Abstract
Macro-autophagy (hereafter referred to as autophagy) delivers cytoplasmic material to the lysosome for degradation, and has been implicated in many cellular processes, including stress, infection, survival, and death. While the regulation and role that autophagy plays in stress, infection, and survival is apparent, the regulation of and role that autophagy has during cell death remains relatively unclear. In this review, we highlight what is known about the role that autophagy can play during physiological cell death, and discuss the implications of better understanding cellular destruction that involves autophagy.
Keywords: cell death, autophagy, apoptosis, necrosis, ATG genes
AUTOPHAGY
The initiation of autophagy starts with the formation of the pre-autophagosomal structure. This pre-autophagosomal structure serves as a nucleation point for the formation of the isolation membrane. The isolation membrane encapsulates cellular material such as proteins and organelles in a double membrane vesicle called an autophagosome [1]. Lysosomes then fuse with the autophagosome forming an autolysosome subsequently enabling their hydrolases to degrade the isolated contents [1]. The autophagic steps can be analyzed by methods that have been previously described [2].
Pioneering genetic studies in yeast led to the identification of the autophagy (ATG) related genes and proteins [3–7]. Significantly, the core autophagy machinery is conserved from yeast to humans, indicating that studies of model organisms can enhance our understanding of how autophagy may function in distinct cellular and physiological contexts in humans. However, the majority of studies have focused on understanding how autophagy functions in the context of nutrient deprivation-triggered induced cell survival, and more work is needed to understand where and how autophagy may contribute in other contexts, such as programmed cell death.
A history of autophagic cell death
In 1973, Schweichel and Merker described three types of physiological cell death that occurred in the developing embryos and fetuses of rats and mice [8]. In type I cell death, isolated dying cells displayed condensation of the nucleus and cytoplasm, and phagocytosis of the dying cells by neighboring cells resulting in the subsequent degradation of the dying cell by the lysosomal system of the engulfing cells. Type I cell death is known as apoptosis [9]. In type II cell death, groups of dying cells were removed in toto with no phagocytosis by neighboring cells. These dying cells contained autophagosomes that isolated regions of their cellular contents, and fused with the dying cell’s own lysosomes to self-degrade the autophagosomes’ contents. Type II cell death would be known as autophagic cell death. Finally, in type III cell death, which is also called necrosis, they observed the swelling of membrane compartments, membrane rupture and “disintegration” of the dying cells with no apparent phagocytosis or lysosomal elements associating with this process [8].
GENETIC SYSTEMS
In recent years, autophagic cell death has been observed in distinct eukaryotic kingdoms from which the studies of genetic model systems have illuminated the roles that autophagy can play in dying cells.
Dictyostelium discoideum
Dictyostelium is a protist that can exist in either a unicellular or multicellular state. During the formation of its multicellular fruiting body, the supportive stalk cells which comprise of approximately 20 percent of all Dictyostelium cells undergo programmed cell death [10]. Having evolutionarily diverged around one billion years ago, Dictyostelium represents one of the most primitive and ancient examples of programmed cell death [11]. Interestingly, Dictyostelium does not possess phagocytes, and no caspases have been found in its genome. Therefore, apoptosis is impossible, and all cell death occurs via autophagy [12].
During starvation, unicellular Dictyostelium begins to aggregate and form a multicellular fruiting body full of viable spores atop a stalk of dead cells. As the stalk cells die they exhibit high levels of autophagy. During this death process, the stalk cells first induce autophagy as a response to starvation, and only after this starvation-induced autophagy is initiated do they receive an additional signal from the differentiation-inducing factor, DIF-1, to promote programmed cell death [13, 14]. Interestingly, sole induction of autophagy or the presence of DIF-1 alone cannot induce cell death [15, 16]. Therefore, autophagy in Dictyostelium appears to be first induced as a starvation response, and only later, along with additional signals, can cell death occur via a mechanism in which autophagy is also necessary [12].
In Dictyostelium, the regulation of starvation-induced autophagy appears to be well conserved as homologues of mTOR complex 1 exist in the Dictyostelium genome. However, the mechanism of how the transition from the use of autophagy for starvation to the use of autophagy for death remains less clear. While it is known that starvation induced autophagy is necessary for cell death to occur in Dictyostelium, as mentioned earlier, another factor, DIF-1, is also necessary. Unfortunately, the DIF-1 signaling pathway has not been fully elucidated. Genetic screens in Dictyostelium have revealed certain genes that are necessary for the completion of autophagic cell death that is triggered though DIF-1, such as iplA (the IP3 receptor) [17]. Interestingly, as described later, the IP3 receptor was also shown to be necessary for autophagic cell death in the salivary glands of Drosophila melanogaster [18]. Therefore, it seems possible that regulation of autophagy during cell death may be evolutionarily conserved.
Arabidopsis thaliana
Unlike metazoans, plants do not exhibit apoptosis because the cell wall of plants prevents the breakdown of cells into apoptotic bodies, and plants do neither have phagocytes nor canonical caspases [19]. However, it should be noted that the activation of caspase-like proteases have been detected during certain types of cell death [20], but the physiological consequences of these metacaspases remains unclear. As such, autophagic cell death is one of the primary means of cell death in plants, and has been observed in Arabidopsis during developmental cell death as well as the pathogen-triggered hypersensitive response [19].
In plants, the tracheary element of the xylem serves as a means of water-conducting vessels. During tracheal development in Arabidopsis, the tracheary elements undergo programmed cell death in order to remove the contents from the center of the tracheal vessel. In Arabidopsis, autophagy has been shown to be activated during tracheary element differentiation in cell culture, and that the autophagy gene, Atg5, was required for cell death during tracheary differentiation [21]. This indicates that autophagic cell death can occur during plant development.
The hypersensitive-response in plants serves as a mechanism to prevent the spread of pathogens, and is characterized by localized cell death around a region of infection. Interestingly, during the Arabidopsis hypersensitive-response, autophagy appears to be able to function in both a pro-survival and pro-death manner depending on the context of the infection [19]. Interestingly, this context-specific dual use of autophagy is similar to the roles that autophagy can play in tumor cells as either a pro-survival or pro-death mechanism. In Arabidopsis, days after infection, loss of autophagy was shown to result in the inability to prevent the spread of cell death in non-infected cells around the site of infection [22], indicating that autophagy is required for survival. Conversely, autophagy has also been shown to be induced in infected cells shortly after infection, and hypersensitive-response cell death was suppressed by autophagy gene mutants [23]. As such, it appears that the use of autophagy as a means to either prevent the spread of disease-induced cell death or as a mechanism to kill uninfected cells depends on disease contexts, and the time after infection.
