Summary
Bacteroidales are the most abundant Gram negative bacteria of the human intestinal microbiota comprising more than half of the bacteria in many individuals. Some of the factors that these bacteria use to establish and maintain themselves in this ecosystem are beginning to be identified. However, ecological competition, especially interference competition where one organism directly harms another, is largely unexplored. To begin to understand the relevance of this ecological principle as it applies to these abundant gut bacteria and factors that may promote such competition, we screened Bacteroides fragilis for the production of antimicrobial molecules. We found that the production of extracellularly secreted antimicrobial molecules is widespread in this species. The first identified molecule, described in this manuscript, contains a membrane attack complex/perforin (MACPF) domain present in host immune molecules that kill bacteria and virally infected cells by pore formation, and mutations affecting key residues of this domain abrogated its activity. This antimicrobial molecule, termed BSAP-1, is secreted from the cell in outer membrane vesicles and no additional proteins are required for its secretion, processing or immunity of the producing cell. This study provides the first insight into secreted molecules that promote competitive interference among Bacteroidales strains of the human gut.
Keywords: Bacteroidales, Bacteroides, Secreted antimicrobial protein, MACPF domain, Intestinal symbiont, Outer membrane vesicles OMV, BSAP
Introduction
The human intestinal microbiota is one of the densest microbial ecosystems on earth comprising more than a hundred different species (Faith et al., 2013). This community of organisms is critical to human health and also has a role in disease (reviewed (Shanahan, 2013)). There has been a great deal of effort devoted to cataloging the composition of this ecosystem not only in healthy individuals (Human Microbiome Project, 2012, Arumugam et al., 2011), but also during disease, and under different dietary conditions (reviewed (Kau et al., 2011). Studies have also addressed the stability of the ecosystem (Zitomersky et al., 2011, Faith et al., 2013) and its resilience following various perturbations (reviewed (Lozupone et al., 2012)). With our greatly increased understanding not only of the microbes present in this ecosystem but also their genetic content, the groundwork has been laid for more intensive efforts to understand how these bacteria interact with one another, not only in interactions where one or more members benefit (Rakoff-Nahoum et al., 2014) but also in competitive interactions. Such analyses are essential for understanding the factors that dictate compositional dynamics and stability within this diverse ecosystem.
In ecology, competitive interactions are categorized as either exploitative, where two individuals compete for the same resources (Case & Gilpin, 1974), or interference, where one individual directly harms another (Schoener, 1983). With the exception of recently described Type VI secretion systems in gut Bacteroidales (Coyne et al., 2014) (Russell et al., 2014) which require cell to cell contact for killing, both of types of competition are largely unexplored among the Bacteroidales. Species that thrive in such dense and diverse microbial communities must have mechanisms to establish themselves, persist, and thwart competitors. As the microbes that colonize the human intestine evolved in a competitive environment where successful species can achieve extremely high densities, the bacterial composition of this ecosystem should be greatly influenced by factors that promote competitive interference, such as the production of antimicrobial molecules.
Secreted antimicrobial proteins of bacteria include small peptide bacteriocins such as the microcins and lantibiotics (reviewed (Cotter et al., 2013)) and larger molecules such as colicins (Cascales et al., 2007) and pyocins (Michel-Briand & Baysse, 2002). These molecules are structurally and functionally diverse and inhibit or kill cells by various mechanisms including nuclease activity, pore formation, enzyme inhibition, and impairment of the cell wall. Some antimicrobial proteins such as group A colicins and R type pyocins are released only upon lysis of the bacterial cell. Other antimicrobial molecules such as microcins and other small peptide bacteriocins of Gram positive bacteria are actively released from growing cells. In most cases, producing strains of both peptide and protein antimicrobials synthesize an immunity protein to protect themselves from the molecule’s toxicity.
Most of what we know about secreted antimicrobial molecules produced by gut bacteria comes from studies of minor components of the human adult microbiota such as Lactobacillus and Bifidobacterium species. These organisms produce numerous types of small peptide bacteriocins, some of which have been demonstrated to have antimicrobial activity against both Gram positive and Gram negative bacteria, including enteric pathogens (reviewed (Cotter et al., 2013)). Research in this field is largely aimed at understanding the diversity and structures of these peptides, their applicability to probiotic design, and potential as antimicrobials against enteric pathogens.
Less is known of the ecological relevance of antimicrobial molecules produced by intestinal bacteria in their natural ecosystem. As Bacteroidales are the most abundant Gram negative bacteria of the intestinal microbiota and also the most temporally stable compared to the abundant Gram positive members of this ecosystem (Faith et al., 2013), it is important to determine the ecological impact of antimicrobial production on community stability, diversity and ecosystem invasion.
Production of antimicrobial proteins by intestinal Bacteroides species has been reported, with the most recent study in 1999 (Papastathopoulou et al., 1997, Farias et al., 1994, Miranda et al., 1993, Avelar et al., 1999). However, no responsible gene was cloned nor was the protein purified, therefore, the identity of these molecules remains unknown. The present study was undertaken to begin to molecularly analyze antimicrobial molecule production in Bacteroides fragilis, a representative species of the abundant gut Bacteroidales. Such analyses will allow for future studies designed at understanding the ecological significance of these antimicrobial molecules in shaping the composition of the microbiota and factors that should be considered in altering the microbiota to bring about human health benefits.
Results and Discussion
Intraspecies growth inhibition among B. fragilis
To begin to address the ability of B. fragilis to secrete antimicrobial molecules that target other B. fragilis strains, we tested 42 strains for their ability to inhibit the growth of type strain NCTC 9343 using the agar overlay spot test (Avelar et al., 1999). We found that 21 of these 42 strains secrete factors that inhibit the growth of NCTC 9343 in this assay (Table S1). To extend these analyses, we tested a panel of ten B. fragilis strains for their ability to secrete molecules that inhibit the growth of each other. These data demonstrate great diversity in both the ability to inhibit and to be inhibited by extracellularly secreted molecules of heterologous strains (Table 1) and suggest that the antimicrobial molecules and the receptors/targets of the sensitive strains are encoded by non-conserved genes of the B. fragilis genome. Strains 638R, NCTC 9343, 379 and CL07T12C05 secrete molecule(s) that inhibit the growth of at least half of the strains tested, whereas three of these ten strains do not secrete any molecules that inhibit the nine heterologous strains tested here. We also tested three Escherichia coli strains and four Vibrio cholerae strains in the agar overlay assay and found that none of the 10 B. fragilis strains secrete factors that inhibit the growth of these Proteobacterial strains (data not shown).
Table 1.
Analysis of growth inhibition among 10 B. fragilis strains by secreted factors
| B. fragilis producer strain (dotted) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| NCTC9343 | 638R | CM11 | 379 | US326 | 12905 | CM13 | CL03T12C07 | CL05T12C13 | CL07T12C05 | ||
|
|
|||||||||||
| Overlay strain | NCTC 9343 |
− | + | − | + | − | − | − | − | − | + |
| 638R |
− | − | − | − | − | − | − | − | − | + | |
| CM11 |
+ | − | − | − | − | − | − | − | − | − | |
| 379 |
− | − | + | − | − | − | − | − | + | − | |
| US326 |
+ | + | − | + | − | − | − | − | − | − | |
| 12905 |
+ | + | − | + | + | − | − | − | − | + | |
| CM13 |
− | + | − | − | − | − | − | − | + | − | |
| CL03T12C07 |
+ | + | − | + | − | − | − | − | + | + | |
| CL05T12C13 |
+ | + | − | + | − | − | − | − | − | + | |
| CL07T12C05 |
− | − | − | − | − | − | − | − | + | − | |
+ sensitive (growth inhibition)
− no growth inhibition
Identification of a genetic region necessary for growth inhibition
We choose to identify the inhibitory molecule(s) of strain 638R as this strain inhibits the growth of six of the ten B. fragilis strains testedand because it is sensitive to erythromycin and tetracycline and, therefore, amenable to genetic manipulation using available Bacteroides genetic tools. We performed transposon mutagenesis with plasmid pYT646b (Tang & Malamy, 2000) and identified a transposon mutant (M26) that abrogated the inhibitory activity against two of the six strains shown in Table 1; (12905 and 9343) (Fig. 1). The other four strains were still inhibited by factor(s) secreted by this transposon mutant.
Figure 1.
