Abstract
Objectives
In adults with unexplained pancreatitis, the yield of complete gene versus select exosome sequencing on mutation detection and distinguishing clinical characteristics associated with mutations requires clarification. We sought to: 1) compare frequency of mutations identified using difference techniques and 2) compare clinical characteristics between adults with and without mutations.
Methods
Cohort study of adults with unexplained pancreatitis who underwent genetic testing between 1/2008-12/2012. We compare probabilities of having a positive mutation with complete gene sequencing versus alternatives, and describe differences in characteristics among patients with and without mutations.
Results
Of 370 patients, 67 (18%) had a genetic mutation; 24 (6%) were high-risk. Mutations were significantly more prevalent with use of complete sequencing (42%) versus other approaches (8%, p<0.0001). The majority (44/67, 66%) with a mutation had no family history. Those with high-risk mutations were more likely to have and family history of chronic pancreatitis (21% v. 4%, p=0.002). Patients with pancreas divisum were more likely to have mutations (27% v. 14%, p=0.0007).
Conclusion
Among individuals with adult-onset pancreatic disease, the probability of finding any mutation, including high-risk, is significantly higher using complete gene sequencing. The impact on patients and providers requires further investigation.
Keywords: genetics, pancreatitis, idiopathic pancreatitis, gene sequencing, pancreas divisum
Introduction
Patients with adult onset, idiopathic chronic pancreatitis (CP) and recurrent acute pancreatitis (RAP) increasingly undergo testing to evaluate for a genetic predisposition. Genes of interest include the cystic fibrosis transmembrane conductance regulator (CFTR), the protease serine 1 (PRSS1), the serine protease inhibitor, Kazal type 1 (SPINK1), and chymotrypsin C (CTRC).1–18 Inheritance patterns may be autosomal dominant (PRSS1), autosomal recessive (CFTR, SPINK1), or complex (e.g., heterozygous CFTR, SPINK1, and CTRC); the latter includes gene-environment and gene-gene (i.e., compound heterozygosity) interactions.1, 4
It is increasingly recognized that a genetic predisposition to pancreatitis is not limited to a pediatric population, as some individuals will develop symptoms during adulthood (age > 18 years). Among small series of adults with CP or RAP, pancreas divisum has been associated with a higher prevalence of genetic mutations.19–21 The clinical significance of a genetic mutation as the sole risk factor or covariate with environmental factors (e.g., alcohol and smoking) requires clarification.22
Our objectives were two-fold. First, while newer assays utilize complete gene sequence analysis, their impact on detecting mutations in an adult population as compared to other methods is unclear. We compare the frequency of positive gene mutations identified using different assays. Second, since genetic testing is expensive and identifying a mutation (even one with unclear clinical significance) may have unintended long-term implications for patients (e.g., insurability, anxiety), we sought to identify clinical characteristics associated with positive genetic mutations. This may inform clinicians of at-risk groups who would be most likely to have a mutation.
Methods
Study Design and Patient Selection
We conducted a retrospective cohort study of all symptomatic adults who underwent pancreas-specific genetic testing between January 2008 and December 2012. Patients were identified from our pancreatobiliary clinical practice at Indiana University Health University Hospital, a tertiary referral center for chronic pancreatitis and other pancreatic disorders. We used an institutional review board-approved clinical database that contains prospectively entered clinical data. We excluded patients age <18 at the time of genetic testing. The study received approval from our local institutional review board prior to the collection of data and none of the authors declare any conflict of interests.
The indications for genetic testing included: idiopathic RAP, idiopathic CP, or symptoms consistent with CP (e.g., steatorrhea responding to oral pancreatic enzymes, abdominal pain consistent with CP). Medical records were abstracted for patient demographics, the presence of pancreatitis-related symptoms including abdominal pain (not associated with an episode of acute pancreatitis), exocrine insufficiency, endocrine insufficiency and age of symptom onset. When available, we recorded specific morphologic abnormalities identified by imaging (i.e., computed tomography (CT), magnetic resonance imaging (MRI), endoscopic ultrasound (EUS), and endoscopic retrograde pancreatography (ERCP)): pancreas divisum, pancreatic calcifications, and main pancreatic duct stricture/dilated main pancreatic duct, among others. We recorded family history of acute pancreatitis, RAP, CP, and pancreatic cancer.