Drosophila melanogaster
During the development of Drosophila from a larva to an adult fly, the steroid hormone 20-hydroxyecdysone (ecdysone) signals for many obsolete larval tissues to undergo programmed destruction. Two of these tissues, the larval midgut and salivary glands, degrade through programmed autophagic cell death.
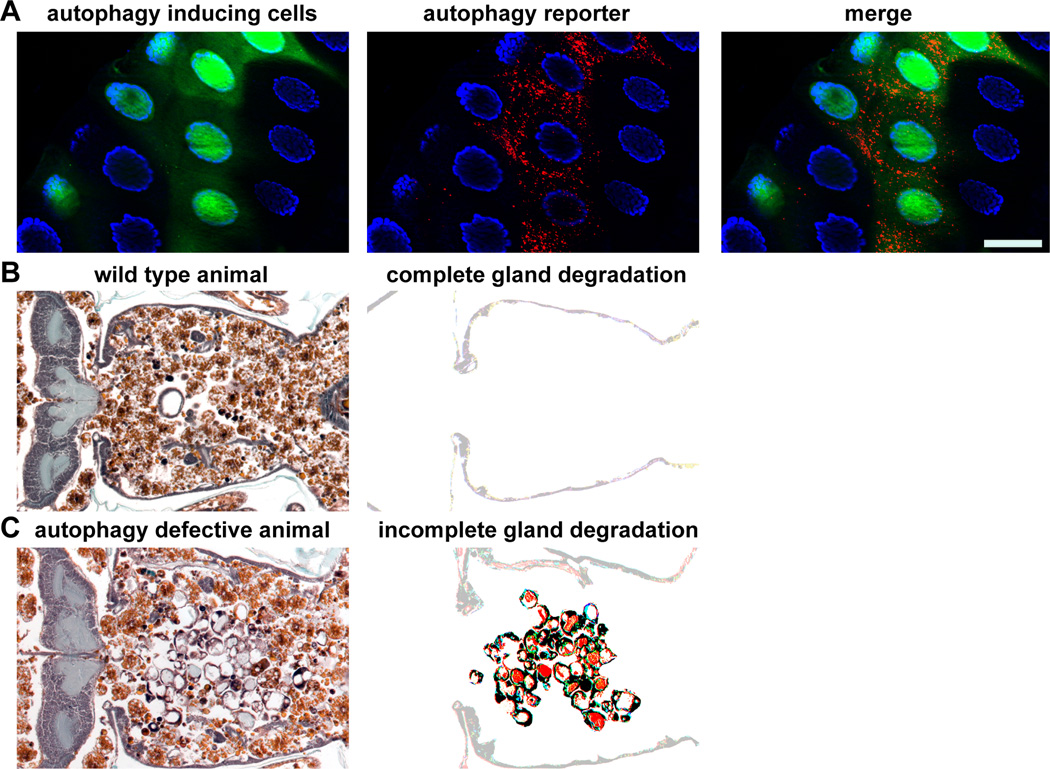
Just prior to its destruction, the larval midgut of Drosophila comprises a large amount of the total volume of larval tissue. At puparium formation, ecdysone signals for the destruction of this tissue, and within four hours, the entire midgut has essentially died [24]. The destruction of this tissue in toto is a classic hallmark of the type II cell death described by Schweichel and Merker [8]. Upon closer examination, high levels of autophagy can be observed during midgut degradation [24], and when autophagy genes are mutated, the midgut fails to degrade [25](Fig. 1). Interestingly, at the time of cell death, midgut cells are TUNEL-positive indicating DNA fragmentation, a sign that caspases are active. However, caspases play no role in the destruction of the midgut as their inhibition does not hinder cell death [25]. Currently, no involvement of phagocytes has ever been observed during midgut cell death. Therefore, autophagy is the sole known cellular process necessary for the self-degradation of the larval midgut during cell death and degradation, and autophagy is important for a programmed cell size reduction and clearance of mitochondria from these dying cells [26].
Figure 1. Autophagy in the larval salivary glands of Drosophila melanogaster is necessary for cell death.
A) A larval salivary gland that expresses an autophagy inducing gene only in green fluorescent protein (GFP, green) cells. Autophagy is visualized by using an autophagy reporter that is expressed in all cells and consists of a mCherry tagged Atg8a protein that localizes to autophagosomes and autolysosomes. Nuclei are stained with DAPI (blue). Scale bar represents 50µm.
B) A histology section of a wild type animal in which the salivary glands degrade normally. Note the lack of any salivary gland material (enhanced in the right image). The anterior of the animal is on the left of the image and the posterior of the animal is on the right of the image.
C) A histology section of an animal in which autophagy is defective. Note the large amount of salivary gland material (enhanced in the right image). The anterior of the animal is on the left of the image and the posterior of the animal is on the right of the image.
Approximately 12 hours after initiation of midgut cell death, another pulse of ecdysone signals for the degradation of the larval salivary glands in toto. Like the midgut, during cell death, salivary gland cells become TUNEL positive, and no phagocytes appear to be involved [27, 28]. Additionally, high levels of autophagy occur, and genetic inhibition of autophagy results in incomplete gland degradation [29, 30]. However, unlike the midgut, in addition to autophagy, caspases are also necessary for salivary gland destruction. Interestingly, these two processes appear to function independently of each other, and both are needed to ensure complete tissue degradation. Therefore, when one process, either autophagy or caspases, is inhibited, partial degradation of the salivary glands still occurs [30]. While caspases and autophagy apparently function independently of each other in the salivary glands, their inductions appear to be linked as genes involved in steroid signaling including the Ecdysone Receptor, nuclear receptor coregulators and the histone H3 lysine trimethyl demethylase dUTX are necessary for the transcription of both caspase and autophagy genes [31, 32].
