Abstract
Components of the prokaryotic clustered regularly interspersed palindromic repeat (CRISPR) loci have recently been repurposed for use in mammalian cells1–6. The Cas9 protein can be programmed with a single guide RNA (sgRNA) to generate site-specific DNA breaks, but there are few known rules governing on-target efficacy of this system7,8. We created a pool of sgRNAs, tiling across all possible target sites of a panel of six endogenous mouse and three endogenous human genes and quantitatively assessed their ability to produce null alleles of their target gene by antibody staining and flow cytometry. We discovered sequence features that improved activity, including a further optimization of the proto-spacer adjacent motif (PAM) of Streptococcus pyogenes Cas9. The results from 1,841 sgRNAs were used to construct a predictive model of sgRNA activity to improve sgRNA design for gene editing and genetic screens. We provide an online tool for the design of highly active sgRNAs for any gene of interest.
When introduced into mammalian cells, the Cas9-sgRNA complex creates sequence specific dsDNA breaks that are repaired by the error-prone non-homologous end joining pathway (NHEJ), often resulting in gene inactivation by the creation of frameshift alleles4–6. Recently, we and others have shown that CRISPR technology can be used for large scale genetic screens in mammalian cells7,9–11. Hit genes from these screens exhibited high concordance between distinct sgRNAs targeting the same gene, higher than the agreement usually seen in RNAi screens. This indicates that the phenotypic consequences of knockout alleles may be more consistent than the varying degrees of knockdown induced by RNAi, and that off-target effects of sgRNAs, while detectable, are rare enough to allow high concordance of multiple sgRNAs targeting the same gene, facilitating the nomination of true positive hits7,9–15.
All genetic CRISPR-based screens published so far have either relied on positive selection for resistance to cytotoxic drugs or negative selection based on the depletion of essential genes7,9–11. While producing promising results, these assays all involved strong selective pressure expected to show robust signal even if only a modest proportion of cells receiving a particular sgRNA experienced full gene inactivation. Many future screens, especially those where screen success is not measured by increased survival or proliferation, will demand a high fraction of sgRNA-treated cells to be complete knockouts, as cells with no knockout alleles, heterozygous knockout, or hypomorphic alleles will dilute assay signals. Studies to date suggest that while sgRNA activity can be quite high, there is significant variability among sgRNAs in their ability to produce null alleles4–7,9,10,16–19. Design criteria to maximize sgRNA efficacy are thus of great utility, both to improve screening libraries and also for smaller scale gene-editing experiments, which often require researchers to first screen multiple sgRNAs for activity.
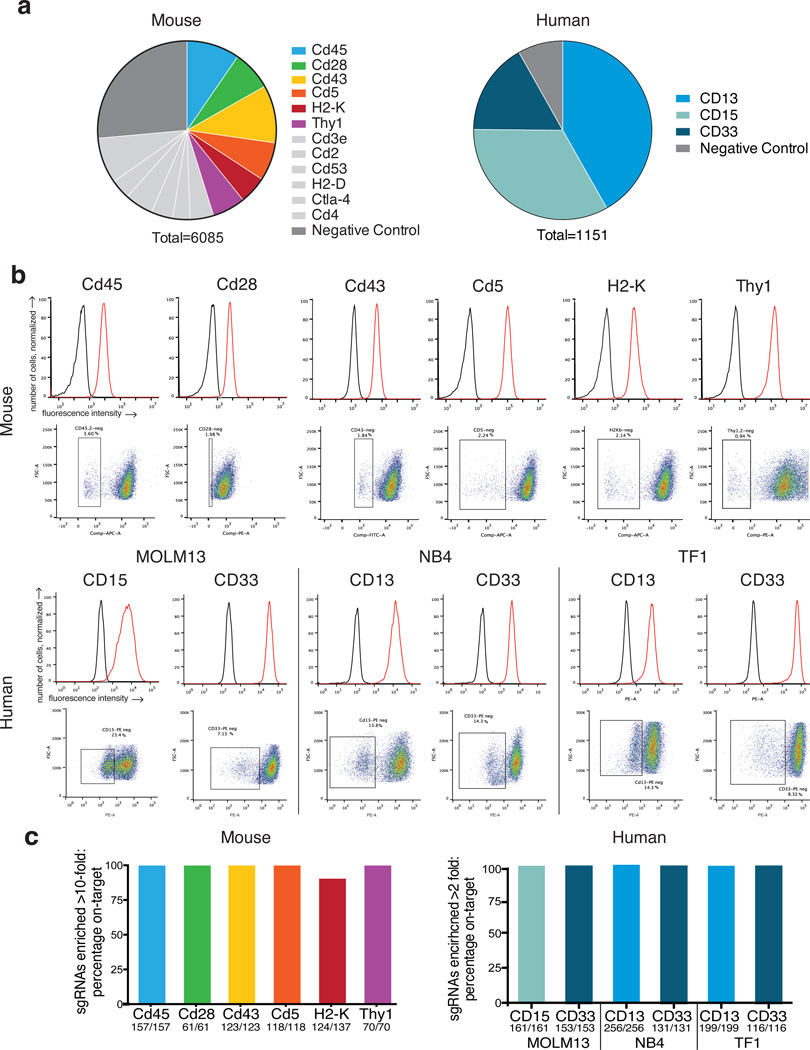
We therefore sought to discover sequence features within and surrounding the target site that predict sgRNA efficacy. To discover generally applicable rules, we tested a wide diversity of sgRNAs against multiple gene targets. Our strategy was to target cell surface markers in a large cell population, delivering one sgRNA per cell, and then isolating complete (biallelic) knockout cells by fluorescence-activated cell sorting (FACS), thereby separating the most active sgRNAs. We designed sgRNAs targeting a panel of mouse genes in all exons and flanking intronic sequence at all 20 nucleotide (nt) target sites that preceded the NGG PAM required by S. pyogenes Cas9, and added a large number of negative control sgRNAs (Fig. 1a, Supplementary Table 1). These sgRNAs were cloned as a pool into a lentiviral vector that simultaneously delivers Cas9, confers puromycin resistance, and expresses a sgRNA, as previously described9. A second pool targeting the coding sequence of three human cell surface markers and also including negative controls was separately cloned into a lentiviral vector that expresses only the sgRNA (Fig. 1a, Supplementary Table 2).