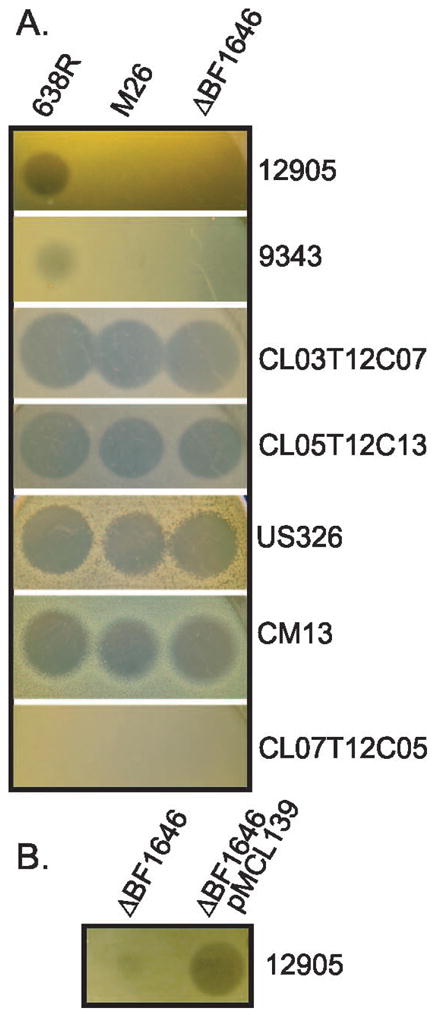
Agar overlay spot assay showing growth inhibition of B. fragilis strains by wild type 638R and mutants. A. Each strain from Table 1 that was growth inhibited by 638R was tested for growth inhibition by transposon mutant M26 and deletion mutant ΔBF1646. Overlaid B. fragilis strains tested for growth inhibition listed vertically. B. BF638R-1646 provided in trans by plasmid pMCL139 restores inhibitory activity to ΔBF1646.
The genetic region corresponding to the insertion site of the transposon is shown in Figure 2. The transposon inserted into BF638R_1646, 450 bp from the start of the gene. Analysis of this genetic region from 56 other B. fragilis strains with sequenced genomes covering this region demonstrated that this gene is present in 21 of these strains but absent in 35. This genetic region occurs as two main types, those similar to strain 638R, and those similar to strain 9343 (Fig. 2). The gene downstream of BF638R_1646 encodes a putative nucleotide deaminase (red) that is highly similar between these 12 B. fragilis strains (94.2–100% identical), whereas the proteins encoded by the genes upstream of BF638R_1646 are less similar (Fig. 2). The proteins encoded upstream of BF638R_1644 are nearly identical in all strains and there is no evidence that the genes of this region are contained on a mobile genetic element. Strain CL07T12C05 is one of the strains where this genetic region is 638R-like, with the orthologous protein, HMPREF1056_01634, identical to BF638R_1646 in all but one amino acid. This strain was included among the panel of strains tested for growth inhibition (Table 1). Both of the strains that are growth inhibited by 638R but not by the M26 mutant are also growth inhibited by strain CL07T12C05, as would be expected if a molecule encoded by this region was responsible for the inhibitory activity.
Figure 2.
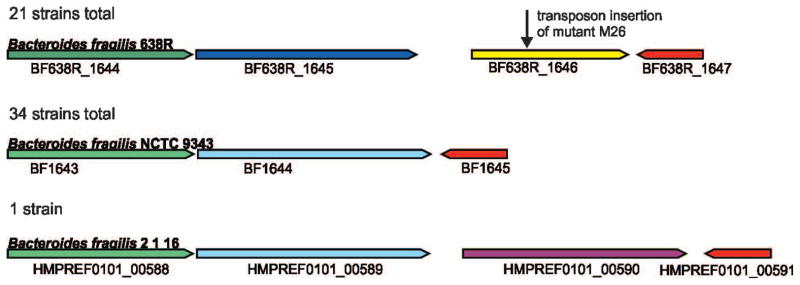
Region of the B. fragilis chromosome surrounding the transposon insertion of mutant M26. The genes in this region were analyzed in all 70 B. fragilis strains with genome sequences; 14 B. fragilis genomes were not included due to this region being incomplete or detectable on multiple contigs. These regions segregated into two different types, those similar to 638R (21 strains) and those similar to 9343 (34 strains), and one outlier. Genes encoding similar proteins are shown in the same color, with the shade distinguishing the genes encoding very similar 638R-like proteins from the 9343-like genes also encoding very similar proteins. The gene immediately upstream of the gene into which the transposon inserted (blue) encodes a putative outer membrane transporter that is 92.3–100% identical within the 638R-like or 9343-like genomes and 57.5–60.4% identical between these two groups. The gene upstream of that encodes a protein of unknown function (green) in which the 638R-like genes are 93.5 – 100% identical to each other, those the 9343-like are 90.3 – 100% identical to each other, whereas the % identity between groups is 71.9 – 86.0%. Although the genetic region of strain 2_1_16 is 9343-like, there is an additional gene, HMPREF0101_00590, encoding a protein of unknown function inserted between the blue and red genes. The gene shown in red encodes a putative nucleotide deaminase which is 94.6 – 100% among all B. fragilis strains.
Analysis of the gene into which the transposon inserted revealed an interesting finding. BF638R_1646 encodes a protein with a membrane attack complex/perforin (MACPF) domain (PF01823), present at amino acids 131 – 322 of the 372 amino acid protein. These domains are present in proteins produced by immune cells that kill bacteria by pore formation. These include complement components (C6, C7, C8α, C8β, C9) that are involved in the membrane attack complex that make pores in the outer membranes of Gram negative bacteria, and perforin produced by cytotoxic T-cell that lyse virally infected cells. BF638R_1646 is also similar to an Arabidopsis thaliana MACPF domain protein annotated as CAD1 for “constitutively activated cell death” that is involved in plant immunity (Morita-Yamamuro et al., 2005). No bacterial molecules with this domain have been shown to have antibacterial activity.
To confirm that the interruption of BF638R_1646 is responsible for the loss of antimicrobial activity, an internal deletion mutant of BF638R_1646 was created (ΔBF1646). Deletion of this gene abrogates the inhibitory activity to the same extent as the transposon mutant M26 (Fig 1A), and addition of pMCL139, containing BF638R_1646, in trans to ΔBF1646 restored this inhibitory activity (Fig. 1B). Therefore, BF638R_1646 is necessary for the 638R secreted antibacterial activity against these two strains.
BF638R_1646 is the only factor necessary for the inhibitory activity
Many bacterially-produced antimicrobial molecules require other proteins for their secretion, modification, or for protection of the producing strain. The genomic context of BF638R_1646 does not indicate an adjacent gene encoding an immunity protein. In addition, the prediction of an N-terminal signal peptidase II (SpII) cleavage site suggests that BF638R_1646 encodes a protein that will be modified with a lipid and secreted by the normal cellular machinery. Therefore, we predicted that if BF638R_1646 was cloned into a heterologous B. fragilis strain that does not produce a BF638R_1646 ortholog, this strain would acquire this antimicrobial activity. To test this prediction, we added pMCL139, containing BF638R_1646, in trans to wild type B. fragilis strain CM13. This tranconjugant was tested for its ability to inhibit the growth of all six of the strains that are inhibited by 638R in the agar overlay assay. CM13 does not normally inhibit the growth of any of the strains analyzed (Table 1); however, it acquires the ability to inhibit strains 12905 and 9343 when BF638R_1646 is supplied in trans (Fig 3A). These analyses revealed another interesting finding. Strains CL03T12C07 and CL05T12C13, which are inhibited by secreted molecules of wild type 638R, transposon mutant M26 and the ΔBF1646 mutant, are also growth inhibited by CM13 synthesizing BF638R_1646. Therefore, there are at least two antimicrobial molecules produced by 638R; BF638R_1646, and another molecule. Strains CM13 and US326, which are inhibited by both wild type 638R and ΔBF1646, are not growth inhibited by CM13 expressing BF638R_1646. Therefore, a molecule other than BF638R_1646 inhibits the growth of these strains, possibly the same second antimicrobial molecule that additionally targets CL03T12C07 and CL05T12C13.
Figure 3.