The core ATG genes are necessary for the induction of autophagy in multiple contexts including starvation and programmed cell death during the life cycle of Drosophila. However, the means of regulating the induction in specific contexts are different. The mechanisms that control autophagy as a starvation response are well understood, and the mTOR complex plays a pivotal role in nutrient sensing and directly regulates the induction of autophagy via ATG proteins [33]. Interestingly, during larval development, mTOR signaling is required for cellular growth in the salivary glands and midgut. Additionally, the Hippo signaling pathway is required for proper growth in the salivary glands. As these processes prevent the induction of autophagy, inhibition of these pathways is necessary to allow for the induction of autophagy during programmed cell death [30, 34, 35]. It is assumed that the arrest of these growth signaling pathways in both of these tissues is because animals stop feeding at this stage in development, but how this dietary information is transmitted to these tissues, if insulin-like peptides are involved, and if steroid signaling regulates this event remains unclear. While growth arrest is critical to allow for the induction of autophagy, the exact mechanisms of how autophagy is induced during cell death remain relatively unknown. For example, specific genes such as the engulfment receptor Draper, the IP3 kinase, IP3K2, the IP3 receptor, and Calmodulin have been shown to regulate autophagy specific during salivary gland degradation [18, 28]. Although steroid signaling may regulate their function, it is unclear how these genes specifically regulate the autophagy machinery. Furthermore, the E1 enzyme ATG7 is not required for autophagy in dying fly midgut cells even though it is required for most other autophagy that has been studied, including starvation-induced autophagy in Drosophila [26]. In order to better understand the regulatory differences in the uses of autophagy further investigation is needed.
Vertebrates
Since its description [8], autophagic cell death has been observed in multiple cell types during development in a variety of vertebrates, including mouse, rat, chick, and frog (reviewed [36]). Unfortunately, our knowledge of autophagic cell death in vertebrates is vague. Therefore, the evidence for autophagic cell death in vertebrates is based almost exclusively on observations in which dying cells showed high levels of autophagy with no evidence of apoptosis. As such, it remains unclear if autophagy actually plays an active role in the death of some or all of these examples. This important gap in our knowledge needs to be overcome through in depth genetic studies of these vertebrates’ dying cells in which autophagic cell death has been reported.
While it is obvious that more complete genetic studies are needed to fully understand the roles autophagy can play in vertebrate programmed cell death, a few studies have attempted to determine the role autophagy may or may not play during mammalian development. For example, embryoid bodies derived from cells lacking ATG genes failed to undergo cavitation, a process that is reminiscent of developmental programmed cell death [37]. However, it was concluded that in this context autophagy is used to signal to clear the cell corpses and not as a means to achieve cell death. It should be noted that this study was done with embyroid bodies, and, may not be a complete representation of what occurs during true animal development. Therefore, further studies are needed.
Although analyses of a role of autophagy in dying animals is lacking, multiple studies in derived cell line suggest the possibility that autophagic cell death occurs in mammals. For example, Ras-induced autophagy was sufficient to trigger caspase-independent cell death [38]. Additionally, activation of autophagy was shown to be sufficient to kill cultured cells when exposed to the autophagy-inducing peptide, Tat-Beclin 1, in a process termed autosis [39]. Moreover, autosis was also observed in vivo during cerebral hypoxic-ischemic injury in rats. While these findings are not in developmental contexts, and are in vitro or in non-physiological conditions, they indicate that autophagy can play an active role during cell death.
AUTOPHAGY AND PROMOTION OF CELL DEATH
Descriptive studies originally suggested that autophagy controls cell death [8, 36]. However, the clear role of autophagy in promoting health and survival in many cell contexts indicates that mechanistic studies are needed to clearly determine the difference in how autophagy can promote cell death [40]. In addition, to be able to modulate autophagy for therapeutic purposes it is useful to know the mechanisms underlying the promotion of cell death by autophagy versus cell health and survival by autophagy. Unfortunately, it remains unclear how autophagy can promote cellular death.
Several possibilities have been proposed for how autophagy may promote cell death, and multiple mechanisms may exist. One possibility is that key survival factors are selectively recruited into autophagosomes for degradation, and multiple studies support this possibility. For example, programmed cell death occurs during fly oogenesis, and the inhibitor of apoptosis Bruce was shown to co-localize with autophagosomes as well as accumulate in autophagy-defective cells [41]. In addition, recruitment of cytoplasmic catalase into autophagosomes was shown to lead to elevated ROS and cell death in L929 cells [42].
An alternative model is that high levels of autophagy may deplete mitochondria and metabolic substrates, and that this could cause a type of metabolic catastrophe that causes cell death. Although it is unclear if this actually occurs, data from flies suggest this possibility. For example, activation of autophagy by mis-expression of the Atg1 kinase is sufficient to kill multiple cell types, including the larval fat body, salivary glands and midgut [26, 30, 43]. Interestingly, this cell death in fat body is delayed by expression of the caspase inhibitor p35, while it is caspase-independent in salivary glands [30, 43].
Recent work in mammalian cells suggests multiple mechanisms for how autophagy may promote cell death. In one case, Ras-triggered cell death was associated with Bcl-2 family member Noxa displacement of Mcl-1 from Beclin1 that led to autophagy-dependent cell death [38]. By contrast, Tat-Beclin 1-triggered autophagic cell death depends on Na(+),K(+)-ATPase function [39], and while it is possible this functions in other dying cells that depend on autophagy, this has never been previously reported. Although it is intriguing to consider such mechanisms, more work needs to be done to be certain these processes function in animals.
CONCLUSIONS
When cells die, apoptosis is often the only form of cell death considered. Autophagic cell death has been shown to play pivotal roles in the development of protists, insects, plants, and potentially mammals. Furthermore, autophagy plays a critical role in immune response-associated cell death of plants. While we have focused on only a few genetic model systems, evidence for autophagic cell death in other organisms in these kingdoms exists. Therefore, the potential prominence of this form of cell death could be greatly under-estimated.
