Abstract
The generation of human induced pluripotent stem cells (iPS) has raised enormous expectations within the biomedical community due to their potential vast implications in regenerative and personalized medicine. However, reprogramming to iPS is still not fully comprehended. Difficulties found in ascribing specific molecular patterns to pluripotent cells (PSCs), and inherent inter-line and intra-line variability between different PSCs need to be resolved. Additionally, and despite multiple assumptions, it remains unclear whether the current in vitro culturing conditions for the maintenance and differentiation of PSCs do indeed recapitulate the developmental processes observed in vivo. As a consequence, basic questions such as what is the actual nature of PSCs remain unanswered and different theories have emerged in regards to the identity of these valuable cell population. Here we discuss on the published theories for defining PSC identity, the implications that the different postulated models have for the reprogramming field as well as speculate on potential future directions that might be opened once a precise knowledge on the nature of PSCs is accomplished.
Introduction
The zygote is a totipotent cell capable of generating trophoectodermal lineages as well as the inner cell mass (ICM). The cells present in the ICM, defined as pluripotent stem cell (PSCs), give rise to all three different germ cell layers of the embryo and ultimately, the adult individual[1,2]. The precise molecular dynamics leading to the formation of PSCs in vivo as well as their fundamental properties have long remained elusive. The ability to generate PSCs in vitro, may bring new light into these questions as well novel strategies for regenerative medicine[1,3–10]μ[11–15].
Stemness: a fixed identity or a dynamic functional equilibrium?
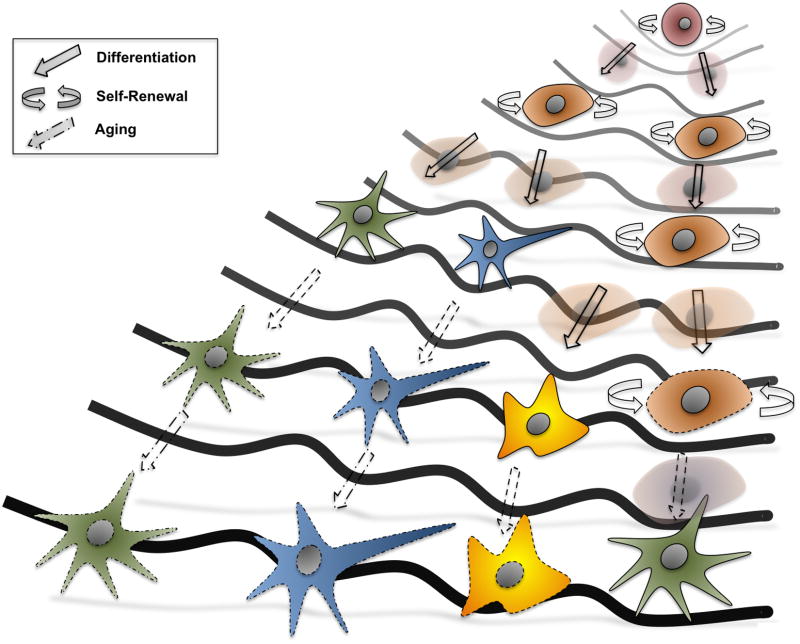
According to the traditional metaphor of embryonic development illustrated by Conrad Waddington, totipotent cells at the top of a hill fall down progressively to appropriate valleys representing different intermediate steps, and ultimately lead to the generation of every other terminally differentiated somatic cell lineage (Figure 1) [16–18].
Figure 1. Revisiting Waddington’s landscape.
The original metaphor of development as depicted by Conrad Waddington presented a simplified representation of cell differentiation to somatic cell lineages. Nowadays, development can be seen as a continuous in where not only adult stem cells can reside in a quiescent state during adulthood, but also where the whole life-span of an individual, including aging processes, is considered.