Figure 1.
sgRNA activity screens in mouse and human cells. (a) Representation of the sgRNA libraries. Colors represent genes assayed by FACS; light gray indicates genes either poorly expressed or not assayed; dark gray indicates targets not found in the mouse or human genomes. (b) Top: Antibody staining in cells (red) compared to unstained cells (black). Bottom: FACS plots indicating the negative population isolated for each cell surface marker after library transduction. (c) Percent of sgRNAs enriched >10-fold (mouse) or >2-fold (human) in the marker-negative population that were on-target.
We transduced EL4 cells, a mouse thymic cell line, with the mouse sgRNA pool. Nine days post transduction we stained cells for each of nine cell surface markers and analyzed by FACS. Endogenous Thy1, H2-K, Cd45, Cd43, Cd28, and Cd5 exhibited good resolution of marker-negative cell populations (Fig. 1b), while Cd2, Cd3e, and Cd53 were poorly expressed and excluded from subsequent analyses (Supplementary Fig. 1). To assess human gene targeting, we prepared three human AML cell lines, MOLM13, NB4, and TF1, by transduction with a vector delivering Cas9 and conferring blasticidin resistance. We confirmed Cas9 activity in these polyclonal lines (Supplementary Fig. 2), then transduced each with the human sgRNA pool and collected marker-negative populations 8 days post transduction. CD15, CD13 and CD33 were evaluated in one, two and three of the cell lines, respectively. For all 12 sorted cell populations, PCR of genomic DNA, followed by next generation sequencing (NGS) identified the sgRNAs that led to complete loss of the protein of interest (Supplementary Tables 2, 3).
We first examined the knockout specificity of the sgRNAs targeting each gene. In the mouse pool we observed between 61 – 157 sgRNAs per gene enriched at least 10-fold in each of the marker-negative cell populations after normalizing the abundance of sgRNAs in each sorted population to their starting abundance in the unsorted population. In the human pool, 116 – 256 sgRNAs per gene were at least 2-fold enriched after similar normalization; a lower threshold was used for this pool because each gene comprises a larger fraction of the overall pool. For 11 of the 12 sorted marker-negative populations, 100% of the highly-active sgRNAs were those that targeted the sorted marker, i.e. the ‘on-target’ sgRNAs, showing the excellent specificity of sgRNAs and of this FACS-based readout (Fig. 1c). For H2-K, all of the 10-fold enriched off-target sgRNAs were included in the pool to target H2-D, a gene not expressed in EL4 cells but highly similar in sequence to H2-K. All of these H2-D-targeting sgRNAs had at least 17 nts of complementarity to H2-K, with 11 differing by only a single nucleotide (Supplementary Table 4). As expected from previous studies on the specificity of sgRNAs, single-base mismatches that preserved activity were more frequent in the 5’ half of the sgRNA sequence14,15,19,20. Many of these off-target sgRNAs scored as likely off-target matches using a widely-used off-target scoring algorithm.14 Several, however, received low off-target scores, suggesting possible room for improvement in off-target predictions.