It is safe to assume that when considering the biomass of the Earth, a large portion of the programmed cell death may occur though the utilization of autophagy. This is supported by our knowledge that plants and insects are dependent on autophagy for a large amount of cell death, and that taxa within these groups represent a large amount of biomass on Earth. It is important to note that plant crops and potentially a large number of pollinators, such as honey bees, likely rely on the use of autophagy during cell death. Additionally, parasitic protists such as certain trichomonads have been shown to undergo autophagic cell death [44]. Furthermore, autophagic cell death has been observed during the development of pests, such as disease transmitting mosquitoes [45]. Therefore, understanding the roles and regulation of autophagy during cell death not only can help crop health and growth, but may also help in the prevention infectious diseases.
Evidence indicates that at least some of the mechanisms that regulate autophagic cell death are conserved between evolutionarily distant species [17, 18]. Furthermore, observations suggest that autophagic cell death occurs during development in higher metazoans such as mammals [36]. As such, a better understanding of autophagic cell death in model organisms may help us understand autophagic cell death in humans. As the apoptotic machinery is often disrupted in diseases, such as cancer, autophagic cell death may offer an alternative mechanism to kill tumor cells. Future studies focused on understanding the regulation and function of autophagy in cell death could significantly advance science, agriculture, and the treatment of disease.
Acknowledgements
Research related to this subject is supported by NIH grants GM079431 and CA159314 to E.H.B.. E.H.B. is an Ellison Medical Foundation Scholar.
References
- 1.Mizushima N, Komatsu M. Autophagy: Renovation of Cells and Tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, Ait-Mohamed O, Ait-Si-Ali S, Akematsu T, Akira S, Al-Younes HM, Al-Zeer MA, Albert ML, Albin RL, Alegre-Abarrategui J, Aleo MF, Alirezaei M, Almasan A, Almonte-Becerril M, Amano A, Amaravadi R, Amarnath S, Amer AO, Andrieu-Abadie N, Anantharam V, Ann DK, Anoopkumar-Dukie S, Aoki H, Apostolova N, Arancia G, Aris JP, Asanuma K, Asare NY, Ashida H, Askanas V, Askew DS, Auberger P, Baba M, Backues SK, Baehrecke EH, Bahr BA, Bai XY, Bailly Y, Baiocchi R, Baldini G, Balduini W, Ballabio A, Bamber BA, Bampton ETBG, Bartholomew CR, Bassham DC, Bast RC, Jr, Batoko H, Bay BH, Beau I, Béchet DM, Begley TJ, Behl C, Behrends C, Bekri S, Bellaire B, Bendall LJ, Benetti L, Berliocchi L, Bernardi H, Bernassola F, Besteiro S, Bhatia-Kissova I, Bi X, Biard-Piechaczyk M, Blum JS, Boise LH, Bonaldo P, Boone DL, Bornhauser BC, Bortoluci KR, Bossis I, Bost F, Bourquin JP, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady NR, Brancolini C, Brech A, Brenman JE, Brennand A, Bresnick EH, Brest P, Bridges D, Bristol ML, Brookes PS, Brown EJ, Brumell JH, Brunetti-Pierri N, Brunk UT, Bulman DE, Bultman SJ, Bultynck G, Burbulla LF, Bursch W, Butchar JP, Buzgariu W, Bydlowski SP, Cadwell K, Cahová M, Cai D, Cai J, Cai Q, Calabretta B, Calvo-Garrido J, Camougrand N, Campanella M, Campos-Salinas J, Candi E, Cao L, Caplan AB, Carding SR, Cardoso SM, Carew JS, Carlin CR, Carmignac V, Carneiro LA, Carra S, Caruso RA, Casari G, Casas C, Castino R, Cebollero E, Cecconi F, Celli J, Chaachouay H, Chae HJ, Chai CY, Chan DC, Chan EY, Chang RC, Che CM, Chen CC, Chen GC, Chen GQ, Chen M, Chen Q, Chen SS, Chen W, Chen X, Chen X, Chen X, Chen YG, Chen Y, Chen Y, Chen YJ, Chen Z, Cheng A, Cheng CH, Cheng Y, Cheong H, Cheong JH, Cherry S, Chess-Williams R, Cheung ZH, Chevet E, Chiang HL, Chiarelli R, Chiba T, Chin LS, Chiou SH, Chisari FV, Cho CH, Cho DH, Choi AM, Choi D, Choi KS, Choi ME, Chouaib S, Choubey D, Choubey V, Chu CT, Chuang TH, Chueh SH, Chun T, Chwae YJ, Chye ML, Ciarcia R, Ciriolo MR, Clague MJ, Clark RS, Clarke PG, Clarke R, Codogno P, Coller HA, Colombo MI, Comincini S, Condello M, Condorelli F, Cookson MR, Coombs GH, Coppens I, Corbalan R, Cossart P, Costelli P, Costes S, Coto-Montes A, Couve E, Coxon FP, Cregg JM, Crespo JL, Cronjé MJ, Cuervo AM, Cullen JJ, Czaja MJ, D'Amelio M, Darfeuille-Michaud A, Davids LM, Davies FE, De Felici M, de Groot JF, de Haan CA, De Martino L, De Milito A, De Tata V, Debnath J, Degterev A, Dehay B, Delbridge LM, Demarchi F, Deng YZ, Dengjel J, Dent P, Denton D, Deretic V, Desai SD, Devenish RJ, Di Gioacchino M, Di Paolo G, Di Pietro C, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dikic I, Dinesh-Kumar SP, Ding WX, Distelhorst CW, Diwan A, Djavaheri-Mergny M, Dokudovskaya S, Dong Z, Dorsey FC, Dosenko V, Dowling JJ, Doxsey S, Dreux M, Drew ME, Duan Q, Duchosal