Abstract
Cancer has been shown to result from the sequential acquisition of genetic alterations in a single lineage of cells. In leukemia, increasing evidence has supported the idea that this accumulation of mutations occurs in self-renewing hematopoietic stem cells (HSCs). These HSCs, containing some, but not all, leukemia-specific mutations have been termed pre-leukemic. Multiple recent studies have sought to understand these pre-leukemic HSCs and determine to what extent they contribute to leukemogenesis. These studies have elucidated patterns in mutation acquisition in leukemia, demonstrated resistance of pre-leukemic cells to standard induction chemotherapy, and identified these pre-leukemic cells as a putative reservoir for the generation of relapsed disease. When combined with decades of research on clonal evolution in leukemia, mouse models of leukemogenesis, and recent massively parallel sequencing-based studies of primary patient leukemia, these studies of pre-leukemic HSCs begin to piece together the evolutionary puzzle of leukemogenesis. These results have broad implications for leukemia treatment, targeted therapies, minimal residual disease monitoring, and early detection screening.
Keywords: Cancer evolution, pre-leukemia
Introduction
Evolution is the stepwise process through which genetic alterations are translated into phenotypic changes, and if advantageous, these phenotypic changes grow to predominate in a population. In the context of leukemia, the phenotypic changes that lead to disease are a block in differentiation and the ability to proliferate without exhaustion. The generation of these phenotypic changes requires multiple genetic events to accumulate in a single lineage of cells, a process that has been shown to take decades in other cancers1,2. Given the low spontaneous mutation rate in hematopoietic cells3, and the absence of hypermutator phenotypes in most leukemias4, the process of leukemogenesis is similarly thought to occur over many years. This hypothesis has led to a model for leukemia evolution whereby mutations accumulate in functionally-normal hematopoietic stem cells (HSCs) during a prolonged “pre-leukemic” phase. These intermediate HSCs harboring some, but not all, leukemia-specific mutations have been termed pre-leukemic HSCs.
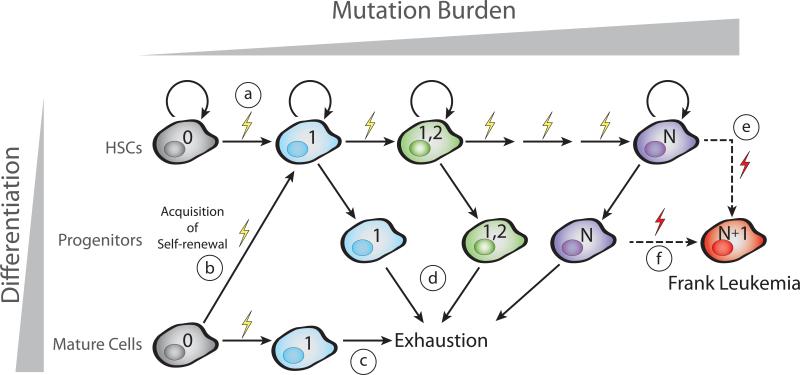
A model for pre-leukemic clonal evolution has been developed from multiple lines of evidence including mouse models of leukemia, targeted analysis of known leukemogenic mutations, and unbiased high-throughput sequencing studies. This model (Figure 1) requires that the first leukemogenic mutation either occurs in a self-renewing cell (A) or confers self-renewal to a more differentiated cell (B). If, instead, a mutation occurred in a differentiated cell but did not confer self-renewal, this mutation would be lost over time due to exhaustion or terminal differentiation (C). Successive mutations accumulate in this mutated self-renewing cell lineage (up to N mutations). These self-renewing cells retain some ability to produce differentiated progeny which are lost to terminal differentiation (D). Eventually, one of these pre-leukemic HSCs (E) or one of their more differentiated progeny (F) acquires an additional mutation (N+1) which leads to the loss of normal HSC functions and the development of frank leukemia. The evolutionary processes that govern the accumulation of mutations in pre-leukemic HSCs are the subject of this review. We will present evidence from multiple subtypes of leukemia supporting this model for pre-leukemic mutation acquisition and discuss the current understanding of clonal evolution that occurs prior to the onset of disease.
Figure 1. Model for pre-leukemic evolution of leukemia.
Sequential acquisition of mutations during clonal evolution is illustrated by changes in color. Multiple scenarios for leukemic evolution exist. The simplest model posits that the first mutation would occur in a normal HSC (A) that retains the ability to self-renew. Alternatively, the first mutation could occur in a more differentiated cell and could confer self-renewal to this previously non-self-renewing cell (B). If, however, the first mutation were to occur in a differentiated cell but not confer self-renewal, this mutation would likely be lost due to exhaustion of this lineage (C). Subsequent mutations accumulate in this self-renewing HSC lineage. At each stage of evolution, these self-renewing cells retain the ability to produce differentiated progeny of both the myeloid and lymphoid lineages (D). Eventually, one or a few additional mutations lead to the generation of a fully leukemic cell. This mutational event could occur in a bonafide HSC (E) or in a progenitor cell (F). The resultant leukemia cell loses the capability to differentiate into multiple cellular lineages.
