Abstract
“Epigenetic Transgenerational Inheritance” (ETI) has been defined as germline (sperm or egg) transmission of epigenetic information between generations in the absence of direct exposures or genetic manipulations. Among reported cases of ETI in mammals, the majority are induced by environmental factors, including environmental toxicants [e.g. agricultural fungicide vinclozolin, plastic additive bisphenol A, pesticide methoxychlor, dioxin, di-(2-ethylhexyl) phthalate, dichlorodiphenyltrichloroethane, and hydrocarbons] and poor nutritional conditions. Although the ETI phenomenon is well established, the underlying mechanism remains elusive. Putative epimutations, including changes in DNA methylation and histone modification patterns, have been reported, but it remains unclear how these epimutations are formed in the first place, and how they are memorized in the germline and then get transmitted to subsequent generations. Based on recent advances in our understanding of regulatory noncoding RNAs (ncRNAs), I propose that ncRNAs are involved in ETI, during both the initial epimutation formation and the subsequent germline transmission of epimutations. ncRNAs can function at epigenetic levels by affecting DNA methylation and histone modifications, thereby changing gene transcriptional activities, which can lead to an altered mRNA transcriptome associated with a disease phenotype. Alternatively, novel or altered ncRNA expression can cause dysregulated post-transcriptional regulation, thus directly affecting the mRNA transcriptome and inducing a disease phenotype. Sperm-borne ncRNAs are potential mediators for epigenetic memory across generations, but they alone may not be sufficient for stable transmission of epimutations across generations. Overall, research on ncRNAs in the context of ETI is urgently needed to shed light on the underlying mechanism of ETI.
Keywords: Gamete, Sperm, Oocyte, Epigenetics, Genetics, Environment, Inheritance, Disease etiology
1. Introduction
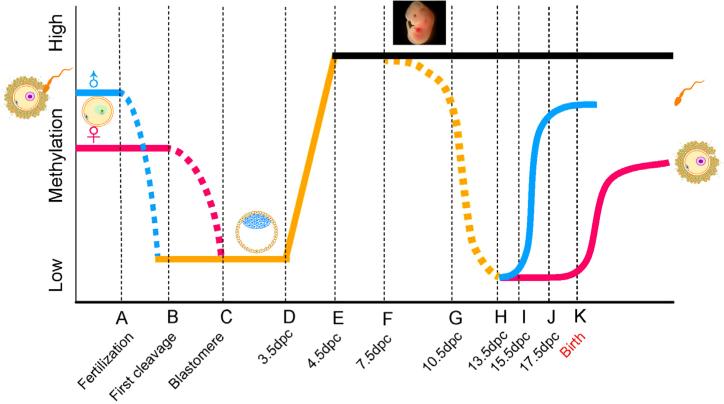
A recent definition refers to “epigenetics” as “the molecular factors/processes around the DNA that regulate genome activity independent of DNA sequence, and these processes are mitotically stable” (Skinner, 2014). Although epigenetic regulation does not involve changes in DNA sequences, it can significantly influence genome activities and gene expression levels, leading to phenotypic changes (Feinberg, 2007). Therefore, it has been widely accepted that epigenomic changes represent aspects of the physiological, or pathophysiological, response to changes in the environment in which an organism lives (Bonasio et al., 2010; Feinberg, 2007). Adverse epigenetic changes can lead to disease conditions, whereas beneficial epigenetic modifications may enhance the physiological fitness. Epigenetic changes in the somatic cells are not passed on to future generations. For organisms of sexual reproduction, these changes must be reflected in the germline in order to transmit to subsequent generations. However, even if epigenetic changes arise in the germline (sperm or oocyte), it has long been believed that the altered epigenetic information would not be passed on to subsequent generations because of the two waves of global epigenetic reprogramming that occur right after fertilization, and during primordial germ cell development in the fetal gonads (Fig. 1) (Bao and Yan, 2012; McCarrey, 2014). In mice, shortly after fertilization, the paternal and maternal genomes undergo active and passive demethylation, respectively; by the time of implantation, the genome of the embryo is largely demethylated. However, soon after implantation, the embryonic genome is rapidly remethylated followed by further methylation changes over the course of organogenesis. The physiological implication of this first wave of reprogramming lies in the erasure of gamete epigenetic marks, including some of those essential for gamete identity, and those potentially gained during parental gametogenesis, so that the new embryonic epigenome can be established to support early development (Seisenberger et al., 2013). When the embryonic primordial germ cells (PGCs) arise and start migrating towards the genital ridges, the PGC genome undergoes global demethylation. Upon arrival in the genital ridges, the PCG genome is largely demethylated; Soon after sexual differentiation in the primitive gonads, both the prospermatogonial, and the oocyte, genomes gain their germlinespecific methylation patterns, which are further modified in a relatively subtle manner during gametogenesis in adulthood. The second wave of reprogramming in PGCs is critical for germ cell fate commitment (Seisenberger et al., 2013). It was previously believed that the two waves of reprogramming events remove the vast majority, if not all, of the methylation marks gained from the parental germline (Seisenberger et al., 2013). However, subsequent studies reveal that neither of the two waves of DNA demethylation is complete as many of the imprinted loci and numerous repetitive sequences [e.g., IAPs (Intracisternal A-particle)] remain methylated after the first wave of reprogramming in the somatic cell lineages, or even after the second wave of reprogramming in the germline (Arney et al., 2001; Mochizuki and Matsui, 2010). Although several DNA-binding factors (e.g. MBD3, ZFP57, ARID4A and ARID4B) have been found essential for the maintenance of several specific genomic imprints (Bartolomei, 2009), the underlying molecular mechanisms remain largely unknown.
