Abstract
Cherubism (OMIM#118400) is a genetic disorder with excessive jawbone resorption caused by mutations in the signaling adaptor protein SH3BP2. Studies on the mouse model for cherubism carrying a P416R knock-in mutation have revealed that mutant SH3BP2 enhances TNF-α production and RANKL-induced osteoclast differentiation in myeloid cells. TNF-α is expressed in human cherubism lesions, which contain a large number of TRAP-positive multinucleated cells, and TNF-α plays a critical role in inflammatory bone destruction in homozygous cherubism mice (Sh3bp2KI/KI). The data suggest a pathophysiological relationship between mutant SH3BP2 and TNF-α-mediated bone loss by osteoclasts. Therefore, we investigated whether P416R mutant SH3BP2 is involved in TNF-α-mediated osteoclast formation and bone loss. Here, we show that bone marrow-derived M-CSF-dependent macrophages (BMMs) from the heterozygous cherubism mutant (Sh3bp2KI/+) mice are highly responsive to TNF-α and can differentiate into osteoclasts independently of RANKL in vitro by a mechanism that involves SYK and PLCγ2 phosphorylation, leading to increased nuclear translocation of NFATc1. The heterozygous cherubism mutation exacerbates bone loss with increased osteoclast formation in a mouse calvarial TNF-α injection model as well as in a human TNF-α transgenic mouse model (hTNFtg). SH3BP2 knockdown in RAW264.7 cells results in decreased TRAP-positive multinucleated cell formation. These findings suggest that the SH3BP2 cherubism mutation can cause jawbone destruction by promoting osteoclast formation in response to TNF-α expressed in cherubism lesions and that SH3BP2 is a key regulator for TNF-α-induced osteoclastogenesis. Inhibition of SH3BP2 expression in osteoclast progenitors could be a potential strategy for the treatment of bone loss in cherubism as well as in other inflammatory bone disorders.
Keywords: SH3BP2, cherubism, TNF-α, osteoclast, arthritis
Introduction
Cherubism (OMIM#118400) is an autosomal dominant craniofacial disorder with disfiguring facial appearance in children due to focal resorption of maxillary and mandibular bones and their replacement with excessively proliferating fibro-osseous tissue masses. The lesions consist mostly of spindle-shaped fibroblastoid cells and a large number of tartrate-resistant acid phosphatase (TRAP)-positive (+) multinucleated giant cells.(1) Our previous studies have revealed that heterozygous gain-of-function mutations in the SH3 domain binding protein 2 (SH3BP2) are responsible for cherubism.(2,3) SH3BP2 is an adaptor protein originally discovered as one of the proteins that bind to the SH3 domain of the protein tyrosine kinase ABL1.(4) SH3BP2 can interact with a variety of proteins including SYK,(5) 14-3-3,(6) VAV,(7) LYN,(8) PLCγ1 and PLCγ2,(5,9) SHP-1,(10,11) BLNK,(12) and SRC (13) in various hematopoietic cell types including T cells, B cells, mast cells, neutrophils, and macrophages as well as in osteoblasts and osteoclasts, indicating that SH3BP2 plays roles in modulating the immune and skeletal system under physiological conditions.(13-15)
Analysis of the P416R knock-in (KI) mouse model for cherubism (equivalent to the most common P418R mutation in cherubism patients) revealed that heterozygous mutants (Sh3bp2KI/+) exhibit systemic osteopenia due to increased osteoclast formation in response to receptor activator of nuclear factor-κB ligand (RANKL). In addition, homozygous mutants (Sh3bp2KI/KI) spontaneously develop severe inflammatory bone loss and joint destruction resulting from systemic macrophage-rich inflammation that overproduces tumor necrosis factor (TNF)-α.(16) More recently, it has been discovered that SH3BP2 interacts with TANKYRASE1 and TANKYRASE2, members of the poly (ADP-ribose) polymerase (PARP) superfamily, and that cherubism mutant SH3BP2 protein undergoes reduced poly (ADP-ribosylation), resulting in decreased proteasomal degradation.(17,18) Therefore, elevated levels of the mutant SH3BP2 protein in Sh3bp2KI/KI myeloid cells lead to enhanced osteoclast formation and TNF-α production in macrophages in a gain-of-function manner. Because elevated levels of wild-type SH3BP2 protein are sufficient to induce enhanced osteoclast formation and TNF-α production by macrophages (16,17), effects shown by Sh3bp2KI/+ and Sh3bp2KI/KI myeloid cells are not specific for the P416R mutation, but are due to the elevated amount of SH3BP2 protein.
In inflammatory bone diseases including rheumatoid arthritis, synovial macrophages and fibroblasts as well as T cells in inflamed joints express a variety of proinflammatory cytokines such as TNF-α, interleukin (IL)-1, IL-6, and IL-17.(19) Among these cytokines, TNF-α is a dominant cytokine that plays a critical role in the promotion of pathological osteoclast formation leading to inflammatory bone destruction.(20-22) Clinical effectiveness of anti-TNF-α treatment for rheumatoid arthritis has demonstrated the essential role of TNF-α in inflammatory bone loss.(23) However, since previous in vitro studies have shown that TNF-α alone does not efficiently induce osteoclast differentiation of bone marrow-derived M-CSF-dependent macrophages (BMMs) as RANKL does,(24-26) TNF-α has been regarded as a cytokine that synergistically potentiates osteoclast differentiation and function in the presence of other cytokines such as RANKL, IL-1 and TGF-β in vitro.(24,27-29) Similarly, in vivo enhancement of osteoclast formation by TNF-α is largely dependent on the stimulation of RANK, the receptor for RANKL, because it has been shown that TNF-α-challenged Rank−/− mice do not exhibit significant signs of bone resorption.(30) Taken together, previous reports implicate that TNF-α is much less potent in inducing osteoclast formation compared to RANKL and that TNF-α alone cannot fully substitute for RANKL both in vitro and in vivo.
Although many advances have been made towards understanding the pathogenesis of inflammatory bone diseases and the role of TNF-α in pathological bone resorption by osteoclasts,(24,27,28,31,32) further investigation is necessary to address the question of which molecules and signaling pathways are involved in the mechanisms that control TNF-α-induced or -assisted osteoclastogenesis. Since TNF-α is expressed in human cherubism lesions (33) and is critically important for the pathogenesis of Sh3bp2KI/KI mice as demonstrated by the rescued inflammatory bone destruction in TNF-α-deficient Sh3bp2KI/KI mice,(16) a pathophysiological link between SH3BP2 and TNF-α-mediated inflammatory bone loss via osteoclasts has been suggested. Therefore, we investigated whether P416R mutant SH3BP2 is involved in TNF-α-mediated osteoclast formation and bone loss.
In the present study, we show that the gain-of-function P416R SH3BP2 mutation potentiates the formation of TNF-α-induced TRAP+ multinucleated cells (MNCs) in the absence of RANK-RANKL interaction in BMM cultures through increased nuclear translocation of NFATc1. We also show that the mutant SH3BP2 exacerbates bone loss in a calvarial TNF-α injection model as well as in transgenic mice expressing human TNF-α (hTNFtg), a model for human rheumatoid arthritis. Furthermore, TRAP+ MNC formation is suppressed in SH3BP2 knockdown RAW264.7 cells. Thus, we demonstrate that SH3BP2 plays a role in TNF-α-induced osteoclastogenesis by modulating the sensitivity of osteoclast progenitors to TNF-α. The data suggest that inhibition of SH3BP2 expression in osteoclast progenitors could be a potential strategy for the treatment of bone loss in inflammatory bone disorders as well as in cherubism.
Methods
Mice
SH3BP2 P416R knock-in mutant mice on C57BL/6 background were previously described.(16) Nfatc1-floxed (Nfatc1fl/fl) mice were kindly provided by Dr. Laurie H. Glimcher. (34) Mx1-Cre and c-Fos-deficient (c-fos−/−) mice on C57BL/6 background were purchased from Jackson Laboratory (Bar Harbor, ME, USA). Human TNF-α-transgenic (hTNFtg) mice were obtained from Taconic (Hudson, NY, USA)(35) and bred with Sh3bp2KI/+ mice under a crossbreeding agreement. To generate Nfatc1-deleted (Nfatc1Δ/Δ) mice, Nfatc1fl/fl mice were crossed with Mx1-Cre mice, and Mx1-Cre/Nfatc1fl/fl mice were injected intraperitoneally with 250 μg of pI:pC (GE Healthcare, Pittsburgh, PA, USA) in PBS every other day for a total of 3 doses starting at postnatal day 10.(34) All procedures were approved by the Institutional Animal Care and Use Committee.
Reagents and antibodies
Recombinant murine M-CSF, TNF-α, and RANKL proteins were purchased from Peprotech (Rocky Hill, NJ, USA). Recombinant mouse osteoprotegerin Fc domain fusion protein (OPG-Fc) was purchased from R&D systems (Minneapolis, MN, USA). Etanercept was purchased from Pfizer (New York, NY, USA). FK506, BAY61-3606, R406, and U73122 were obtained from Sigma-Aldrich (St. Louis, MO, USA), Millipore (Billerica, MA, USA), Selleck chemicals (Houston, TX, USA), and Calbiochem (Billerica, MA, USA), respectively. Antibodies for Western blot were obtained from Cell Signaling Technology (Danvers, MA, USA), Santa Cruz Biotechnology (Santa Cruz, CA, USA), GeneTex (Irvin, CA, USA), Abcam (Cambridge, MA, USA), Millipore, and Sigma-Aldrich.
Immunohistochemistry
Paraffin embedded cherubism lesions were sectioned (6 μm) and rehydrated. Antigen retrieval was performed using antigen unmasking solution (Vector Laboratories, Burlingame, CA, USA), and endogenous peroxidase was blocked with 3% H2O2. After blocking with normal horse serum, the sections were incubated overnight at 4°C with either mouse anti-TNF-α (P/T2, Abcam)(33) or mouse anti-CD14 (Clone 7, Thermo Scientific, Waltham, MA, USA) antibodies. After washing with PBS, sections were incubated with biotinylated anti-mouse IgG antibody for 10 min and treated with Vectastain elite ABC kit (Vector Laboratories). TNF-α and CD14 were visualized using DAB and VIP peroxidase reagents (ImmPACT DAB and VIP, Vector Laboratories), respectively. The sections were counterstained with Mayer's hematoxylin. As negative controls, specimens were incubated with normal mouse IgG in place of primary antibodies.
Osteoclast differentiation and resorption assay
Bone marrow cells were isolated from long bones of 6 to 10-week-old female mice.(16) Non-adherent cells were collected, seeded on 48-well plates at a density of 2.1 × 105 cells/well, and incubated in α-MEM supplemented with 10% FBS containing M-CSF (25 ng/ml). After 2 days, bone marrow derived M-CSF-dependent macrophages (BMMs) were stimulated with TNF-α or RANKL for additional 3–4 days in the presence of M-CSF (25 ng/ml). For fetal liver cell culture, fetal livers (E13.5) were dissociated in α-MEM, and non-adherent cells were treated as described above. Culture medium was changed every 2 days. TRAP+ MNC formation (3 and more nuclei) was visualized by TRAP staining (Sigma-Aldrich) and counted (n = 4–6 wells/group). Biochemical assay for TRAP activity in the culture supernatant was performed as described.(34,36) OPG-Fc and etanercept were used to neutralize RANKL and TNF-α, respectively. To block calcineurin, SYK, PLC, and NF-κB pathways, we used FK506, SYK inhibitors (BAY61-3606, R406), U73122, or NEMO-binding domain peptide,(37) respectively. For resorption assay, non-adherent bone marrow cells were plated at a density of 8.5 × 104 cells/well (96-well plates) and cultured in the presence of M-CSF (25 ng/ml) and TNF-α (100 ng/ml) for 7 to 21 days. After removing the cells with 1M NH4OH, resorption areas were visualized with toluidine blue or von Kossa staining followed by quantification with analySIS (Soft Imaging System GmbH, Munster, Germany) attached to a Nikon NE600 reflective microscope.