Early Evidence for Pre-Leukemic HSCs
The earliest evidence for the existence of pre-leukemic HSC in human leukemia was gleaned from rigorous clinical studies of both adult and pediatric leukemia. Clonality of AML was first studied in female patients heterozygous for X-chromosome-linked glucose-6-phosphate dehydrogenase (G6PD) gene variants. In each of these patients, the leukemic blast cells expressed only a single allele of G6PD, indicating that the malignant clone was derived from a single cell. In a subset of these patients, circulating erythrocytes and/or platelets were observed to express only the leukemic G6PD allele, suggesting clonal dominance of a pre-leukemic clone contributing to erythropoiesis and thrombopoiesis. Moreover, some patients exhibited partial or complete clonal expression of G6PD in the hematopoietic system even during remission5,6. Additional studies of Epstein-Barr virus transformed B lymphoid cell lines from patients heterozygous for G6PD variants showed that certain AML patients exhibited a significant skewing in G6PD allotypes even in B lymphoid cells7. Cumulatively, these studies provided the first evidence that early leukemogenic mutations arise in multipotent hematopoietic cells and can provide a clonal advantage.
Later studies of adult AML with the recurrent t(8;21) translocation involving the runt-related transcription factor 1 (AML1/RUNX1) and the core-binding factor alpha subunit 2 (ETO/CBFA2T1) genes showed detectable levels of this translocation in normal myeloid cells from patients in long-term remission8,9. Subsequent research isolated highly-purified HSC from t(8;21) patients in long-term remission and demonstrated the presence of the fusion transcript in single-cell derived myeloid and erythroid colonies10. This work provided the first direct demonstration that HSCs could harbor leukemogenic mutations and was the first evidence that pre-leukemic cells could persist during remission.
Extensive work on cases of pediatric leukemia, and in particular twin studies, led to advances in our understanding of pre-leukemic HSC and the clonal progression of this disease. Early research on twin pairs with concordant AML showed that, in some cases, both individuals of the pair harbored the same non-constitutive chromosomal rearrangement of the mixed lineage leukemia (MLL) gene in their leukemic cells11. This provided the first evidence supporting an in utero clonal origin of these pediatric leukemias. This phenomenon was observed in multiple subtypes of leukemia harboring multiple different recurrent chromosomal translocations12–20. This work was complemented by studies of twins with discordant disease status showing the presence of the leukemic TEL-AML1 translocation in normal differentiated cells of the non-leukemic twin21. Moreover, introduction of the TEL-AML1 fusion gene into normal cord blood HSCs did not lead to an overt leukemia, but rather a pre-leukemic phase resembling that of the non-leukemic twin.
Additional studies of chronic leukemias and myeloproliferative neoplasms have also implicated HSCs as a primary reservoir for accumulation of leukemic mutations. The canonical BCR-ABL translocation causing chronic myeloid leukemia (CML) has been detected in differentiated lymphoid cells in chronic phase patients22–26, as well as directly in HSCs27,28. Additional events occurring at the level of the granulocyte-macrophage progenitor are thought to lead to progression to blast-crisis in these patients29. Intriguingly, the clinical use of inhibitors targeting the BCR-ABL kinase has led to durable long-term remissions in CML; however, these patients often have detectable BCR-ABL translocations in their normally-functioning HSCs despite the apparent clearance of all leukemic cells30–32. Moreover, many of these patients will develop recurrent disease if anti-BCR-ABL therapy is discontinued33,34, providing support for the idea that these pre-leukemic HSC constitute a cellular reservoir that is poised to generate recurrent disease. Similarly, research on chronic lymphocytic leukemia (CLL) has demonstrated that the propensity to generate clonal B cells is a cell-intrinsic property that has already been acquired at the level of the HSC. HSCs isolated from CLL patients showed lymphoid-lineage gene priming and restricted mono- or oligo-clonal B cell differentiation much like that observed in monoclonal B cell lymphocytosis, a precursor condition to frank CLL35. This work suggests that, even in mature lymphoid tumors such as CLL, HSCs can be involved in leukemogenesis and act as cellular reservoirs for mutation acquisition. Intriguingly, the molecular identity of pre-leukemic mutations in CLL remains to be determined. Additional work investigating myeloproliferative neoplasms, such as polycythemia vera, has shown that the hallmark mutation of this disease, a valine to phenylalanine mutation at amino acid 617 in the Janus Kinase 2 protein, occurs in hematopoietic stem cells, leading to a predisposition towards and autonomous production of erythroid lineage cells 36.
Collectively, these studies provided early evidence supporting a model in which leukemic mutations accumulate in HSCs that retain their ability to self-renew and generate differentiated progeny. These early studies have been supported both by mouse models of leukemogenesis and by recent human genetic studies that make use of high-throughput DNA sequencing to understand the clonal architecture of leukemia.
Evidence for Pre-Leukemic HSCs from Mouse Models of Leukemia
The application of high-throughput sequencing in the investigation of human AML has led to the identification of recurrent leukemogenic mutations in DNA methyltransferase 3A (DNMT3A), Ten-eleven translocation 2 (TET2), Isocitrate Dehydrogenase 1 and 2 (IDH1/2), and many other genes, including previously identified mutations in FMS-like tyrosine kinase-3 (FLT3). Mouse models of these mutations have provided key insights into their effects on hematopoietic stem and progenitor cells, and have allowed for combinatorial experiments that demonstrate cooperativity between various mutations. Early studies of mice with internal tandem duplication (ITD) mutations in FLT3 demonstrated a robust myeloproliferative phenotype reminiscent of chronic myelomonocytic leukemia37. While these mice had significantly higher numbers of differentiated monocytes and granulocytes, the stem cell potential of the HSC compartment was dramatically reduced as demonstrated by a loss of spleen colony forming unit ability. These results were later corroborated by a second mouse model of FLT3-ITD mutation which showed a transient overproliferation within the normally quiescent long-term HSC compartment that led to a rapid depletion of these self-renewing cells38. Together, these studies demonstrate that the FLT3-ITD mutation impairs HSC function and self-renewal, and suggest it would likely be a late progressor event in leukemogenesis (as in Figure 1E, F).