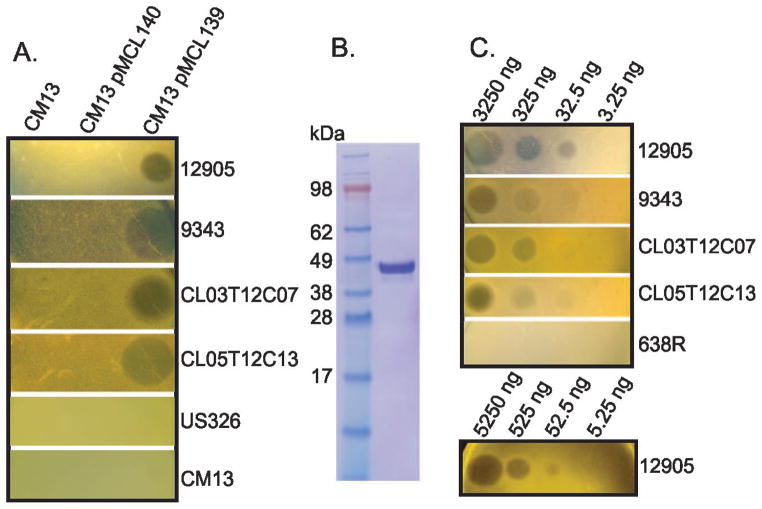
Growth inhibitory activity of BSAP-1 does not require additional factors. A. Agar overlay spot assay showing that addition of BF638R_BF1646 (pMCL139), but not vector alone (pMCL140), to strain CM13 confers the growth inhibitory activity to this strain. B. Coomassie-stained gel of See-Blue Pus2 markers (Life Technologies, lane 1) or purified His-BSAP (lane 2). C. Agar overlay spot assay with two different purifications of His-BSAP-1 showing the potent ability of this protein to inhibit the growth of sensitive strains. Total amount of protein added to the plate in 5 μl volumes is shown in figure.
To further confirm that this single protein encoded by BF638R_1646 is responsible for the growth inhibition of these four strains, we made an N-terminal His-tagged protein by fusing the His-tag with BF638R_1646 at the SpII cleavage site to make a soluble protein (Fig 3B). In agar overlay assays, this purified fusion protein demonstrated very potent inhibitory activity when tested against all four strains shown to be sensitive to BF638R_1646. As little as 32.5 ng of protein produced a visible zone of inhibition in the agar overlay assay against all sensitive strains (Fig. 3C). These collective data demonstrate that this single protein is all that is required for the inhibitory activity and it requires no immunity protein to protect the producing cell or additional factors for its processing and secretion. This protein was named BSAP-1 for Bacteroidales secreted antimicrobial protein 1.
Bactericidal versus bacteriostatic assays
The agar overlay assay demonstrates that BSAP-1 inhibits the growth of the sensitive strains tested, but does not reveal whether it has bactericidal or bacteriostatic activity. Two different assays were performed to address this question. In the first, sensitive strain 12905 was directly added to plates containing purified His-BSAP-1 and 12905 viability was quantified after overnight incubation. For the second assay, 12905 was incubated with 638R or ΔBF1646 and the viability of 12905 was quantified following overnight co-culture.
In the first assay, 10-fold dilutions containing between 102 – 107 cfu of strain 12905 were added in 5μl volumes to BHIS plates containing purified His-BSAP-1 (49 μg protein exp 1, or 75 μg protein exp 2) or with PBS as a control. One set of plates was allowed to grow for several days to visualize bacterial growth, and the other set was harvested after overnight incubation to quantify viable 12905. Visible growth inhibition of 12905 is evident on His-BSAP-1 containing plates, even when 105 bacteria were added (Fig. 4). As expected, His-BSAP-1 did not inhibit the growth of strain 638R. The spots corresponding to the addition of 103, 104 and 105 cfu of 12905 were extracted after overnight incubation and serially diluted to quantify bacteria. The results demonstrate that there is no substantial increase or decrease in the cfu of 12905 after overnight incubation with His-BSAP-1 compared to the number of cfu added to the plate (Table 2). In contrast, when 12905 is added to a plate with PBS, there is a 6.6 – 8.8 fold increase in cfu. These data demonstrate that 12905 remains viable after overnight incubation with His-BSAP-1, but does not replicate, suggesting that BSAP-1 may be bacteriostatic.
Figure 4.
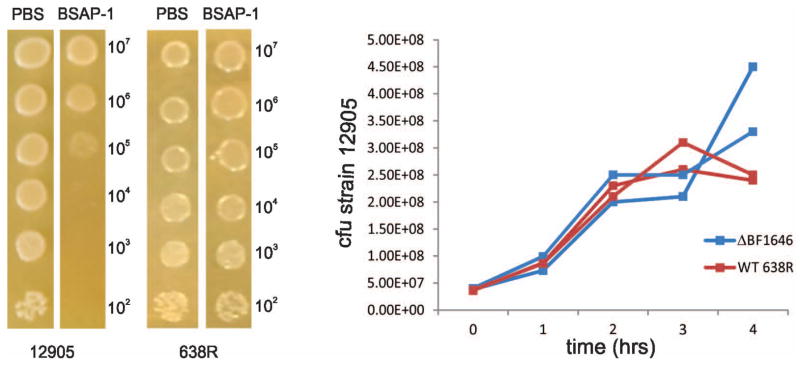
BSAP-1 inhibition of 12905 growth A. Addition of strain 12905 or 638R in 10-fold dilutions to plates containing His-BSAP-1 or PBS. Total cfu of 12905 added to plates is shown on the right and those recovered after overnight incubation is shown in Table 2. B. Broth co-culture experiments of strain 12905 with either wild type 638R or ΔBF1646. The cfus of sensitive strain 12905 are reported.
Table 2.
Quantification of 12905 survival/growth following incubation with purified BSAP-1
| Overnight incubation with purified BSAP-1
or PBS | |||
|---|---|---|---|
| cfu 12905 added | Incubated with BSAP-1 | Incubated with PBS | |
|
| |||
| cfu 12905 retrieved | cfu 12905 retrieved | ||
| Experiment 1 | 1.0 × 105 | 3.5 × 105 | 8.8 × 105 |
| 1.0 × 104 | 9.5 × 103 | 6.7 × 104 | |
| 1.0 × 103 | 1.0 × 103 | 6.6 × 103 | |
| Experiment 2 | 1.0 × 105 | 9.5 × 104 | 1.8 × 106 |
| 1.0 × 104 | 9.0 × 103 | 1.4 × 105 | |
| 1.0 × 103 | 1.1 × 103 | 5.7 × 103 | |
For the co-culture assays, strain 12905 was incubated overnight on BHIS plates with an excess of either 638R or ΔBF1646 to allow for maximal contact of sensitive strain 12905 with secreted BSAP-1. The ratios of bacteria for each experiment are listed in the experimental results section and ranged from 1:12.8 to 1:19.6. Following overnight co-culture, the surviving12905 were quantified by plating onto BHIS with erythromycin (Table 3). Co-culture of 12905 with 638R significantly reduced the cfu of 12905 compared to the number that were added at the start of the co-culture (160-fold average decrease) p-value 0.0076; whereas co-culture with ΔBF1646 allows for robust growth of strain 12905 with more than a 2 log increase for all experiments, p-value 0.0077. These data demonstrate that BSAP-1 is bactericidal rather than bacteriostatic as would be expected for an antimicrobial molecule with a MACPF domain. The differences in the results of the co-culture assays compared to assays with the purified protein could be due to amount of the purified BSAP-1 that was added to the plate. A total of 49 μg or 75 μg of BSAP-1 was spread on the plate and was likely insufficient to completely kill the bacteria that were added. In addition, BSAP-1 activity may not have been stable during the overnight incubation at 37°C. In contrast, co-culture with 638R would yield a continuous supply of BSAP-1, revealing the true bactericidal activity of the molecule.
Table 3.