Immunofluorescent staining
TNF-α-treated non-adherent bone marrow cells (1.8 × 105 cells/well in 8-well chamber slides) were fixed in 4% PFA, permeabilized with 0.2% Triton X-100, blocked in 2% normal goat serum/2.5% BSA/PBS, and incubated with anti-NFATc1 antibody (7A6, Santa Cruz). NFATc1 was detected by Alexa Fluor-555 conjugated goat anti-mouse IgG antibody. Actin and nuclei were co-stained with Alexa Fluor-488 conjugated phalloidin (Life Technologies, Grand Island, NY, USA) and DAPI, respectively. Fluorescent images of cells were acquired with a TCS SP5 II confocal microscope (Leica, Buffalo Grove, IL, USA). To quantify nuclear localization of NFATc1, three images per each group were taken with a 4X objective on a Nikon TE800 fluorescent microscope. Cells were considered positive for NFATc1 nuclear localization when the fluorescence intensity of NFATc1 in nuclei exceeded that in cytoplasm. Numbers of DAPI-and NFATc1-positive nuclei were counted using ImageJ (NIH), and the percentages of NFATc1-positive nuclei per total nuclei were calculated as described.(38) Approximately 1000 nuclei in each group were analyzed.
Real-time quantitative PCR (qPCR)
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized using High Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Carlsbad, CA, USA). qPCR reactions were performed using Absolute Blue QPCR Master Mixes (Thermo Scientific) with StepOne Plus system (Applied Biosystems). Gene expression levels relative to Hprt were calculated by ΔΔCt method and were normalized to baseline controls as indicated in each experiment. qPCR primers used in this study are listed in Supplemental Table 1. All qPCR reactions yielded products with single peak dissociation curves.
Immunoprecipitation and Western blot
Cells were washed with ice-cold PBS and lysed with lysis buffer (25 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA, 10% glycerol, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 0.7 mM β-glycerophosphate) with protease inhibitor and phosphatase inhibitor cocktails (Sigma-Aldrich). After pre-clearing with protein A/G-Plus-Agarose (Santa Cruz Biotechnology), approximately 200–300 μg of total protein was incubated with 1–2 μg of indicated primary antibodies at 4°C for 2 hours, then incubated with 20 μl of protein A/G-Plus-Agarose for additional 2 hours. After washing three times with lysis buffer, the agarose was resuspended in Laemmli loading buffer and boiled for 5 min. For the preparation of Triton X-100-solubilized protein, cells were washed with ice-cold PBS and lysed with lysis buffer. For nuclear and cytoplasmic fractionation, cells were lysed on ice in cytoplasmic lysis buffer (10 mM HEPES (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.05% Igepal), and nuclei were sedimented by centrifugation and lysed in nuclear lysis buffer (2% SDS, 2 M urea, 8% sucrose, 20 mM sodium β-glycerophosphate, 1 mM NaF, and 5 mM Na2VO4). Five μg of TritonX-100-solubilized protein, 1 μg of nuclear protein and 4 μg of cytoplasmic protein were resolved by SDS-PAGE and transferred to nitrocellulose membranes. After blocking with 5% skim milk in TBST buffer, membranes were incubated with primary antibodies followed by incubation with appropriate HRP-conjugated species-specific secondary antibodies (Cell Signaling Technology). Bands were detected using SuperSignal West Dura or Femto chemiluminescent substrate (Thermo Scientific) and visualized by ImageQuant LAS-4000 (GE Healthcare). Actin, HSP90, and nuclear matrix protein p84 were used as loading controls.
Retrovirus production and SH3BP2 overexpression in RAW264.7 cells
C-terminal FLAG-tagged wild-type and P416R Sh3bp2 cDNA fragments were cloned into pMXs-IP vector.(39) 293 GPG cells were transfected using FuGENE HD (Roche, Alameda, CA, USA). After 48 hours, supernatants were collected and used for infection. RAW264.7 cells were cultured in medium containing retrovirus and polybrene (8 μg/ml) for 2 days. Infected RAW264.7 cells were selected with puromycin (10 μg/ml), re-seeded on 48-well plates at a density of 2000 cells/well, and stimulated with TNF-α (100 ng/ml) for 4–5 days. For resorption assays, infected RAW264.7 cells were plated at a density of 1.0 × 103 cells/well in DMEM/10% FBS on Osteo-Assay Surface plates (Corning Life Sciences, Lowell, MA). After 8 days of culture with TNF-α (100 ng/ml) at 10% CO2, the cells were removed with 10% bleach solution, and wells were stained with 5% silver nitrate solution. Resorbed area/well was quantified using ImageJ.
TNF-α calvarial injection
Ten-week-old Sh3bp2KI/+ and Sh3bp2+/+ male mice were subjected to daily supracalvarial injections with TNF-α (1.5 μg/mouse/day) or PBS for 5 days.(40) Mice were euthanized 24 hours after the last injection, and the calvariae were fixed in 4% paraformaldehyde (PFA) for 2 days. For RNA collection, calvariae were crushed under liquid nitrogen conditions using a tissue pulverizer and immediately stored in TRIzol reagent.
Micro-computed tomographic (microCT) analysis
PFA-fixed calvariae and hind limbs were immersed in 70% ethanol and scanned with a vivaCT 40 (Scanco Medical AG, Bassersdorf, Switzerland) with an X-ray energy of 55 kVp (145 μA), a voxel resolution of 15 μm, and an integration time of 200 ms. Trabecular and cortical bone properties of tibia and talus bone volume were analyzed using Scanco bone evaluation software. The talus bone volumes were evaluated for a quantitative measurement of bone erosion.(41) For three-dimensional (3D) reconstruction of calvarial and hind paw bones, threshold was set to 300. The region of trabecular bone analysis comprised 70 slices of secondary spongiosa beginning just beneath primary spongiosa of the tibia; the region of cortical bone analysis comprised 30 slices of midshaft (1 mm proximal to the tibio-fibular junction) of the tibia. All microCT parameters were described according to American Society for Bone and Mineral Research (ASBMR) guidelines.(42)
Histomorphometry
Bone samples fixed in 4% PFA in PBS were decalcified for 4 weeks in 0.5 M EDTA (pH 7.2) at 4°C. Six μm paraffin sections were stained with hematoxylin and eosin (H&E), and osteoclasts were visualized by TRAP staining. Methyl green was used for counterstaining. Histomorphometric measurements were performed in a blinded manner using the OsteoMeasure analysis system (OsteoMetrics Inc., Atlanta, GA, USA) interfaced with Nikon E800 microscope. Eroded surface per bone surface (ES/BS), number of osteoclasts per bone surface (N.Oc/BS), and osteoclast surface per bone surface (Oc.S/BS) were determined. The terminology and units used are those recommended by the Histomorphometry Nomenclature Committee of the ASBMR.(43)
Evaluation of TNF-α-mediated arthritis and bone loss
Sh3bp2KI/+ mice were crossed with hTNFtg mice, and inflammation and bone loss were evaluated as described.(25) Arthritis severity was assessed once a week in a blinded manner using the following criteria: 0 = Normal; 1 = Mild erythema or swelling of the wrist or ankle or erythema and swelling of any severity for 1 digit; 2 = More than three inflamed digits or moderate erythema and swelling of the ankle or wrist; 3 = Severe erythema and swelling inflammation of wrist or ankle; 4 = Complete erythema and swelling of the wrist and ankle including all digits. Each limb was graded, giving a maximum score of 16. After fixation, hind limb samples were subjected to microCT analysis. Tibio-talar joints and tibiae were analyzed to determine focal and systemic bone loss, respectively. Severity of inflammation around ankle joints was evaluated on H&E staining sections using the following criteria: 0 = normal, 1 = mild diffuse inflammatory infiltrates, 2 = moderate inflammatory infiltrates, 3 = marked inflammatory infiltrates, 4 = severe inflammation with pannus formation.
ELISA assay for human and mouse TNF-α
Human and mouse TNF-α concentrations in serum collected from 16-week-old hTNFtg mice were measured with DuoSet ELISA Development kits (R&D).
SH3BP2 knockdown in RAW264.7 cells
GIPZ lentiviral shRNAmir (mouse SH3BP2 shRNA and non-silencing shRNA) were obtained from Thermo Scientific. Clone numbers are: #1 (V3LMM_444790), #2 (V3LMM_444792), non-silencing (RHS4348). RAW264.7 cells were transduced with the lentiviral particles at a multiplicity of infection (MOI) of 30 diluted in DMEM containing 8 μg/ml of polybrene (Sigma-Aldrich). After 6-hour incubation, equal amount of DMEM/20%FBS was added to each well. The transfection medium was removed after 2 days and replaced with complete culture medium. RAW264.7 cells expressing SH3BP2 shRNA were selected with puromycin (10 μg/ml). To evaluate osteoclastogenesis, the infected and non-infected cells were seeded on 48-well plates at a density of 2000 cells/well and stimulated with TNF-α (100 ng/ml) for 5 days. Knockdown efficiency was confirmed by Western blot. Resorption assay was performed as described in the retroviral SH3BP2 overexpression section.
Statistical analysis
All results are given as mean ± SD. Statistical analysis was performed by the two-tailed unpaired Student's t test to compare two groups and by one-way ANOVA (Tukey post-hoc test) to compare three or more groups using GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA) and SPSS Statistics 20 (IBM, Armonk, NY, USA). P values less than 0.05 were considered statistically significant.
Results
TNF-α- and CD14-positive cells in human cherubism lesions
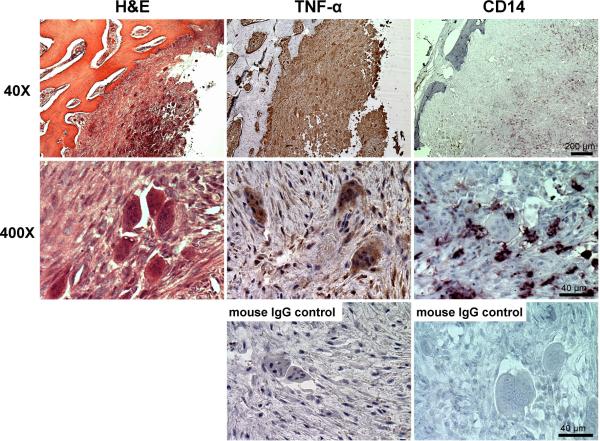
To investigate the involvement of TNF-α and osteoclast precursors in human cherubism lesions, we performed immunohistochemical analyses for TNF-α and CD14, a marker for monocytes/macrophages. Cherubism lesions contained a number of multinucleated giant cells embedded in fibrous stromal cells (Fig. 1, left). Both giant cells and fibrous cells were positive for TNF-α (Fig. 1, middle), which is consistent with a previous report by Hero et al.(33) CD14+ monocytes, which have the potential to differentiate into osteoclasts,(44,45) were also present (Fig. 1, right). These findings suggest that in human cherubism lesions osteoclast precursor cells are constantly exposed to TNF-α and indicate a pathological link between TNF-α and TRAP+ MNC formation in cherubism lesions.(3,46) Therefore, we hypothesized that mutant SH3BP2 enhances TNF-α-induced osteoclastogenesis.
Fig. 1. TNF-α- and CD14-positive cells in human cherubism lesions.
H&E (left) staining and immunohistochemical analysis of human cherubism jaw lesion (family A (2,3)) by specific antibodies against TNF-α (middle) and CD14 (right). As controls, tissue specimens were incubated with normal mouse IgG in place of the specific primary antibodies.