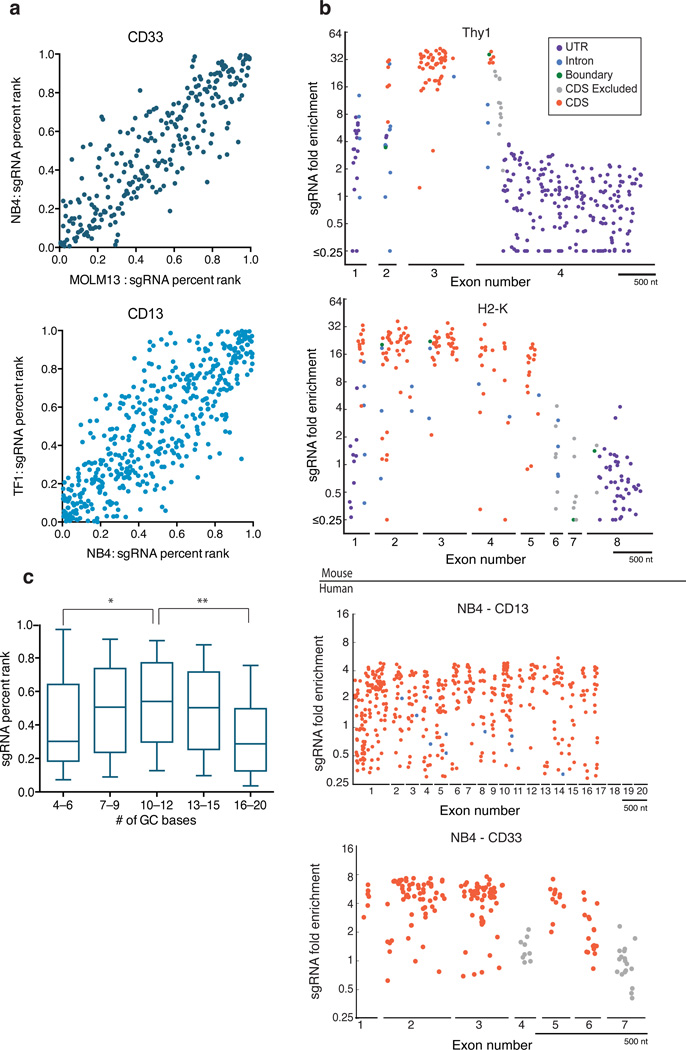
We next examined the consistency across cell lines of the activity of sgRNAs targeting CD13 or CD33. We observed strongly correlated sgRNA activity in four pairwise cross-cell-line comparisons, suggesting that relative levels of sgRNA activity can generalize across cellular contexts (Fig. 2a, Supplementary Fig. 3). To further validate the results obtained in pooled screening format, we re-tested the activity of 17 sgRNAs targeting three genes in arrayed format and observed good correspondence between these two assays (Supplementary Fig. 4a, Supplementary Table 5). We then examined the spectrum of DNA lesions caused by the 17 sgRNAs. As expected, we found that frameshift alleles were more common for the sgRNAs that were more enriched in the marker negative populations (Supplementary Fig. 4b).
Figure 2.
Features of sgRNA activity. (a) sgRNA concordance across cell lines. Pairwise comparison between cell lines of sgRNA percent-rank (see Methods for percent-rank calculation) for sgRNAs targeting CD13 or CD33; Spearman rank correlation of 0.87 and 0.80, respectively. (b) Activity maps of sgRNA by cut site position. Exons and 100 nts of flanking intron are represented as lines on the x-axis with gaps marking the remaining intronic sequence. sgRNAs excluded from activity modeling are indicated in gray. Boundary sgRNAs (green) are those where the cut site, between nts 17 and 18, falls between annotated regions (e.g. CDS/intron). All sgRNAs with fold enrichment ≤ 0.25 are grouped at the bottom of the y-axis. Scale bar indicates 500nt of sequence. (c) Activity as a function of G/C content for the 1,841 CDS-targeting sgRNAs analyzed. The top, middle and bottom lines of the box represent the 25th, 50th, and 75th percentiles, respectively; the whiskers represent the 10th and 90th percentiles. p* = 0.0003, p** = 3 × 10−11, Kolmogorov-Smirnov test.
For all nine target genes we annotated each sgRNA by the location of its cut site to determine how the position of a target site within the gene relates to its efficacy (Fig. 2b; Supplementary Fig. 5; Supplementary Table 6). Some exons contained no active sgRNA targets, suggesting that these exons were not expressed in the assayed cell line (Supplementary Fig. 5). As expected, we observed diminished activity of sgRNAs targeting close to the C’-terminus, since frameshift mutations close to the end of a protein are less likely to disrupt expression (Supplementary Fig. 6). Gene-specific patterns also emerged; for example, the N’-terminus of CD15 was a less-effective target site, perhaps reflecting local chromatin structure. These results show that while a wide-range of the CDS is generally suitable as a target site, exceptions could arise from gene-specific features. In a library-design context, targeting more than one site per gene should help to compensate for gene-specific limitations.
We next examined the activity of sgRNAs targeting non-coding regions in the mouse pool. We saw >10-fold enrichment among the knockout cells for 55% of sgRNAs with an expected cut site exactly at the exon - intron boundary. Activity quickly decreased as a function of distance to the nearest CDS: only 2 out of 50 sgRNAs with an expected cut site 6 nts or farther from the CDS showed greater than 10-fold enrichment (Supplementary Fig. 7). Finally, we observed that sgRNAs targeting the 5’ and 3’ UTRs were highly ineffective: 1 of 119 5’UTR-targeted sgRNAs and 0 of 1,044 3’UTR-targeted sgRNAs were enriched at least 10-fold in the target gene-negative cell population. These results suggest that sgRNAs should generally be designed to target the CDS, although target sites that disrupt splicing can be efficacious and may be particularly useful when it is desirable to re-introduce the CDS, such as for phenotypic rescue experiments.