Fig. 1.
Schematic illustration showing the two global reprogramming events during murine development. Relative genomic methylation levels are represented on the vertical axis, whereas key stages of preimplantation and fetal germ cell development are shown on the horizontal axis. Blue and red lines denote the paternal and the maternal genomes, respectively. Dash lines represent the progression of demethylation. “dpc” stands for “days post-cotium”. This figure is adapted from (Bao and Yan, 2012).
Because of these reprogramming events, it has been widely accepted that epigenetic changes acquired during the lifetime of one individual would be completely erased and reset in the germline, and thus, there would be no chance for those epigenetic changes to be transmitted to next generations (McCarrey, 2014). It was furthermore believed that even if some of the acquired epigenetic information were to be transmitted to the immediate next generation, the reprogramming events would eventually erase those epigenetic marks in subsequent generations (McCarrey, 2014). Therefore, the concept of transgenerational epigenetic inheritance (ETI) has been controversial (McCarrey, 2014; Skinner, 2011). However, increasing lines of evidence suggest that certain induced pheno-types indeed can be passed on to subsequent generations through the germline, and that this non-Mendelian transmission of pheno-type is most likely mediated by epigenetic mechanisms.
2. Environmental epigenetic transgenerational inheritance (EETI)
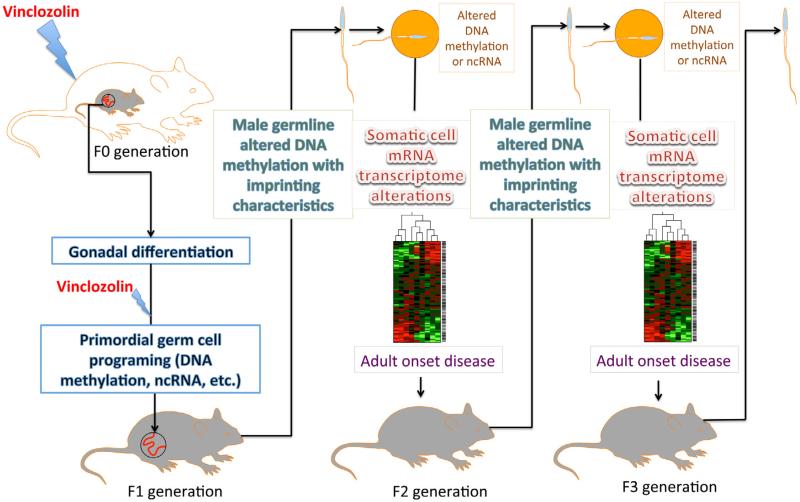
Environmental epigenetic transgenerational inheritance (EETI) refers to the phenomenon in which exposure of the gestating mother (F0), along with the developing fetus (F1), to environmental insults, such as endocrine disruptors, toxicants, poor nutrition, etc., causes epimutations that are transmitted to the F3 generation, and beyond, through the germline, in the absence of continued exposure (Cuzin et al., 2008; Gluckman et al., 2007; Skinner, 2011; Skinner and Guerrero-Bosagna, 2009; Whitelaw and Whitelaw, 2008). The Skinner lab first reported, in 2005, that exposure of gestating female rats to vinclozolin, an environmental endocrine disruptor, during the time window (E8.5–E14.5) of global demethylation in the PGCs in the fetal rat gonads (F1) causes a phenotype characterized by increased incidence of adult onset diseases, including disrupted spermatogenesis, prostate or kidney diseases, immune abnormalities, and tumorigenesis, in not only F1 and F2 generations, but also the F3 generation that is never directly exposed to vinclozolin (Anway et al., 2005) (Fig. 2). More interestingly, the phenotype can only be transmitted through the paternal germline, i.e., sperm. This non-Mendelian inheritance and lack of segregation suggest that epigenetic mechanisms are responsible for the transgenerational propagation of the initial effects (Anway et al., 2005; Skinner, 2008). Subsequent studies demonstrate that this transgenerational effect is associated with epimutations manifested as altered DNA methylation patterns, termed differential DNA methylation regions (DMRs), which are propagated specifically through the male germline (Anway et al., 2008; Clement et al., 2010; Guerrero-Bosagna et al., 2010; Skinner et al., 2008). These studies imply that the epimutations caused by the initial vinclozolin exposure may possess features similar to those of imprinted genes, thus allowing them to avoid reprogramming and to achieve transgenerational propagation. Moreover, recent data from the Skinner lab demonstrate that sperm epimutations subsequently alter the epigenomes and transcriptomes of developing Sertoli cells or granulosa cells, which may be responsible for the adult onset spermatogenic and ovarian disruptions leading to fertility defects (Nilsson et al., 2012; Skinner et al., 2012).
Fig. 2.