Increased TNF-α-induced osteoclast formation in Sh3bp2KI/+ BMMs
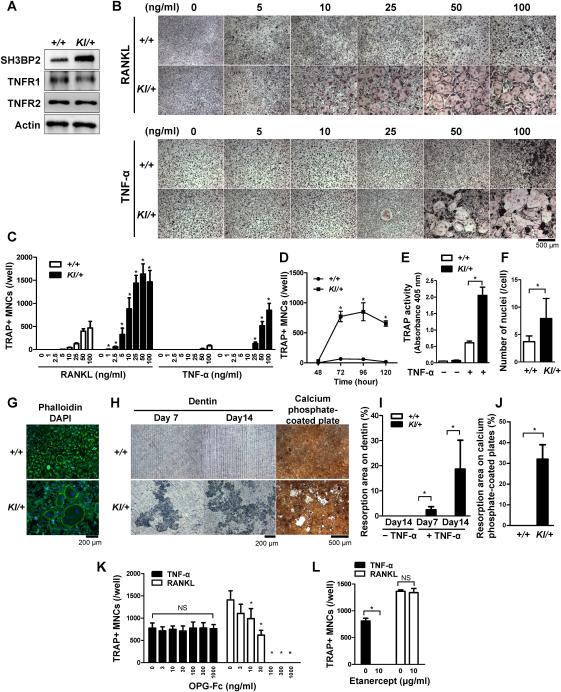
Here we use heterozygous Sh3bp2KI/+ mice for further analyses, since spontaneous development of systemic inflammation in homozygous Sh3bp2KI/KI mice results in short lifespan (16) which could hinder future in vivo experiments (as described in Fig. 7 and 8) and because cherubism occurs as an autosomal dominant disease in humans with a heterozygous SH3BP2 mutation.(3) To examine whether the P416R SH3BP2 mutation regulates TNF-α-induced osteoclastogenesis of osteoclast precursors, we performed in vitro osteoclast differentiation assays using BMMs. The expression level of SH3BP2 protein was higher in Sh3bp2KI/+ BMMs than in Sh3bp2+/+ BMMs as reported by Levaot et al.,(13) and both TNF-α receptors (TNFR1 and TNFR2) were equally expressed in Sh3bp2KI/+ and Sh3bp2+/+ BMMs (Fig. 2A). As we have previously reported,(16) Sh3bp2KI/+ BMMs formed more TRAP+ MNCs than Sh3bp2+/+ BMMs in response to RANKL (Fig. 2B, upper panels). Sh3bp2KI/+ BMMs stimulated with TNF-α formed more TRAP+ MNCs than Sh3bp2+/+ BMMs (Fig. 2B, lower panels), which was confirmed by the quantitative measurements of the number of TRAP+ MNCs (Fig. 2C). TNF-α consistently induced 12 to 37 times more TRAP+ MNC formation in Sh3bp2KI/+ BMMs throughout the culture period (Fig. 2D). Homozygous Sh3bp2KI/KI BMMs formed similar numbers of TRAP+ MNCs compared to heterozygous Sh3bp2KI/+ BMMs (data not shown). M-CSF stimulation alone did not induce TRAP+ MNC formation in Sh3bp2+/+, Sh3bp2KI/+, and Sh3bp2KI/KI BMM cultures (data not shown). TRAP activity in culture medium as well as the number of nuclei per TRAP+ MNC were significantly increased in Sh3bp2KI/+ BMM cultures (Fig. 2E, 2F). Sh3bp2KI/+ TRAP+ MNCs formed actin rings (Fig. 2G) and exhibited higher dentine and calcium phosphate resorption activity compared to Sh3bp2+/+ BMMs after TNF-α stimulation (Fig. 2H, 2I, and 2J). These results indicate that Sh3bp2KI/+ BMMs stimulated with TNF-α can differentiate into fully functional osteoclasts which can resorb mineralized matrices. In agreement with a previous report by Kobayashi et al.,(27) Sh3bp2+/+ TRAP+ MNCs induced by TNF-α did not form resorption pits (Fig. 2H, 2I, and 2J).
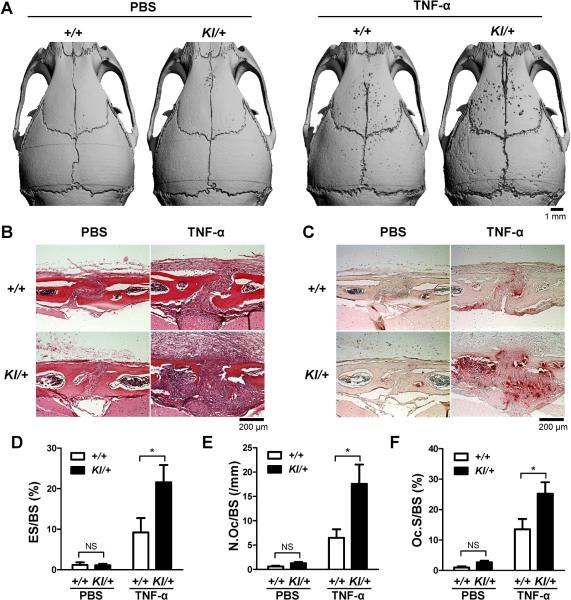
Fig. 7. In vivo effect of TNF-α on osteoclast formation in SH3BP2 cherubism mutant mice.
(A–F) Ten-week-old male Sh3bp2+/+ and Sh3bp2KI/+ mice were subcutaneously injected with either PBS or mouse recombinant TNF-α protein (1.5 μg/mouse/day) for 5 days over the calvarial bone (n = 5–8/group). Calvarial bone samples were harvested 24 hours after the last injection. (A) Representative microCT images of calvarial bone erosion after TNF-α injection (right panel). (B) H&E staining of calvarial tissue sections. (C) TRAP staining of calvarial tissues. (D–F) Histomorphometric analysis. (D) Eroded surface per bone surface (ES/BS), (E) number of osteoclasts per bone surface (N.Oc/BS), and (F) osteoclast surface per bone surface (Oc.S/BS) at the sagittal suture (100−200 μm posterior from bregma). +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+. Data are presented as mean ± SD. * P < 0.05, NS: not significant.
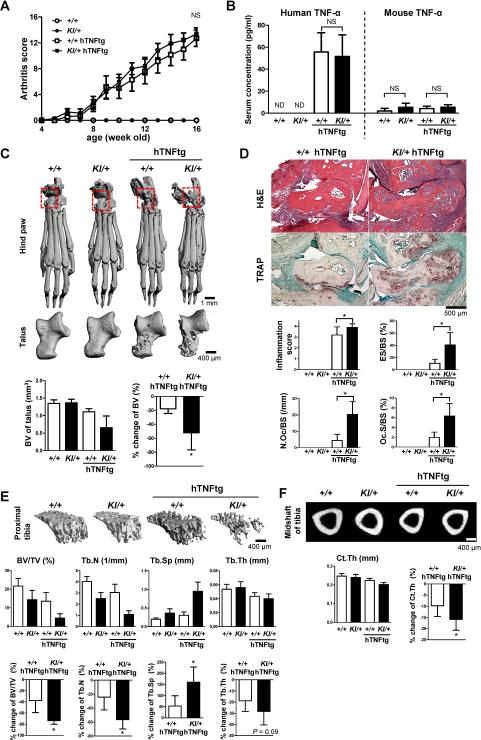
Fig. 8. P416R SH3BP2 cherubism mutation exacerbates bone loss in a TNF-α-mediated arthritis model.
Sh3bp2+/+ and Sh3bp2KI/+ mice were crossed with human TNF-α transgenic (hTNFtg) mice. Joint inflammation was monitored until the age of 16 weeks. Serum and bone samples were collected and subjected to ELISA, microCT and histological analysis. (A) Changes in clinically assessed joint inflammation scores in Sh3bp2+/+ (n = 11), Sh3bp2KI/+ (n = 11), Sh3bp2+/+/hTNFtg (n = 8), and Sh3bp2KI/+/hTNFtg (n = 10) male mice. (B) Serum concentrations of human and mouse TNF-α. ND: not detectable. (C) Representative microCT images of hind paws and talus bones. Bone volumes (BV) of talus and % change of the BV of talus are shown. (D) H&E (upper) and TRAP staining (lower) images of tibio-talar joints. Inflammation score, eroded surface per bone surface (ES/BS), number of osteoclasts per bone surface (N.Oc/BS), and osteoclast surface per bone surface (Oc.S/BS) were quantitated. (E) Representative microCT images of trabecular bone of proximal tibia (top). Bone volume per total volume (BV/TV), trabecular number (Tb.N), trabecular separation (Tb.Sp), trabecular thickness (Tb.Th) in trabecular bone of proximal tibia (middle), and % change of the parameters (bottom). (F) Representative microCT images of midshaft of tibia (top). Cortical thickness (Ct.Th) of midshaft of tibia (bottom left) and % change of the Ct.Th (bottom right). (D). Data are presented as mean ± SD. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+. * P < 0.05, NS: not significant.
Fig. 2. P416R SH3BP2 cherubism mutation enhances TNF-α-induced osteoclast differentiation independently of RANKL in vitro.
Bone marrow cells were isolated from Sh3bp2+/+ and Sh3bp2KI/+ mice. Non-adherent bone marrow cells were seeded at a density of 2.1 × 105/well on 48-well plates. After 2-day preculture with M-CSF (25 ng/ml), BMMs were stimulated with RANKL or TNF-α for 72 or 96 hours, respectively. (A) Western blot analysis for SH3BP2, TNFR1, and TNFR2 in Sh3bp2+/+ and Sh3bp2KI/+ BMMs after 2-day preculture with M-CSF. (B) TRAP staining of BMMs stimulated with either RANKL or TNF-α at indicated concentrations. (C) Quantitation of TRAP-positive MNCs (TRAP+ MNCs) per well after RANKL or TNF-α stimulation. (D) Number of TRAP+ MNCs. BMMs were stimulated with TNF-α (100 ng/ml). (E) TRAP activity in the culture supernatant after 96-hour treatment with or without TNF-α (100 ng/ml). (F) Number of nuclei per TRAP+ MNCs. BMMs were stimulated with TNF-α (100 ng/ml) for 96 hours. (G) Phalloidin and DAPI staining of TNF-α-stimulated BMMs. Cells were fixed 96 hours after TNF-α treatment (100 ng/ml). Actin and nuclei were visualized with phalloidin (green) and DAPI (blue), respectively. (H–J) Resorption assays. BMMs were cultured with TNF-α (100 ng/ml) on dentin slices for 14 days and on calcium phosphate-coated plates for 21 days. After removal of the cells, resorption areas were visualized by toluidine blue or von Kossa staining. Resorbed areas (%) on dentin slices (I) and on calcium phosphate-coated plates (J) were quantified (n = 3–4/group). (K, L) Quantitation of TRAP+ MNCs. Sh3bp2KI/+ BMMs were stimulated with TNF-α (100 ng/ml) for 96 hours or RANKL (50 ng/ml) for 72 hours in the presence of (K) osteoprotegerin-Fc fusion protein (OPG-Fc) or (L) soluble TNFR2-Fc fusion protein (etanercept). +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+. Data are presented as mean ± SD. * P < 0.05, NS: not significant.
Next, to test if TNF-α induces osteoclast differentiation of Sh3bp2KI/+ BMMs independently of RANKL, recombinant murine osteoprotegerin (OPG-Fc) was added to the BMM cultures. OPG-Fc completely failed to inhibit TNF-α-induced osteoclastogenesis in Sh3bp2KI/+ BMM cultures, while RANKL-induced osteoclastogenesis was suppressed in a dose-dependent manner (Fig. 2K), confirming that TNF-α stimulation of Sh3bp2KI/+ BMMs can induce osteoclast formation independently of RANKL in vitro. In contrast, etanercept, a TNF-α inhibitor, effectively blocked TNF-α-induced osteoclastogenesis in Sh3bp2KI/+ BMM cultures (Fig. 2L). Taken together, these data demonstrate that the P416R mutant SH3BP2, which acts in a gain-of-function manner, potentiates TNF-α-induced formation of TRAP+ MNCs independently of RANKL and that Sh3bp2KI/+ TRAP+ MNCs induced by TNF-α are functionally mature osteoclasts.