Quantification of 12905 survival/growth following co-culture with 638R or ΔBF1646
| Overnight co-culture on BHIS plates of
12905 with 638R or ΔBSAP-1 | |||
|---|---|---|---|
| Experiment | cfu 12905 added to co-culture | Co-cultured with 638R | Co-cultured with ΔBF1646 |
|
| |||
| cfu 12905 retrieveda | cfu 12905 retrievedb | ||
| 1 | 1.5 × 106 | 1.2 × 106 | 5.0 × 108 |
| 2 | 4.6 × 105 | 1.5 × 103 | 5.1 × 107 |
| 3 | 4.6 × 105 | 5.5 × 103 | 5.5 × 107 |
| 4 | 4.6 × 105 | 2.0 × 103 | 7.2 × 107 |
| 5 | 4.6 × 105 | 2.0 × 103 | 7.6 × 107 |
| 6 | 8.7 × 105 | 6.5 × 103 | 1.0 × 108 |
| 7 | 8.7 × 105 | 9.0 × 103 | 9.5 × 107 |
| 8 | 8.7 × 105 | 4.3 × 103 | 9.2 × 107 |
| 9 | 8.7 × 105 | 5.5 × 103 | 1.2 × 108 |
statistically significant decrease from initial cfu. p-value 0.0076
statistically significant increase from initial cfu. p-value 0.0077
The co-culture experiments were repeated with bacteria grown in liquid culture. In support of the concentration dependent effect of BSAP-1, growth inhibition of sensitive strain 12905 was only detected in co-culture with BSAP-1 producing strain 638R at later time points (Fig. 4B). In both experimental co-cultures with strain 638R, cfu of 12905 began to decrease after three hours; whereas the cfu of 12905 incubated with ΔBF1646 continued to increase.
Site-directed mutations in the MACPF domain
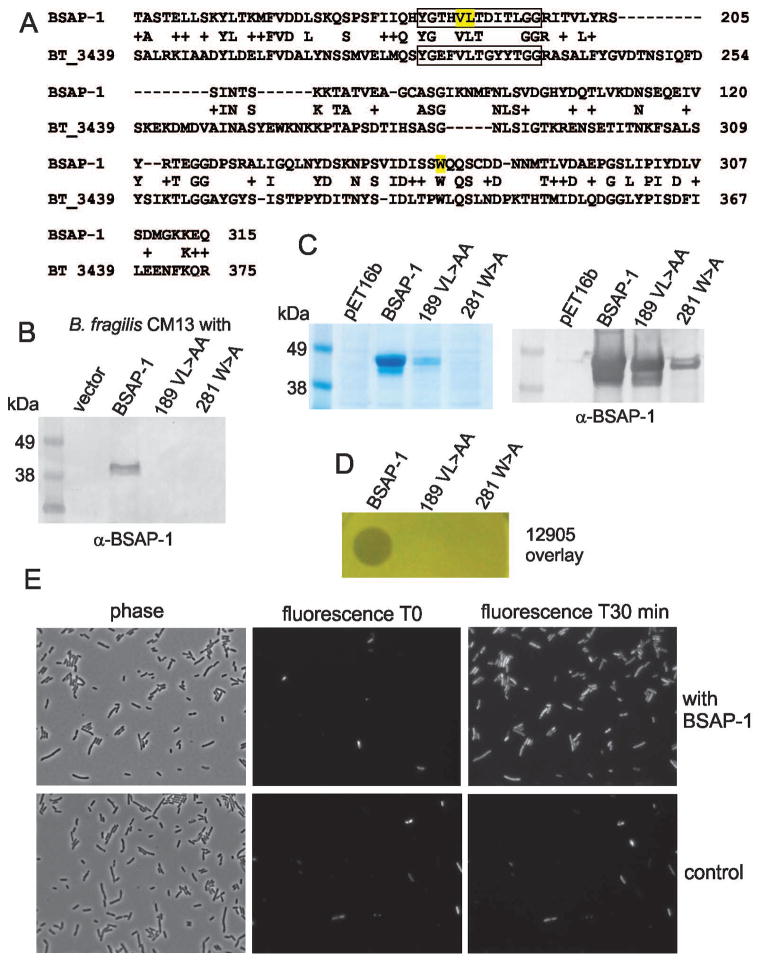
To determine whether the MACPF domain of BSAP-1 is necessary for its killing, we made site-directed mutations affecting critical residues of this domain. A MACPF protein of unknown function from Bacteroides thetaiotaomicron (BT-3439) was previously structured with its critical residues mapped in relation to other MACPF containing proteins (Xu et al., 2010). Therefore, we aligned the primary sequences of the MACPF domains of BSAP-1 and BT_3439 to identify these critical residues in BSAP-1 (Fig 5A). The MACPF motif Y/W-G-T/S-H-F/Y-X6-GG (Ponting, 1999) of both BT_3439 and BSAP-1 are boxed, (Fig 5A) with the MACPF motif of BSAP-1 conforming better to this consensus than BT_3439. Based on the conservation of the VL residues between these two proteins in this critical motif, they were selected for mutagenesis by converting to AA. In addition, a highly conserved tryptophan residue contained within a common helix conserved in all known MACPFs was also targeted by making alterations to change the residue to A (Fig 5A). These mutant proteins were synthesized in B. fragilis CM13, to determine if they confer upon this strain the ability to kill BSAP-1 sensitive strains. In addition, recombinant mutant proteins were synthesized with an N-terminal His-tag and purified from E. coli to assess killing activity of the purified protein. We found that alteration of the BSAP-1 MACPF domain at either of these two sites completely abolished the ability of B. fragilis to synthesize and secrete BSAP-1 (Fig 5B). In addition, alteration of either of these critical MACPF sites resulted in proteins that were not present in the soluble fraction of E. coli lysates under standard induction conditions, suggesting that the altered proteins were improperly folded. To circumvent this caveat, we performed the induction at room temperature in an attempt to prevent inclusion body formation. Induction at room temperature allowed a portion of the VL>AA mutant protein to be isolated from the soluble phase, but extremely low levels of the W>A mutant protein was in the soluble phase (Fig 5C). The purified mutant proteins were concentrated 10-fold, resulting in a concentration for the VL>AA protein half that of the wild type His-BSAP-1 concentration. Therefore, for the agar overlay assay, the mutant proteins were applied twice in the same area and allowed to dry between each application. As shown in figure 5D, the alteration of the critical MACPF residues of the MACPF domain completely abrogated the killing activity of the protein. These data demonstrate that the MACPF domain is critical for BSAP-1 activity and that alteration of critical residues of this domain of BSAP-1 severely affect its solubility and functionality.
Figure 5.
MACPF domain is necessary for BSAP-1 activity. A. Alignment of the MACPF domains of BSAP-1 and BT_3439 of B. thetaiotaomicron VPI-5482. A conserved MACPF motif is boxed with residues altered by site-directed mutagenesis highlighted in yellow. B. Western blot of supernatants of B. fragilis CM13 containing the gene for WT BSAP-1, or BSAP-1 site-directed mutant genes, probed with antiserum to BSAP-1. C. Coomassie-stained gel (left panel) or western immunoblot (right panel) showing the purification of His-tagged BSAP-1 or BSAP-1 site-directed mutants from BL21/DE3 induced at room temperature. The coomassie-stained gel shows the unconcentrated mutant proteins following purification whereas the western blot shows reactivity when the mutant protein samples were each concentrated 10-fold. The blot was probed with antiserum to BSAP-1. D. Analysis of the killing of strain 12905 in an agar overlay by purified BSAP-1 or concentrated BSAP-1 site-directed mutants E. Microscopic analysis of PI incorporation by strain 12905 exposed to BSAP-1 contained in culture supernatant or control supernatant. Fluorescence images are shown at the starting point and 30 minutes later.
BSAP-1 allows rapid PI internalization
MACPF proteins can oligomerize and form pores in susceptible cells (Tschopp et al., 1986, Rosado et al., 2007). To analyze if BSAP-1 causes membrane permeability in strain 12905, we used fluorescence microscopy to monitor prodidium iodide (PI) uptake. PI does not penetrate intact membranes but when membranes are compromised, PI enters cells, binds to DNA, and fluoresces. Using a modified PI infused agar pad method where bacteria are added and maintained under anaerobic conditions during visualization, we detected a rapid PI labeling of bacteria exposed to BSAP-1. All bacteria in the field were labeling within 30 min; however, there was almost no change in the fluorescence of bacteria exposed to the control sample (Fig. 5E).