Schematic presentation of vinclozolin-induced transgenerational epigenetic inheritance of the adult onset disease phenotype. When a gestating rat (F0) is exposed to vinclozolin in the time window of primordial germ cell (PGC) reprogramming (E10.5–E13.5), epimutations [e.g. altered DNA methylation leading to the formation of differential methylation regions (DMRs)] arise and persist throughout spermatogenesis in F1 and also the post-fertilization global reprogramming in F2 embryos. These epimutations can directly cause aberrant expression of many mRNA genes, or indirectly affect mRNA expression by altering ncRNA expression. Dysregulated mRNA transcriptome results in different adult onset diseases in various organs. Epimutations and the disease phenotype can then be stably transmitted through the male germline, to not only the exposed generations (F1 and F2), but also subsequent generations that have never been exposed to vinclozolin (F3, F4, and beyond).
Since the report of vinclozolin-induced EETI in rats (Anway et al., 2005) (Fig. 2), similar phenomena have been reported in other species, including plants (Schmitz et al., 2011), worms (Greer et al., 2011), insects (Ruden and Lu, 2008), mice (Anway et al., 2005; Doyle et al., 2013) and humans (Pembrey, 2010), when exposed to various environmental factors, e.g., poor nutrition (Burdge et al., 2011; Hoile et al., 2011), cell culture conditions (Mahsoudi et al., 2007), vinclozolin (Anway et al., 2005), bisphenol A (Salian et al., 2009), dioxin (Bruner-Tran and Osteen, 2011), di-(2-ethylhexyl) phthalate (DEHP) (Doyle et al., 2013; Manikkam et al., 2013), dithiothreitol (DTT) (Kabasenche and Skinner, 2014; Skinner et al., 2013) and alcohol (Ramsay, 2010). Abnormalities manifested in the F1 (developing fetus) and F2 (progeny of F1) generations are not surprising because the F1s are directly exposed to the environmental factors, and F2s are derived from the sperm or eggs of F1s, which are also directly exposed during the fetal development (Fig. 2). However, observation of the same phenotype in the F3 generation, and beyond, is unexpected because these subsequent generations were never directly exposed (Daxinger and Whitelaw, 2012; Skinner, 2011; Whitelaw and Whitelaw, 2008). As discussed above; the two waves of reprogramming during post-fertilization and PGC development (Fig. 1) would erase the environmentally induced epimutations, if they were not protected through some means.
These data clearly demonstrate connections between DMRs, the altered mRNA transcriptome, and the adult onset disease pheno-type, which may provide some mechanistic insights into the EETI phenomenon. However, many questions remain unanswered, e.g.: How are specific genomic regions recognized and targeted for methylation to form DMRs? How do limited initial epigenetic changes induce alterations in expression profiles of many genes located outside those DMRs? How do those epimutations avoid correction by the global epigenetic reprogramming during post-fertilization and PGC development? How are those epimutations memorized by the germ cells and then transmitted to subsequent generations? Increasing lines of evidence suggest that the answers may lie in noncoding RNAs (ncRNAs).
3. Involvement of noncoding RNAs in the initial formation of epimutations
The majority of the noncoding regions of the mammalian genome are transcribed and processed into small and large noncoding RNAs (ncRNAs) (Cech and Steitz, 2014). ncRNAs can be divided into two main groups based on size; those with a size of >200 nt are called large noncoding RNAs (lncRNAs), while ncRNAs <200 nt represent small noncoding RNAs (sncRNAs). Accumulating data have shown that ncRNAs, especially large intergenic noncoding RNAs (lincRNAs), can function as epigenetic regulators by interacting directly with epigenetic factors involved in chromatin remodeling, DNA methylation, histone modifications or epigenetic memory (e.g. genomic imprinting and paramutations), thus achieving regulation of complex gene networks (Orom and Shiekhattar, 2013). Alternatively, lincRNAs can indirectly regulate epigenetic states by affecting transcriptional or translational activity, or by affecting the stability of mRNAs encoding epigenetic factors (Aravin and Hannon, 2008; Clark and Mattick, 2011; Costa, 2008; Esteller, 2011; Kaikkonen et al., 2011; Mattick, 2007, 2011; Morris, 2009). Therefore, ncRNAs, in theory, should have multiple molecular means to participate in EETI.
First, some of the long noncoding RNAs (lncRNAs), especially lincRNAs, are transcribed from sites within, or close to, the genomic regions that are destined to be silenced (Esteller, 2011; Gupta et al., 2010; Khalil et al., 2009; Tsai et al., 2010). These lincRNAs can serve as “sequence guides”, attracting chromatin remodeling complexes, or other epigenetic machineries, to achieve regional silencing through DNA methylation, or repressive histone modifications (Gupta et al., 2010; Khalil et al., 2009; Tsai et al., 2010). For example, the methylation of H19, a paternally imprinted gene, requires another lincRNA that is transcribed from the H19 locus (Gabory et al., 2010; Kwong et al., 2006). This lincRNA is essential for maintaining the hypermethylation status of the paternal copy of the H19 gene. Recently, it has also been demonstrated that the differential expression patterns in the HOXC gene cluster depend upon the expression of a lincRNA, called HOTAIR (Gupta et al., 2010; Rinn et al., 2007; Tsai et al., 2010). HOTAIR is transcribed in a region close to the HOXC gene cluster, and it enables the tethering of two distinct repressive complexes to chromatin for coupled H3K27 methylation and H3K4 demethylation (Gupta et al., 2010; Tsai et al., 2010). In this way, the HOXC gene cluster displays a wide variety of expression profiles in multiple organs of the body during development. Additionally, lincRNA-induced epigenetic silencing through chromatin remodeling may not have to occur in a sequence-specific manner, e.g., lncRNA XIST-mediated X chromosome inactivation (XCI) (Ng et al., 2007). XIST is expressed on the targeted X chromosome; it attracts and binds polycomb group protein complexes (PcGs), which, in turn, trimethylate H3K27 in cis, thus initiating the panchromosomal epigenetic silencing (Ponting et al., 2009). The fact that ~20% of lincRNAs expressed in human cells are bound by PcGs suggests that a similar mechanism may be utilized by other genes or gene clusters (Khalil et al., 2009). Therefore, it is conceivable that some lincRNAs may be involved in the initial formation of epimutations, caused by environmental exposures, possibly during the global epigenetic reprogramming that occurs in PGCs, or during gametogenesis.