Definitions
Acute pancreatitis was defined as acute abdominal pain associated with increased serum pancreatic enzyme levels (amylase and/or lipase) > 3 times the upper limit of normal value or imaging changes consistent with acute pancreatitis;23 documentation of such findings were required in the medical records. RAP was defined as two or more discrete episodes of documented acute pancreatitis.24 Patients were diagnosed with CP based on standard radiographic criteria, including the presence of pancreatic calcifications on CT, EUS, or ERCP; pancreatic ductal abnormalities on pancreatography obtained by magnetic resonance cholangiopancreatography (MRCP) or ERCP (using the Cambridge classification); ≥5 endosonographic criteria on EUS; marked pancreatic atrophy on CT or MRCP; and exocrine insufficiency based on a history of steatorrhea.25–28 A diagnosis of pancreas divisum required confirmation via ERCP or MRCP.29–31
Genetic Testing
The decision to perform genetic testing was always at the discretion of the treating physician. During the study period, we used several third party assays for genetic testing (table 1). Since 2011, we have used complete gene sequence analysis (Ambry Genetics) for all of our patients. Prior to 2011, several other entities were used: CFTR testing by other entities (Genzyme and Quest Diagnostics) used enzymatic amplification of known mutations (97 and 32, respectively); PRSS1 testing by other entities (Mayo Clinic and University of Pittsburgh Medical Center) employed gene sequence analysis for a smaller portion of the gene where mutations were known to impact pancreatitis risk (table 1). For descriptive purposes in this study, “complete gene sequencing” refers to assays conducted by Ambry Genetics.
Table 1.
Summary of modalities for genetic testing used during the study period
| Genetic Test | CFTR | PRSS1 | SPINK1 | CTRC |
|---|---|---|---|---|
| Ambry Genetics (Aliso Viejo, CA, USA) | DNA sequencing of 5′ UTR, 27 exons, all introns, polyT status and TG repeats | DNA sequencing of 5′ UTR, exons 1–5 and all introns | DNA sequencing of 5′ UTR, exons 1–4 and all introns | DNA sequencing of 5′ UTR, exons 1–8 and all introns |
| Genzyme Corporation (Cambridge, MA, USA) | Enzymatic amplification of DNA for 97 mutations | |||
| Quest Diagnostics (Madison, NJ, USA) | Enzymatic amplification of DNA for 32 mutations | |||
| Mayo Clinic (Rochester, MN, USA) | DNA sequencing of exons 2 and 3 | |||
| University of Pittsburgh Medical Center (Pittsburgh, PA, USA) | DNA sequencing of exons 2 and 3 |
5′ UTR = 5′ untranslated region
We defined a positive genetic test as a mutation detected for any of the 4 genes tested. We also present the frequency of high-risk mutations32–34, which we defined as: 1) a single copy mutation of PRSS1; 2) homozygous mutations of CFTR, SPINK1 or CTRC; or 3) compound heterozygous mutations of CFTR, SPINK1 and/or CTRC.
Analysis Plan
The cohort was dichotomized by the presence of any genetic mutation. Given the clinical significance of individuals having mutations known to have high risk for leading to pancreatic disease, we also present the characteristics of this subgroup. We used descriptive statistics (mean ± standard deviation, proportion with 95% confidence intervals) to present patient and phenotypic characteristics. To examine differences between study populations (those with any genetic mutation versus all others and those with a high-risk mutation versus all others), we used comparative statistics (Student’s t test or Fisher’s exact test) where appropriate.
Results
Prevalence of pancreas-specific genetic mutations
During the five-year study period, we identified 370 symptomatic adults who underwent genetic testing through the adult pancreatobiliary practice at Indiana University. Of these, 67 (18.1%) had a genetic mutation, 24 (6.5%) of which were high-risk (figure 1). A per-patient summary of mutations is available (supplementary table). Among patients who underwent testing for CFTR (n=359), 109 (30.4%) were via complete gene sequencing (Ambry), 249 (69.4%) were via enzymatic amplification for 97 mutations (Genzyme), and 1 (0.3%) was via enzymatic amplification for 32 mutations (Quest). For PRSS1 mutations (n=267), 106 (39.7%) were via complete gene sequencing (Ambry) and 161 (60.3%) were via DNA sequencing of exons 2 and 3 (Mayo Clinic and University of Pittsburgh). All SPINK1 (n=107) and CTRC (n=94) assays were performed using complete gene sequencing (Ambry).
Figure 1.