In contrast, somatic loss-of function mutations in the TET2 gene have been shown to lead to an increase in stem cell self-renewal39,40. In vitro studies of hematopoietic cells demonstrated increased serial replating upon knockdown of TET2. Moreover, a mouse model for hematopoietic-specific conditional loss of TET2 showed progressive defects in the bone marrow and extramedullary hematopoiesis39. Specifically, both the number and re-populating ability of HSCs was increased in TET2−/− mice compared to wildtype controls, and TET2−/− HSC exhibited a strong advantage upon competitive transplantation39,40. These data underscore a role for TET2 mutation in the maintenance of HSC self-renewal and expansion, and suggest TET2 mutations could occur as an early event in leukemogenesis (as in Figure 1A).
Similar studies have analyzed the effects of loss of DNMT3A on the hematopoietic system41. Conditional ablation of DNMT3A led to increased impairment of HSC differentiation through multiple rounds of serial transplantation. This impaired differentiation was accompanied by a simultaneous expansion in the number of bone marrow HSC. These HSC changes were shown to be a consequence of alterations in CpG methylation at key HSC-specific genes, leading to the hypothesis that wildtype DNMT3A is a critical participant in the silencing of HSC regulatory genes during differentiation. Thus, much like mutation of TET2, loss of DNMT3A function leads to an increase in HSC self-renewal and expansion, and implicates such mutations as potential early players in the evolution of leukemia (as in Figure 1A).
Unlike mutation of TET2 or DNMT3A, mouse models of IDH1 mutation have not shown an effect on HSC self-renewal or expansion. Instead, knock-in of the IDH1(R132H) mutation in myeloid lineage cells led to an increase in lineage-restricted hematopoietic progenitors, a decrease in bone marrow cellularity, and extramedullary hematopoiesis42. However, knock-in of this mutation into all hematopoietic cells, including HSC, did not result in overt hematologic abnormalities, indicating a more subtle effect on HSC functions. These results do not preclude a role for IDH1 mutation early in leukemic evolution, but do suggest that IDH1 mutation does not markedly disrupt HSC function.
Overall, the use of mouse models to study leukemic mutations has provided key insights into the effects of these mutations on the hematopoietic system, and HSC in particular. Certain mutations (e.g. DNMT3A, TET2) seem to play clear roles in HSC maintenance and expansion while others (e.g. FLT3) seem to antagonize this process in favor of dysregulated proliferation. Additional studies based on high-throughput sequencing analysis of primary patient samples have augmented these model systems and provided substantial evidence supporting the pre-leukemic evolution model of leukemogenesis.
Evidence for Pre-Leukemic HSCs from Massively Parallel Sequencing Data
The advent of high-throughput sequencing has revolutionized the study of cancer and clonal evolution. Hundreds of leukemia samples have been sequenced and the recurrent mutations associated with each subset of the disease have been identified, allowing for tracking of these mutations through time and the inference of clonal evolutionary patterns. Pioneering early work using SNP arrays to investigate pediatric ALL showed that the majority of cases of relapsed disease originate from clones antecedent to the diagnosis clone43. However, these relapsed leukemias shared some, but not all, of the clonal mutations found in the leukemia at diagnosis, implicating the putative involvement of preleukemic HSC in this evolutionary progression. In normal individuals, analysis of mutation acquisition in HSC over time was accomplished through whole exome sequencing (WXS) of single HSC-derived colonies, which showed that the total number of mutations in individual HSC increases with age44. While the majority of these mutations are likely passenger mutations, these results highlight the ability of these cells to accumulate multiple genetic lesions over time. Additionally, these data serve to underscore the notion that diseases such as AML are often diseases of the elderly, and this may be a consequence of the long time period required to acquire multiple mutations in a single cell lineage without apparent genomic instability. In line with these observations, exome sequencing of multiple elderly women with evidence of clonal hematopoiesis based on X-inactivation identified recurrent somatic mutations in the TET2 gene45. Subsequent analysis of 182 additional elderly women with clonal hematopoiesis determined that >5% of these individuals had mutations in TET2. While these individuals had no clinical evidence of hematologic malignancies, the presence of recurrent somatic mutations in a known leukemia driver in the setting of clonal hematopoiesis indicates that acquisition of such mutations at the level of the HSC could be causative in the early evolution of leukemia. Collectively, this research laid the groundwork for additional studies focused on understanding the clonal evolution events that precede the onset of frank leukemia through the direct investigation of pre-leukemic HSCs.
Substantial advances in stem cell biology have further facilitated the study of pre-leukemic HSCs. In particular, the discovery of leukemia-specific antigens46 has enabled the prospective isolation of residual normal HSC from leukemia samples. In particular, combinations of the leukemia-specific markers CD47, CD99, and the T-cell immunoglobulin and mucin domain-containing protein 3 (TIM3) have been used to isolate residual normal HSC by fluorescence activated cell sorting (FACS)47,48. These residual normal HSC are capable of forming both myeloid and erythroid colonies in methylcelluose, and are capable of producing both CD33+ myeloid and CD19+ lymphoid cells in long-term xenotransplantation studies. Together, these results indicate that, in the best stem cell assays available, these cells are bona fide HSCs. According to the model described above (Figure 1), this residual normal HSC population should contain individual cells harboring leukemia-specific mutations, termed preleukemic HSC. The first sequencing studies of these residual HSCs provided strong support for this model, by determining that in diagnostic samples from AML patients harboring a FLT3-ITD mutation, some, but not all, leukemia-specific mutations could be detected in prospectively isolated residual HSCs49. Moreover, by targeted genotyping of single HSC-derived colonies, the order of mutation acquisition was determined in a handful of these cases. This study validated the model described here and was the first study to isolate pre-leukemic HSC in AML.