But what is the nature of the cells at the top of the Waddington landscape? Pluripotent stem cells are considered to represent a naïve ground state that lack expression of genes and protein products typical of more differentiated lineages[8]. Assuming such ground state, differentiation proceeds upon targeted activation of specific gene programs defining specific somatic cellular identities. It is therefore assumed that different pluripotent stem cell lines, whether embryonic stem cells (ESCs) or induced pluripotent stem cells (iPSCs), should present a common molecular signature that defines their pluripotent identity. An alternative theory postulates that pluripotent stem cells could be viewed as a defined functional cellular state, where differential molecular programs (e.g variations in marker expression) can be observed, including the expression of the so-called lineage markers. Assuming lineage marker expression, differentiation of pluripotent cells can be explained by a progressive switching off of certain markers while maintaining, or even potentiating, the expression of other lineage genes for defining final cellular identity[19].
Efforts at defining PSC identity at the molecular level seem to suggest that dynamic transitions occur between different cellular states[19–24]. In such a scenario, differential marker expression can be observed between PSCs despite all different lines presenting functional pluripotent differentiation properties. Nonetheless, evidence for and against one model (that PSCs represent a fixed identity) or another (that PSCs dynamically transit between different functional states in where varying expression of markers can be observed), derive mainly from in vitro experiments. These observations might be influenced or altered by the artificiality of the in vitro systems themselves, and raise the question of which of the two models, if any, apply during in vivo development. For example, it is well known the inter-line and intra-line variability observed depending of the culture system utilized. Culturing conditions for pluripotent cells were originally optimized for the stabilization of an undifferentiated pluripotent state, a stable proliferative phase that may rarely occur during early embryonic development. Additionally, inter-laboratory variability in terms of culturing conditions has also been shown to affect PSC molecular signatures. Small and somewhat uncontrollable variability during PSC culturing (e.g.: the relative activity of growth factors; the rates of binding to their receptors or even small batch-to-batch differences in common reagents used for maintaining PSCs) might additionally affect the patterns of expression of a given set of genes[22,23,25]. Indeed, it is quite clear to the experienced stem cell researcher that despite culture optimization, spontaneous differentiation in pluripotent stem cell cultures occurs relatively frequently. Therefore, it is unclear whether such dynamic variations might be also present during embryogenesis in vivo and whether dynamic expression of “lineage markers” occurs in vivo as opposed to the presence of an invariable and discrete pluripotent cell identity.
Dynamic transitions in biology are not atypical. Examples of dynamic transitions between metastable cellular states have been previously reported when attempting at defining cancer stem cells[26]. As an example, putative stem cells were first isolated from Glioblastoma multiforme (GBMs) on the bases of CD133 expression. CD133 rapidly emerged as a “marker” of glioma stem cells and CD133+ isolated from the tumor mass did seem to fulfill all functional criteria of TICs. CD133+ cells were found not only to self-renew but also to bear differentiation potential. Thus, convincingly demonstrating the “stem-like” nature of the isolated cells. Yet, it was soon after demonstrated that CD133-cells bore the same properties, therefore, questioning the presence of individualized discrete cancer stem cell identities[27–32]. In line with these observations, stem cell-like signatures have been shown to arise during cancer progression, raising the possibility that differentiated cancer cells could transit between two dedifferentiated stem-like states[26,33,34]. Whereas cancer stem cells share defining properties with non-malignant stem cells, namely multilineage differentiation and self-renewing potential, it remains unclear whether such transitions occur in pluripotent cells in vivo and further investigations are required to answer these points.