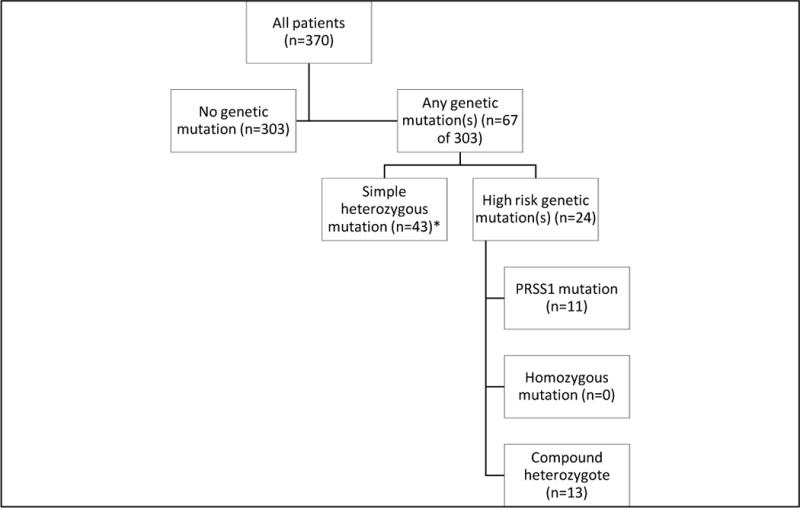
Schema: Adult individuals who underwent genetic testing for pancreas-specific mutations
* Defined as a heterozygous mutation of CFTR, CTRC or SPINK1
The probability of a detecting any mutation and a high-risk mutation significantly increased when complete gene sequencing was performed (table 2). Patients tested by complete gene sequencing had a mutation detected in 42.2% of cases as compared to 8.1% when using any other method (p<0.0001). Similarly, patients having a high-risk mutation were identified in 18.3% of cases using complete gene sequencing versus 1.5% when using other modalities (p<0.001). These differences remained significant for the subgroup of individuals tested for PRSS1 or CFTR mutations.
Table 2.
Probability of an abnormal genetic test
| Gene mutation | Proportion with mutation | P value¥¥¥ | |
|---|---|---|---|
| Complete Gene Sequencing (n=109) N, (%) |
Other tests (n=261) N, (%) |
||
| Any mutation | 46 (42.2%) | 21 (8.1%) | < 0.001 |
| High-risk mutation | 20 (18.3%) | 4 (1.5%) | < 0.001 |
| PRSS1 (n=266)¥ | 8 (7.5%) | 3 (1.9%) | 0.029 |
| SPINK1 (n=107)¥ | 15 (14.0%) | N/A¥¥ | |
| CFTR (n=359)¥ | 29 (26.6%) | 20 (8.0%) | <0.0001 |
| CTRC (n=94)¥ | 7 (7.5%) | N/A¥¥ | |
No patients were found to have a homozygous mutation
All patients who underwent testing for SPINK1 and CTRC mutations utilized complete gene sequencing.
Fisher’s exact test
Clinical history
The majority (354 of 370, 96%) of patients were Caucasian. There were no significant differences in patient characteristics of those with and without genetic mutations (table 3). Age of presentation was younger for individuals with high-risk mutations (32.3±15.4, p=0.05 vs. all others), although those having no mutations also presented with an average age < 40 (39.5±16.1). Patients with high-risk mutations were more likely to report a family history of chronic pancreatitis (20.8%, p=0.002 vs. all others), but there was no significant difference between those having any mutation (7.6%) and no mutation (3.7%, p=0.18). There was a greater likelihood of having a family history of acute pancreatitis among high-risk individuals, but this did not reach statistical significance (p=0.17). The prevalence of a family history of pancreatic cancer was similar in all groups. Importantly, 44 of the 67 (65.7%) patients with a mutation had no family history of acute pancreatitis, chronic pancreatitis or pancreatic cancer. Among those with high-risk mutations (n=24), 14 (58.3%) had no family history.
Table 3.
Patient Characteristics
| Variable | No mutation (n=303) | Any mutation (n=67) | High-risk mutation¥ (n=24) | P value (no vs. any mutation) | P value (high-risk mutation vs. all others) |
|---|---|---|---|---|---|
| Age of symptom onset (mean, SD) | 39.5 (16.1) | 35.5 (17.5) | 32.3 (15.4) | 0.08 | 0.05 |
| Age of presentation (mean, SD) | 44.5 (15.5) | 43.0 (16.8) | 42.5 (13.9) | 0.48 | 0.57 |
| Male sex, n (%) | 102 (33.7) | 25 (37.3) | 8 (33.3) | 0.57 | 1.0 |
| BMI (mean, SD) | 27.8 (6.7) | 26.8 (6.9) | 27.1 (6.1) | 0.30 | 0.69 |
|
Smoking status, n (%) • Current • Former • Never |
93 (31.5) 55 (18.6) 147 (49.8) |
22 (33.9) 11 (16.9) 32 (49.2) |
12 (52.2) 2 (8.7) 9 (39.1) |
0.91 | 0.10 |
|
Alcohol use, n (%) • Current • Former • Never |
61 (20.6) 91 (30.7) 144 (48.7) |
10 (15.4) 27 (41.5) 28 (43.1) |
5 (21.7) 11 (47.8) 7 (30.4) |
0.23 | 0.18 |
| Family History, n (%) | |||||
| • Acute Pancreatitis | 51 (17.2) | 16 (24.2) | 7 (29.2) | 0.22 | 0.17 |
| • Chronic Pancreatitis | 11 (3.7) | 5 (7.6) | 5 (20.8) | 0.18 | 0.002 |
| • Pancreatic Cancer | 28 (9.4) | 7 (10.6) | 3 (12.5) | 0.82 | 0.72 |
Defined as any PRSS1 mutation, compound heterozygote mutation, or homozygous mutation for SPINK1, CFTR or CTRC gene.