BSAP-1 is released in OMV
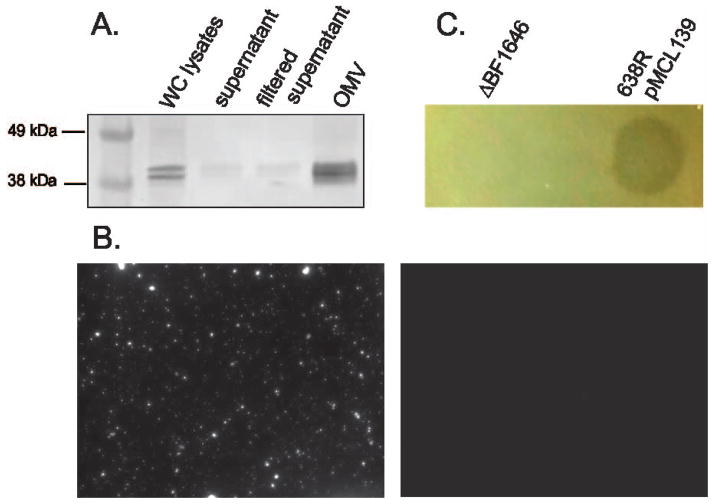
In Bacteroides species, lipoproteins are mainly targeted to the outer surface of the bacterium (Shipman et al., 1999, Fletcher et al., 2009). B. fragilis has been shown to release outer membrane vesicles (OMV), which contain proteins with enzymatic activities (Patrick et al., 1996, Rakoff-Nahoum et al., 2014, Elhenawy et al., 2014) and capsular polysaccharide that produce immunomodulatory effects in the host (Shen et al., 2012). Both B. fragilis and Bacteroides thetaiotaomicron, preferentially package acidic proteins into OMV (Elhenawy et al., 2014). Pseudomonas aeruginosa has been shown to release outer membrane vesicles containing hydrolytic enzymes that lyse heterologous strains (Kadurugamuwa & Beveridge, 1996); providing precedence for release of antimicrobial molecules in OMVs To determine if BSAP-1 is released in OMVs, we used the purified recombinant His-tagged protein to raise antibodies in rabbits to track this protein. We purified OMVs from 638R pMCL139 and performed Western blot analysis as well as fluorescently labeling the OMV fraction using BSAP-1 antiserum. These analyses demonstrate that BSAP-1 is released from the bacterial surface in OMVs rather than in soluble form (Fig. 6A,B) as would be expected for a protein containing a lipid moiety and consistent with its acidic pI (6.36 without the signal sequence). We also performed the agar spot overlay assay by dotting purified OMV from 638R pMCL139 and found that the OMV fraction itself inhibited the growth of 12905 in this assay (Fig. 6C).
Figure 6.
BSAP-1 is secreted extracellularly in outer membrane vesicles. A. Western immunoblot of 638R pMCL139 whole cell lysate, supernatant, or OMV fraction probed with polyclonal antiserum to His-BSAP-1. B. Immunofluorescence analysis of OMV fraction of 638R pMCL139 (left) or ΔBF1646 (right) using antiserum to His-BSAP-1. C. Agar spot overlay assay showing growth inhibition of strain 12905 by purified OMVs from 638R pMCL139, but not ΔBF1646. OMVs were harvested at mid-log (OD600 = 0.6) and 50 μl volumes containing 50 μg (ΔBF1646) or 52 μg (638R pMCL139) of total protein were added in 50 μl volumes to plates prior to overlay.
MACPF proteins are widely distributed in Bacteroidetes
As most functionally described MACPF domain-containing proteins are produced by eukaryotes, we searched all available complete and draft bacterial genomes for genes encoding proteins with a MACPF domain (pfam01823). Of the 15,482 non-Bacteroidetes bacterial genomic sequences available, 161 MACPF containing proteins were identified. Of these, the majority (121) are encoded by genomes of Chlamydia species. The Chlamydia MACPF containing proteins are highly similar to each other and are of undescribed function (Ponting, 1999, Taylor et al., 2010). The remaining 40 MACPF-containing proteins of non-Bacteroidetes, non-Chlamydia species are sporadically distributed among eight bacterial phyla (Table S2).
The 466 sequenced Bacteroidetes/Chlorobi strains collectively encode 163 proteins containing a MACPF domain, more than are encoded by the sequenced bacterial genomes of all other phyla combined. Although these proteins all contain the MACPF domain, there is extensive heterogeneity in their sequences and they range in size from 187 amino acids to 1111 amino acids. Figure S1 shows a cladogram of the MACPF proteins encoded by Bacteroidetes genomes. Some of these MACPF contain additional domains such as ankyrin domains and carbohydrate binding modules. In addition, diverse Bacteroidetes strains from the Flavobacteridales and Sphingobacteriales that live in soil and marine environments also encode MACPF-domain containing proteins (blue font in Fig. S1). Therefore, with the exception of Chlamydia, bacterially produced MACPF domain-containing proteins are largely encoded by Bacteroidetes genomes and may provide an ecological advantage to these bacteria in their natural microbial communities. These 163 Bacteroidetes-encoded MACPF proteins were analyzed for the presence of an SpII signal peptide (cleavage site) using LipoP (Juncker et al., 2003). 131 of the 163 proteins are predicted to have a signal sequence with an SpII cleavage site and therefore may also be released by these diverse Bacteroidetes species into their surroundings in OMVs. In addition, 125 of the 163 mature proteins (signal sequences removed) are acidic, another factors shown to be important for packaging into OMV (Elhenawy et al., 2014). It is interesting to note that BSAP-1 is encoded only by genomes of B. fragilis strains, whereas other MACPF-containing proteins that are very similar or identical to each other are synthesized by various Bacteroidales species. We recently described that one of these genes, present in four different Bacteroidales species (highlighted in yellow in Fig S1), is carried on an integrative conjugative elements that was transferred between these coresident Bacteroidales species within the gut of a human (Coyne et al., 2014).
We chose five of these MACPF proteins (highlighted in red in Fig S1) for analysis of antimicrobial activity. B. fragilis BF638R_2714 was chosen because it is the only other MACPF domain containing protein of strain 638R and would allow us to determine if this gene encodes the additional antimicrobial activity detected in this strain. BACUNI_00959 of B. uniformis ATCC 8492 was chosen because it encodes the closest ortholog to the BSAP-1 group (54% similarity over its length). As the structure of BT_3439 has previously been elucidated (Xu et al., 2010), we analyzed all three MACPF-containing proteins from the well-studied B. thetaiotaomicron type strain VPI 5482 (BT_3120, BT_3437, and BT_3439). BT_3120 and BT_3439 are not contained in Fig. S1 as they scored below the gathering threshold bit-score for the MACPF pfam; however, structural and domain architecture analyses demonstrate that these proteins have MACPF domains (Xu et al., 2010). These five genes were cloned into the Bacteroides expression vector pMCL140 and placed into the native background strain to allow for detection of activity when these genes are expressed from a constitutive promoter. The constructs were also placed in trans in B. fragilis CM13 to determine whether any detected activity in the native strain is due to the product of the cloned gene rather than a different product of the host strain. The panel of strains tested for sensitivity in the agar spot overlay assay is shown in Table S4. Expression of the second MACPF-encoding gene of B. fragilis 638R did not confer to CM13 the ability to inhibit the growth of any of the four B. fragilis strains growth inhibited by the ΔBF1646 mutant (Table S4). Therefore, this second MACPF-domain protein of 638R is not the additional antimicrobial factor that inhibits the growth of these B. fragilis stains. The B. thetaiotaomicron type strain was able to inhibit the growth of B. fragilis 12905, but this activity was not attributed to any of its MACPF-containing proteins. This finding demonstrates that secreted antimicrobial factors are not limited to intraspecies competitive interactions and that interspecies competitions are also likely to be important in the gut ecosystem. This B. thetaiotaomicron strain only slightly inhibited the growth (visible but turbid zone) of one of the other 19 B. thetaiotaomicron strains tested in this assay. The B. uniformis type strain slightly inhibited the growth of 12905 and three of the other B. uniformis strains tested, but this activity could not be attributed to BACUNI_00959 as CM13 containing this gene did not inhibit the growth of these strains. Therefore, growth inhibitory activity could not be demonstrated for any of these five MACPF domain-containing proteins.