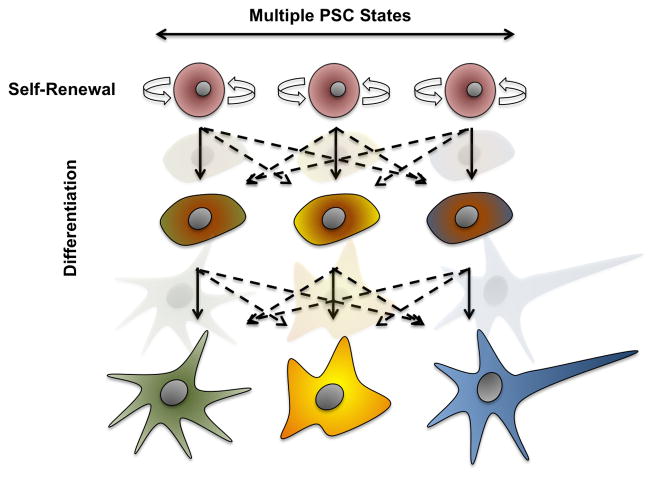
Contrary to cancer biology, the differentiation of stem cells to defined somatic lineages was traditionally considered an irreversible process, however, reprogramming to iPSCs has demonstrated the reversibility of somatic identity to a pluripotent state[5,6]. Since reprogramming occurs upon the forceful induction of given genes and/or epigenetic modifications this may not necessarily represent a natural process. Nevertheless, spontaneous reprogramming does indeed occur and transitions between differentiated somatic cells and dedifferentiated states can be observed in situations other than cancer. Regenerative organisms generate, as a response to injury, a dedifferentiated cellular mass, called blastema, which is able to re-differentiate into all appropriate lost tissues and cell lineages[35–38]. More recently, the concept of dedifferentiation to a plastic stem cell state has also been demonstrated in mammals as a response to injury[39,40]. If a equilibrium between multiple undifferentiated pluripotent states indeed exists in vivo, then a flattened area in where multiple PSCs, solely defined in terms of functionality, should be considered (Figure 2). This situation will raise additional questions such as whether differentiation of all major somatic tissues proceeds in an oligoclonal manner in vivo. If this was true, it might well be that certain populations of pluripotent cells present bias differentiation potential towards specific somatic lineages due to small variance in the expression of certain “lineage” genes (e.g.: the relative levels of defined growth factor receptors), as opposed to monoclonal differentiation where all pluripotent cells present the same identity and relative ability to contribute to all differentiated lineages.
Figure 2. Pluripotent Stem Cell identity: a fixed entity or a compendium of functional states?
Unequivocally, development starts by differentiation of a single cell, the zygote. However, PSCs contained in the Inner Cell Mass (ICM) of a differentiated blastocyst might not only present different molecular signatures but also, potentially transit dynamically between different pluripotent states. Dynamic transitions might indeed not only hamper efforts at defining PSC identity and molecular profiles but might additionally lead to differentiation bias in vivo. It is therefore tempting to speculate that should PSC dynamically change in terms of their genetic and epigenetic programs, oligoclonal differentiation might indeed be responsible for the development of an adult organism. However, current in vitro settings favoring differentiation to multiple lineages for the assessment of pluripotency alongside our inability to precisely model the developing embryonic niche in vitro still compromises our ability to question the models presented here.
Defining the nature of pluripotent cells will ultimately influence current approaches for understanding stem cell biology. Most importantly, precise knowledge on the nature of PSCs and how differentiation actually occurs might open new venues for a better understanding of reprogramming and differentiation leading to the development of regenerative medicine strategies for the treatment of human disease and injury.
Reprogramming by lineage specifiers
In 2006, Takahashi and Yamanaka were able to identify specific factors able to dedifferentiate somatic cell lineages to a pluripotent state[6]. Following these initial studies the search for reprogramming factors focused on genes enriched in ESCs, that could, upon overexpression, drive the conversion of somatic cells to a defined pluripotent state. This has proven to be a successful approach and multiple factors typically expressed in ESCs have been used for the derivation of iPSCs[37,41–45]. Such an approach inherently recognizes pluripotency as a fixed identity where Transcription Factors (TFs) can impose a “pluripotent” genetic program in somatic cells eventually overcoming differentiated gene signatures.