To identify sequence features of sgRNAs that correlated with activity, we focused on the subset of sgRNAs targeting the CDS. We eliminated all sgRNAs in broadly ineffective target regions, e.g. due to proximity to the C’-terminus or apparent lack of exon expression, resulting in a set of 1,841 sgRNAs which were normalized by percent-rank within each gene (Supplementary Table 7). We examined target strand as a function of activity and saw no statistically-significant difference in contrast to a previously observed slight preference for the antisense strand (Supplementary Fig. 8)7. Additionally, we observed that sgRNAs with low or high G/C content tended to be less active (Fig. 2c), as previously reported7,8.
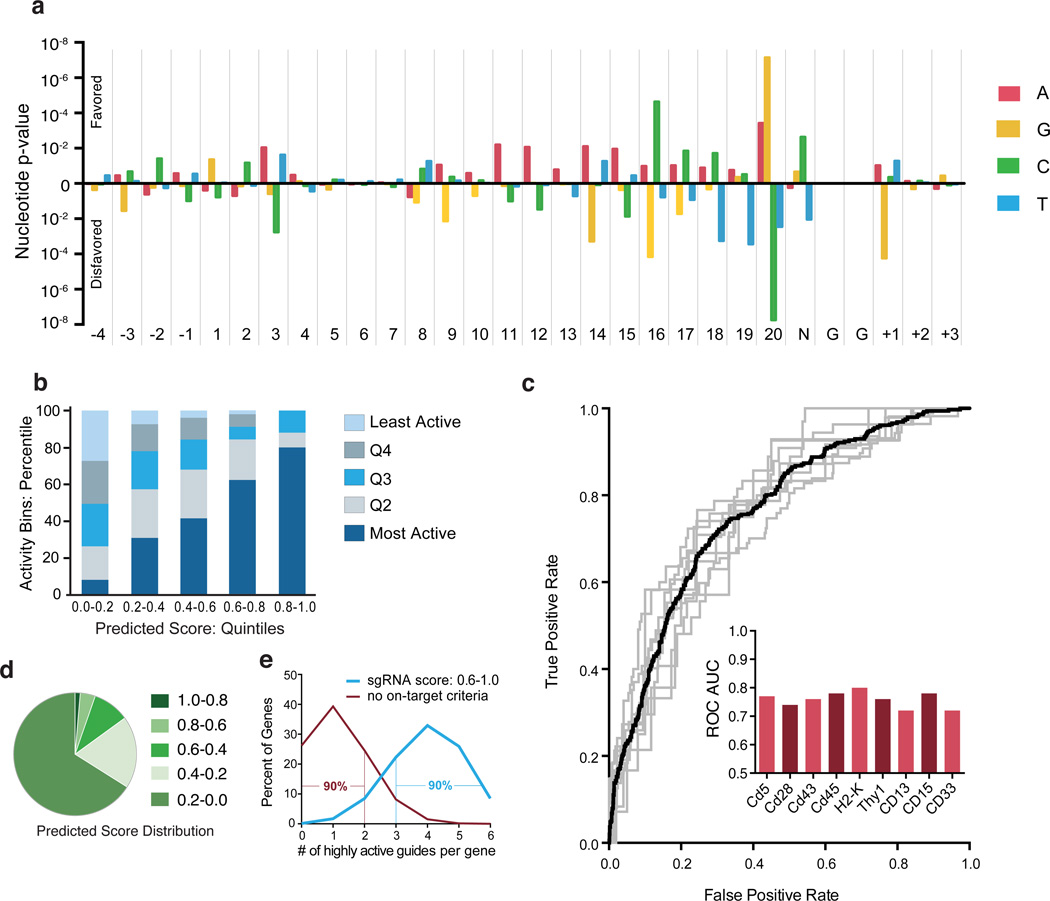
We next examined nucleotide preferences for active sgRNAs at every position across the length of the sgRNA and flanking target sequence. Specifically, we looked for statistical enrichment or depletion of sgRNAs with a given sequence feature among the top 20% most active of all sgRNAs for the same gene target, as these high-activity sgRNAs are of most interest (Fig. 3a, Supplementary Table 8). Within the sgRNA sequence, the most significant differences appeared at position 20, the nucleotide immediately adjacent to the PAM; in agreement with previous observations, we see that guanine is strongly preferred, and in our data, cytosine is strongly unfavorable7,8. Additionally, we see a preference for cytosine and against guanine at position 16. This is the last nucleotide of the seed region defined by a recent genome-wide analysis of Cas9 binding affinity20. In further agreement with Wang et al.7, there was a consistent preference for adenine in the middle of the sgRNA, and cytosine was disfavored at position 3.
Figure 3.