Follow up studies of this work expanded these conclusions to a more diverse cohort of AML patients representing the heterogeneity seen in this disease, further validating the model that leukemogenic mutations are serially acquired in self-renewing HSC 50,51. Notably, these studies characterized the contribution of pre-leukemic HSCs to lymphoid cells at diagnosis and found that, in multiple genetic subtypes of AML, pre-leukemic HSCs contribute to lymphopoiesis. Together, these studies demonstrated that residual HSCs contain pre-leukemic cells and showed that HSCs can harbor multiple leukemogenic driver mutations and still retain normal differentiation in the context of xenotransplantation. Moreover, these results showed that some pre-leukemic mutations appear to confer a growth advantage, enabling pre-leukemic HSC to outcompete unmutated HSC. This initial isolation and genetic characterization of pre-leukemic HSCs in AML proved that, at least in some cases, leukemia evolves from sequential acquisition of mutations in functionally-normal HSCs.
This work with residual HSC from diagnosis led to the hypothesis that pre-leukemic HSCs may survive standard induction chemotherapy regimens and persist in remission. Sequencing studies of AML patient-matched diagnosis and remission samples showed that all mutations identified to be preleukemic at diagnosis were also found in both mature hematopoietic cells and immature HSPCs during remission50,51. Importantly, none of the leukemic mutations absent from pre-leukemic HSC were found to persist during remission. Moreover, in a subset of these patients where matched relapse disease samples were available, all pre-leukemic mutations were found in the relapse clone50. Together, these data indicate that pre-leukemic HSCs are capable of surviving induction chemotherapy and can contribute to hematopoiesis during remission. The presence of these cells during remission additionally highlights the potential for pre-leukemic HSCs to participate in the generation of relapsed disease. As these cells contain some, but not all of the mutations necessary to generate leukemia, they are poised to acquire a small number of mutations and generate relapsed disease. Formal proof of this has not been provided, but would require exclusion of the possibility that leukemic populations emerging at relapse were not present during remission. Successful demonstration of a role for pre-leukemic cells in the generation of relapsed disease would have substantial implications for how leukemia is treated and would identify pre-leukemic HSCs as an important and novel therapeutic target.
Patterns of Mutation Acquisition in Leukemia
The study of pre-leukemic HSC has major implications for the order of acquisition of mutations in this disease. The strongest evidence for patterns of mutations in leukemia has come from large scale sequencing efforts in AML showing that certain mutations often co-occur (e.g. DNMT3A, NPM1, and FLT3) or are mutually exclusive (e.g. TET2 and IDH1/2). These patterns imply that there may also be patterns in the temporal acquisition of mutations. If, for example, mutation of a certain gene is frequently found in pre-leukemic HSCs, this mutation would frequently be an early event in leukemogenesis. Conversely, if mutation of a certain gene is never or rarely seen in pre-leukemic HSCs, this mutation would be considered a late event. Characterization of pre-leukemic mutations in 16 patients by WXS has identified such statistically significant patterns. In particular, mutations in genes that affect the epigenome through processes such as DNA methylation, histone modification, or regulation of chromatin topology occurred statistically significantly earlier in leukemogenesis 50. Moreover, mutations in genes that lead to activated signaling and increases in proliferation, such as those in FLT3 and RAS, were found to be almost exclusively late events50. These results were validated in a larger cohort of patients by targeted deep sequencing. Similar studies investigating patients with mutations in DNMT3A showed that in 12 of 15 patients (70.5%), the DNMT3A mutation was preleukemic51. In these same patients, mutations in NPM1 and FLT3 were never found to be pre-leukemic. Additional studies focusing on the persistence and loss of mutations between diagnosis and relapse have come to strikingly similar conclusions. Notably, mutations in genes such as IDH1/2 and DNMT3A appear highly stable between diagnosis and relapse (i.e. occur early) while mutations in genes such as FLT3 and RAS appear to be gained or lost at relapse (i.e. occur late) 52. Intriguingly, this study indicated that mutations in NPM1 were almost always retained between diagnosis and relapse, a result that is contrasted by the absence of this mutation in pre-leukemic HSCs as mentioned above. This contrast identifies an important avenue for future research – how do the patterns identified in mutation acquisition correlate with disease progression and what are the therapeutic implications for a mutation to be pre-leukemic versus late? Together, these studies indicate that patterns of mutation acquisition exist in leukemia – a discovery that has significant implications for the development of effective targeted therapies.
Implications for Patient Survival, Therapy Development, and MRD Detection
The existence of pre-leukemic HSC and the elucidation of patterns of mutation acquisition in leukemia have important clinical implications. Importantly, pre-leukemic, early mutations would be present in all leukemia cells, while mutations present in leukemic subclones would be the latest mutations. Thus, therapies aimed at targeting every leukemic cell should be designed against the earliest pre-leukemic mutations. However, it remains possible that the earliest leukemic mutations function via a “hit-and-run” mechanism, where they are required to initiate disease, but not to maintain it. If this were the case, targeting of these early mutations would not affect the leukemic cells. Recent studies have begun to investigate this question using mouse models of AML53. Using a transgenic mouse expressing the FLT3-ITD oncogene and a repressible version of mutant IDH2, the authors show that silencing of mutant IDH2 in frankly leukemic cells leads to a reversal of the differentiation block caused by IDH2 mutation. These results suggest that therapeutic targeting of early mutations, such as IDH2, will have functional effects on leukemic cells. It remains to be seen whether this phenomenon can be recapitulated in primary patient AML cells, and whether targeted therapies directed towards preleukemic mutations will be effective.