Presenting symptoms (abdominal pain, steatorrhea, diabetes, and weight loss) were similar between patients having no mutation and any mutation, including those with a high-risk mutation (table 4). Patients with any mutation (67.2%) were more likely to present with recurrent acute pancreatitis compared to those without a mutation (47.5%, p=0.004). There was no significant difference in the rate of CP when comparing patients with no genetic mutation with any genetic mutations (p=0.12) or a high-risk mutation (p=0.13). The prevalence of moderate/severe CP and having a history of a single episode of acute pancreatitis was similar among all populations.
Table 4.
Clinical presentation
| Variable | No mutation (n=303) | Any mutation (n=67) | High-risk mutation¥ (n=24) | P value (no v. any mutation) | P value (high-risk v. all others) |
|---|---|---|---|---|---|
| Acute Pancreatitis | 210 (69.3) | 53 (79.1) | 20 (83.3) | 0.14 | 0.24 |
| Recurrent Acute pancreatitis | 144 (47.5) | 45 (67.2) | 17 (70.8) | 0.004 | 0.06 |
| Chronic pancreatitis | 184 (60.9) | 48 (71.6) | 19 (79.2) | 0.12 | 0.125 |
| • Moderate/Severe Chronic Pancreatitis |
93 (30.7) | 27 (40.9) | 10 (41.7) | 0.11 | 0.22 |
| Symptoms | |||||
| • Abdominal pain | 261 (86.1) | 58 (86.6) | 20 (83.3) | 1.0 | 0.76 |
| • Steatorrhea | 82 (27.1) | 14 (20.9) | 5 (20.8) | 0.36 | 0.64 |
| • Diabetes | 68 (22.4) | 10 (14.9) | 4 (16.7) | 0.19 | 0.80 |
| • Weight loss | 131 (43.2) | 29 (43.3) | 12 (50.0) | 1.0 | 0.53 |
n (%) for all variables
Fisher’s exact test was used for all comparisons.
Defined as any PRSS1 mutation, compound heterozygote mutation, or homozygous mutation for SPINK1, CFTR or CTRC gene.
Phenotypic characteristics
Despite similarities in age of symptom onset, age of presentation, and clinical history, there were important phenotypic differences between those with and without a pancreas-specific genetic mutation (table 5). In the entire cohort, there were 59 (15.9%) patients with pancreas divisum, of who 48 (81.4%) were diagnosed by ERCP and 11 (18.6%) by MRCP. Patients without a mutation were less likely to have pancreas divisum (13.6%) compared to 26.9% of those with any mutation (p=0.01) and 33.3% of those with a high-risk mutation (p=0.04). This remained significant when the analysis was limited to the subgroup (n=279) that underwent ERCP. Pancreatic calcifications and main duct strictures were more frequently diagnosed among patients with genetic mutations, particularly those classified as high risk.
Table 5.
Phenotypic Characteristics
| Variable | No mutation (n=303) | Any mutation (n=67) | High-risk mutation¥ (n=24) | P value (no v. any mutation) | P value (high-risk v. all others) |
|---|---|---|---|---|---|
| Pancreas divisum | 41 (13.6) | 18 (26.9) | 8 (33.3) | 0.01 | 0.04 |
| • Confirmed by ERCP (n=279)¥ | 32 (14.2) | 14 (26.4) | 6 (35.3) | 0.04 | 0.04 |
| Pancreatic calcifications | 41 (18.1) | 14 (26.4) | 7 (41.2) | 0.18 | 0.05 |
| Pancreatic duct stricture | 46 (20.4) | 16 (30.2) | 8 (47.1) | 0.14 | 0.029 |
n (%) for all variables
Fisher’s exact test was used for all comparisons.
279 of 370 patients underwent ERCP as part of their diagnostic and treatment plan. These proportions represent the subgroup that underwent ERCP.