Model of sgRNA activity. (a) p-values of observing the conditional probability of a guide with a percent-rank activity of >0.8 under the null distribution examined at every position including the 4 nt upstream of the sgRNA target site, the 20 nt of sgRNA complementarity, the PAM, and the 3 nt downstream of the sgRNA target sequence. p-values were calculated from the binomial distribution with a baseline probability of 0.2 using 1,841 CDS-targeting guides. (b) Performance evaluation of sgRNA activity prediction scores based on nucleotide features. Scores for 1,841 sgRNAs are divided by quintile (x-axis) and experimentally-determined activity within each prediction group is assessed by sgRNA percent rank, and also binned by quintile (y-axis). (c) Performance validation of sgRNA prediction algorithm. The model was trained on all possible combinations of 8 genes and tested individually on the remaining held-out gene. Each gray line indicates the ROC curve for a held-out gene. The black line is the mean ROC curve. The bar graph inset indicates the Area Under the Curve (AUC) for each gene. (d) Distribution of 1,841 sgRNAs across predicted score quintiles. (e) Simulation of the fraction of most-active sgRNAs, arbitrarily defined as the top 20% of sgRNA for a gene, in hypothetical libraries with 6 sgRNAs per gene. For a library designed with no on-target criteria (null, in red) the values are simply the binominal expansion of 0.2. For the hypothetical library that incorporates sgRNA scoring rules to enrich for highly-active sgRNAs (blue), the model predicts that the top two quintiles of scores (0.6 – 1.0) contain 66.3% of most-active sgRNAs, and thus the values are the binomial expansion of 0.663.
Notably, we also observed a preference in the variable nucleotide of the PAM, where cytosine was favored and thymine was disfavored. The preference for cytosine at this position has also recently been observed in zebrafish8. The bias against thymine towards the 3’ end of the 20 nt sgRNA target site observed by us and others has previously been explained from the perspective of sgRNA expression, as RNA polymerase III terminates at U-rich regions and the transcript sequence immediately downstream of the 20 nt targeting sequence is U-rich7,20. This mechanism cannot be extended to explain the bias against thymine in the PAM, however, as this thymine is a feature of the DNA target site and is not included in the sgRNA transcript. Additionally, we observed a strong bias against guanine immediately 3’ of the PAM suggesting that an extended PAM sequence of CGGH is optimal for the use of S. pyogenes Cas9 to engineer dsDNA breaks in mammalian cells. Indeed, 39% of targets with a CGGT PAM were in the highest-activity quintile, compared to only 11% in the lowest quintile. Conversely, 42% of targets with the least-optimal PAM sequence of TGGG were in the lowest-activity quintile while only 8% were in the highest quintile.
We built a predictive model for sgRNA activity by training a logistic regression classifier to discriminate the highest-activity quintile of sgRNAs for each gene using sequence features. We used the data from all nine mouse and human genes to determine sequence feature weights for activity predictions (Supplementary Table 9). The quintile of highest scores was 80% comprised of the highest-activity sgRNAs and contained the fewest low-activity sgRNAs (Fig. 3b). Conversely, the lowest-score quintile contained the most low-activity sgRNAs and the smallest fraction of high-activity sgRNAs. We provide a simple web tool using this model to generate sgRNA scores for any sequence of interest (http://www.broadinstitute.org/rnai/public/analysis-tools/sgrna-design).
To ensure that this model generalizes across genes, we first cross-validated by training on eight genes while holding out the remaining gene, and the model accurately predicted activities for all nine held-out genes (Fig. 3c). Similarly, base preferences determined from the 959 sgRNAs in the mouse pool alone closely converge to the preferences obtained using the full 1,841-sgRNA dataset (Supplementary Fig. 9). Notably, the nine genes span a broad range of G/C content and length, and do not share any appreciable sequence homology, consistent with the observation of no cross-reactivity of sgRNAs among these genes (Fig. 1c). These analyses suggest that the dataset is large enough for the model to converge on a consistent pattern of base preferences.
We further validated the generalizability of the model against a set of 1,278 sgRNAs targeting 414 genes, using data from an earlier screen for viability effects in A375 cells, a human melanoma line9. We examined functional categories previously established to be most highly enriched for essential genes in all cell types (e.g. proteasome, ribosome, etc.) and analyzed the subset of genes that, in this viability screen, had multiple targeting sgRNAs that were depleted over time9,12,13. We then compared the predicted-efficacy scores for the sgRNAs targeting these 414 genes to their observed depletion in the screen. Similar to our observations for the FACS protein knockout assay, we saw that the highest quintile of predicted scores was comprised of the greatest proportion of high-activity sgRNAs, while the lowest-score quintile had the most low-activity sgRNAs (Supplementary Fig. 10, Supplementary Table 10). This prediction of activity for 1,278 sgRNAs targeting 414 genes, together with the high consistency observed in the base preferences across all sgRNAs for 9 genes, show that the model presented here generalizes widely to predict highly-active sgRNAs.
For screening approaches, a library of potent sgRNAs that provides good genome coverage is of primary importance, and we were thus more concerned with correctly identifying the highest activity sgRNAs than accurately modeling the activity of all sgRNAs. As a result, the scoring system presented here stringently scores predicted activity: only 5% of sgRNAs received a score of 0.6 or greater, while the majority of sgRNAs, including many sgRNAs that were experimentally highly-active, received scores of < 0.2 (Fig. 3d). Accordingly, the most powerful application of this model is as a sgRNA design tool, i.e. to select a few of the highest-scoring sgRNAs in order to obtain those most likely to be highly effective. Existing genome-wide libraries, while designed to avoid off-target sites, have not incorporated any criteria to enhance on-target activity7,9–11. A library with, for example, 6 sgRNAs per gene designed without any on-target activity criteria would contain 2 or fewer sgRNAs in the highest quintile of activity for 90% of genes, while a library designed with the criteria for enhanced activity presented here would have at least 3 highest-activity quintile sgRNAs for 90% of genes (Fig. 3e).