More recently, it has been demonstrated that so-called pluripotent factors act as pioneer TFs whose initial activities do not depend on their direct binding to stem cell gene promoters but rather on their global activity and somewhat unspecific opening the chromatin. Upon binding, pioneer TFs expose specific gene promoters, facilitating and ultimately driving the binding to specific promoters[46–48]. Contrary to trans-reprogramming processes, pioneer activities acting over the chromatin are referred to as cis-reprogramming events. Cis-reprogramming events can indeed explain why the initial phase of reprogramming occurs as a stochastic process[49], as it would imply a randomized series of events that could vary from experiment to experiment until chromatin structures have been modified and specific promoters are readily accessible to defined TFs.
Two elegant reports have shed new light on the actual molecular events underlying reprogramming[49,50]. First, the report by Buganim et al convincingly demonstrates that a first phase of reprogramming occurs randomly, or stochastically, until the expression of endogenous Sox2 is accomplished. Once Sox2 is expressed, and presumably genome-wide chromatin modifications have occurred, a hierarchical phase of gene expression occurs[49]. Another report by the Hochedlinger’s laboratory, further demonstrated the suitability of population level approaches to investigate the mechanisms of reprogramming irrespectively of specific cell identities. In this case, it was observed that the first days of reprogramming lead to global chromatin modifications in line with the pioneer activity of reprogramming factors. Following this first wave of chromatin remodeling, a gradual phase of gene expression changes ultimately lead to the activation of endogenous pluripotent machinery and the formation of iPSCs[50,51].
In line with the theoretical framework provided by these reports, three different recent manuscripts have reported on the generation of iPSCs by using somewhat unconventional strategies (Figure 3)[12,13,52]. Deng and colleagues have shown that critical compounds indispensable for chemical reprogramming, such as DNzep (an inhibitor of the lysine methyltransferase EZH2 (KMT6) that catalyzes trimethylation of lysine 27 on histone H3), target the chromatin. It is possible that by remodeling the chromatin some of the compounds used for chemical reprogramming mimic the pioneer activities of reprogramming TFs[52]. It is however unclear how, and which TFs, drive trans-reprograming events during the second phase of chemical reprogramming once the chromatin has been opened. On support for the need for pioneer activities, the Deng’s laboratory first reported on the reprogramming of murine fibroblasts to iPSCs by using genes typical of differentiated lineages, the so-called lineage specifiers[13]. In this case, it was found that several mesendodermal and ectodermal linage genes sufficed for the replacement of Oct4 and Sox2 respectively. The findings that lineage genes could “balance” differentiation forces and allow for the generation of iPSCs resulted in the hypothesis of a seesaw model for reprogramming. It was speculated that replacement of pluripotency factors was possible due to their actual activities in lineage specification[10,53]. In support of this hypothesis, reprogramming of Neural Progenitor Cells (NPCs), an ectodermal-derived cell population characterized by high levels of Sox2 expression, could indeed be accomplished by sole overexpression of Oct4[54]. By considering pluripotency factors as lineage specifiers, it was further speculated that the establishment of a functional equilibrium of counteracting differentiation forces might suffice for the generation of iPSCs[11,13,14]. Conceptually similar to the report by the Deng laboratory, we have reported on the used of lineage specifiers for the reprogramming of human somatic fibroblasts[12]. Interestingly, we observed that even though highly dynamic, expression of genes typically ascribed to differentiated lineages could be readily observed in undifferentiated pluripotent stem cells, albeit at very low levels. Contrary to the reports from Deng in murine cells, we found a positive loop between GATA3 and Oct4 expression that further complicates our understanding of how reprogramming with lineage specifiers occurs in human cells. The fact is that whereas Oct4 is a driver of mesendodermal genes, one of which is actually GATA3[53], GATA3 also acts alongside cdx2 early on during development for the formation of trophoectodermal lineages. Therefore, it remains obscure whether reprogramming of human cells by lineage specifiers occurs due to pioneer TF activities, by specific balancing of counteracting differentiation forces, or simply by establishing a discrete stem cell identity that might appear, yet been overseen, during human embryonic development[12]. Last, it would be of interest to consider the use of lineage specifiers for in vivo reprogramming. If lineage specifiers demonstrate suitable for in vivo reprogramming, their use might indeed allow for a more efficient and controllable dedifferentiation process minimizing the risk for acquisition of neoplastic properties[55].