Discussion
Genetic mutations among adults with unexplained pancreatic disease
Consistent with previous studies, the prevalence of genetic mutations in a cohort of adult individuals with pancreatic disease undergoing genetic testing is substantial (18.1%),1–3, 5, 6, 8–11, 13–18 though high-risk mutations are identified less frequently (6.5%). Importantly, use of a test that sequences the entire gene(s) of interest identified significantly (greater than 5-fold) more mutations, including those classified as high-risk (greater than 12-fold). Prior studies using genetic sequencing of either the entire gene or a large percentage of it have also shown high rates of finding genetic mutations with a 45–49% rate of mutations found in the CFTR, PRSS1 and SPINK1 genes in similar patient populations.1, 5, 35 Alternative analytics for mutations such as high resolution DNA melting may provide rapid and accurate diagnostics at a lower cost.36 Other genetic testing modalities utilized in our cohort either sequenced a smaller portion of the gene or used other techniques such as enzymatic amplification of predefined mutations. The rate of detected mutations in our cohort likely would have been higher if all individuals had undergone gene sequence analysis of all 4 genes. The clinical significance and impact of identifying many of these mutations will require further clarification and not all mutations identified to date have been associated with the development of pancreatic disease. Specifically, counseling of patients and relatives, screening for pancreatic cancer and impact of potential therapies (both endoscopic and surgical) in patients with genetic mutations and pancreatic disease such as chronic pancreatitis will require further investigation. Furthermore, the pathogenic significance of some mutations (e.g., c.592-24C>T) is unknown; future studies may impact the classification of high-risk and unknown risk mutations presented in this paper.
Clinical factors do not distinguish adults with pancreas-specific genetic mutations
Age of symptom onset, age of clinical presentation, and presenting symptoms were not associated with having a genetic mutation; those having a high-risk mutation developed overt symptoms in their early 30s (mean age 32) while those with no mutation were in their late 30s (mean age 39). Since we limited our analysis to individuals who underwent genetic testing, our study is susceptible to a selection bias so this observation should be interpreted with caution: the treating physician suspected a genetic abnormality based on family history, age, and/or idiopathic nature of their illness. Nevertheless, these demographic and clinical factors were similar among those with and without mutations. Further, the absence of a family history in more than half of individuals with a mutation (including those deemed high-risk) highlights their variable penetrance: autosomal dominant (minority), autosomal recessive (one parent with two mutations or two parents each with a single mutation), and complex genetic-environment interactions. Our cohort represents a “real-world” adult population that would be considered for genetic testing.
Family history is not a prerequisite for pancreas-specific genetic mutations
Prior studies have shown that 4% of patients with familial (non-autosomal dominant) CP have a mutation in the CTRC gene14 and up to 86% have a mutation in the SPINK1 gene12. While patients with high-risk mutations in our cohort were more likely to have a family history of CP (20.8%), the majority of individuals with high-risk or any mutations had no known family history of chronic pancreatitis; therefore, the absence of a family history should not dissuade the clinician from pursuing a genetic evaluation.
Previous reports have shown that patients with RAP have an increased rate of mutations in the CFTR gene, ranging from 12-32%1, 15, 16, as well as an increased rate of compound heterozygosity1. Numerous studies using a variety of methods for genetic testing have reported more mutations in patients with CP, including CFTR (18–44%),1, 4–7, 17 PRSS1 (2%),1 SPINK1 (5–23%),1, 5, 6, 10–12, 18, 37, 38 and CTRC (3–12%).13, 14. In our cohort, 67% of patients with a mutation had RAP (71% of high-risk patients). We also found that patients with a high-risk genetic mutation had a 79% probability of having chronic pancreatitis. Thus, our findings are in agreement with prior studies that the presence of RAP or CP as opposed to a single attack of acute pancreatitis alone increases the chances of having a positive genetic test.
Pancreas divisum is more prevalent among patients with genetic abnormalities
Consistent with previous studies, we found a higher incidence of mutations among individuals with pancreas divisum. Previous studies reported high rates of CFTR (47–70%), PRSS1 (16%), and SPINK1 (16%) mutations among patients with CP and pancreas divisum.19–21 Therefore, pancreas divisum is an important phenotypic clue that a patient is at-risk for a genetic mutation (although the majority of patients with divisum were still negative for a mutation). Prior studies evaluating the effectiveness of minor papilla endotherapy in the setting of both RAP and pancreas divisum suggest that therapy is more beneficial in patients that have not yet developed chronic pancreatitis.39, 40 It should be noted that genetic testing was not evaluated in these studies and future work evaluating RAP and pancreas divisum should consider whether the presence of a genetic mutation correlates with response to minor papilla endotherapy.