Local chromatin structure has recently been identified as a major factor affecting the ability of Cas9 to find the PAM and begin to bind DNA with the seed region of the sgRNA20,21. The sequence features we find to promote activity apply across different cellular contexts, suggesting that some of the steps involved in Cas9-based DNA targeting are governed by intrinsic features of the target site and sgRNA sequence. We speculate that, even with optimal design rules, certain cellular contexts or sequence properties may render some genes difficult to target efficiently with current CRISPR technology; in these cases, RNAi technology might provide a better option for probing gene function.
Here we quantitatively assayed the activity of thousands of sgRNAs to uncover sequence features that modulate the ability of Cas9 to bind DNA, cleave the target site, and result in a null allele. Similar approaches were previously applied to RNAi knockdown22,23. We used a direct measurement of target protein levels to categorize sgRNA activity rather than phenotypic outcomes that generally do not distinguish biallelic inactivation from haploinsufficiency. Indeed, the large number of sgRNAs combined with the quantitative assay for protein knockout efficacy may have allowed detection of preferences in the PAM region that had not been observed previously, and provide a quantitative measure of the base preferences throughout the target site4–6,16,20,24. We found sequence features that are predictive of sgRNA activity, developed a quantitative model based on these features to optimize sgRNA activity prediction, and created a tool to use this model for sgRNA design. The sequence feature model generalized to each of 9 distinct genes and 4 cell lines of both human and mouse origin, and also to sgRNA activities for 414 hit genes in a genome-wide proliferation screen. By incorporating additional datasets, activity readouts, and modeling approaches, it will be possible to further refine these activity predictions, to determine which mechanistic steps drive sequence preferences and to identify other factors that influence activity. The sequence features shown here that correlate to on-target activity of the Cas9-sgRNA complex will enable more effective application of CRISPR technology to edit the genome and probe gene function.
METHODS
Library design
For a collection of mouse cell surface markers, as well as negative control targets, we identified all sgRNA target sites preceding the NGG PAM sequences on the plus and minus strands of DNA for all exons, including 25 nts of flanking intronic sequence, as annotated in the Ensembl Genome Browser. To all 20 nt sgRNA sequences we prepended a G to allow for proper transcription initiation by RNA polymerase III (Supplementary Table 1). For the human cell surface markers, we limited our design to coding sequence sgRNAs.
Library creation
The mouse library creation was as described9. Briefly, oligonucleotides (Agilent) were PCR amplified (Pfusion, New England Biolabs) and the resulting PCR product was column cleaned (Qiagen PCR Purification) and cloned by Gibson assembly (New England Biolabs) into pXPR_001 (e.g. lentiCRISPR, Addgene plasmid 49535). The assembly was column cleaned (Qiagen PCR Purification), electroporated into E. coli (Lucigen) and grown at 37°C for 16 hours. Colonies were harvested and DNA was prepared (Qiagen, Endotoxin Free MaxiPrep). Virus production was as described9,13. Centrifugation at 100,000 × g for 2 hours was used to concentrate the virus, followed by resuspension in DMEM + 10% FBS at 4°C overnight. For the human library, pairs of oligonucleotides (IDT) with BsmBI-compatible overhangs were ordered, individually annealed, and then ligated as a pool into pXPR_003 (e.g. lentiGuide, Addgene plasmid 52963).
Cell culture and lentiviral infection
EL4 cells were maintained in DMEM + 10% FBS + 50 µM 2-mercaptoethanol and were obtained from ATCC. MOLM14 and NB4 were maintained in RPMI + 10% FBS. Both were obtained from the Cancer Cell Line Encyclopedia (CCLE). TF1 were maintained in RPMI + 10% FBS + 2 ng/mL GM-CSF (Invitrogen) and were obtained from CCLE.
Screen Infection Conditions
Mouse EL4 cells
Optimal infection conditions to achieve 30–50% infection efficiency, corresponding to an MOI <1 were determined by infecting at multiple virus volumes (eg. 150 µL, 300 µL, 500 µL and 1 mL) and then 48h post infection, splitting an equal number of cells from each infection volume into 2 wells of a 6-well dish, one containing complete medium supplemented with 2 µg/mL puromycin, the other containing complete medium only. The ratio of live cells 2 days after puromycin addition is the infection efficiency of that viral dose. Cells were then infected in 12-well plate format such that each well contained 1×106 cells, 140 µL of ultracentrifuge-concentrated virus, and complete media to a final volume of 2 mL supplemented with 4 µg/mL polybrene. After centrifugation for 2 hours at 1000 × g, 2 mL of complete media was added per well. The following day, cells were split out of the 12-well dish and cultured for an additional 24 hours prior to addition of puromycin (2 µg/mL).