Figure 3. Reprogramming and the nature of pluripotent stem cells.
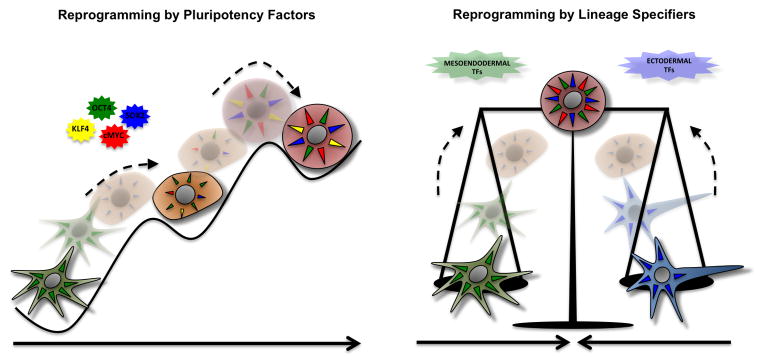
Defining the actual nature of PSCs presents interesting possibilities for reprogramming. On the one hand (left side), if pluripotency represents a discrete and molecularly defined identity, identification of “pluripotent factors” defining a specific pluripotent signature should unequivocally allow for the reprogramming of somatic cells and expectably, a finite number of pluripotent factors should be found. On the contrary (right side), if pluripotency is defined as a balance of counter-acting differentiation forces, there might be un infinite number of combinations and variables such as “differentiation strength” to be taking into account for the reprogramming of somatic cells to iPSCs. Should this be the case, reprogramming strategies should additionally carefully considered the initial somatic cell types and molecularly different types of PSCs might be obtained while still functionally presenting pluripotent properties.
In summary, it remains unclear the precise mechanisms that drive the reprogramming of somatic cells as well as the actual nature of the pluripotent state. However, the presence of lineage markers in undifferentiated PSCs and their use for generating iPSCs blurs traditional cell marker definitions. Ultimately, a profound knowledge of the molecular mechanisms underlying reprogramming and a detailed characterization of the driving forces behind this process might allow for the design of strategies aiming towards the generation of higher quality stem cells, reversion of aging, or even induction of a pro-regenerative state in vivo.
Acknowledgments
We thank M. Schwarz for administrative support. I.S.M. was partially supported by a Nomis Foundation postdoctoral fellowship. A.O. was partially supported by an NIH T32 Cancer Training Grant. Work in the laboratory of J.C.I.B. was supported by grants from NIH (5U01HL107442), CIRM, the G. Harold and Leila Y. Mathers Charitable Foundation and The Leona M. and Harry B. Helmsley Charitable Trust (2012-PG-MED002).