Summary
Our study is limited by its retrospective design and potential for detection or recall bias, particularly when looking at issues such as family history. In addition, our study population included individuals with pancreatic disease who underwent genetic testing; therefore, the treating physician suspected a genetic abnormality in all cases and so our results do not represent the prevalence of mutations in a general population of adults with pancreatic disease. Nevertheless, our observations are important since the sampling frame represents a “real-world” cohort of adults who are likely to undergo genetic testing. A history of RAP, CP and pancreas divisum increase the likelihood of a high-risk mutation, whereas the absence of a family history of RAP or CP does not rule out its possibility. Finally, the prevalence of both high-risk and any pancreas-specific mutation significantly increased with the use of complete gene sequence analysis. The clinical implications of this increase require further investigation.
In conclusion, patient characteristics that are associated with a pancreas-specific genetic mutation include a personal history of RAP, CP, family history of CP, and the presence of pancreas divisum. However, the majority of adults with a positive test had no significant family history. We have also shown that complete gene sequence analysis increases the probability of all and high-risk genetic mutations, keeping in mind that not all mutations have been characterized in terms of their clinical significance or association with pancreatitis. The potential clinical implications of identifying a genetic mutation are several: response to medical, surgical or endoscopic therapy may differ in this population, and physicians should push more aggressively for lifestyle modifications including smoking and alcohol cessation. Future prospective studies should evaluate differences in outcomes of individuals with pancreatic disease and genetic mutations, and specific medical therapies that compensate for genetic abnormalities are awaited.
Supplementary Material
Acknowledgments
Sources of funding: GC is supported by a grant (K23DK095148) from the NIH.
Abbreviations
- CP
chronic pancreatitis
- RAP
recurrent acute pancreatitis
- CFTR
cystic fibrosis transmembrane conductance regulator
- PRSS1
protease serine 1
- SPINK1
serine protease inhibitor, Kazal type 1
- CTRC
chymotrypsin C
- CT
computed tomography
- MRI
magnetic resonance imaging
- EUS
endoscopic ultrasound
- ERCP
endoscopic retrograde cholangiopancreatography
- MRCP
magnetic resonance cholangiopancreatography
- 5′ UTR
5′ untranslated region
Footnotes
Conflicts of interest: For the remaining authors none are declared.
References
- 1.Keiles S, Kammesheidt A. Identification of CFTR, PRSS1, and SPINK1 mutations in 381 patients with pancreatitis. Pancreas. 2006;33:221–7. doi: 10.1097/01.mpa.0000232014.94974.75. [DOI] [PubMed] [Google Scholar]
- 2.Truninger K, Malik N, Ammann RW, et al. Mutations of the cystic fibrosis gene in patients with chronic pancreatitis. Am J Gastroenterol. 2001;96:2657–61. doi: 10.1111/j.1572-0241.2001.04047.x. [DOI] [PubMed] [Google Scholar]
- 3.Pezzilli R, Morselli-Labate AM, Mantovani V, et al. Mutations of the CFTR gene in pancreatic disease. Pancreas. 2003;27:332–6. doi: 10.1097/00006676-200311000-00011. [DOI] [PubMed] [Google Scholar]
- 4.Noone PG, Zhou Z, Silverman LM, et al. Cystic fibrosis gene mutations and pancreatitis risk: relation to epithelial ion transport and trypsin inhibitor gene mutations. Gastroenterology. 2001;121:1310–9. doi: 10.1053/gast.2001.29673. [DOI] [PubMed] [Google Scholar]
- 5.Weiss FU, Simon P, Bogdanova N, et al. Complete cystic fibrosis transmembrane conductance regulator gene sequencing in patients with idiopathic chronic pancreatitis and controls. Gut. 2005;54:1456–60. doi: 10.1136/gut.2005.064808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Audrezet MP, Chen JM, Le Marechal C, et al. Determination of the relative contribution of three genes-the cystic fibrosis transmembrane conductance regulator gene, the cationic trypsinogen gene, and the pancreatic secretory trypsin inhibitor gene-to the etiology of idiopathic chronic pancreatitis. Eur J Hum Genet. 2002;10:100–6. doi: 10.1038/sj.ejhg.5200786. [DOI] [PubMed] [Google Scholar]