Second, a subclass of sncRNAs that are associated with MIWI2 are exclusively expressed in PGCs, and thus, called MIWI2-piRNAs (or pre-pachytene piRNAs) (Aravin et al., 2008; Kuramochi-Miyagawa et al., 2008). MIWI2-piRNAs are mainly derived from repetitive sequences (e.g. transposons) (Aravin et al., 2008; Kuramochi-Miyagawa et al., 2008). These piRNAs are required for remethylation of transposons, like LINE1 elements, during the latter half (E14.5– E18.5) of reprogramming in male germ cells (Aravin et al., 2008; O’Donnell and Boeke, 2007), and for the maintenance of the hypermethylation status of at least one of several known imprinted genes, Rasgrf1 (Watanabe et al., 2011). In the MIWI2 KO mice, the activity of LINE1 transposons is drastically elevated, and the methylation pattern of Rasgrf1 is massively altered (Aravin et al., 2008; Bao et al., 2014; Kuramochi-Miyagawa et al., 2008; Zheng et al., 2010). Therefore, it is also possible that the initial epimutations induced by the environmental factors may involve some of the MIWI2-piRNAs.
Supporting these hypotheses, studies have shown that expression of lincRNAs and piRNAs occurs prior to heterochromatin formation, hypermethylation of DNA or repressive histone modifications (Kaikkonen et al., 2011; Morris, 2009). More importantly, these ncRNAs are expressed either from, or close to, the regions or loci that are targeted for silencing (Kaikkonen et al., 2011; Morris, 2009). These facts suggest that ncRNAs may serve as “sequence guides” that direct chromatin modifying protein complexes to the targeted regions/loci for epigenetic modifications. Therefore, it would be interesting to examine whether an environmental exposures induce the expression of unique lncRNAs and piRNAs from regions within, or proximal to, the exposure-specific DMRs in both somatic and germ cells.
Third, the DMRs identified by the Skinner lab (Skinner et al., 2012) contain numerous miRNAs, endo-siRNAs and pachytene/MIWI2-piRNAs, all of which mainly function by regulating mRNA stability and translational efficiency (Kaikkonen et al., 2011). Another group of ncRNAs, including promoter-associated RNAs (PARs) and enhancer RNAs (eRNAs), have been determined to bind the promoter regions of mRNA genes, and function as transcriptional activators by interacting with the transcriptional machinery (e.g. transcription factors, RNA Pol II, RNA-binding proteins, etc.) (Kaikkonen et al., 2011). If a DMR contains ncRNAs that regulate gene expression at transcriptional (PARs and eRNAs) or post-transcriptional (miRNAs, endo-siRNAs, and pachytene piRNAs) levels, then the implicated ncRNAs could have effects on the expression of numerous target genes, which could be encoded by genes distributed throughout the genome.
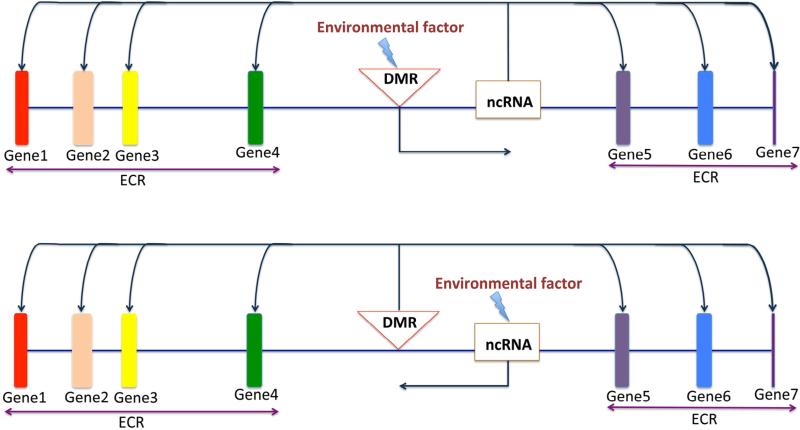
Alternatively, ncRNAs can also be involved in the distal effects of DMRs on the expression of multiple genes constituting an epi-genetic control region (ECR), through the following two potential mechanisms: First, environmental factors (e.g. vinclozolin) cause primary DNA methylation changes (DMRs) which, in turn, affect the expression of ncRNAs that are adjacent to the DMRs (Fig. 3, upper panel). Altered ncRNA expression then causes disrupted expression of many of its target genes, even those located distally. Second, environmental factors (e.g. vinclozolin) can directly affect the production of ncRNAs, especially large intergenic noncoding RNAs (lincRNAs), which are essential for sequence-specific DNA methylation and chromatin remodeling. Aberrant ncRNA production leads to altered DNA methylation patterns generally manifesting in DMRs, which, in turn, affect the expression of multiple mRNA genes located throughout the genome (Fig. 3, lower panel). Therefore, mapping ncRNAs located within, or proximal to, the DMRs would allow for the dissection of the relationship between changes in ncRNAs and alterations in the mRNA transcriptome.