In addition to targeting the earliest mutations with the hope of eliminating every frankly leukemic cell, it may be important to target these early mutations to eradicate the pre-leukemic HSCs as well. As stated above, pre-leukemic HSCs persist in remission and harbor some, but not all of the leukemia-specific mutations, and are therefore poised to acquire additional mutations and, potentially, generate relapsed disease10,50,51. One implication is that in order to achieve more durable, and eventually, permanent remissions, therapies may need to eradicate the pre-leukemic HSCs as well as the frankly leukemic cells. Moreover, therapies used to eradicate leukemic cells may differ from therapies needed to eradicate pre-leukemic HSCs given the highly divergent cellular contexts. For example, ABL-tyrosine kinase inhibitors are effective at eliminating BCR-ABL progenitors in chronic myeloid leukemia (CML), but are ineffective at eradicating the BCR-ABL leukemic stem cell compartment54. This example illustrates that the efficacy of a molecularly targeted therapy can vary between different cellular contexts. While the contribution of pre-leukemic HSCs to relapsed disease has not been proven, it is possible that, as our ability to eradicate leukemic cells increases, the contribution of pre-leukemic HSCs to relapsed disease will become more and more apparent. More specifically, if we are able to fully eradicate leukemia cells, then relapsed disease may no longer originate from outgrowth of leukemic subclones or from therapy-resistant leukemia cells, but rather from further evolution of pre-leukemic HSC. As the duration of remissions increases, so does the time during which a pre-leukemic cell could acquire additional mutations. Increased eradication of leukemic cells resulting in more durable remissions may unveil the contribution of pre-leukemic HSCs to relapsed disease. As some pre-leukemic cells are capable of harboring multiple recurrent driver mutations49,50, the re-evolution of disease from a pre-leukemic cell may not require the acquisition of multiple genetic aberrations. As an example, studies of patient-matched diagnosis and relapsed AML have shown that patients whose relapsed disease is more divergent from their disease at diagnosis have an increased duration of relapse-free survival52. Conceptually, a more divergent relapsed disease indicates a further step back in evolutionary time, potentially all the way to a pre-leukemic cell, which may take longer to accumulate new mutations that lead to relapse. Despite this increase in relapse-free survival, these patients ultimately still relapse, underscoring the potential importance of therapeutically targeting pre-leukemic HSCs.
Another important implication of the persistence of pre-leukemic mutations in HSCs during remission is the contribution of these cells to minimal residual disease (MRD) monitoring. MRD, defined as the presence of small numbers of leukemic cells in a patient after treatment, is one of the primary causes of relapse in leukemia. MRD monitoring is designed to detect leukemic cells and can employ different modalities, such as flow cytometry or nucleic acid-based mutation detection. However, in the case of MRD monitoring with a pre-leukemic mutation, pre-leukemic HSCs and/or their differentiated progeny would be captured as if they were fully-leukemic cells. Therefore, the apparent MRD burden would be higher than the actual level of frankly leukemic cells. Mutations in NPM1 have been used with efficacy in MRD monitoring in patients with AML55, and this may be due to the fact that they have not been identified as pre-leukemic mutations 50,51. These results identify a paradox in the monitoring of MRD – the ideal markers of MRD are those that are only present in all frank leukemia cells and not preleukemic cells, but such markers are the most likely to be divergent between diagnosis and relapse. Currently, the data support the use of NPM1 as a marker for MRD in the clinical setting. In the future, it will be important to understand whether patients with higher degrees of pre-leukemic burden during remission have a shorter overall time-to-relapse than patients with low or undetectable pre-leukemic burden. Such a scenario may make monitoring of pre-leukemic burden during remission clinically relevant.
Conclusions and Future Directions
Pre-leukemic HSCs in leukemia represent an important scientific and clinical entity. The study of their genetics has led to an understanding of patterns of mutation acquisition, which has substantial implications for the treatment of leukemia. Mutations altering genes involved in regulation of the epigenome appear to occur significantly earlier in AML evolution. As such, these mutations represent ideal targets for therapeutic intervention as they would be present in every leukemic cell and not just a minor subclone. Conversely, mutations that lead to activated signaling have been identified as late events and may be poor choices for therapeutic intervention as they are more likely to be present only in subclones. Moreover, the patterns of mutation acquisition observed in AML may also extend to other hematologic malignancies. Further studies investigating pre-leukemic cells in other diseases will shed light on this question and indicate whether the patterns observed in AML are general patterns of tumorigenesis or specific to the biology of AML.