- 7.Cohn JA, Friedman KJ, Noone PG, et al. Relation between mutations of the cystic fibrosis gene and idiopathic pancreatitis. N Engl J Med. 1998;339:653–8. doi: 10.1056/NEJM199809033391002. [DOI] [PubMed] [Google Scholar]
- 8.Witt H, Luck W, Becker M. A signal peptide cleavage site mutation in the cationic trypsinogen gene is strongly associated with chronic pancreatitis. Gastroenterology. 1999;117:7–10. doi: 10.1016/s0016-5085(99)70543-3. [DOI] [PubMed] [Google Scholar]
- 9.Pfutzer RH, Whitcomb DC. Trypsinogen mutations in chronic pancreatitis. Gastroenterology. 1999;117:1507–8. doi: 10.1016/s0016-5085(99)70312-4. [DOI] [PubMed] [Google Scholar]
- 10.Schneider A, Barmada MM, Slivka A, et al. Clinical characterization of patients with idiopathic chronic pancreatitis and SPINK1 Mutations. Scand J Gastroenterol. 2004;39:903–4. doi: 10.1080/00365520410006710. [DOI] [PubMed] [Google Scholar]
- 11.Drenth JP, te Morsche R, Jansen JB. Mutations in serine protease inhibitor Kazal type 1 are strongly associated with chronic pancreatitis. Gut. 2002;50:687–92. doi: 10.1136/gut.50.5.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Threadgold J, Greenhalf W, Ellis I, et al. The N34S mutation of SPINK1 (PSTI) is associated with a familial pattern of idiopathic chronic pancreatitis but does not cause the disease. Gut. 2002;50:675–81. doi: 10.1136/gut.50.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masson E, Chen JM, Scotet V, et al. Association of rare chymotrypsinogen C (CTRC) gene variations in patients with idiopathic chronic pancreatitis. Hum Genet. 2008;123:83–91. doi: 10.1007/s00439-007-0459-3. [DOI] [PubMed] [Google Scholar]
- 14.Rosendahl J, Witt H, Szmola R, et al. Chymotrypsin C (CTRC) variants that diminish activity or secretion are associated with chronic pancreatitis. Nat Genet. 2008;40:78–82. doi: 10.1038/ng.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavestro GM, Zuppardo RA, Bertolini S, et al. Connections between genetics and clinical data: Role of MCP-1, CFTR, and SPINK-1 in the setting of acute, acute recurrent, and chronic pancreatitis. Am J Gastroenterol. 2010;105:199–206. doi: 10.1038/ajg.2009.611. [DOI] [PubMed] [Google Scholar]
- 16.Ockenga J, Stuhrmann M, Ballmann M, et al. Mutations of the cystic fibrosis gene, but not cationic trypsinogen gene, are associated with recurrent or chronic idiopathic pancreatitis. Am J Gastroenterol. 2000;95:2061–7. doi: 10.1111/j.1572-0241.2000.02228.x. [DOI] [PubMed] [Google Scholar]
- 17.Sharer N, Schwarz M, Malone G, et al. Mutations of the cystic fibrosis gene in patients with chronic pancreatitis. N Engl J Med. 1998;339:645–52. doi: 10.1056/NEJM199809033391001. [DOI] [PubMed] [Google Scholar]
- 18.Witt H, Luck W, Hennies HC, et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000;25:213–6. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- 19.Bertin C, Pelletier AL, Vullierme MP, et al. Pancreas divisum is not a cause of pancreatitis by itself but acts as a partner of genetic mutations. Am J Gastroenterol. 2012;107:311–7. doi: 10.1038/ajg.2011.424. [DOI] [PubMed] [Google Scholar]
- 20.Garg PK, Khajuria R, Kabra M, et al. Association of SPINK1 gene mutation and CFTR gene polymorphisms in patients with pancreas divisum presenting with idiopathic pancreatitis. J Clin Gastroenterol. 2009;43:848–52. doi: 10.1097/MCG.0b013e3181a4e772. [DOI] [PubMed] [Google Scholar]
- 21.Choudari CP, Imperiale TF, Sherman S, et al. Risk of pancreatitis with mutation of the cystic fibrosis gene. Am J Gastroenterol. 2004;99:1358–63. doi: 10.1111/j.1572-0241.2004.30655.x. [DOI] [PubMed] [Google Scholar]
- 22.Whitcomb DC. Framework for interpretation of genetic variations in pancreatitis patients. Front Physiol. 2012;3:440. doi: 10.3389/fphys.2012.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banks PA, Bollen TL, Dervenis C, et al. Classification of acute pancreatitis–2012: revision of the Atlanta classification and definitions by international consensus. Gut. 2013;62:102–11. doi: 10.1136/gutjnl-2012-302779. [DOI] [PubMed] [Google Scholar]