Footnotes
Competing interests
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- *1.Evans M. Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat Rev Mol Cell Biol. 2011;12:680–686. doi: 10.1038/nrm3190. This review provides an excellent first-person overview of the key steps leading to the establishment of the current stem cell reprogramming field by one of his most notable historical contributors. [DOI] [PubMed] [Google Scholar]
- 2.Izpisua Belmonte JC. Reprogramming development and aging: cell differentiation as a malleable process. Curr Opin Cell Biol. 2012;24:713–715. doi: 10.1016/j.ceb.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 4.Kaufman MH, Robertson EJ, Handyside AH, Evans MJ. Establishment of pluripotential cell lines from haploid mouse embryos. J Embryol Exp Morphol. 1983;73:249–261. [PubMed] [Google Scholar]
- **5.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. This paper describes for the first time the reprogramming of human cells to induced Pluripotent Stem Cells (iPSCs) by overexpression of 4 core reprogramming factors. This manuscript represents the first time in where iPSCs were generated. [DOI] [PubMed] [Google Scholar]
- **6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. By using a funnel strategy this manuscript describes the identification of key factors typically expressed in embryonic stem cells, which allow for the conversion of murine somatic cells to an embryonic-like phenotype. This paper describes for the first time the reprogramming of human cells to induced Pluripotent Stem Cells by overexpression of embryonic stem cell factors. [DOI] [PubMed] [Google Scholar]
- **7.Tachibana M, Amato P, Sparman M, Gutierrez NM, Tippner-Hedges R, Ma H, Kang E, Fulati A, Lee H-S, Sritanaudomchai H, et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell. 2013;153:1228–1238. doi: 10.1016/j.cell.2013.05.006. This manuscript describes the generation of embryonic stem cell lines by transfer of somatic human nuclei to enucleated oocytes. This report represents the first demonstration after decades of work in other mammals that therapeutic cloning and derivation of human patient-specific Embryonic Stem Cell lines is possible. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Welling M, Geijsen N. Uncovering the true identity of naïve pluripotent stem cells. Trends Cell Biol. 2013;23:442–448. doi: 10.1016/j.tcb.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 10.Loh KM, Lim B. A precarious balance: pluripotency factors as lineage specifiers. Cell Stem Cell. 2011;8:363–369. doi: 10.1016/j.stem.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Shu J, Deng H. Lineage specifiers: new players in the induction of pluripotency. Genomics Proteomics Bioinformatics. 2013;11:259–263. doi: 10.1016/j.gpb.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **12.Montserrat N, Nivet E, Sancho-Martinez I, Hishida T, Kumar S, Miquel L, Cortina C, Hishida Y, Xia Y, Esteban CR, et al. Reprogramming of human fibroblasts to pluripotency with lineage specifiers. Cell Stem Cell. 2013;13:341–350. doi: 10.1016/j.stem.2013.06.019. Contrary to previous reports Oct4, a core reprogramming factor thought to be indispensable for the reprogramming of human somatic cells, was found to be replaceable. This paper describes for the first time the reprogramming of human fibroblasts to iPSCs by the use of genes typically involved in lineage specification. [DOI] [PubMed] [Google Scholar]
- **13.Shu J, Wu C, Wu Y, Li Z, Shao S, Zhao W, Tang X, Yang H, Shen L, Zuo X, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–975. doi: 10.1016/j.cell.2013.05.001. This paper describes the generation of murine iPSCs upon balancing of counteracting differentiation forces with lineage specifiers. The manuscript also reports on the role of different reprogramming factors in lineage specification and postulates a seesaw model of pluripotency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-David U, Nissenbaum J, Benvenisty N. New balance in pluripotency: reprogramming with lineage specifiers. Cell. 2013;153:939–940. doi: 10.1016/j.cell.2013.04.051. [DOI] [PubMed] [Google Scholar]
- 15.Liu K, Song Y, Yu H, Zhao T. Understanding the roadmaps to induced pluripotency. Cell Death Dis. 2014;5:e1232. doi: 10.1038/cddis.2014.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Zhang K, Xu L, Wang E. Quantifying the Waddington landscape and biological paths for development and differentiation. [no date], [no volume] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iovino N, Cavalli G. Rolling ES cells down the Waddington landscape with Oct4 and Sox2. Cell. 2011;145:815–817. doi: 10.1016/j.cell.2011.05.027. [DOI] [PubMed] [Google Scholar]