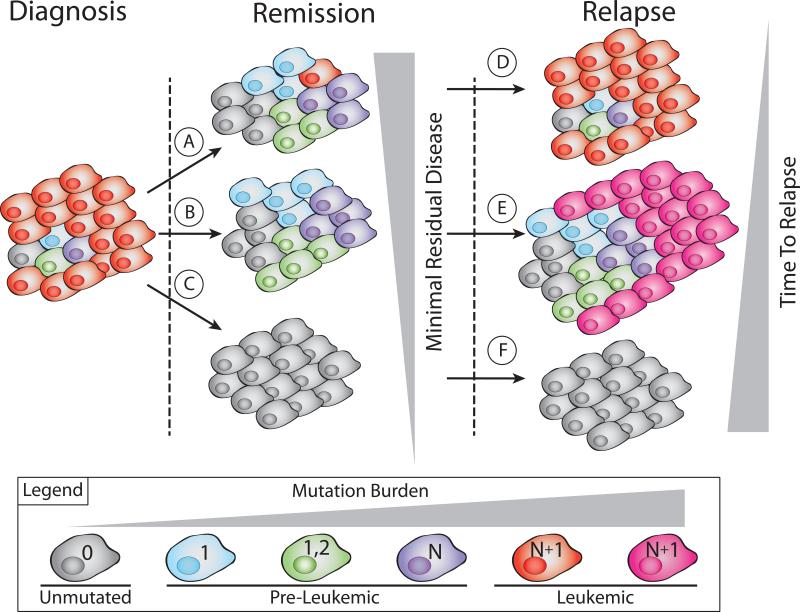
Here we summarized careful sequencing studies of the clonal evolution of these pre-leukemic cells that have provided key insights into the putative mechanisms of relapsed disease. Importantly, identification of pre-leukemic HSCs and differentiated cells derived from pre-leukemic HSCs during remission has supported multiple models of relapse (Figure 2). In many patients, our current standard chemotherapy regimens do not fully eradicate all leukemia cells (Figure 2A). The leukemic cells that survive this therapy rebound and eventually present as relapsed disease (Figure 2D). Multiple sequencing studies have indicated that these patients who are classified as having detectable MRD tend to relapse more quickly and often present with relapsed disease that is not genetically divergent from the disease at diagnosis. We hypothesize another avenue of relapse whereby all leukemic cells are eliminated with standard chemotherapy regimens (Figure 2B), but over time, therapy refractory preleukemic cells undergo further mutation evolution and seed genetically divergent relapsed disease (Figure 2E). As targeted therapies are developed and our ability to eliminate leukemic cells increases, we believe that it will become necessary to additionally target the pre-leukemic cells, which may require different therapies. In such a scenario, all pre-leukemic and frankly leukemic cells could be eradicated from a patient (Figure 2C), which would result in a very durable remission, and potentially, long-term cure of the disease (Figure 2F). Future investigation of the biology of pre-leukemic HSCs and how they differ from both unmutated HSCs and frankly leukemic cells will be critical for determining their contribution to disease progression and ways in which these cells can be therapeutically targeted. While the persistence of these cells in remission indicates their potential to be involved in the generation of relapsed disease, formal proof of this possibility will identify these pre-leukemic HSCs as important therapeutic targets and may eventually lead to durable remissions and eventual cures.
Figure 2. Mechanisms of relapse resulting from varying levels of leukemia cell eradication.
Multiple distinct clinical scenarios exist after initial treatment of leukemia. If there is incomplete eradication of leukemia cells (A), the patient would be considered to have some minimal residual disease, and these residual leukemia cells would be able to proliferate and cause relapsed disease with short latency that is genetically similar to the disease at diagnosis. Alternatively, complete eradication of leukemia cells but incomplete targeting of pre-leukemic cells (B) could result in low or undetectable minimal residual disease. However, it remains possible for these pre-leukemic cells to acquire further genetic alterations leading to a genetically divergent relapsed disease with a longer latency (E). The optimal therapeutic situation would be one in which all leukemic and pre-leukemic cells are eradicated (C). Such a scenario would result in the most durable remissions, and potentially, long-term cure of AML (F).
Acknowledgements
M.R.C.Z. is supported by the Smith Fellowship, the NSF GRFP, and the NIH F31 Pre-doctoral fellowship. R.M. holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund and is a New York Stem Cell Foundation Robertson Investigator.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones S, Chen W-D, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4283–8. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araten DJ, Golde DW, Zhang RH, Thaler HT, Gargiulo L, Notaro R, et al. A quantitative measurement of the human somatic mutation rate. Cancer Res. 2005;65:8111–7. doi: 10.1158/0008-5472.CAN-04-1198. [DOI] [PubMed] [Google Scholar]
- 4.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio S. a. J. R., Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013 doi:10.1038/nature12477. [Google Scholar]
- 5.Fialkow PJ, Singer JW, Raskind WH,W,AJ, Jacobson RJ, Bernstein ID, et al. Clonal Development, Stem-cell Differentiation, and Clinical Remissions in Acute Non-lymphocytic Leukemia. N. Engl. J. Med. 1987;317:468–473. doi: 10.1056/NEJM198708203170802. [DOI] [PubMed] [Google Scholar]
- 6.Fialkow PJ, Janssen JW, Bartram CR. Clonal remissions in acute nonlymphocytic leukemia: evidence for a multistep pathogenesis of the malignancy. Blood. 1991;77:1415–7. [PubMed] [Google Scholar]
- 7.Ferraris a M., Raskind WH, Bjornson BH, Jacobson RJ, Singer JW, Fialkow PJ. Heterogeneity of B cell involvement in acute nonlymphocytic leukemia. Blood. 1985;66:342–4. [PubMed] [Google Scholar]
- 8.Kwong YL, Wong KF, Chan V, Chan CH. Persistence of AMLI Rearrangement in Peripheral Blood Cells in t(8;21). Cancer Genet Cytogenet. 1996;54:151–154. doi: 10.1016/0165-4608(95)00282-0. [DOI] [PubMed] [Google Scholar]
- 9.Miyamoto T, Nagafuji K, Akashi K, Harada M, Kyo T, Akashi T, et al. Persistence of multipotent progenitors expressing AML1/ETO transcripts in long-term remission patients with t(8;21) acute myelogenous leukemia. Blood. 1996;87:4789–96. [PubMed] [Google Scholar]