Several identified MACPF domain-containing proteins do not have lytic activity. Complement components C6, C7, C8α, C8β and C9 all have MACPF domains, however, only C8α and C9 are capable of lysing cells. In addition, the pores formed by perforin oligomerization mediate the delivery of cytotoxic proteases (granzymes) that contribute to cell death (Bolitho et al., 2007, Pipkin & Lieberman, 2007). The crystal structures of two bacterially encoded MACPF proteins have been elucidated. The structure of The Plu-MACPF of the plant pathogen Photorhabdus luminescens revealed that MACPF proteins have structural similarity with pore-forming cholesterol-dependent cytolysins (CDCs) of Gram-positive bacteria (Rosado et al., 2007). The B. thetaoiotaomicron MACPF-domain containing protein BT_3439 contains two additional domains resembling ribonuclease H and interleukin 8 (Xu et al., 2010). It is speculated that 16 copies of this molecule oligomerize to form a porin-like transmembrane pore and that the MACPF/CDC fold of BT_3439 may be used for nonlytic purposes including protein secretion or nutrient uptake (Xu et al., 2010). It is likely that the diverse MACPF domain containing proteins of these Bacteroidetes species have varied functions. The prevalence of MACPF domain-containing proteins, particularly within the phylum Bacteroidetes, warrants their continued investigation.
This is the first study to characterize a secreted antimicrobial molecule of the abundant gut Bacteroidales and is the first bacterially produced MACPF domain-containing protein shown to have antimicrobial activity. We found that a single B. fragilis strain can produce multiple secreted antimicrobial proteins, each targeting a specific set of strains. These data suggest not only that secreted antimicrobial proteins are encoded by nonconserved regions of the chromosome, but so too are the receptor/targets of the sensitive strains. This is in contrast to the well described colicins and microcins of E. coli that bind a conserved surface receptor of the species. In addition, our analyses show that this BSAP does not require an immunity protein to protect the producing strain, and it is secreted from actively growing cell in OMVs.
It will be important to determine how prevalent the production of secreted antimicrobial proteins is among the intestinal Bacteroidales and if Bacteroidales members co-resident in a human gut microbiota produce antimicrobial molecules that target each other, similar to the rock paper scissor scenario (Kerr et al., 2002) shown to contribute to E. coli strain diversity in a mouse model (Kirkup & Riley, 2004). Alternatively, in a stable ecosystem, these battles may have been fought and reconciled, or may occasionally be waged if antimicrobial producing strains are introduced into an established ecosystem with a corresponding sensitive strain.
Studies are beginning to reveal the great complexity of interactions that occur among gut Bacteroidales strains/species. Such interactions can be categorized either by their effects on other bacterial members of the ecosystem (competitive or beneficial) or by the mode of delivery of the product (contact dependent versus secreted). Contact dependent interactions include transfer of mobile genetic elements by conjugation, which we showed occurs extensively between co-resident Bacteroidales species in the human gut (Coyne et al., 2014). Another type of contact dependent interaction that occurs among the Bacteroidales is type VI secretion system (T6SS) mediated killing (Coyne et al., 2014) (Russell et al., 2014). It will be interesting to determine the impact of T6SS mediated killing verses killing by secreted BSAPs and what factors trigger each type of killing system.
Interactions dependent on secreted factors include not only antagonistic interactions mediated by BSAPs, but also interactions that benefit one or more members. Bacteroidales species secrete glycoside hydrolases and polysaccharide lyases in OMVs (Patrick et al., 1996, Rakoff-Nahoum et al., 2014, Elhenawy et al., 2014), which allow other species to utilize plant polysaccharides that would otherwise be inaccessible to them (Rakoff-Nahoum et al., 2014). Therefore, OMVs are the vehicles of both competitive and beneficial interactions between Bacteroidales strains/species.
We do not yet know how these processes are regulated and under what conditions in the gut a particular interaction may be favored over another. It is possible that if nutritional sources are limited, or if bacteria sense other stresses, competitive interactions would be favored (Cornforth & Foster, 2013). Deciphering the significance of each of these interactions, how they are regulated, and how they contribute to the composition of our gut microbiota will be important areas of continued study within this human health related ecosystem.
Experimental Procedures
All primers used in this study are listed in Table S5.
Bacterial strains and growth conditions
Bacteroides strains used in this study were described previously (Comstock et al., 2000, Zitomersky et al., 2011). All Bacteroides were grown in supplemented basal medium (Pantosti et al., 1991), or on supplemented brain heart infusion plates. Antibiotics (5 μg/ml erythromycin or 2 μg/ml tetracycline) were added where indicated. Escherichia coli strains were grown in L broth or L plates with antibiotics added where appropriate; ampicillin 100 μg/ml, trimethoprim 100 μg/ml, kanamycin 50 μg/ml. E. coli strains HB101, DH5α, E2348/69, and V. cholerae strains O395, N16961, V52 and C6706 were tested for sensitivity to secreted antimicrobial products of B. fragilis strains in the agar overlay assay under both aerobic and anaerobic conditions.
Agar spot test for growth inhibition analysis
The ability of one Bacteroides strain to inhibit the growth of another was assayed using the agar spot test (Avelar et al., 1999). In brief, Bacteroides strains were resuspended from a plate into PBS at a density of approximately 1010/ml and 2 μl volumes were spotted on a BHIS plate and grown anaerobically at 37°C overnight. The bacteria were removed with a swab and the residual bacteria remaining on the plate were killed by exposing to chloroform vapor for 15 min. Strains to be tested for growth inhibition were grown to an OD600 of 0.6, and then 100 μl was mixed with 4 ml top agar and overlaid onto the chloroform-treated plate. The zones of inhibition were analyzed after overnight incubation at 37°C anaerobically.
Transposon mutagenesis and deletion of BF638R_1646
Random mutagenesis of B. fragilis 638R was performed using the transposon containing plasmid pYT646b as described (Tang & Malamy, 2000) using tetracycline selection. Transposon mutants were screened for loss of inhibitory activity using the agar spot test with B. fragilis 12905 as the overlay strain. The transposon insertion site of M26, which lost the ability to inhibit the growth of strain 12905, was identified by cloning the junction, taking advantage of the β-lactamase gene and E. coli origin of replication contained at the end of the transposon. The chromosomal DNA was digested with HindIII followed by dilute ligation, transformed into E. coli DH5α and plated on ampicillin plates. The junctional DNA from the resulting plasmid was identified by DNA sequencing using a primer directed out of the transposon.
A deletion of BF638R_1646 was constructed such that 1025 bp of 1119 bp gene was removed. DNA segments upstream and downstream of the region to be deleted were PCR amplified and the PCR products were digested with BamHI and MluI and cloned by three way ligation into the BamHI site of pNJR6 (Stevens et al., 1990). The resulting plasmid was conjugally transferred into wild-type B. fragilis 638R using helper plasmid R751and cointegrates were selected by erythromycin resistance. Double cross outs were screened by PCR for the mutant genotype.
Cloning and expression of BF638R_1646 and His-BF638R_1646
BF638R_1646 was PCR amplified using primers with BamHI sites, the PCR product was digested with BamHI and cloned into the BamHI site of Bacteroides expression vector pMCL140 (Chatzidaki-Livanis et al., 2010) and screened for orientation. This yielded plasmid pMCL139, and was added in trans to B. fragilis strain CM13 or 638R by conjugal mating using helper plasmid RK231.
An N-terminal His-tagged fusion of BF638R_1646 was constructed by cloning the PCR amplified BF638R_1646 into the NdeI site of pET16b (Novagen) and screening for orientation. The resulting recombinant protein, His-BSAP-1, was induced using the standard IPTG protocol with 40μg/ml IPTG for 2.5 hrs. His-BSAP-1 was purified using the ProBond Purification System (Life Technologies). The eluted protein was dialyzed in PBS. Protein concentration was determined using the BCA kit (Thermo Scientific).
Site-directed mutations in BF638R_1646
Site-directed mutagenesis of BF638R_1646 in plasmid pMCL139 (for expression in B. fragilis) or plasmid pMCL189 (for His-tagged expression in E. coli) was carried out with the QuikChange XL Kit (Agilent Technologies) with an elongation time of 13 min with the primers listed in Table S5. All mutations were confirmed by sequencing of the entire gene and the promoter region. In order to get soluble mutant proteins, the induction of both the His-tagged wild type protein and the mutant His-tagged proteins was performed at room temperature. Purification of the His-tagged proteins occurred as described above. Purified mutant His-tagged proteins were concentrated using Amicon Ultra centrifugal filters.
Cloning of additional MACPF-encoded proteins
BT_3120, BT_3437, BT_3439, BACUNI_00959 and BF638R_2714 were PCR amplified and cloned into the BamHI site of Bacteroides expression vector pMCL140 (Chatzidaki-Livanis et al., 2010) and screened for orientation. This yielded plasmids pMCL176, pMCL175, pMCL174, pMCL178, and pMCL177, respectively, which were added in trans to B. fragilis strain CM13 or the wild type strain by conjugal mating using helper plasmid RK231.