Increased mRNA expression levels of osteoclast-associated genes and NFATc1 nuclear translocation in TNF-α-stimulated Sh3bp2KI/+ BMMs
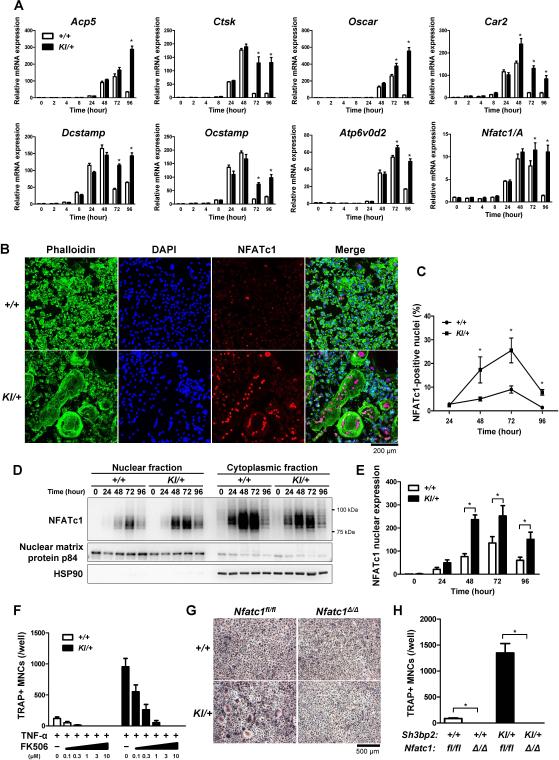
Next, we measured mRNA expression levels of genes that are associated with osteoclast differentiation and function, such as acid phosphatase 5 (Acp5), cathepsin K (Ctsk), osteoclast-associated receptor (Oscar), carbonic anhydrase II (Car2), dendritic cell-specific transmembrane protein (Dcstamp), osteoclast stimulatory transmembrane protein (Ocstamp), and the d2 isoform of the vacuolar (H+) ATPase V0 domain (Atp6v0d2). Real-time PCR analysis revealed that TNF-α stimulation induces increased expression of osteoclast-associated genes in Sh3bp2KI/+ BMMs primarily at 72 to 96 hours after TNF-α stimulation compared to those in Sh3bp2+/+ BMMs (Fig. 3A).
Fig. 3. P416R SH3BP2 mutation augments osteoclast-associated genes expression and nuclear translocation of NFATc1.
(A) Quantitative-PCR analysis of osteoclast associated genes. Bone marrow cells were isolated from Sh3bp2+/+ and Sh3bp2KI/+ mice. After 2-day preculture with M-CSF (25 ng/ml), BMMs were stimulated with TNF-α (100 ng/ml) for 96 hours. RNA samples were collected at indicated time points. Gene expression levels relative to Hprt were calculated and normalized to the expression level of Sh3bp2+/+ BMMs at 0 hour. The data are representative of three independent experiments. (B) Immunofluorescent staining of actin, nuclei, and NFATc1 visualized by phalloidin, DAPI, and anti-NFATc1 antibody (clone: 7A6), respectively. BMMs were fixed 72 hours after TNF-α (100 ng/ml) treatment. (C) Quantitation of NFATc1-positive nuclei. The percentage of NFATc1-positive nuclei per total nuclei was measured at the indicated time points. (D) Western blot analysis for NFATc1. NFATc1 expression levels in nuclear and cytoplasmic fractions were determined at the indicated time points. (E) Relative NFATc1 expression. Ratio of NFATc1 to nuclear matrix protein p84 in nuclear fractions were calculated and normalized to the ratio of Sh3bp2+/+ at 0 hour (n = 3). (F) Quantitation of TRAP+ MNCs. BMMs were stimulated with TNF-α (100 ng/ml) for 96 hours in the presence of FK506. (G, H) TRAP staining images and quantitation of TRAP+ MNCs. Sh3bp2KI/+ mice were crossed with Nfatc1-floxed (Nfatc1fl/fl) Mx1-Cre mice. Nfatc1 gene was deleted in hematopoietic cells by the injection of pI:pC. Nfatc1-deleted (Nfatc1Δ/Δ) bone marrow cells were isolated and stimulated with TNF-α (100 ng/ml) for 96 hours. Data are presented as mean ± SD. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+. * P < 0.05, NS: not significant.
Nuclear factor of activated T-cells cytoplasmic 1 (NFATc1) isoform A (NFATc1/A) is a master regulator of the osteoclast transcriptome, promoting the expression of numerous genes required for osteoclast differentiation and bone resorption.(47,48) Since the expression of osteoclast-associated genes is primarily under the control of NFATc1 in RANKL-induced osteoclasts,(34) increased expression of such genes in TNF-α-stimulated Sh3bp2KI/+ BMMs suggested that the P416R SH3BP2 mutation potentiates TNF-α induction of osteoclastogenesis through robust induction of NFATc1 similar to autoamplification,(48) a mechanism of NFATc1 induction by NFATc1 itself, in RANKL-induced osteoclastogenesis. As expected, Nfatc1/A mRNA expression was significantly increased in TNF-α-stimulated Sh3bp2KI/+ BMMs compared to Sh3bp2+/+ BMMs at 72 to 96 hours after TNF-α stimulation due to the sustained Nfatc1/A expression, but was transient in Sh3bp2+/+ BMMs (Fig. 3A).
Osteoclast differentiation is regulated by several other transcription factors such as Nfkb1, Nfkb p65, c-fos, c-jun, PU.1, and Mitf.(19) mRNA expression of these transcription factors were comparable between Sh3bp2KI/+ and Sh3bp2+/+ BMMs stimulated with TNF-α (Supplemental Fig. 1). Expression of other regulators of RANKL-induced osteoclast differentiation (Irf8, Mafb, Bcl6, Blimp1)(49-52) as well as Rbpj, a nuclear DNA-binding protein involved in TNF-α-induced osteoclastogenesis,(26) were not different between Sh3bp2KI/+ and Sh3bp2+/+ BMMs (Supplemental Fig. 1). Collectively, these results indicate that expression of osteoclast-associated genes that are controlled by NFATc1 is increased in TNF-α stimulated Sh3bp2KI/+ BMMs.
TNF-α induces NFATc1 nuclear localization in human macrophages.(53) Therefore, increased Nfatc1/A mRNA expression in Sh3bp2KI/+ BMMs led us to investigate the NFATc1 nuclear translocation in Sh3bp2KI/+ BMMs in response to TNF-α. Immunofluorescent staining revealed that NFATc1 nuclear localization is observed predominantly in giant cells (Fig. 3B) and that the percentages of NFATc1+ nuclei were greater in Sh3bp2KI/+ BMM cultures (25.5 ± 5.2% in Sh3bp2KI/+ vs. 9.0 ± 1.5% in Sh3bp2+/+ BMMs at 72 hours) (Fig. 3C). Consistent with this observation, nuclear NFATc1 protein levels were elevated 1.9 to 3.1-fold in TNF-α-stimulated Sh3bp2KI/+ BMMs at 48 to 96 hours after TNF-α stimulation compared with those in Sh3bp2+/+ BMMs (Fig. 3D, left, and Fig. 3E). To confirm whether NFATc1 is located downstream of mutant SH3BP2, Sh3bp2KI/+ BMMs were stimulated with TNF-α in the presence of a calcineurin inhibitor FK506. FK506 inhibited the formation of TRAP+ MNCs in a dose-dependent manner (Fig. 3F). Furthermore, Nfatc1-deleted (Nfatc1Δ/Δ) Sh3bp2KI/+ BMMs cultured with TNF-α did not form TRAP+ MNCs. This indicates that NFATc1 is downstream of SH3BP2 in BMMs stimulated with TNF-α and necessary for the increased TRAP+ MNC formation (Fig. 3G, 3H), which is consistent with the requirement of NFATc1 in RANKL-induced osteoclast differentiation of Sh3bp2KI/+ and Sh3bp2KI/KI BMMs.(34)
It has been shown that nuclear translocation of NF-κB family transcription factors and c-Fos precedes the robust induction of NFATc1.(19,48) To examine whether P416R mutant SH3BP2 enhances NF-κB and c-Fos activation prior to the induction of NFATc1, we determined the protein levels in the nucleus. Immunoblot analysis revealed that TNF-α induces nuclear localization of NF-κB (p50, p52, and p65) and c-Fos, but expression levels of the transcription factors in the nuclear fraction were comparable between TNF-α-stimulated Sh3bp2KI/+ and Sh3bp2+/+ BMMs (Supplemental Fig. 2). On the other hand, inhibition of NF-κB or c-Fos function suppressed TNF-α-induced TRAP+ MNC formation in Sh3bp2KI/+ BMMs (Supplemental Fig. 3A-D). Collectively, these data indicate that the P416R SH3BP2 mutation promotes TRAP+ MNC formation via increased NFATc1 nuclear translocation without affecting NF-κB and c-Fos expressions, but NF-κB and c-Fos are necessary for the induction of TRAP+ MNCs.
P416R mutant SH3BP2 increases SYK and PLCγ2 phosphorylation and SYK-dependent pathway is required for increased NFATc1 nuclear localization in Sh3bp2KI/+ BMMs
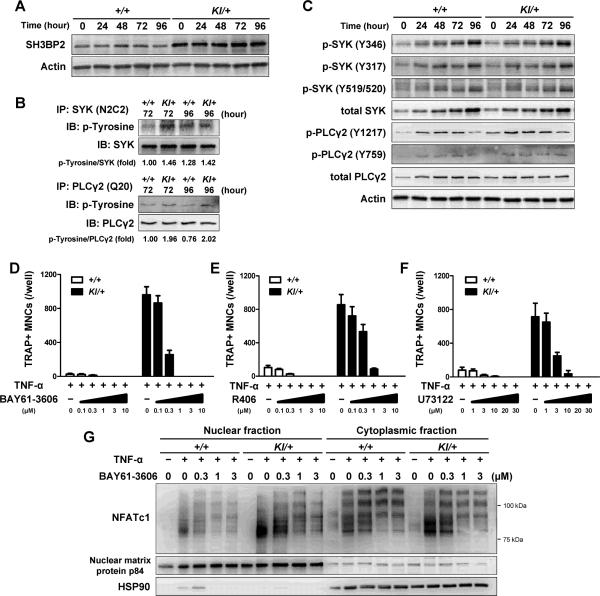
Having shown that the P416R mutation enhances TNF-α-induced osteoclast formation, we moved to the analysis of the mechanism by which mutant SH3BP2 modulates the signaling pathway downstream of TNF-α receptors in BMMs. The protein levels of SH3BP2 were constantly high in Sh3bp2KI/+ BMMs before and after TNF-α stimulation (Fig. 4A). In COS-7 and DT40 B cell lines, SYK phosphorylates SH3BP2,(8,12,54) and they cooperatively control downstream signaling events leading to the activation of NFAT,(12) suggesting the possibility that SYK activation is involved in the increased TRAP+ MNC formation of Sh3bp2KI/+ BMMs. Therefore, we examined whether phosphorylation levels of SYK are altered in Sh3bp2KI/+ BMMs treated with TNF-α. We found that total tyrosine phosphorylation of SYK in Sh3bp2KI/+ BMMs was increased at 72 to 96 hours after TNF-α stimulation (Fig. 4B, upper panels). Since elevated phosphorylation of Y346 in SYK has been reported in RANKL-stimulated Sh3bp2KI/+ BMMs,(16) we next investigated the phosphorylation levels of the specific tyrosine residues in SYK. TNF-α did induce SYK expression and phosphorylation of Y317, Y346, Y519/520 in both Sh3bp2KI/+ and Sh3bp2+/+ BMMs at comparable phosphorylation levels (Fig. 4C), indicating that phosphorylation of tyrosine residues other than Y317, Y346, Y519/520 are elevated in Sh3bp2KI/+ BMMs compared to Sh3bp2+/+ BMMs.