MA, Duff K, Dugail I, Durbeej M, Duszenko M, Edelstein CL, Edinger AL, Egea G, Eichinger L, Eissa NT, Ekmekcioglu S, El-Deiry WS, Elazar Z, Elgendy M, Ellerby LM, Eng KE, Engelbrecht AM, Engelender S, Erenpreisa J, Escalante R, Esclatine A, Eskelinen EL, Espert L, Espina V, Fan H, Fan J, Fan QW, Fan Z, Fang S, Fang Y, Fanto M, Fanzani A, Farkas T, Farré JC, Faure M, Fechheimer M, Feng CG, Feng J, Feng Q, Feng Y, Fésüs L, Feuer R, Figueiredo-Pereira ME, Fimia GM, Fingar DC, Finkbeiner S, Finkel T, Finley KD, Fiorito F, Fisher EA, Fisher PB, Flajolet M, Florez-McClure ML, Florio S, Fon EA, Fornai F, Fortunato F, Fotedar R, Fowler DH, Fox HS, Franco R, Frankel LB, Fransen M, Fuentes JM, Fueyo J, Fujii J, Fujisaki K, Fujita E, Fukuda M, Furukawa RH, Gaestel M, Gailly P, Gajewska M, Galliot B, Galy V, Ganesh S, Ganetzky B, Ganley IG, Gao FB, Gao GF, Gao J, Garcia L, Garcia-Manero G, Garcia-Marcos M, Garmyn M, Gartel AL, Gatti E, Gautel M, Gawriluk TR, Gegg ME, Geng J, Germain M, Gestwicki JE, Gewirtz DA, Ghavami S, Ghosh P, Giammarioli AM, Giatromanolaki AN, Gibson SB, Gilkerson RW, Ginger ML, Ginsberg HN, Golab J, Goligorsky MS, Golstein P, Gomez-Manzano C, Goncu E, Gongora C, Gonzalez CD, Gonzalez R, González-Estévez C, González-Polo RA, Gonzalez-Rey E, Gorbunov NV, Gorski S, Goruppi S, Gottlieb RA, Gozuacik D, Granato GE, Grant GD, Green KN, Gregorc A, Gros F, Grose C, Grunt TW, Gual P, Guan JL, Guan KL, Guichard SM, Gukovskaya AS, Gukovsky I, Gunst J, Gustafsson AB, Halayko AJ, Hale AN, Halonen SK, Hamasaki M, Han F, Han T, Hancock MK, Hansen M, Harada H, Harada M, Hardt SE, Harper JW, Harris AL, Harris J, Harris SD, Hashimoto M, Haspel JA, Hayashi S, Hazelhurst LA, He C, He YW, Hébert MJ, Heidenreich KA, Helfrich MH, Helgason GV, Henske EP, Herman B, Herman PK, Hetz C, Hilfiker S, Hill JA, Hocking LJ, Hofman P, Hofmann TG, Höhfeld J, Holyoake TL, Hong MH, Hood DA, Hotamisligil GS, Houwerzijl EJ, Høyer-Hansen M, Hu B, Hu CA, Hu HM, Hua Y, Huang C, Huang J, Huang S, Huang WP, Huber TB, Huh WK, Hung TH, Hupp TR, Hur GM, Hurley JB, Hussain SN, Hussey PJ, Hwang JJ, Hwang S, Ichihara A, Ilkhanizadeh S, Inoki K, Into T, Iovane V, Iovanna JL, Ip NY, Isaka Y, Ishida H, Isidoro C, Isobe K, Iwasaki A, Izquierdo M, Izumi Y, Jaakkola PM, Jäättelä M, Jackson GR, Jackson WT, Janji B, Jendrach M, Jeon JH, Jeung EB, Jiang H, Jiang H, Jiang JX, Jiang M, Jiang Q, Jiang X, Jiang X, Jiménez A, Jin M, Jin S, Joe CO, Johansen T, Johnson DE, Johnson GV, Jones NL, Joseph B, Joseph SK, Joubert AM, Juhász G, Juillerat-Jeanneret L, Jung CH, Jung YK, Kaarniranta K, Kaasik A, Kabuta T, Kadowaki M, Kagedal K, Kamada Y, Kaminskyy VO, Kampinga HH, Kanamori H, Kang C, Kang KB, Kang KI, Kang R, Kang YA, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kanthasamy A, Karantza V, Kaushal GP, Kaushik S, Kawazoe Y, Ke PY, Kehrl JH, Kelekar A, Kerkhoff C, Kessel DH, Khalil H, Kiel JA, Kiger AA, Kihara A, Kim DR, Kim DH, Kim DH, Kim EK, Kim HR, Kim JS, Kim JH, Kim JC, Kim JK, Kim PK, Kim SW, Kim YS, Kim Y, Kimchi A, Kimmelman AC, King JS, Kinsella TJ, Kirkin V, Kirshenbaum LA, Kitamoto K, Kitazato K, Klein L, Klimecki WT, Klucken J, Knecht E, Ko BC, Koch JC, Koga H, Koh JY, Koh YH, Koike M, Komatsu M, Kominami E, Kong HJ, Kong WJ, Korolchuk VI, Kotake Y, Koukourakis MI, Kouri Flores JB, Kovács AL, Kraft C, Krainc D, Krämer H, Kretz-Remy C, Krichevsky AM, Kroemer G, Krüger R, Krut O, Ktistakis NT, Kuan CY, Kucharczyk R, Kumar A, Kumar R, Kumar S, Kundu M, Kung HJ, Kurz T, Kwon HJ, La Spada AR, Lafont F, Lamark T, Landry J, Lane JD, Lapaquette P, Laporte JF, László L, Lavandero S, Lavoie JN, Layfield R, Lazo PA, Le W, Le Cam L, Ledbetter DJ, Lee AJ, Lee BW, Lee GM, Lee J, Lee JH, Lee M, Lee MS, Lee SH, Leeuwenburgh C, Legembre P, Legouis R, Lehmann M, Lei HY, Lei QY, Leib DA, Leiro J, Lemasters JJ, Lemoine A, Lesniak MS, Lev D, Levenson VV, Levine B, Levy E, Li F, Li JL, Li L, Li S, Li W, Li XJ, Li YB, Li YP, Liang C, Liang Q, Liao YF, Liberski PP, Lieberman A, Lim HJ, Lim KL, Lim K, Lin CF, Lin FC, Lin J, Lin JD, Lin K, Lin WW, Lin WC, Lin YL, Linden R, Lingor P, Lippincott-Schwartz J, Lisanti MP, Liton PB, Liu B, Liu CF, Liu K, Liu L, Liu QA, Liu W, Liu YC, Liu Y, Lockshin RA, Lok CN, Lonial S, Loos B, Lopez-Berestein G, López-Otín C, Lossi L, Lotze MT, Lőw P, Lu B, Lu B, Lu B, Lu Z, Luciano F, Lukacs NW, Lund AH, Lynch-Day MA, Ma Y, Macian F, MacKeigan JP, Macleod KF, Madeo F, Maiuri L, Maiuri MC, Malagoli D, Malicdan MC, Malorni W, Man N, Mandelkow EM, Manon S, Manov I, Mao K, Mao X, Mao Z, Marambaud P, Marazziti D, Marcel YL, Marchbank K, Marchetti P, Marciniak SJ, Marcondes M, Mardi M, Marfe G, Mariño G, Markaki M, Marten MR, Martin SJ, Martinand-Mari C, Martinet W, Martinez-Vicente M, Masini M, Matarrese P, Matsuo S, Matteoni R, Mayer A, Mazure