Growth inhibitory analyses using purified His-BSAP-1
Agar overlay assays using purified His-BSAP-1 were performed the same as the regular agar spot assays except that dilutions of the purified protein in PBS were added to the BHIS plates in 5 μl volumes, allowed to dry and then overlaid with strains as described.
In separate experiments, 150 μl volumes of purified His-BSAP-1 containing either 49 μg of protein (experiment 1) or 75 μg of protein (experiment 2) or PBS as a control, were added to BHIS plates and allowed to dry. Dilutions of bacteria as indicated in PBS were added directly to these plates in 5 μl volumes and incubated overnight. After overnight growth, the area where the bacteria were applied were excised, added to PBS and mixed until completely dispersed, and the number of viable bacteria was enumerated by plate count. Duplicate plates were incubated for several days to visualize bacterial growth.
Propidium iodode labeling
Supernatants of overnight cultures of 638R pMCL139 and 638RΔ1646 grown without antibiotics were filtered through a 0.22μm filter and mixed with and equal volume of log phase grown 12905. Two μl of the mixtures were placed on 0.8% agarose pads containing propidium iodide (Molecular Probes) and sealed under anaerobic conditions. Bacteria were immediately imaged using a time-lapse Axioplan 2 imaging system (Zeiss) at 1000x (540/625 channel).
Co-culture experiments
For co-culture experiments on BHIS plates, a 10μl aliquot of log phase 12905, containing bacteria as listed in Table 3, was mixed with 90μl aliquots of log phase 638R or ΔBF1646 in excess with the following ratios for the various experiments (12905:638R or ΔBF1646) exp 1, 1:18; exps 2 – 5, 1:12.8, 1:19.6; exps 6 – 9, 1:14.5, 1:19.5. 10μl aliquots of the above mixtures were spotted on BHIS plates and incubated anaerobically at 37°C overnight. The spots were excised, suspended in PBS and serial dilutions were plated to erythromycin plates to quantify 12905, which is resistant whereas 638R is erythromycin sensitive. Statistical analyses were performed using the Wilcoxon signed-rank test.
For co-culture experiments in broth, a 500μl aliquot of log phase 12905 and a 500μl aliquot of 638R or ΔBF1646 were added to 9 ml of fresh medium and incubated anaerobically at 37°C. Cfus of 12905 were monitored over time by plate counts under erythromycin selection.
Western immunoblot analyses and preparation of anti-His-BSAP-1 antiserum
For Western immunoblot analyses, bacteria or cellular fractions were boiled in LDS sample buffer and subjected to electrophoresis using NuPAGE 4 to 12% gradient sodium dodecyl sulfate-polyacrylamide gels with MES buffer (Life Technologies). The contents of the gels were transferred to PVDF membranes. Antiserum to purified His-BSAP-1 was prepared in rabbits by Lampire Biologicals using the EXPRESS-LINE polyclonal antiserum protocol. Alkaline phosphatase-labeled anti-rabbit IgG secondary antibody (Pierce) was the secondary antibody, and the membranes were developed with BCIP/NBT (KPL, Gaithersburg, MD).
Outer membrane vesicle (OMV) preparation and labeling
OMVs from B. fragilis 638R pMCL139 were prepared essentially as described (Patrick et al., 1996). In brief, the supernatants of bacterial cultures grown to OD600 of 0.6 were collected and filtered through a 0.45μm filter to remove any remaining bacteria. The filtered supernatants were ultracentrifuged at 131,000 × g at 10°C for 1hr to pellet the OMVs. The OMVs were washed in 10 ml PBS and ultracentrifuged as before. For immunolabeling, the washed OMVs were incubated with anti-His-BSAP-1 antiserum (1:100) for 1 hr, ultracentrifuged and washed and then incubated with Alexa-488 goat-anti-rabbit IgG for 1hr and washed by ultracentrifugation. The OMV were visualized using an Axioplan 2 imaging system (Zeiss) at 1000x. For agar spot overlay analyses, washed OMVs from the supernatant of 15 ml of 638R pMCL139 or ΔBF1646 grown to mid-log (OD600 0.6) containing 52 μg or 50 μg of total protein respectively, were spotted in 50 μl aliquots on a BHIS plate, allowed to dry and then overlaid with strain 12905.
Identification of MACPF-containing proteins encoded by bacterial genomes
Complete and draft bacterial genomes available from GenBank (Benson et al., 2013) (at ftp.ncbi.nih.gov/genbank/genomes/Bacteria and /Bacteria_DRAFT, respectively) and RefSeq (at ftp.ncbi.nih.gov/genomes/Bacteria and /Bacteria_DRAFT, respectively) as of June 8, 2014 were retrieved if the submission included the proteome in fasta format (as an .faa file). Where multiple genomes had been assigned the same NCBI unique id number (UID) or the same NCBI taxonomic ID, one genome from the duplicated group was selected. The genome set thus compiled comprises 15,948 genomes (8,121 RefSeq genomes (2,695 complete and 5,426 draft) and 7,827 GenBank genomes (49 complete and 7,778 draft)). The profile hidden Markov model (HMM) for the MAC/Perforin domain (PF01823.14) was retrieved from the library of Pfam-A HMMs maintained by the Wellcome Trust Sanger Institute as part of Pfam version 27.0 (Punta et al., 2012). A custom Perl script was written to compare this HMM profile to all 55,185,390 protein sequences using version v3.1b1 of the Windows hmmsearch program contained in the HMMER 3.1 suite (Eddy, 2011). Proteins found to have a full sequence bit score that equaled or exceeded the gathering threshold bit score cut-off for the profile HMM (20.70) as a result of this search were enumerated and retained as MACPF-containing proteins (Table S2). Each of the MACPF-containing proteins was additionally scanned for the presence of an SpII cleavage site (signal sequence) using the Linux version 1.0a of LipoP (Juncker et al., 2003) under Fedora 15.
Generation of the phylogenetic tree
The 163 protein sequences from the Bacteroidetes phylum found to have a significant match to the PFam MAC/Perforin domain (PF01823.14) were consolidated into 71 groups using version 2.2.26 of blastclust from NCBI setting a similarity threshold (-S, percent of identical residues) of 99%, a minimum length coverage (-L) of 1.0, and with the alignment length threshold enforced on only one member of a sequence pair (-b F). A representative member of each cluster was used in the generation of the tree; all members of the clusters are listed in Table S4.
The set of representative sequences was aligned using CLUSTALW (Larkin et al., 2007) as implemented in the MEGA6 program (Tamura et al., 2013)._Inference of evolutionary relationships was performed using the Neighbor-Joining method (Saitou & Nei, 1987), evolutionary distances were computed using the JTT matrix-based method (Jones et al., 1992) and the reliability of the phylogeny was estimated using the bootstrap test (Felsenstein, 1985) with 1,000 replicates. Evolutionary distances are shown as the number of amino acid substitutions per site. During the analysis, ambiguous positions were removed for each sequence pair, yielding a total of 1,149 positions in the Bacteroidetes datasets, respectively.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grants AI081843 and AI093771 from the National Institute of Allergy and Infectious Diseases.
Footnotes
The authors declare no conflict of interest.