Fig. 3.
Two potential mechanisms by which ncRNAs are involved in distal effects of differential DNA methylation regions (DMRs) on expression of multiple genes constituting an epigenetic control region (ECR). Upper panel: Environmental factors (e.g. vinclozolin) cause primary DNA methylation changes (DMRs), which, in turn, affect the expression of ncRNAs that are adjacent to the DMRs. Altered ncRNA expression then affects the expression of their target genes located distally. Lower panel: Alternatively, environmental factors (e.g. vinclozolin) can directly affect the production of ncRNAs, especially those large intergenic noncoding RNAs (lincRNAs), which are essential for sequence-specific DNA methylation (e.g. H19 lincRNA for the imprinting of H19 locus) and chromatin remodeling (e.g. HOTAIR for the epigenetic control of the Hox gene cluster). Aberrant ncRNA production leads to altered DNA methylation patterns manifested as DMRs which, in turn, affect expression of multiple mRNA genes located throughout the genome.
4. Sperm-borne ncRNAs as the epigenetic memory
The paternal transmission of epimutations, resulting from the in utero exposure of an F0 mother to vinclozolin, through the mother's descendants, suggests that the epimutations are ingrained into the “epigenetic memory” of the male germline, and that this information can be transmitted via sperm. The sperm epigenome includes the entire methylome, which is largely established during fetal germ cell development, and further completed during spermatogenesis (Carrell and Hammoud, 2010; Jenkins and Carrell, 2011; Talbert and Henikoff, 2010). Sperm chromatin is protamine-dominant, and only a small amount of histones are retained. Recent data suggest that sperm retained histones exist in various forms of modifications, and are associated with specific genomic regions that encode early developmental genes or sncRNAs (Anway et al., 2005; Brykczynska et al., 2010; Hammoud et al., 2009; Ihara et al., 2014; Molaro et al., 2011). In addition, sperm-borne RNAs, including RNAs that are associated with the sperm plasma membrane and sperm chromatin, also represent an integral part of the sperm epigenome because these RNA molecules can be delivered into the oocyte during fertilization (Ostermeier et al., 2002, 2004). The fact that the use of “membrane-free” sperm heads yields similar fertilization and birth rates through intracytoplasmic sperm injection (ICSI), as compared to unaltered sperm, suggests that the RNAs located in parts other than the head are not critically required for fertilization and subsequent embryonic development (Yan et al., 2008; Yanagimachi, 2005).
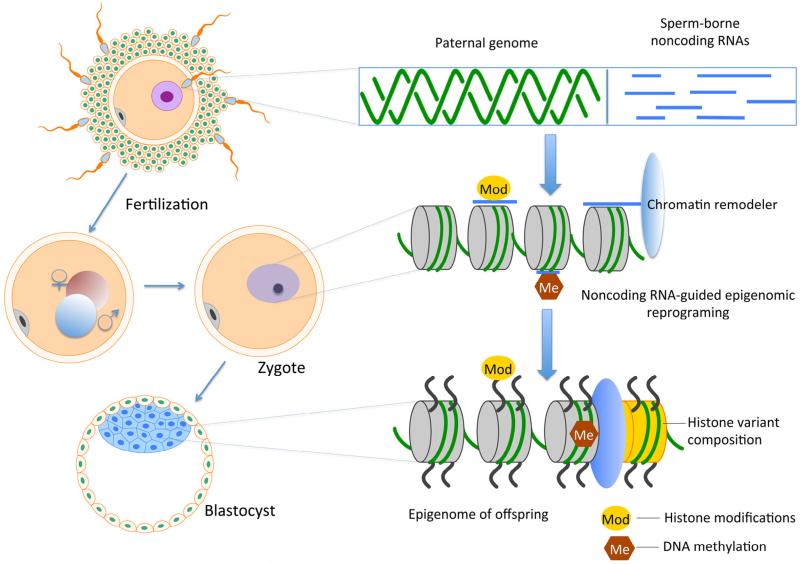
Soon after fertilization, the paternal genome undergoes reprogramming, which involves genome-wide demethylation, chromatin decondensation and histone restoration (Reik et al., 2001; Wilkins, 2005). Epigenetic reprogramming events in the preimplantation embryo, along with the subsequent germline-specific reprogramming events, would be expected to correct all acquired epimutations, unless those epimutations were to adopt an imprinting-like mechanism, thus persisting in the germline. Alternatively, sperm-borne RNAs, especially ncRNAs, may function as templates or sequence guides, to induce similar epimutations once released into oocytes during fertilization (Fig. 4). Sperm-borne RNAs, both large (mRNAs and lncRNAs) and small (sncRNAs), have been confirmed in many mammalian species (Krawetz et al., 2011; Lalancette et al., 2008; Martins and Krawetz, 2005; Miller et al., 2005; Ostermeier et al., 2002, 2004; Peng et al., 2012; Yan et al., 2008). These RNA molecules are delivered to the oocyte during fertilization, but their physiological roles remain elusive (Ostermeier et al., 2004). Previous studies suggest that sperm-borne RNAs may contribute to paramutational effects (i.e., one allele affects the expression of the other allele of a gene) (Rassoulzadegan et al., 2006). In plants, sperm-borne mRNAs and ncRNAs can serve as templates for modifying DNA sequences (Alleman et al., 2006; Lolle et al., 2005; Storici et al., 2007). However, this dramatic effect has not been observed in animal kingdom. Nevertheless, given that the epimutations induced by vinclozolin are selectively transmitted through the male germline, it is plausible to hypothesize that the RNA contents associated with sperm chromatin may contribute to this transgenerational inheritance (Fig. 4). Therefore, it would be interesting to determine changes in sperm-borne ncRNA profiles, in exposed sperm, across three or more generations, and to explore the effects of altered sperm ncRNA contents on the formation of specific DMRs and/or the disruption of mRNA expression in preimplantation embryos and adults. Similar work should also be conducted on oocytes because oocyte-mediated transgenerational epigenetic inheritance has been documented (Skinner, 2014; Skinner et al., 2013; Waterland et al., 2007), and similar mechanisms may function in the female germline to promote transgenerational epigenetic inheritance.