NM, McConkey DJ, McConnell MJ, McDermott C, McDonald C, McInerney GM, McKenna SL, McLaughlin B, McLean PJ, McMaster CR, McQuibban GA, Meijer AJ, Meisler MH, Meléndez A, Melia TJ, Melino G, Mena MA, Menendez JA, Menna-Barreto RF, Menon MB, Menzies FM, Mercer CA, Merighi A, Merry DE, Meschini S, Meyer CG, Meyer TF, Miao CY, Miao JY, Michels PA, Michiels C, Mijaljica D, Milojkovic A, Minucci S, Miracco C, Miranti CK, Mitroulis I, Miyazawa K, Mizushima N, Mograbi B, Mohseni S, Molero X, Mollereau B, Mollinedo F, Momoi T, Monastyrska I, Monick MM, Monteiro MJ, Moore MN, Mora R, Moreau K, Moreira PI, Moriyasu Y, Moscat J, Mostowy S, Mottram JC, Motyl T, Moussa CE, Müller S, Muller S, Münger K, Münz C, Murphy LO, Murphy ME, Musarò A, Mysorekar I, Nagata E, Nagata K, Nahimana A, Nair U, Nakagawa T, Nakahira K, Nakano H, Nakatogawa H, Nanjundan M, Naqvi NI, Narendra DP, Narita M, Navarro M, Nawrocki ST, Nazarko TY, Nemchenko A, Netea MG, Neufeld TP, Ney PA, Nezis IP, Nguyen HP, Nie D, Nishino I, Nislow C, Nixon RA, Noda T, Noegel AA, Nogalska A, Noguchi S, Notterpek L, Novak I, Nozaki T, Nukina N, Nürnberger T, Nyfeler B, Obara K, Oberley TD, Oddo S, Ogawa M, Ohashi T, Okamoto K, Oleinick NL, Oliver FJ, Olsen LJ, Olsson S, Opota O, Osborne TF, Ostrander GK, Otsu K, Ou JH, Ouimet M, Overholtzer M, Ozpolat B, Paganetti P, Pagnini U, Pallet N, Palmer GE, Palumbo C, Pan T, Panaretakis T, Pandey UB, Papackova Z, Papassideri I, Paris I, Park J, Park OK, Parys JB, Parzych KR, Patschan S, Patterson C, Pattingre S, Pawelek JM, Peng J, Perlmutter DH, Perrotta I, Perry G, Pervaiz S, Peter M, Peters GJ, Petersen M, Petrovski G, Phang JM, Piacentini M, Pierre P, Pierrefite-Carle V, Pierron G, Pinkas-Kramarski R, Piras A, Piri N, Platanias LC, Pöggeler S, Poirot M, Poletti A, Poüs C, Pozuelo-Rubio M, Prætorius-Ibba M, Prasad A, Prescott M, Priault M, Produit-Zengaffinen N, Progulske-Fox A, Proikas-Cezanne T, Przedborski S, Przyklenk K, Puertollano R, Puyal J, Qian SB, Qin L, Qin ZH, Quaggin SE, Raben N, Rabinowich H, Rabkin SW, Rahman I, Rami A, Ramm G, Randall G, Randow F, Rao VA, Rathmell JC, Ravikumar B, Ray SK, Reed BH, Reed JC, Reggiori F, Régnier-Vigouroux A, Reichert AS, Reiners JJ, Jr, Reiter RJ, Ren J, Revuelta JL, Rhodes CJ, Ritis K, Rizzo E, Robbins J, Roberge M, Roca H, Roccheri MC, Rocchi S, Rodemann HP, Rodríguez de Córdoba S, Rohrer B, Roninson IB, Rosen K, Rost-Roszkowska MM, Rouis M, Rouschop KM, Rovetta F, Rubin BP, Rubinsztein DC, Ruckdeschel K, Rucker EB, 3rd, Rudich A, Rudolf E, Ruiz-Opazo N, Russo R, Rusten TE, Ryan KM, Ryter SW, Sabatini DM, Sadoshima J, Saha T, Saitoh T, Sakagami H, Sakai Y, Salekdeh GH, Salomoni P, Salvaterra PM, Salvesen G, Salvioli R, Sanchez AM, Sánchez-Alcázar JA, Sánchez-Prieto R, Sandri M, Sankar U, Sansanwal P, Santambrogio L, Saran S, Sarkar S, Sarwal M, Sasakawa C, Sasnauskiene A, Sass M, Sato K, Sato M, Schapira AH, Scharl M, Schätzl HM, Scheper W, Schiaffino S, Schneider C, Schneider ME, Schneider-Stock R, Schoenlein PV, Schorderet DF, Schüller C, Schwartz GK, Scorrano L, Sealy L, Seglen PO, Segura-Aguilar J, Seiliez I, Seleverstov O, Sell C, Seo JB, Separovic D, Setaluri V, Setoguchi T, Settembre C, Shacka JJ, Shanmugam M, Shapiro IM, Shaulian E, Shaw RJ, Shelhamer JH, Shen HM, Shen WC, Sheng ZH, Shi Y, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shintani T, Shirihai OS, Shore GC, Sibirny AA, Sidhu SB, Sikorska B, Silva-Zacarin EC, Simmons A, Simon AK, Simon HU, Simone C, Simonsen A, Sinclair DA, Singh R, Sinha D, Sinicrope FA, Sirko A, Siu PM, Sivridis E, Skop V, Skulachev VP, Slack RS, Smaili SS, Smith DR, Soengas MS, Soldati T, Song X, Sood AK, Soong TW, Sotgia F, Spector SA, Spies CD, Springer W, Srinivasula SM, Stefanis L, Steffan JS, Stendel R, Stenmark H, Stephanou A, Stern ST, Sternberg C, Stork B, Strålfors P, Subauste CS, Sui X, Sulzer D, Sun J, Sun SY, Sun ZJ, Sung JJ, Suzuki K, Suzuki T, Swanson MS, Swanton C, Sweeney ST, Sy LK, Szabadkai G, Tabas I, Taegtmeyer H, Tafani M, Takács-Vellai K, Takano Y, Takegawa K, Takemura G, Takeshita F, Talbot NJ, Tan KS, Tanaka K, Tanaka K, Tang D, Tang D, Tanida I, Tannous BA, Tavernarakis N, Taylor GS, Taylor GA, Taylor JP, Terada LS, Terman A, Tettamanti G, Thevissen K, Thompson CB, Thorburn A, Thumm M, Tian F, Tian Y, Tocchini-Valentini G, Tolkovsky AM, Tomino Y, Tönges L, Tooze SA, Tournier C, Tower J, Towns R, Trajkovic V, Travassos LH, Tsai TF, Tschan MP, Tsubata T, Tsung A, Turk B, Turner LS, Tyagi SC, Uchiyama Y, Ueno T, Umekawa M, Umemiya-Shirafuji R, Unni VK, Vaccaro MI, Valente EM, Van den Berghe G, van der Klei IJ, van Doorn W, van Dyk LF, van Egmond M, van Grunsven LA, Vandenabeele P, Vandenberghe WP, Vanhorebeek I, Vaquero EC, Velasco G, Vellai T, Vicencio JM, Vierstra RD, Vila M, Vindis C, Viola G, Viscomi