Fig. 4. P416R SH3BP2 cherubism mutation increases SYK and PLCγ2 phosphorylation during TNF-α-induced osteoclastogenesis.
Bone marrow cells were isolated from Sh3bp2+/+ and Sh3bp2KI/+ mice. After 2-day preculture with M-CSF (25 ng/ml), cells were stimulated with TNF-α (100 ng/ml). Protein samples were collected at the indicated time points. (A) Western blot analysis for SH3BP2 in Sh3bp2+/+ and Sh3bp2KI/+ BMMs stimulated with TNF-α. (B) Immunoprecipitation and Western blot analysis for the phosphorylation of SYK and PLCγ2. SYK and PLCγ2 proteins were immunoprecipitated using anti-SYK (N2C2) and anti-PLCγ2 (Q20) antibodies, respectively, and probed by anti-phosphotyrosine antibody (clone: 4G10). Intensities of bands were quantified, and ratio of phosphotyrosine to SYK or to PLCγ2 were calculated and normalized to that of Sh3bp2+/+ samples at 72 hours. (C) Western blot analysis for phosphorylated SYK and PLCγ2. (D–F) Quantitation of TRAP+ MNCs. BMMs were stimulated with TNF-α (100 ng/ml) for 96 hours in the presence of SYK inhibitors (BAY61-3606 (D) and R406 (E)), PLC inhibitor (U73122 (F)), and DMSO (as control). (G) NFATc1 protein levels in nuclear and cytoplasmic fractions. Protein samples were collected at 72 hours after TNF-α (100 ng/ml) treatment in the presence of BAY61-3606. Data are presented as mean ± SD. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+.
In RANKL-induced osteoclastogenesis, activated PLCγ2 is a key downstream effector of SYK, and this pathway is crucial for robust NFATc1 induction.(19,55) We also found that tyrosine phosphorylation levels of PLCγ2 are approximately 2-fold higher in Sh3bp2KI/+ BMMs than in Sh3bp2+/+ BMMs after TNF-α treatment (Fig. 4B, lower panels). We observed that TNF-α induces PLCγ2 expression in both Sh3bp2KI/+ and Sh3bp2+/+ BMMs and that the phosphorylation levels of the Y1217 are constantly increased in Sh3bp2KI/+ BMMs compared to Sh3bp2+/+ BMMs (Fig. 4C). Expression and phosphorylation patterns of other pathways important for osteoclast differentiation including NF-κB and MAP kinases (ERK, p38, JNK) were comparable between Sh3bp2KI/+ and Sh3bp2+/+ BMMs stimulated with TNF-α (Supplemental Fig. 4A). Expression of TRAF family proteins and phosphorylation of SRC, LYN, FYN, VAV1, RAC, PLCγ1, and AKT, which are potentially involved in SH3BP2-mediated signaling pathway, were similar in Sh3bp2KI/+ and Sh3bp2+/+ BMMs stimulated with TNF-α (Supplemental Fig. 4B).
To examine the involvement of SYK- and PLC-mediated pathways in TNF-α-induced osteoclastogenesis, SYK and PLC inhibitors were tested. Two SYK inhibitors (BAY61-3606 and R406) as well as a PLC inhibitor (U73122) suppressed the formation of TRAP+ MNCs in Sh3bp2KI/+ BMM cultures treated with TNF-α in a dose-dependent manner (Fig. 4D, 4E, and 4F). Furthermore, treatment with BAY61-3606 suppressed the nuclear translocation of NFATc1 in Sh3bp2KI/+ BMMs (Fig. 4G). These results suggest that mutant SH3BP2 increases NFATc1 nuclear translocation through mechanisms that involve the SYK-PLCγ2 pathway.
SH3BP2 overexpression in RAW264.7 cells enhances TNF-α-induced TRAP+ MNC formation and resorption function
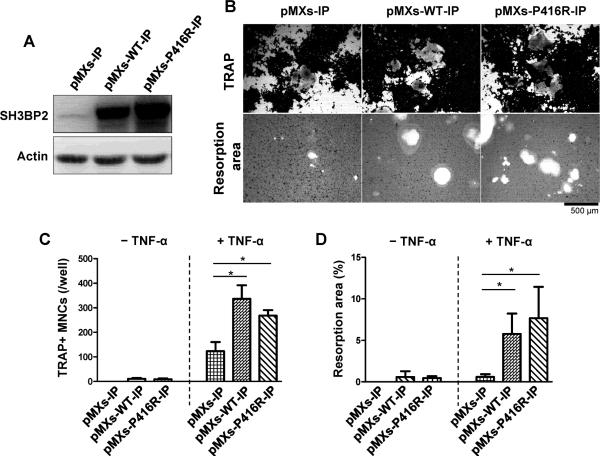
We next asked which factor, mutation-specific function or expression level of SH3BP2, is critical in the increased formation of TRAP+ MNCs in response to TNF-α. We overexpressed P416R mutant or wild-type SH3BP2 in the RAW264.7 osteoclast precursor cell line by retroviral transfection (Fig. 5A). Overexpression of wild-type SH3BP2 augmented TNF-α-induced TRAP+ MNC formation comparable to that of mutant SH3BP2 (Fig. 5B upper panels and 5C). Moreover, overexpression of wild-type SH3BP2 increased mineralized matrix resorption of the RAW264.7 cells comparable to mutant SH3BP2 (Fig. 5B lower panels and 5D). These results demonstrate that increased wild-type SH3BP2 expression can also enhance the sensitivity of osteoclast precursors to TNF-α and that the increased sensitivity is not due to specific properties of the SH3BP2 mutation, but is determined by the total amount of SH3BP2 protein, which is in agreement with previous reports.(16,17)
Fig. 5. Overexpression of SH3BP2 in RAW264.7 cells enhances TNF-α-induced TRAP+ MNC formation and resorption function.
RAW264.7 cells were infected with pMXs-WT-IP or pMXs-P416R-IP retrovirus to overexpress wild-type or P416R mutant SH3BP2, respectively. Empty retrovirus (pMXs-IP) was used as a control. After selection with puromycin (10 μg/ml), cells were cultured with or without TNF-α (100 ng/ml) for 120 hours. (A) Western blot analysis for SH3BP2. Total cell lysates were subjected to Western blot analysis for SH3BP2 expression. (B) Infected RAW264.7 cells were stimulated with TNF-α (100 ng/ml) for 120 hours, followed by TRAP staining (upper panels). Infected RAW264.7 cells were stimulated with TNF-α (100 ng/ml) on calcium phosphate-coated plates for 8 days. After removal of the cells, resorption areas were visualized by von Kossa staining (lower panels). (C) Quantitation of TRAP+ MNCs (from B). (D) Percentages of resorption area per total area were determined (from B). Data are presented as mean ± SD. * P < 0.05.
Comparison of the efficacy of TNF-α and RANKL on Sh3bp2KI/+ osteoclastogenesis
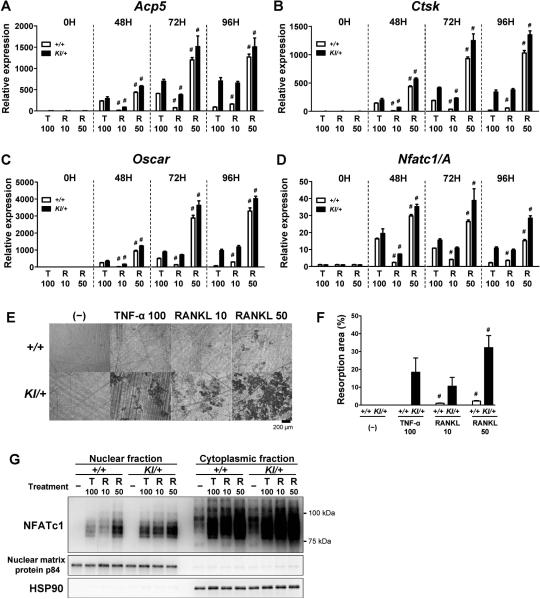
We examined how potently TNF-α induces osteoclastogenesis compared to RANKL in Sh3bp2KI/+ BMMs. BMMs were treated with TNF-α (100 ng/ml) or RANKL (10 and 50 ng/ml), and osteoclast-associated gene expression, bone-resorbing capacity, and nuclear localization of NFATc1 were compared between TNF-α and RANKL stimulation. We found that the expression levels of osteoclast-associated genes in Sh3bp2KI/+ BMMs in response to TNF-α (100 ng/ml) were equal to or greater than those induced by 10 ng/ml of RANKL (Fig. 6A−D). Similarly, bone resorption activity of TNF-α-stimulated Sh3bp2KI/+ BMMs was comparable to that of 10 ng/ml of RANKL (Fig. 6E, 6F). Finally, in Sh3bp2KI/+ BMMs, nuclear localization of NFATc1 at 72 hours after TNF-α stimulation was comparable to that induced by 10 ng/ml RANKL (Figure 6G). Induction of osteoclast-associated gene expression, resorption activity, and NFATc1 nuclear localization in Sh3bp2KI/+ BMMs by 100 ng/ml TNF-α was greater than that in Sh3bp2+/+ BMMs by 10 ng/ml RANKL, but not as effective as 50 ng/ml RANKL (except for resorption activity). These data suggest that 100 ng/ml TNF-α is able to induce osteoclast differentiation and function in Sh3bp2KI/+ BMMs as efficiently as 10 ng/ml RANKL in Sh3bp2KI/+ BMMs, and with higher efficiency than 10 ng/ml RANKL in Sh3bp2+/+ BMMs.
Fig. 6. Comparison of the effects of TNF-α and RANKL on osteoclastogenesis.
(A–D) Quantitative PCR of osteoclast-associated genes. BMMs were stimulated with TNF-α (100 ng/ml) and RANKL (10 and 50 ng/ml). Gene expression levels of Acp5 (A), Ctsk (B), Oscar (C), and Nfatc1/A (D) were determined. Gene expression levels relative to Hprt were calculated and normalized to the expression level of Sh3bp2+/+ BMMs at 0 hour. (E, F) Resorption assay. BMMs were cultured with TNF-α or RANKL at indicated concentrations for 14 days on dentin slices. After removal of the cells, resorption areas were visualized by toluidine blue (E). Resorbed areas per total surface area were quantified (F). (G) Western blot analysis for NFATc1. BMMs were stimulated with TNF-α (100 ng/ml) or RANKL (10 and 50 ng/ml) for 72 hours. NFATc1 expression levels in nuclear and cytoplasmic fractions were determined. Data are presented as mean ± SD. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+. # P < 0.05 compared to TNF-α-stimulated Sh3bp2KI/+ BMMs at each time point.