Human MOLM13, NB4 and TF1 cells
MOLM13 and NB4 cells were transduced at MOI <1 with lentivirus prepared from pLX_TRC311-Cas9, and selected with 5 µg/mL blasticidin for 9 days. TF1 cells were transduced at a MOI <1 with lentivirus prepared from pXPR_101 (e.g. lentiCas9-blast, Addgene plasmid 52962), and selected with 5 µg/mL blasticidin for 14 days as well as for 2 days directly preceding infection with the human sgRNA library. Infection efficiency for the human sgRNA library was similarly determined to achieve an infection efficiency corresponding to an MOI <1. Briefly, 4 wells of each cell line at 2.5×106 cells per well in 2 mL media and 1 mL virus (MOLM13 & NB4) or 300 µL virus (TF1) supplemented with 4 µg/mL polybrene were centrifuged for 2 hours at 1000 × g then 2 mL media was added per well. 24-hours post infection, cells were split out of the 12-well plates and 48-hours post infection 2 µg/mL puromycin was added and maintained until analysis by FACS.
Cas9 activity assay
Cas9 expressing MOLM13, NB4 and TF1 cell lines were transduced with pXPR-011 (Addgene plasmid 59702) at an MOI ~1. Briefly, cells were infected in 24-well plate format, with each well containing 2×105 cells, 100 µL virus and 300 µL of media supplemented with 4 µg/mL polybrene. 48 hours post infection, 2 µg/mL puromycin was added and cells were selected for 3 days. Parental lines transduced with XPR-011 only were maintained in parallel with Cas9 and pXPR-011 expressing cell lines; samples from both were analyzed on a BD-LSRFortessa X-20 ten days post infection. Active Cas9-expressing lines will result in a reduction in GFP when transfected with pXPR-011 as this vector delivers both GFP and a guide targeting GFP. Because GFP is downstream of puromycin and after a 2A site, abrogation of GFP will have no impact on puromycin resistance (Supplementary Fig 2).
FACS
Human and mouse cell surface markers were selected the basis of homogeneity of expression as assessed by antibody staining profiles. Only cell lines which showed expression of a particular cell surface marker in >98% cells were chosen for analysis.
EL4 cells were independently stained and sorted on a FACS Aria flow cytometer 8 days post transduction. Antibodies used in this study included: eBioscience 17-5958-80 Anti-Mouse MHC Class I (H-2Kb) APC; eBioscience 12-0281-81 Anti-Mouse CD28 PE; eBioscience 11-0021-81 Anti-Mouse CD2 FITC; eBioscience 11-0431-81 Anti-Mouse CD43 FITC; eBioscience 17-0031-81 Anti-Mouse CD3e APC; eBioscience 17-0051-81 Anti-Mouse CD5 APC; Biolegend 124705 FITC anti-mouse CD53; BD Biosciences 561974 APC conjugated anti-CD90.2 (Thy1.2); BD Biosciences 560695 PE conjugated anti CD45.2
MOLM13, NB4 and TF1 cells were stained and sorted on a BD-FACS Aria II 8 days post transduction with the human sgRNA library. Antibodies used in this study included: BD Pharmingen 555450 CD33-PE; BD Pharmingen 555394 CD13-PE; BD Pharmingen 562371 CD15-PE.
Arrayed Guide Activity Assays
EL4 cells expressing Cas9 (pXPR_101) were infected with 17 sgRNAs targeting three cell surface markers. Nine days post infection 1×106 cells were isolated, genomic DNA extracted (DNeasy Blood and Tissue Kit, Qiagen) and sequencing performed on an Illumina MiSeq. Eleven days post sgRNA transduction, 2×105 cells from each sgRNA-infected population were stained with their corresponding antibodies and analyzed on an Accuri C6 Flow Cytometer for gene knockout. Reads were classified as a) wildtype, b) mismatch, c) indels producing a frameshift, and d) in-frame indels. The percentage of frameshift alles (Supplementary Fig. 4b) was calculated as the number of reads in category c divided by the total number of reads.
Genomic DNA isolation and sequencing
Genomic DNA was isolated (DNeasy Blood and Tissue Kit, Qiagen); cell populations < 1 million were supplemented with 1 µg of yeast RNA before purification. PCR was performed using as previously described, with the exception of using NEBNext High-Fidelity 2× PCR Master Mix9. Samples were sequenced on an Illumina HiSeq 2500 or MiSeq.
Data processing and analysis
Illlumina sequencing reads were processed by counting the number of unique reads for each sgRNA in each experimental condition (Supplementary Tables 2, 3). With each sample for each sgRNA, “Reads per Million” was determined by dividing the number of reads for an individual sgRNA by the total number of sgRNA reads in that sample, multiplying by one million, adding one, and then log2 transforming. Multiple samples for the same experimental condition were then averaged.
For analysis of pooled screening data, log-fold-change values were calculated for each sgRNA by subtracting the abundance in the unsorted population from the abundance in the marker-negative population. We excluded all sgRNAs that had a run of 4 or more thymidines, as this would be expected to cause premature transcription termination. We also excluded all sgRNAs that were present at less than 32 reads per million in the unsorted population. We visually examined activity maps as a function of cut site position to exclude from our predictive model any sgRNAs that targeted areas of generally low activity, even if that meant excluding some outlier sgRNAs with high activity, in order to ensure that we were not contaminating our modeling dataset with sequences erroneously assigned as low activity by virtue of their target site rather than their intrinsic potential efficacy.