- 10.Miyamoto T, Weissman IL, Akashi K. AML1/ETO-expressing nonleukemic stem cells in acute myelogenous leukemia with 8;21 chromosomal translocation. Proc. Natl. Acad. Sci. U. S. A. 2000;97:7521–6. doi: 10.1073/pnas.97.13.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ford AM, Ridge SA, Cabrera ME, Mahmoud H, Steel CM, Chan LC, et al. In utero rearrangements in the trithorax-related oncogene in infant leukaemias. NatureNature. 1993;363:358–360. doi: 10.1038/363358a0. [DOI] [PubMed] [Google Scholar]
- 12.Wiemels JL, Cazzaniga G, Daniotti M, Eden OB, Addison GM, Masera G, et al. Prenatal origin of acute lymphoblastic leukaemia in children. Lancet. 1999;354:1499–503. doi: 10.1016/s0140-6736(99)09403-9. [DOI] [PubMed] [Google Scholar]
- 13.Zuna J, Burjanivova T, Mejstrikova E, Zemanova Z, Muzikova K, Meyer C, et al. Covert Preleukemia Driven by MLL Gene Fusion. 2009;107:98–107. doi: 10.1002/gcc.20622. [DOI] [PubMed] [Google Scholar]
- 14.Zuna J, Muzikova K, Ford AM, Maia AT, Krejci O, Tousovska K, et al. Pre-natal, Clonal Origin of Acute Lymphoblastic Leukaemia in Triplets. Leuk. Lymphoma. 2003;44:2099–2102. doi: 10.1080/1042819031000123393. [DOI] [PubMed] [Google Scholar]
- 15.Van Delft FW, Horsley S, Colman S, Anderson K, Bateman C, Kempski H, et al. Clonal origins of relapse in ETV6-RUNX1 acute lymphoblastic leukemia. Blood. 2011:6247–6254. doi: 10.1182/blood-2010-10-314674. doi:10.1182/blood-2010-10-314674. [DOI] [PubMed] [Google Scholar]
- 16.Ford a M., Bennett C. a, Price CM, Bruin MC, Van Wering ER, Greaves M. Fetal origins of the TEL-AML1 fusion gene in identical twins with leukemia. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4584–8. doi: 10.1073/pnas.95.8.4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gale KB, Ford a M., Repp R, Borkhardt a, Keller C, Eden OB, et al. Backtracking leukemia to birth: identification of clonotypic gene fusion sequences in neonatal blood spots. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13950–4. doi: 10.1073/pnas.94.25.13950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maia AT, Tussiwand R, Cazzaniga G, Rebulla P, Colman S, Biondi A, et al. Identification of preleukemic precursors of hyperdiploid acute lymphoblastic leukemia in cord blood. Genes. Chromosomes Cancer. 2004;40:38–43. doi: 10.1002/gcc.20010. [DOI] [PubMed] [Google Scholar]
- 19.Greaves M. Pre-natal Origins of Childhood Leukemia. Rev Clin Exp Hematol. 2003 [PubMed] [Google Scholar]
- 20.Horsley SW, Colman S, Mckinley M, Bateman CM, Jenney M, Chaplin T, et al. Genetic Lesions in a Preleukemic Aplasia Phase in a Child with Acute Lymphoblastic Leukemia. 2008;340:333–340. doi: 10.1002/gcc.20537. [DOI] [PubMed] [Google Scholar]
- 21.Hong D, Gupta R, Ancliff P, Atzberger A, Brown J, Soneji S, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008;319:336–9. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 22.Knuutila S, Teerenhovi L, Larramendy ML, Elonen E, Franssila KO, Nylund SJ, et al. Cell lineage involvement of recurrent chromosomal abnormalities in hematologic neoplasms. Genes. Chromosomes Cancer. 1994;10:95–102. doi: 10.1002/gcc.2870100204. [DOI] [PubMed] [Google Scholar]
- 23.Jonas D, Lübbert M, Kawasaki ES, Henke M, Bross KJ, Mertelsmann R, et al. Clonal analysis of bcr-abl rearrangement in T lymphocytes from patients with chronic myelogenous leukemia. Blood. 1992;79:1017–23. [PubMed] [Google Scholar]
- 24.Haferlach T, Winkemann M, Nickenig C, Meeder M, Ramm-Petersen L, Schoch R, et al. Which compartments are involved in Philadelphia-chromosome positive chronic myeloid leukaemia? An answer at the single cell level by combining May-Grünwald-Giemsa staining and fluorescence in situ hybridization techniques. Br. J. Haematol. 1997;97:99–106. doi: 10.1046/j.1365-2141.1997.9662656.x. [DOI] [PubMed] [Google Scholar]
- 25.Nitta M, Kato Y, Strife A, Wachter M, Fried J, Perez A, et al. Incidence of involvement of the B and T lymphocyte lineages in chronic myelogenous leukemia. Blood. 1985:1053–1061. [PubMed] [Google Scholar]
- 26.Juneja HS, Weiner R. Presence of the Philadelphia Chromosome ( Ph 1 ) in Pokeweed Mitogen Stimulated Lymphocytes during Chronic Phase of. Cancer Genet Cytogenet. 1981;4:39–44. doi: 10.1016/0165-4608(81)90006-6. [DOI] [PubMed] [Google Scholar]
- 27.Fialkow PJ, Jacobson RJ, Papayannopoulou T. Chronic myelocytic leukemia: clonal origin in a stem cell common to the granulocyte, erythrocyte, platelet and monocyte/macrophage. Am. J. Med. 1977;63:125–30. doi: 10.1016/0002-9343(77)90124-3. [DOI] [PubMed] [Google Scholar]
- 28.Bedi a, Zehnbauer B. a, Collector MI, Barber JP, Zicha MS, Sharkis SJ, et al. BCR-ABL gene rearrangement and expression of primitive hematopoietic progenitors in chronic myeloid leukemia. Blood. 1993;81:2898–902. [PubMed] [Google Scholar]