In vivo effect of TNF-α on osteoclast formation in SH3BP2 cherubism mutant mice
To determine whether the P416R SH3BP2 mutation plays a role in TNF-α-mediated TRAP+ MNC formation in vivo, we conducted TNF-α injection experiments on calvarial bone of Sh3bp2KI/+ mice.(40) Ten-week-old male Sh3bp2KI/+ and control Sh3bp2+/+ mice were injected with TNF-α (1.5 μg/day) or PBS subcutaneously over the calvariae for 5 days. MicroCT analysis showed that the number of erosion pits on calvarial bone surface in TNF-α-injected Sh3bp2KI/+ mice is increased compared to Sh3bp2+/+ mice (Fig. 7A), and calvarial bone destruction was more severe in TNF-α-injected Sh3bp2KI/+ mice (Fig. 7B). Consistent with these results, increased numbers of TRAP+ MNCs were observed in TNF-α-injected Sh3bp2KI/+ mice (Fig. 7C). Histomorphometrical measurements of eroded surface per bone surface (ES/BS), number of osteoclasts per bone surface (N.Oc/BS), and osteoclast surface per bone surface (Oc.S/BS) at the sagittal suture confirmed increased osteoclast formation in TNF-α-injected Sh3bp2KI/+ mice (Fig. 7D, 7E, and 7F). The Rankl and Opg mRNA expression ratio in the calvarial bone tissues injected with TNF-α was comparable between Sh3bp2KI/+ and Sh3bp2+/+ mice (Supplemental Fig. 5A-C). Collectively, these data indicate that Sh3bp2KI/+ mice are more susceptible to TNF-α stimulation than Sh3bp2+/+ mice, resulting in increased osteoclast formation and inflammatory bone loss.
hTNFtg mice in Sh3bp2KI/+ background exhibit increased rate of bone loss
We next questioned whether the gain-of-function P416R mutation enhances TNF-α-driven inflammatory bone loss in a rheumatoid arthritis model. The Sh3bp2KI/+ mice were crossed with transgenic mice expressing human TNF-α (hTNFtg), which spontaneously develop arthritis and systemic bone loss.(35) Swelling of the paws was assessed in Sh3bp2+/+ (n = 11), Sh3bp2KI/+ (n = 11), Sh3bp2+/+/hTNFtg (n = 8), and Sh3bp2KI/+/hTNFtg (n = 10) male mice until age of 16 weeks when serum and hind limbs samples were collected. Both Sh3bp2KI/+/hTNFtg and Sh3bp2+/+/hTNFtg mice developed severe paw swelling (arthritis), and the severity of arthritis was similar in both double mutants (Fig. 8A). Serum levels of human and mouse TNF-α were not different between Sh3bp2KI/+/hTNFtg and Sh3bp2+/+/hTNFtg mice (Fig. 8B), indicating that osteoclast progenitor cells in both mutant lines are exposed to equivalent amount of TNF-α.
MicroCT images of hind paws and talus bones showed severe joint destruction in Sh3bp2KI/+/hTNFtg mutants. MicroCT analysis revealed that Sh3bp2KI/+/hTNFtg mutants exhibit more severe focal bone loss as shown by 2.9-fold greater reduction rate of talus bone volume in Sh3bp2KI/+/hTNFtg mutants (Fig. 8C). Histological analysis of ankle joints showed an increased inflammatory bone destruction (Fig. 8D, H&E) and greater numbers of osteoclast formation on the surface of talus bones in Sh3bp2KI/+/hTNFtg mice than in Sh3bp2+/+/hTNFtg mice (Fig. 8D, TRAP). Inflammation score of ankle joints, ES/BS, N.Oc/BS, and Oc.S/BS were all increased in Sh3bp2KI/+/hTNFtg mice compared to Sh3bp2+/+/hTNFtg mice (Fig. 8D). Further microCT analysis revealed that Sh3bp2KI/+/hTNFtg mice exhibit approximately 2-fold greater reduction rate of systemic bone mass as represented by trabecular bone volume per total volume (BV/TV), trabecular bone number (Tb.N), trabecular bone separation (Tb.Sp), and trabecular thickness (Tb.Th) at proximal tibia (Fig. 8E). The reduction rate of cortical bone thickness (Ct.Th) at the midshaft of tibiae was also 1.6-fold increased (Fig. 8F). These results indicate that the P416R mutation promotes osteoclast formation and subsequent bone erosion and systemic bone loss in response to TNF-α in hTNFtg mice.
Decreased SH3BP2 levels suppress TNF-α induction of TRAP+ MNC formation and mineralized matrix-resorbing capacity in RAW264.7 cells
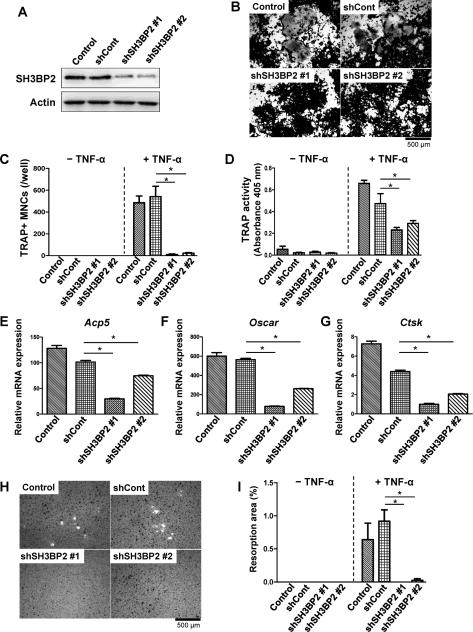
Since overexpression of wild-type SH3BP2 increased the TRAP+ MNC formation and mineralized matrix-resorbing capacity of RAW264.7 cells in response to TNF-α (Fig. 5), we investigated whether reduced SH3BP2 expression affects the TNF-α-induced TRAP+ MNC formation and the resorption capacity. Lentiviruses that overexpress shRNA against Sh3bp2 were used to knockdown SH3BP2 in RAW264.7 cells. Two independent SH3BP2 knockdown cell lines, non-silencing shRNA-infected cells, and non-infected parent RAW264.7 cells (Fig. 9A) were stimulated with TNF-α for 5 days. The two knockdown cell lines formed smaller numbers of TRAP+ MNCs in response to TNF-α compared to the non-infected cells and non-silencing shRNA infected cells (Fig. 9B, 9C), and TRAP activity in the culture supernatant was decreased in both knockdown cell lines (Fig. 9D). Consistent with suppressed formation of TRAP+ MNCs, mRNA expression of osteoclast-associated genes such as Acp5, Oscar, and Ctsk was decreased in the knockdown cells (Fig. 9E, 9F, and 9G). Furthermore, SH3BP2 knockdown in the RAW264.7 cells suppressed TNF-α-induced resorption of mineralized matrix (Fig. 9H, 9I). These results demonstrate that reduced level of SH3BP2 results in less potential for osteoclast progenitors to differentiate into mature osteoclasts in response to TNF-α.
Fig. 9. SH3BP2 knockdown suppresses TNF-α-induced TRAP+ MNC formation and resorption function in RAW264.7 cells.
RAW264.7 cells were infected with non-silencing (shCont) and SH3BP2 shRNA (shSH3BP2 #1 and #2) lentiviruses. After puromycin selection, the infected RAW264.7 cells and non-infected RAW264.7 cells (Control) were stimulated with TNF-α (100 ng/ml) for 120 hours. (A) Western blot analysis for SH3BP2. Total cell lysates were subjected to the Western blot analysis for SH3BP2 expression. (B) TRAP staining, (C) quantitation of TRAP+ MNCs, and (D) TRAP activity in the culture supernatant were determined 120 hours after TNF-α treatment. (E–G) Osteoclast-associated gene expression. RNA samples were collected 96 hours after TNF-α treatment and subjected to qPCR analysis for Acp5, Oscar, and Ctsk mRNA expression. Expression levels relative to Hprt were calculated and normalized to the expression level of non-stimulated shCont cells. (H, I) Infected RAW264.7 cells were stimulated with TNF-α (100 ng/ml) on calcium phosphate-coated plates for 8 days. After removal of the cells, resorption area was visualized by von Kossa staining (H). Percentages of resorption area per total area were determined (I). The data are representative of two independent experiments. Data are presented as mean ± SD. * P < 0.05.
Discussion
Under inflammatory conditions, proinflammatory cytokines such as TNF-α and IL-1 can cooperate with RANKL and synergistically potentiate bone resorption by osteoclasts.(22,29) TNF-α is known as a dominant proinflammatory cytokine that plays a critical role in the promotion of osteoclastogenesis leading to inflammatory bone resorption.(22) Therefore, obtaining insights into the mechanisms that modulate inflammatory cytokine-induced or -assisted osteoclast differentiation and function will propel the development of novel treatments for inflammatory bone diseases. Our data suggest that SH3BP2 plays a role in TNF-α-induced osteoclastogenesis.
Our in vitro studies showed that the P416R cherubism gain-of-function mutation in SH3BP2 enhances TNF-α induction of osteoclast formation comparable to the level of RANKL stimulation of wild-type BMMs (Fig. 2C), although previous cell culture studies have shown that TNF-α is less potent in inducing osteoclast differentiation of BMMs than RANKL.(24-26) This result demonstrates that SH3BP2 regulates TNF-α-induced osteoclastogenesis by modulating the sensitivity of osteoclast precursor cells to TNF-α in vitro. We further showed that SH3BP2 gain-of-function exacerbates bone volume loss in hTNFtg mice, a mouse model for human rheumatoid arthritis, demonstrating that SH3BP2 serves as a regulatory protein for TNF-α-mediated inflammatory bone loss in vivo.
Interestingly, overexpression of wild-type SH3BP2 increased the TRAP+ MNC formation and mineralized matrix-resorbing function of RAW264.7 cells in response to TNF-α (Fig. 5). In contrast, knockdown of wild-type SH3BP2 decreased the TRAP+ MNC formation and mineralized matrix resorption (Fig. 9). These results suggest that wild-type SH3BP2 plays a role in TNF-α-induced osteoclastogenesis and that SH3BP2 could be a potential therapeutic target of the osteoclast-driven bone loss in inflammatory diseases that involve TNF-α. Taking into account the previous findings that SH3BP2 is important for RANKL-induced osteoclastogenesis,(13,16,34,56) our results imply that SH3BP2 controls both physiological and pathological bone remodeling by osteoclasts through mechanisms that modulate the sensitivities of osteoclast progenitors to RANKL and TNF-α, respectively.
In addition, our data suggest that TNF-α could be an inducer of TRAP+ MNC formation in cherubism lesions. Given the fact that cherubism lesions contain CD14+ osteoclast progenitor cells as well as TNF-α-expressing stromal cells (Fig. 1), it is reasonable to assume that formation of TRAP+ MNCs embedded in the cherubism lesions is attributed to the increased responsiveness of mutant osteoclast progenitors to TNF-α as well as to RANKL in the lesions.(57,58) This concept is supported by our finding that BMMs from Sh3bp2KI/+ mice responded to TNF-α (100 ng/ml) at a comparable level to RANKL (10 ng/ml) in in vitro osteoclastogenesis (Fig. 6).
Consistent with Kobayashi et al.,(27) wild-type (Sh3bp2+/+) TRAP+ MNCs induced by TNF-α did not form resorption pits in our resorption assays (Fig. 2H, 2I, and 2J), suggesting that osteoclasts induced solely by TNF-α might lack or have significantly lower bone-resorbing activity (quiescent osteoclast (27)), compared to TRAP+ MNCs induced by RANKL. Interestingly, we found that Sh3bp2KI/+ TRAP+ MNCs induced only by TNF-α are able to form resorption pits (fully functional osteoclasts). Kobayashi et al. also demonstrated that IL-1α treatment together with TNF-α is able to induce bone resorption activity in wild-type TRAP+ MNCs. (27) Therefore, formation of the bone resorbing TRAP+ MNCs from TNF-α-stimulated Sh3bp2KI/+ BMMs is likely due to the involvement of mutant SH3BP2 in enhanced production of inflammatory cytokines other than TNF-α (59) that induce the formation of bone-resorbing TRAP+ MNCs synergistically with TNF-α in the BMM cultures.