- 18.Morris R, Sancho-Martinez I, Sharpee TO, Belmonte JCI. pnas org. Mathematical approaches to modeling development and reprogramming. [no date], [no volume] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zipori D. The nature of stem cells: state rather than entity. Nat Rev Genet. 2004;5:873–878. doi: 10.1038/nrg1475. [DOI] [PubMed] [Google Scholar]
- 20.MacArthur BD, Lemischka IR. Statistical mechanics of pluripotency. Cell. 2013;154:484–489. doi: 10.1016/j.cell.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 21.Lu R, Markowetz F, Unwin RD, Leek JT, Airoldi EM, MacArthur BD, Lachmann A, Rozov R, Ma’ayan A, Boyer LA, et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature. 2009;462:358–362. doi: 10.1038/nature08575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang G, Zhang Y. Genetic and epigenetic variations in iPSCs: potential causes and implications for application. Cell Stem Cell. 2013;13:149–159. doi: 10.1016/j.stem.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cahan P, Daley GQ. Origins and implications of pluripotent stem cell variability and heterogeneity. Nat Rev Mol Cell Biol. 2013;14:357–368. doi: 10.1038/nrm3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris R, Sancho-Martinez I, Sharpee TO, Izpisua Belmonte JC. Mathematical approaches to modeling development and reprogramming. Proceedings of the National Academy of Sciences. 2014;111:5076–5082. doi: 10.1073/pnas.1317150111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki A, Raya A, Kawakami Y, Morita M, Matsui T, Nakashima K, Gage FH, Rodríguez-Esteban C, Izpisua Belmonte JC. Maintenance of embryonic stem cell pluripotency by Nanog-mediated reversal of mesoderm specification. Nat Clin Pract Cardiovasc Med. 2006;3 (Suppl 1):S114–22. doi: 10.1038/ncpcardio0442. [DOI] [PubMed] [Google Scholar]
- *26.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. This manuscript describes the dynamic transitions underlying cancer stem cell formation and postulates a dynamic equilibrium mode for driving transitions towards a stem cell state. [DOI] [PubMed] [Google Scholar]
- 27.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 28.Choi SA, Lee JY, Phi JH, Wang K-C, Park C-K, Park S-H, Kim S-K. Identification of brain tumour initiating cells using the stem cell marker aldehyde dehydrogenase. Eur J Cancer. 2014;50:137–149. doi: 10.1016/j.ejca.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Gilbertson RJ, Rich JN. Making a tumour’s bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer. 2007;7:733–736. doi: 10.1038/nrc2246. [DOI] [PubMed] [Google Scholar]
- 30.Visvader JE, Lindeman GJ. Cancer stem cells: current status and evolving complexities. Cell Stem Cell. 2012;10:717–728. doi: 10.1016/j.stem.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Medema JP. Cancer stem cells: the challenges ahead. Nat Cell Biol. 2013;15:338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 33.Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proceedings of the National Academy of Sciences. 2011;108:7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **34.Chaffer CL, Marjanovic ND, Lee T, Bell G, Kleer CG, Reinhardt F, D’Alessio AC, Young RA, Weinberg RA. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell. 2013;154:61–74. doi: 10.1016/j.cell.2013.06.005. This manuscript describes the reprogramming of differentiated cancer cells to stem cell-like phenotypes by modulation of the chromatin. It provides the first demonstration that ZEB1 can drive the dedifferentiation of breast cancer cells to a breast cancer stem cell phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sánchez Alvarado A. Developmental biology: A cellular view of regeneration. Nature. 2009;460:39–40. doi: 10.1038/460039a. [DOI] [PubMed] [Google Scholar]
- *36.Sánchez Alvarado A, Yamanaka S. Rethinking differentiation: stem cells, regeneration, and plasticity. Cell. 2014;157:110–119. doi: 10.1016/j.cell.2014.02.041. This review provides one of the most provocative overviews on the field of development, regeneration and reprogramming. The manuscript also provides compelling arguments on the differences between embryonic development and development as a whole continuous process and proposes aging as aprt of the developmental program of an organism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eguizabal C, Montserrat N, Veiga A, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation, and reprogramming: future directions in regenerative medicine. Semin Reprod Med. 2013;31:82–94. doi: 10.1055/s-0032-1331802. [DOI] [PubMed] [Google Scholar]