- 29.Jamieson CHM, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 2004;351:657–67. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 30.Chu S, McDonald T, Lin A, Chakraborty S, Huang Q, Snyder DS, et al. Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood. 2011;118:5565–72. doi: 10.1182/blood-2010-12-327437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chomel J-C, Bonnet M-L, Sorel N, Bertrand A, Meunier M-C, Fichelson S, et al. Leukemic stem cell persistence in chronic myeloid leukemia patients with sustained undetectable molecular residual disease. Blood. 2011;118:3657–60. doi: 10.1182/blood-2011-02-335497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. 2011;121 doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahon F-X, Réa D, Guilhot J, Guilhot F, Huguet F, Nicolini F, et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol. 2010;11:1029–35. doi: 10.1016/S1470-2045(10)70233-3. [DOI] [PubMed] [Google Scholar]
- 34.Ross DM, Branford S, Seymour JF, Schwarer AP, Arthur C, Yeung DT, et al. Safety and efficacy of imatinib cessation for CML patients with stable undetectable minimal residual disease: results from the TWISTER study. Blood. 2013;122:515–22. doi: 10.1182/blood-2013-02-483750. [DOI] [PubMed] [Google Scholar]
- 35.Kikushige Y, Ishikawa F, Miyamoto T, Shima T, Urata S, Yoshimoto G, et al. Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell. 2011;20:246–59. doi: 10.1016/j.ccr.2011.06.029. [DOI] [PubMed] [Google Scholar]
- 36.Jamieson CHM, Gotlib J, Durocher J. a, Chao MP, Mariappan MR, Lay M, et al. The JAK2 V617F mutation occurs in hematopoietic stem cells in polycythemia vera and predisposes toward erythroid differentiation. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6224–9. doi: 10.1073/pnas.0601462103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee BH, Tothova Z, Levine RL, Anderson K, Buza-Vidas N, Cullen DE, et al. FLT3 mutations confer enhanced proliferation and survival properties to multipotent progenitors in a murine model of chronic myelomonocytic leukemia. Cancer Cell. 2007;12:367–80. doi: 10.1016/j.ccr.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chu SH, Heiser D, Li L, Kaplan I, Collector M, Huso D, et al. FLT3-ITD Knockin Impairs Hematopoietic Stem Cell Quiescence/Homeostasis, Leading to Myeloproliferative Neoplasm. Cell Stem Cell. 2012;11:346–58. doi: 10.1016/j.stem.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 Loss Leads to Increased Hematopoietic Stem Cell Self-Renewal and Myeloid Transformation. Cancer Cell. 2011:1–14. doi: 10.1016/j.ccr.2011.06.001. doi:10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quivoron C, Couronné L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 41.Challen G. a, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2011:1–11. doi: 10.1038/ng.1009. doi:10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasaki M, Knobbe CB, Munger JC, Lind EF, Brenner D, Brüstle A, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 1. 2012 doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mullighan CG, Phillips L. a., Su X, Ma J, Miller CB, Shurtleff S, et al. Genomic Analysis of the Clonal Origins of Relapsed Acute Lymphoblastic Leukemic. 2008;28:1377–1380. doi: 10.1126/science.1164266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The Origin and Evolution of Mutations in Acute Myeloid Leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 2012;44:1179–81. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Majeti R. Monoclonal antibody therapy directed against human acute myeloid leukemia stem cells. Oncogene. 2010:1–11. doi: 10.1038/onc.2010.511. doi:10.1038/onc.2010.511. [DOI] [PubMed] [Google Scholar]
- 47.Majeti R, Chao MP, Alizadeh A. a, Pang WW, Jaiswal S, Gibbs KD, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138:286–99. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jan M, Chao MP, Cha AC, Alizadeh A. a, Gentles AJ, Weissman IL, et al. Prospective separation of normal and leukemic stem cells based on differential expression of TIM3, a human acute myeloid leukemia stem cell marker. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5009–14. doi: 10.1073/pnas.1100551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med. 2012;4:149ra118. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corces-Zimmerman MR, Hong W-J, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. U. S. A. 2014;111:2548–53. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014 doi: 10.1038/nature13038. doi:10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kronke J, Bullinger L, Teleanu V, Tschurtz F, Gaidzik VI, Kuhn MWM, et al. Clonal evolution in relapsed NPM1 mutated acute myeloid leukemia. Blood. 2013:100–108. doi: 10.1182/blood-2013-01-479188. doi:10.1182/blood-2013-01-479188. [DOI] [PubMed] [Google Scholar]
- 53.Kats LM, Reschke M, Taulli R, Pozdnyakova O, Burgess K, Bhargava P, et al. Proto-Oncogenic Role of Mutant IDH2 in Leukemia Initiation and Maintenance. Cell Stem Cell. 2014;14:329–341. doi: 10.1016/j.stem.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jamieson CH. Chronic myeloid leukemia stem cells. Hematology Am. Soc. Hematol. Educ. Program. 2008:436–42. doi: 10.1182/asheducation-2008.1.436. doi:10.1182/asheducation-2008.1.436. [DOI] [PubMed] [Google Scholar]
- 55.Schnittger S, Kern W, Tschulik C, Weiss T, Dicker F, Falini B, et al. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood. 2009;114:2220–31. doi: 10.1182/blood-2009-03-213389. [DOI] [PubMed] [Google Scholar]