Our results demonstrate that the mutant SH3BP2 enhances TNF-α-induced TRAP+ MNC formation via induction of NFATc1. In RANKL-induced osteoclast formation, NFATc1 induction is required for the full differentiation and activation of osteoclasts. Initiation of this induction is dependent on both the TRAF6-NF-κB and c-Fos pathways.(47,60) However, nuclear expressions of NF-κB (p50, p52, p65) and c-Fos in TNF-α-stimulated BMMs were not affected by the gain-of-function of SH3BP2 (Supplemental Fig. 2), suggesting that mutant SH3BP2 regulates TNF-α-induced TRAP+ MNC formation by exclusively modulating robust and sustained NFATc1 induction and nuclear translocation mechanism following TNF-α stimulation,(53) which is similar to the NFATc1 autoamplification mechanism in RANKL-induced osteoclastogenesis.(48) Sustained expression of Nfatc1/A and osteoclast marker genes (Fig. 3A) further suggests that elevated levels of SH3BP2 protein might increase the survival of TRAP+ MNCs under TNF-α stimulation.(61)
RBPJ-Blimp1-IRF8 is an important axis for the regulation of TNF-α-induced osteoclast differentiation through mechanisms that also control NFATc1 expression.(26,49) Our Western blotting analysis with Sh3bp2KI/+ BMMs did not show marked differences in the expression of IRF8 in the nucleus following TNF-α stimulation (Supplemental Fig. 2), suggesting that the mutant SH3BP2 is unlikely to control TNF-α-induced TRAP+ MNC formation by mechanisms that involve the RBPJ-Blimp1-IRF8 axis. Mafb, Bcl6, and NF-κB p100, which are negative transcription factors for osteoclastogenesis,(25,50,51) are not likely to be involved in the SH3BP2 control of TNF-α-induced TRAP+ MNC formation either (Supplemental Fig. 1 and Supplemental Fig. 2).
SYK activation precedes NFATc1 induction in RANKL-induced osteoclastogenesis,(19) and the activation of SYK plays a critical role in differentiation and function of osteoclasts by associating with phosphorylated immunoreceptor tyrosine-based activation motifs (ITAMs) within FcRγ and DAP12 adaptors,(62) where PLCγ2 mediates the activation signal to induce robust expression of NFATc1.(19,55) In fact, SYK-, PLCγ2-, and NFATc1-deficient osteoclast progenitors are all defective in osteoclast development.(34,62-64) Increased phosphorylation of SYK was observed in Sh3bp2KI/+ BMMs stimulated with TNF-α compared to Sh3bp2+/+ BMMs (Fig. 4B). Interestingly, phosphorylation levels of tyrosine 346 (Y346) in SYK, which are elevated in RANKL-stimulated Sh3bp2KI/+ BMMs compared to Sh3bp2+/+ BMMs,(16) were comparable between TNF-α-simulated Sh3bp2KI/+ and Sh3bp2+/+ BMMs (Fig. 4C). This result indicates that the signaling pathway through which mutant SH3BP2 regulates TNF-α-induced osteoclast differentiation is different from the RANKL-stimulated pathway, although increased NFATc1 induction is the common final consequence of Sh3bp2KI/+ BMM stimulation by the two different cytokines.(34) It has been reported that TNF-α rapidly (within 30 seconds) activates SYK in human T cells (Jurkat) and myeloid cells (U937 and KBM-5) following the recruitment of SYK in the TNFR1 and TNFR2 complex, but how TNF-α stimulation activates SYK is not clear.(65) Because SH3BP2 associates with SYK (5,56,66) and P416R mutant SH3BP2 increases SYK phosphorylation in Sh3bp2KI/+ BMMs, it is evident that SH3BP2 mediates TNF-α-induced SYK activation. Nevertheless, SH3BP2 is unlikely to phosphorylate SYK directly, since SH3BP2 is an adaptor protein with no kinase activity. Therefore, identification of a kinase (including SYK itself) that phosphorylates SYK in cooperation with SH3BP2 upon TNF-α stimulation would advance our understanding of the SH3BP2 function and allow us to decipher the differences in signaling pathway between TNF-α- and RANKL-induced osteoclastogenesis.
PLCγ2 regulates RANKL-induced osteoclastogenic transcriptional activation by two distinct mechanisms: adaptor function that mediates GAB2 phosphorylation and enzyme function that activates calcium signaling.(47,64,67) Y1217 of PLCγ2 was hyperphosphorylated in Sh3bp2KI/+ BMMs stimulated with TNF-α (Fig. 4C), which was also the case in RAW264.7 cells overexpressing human P418R mutant SH3BP2 (equivalent to P416R in mice).(68) Therefore, it can be speculated that increased activation of PLCγ2 by Y1217 hyperphosphorylation (69) leads to either modulated adaptor function of PLCγ2 or elevated Ca2+-calcineurin signaling in TNF-α-stimulated Sh3bp2KI/+ BMMs.
We have previously reported that cherubism mutant SH3BP2 controls both inflammation and osteoclastogenesis by two distinct pathways, which are mediated through ERK and SYKNFATc1, respectively.(16,34) In the mouse model, spontaneous inflammation is observed only in homozygous Sh3bp2KI/KI mice, in which SH3BP2 expression is even higher than in heterozygous Sh3bp2KI/+ mice.(17) In this current study, we found that the heterozygous P416R SH3BP2 mutation does not significantly influence the arthritis scores (inflammation), but it affects the focal and systemic bone loss in the hTNFtg model (Fig. 8). We speculate that this is because the increased amount of SH3BP2 protein in heterozygous Sh3bp2KI/+ macrophages is not sufficient to enhance TNF-α production and TNF-α-mediated inflammation in the hTNFtg model but adequate for enhancing osteoclastogenesis. The dissociation of inflammation from bone loss indicates the diverse function of SH3BP2 in modulating immune and skeletal homeostasis, which presumably depends on cellular levels of SH3BP2 protein.
The RANKL-RANK pathway is critically important for inflammatory bone loss, because TNF-α induces RANKL expression in stromal cells and RANK expression in osteoclast progenitors synergistically with RANKL.(20-22,70) Since SH3BP2 regulates both RANKL- and TNF-α-induced osteoclast formation in vitro, it is reasonable to conclude that exacerbation of bone loss in Sh3bp2KI/+/hTNFtg mice is due to the accumulative effects of osteoclastic bone resorption controlled by SH3BP2 downstream of both RANK and TNF-α receptors.
In summary, we demonstrated that the cherubism mutation in SH3BP2 enhances TNF-α-induced osteoclast formation in vitro and TNF-α-mediated osteoclast formation in vivo, resulting in increased bone loss and destruction. We also showed that wild-type SH3BP2 knockdown in RAW264.7 cells exhibit decreased TRAP+ MNC formation in response to TNF-α. SH3BP2 inhibition in osteoclast precursors, which could prevent TNF-α-mediated inflammatory bone loss, may benefit not only cherubism patients but also individuals with inflammatory bone diseases including rheumatoid arthritis that impair both the immune and the skeletal system.
Supplementary Material
Supplemental Fig. 1. Osteoclast-associated gene expression in TNF-α-stimulated Sh3bp2+/+ and Sh3bp2KI/+ bone marrow cells. BMMs from Sh3bp2+/+ and Sh3bp2KI/+ mice were stimulated with TNF-α (100 ng/ml) for 96 hours. RNA samples were collected at the indicated time points and subjected to qPCR analysis. Gene expression levels relative to Hprt were calculated and normalized to the expression of Sh3bp2+/+ at 0 hour. The data are representative of three independent experiments. Data are presented as mean ± SD. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+. * P < 0.05.
Supplemental Fig. 2. Nuclear translocation of transcription factors during TNF-α-induced TRAP+ MNC formation in Sh3bp2+/+ and Sh3bp2KI/+ BMMs. BMMs from Sh3bp2+/+ and Sh3bp2KI/+ mice were stimulated with TNF-α (100 ng/ml). Nuclear and cytoplasmic protein samples were collected at indicated time points and subjected to Western blot analysis for indicated proteins. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+.
Supplemental Fig. 3. Effect of the inhibition of NF-κB- and c-Fos-mediated signaling pathways on TNF-α-induced TRAP+ MNC formation. (A, B) TRAP staining images and quantitation of TRAP+ MNCs. BMMs from Sh3bp2+/+ and Sh3bp2KI/+ mice were stimulated with TNF-α (100ng/ml) in the presence of wild-type and mutant forms of NEMO-binding domain (NBD) peptides and vehicle (DMSO) for 96 hours. (C, D) TRAP staining images and quantitation of TRAP+ MNCs. Sh3bp2KI/+ mice were crossed with c-Fos-deficient (c-fos−/−) mice. Fetal liver cells from c-Fos-deficient Sh3bp2KI/+ mice (Sh3bp2KI/+/c-fos−/−) were stimulated with TNF-α (100 ng/ml) for 96 hours. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+. Data are presented as mean ± SD. *P < 0.05.
Supplemental Fig. 4. Intracellular signaling pathways during TNF-α-induced TRAP+ MNC formation. (A, B) BMMs from Sh3bp2+/+ and Sh3bp2KI/+ mice were stimulated with TNF-α (100 ng/ml). Protein samples were collected at indicated time points and subjected to Western blot analysis for indicated proteins. Expression levels were examined in (A) short-term culture and (B) long-term culture. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+.
Supplemental Fig. 5. Rankl and Opg mRNA expression in TNF-α-injected calvarial tissues. (A–C) Ten-week-old male Sh3bp2+/+ and Sh3bp2KI/+ mice were subcutaneously injected with either PBS or TNF-α (1.5 μg/mouse/day) over calvariae for 5 days. Mice were euthanized 24 hours after last injection. Gene expression of Rankl and Opg and their ratio. RNA samples were isolated from mouse calvarial tissues and subjected to qPCR analysis (n = 3/group). Gene expression levels relative to Hprt were calculated and normalized to the expression levels of PBS-injected Sh3bp2+/+ control. Data are presented as mean ± SD. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+. * P < 0.05, NS: not significant.
Acknowledgments
We would like to thank Drs. L. Bonewald, M. Johnson, J. Gorski, S. Dallas, and members of Bone Biology Research Program at the University of Missouri-Kansas City (UMKC), School of Dentistry for critical suggestions. We are grateful to M. Ueki and S. Ueki for technical assistance. We are also indebted to the staff at the UMKC Laboratory Animal Research Core. This work was supported by a grant from the NIH (R01DE020835) to YU. TM is a recipient of 2008 Japan Rheumatism Foundation Rheumatology Traveling Fellowship and 2011 ASBMR Young Investigator Award. We acknowledge the use of a confocal microscope in the UMKC, School of Dentistry Confocal Microscopy Core supported by the UMKC Office of Research Services, UMKC Center of Excellence in Dental and Musculoskeletal Tissues, and NIH grant S10RR027668.
Footnotes
Authors’ roles: Study design: TM and YU. Study conduct and data collection: TM, SI, RI, RG. Data analysis: TM, TY, MK. Data interpretation: TM, YLL, RR, MB, ER, and YU. Drafting manuscript: TM and YU. YU supervised the overall study and wrote the final manuscript. TM and YU take responsibility for the integrity of data analysis.
Conflict of Interests
All authors state that they have no conflict of interest.