Fig. 4.
Schematic illustration of a potential mechanism by which sperm-borne ncRNAs mediate transgenerational epigenetic inheritance. In this model, sperm-borne ncRNAs are released during paternal chromatin decondensation. In the subsequent reprogramming of the preimplantation embryos, these paternal ncRNAs may function as the “sequence guide” to direct epigenetic machineries for DNA methylation, histone modification and chromatin remodeling to set up the epigenome of the early embryo. Some, if not all, of these ncRNA-mediated epigenetic marks may possess features of those imprinted loci, and thus, can avoid erasure during the 2nd round of reprogramming in the male germline, leading to transgenerational inheritance. The key in this model is that the ncRNAs are required for reprogramming.
In summary, ncRNAs might be the missing link between DNA sequences and their ultimate epigenetic states. ncRNAs can either act directly at epigenetic levels by affecting epigenetic modifications of the genome, or function to control the expression of epigenetic modulators and thus, indirectly induce epigenetic changes. ncRNAs exist and function in both somatic and germ cells; ncRNA changes in somatic cells lead to altered mRNA transcriptomes, and could potentiate disease phenotypes, whereas germline ncRNAs may mediate the transmission of epigenetic memory. These hypotheses may, or may not, be proven true, but are worth investigation, as elucidation of the underlying mechanisms of ETI would have a profound impact on our understanding of disease etiology.
Highlights.
ncRNAs may be involved in epigenetic transgenerational inheritance.
ncRNAs can function at epigenetic levels by affecting DNA methylation and histone modifications.
Novel or altered ncRNA expression can cause dysregulated post-transcriptional regulation.
Sperm-borne ncRNAs are potential mediators for epigenetic memory.
Acknowledgments
I would like to apologize to the authors whose relevant work was not cited due to the space limitation. Research in the Yan lab is supported by the NIH grants (HD060858, HD071736 and HD074573 to W. Y.). Dr. Daniel Oliver and Mr. Andrew Schuster are acknowledged for critical reading and text editing.
References
- Alleman M, Sidorenko L, McGinnis K, Seshadri V, Dorweiler JE, White J, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anway MD, Rekow SS, Skinner MK. Comparative anti-androgenic actions of vinclozolin and flutamide on transgenerational adult onset disease and spermatogenesis. Reprod. Toxicol. 2008;26:100–106. doi: 10.1016/j.reprotox.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb. Symp. Quant. Biol. 2008;73:283–290. doi: 10.1101/sqb.2008.73.058. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arney KL, Erhardt S, Drewell RA, Surani MA. Epigenetic reprogramming of the genome – from the germ line to the embryo and back again. Int. J. Dev. Biol. 2001;45:533–540. [PubMed] [Google Scholar]
- Bao J, Yan W. Male germline control of transposable elements. Biol. Reprod. 2012;86:162, 1–14. doi: 10.1095/biolreprod.111.095463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Zhang Y, Schuster AS, Ortogero N, Nilsson EE, Skinner MK, et al. Conditional inactivation of Miwi2 reveals that MIWI2 is only essential for prospermatogonial development in mice. Cell Death Differ. 2014;21:783–796. doi: 10.1038/cdd.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS. Genomic imprinting: employing and avoiding epigenetic processes. Genes Dev. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruner-Tran KL, Osteen KG. Developmental exposure to TCDD reduces fertility and negatively affects pregnancy outcomes across multiple generations. Reprod. Toxicol. 2011;31:344–350. doi: 10.1016/j.reprotox.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, et al. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 2010;17:679–687. doi: 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- Burdge GC, Hoile SP, Uller T, Thomas NA, Gluckman PD, Hanson MA, et al. Progressive, transgenerational changes in offspring phenotype and epigenotype following nutritional transition. PLoS ONE. 2011;6:e28282. doi: 10.1371/journal.pone.0028282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrell DT, Hammoud SS. The human sperm epigenome and its potential role in embryonic development. Mol. Hum. Reprod. 2010;16:37–47. doi: 10.1093/molehr/gap090. [DOI] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Clark MB, Mattick JS. Long noncoding RNAs in cell biology. Semin. Cell Dev. Biol. 2011;22:366–376. doi: 10.1016/j.semcdb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Clement TM, Savenkova MI, Settles M, Anway MD, Skinner MK. Alterations in the developing testis transcriptome following embryonic vinclozolin exposure. Reprod. Toxicol. 2010;30:353–364. doi: 10.1016/j.reprotox.2010.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FF. Non-coding RNAs, epigenetics and complexity. Gene. 2008;410:9–17. doi: 10.1016/j.gene.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Cuzin F, Grandjean V, Rassoulzadegan M. Inherited variation at the epigenetic level: paramutation from the plant to the mouse. Curr. Opin. Genet. Dev. 2008;18:193–196. doi: 10.1016/j.gde.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat. Rev. Genet. 2012;13:153–162. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- Doyle TJ, Bowman JL, Windell VL, McLean DJ, Kim KH. Transgenerational effects of di-(2-ethylhexyl) phthalate on testicular germ cell associations and spermatogonial stem cells in mice. Biol. Reprod. 2013;88:112. doi: 10.1095/biolreprod.112.106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Gabory A, Jammes H, Dandolo L. The H19 locus: role of an imprinted non-coding RNA in growth and development. Bioessays. 2010;32:473–480. doi: 10.1002/bies.200900170. [DOI] [PubMed] [Google Scholar]