Off-target scores (Supplementary Table 4) were obtained from crispr.mit.edu, accessed April 24, 2014. Data from Hsu et al. were used to calculate the score, as described on the webserver14.
sgRNA Activity Predictive Model
Within each gene, passing sgRNAs were first ranked, with the best shRNA receiving the rank of 1. This number was then divided by the total number of sgRNAs, which was then subtracted from 1 to determine a percent-rank. This results in the worst sgRNA for a gene receiving a percent-rank of 0, while the best sgRNA will have a percent-rank approaching 1. Percent-rank values were averaged for genes that were assayed in more than one cell line.
The features used for prediction were the individual nucleotides and all pairs of adjacent nucleotides indexed by position in the 30 mer target site. We also included the count of Gs and Cs in the 20 nt of the sgRNA. Because of an observed non-linear dependence between G/C content and efficacy, two G/C-count features were also incorporated: one for deviations below ten and one for deviations above ten. To allow for independent weights for each nucleotide feature, the nucleotide feature space was represented with one-hot encoding.
Because the full set of features – with 120 single nucleotide features, 464 dinucleotide features, and the two G/C-count features – was over-determined, we incorporated feature selection to choose a subset of features with the best generalization error. An L1-regularized linear support vector machine (SVM) implemented in the python module scikit-learn was used to generate sets of features as a function of the L1-norm penalty. Given the set of features from the SVM, a logistic regression classifier was trained to discriminate the top quintile of sgRNAs for each gene from the remainder. We cross-validated the model by training on the data for eight genes and predicting on the data for the remaining gene. The feature selection step was run in a nested stratified cross validation loop on the training data in which each fold excluded an equal proportion of the sgRNAs for each of the eight training genes. The L1-norm penalty was chosen to maximize the average holdout AUC in the nested loop. We also used leave-one-sgRNA-out cross validation to measure the performance of the model, though leave-one-gene-out is a more realistic measure of generalization performance. After validation, we trained a final model using all available data (Supplementary Table 9), which used only 72 of the 586 features, including both GC-count features.
The model weights presented in Supplementary Table 9 can be used to easily compute the sgRNA score. A guide necessarily only has a subset of all the features, indicated via one-hot encoding as binary variables. Let the model weights for the features i for a particular guide sj be wii, the intercept int. Then the sgRNA score f(sj) is given via logistic regression as:
Model scores f(sj) will fall into the range [0,1], and higher values predict higher activity.
Analysis of A375 viability data
For analysis of A375 screening data for lethal sgRNAs, we were interested in generating a set of sgRNAs with as few false positives as possible, and were less interested in capturing all true positives. Starting from a library of 64,751 sgRNAs, we applied numerous filters to improve data quality: a) removed any sgRNAs present at fewer than 8 reads per million at the early time point; b) removed any sgRNAs containing a run of 4 or more thymidines; c) examined only sgRNAs targeting genes in sets already well-established to be essential for viability9,25 d) required at least two sgRNAs targeting the gene to remain in the dataset. This generated a list of 1278 sgRNAs (Supplementary Table 10). We subtracted the gene average depletion from the depletion caused by the individual sgRNA, to produce a gene-normalized activity for each sgRNA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michael Waring (Ragon Institute, Cambridge, MA) for expert assistance with flow cytometry; Sefi Rosenbluh (Broad Institute) for sharing the pLX_311-Cas9 vector; Tamara Mason (Broad Institute) for Illumina sequencing advice and execution; Doug Alan, Adam Brown, Mark Tomko, Matt Greene, and Tom Green (Broad Institute) for assistance in building the sgRNA design website; Jennifer Listgarten and Nicolo Fuso (Microsoft Research) for a critical analysis of the sgRNA activity prediction model.
Z.T. is supported by NIH 5T32CA009172-39.
Footnotes
Accession codes
SRA: SRP045596
AUTHOR CONTRIBUTIONS
E.H., D.G., Z.T., and J.D. designed experiments; E.H., M.S., D.G., and Z.T. executed experiments; E.H. and J.D. analyzed experimental results; M.H. and J.D. analyzed sequencing data and developed analysis tools; I.S. developed the sgRNA scoring model; E.H., D.R., and J.D. wrote the manuscript with help from other authors; B.E., R.X. and D.R. supervised the research. J.D. is a Merkin Institute Fellow.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Barrangou R, et al. CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 3.Sapranauskas R, et al. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Research. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471–e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cong L, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mali P, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gagnon JA, et al. Efficient Mutagenesis by Cas9 Protein-Mediated Oligonucleotide Insertion and Large-Scale Assessment of Single-Guide RNAs. PLoS ONE. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koike-Yusa H, Li Y, Tan E-P, Del Castillo Velasco-Herrera M, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y, et al. High-throughput screening of a CRISPR-Cas9 library for functional genomics in human cells. Nature. 2014:1–16. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 12.Luo B, et al. Highly parallel identification of essential genes in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2008;105:20380–20385. doi: 10.1073/pnas.0810485105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Research. 2014;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinek M, et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang H, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol. 2014 doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the cas9 endonuclease. Nat Biotechnol. 2014:1–9. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds A, et al. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 23.Fellmann C, et al. Functional Identification of Optimized RNAi Triggers Using a Massively Parallel Sensor Assay. Mol. Cell. 2011;41:733–746. doi: 10.1016/j.molcel.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.