MT, Voitsekhovskaja OV, von Haefen C, Votruba M, Wada K, Wade-Martins R, Walker CL, Walsh CM, Walter J, Wan XB, Wang A, Wang C, Wang D, Wang F, Wang F, Wang G, Wang H, Wang HG, Wang HD, Wang J, Wang K, Wang M, Wang RC, Wang X, Wang X, Wang YJ, Wang Y, Wang Z, Wang ZC, Wang Z, Wansink DG, Ward DM, Watada H, Waters SL, Webster P, Wei L, Weihl CC, Weiss WA, Welford SM, Wen LP, Whitehouse CA, Whitton JL, Whitworth AJ, Wileman T, Wiley JW, Wilkinson S, Willbold D, Williams RL, Williamson PR, Wouters BG, Wu C, Wu DC, Wu WK, Wyttenbach A, Xavier RJ, Xi Z, Xia P, Xiao G, Xie Z, Xie Z, Xu DZ, Xu J, Xu L, Xu X, Yamamoto A, Yamamoto A, Yamashina S, Yamashita M, Yan X, Yanagida M, Yang DS, Yang E, Yang JM, Yang SY, Yang W, Yang WY, Yang Z, Yao MC, Yao TP, Yeganeh B, Yen WL, Yin JJ, Yin XM, Yoo OJ, Yoon G, Yoon SY, Yorimitsu T, Yoshikawa Y, Yoshimori T, Yoshimoto K, You HJ, Youle RJ, Younes A, Yu L, Yu L, Yu SW, Yu WH, Yuan ZM, Yue Z, Yun CH, Yuzaki M, Zabirnyk O, Silva-Zacarin E, Zacks D, Zacksenhaus E, Zaffaroni N, Zakeri Z, Zeh HJ, 3rd, Zeitlin SO, Zhang H, Zhang HL, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang MY, Zhang XD, Zhao M, Zhao YF, Zhao Y, Zhao ZJ, Zheng X, Zhivotovsky B, Zhong Q, Zhou CZ, Zhu C, Zhu WG, Zhu XF, Zhu X, Zhu Y, Zoladek T, Zong WX, Zorzano A, Zschocke J, Zuckerbraun B. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 4.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, Wolf DH. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- 5.Harding TM, Morano KA, Scott SV, Klionsky DJ. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J Cell Bio. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harding TM, Hefner-Gravink A, Thumm M, Klionsky DJ. Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein. J Biol Chem. 1996;271:17621–17624. doi: 10.1074/jbc.271.30.17621. [DOI] [PubMed] [Google Scholar]
- 7.Klionsky DJ, Cregg JM, Dunn WAJ, Emr SD, Sakai Y, Sandoval IV, Sibirny A, Subramani S, Thumm M, Veenhuis M, Ohsumi Y. A unified nomenclature for yeast autophagy-related genes. Dev Cell. 2003;5:539–545. doi: 10.1016/s1534-5807(03)00296-x. [DOI] [PubMed] [Google Scholar]
- 8.Schweichel J-U, Merker H-J. The morphology of various types of cell death in prenatal tissues. Teratology. 1973;7:253–266. doi: 10.1002/tera.1420070306. [DOI] [PubMed] [Google Scholar]
- 9.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittingham WF, Raper KB. Non-viability of stalk cells in Dictyostelium. Proc Natl Acad Sci U S A. 1960;46:427–438. doi: 10.1073/pnas.46.5.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 12.Giusti C, Tresse E, Luciani MF, Golstein P. Autophagic cell death: analysis in Dictyostelium. Biochim Biophys Acta. 2009;1793:1422–1431. doi: 10.1016/j.bbamcr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 13.de Chastellier C, Ryter A. Changes of the cell surface and of the digestive apparatus of Dictyostelium discoideum during the staruation period triggering aggregation. J Cell Biol. 1977;75:218–236. doi: 10.1083/jcb.75.1.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornillon S, Foa C, Davoust J, Buonavista N, Gross JD, Golstein P. Programmed cell death in Dictyostelium. J Cell Sci. 1994;107:2691–2704. doi: 10.1242/jcs.107.10.2691. [DOI] [PubMed] [Google Scholar]
- 15.Town CD, Gross JD, Kay RR. Cell differentiation without morphogenesis in Dictyostelium discoideum. Nature. 1976;262:717–719. doi: 10.1038/262717a0. [DOI] [PubMed] [Google Scholar]
- 16.Levraud JP, Adam M, Luciani MF, de Chastellier C, Blanton RL, Golstein P. Dictyostelium cell death: early emergence and demise of highly polarized paddle cells. J Cell Biol. 2003;160:1105–1114. doi: 10.1083/jcb.200212104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lam D, Kosta A, Luciani MF, Golstein P. The inositol 1,4,5-trisphosphate receptor is required to signal autophagic cell death. Mol Biol Cell. 2008;19:691–700. doi: 10.1091/mbc.E07-08-0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson C, Ambros V, Baehrecke EH. miR-14 regulates autophagy during developmental cell death by targeting ip3-kinase 2. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.09.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofius D, Munch D, Bressendorff S, Mundy J, Petersen M. Role of autophagy in disease resistance and hypersensitive response-associated cell death. Cell Death Differ. 2011;18:1257–1262. doi: 10.1038/cdd.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsiatsiani L, Timmerman E, De Bock PJ, Vercammen D, Stael S, van de Cotte B, Staes A, Goethals M, Beunens T, Van Damme P, Gevaert K, Van Breusegem F. The Arabidopsis metacaspase9 degradome. Plant Cell. 2013;25:2831–2847. doi: 10.1105/tpc.113.115287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon SI, Cho HJ, Jung JH, Yoshimoto K, Shirasu K, Park OK. The Rab GTPase RabG3b functions in autophagy and contributes to tracheary element differentiation in Arabidopsis. Plant J. 2010;64:151–164. doi: 10.1111/j.1365-313X.2010.04315.x. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Hofius D, Schultz-Larsen T, Joensen J, Tsitsigiannis DI, Petersen NH, Mattsson O, Jørgensen LB, Jones JD, Mundy J, Petersen M. Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell. 2009;137:773–783. doi: 10.1016/j.cell.2009.02.036. [DOI] [PubMed] [Google Scholar]