- Gluckman PD, Hanson MA, Beedle AS. Non-genomic transgenerational inheritance of disease risk. Bioessays. 2007;29:145–154. doi: 10.1002/bies.20522. [DOI] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, et al. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature. 2011;479:365–371. doi: 10.1038/nature10572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS ONE. 2010;5:e13100. doi: 10.1371/journal.pone.0013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoile SP, Lillycrop KA, Thomas NA, Hanson MA, Burdge GC. Dietary protein restriction during F0 pregnancy in rats induces transgenerational changes in the hepatic transcriptome in female offspring. PLoS ONE. 2011;6:e21668. doi: 10.1371/journal.pone.0021668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M, Meyer-Ficca ML, Leu NA, Rao S, Li F, Gregory BD, et al. Paternal poly (ADP-ribose) metabolism modulates retention of inheritable sperm histones and early embryonic gene expression. PLoS Genet. 2014;10:e1004317. doi: 10.1371/journal.pgen.1004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, Carrell DT. The paternal epigenome and embryogenesis: poising mechanisms for development. Asian J. Androl. 2011;13:76–80. doi: 10.1038/aja.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabasenche WP, Skinner MK. DDT, epigenetic harm, and transgenerational environmental justice. Environ. Health. 2014;13:62. doi: 10.1186/1476-069X-13-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaikkonen MU, Lam MT, Glass CK. Non-coding RNAs as regulators of gene expression and epigenetics. Cardiovasc. Res. 2011;90:430–440. doi: 10.1093/cvr/cvr097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. U.S.A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawetz SA, Kruger A, Lalancette C, Tagett R, Anton E, Draghici S, et al. A survey of small RNAs in human sperm. Hum. Reprod. 2011;26:3401–3412. doi: 10.1093/humrep/der329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong WY, Miller DJ, Ursell E, Wild AE, Wilkins AP, Osmond C, et al. Imprinted gene expression in the rat embryo-fetal axis is altered in response to periconceptional maternal low protein diet. Reproduction. 2006;132:265–277. doi: 10.1530/rep.1.01038. [DOI] [PubMed] [Google Scholar]
- Lalancette C, Miller D, Li Y, Krawetz SA. Paternal contributions: new functional insights for spermatozoal RNA. J. Cell. Biochem. 2008;104:1570–1579. doi: 10.1002/jcb.21756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolle SJ, Victor JL, Young JM, Pruitt RE. Genome-wide non-Mendelian inheritance of extra-genomic information in Arabidopsis. Nature. 2005;434:505–509. doi: 10.1038/nature03380. [DOI] [PubMed] [Google Scholar]
- Mahsoudi B, Li A, O’Neill C. Assessment of the long-term and transgenerational consequences of perturbing preimplantation embryo development in mice. Biol. Reprod. 2007;77:889–896. doi: 10.1095/biolreprod.106.057885. [DOI] [PubMed] [Google Scholar]
- Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS ONE. 2013;8:e55387. doi: 10.1371/journal.pone.0055387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins RP, Krawetz SA. RNA in human sperm. Asian J. Androl. 2005;7:115–120. doi: 10.1111/j.1745-7262.2005.00048.x. [DOI] [PubMed] [Google Scholar]
- Mattick JS. A new paradigm for developmental biology. J. Exp. Biol. 2007;210:1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- Mattick JS. The central role of RNA in human development and cognition. FEBS Lett. 2011;585:1600–1616. doi: 10.1016/j.febslet.2011.05.001. [DOI] [PubMed] [Google Scholar]
- McCarrey JR. Distinctions between transgenerational and nontransgenerational epimutations. Mol. Cell. Endocrinol. 2014 doi: 10.1016/j.mce.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Miller D, Ostermeier GC, Krawetz SA. The controversy, potential and roles of spermatozoal RNA. Trends Mol. Med. 2005;11:156–163. doi: 10.1016/j.molmed.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Matsui Y. Epigenetic profiles in primordial germ cells: global modulation and fine tuning of the epigenome for acquisition of totipotency. Dev. Growth Differ. 2010;52:517–525. doi: 10.1111/j.1440-169X.2010.01190.x. [DOI] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, et al. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris KV. Long antisense non-coding RNAs function to direct epigenetic complexes that regulate transcription in human cells. Epigenetics. 2009;4:296–301. doi: 10.4161/epi.4.5.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K, Pullirsch D, Leeb M, Wutz A. Xist and the order of silencing. EMBO Rep. 2007;8:34–39. doi: 10.1038/sj.embor.7400871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson E, Larsen G, Manikkam M, Guerrero-Bosagna C, Savenkova MI, Skinner MK. Environmentally induced epigenetic transgenerational inheritance of ovarian disease. PLoS ONE. 2012;7:e36129. doi: 10.1371/journal.pone.0036129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Shiekhattar R. Long noncoding RNAs usher in a new era in the biology of enhancers. Cell. 2013;154:1190–1193. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier GC, Dix DJ, Miller D, Khatri P, Krawetz SA. Spermatozoal RNA profiles of normal fertile men. Lancet. 2002;360:772–777. doi: 10.1016/S0140-6736(02)09899-9. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature. 2004;429:154. doi: 10.1038/429154a. [DOI] [PubMed] [Google Scholar]