- 24.Cote GA, Imperiale TF, Schmidt SE, et al. Similar efficacies of biliary, with or without pancreatic, sphincterotomy in treatment of idiopathic recurrent acute pancreatitis. Gastroenterology. 2012;143:1502–1509 e1. doi: 10.1053/j.gastro.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Axon AT, Classen M, Cotton PB, et al. Pancreatography in chronic pancreatitis: international definitions. Gut. 1984;25:1107–12. doi: 10.1136/gut.25.10.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Catalano MF, Sahai A, Levy M, et al. EUS-based criteria for the diagnosis of chronic pancreatitis: the Rosemont classification. Gastrointest Endosc. 2009;69:1251–61. doi: 10.1016/j.gie.2008.07.043. [DOI] [PubMed] [Google Scholar]
- 27.Sugiyama M, Haradome H, Atomi Y. Magnetic resonance imaging for diagnosing chronic pancreatitis. J Gastroenterol. 2007;42(Suppl 17):108–12. doi: 10.1007/s00535-006-1923-x. [DOI] [PubMed] [Google Scholar]
- 28.Schlaudraff E, Wagner HJ, Klose KJ, et al. Prospective evaluation of the diagnostic accuracy of secretin-enhanced magnetic resonance cholangiopancreaticography in suspected chronic pancreatitis. Magn Reson Imaging. 2008;26:1367–73. doi: 10.1016/j.mri.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Mosler P, Akisik F, Sandrasegaran K, et al. Accuracy of magnetic resonance cholangiopancreatography in the diagnosis of pancreas divisum. Dig Dis Sci. 2012;57:170–4. doi: 10.1007/s10620-011-1823-7. [DOI] [PubMed] [Google Scholar]
- 30.Lai R, Freeman ML, Cass OW, et al. Accurate diagnosis of pancreas divisum by linear-array endoscopic ultrasonography. Endoscopy. 2004;36:705–9. doi: 10.1055/s-2004-825663. [DOI] [PubMed] [Google Scholar]
- 31.Quest L, Lombard M. Pancreas divisum: opinio divisa. Gut. 2000;47:317–9. doi: 10.1136/gut.47.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitcomb DC. Genetic risk factors for pancreatic disorders. Gastroenterology. 2013;144:1292–302. doi: 10.1053/j.gastro.2013.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitcomb DC. Genetics of alcoholic and nonalcoholic pancreatitis. Curr Opin Gastroenterol. 2012;28:501–6. doi: 10.1097/MOG.0b013e328356e7f3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider A, Larusch J, Sun X, et al. Combined bicarbonate conductance-impairing variants in CFTR and SPINK1 variants are associated with chronic pancreatitis in patients without cystic fibrosis. Gastroenterology. 2011;140:162–71. doi: 10.1053/j.gastro.2010.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang MC, Chang YT, Wei SC, et al. Spectrum of mutations and variants/haplotypes of CFTR and genotype-phenotype correlation in idiopathic chronic pancreatitis and controls in Chinese by complete analysis. Clin Genet. 2007;71:530–9. doi: 10.1111/j.1399-0004.2007.00813.x. [DOI] [PubMed] [Google Scholar]
- 36.Montgomery J, Wittwer CT, Kent JO, et al. Scanning the cystic fibrosis transmembrane conductance regulator gene using high-resolution DNA melting analysis. Clin Chem. 2007;53:1891–8. doi: 10.1373/clinchem.2007.092361. [DOI] [PubMed] [Google Scholar]
- 37.Sandhu B, Vitazka P, Ferreira-Gonzalez A, et al. Presence of SPINK-1 variant alters the course of chronic pancreatitis. J Gastroenterol Hepatol. 2011;26:965–9. doi: 10.1111/j.1440-1746.2011.06713.x. [DOI] [PubMed] [Google Scholar]
- 38.Teich N, Bauer N, Mossner J, et al. Mutational screening of patients with nonalcoholic chronic pancreatitis: identification of further trypsinogen variants. Am J Gastroenterol. 2002;97:341–6. doi: 10.1111/j.1572-0241.2002.05467.x. [DOI] [PubMed] [Google Scholar]
- 39.Lehman GA, Sherman S, Nisi R, et al. Pancreas divisum: results of minor papilla sphincterotomy. Gastrointest Endosc. 1993;39:1–8. doi: 10.1016/s0016-5107(93)70001-2. [DOI] [PubMed] [Google Scholar]
- 40.Chacko LN, Chen YK, Shah RJ. Clinical outcomes and nonendoscopic interventions after minor papilla endotherapy in patients with symptomatic pancreas divisum. Gastrointest Endosc. 2008;68:667–73. doi: 10.1016/j.gie.2008.01.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