References
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, Meta HITC, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Merieux A, Melo Minardi R, M’Rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avelar KE, Pinto LJ, Antunes LC, Lobo LA, Bastos MC, Domingues RM, Ferreira MC. Production of bacteriocin by Bacteriodes fragilis and partial characterization. Lett Appl Microbiol. 1999;29:264–268. doi: 10.1046/j.1365-2672.1999.00603.x. [DOI] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41:D36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolitho P, Voskoboinik I, Trapani JA, Smyth MJ. Apoptosis induced by the lymphocyte effector molecule perforin. Current opinion in immunology. 2007;19:339–347. doi: 10.1016/j.coi.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Cascales E, Buchanan SK, Duche D, Kleanthous C, Lloubes R, Postle K, Riley M, Slatin S, Cavard D. Colicin biology. Microbiol Mol Biol Rev. 2007;71:158–229. doi: 10.1128/MMBR.00036-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case TJ, Gilpin ME. Interference competition and niche theory. Proc Natl Acad Sci U S A. 1974;71:3073–3077. doi: 10.1073/pnas.71.8.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzidaki-Livanis M, Weinacht K, Comstock L. Trans locus inhibitors limit concomitant polysaccharide synthesis in the human gut symbiont Bacteroides fragilis. Proc Natl Acad Sci U S A. 2010;107:11976–11980. doi: 10.1073/pnas.1005039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comstock LE, Pantosti A, Kasper DL. Genetic diversity of the capsular polysaccharide C biosynthesis region of Bacteroides fragilis. Infect Immun. 2000;68:6182–6188. doi: 10.1128/iai.68.11.6182-6188.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol. 2013;11:285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- Cotter PD, Ross RP, Hill C. Bacteriocins - a viable alternative to antibiotics? Nat Rev Microbiol. 2013;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- Coyne M, Zitomersky N, McGuire A, Earl A, Comstock L. Evidence of extensive DNA transfer between Bacteroidales species within the human gut. mBio. 2014;5:e1305–1314. doi: 10.1128/mBio.01305-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. Accelerated Profile HMM Searches. PLoS computational biology. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhenawy W, Debelyy MO, Feldman MF. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio. 2014;5:e00909–00914. doi: 10.1128/mBio.00909-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farias LM, Totola AH, Miranda CM, Carvalho MA, Damasceno CA, Tavares CA, Cisalpino EO, Vieira EC. Extraction, partial purification and characterization of a bacteriocin (fragicilin) produced by a strain of Bacteroides fragilis isolated from Callithrix penicillata. Res Microbiol. 1994;145:9–16. doi: 10.1016/0923-2508(94)90062-0. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell. 2009;137:321–331. doi: 10.1016/j.cell.2009.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Computer applications in the biosciences : CABIOS. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Beveridge TJ. Bacteriolytic effect of membrane vesicles from Pseudomonas aeruginosa on other bacteria including pathogens: conceptually new antibiotics. J Bacteriol. 1996;178:2767–2774. doi: 10.1128/jb.178.10.2767-2774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr B, Riley MA, Feldman MW, Bohannan BJ. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- Kirkup BC, Riley MA. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature. 2004;428:412–414. doi: 10.1038/nature02429. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel-Briand Y, Baysse C. The pyocins of Pseudomonas aeruginosa. Biochimie. 2002;84:499–510. doi: 10.1016/s0300-9084(02)01422-0. [DOI] [PubMed] [Google Scholar]
- Miranda CM, Farias LM, Carvalho MA, Damasceno CA, Totola AH, Tavares CA, Cisalpino EO, Vieira EC. Purification and partial characterization of a bacteriocin isolated from Bacteroides ovatus H47. Can J Microbiol. 1993;39:169–174. doi: 10.1139/m93-023. [DOI] [PubMed] [Google Scholar]
- Morita-Yamamuro C, Tsutsui T, Sato M, Yoshioka H, Tamaoki M, Ogawa D, Matsuura H, Yoshihara T, Ikeda A, Uyeda I, Yamaguchi J. The Arabidopsis gene CAD1 controls programmed cell death in the plant immune system and encodes a protein containing a MACPF domain. Plant & cell physiology. 2005;46:902–912. doi: 10.1093/pcp/pci095. [DOI] [PubMed] [Google Scholar]
- Pantosti A, Tzianabos AO, Onderdonk AB, Kasper DL. Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infect Immun. 1991;59:2075–2082. doi: 10.1128/iai.59.6.2075-2082.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papastathopoulou A, Bezirtzoglou E, Legakis NJ. Bacterioides fragilis: production and sensitivity to bacteriocins. Anaerobe. 1997;3:203–206. doi: 10.1006/anae.1997.0106. [DOI] [PubMed] [Google Scholar]
- Patrick S, McKenna JP, O’Hagan S, Dermott E. A comparison of the haemagglutinating and enzymic activities of Bacteroides fragilis whole cells and outer membrane vesicles. Microb Pathog. 1996;20:191–202. doi: 10.1006/mpat.1996.0018. [DOI] [PubMed] [Google Scholar]
- Pipkin ME, Lieberman J. Delivering the kiss of death: progress on understanding how perforin works. Current opinion in immunology. 2007;19:301–308. doi: 10.1016/j.coi.2007.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. Chlamydial homologues of the MACPF (MAC/perforin) domain. Curr Biol. 1999;9:R911–913. doi: 10.1016/s0960-9822(00)80102-5. [DOI] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakoff-Nahoum S, Coyne MJ, Comstock LE. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol. 2014;24:40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado CJ, Buckle AM, Law RH, Butcher RE, Kan WT, Bird CH, Ung K, Browne KA, Baran K, Bashtannyk-Puhalovich TA, Faux NG, Wong W, Porter CJ, Pike RN, Ellisdon AM, Pearce MC, Bottomley SP, Emsley J, Smith AI, Rossjohn J, Hartland EL, Voskoboinik I, Trapani JA, Bird PI, Dunstone MA, Whisstock JC. A common fold mediates vertebrate defense and bacterial attack. Science. 2007;317:1548–1551. doi: 10.1126/science.1144706. [DOI] [PubMed] [Google Scholar]
- Russell AB, Wexler AG, Harding BN, Whitney JC, Bohn AJ, Goo YA, Tran BQ, Barry NA, Zheng H, Peterson SB, Chou S, Gonen T, Goodlett DR, Goodman AL, Mougous JD. A Type VI Secretion-Related Pathway in Bacteroidetes Mediates Interbacterial Antagonism. Cell Host Microbe. 2014;16:227–236. doi: 10.1016/j.chom.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schoener TW. Field experiments on interspecific competition. Am Nat. 1983;122:240–285. [Google Scholar]
- Shanahan F. The colonic microbiota in health and disease. Curr Opin Gastroenterol. 2013;29:49–54. doi: 10.1097/MOG.0b013e32835a3493. [DOI] [PubMed] [Google Scholar]
- Shen Y, Giardino Torchia ML, Lawson GW, Karp CL, Ashwell JD, Mazmanian SK. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe. 2012;12:509–520. doi: 10.1016/j.chom.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipman JA, Cho KH, Siegel HA, Salyers AA. Physiological characterization of SusG, an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron. J Bacteriol. 1999;181:7206–7211. doi: 10.1128/jb.181.23.7206-7211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens AM, Shoemaker NB, Salyers AA. The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. J Bacteriol. 1990;172:4271–4279. doi: 10.1128/jb.172.8.4271-4279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular biology and evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YP, Malamy MH. Isolation of Bacteroides fragilis mutants with in vivo growth defects by using Tn4400’, a modified Tn4400 transposition system, and a new screening method. Infect Immun. 2000;68:415–419. doi: 10.1128/iai.68.1.415-419.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LD, Nelson DE, Dorward DW, Whitmire WM, Caldwell HD. Biological characterization of Chlamydia trachomatis plasticity zone MACPF domain family protein CT153. Infect Immun. 2010;78:2691–2699. doi: 10.1128/IAI.01455-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp J, Masson D, Stanley KK. Structural/functional similarity between proteins involved in complement- and cytotoxic T-lymphocyte-mediated cytolysis. Nature. 1986;3:831–834. doi: 10.1038/322831a0. [DOI] [PubMed] [Google Scholar]
- Xu Q, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Cai X, Carlton D, Chen C, Chiu HJ, Clayton T, Das D, Deller MC, Duan L, Ellrott K, Farr CL, Feuerhelm J, Grant JC, Grzechnik A, Han GW, Jaroszewski L, Jin KK, Klock HE, Knuth MW, Kozbial P, Krishna SS, Kumar A, Lam WW, Marciano D, Miller MD, Morse AT, Nigoghossian E, Nopakun A, Okach L, Puckett C, Reyes R, Tien HJ, Trame CB, van den Bedem H, Weekes D, Wooten T, Yeh A, Zhou J, Hodgson KO, Wooley J, Elsliger MA, Deacon AM, Godzik A, Lesley SA, Wilson IA. Structure of a membrane-attack complex/perforin (MACPF) family protein from the human gut symbiont Bacteroides thetaiotaomicron. Acta crystallographica Section F, Structural biology and crystallization communications. 2010;66:1297–1305. doi: 10.1107/S1744309110023055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitomersky NL, Coyne MJ, Comstock LE. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect Immun. 2011;79:2012–2020. doi: 10.1128/IAI.01348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.