- Pembrey ME. Male-line transgenerational responses in humans. Hum. Fertil. 2010;13:268–271. doi: 10.3109/14647273.2010.524721. [DOI] [PubMed] [Google Scholar]
- Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, et al. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res. 2012;22:1609–1612. doi: 10.1038/cr.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- Ramsay M. Genetic and epigenetic insights into fetal alcohol spectrum disorders. Genome Med. 2010;2:27. doi: 10.1186/gm148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruden DM, Lu X. Hsp90 affecting chromatin remodeling might explain transgenerational epigenetic inheritance in Drosophila. Curr. Genomics. 2008;9:500–508. doi: 10.2174/138920208786241207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salian S, Doshi T, Vanage G. Impairment in protein expression profile of testicular steroid receptor coregulators in male rat offspring perinatally exposed to bisphenol A. Life Sci. 2009;85:11–18. doi: 10.1016/j.lfs.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O'Malley RC, Urich MA, Libiger O, et al. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–373. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos. Trans. R. Soc. Lond. B. Biol Sci. 2013;368:20110330. doi: 10.1098/rstb.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK. What is an epigenetic transgenerational phenotype? F3 or F2. Reprod. Toxicol. 2008;25:2–6. doi: 10.1016/j.reprotox.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK. Environmental epigenetic transgenerational inheritance and somatic epigenetic mitotic stability. Epigenetics. 2011;6:838–842. doi: 10.4161/epi.6.7.16537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK. Endocrine disruptor induction of epigenetic transgenerational inheritance of disease. Mol. Cell. Endocrinol. 2014 doi: 10.1016/j.mce.2014.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Guerrero-Bosagna C. Environmental signals and transgenerational epigenetics. Epigenomics. 2009;1:111–117. doi: 10.2217/epi.09.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS ONE. 2008;3:e3745. doi: 10.1371/journal.pone.0003745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Mohan M, Haque MM, Zhang B, Savenkova MI. Epigenetic transgenerational inheritance of somatic transcriptomes and epigenetic control regions. Genome Biol. 2012;13:R91. doi: 10.1186/gb-2012-13-10-r91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Guerrero-Bosagna C, Haque M, Nilsson E, Bhandari R, McCarrey JR. Environmentally induced transgenerational epigenetic reprogramming of primordial germ cells and the subsequent germ line. PLoS ONE. 2013;8:e66318. doi: 10.1371/journal.pone.0066318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Manikkam M, Tracey R, Guerrero-Bosagna C, Haque M, Nilsson EE. Ancestral dichlorodiphenyltrichloroethane (DDT) exposure promotes epigenetic transgenerational inheritance of obesity. BMC Med. 2013;11:228. doi: 10.1186/1741-7015-11-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F, Bebenek K, Kunkel TA, Gordenin DA, Resnick MA. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. Histone variants –ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science. 2011;332:848–852. doi: 10.1126/science.1203919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA, Travisano M, Tahiliani KG. Diet-induced hypermethylation at agouti viable yellow is not inherited transgenerationally through the female. FASEB J. 2007;21:3380–3385. doi: 10.1096/fj.07-8229com. [DOI] [PubMed] [Google Scholar]
- Whitelaw NC, Whitelaw E. Transgenerational epigenetic inheritance in health and disease. Curr. Opin. Genet. Dev. 2008;18:273–279. doi: 10.1016/j.gde.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Wilkins JF. Genomic imprinting and methylation: epigenetic canalization and conflict. Trends Genet. 2005;21:356–365. doi: 10.1016/j.tig.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Yan W, Morozumi K, Zhang J, Ro S, Park C, Yanagimachi R. Birth of mice after intracytoplasmic injection of single purified sperm nuclei and detection of messenger RNAs and microRNAs in the sperm nuclei. Biol. Reprod. 2008;78:896–902. doi: 10.1095/biolreprod.107.067033. [DOI] [PubMed] [Google Scholar]
- Yanagimachi R. Intracytoplasmic injection of spermatozoa and spermatogenic cells: its biology and applications in humans and animals. Reprod. Biomed. Online. 2005;10:247–288. doi: 10.1016/s1472-6483(10)60947-9. [DOI] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc. Natl Acad. Sci. U.S.A. 2010;107:11841–11846. doi: 10.1073/pnas.1003953107. [DOI] [PMC free article] [PubMed] [Google Scholar]