- 38.Holmberg J, Perlmann T. Maintaining differentiated cellular identity. Nat Rev Genet. 2012;13:429–439. doi: 10.1038/nrg3209. [DOI] [PubMed] [Google Scholar]
- 39.Stange DE, Koo B-K, Huch M, Sibbel G, Basak O, Lyubimova A, Kujala P, Bartfeld S, Koster J, Geahlen JH, et al. Differentiated Troy+ chief cells act as reserve stem cells to generate all lineages of the stomach epithelium. Cell. 2013;155:357–368. doi: 10.1016/j.cell.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **40.Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, et al. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503:218–223. doi: 10.1038/nature12777. By using lineage tracing approaches this manuscript demonstrates for the first time the reversibility of the differentiated state in the absence of exogenous genetic manipulation as a reponse to injury in mammalian cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sancho-Martinez I, Baek SH, Izpisua Belmonte JC. Lineage conversion methodologies meet the reprogramming toolbox. Nat Cell Biol. 2012;14:892–899. doi: 10.1038/ncb2567. [DOI] [PubMed] [Google Scholar]
- 43.Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Theunissen TW, Jaenisch R. Molecular Control of Induced Pluripotency. Cell Stem Cell. 2014;14:720–734. doi: 10.1016/j.stem.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46*.Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell. 2012;151:994–1004. doi: 10.1016/j.cell.2012.09.045. This paper provides the first comprehensive study on the molecular role that the so-called pluripotency factors play in modulating the epigenome during reprogramming. It establishes the role of the reprogramming factors as “pioneer factors” during the initial steps of reprogramming and therefore provides the basic understanding on the molecular control of cell dedifferentiation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soufi A, Zaret KS. Understanding impediments to cellular conversion to pluripotency by assessing the earliest events in ectopic transcription factor binding to the genome. Cell Cycle. 2013;12:1487–1491. doi: 10.4161/cc.24663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49**.Buganim Y, Faddah DA, Cheng AW, Itskovich E, Markoulaki S, Ganz K, Klemm SL, van Oudenaarden A, Jaenisch R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell. 2012;150:1209–1222. doi: 10.1016/j.cell.2012.08.023. This manuscript represents the first comprehensive study describing the molecular dynamics underlying iPSC generation at the single-cell level over time. It additionally demonstrates the distinct phases of reprogramming as well as establishes a hierarchical phase of gene activation during late reprogramming events. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50**.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. This report represents the first comprehensive description of the different molecular changes underlying murine cell reprogramming at the population level. It describes and correlates changes occurring at the transcriptome level as well as those at the epigenetic level and clearly demonstrates that two major waves of reprogramming events underlie the conversion of somatic murine cells to pluripotent cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sancho-Martinez I, Izpisua Belmonte JC. Stem cells: Surf the waves of reprogramming. Nature. 2013;493:310–311. doi: 10.1038/493310b. [DOI] [PubMed] [Google Scholar]
- 52.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 53.Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, Piao Y, Mehta S, Yee S, Nakatake Y, et al. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell. 2009;5:420–433. doi: 10.1016/j.stem.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54**.Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Araúzo-Bravo MJ, Ruau D, Han DW, Zenke M, et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. This paper demonstrates for the first time the suitability of using somatic cells with high endogenous levels of pluripotent factors for the simplification of the reprogramming process. It demonstrates for the first time that Neural Progenitor Cells can be reprogrammed solely upon Oct4 overexpression and additionally provides indirect evidence on the role that balancing counteracting differentiation forces might play in the acquisition of pluripotency. [DOI] [PubMed] [Google Scholar]
- 55.Ohnishi K, Semi K, Yamamoto T, Shimizu M, Tanaka A, Mitsunaga K, Okita K, Osafune K, Arioka Y, Maeda T, et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156:663–677. doi: 10.1016/j.cell.2014.01.005. [DOI] [PubMed] [Google Scholar]