- 24.Lee C-Y, Cooksey BAK, Baehrecke EH. Steroid regulation of midgut cell death during Drosophila development. Dev Biol. 2002;250:101–111. doi: 10.1006/dbio.2002.0784. [DOI] [PubMed] [Google Scholar]
- 25.Denton D, Shravage B, Simin R, Mills K, Berry DL, Baehrecke EH, Kumar S. Autophagy, not apoptosis, is essential for midgut cell death in Drosophila. Curr Biol. 2009;19:1741–1746. doi: 10.1016/j.cub.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang T-K, Shravage BV, Hayes SD, Powers CM, Simin RT, Harper JW, Baehrecke EH. Uba1 functions in Atg7- and Atg3-independent autophagy. Nat Cell Biol. 2013;15:1067–1078. doi: 10.1038/ncb2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin DN, Baehrecke EH. Caspases function in autophagic cell death in Drosophila . Development. 2004;131:275–284. doi: 10.1242/dev.00933. [DOI] [PubMed] [Google Scholar]
- 28.McPhee CK, Logan MA, Freeman MR, EH B. Activation of autophagy during cell death requires the engulfment receptor Draper. Nature. 2010;465:1093–1096. doi: 10.1038/nature09127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee C-Y, Baehrecke EH. Steroid regulation of autophagic programmed cell death during development. Development. 2001;128:1443–1455. doi: 10.1242/dev.128.8.1443. [DOI] [PubMed] [Google Scholar]
- 30.Berry DL, Baehrecke EH. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ihry RJ, Bashirullah A. Genetic control of specificity to steroid-triggered responses in Drosophila. Genetics. 2014;196:767–780. doi: 10.1534/genetics.113.159707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Denton D, Aung-Htut MT, Lorensuhewa N, Nicolson S, Zhu W, Mills K, Cakouros D, Bergmann A, Kumar S. UTX coordinates steroid hormone-mediated autophagy and cell death. Nat Commun. 2013;4:2916. doi: 10.1038/ncomms3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Dutta S, Baehrecke EH. Warts is required for PI3K-regulated growth arrest, autophagy, and autophagic cell death in Drosophila. Curr Biol. 2008;18:1466–1475. doi: 10.1016/j.cub.2008.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denton D, Chang TK, Nicolson S, Shravage B, Simin R, Baehrecke EH, Kumar S. Relationship between growth arrest and autophagy in midgut programmed cell death in Drosophila. Cell Death Differ. 2012;19:1299–1307. doi: 10.1038/cdd.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clarke PGH. Developmental cell death: morphological diversity and multiple mechanisms. Anat Embryol. 1990;181:195–213. doi: 10.1007/BF00174615. [DOI] [PubMed] [Google Scholar]
- 37.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, Hogan RN, Gilpin C, Levine B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 38.Elgendy M, Sheridan C, Brumatti G, Martin SJ. Oncogenic Ras-induced expression of Noxa and Beclin-1 promotes autophagic cell death and limits clonogenic survival. Mol Cell. 2011;42:23–35. doi: 10.1016/j.molcel.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 39.Liu Y, Shoji-Kawata S, Sumpter RMJ, Wei Y, Ginet V, Zhang L, Posner B, Tran KA, Green DR, Xavier RJ, Shaw SY, Clarke PG, Puyal J, Levine B. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc Natl Acad Sci U S A. 2013;110:20364–20371. doi: 10.1073/pnas.1319661110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nezis IP, Shravage BV, Sagona AP, Lamark T, Bjørkøy G, Johansen T, Rusten TE, Brech A, Baehrecke EH, Stenmark H. Autophagic degradation of dBruce controls DNA fragmentation in nurse cells during late Drosophila melanogaster oogenesis. J Cell Biol. 2010;190:523–531. doi: 10.1083/jcb.201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu L, Wan F, Dutta S, Welsh S, Liu Z, Freundt E, Baehrecke EH, Lenardo MJ. Autophagic programmed cell death by selective catalase degradation. Proc Natl Acad Sci USA. 2006;103:4952–4957. doi: 10.1073/pnas.0511288103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol. 2007;17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mariante RM, Vancini RG, Benchimol M. Cell death in trichomonads: new insights. Histochem Cell Biol. 2006;125:545–556. doi: 10.1007/s00418-005-0098-5. [DOI] [PubMed] [Google Scholar]
- 45.Fernandes KM, Neves CA, Serrão JE, Martins GF. Aedes aegypti midgut remodeling during metamorphosis. Parasitol Int. 2014;63:506–512. doi: 10.1016/j.parint.2014.01.004. [DOI] [PubMed] [Google Scholar]