Reference
- 1.Southgate J, Sarma U, Townend JV, Barron J. Flanagan AM Study of the cell biology and biochemistry of cherubism. J Clin Pathol. 1998;51(11):831–7. doi: 10.1136/jcp.51.11.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tiziani V, Reichenberger E, Buzzo CL, Niazi S, Fukai N, Stiller M, et al. The gene for cherubism maps to chromosome 4p16. Am J Hum Genet. 1999;65(1):158–66. doi: 10.1086/302456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueki Y, Tiziani V, Santanna C, Fukai N, Maulik C, Garfinkle J, et al. Mutations in the gene encoding c-Abl-binding protein SH3BP2 cause cherubism. Nat Genet. 2001;28(2):125–6. doi: 10.1038/88832. [DOI] [PubMed] [Google Scholar]
- 4.Ren R, Mayer BJ, Cicchetti P. Baltimore D Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259(5098):1157–61. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 5.Deckert M, Tartare-Deckert S, Hernandez J, Rottapel R, Altman A. Adaptor function for the Syk kinases-interacting protein 3BP2 in IL-2 gene activation. Immunity. 1998;9(5):595–605. doi: 10.1016/s1074-7613(00)80657-3. [DOI] [PubMed] [Google Scholar]
- 6.Foucault I, Liu YC, Bernard A, Deckert M. The chaperone protein 14-3-3 interacts with 3BP2/SH3BP2 and regulates its adapter function. J Biol Chem. 2003;278(9):7146–53. doi: 10.1074/jbc.M209509200. [DOI] [PubMed] [Google Scholar]
- 7.Foucault I, Le Bras S, Charvet C, Moon C, Altman A, Deckert M. The adaptor protein 3BP2 associates with VAV guanine nucleotide exchange factors to regulate NFAT activation by the B-cell antigen receptor. Blood. 2005;105(3):1106–13. doi: 10.1182/blood-2003-08-2965. [DOI] [PubMed] [Google Scholar]
- 8.Maeno K, Sada K, Kyo S, Miah SM, Kawauchi-Kamata K, Qu X, et al. Adaptor protein 3BP2 is a potential ligand of Src homology 2 and 3 domains of Lyn protein-tyrosine kinase. J Biol Chem. 2003;278(27):24912–20. doi: 10.1074/jbc.M301201200. [DOI] [PubMed] [Google Scholar]
- 9.Jevremovic D, Billadeau DD, Schoon RA, Dick CJ, Leibson PJ. Regulation of NK cell-mediated cytotoxicity by the adaptor protein 3BP2. J Immunol. 2001;166(12):7219–28. doi: 10.4049/jimmunol.166.12.7219. [DOI] [PubMed] [Google Scholar]
- 10.Yu Z, Maoui M, Zhao ZJ, Li Y, Shen SH. SHP-1 dephosphorylates 3BP2 and potentially downregulates 3BP2-mediated T cell antigen receptor signaling. FEBS J. 2006;273(10):2195–205. doi: 10.1111/j.1742-4658.2006.05233.x. [DOI] [PubMed] [Google Scholar]
- 11.Chihara K, Nakashima K, Takeuchi K, Sada K. Association of 3BP2 with SHP-1 regulates SHP-1-mediated production of TNF-alpha in RBL-2H3 cells. Genes Cells. 2011;16(12):1133–45. doi: 10.1111/j.1365-2443.2011.01557.x. [DOI] [PubMed] [Google Scholar]
- 12.Shukla U, Hatani T, Nakashima K, Ogi K, Sada K. Tyrosine phosphorylation of 3BP2 regulates B cell receptor-mediated activation of NFAT. J Biol Chem. 2009;284(49):33719–28. doi: 10.1074/jbc.M109.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levaot N, Simoncic PD, Dimitriou ID, Scotter A, La Rose J, Ng AH, et al. 3BP2-deficient mice are osteoporotic with impaired osteoblast and osteoclast functions. J Clin Invest. 2011;121(8):3244–57. doi: 10.1172/JCI45843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G, Dimitriou ID, La Rose J, Ilangumaran S, Yeh WC, Doody G, et al. The 3BP2 adapter protein is required for optimal B-cell activation and thymus-independent type 2 humoral response. Mol Cell Biol. 2007;27(8):3109–22. doi: 10.1128/MCB.01014-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de la Fuente MA, Kumar L, Lu B, Geha RS. 3BP2 deficiency impairs the response of B cells, but not T cells, to antigen receptor ligation. Mol Cell Biol. 2006;26(14):5214–25. doi: 10.1128/MCB.00087-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueki Y, Lin CY, Senoo M, Ebihara T, Agata N, Onji M, et al. Increased myeloid cell responses to M-CSF and RANKL cause bone loss and inflammation in SH3BP2 “cherubism” mice. Cell. 2007;128(1):71–83. doi: 10.1016/j.cell.2006.10.047. [DOI] [PubMed] [Google Scholar]
- 17.Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, et al. Loss of Tankyrase-Mediated Destruction of 3BP2 Is the Underlying Pathogenic Mechanism of Cherubism. Cell. 2011;147(6):1324–39. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guettler S, LaRose J, Petsalaki E, Gish G, Scotter A, Pawson T, et al. Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell. 2011;147(6):1340–54. doi: 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 19.Takayanagi H. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat Rev Immunol. 2007;7(4):292–304. doi: 10.1038/nri2062. [DOI] [PubMed] [Google Scholar]
- 20.Romas E, Gillespie MT, Martin TJ. Involvement of receptor activator of NFkappaB ligand and tumor necrosis factor-alpha in bone destruction in rheumatoid arthritis. Bone. 2002;30(2):340–6. doi: 10.1016/s8756-3282(01)00682-2. [DOI] [PubMed] [Google Scholar]
- 21.Nanes MS. Tumor necrosis factor-alpha: molecular and cellular mechanisms in skeletal pathology. Gene. 2003;321:1–15. doi: 10.1016/s0378-1119(03)00841-2. [DOI] [PubMed] [Google Scholar]
- 22.Hardy R, Cooper MS. Bone loss in inflammatory disorders. J Endocrinol. 2009;201(3):309–20. doi: 10.1677/JOE-08-0568. [DOI] [PubMed] [Google Scholar]
- 23.Redlich K, Smolen JS. Inflammatory bone loss: pathogenesis and therapeutic intervention. Nat Rev Drug Discov. 2012;11(3):234–50. doi: 10.1038/nrd3669. [DOI] [PubMed] [Google Scholar]
- 24.Lam J, Takeshita S, Barker JE, Kanagawa O, Ross FP, Teitelbaum SL. TNF-alpha induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J Clin Invest. 2000;106(12):1481–8. doi: 10.1172/JCI11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yao Z, Xing L, Boyce BF. NF-kappaB p100 limits TNF-induced bone resorption in mice by a TRAF3-dependent mechanism. J Clin Invest. 2009;119(10):3024–34. doi: 10.1172/JCI38716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao B, Grimes SN, Li S, Hu X, Ivashkiv LB. TNF-induced osteoclastogenesis and inflammatory bone resorption are inhibited by transcription factor RBP-J. J Exp Med. 2012;209(2):319–34. doi: 10.1084/jem.20111566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi K, Takahashi N, Jimi E, Udagawa N, Takami M, Kotake S, et al. Tumor necrosis factor alpha stimulates osteoclast differentiation by a mechanism independent of the ODF/RANKL-RANK interaction. J Exp Med. 2000;191(2):275–86. doi: 10.1084/jem.191.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim N, Kadono Y, Takami M, Lee J, Lee SH, Okada F, et al. Osteoclast differentiation independent of the TRANCE-RANK-TRAF6 axis. J Exp Med. 2005;202(5):589–95. doi: 10.1084/jem.20050978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei S, Kitaura H, Zhou P, Ross FP, Teitelbaum SL. IL-1 mediates TNF-induced osteoclastogenesis. J Clin Invest. 2005;115(2):282–90. doi: 10.1172/JCI23394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Sarosi I, Yan XQ, Morony S, Capparelli C, Tan HL, et al. RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. Proc Natl Acad Sci U S A. 2000;97(4):1566–71. doi: 10.1073/pnas.97.4.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fuller K, Murphy C, Kirstein B, Fox SW, Chambers TJ. TNFalpha potently activates osteoclasts, through a direct action independent of and strongly synergistic with RANKL. Endocrinology. 2002;143(3):1108–18. doi: 10.1210/endo.143.3.8701. [DOI] [PubMed] [Google Scholar]
- 32.Azuma Y, Kaji K, Katogi R, Takeshita S, Kudo A. Tumor necrosis factor-alpha induces differentiation of and bone resorption by osteoclasts. J Biol Chem. 2000;275(7):4858–64. doi: 10.1074/jbc.275.7.4858. [DOI] [PubMed] [Google Scholar]
- 33.Hero M, Suomalainen A, Hagstrom J, Stoor P, Kontio R, Alapulli H, et al. Anti-tumor necrosis factor treatment in cherubism - Clinical, radiological and histological findings in two children. Bone. 2013;52(1):347–53. doi: 10.1016/j.bone.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Aliprantis AO, Ueki Y, Sulyanto R, Park A, Sigrist KS, Sharma SM, et al. NFATc1 in mice represses osteoprotegerin during osteoclastogenesis and dissociates systemic osteopenia from inflammation in cherubism. J Clin Invest. 2008;118(11):3775–89. doi: 10.1172/JCI35711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayward MD, Jones BK, Saparov A, Hain HS, Trillat AC, Bunzel MM, et al. An extensive phenotypic characterization of the hTNFalpha transgenic mice. BMC Physiol. 2007;7:13. doi: 10.1186/1472-6793-7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janckila AJ, Takahashi K, Sun SZ, Yam LT. Naphthol-ASBI phosphate as a preferred substrate for tartrate-resistant acid phosphatase isoform 5b. J Bone Miner Res. 2001;16(4):788–93. doi: 10.1359/jbmr.2001.16.4.788. [DOI] [PubMed] [Google Scholar]
- 37.Jimi E, Aoki K, Saito H, D'Acquisto F, May MJ, Nakamura I, et al. Selective inhibition of NF-kappa B blocks osteoclastogenesis and prevents inflammatory bone destruction in vivo. Nat Med. 2004;10(6):617–24. doi: 10.1038/nm1054. [DOI] [PubMed] [Google Scholar]
- 38.Tanabe N, Wheal BD, Kwon J, Chen HH, Shugg RP, Sims SM, et al. Osteopontin signals through calcium and nuclear factor of activated T cells (NFAT) in osteoclasts: a novel RGD-dependent pathway promoting cell survival. J Biol Chem. 2011;286(46):39871–81. doi: 10.1074/jbc.M111.295048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kitamura T. New experimental approaches in retrovirus-mediated expression screening. Int J Hematol. 1998;67(4):351–9. doi: 10.1016/s0925-5710(98)00025-5. [DOI] [PubMed] [Google Scholar]
- 40.Kitaura H, Zhou P, Kim HJ, Novack DV, Ross FP, Teitelbaum SL. M-CSF mediates TNF-induced inflammatory osteolysis. J Clin Invest. 2005;115(12):3418–27. doi: 10.1172/JCI26132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Proulx ST, Kwok E, You Z, Papuga MO, Beck CA, Shealy DJ, et al. Longitudinal assessment of synovial, lymph node, and bone volumes in inflammatory arthritis in mice by in vivo magnetic resonance imaging and microfocal computed tomography. Arthritis Rheum. 2007;56(12):4024–37. doi: 10.1002/art.23128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res. 2010;25(7):1468–86. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 43.Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, et al. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res. 2013;28(1):2–17. doi: 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicholson GC, Malakellis M, Collier FM, Cameron PU, Holloway WR, Gough TJ, et al. Induction of osteoclasts from CD14-positive human peripheral blood mononuclear cells by receptor activator of nuclear factor kappaB ligand (RANKL). Clin Sci (Lond) 2000;99(2):133–40. [PubMed] [Google Scholar]
- 45.Sorensen MG, Henriksen K, Schaller S, Henriksen DB, Nielsen FC, Dziegiel MH, et al. Characterization of osteoclasts derived from CD14+ monocytes isolated from peripheral blood. J Bone Miner Metab. 2007;25(1):36–45. doi: 10.1007/s00774-006-0725-9. [DOI] [PubMed] [Google Scholar]
- 46.Chomette G, Auriol M, Guilbert F, Vaillant JM. Cherubism. Histo-enzymological and ultrastructural study. Int J Oral Maxillofac Surg. 1988;17(4):219–23. doi: 10.1016/s0901-5027(88)80043-2. [DOI] [PubMed] [Google Scholar]
- 47.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, et al. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell. 2002;3(6):889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 48.Asagiri M, Sato K, Usami T, Ochi S, Nishina H, Yoshida H, et al. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J Exp Med. 2005;202(9):1261–9. doi: 10.1084/jem.20051150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao B, Takami M, Yamada A, Wang X, Koga T, Hu X, et al. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat Med. 2009;15(9):1066–71. doi: 10.1038/nm.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim K, Kim JH, Lee J, Jin HM, Kook H, Kim KK, et al. MafB negatively regulates RANKL-mediated osteoclast differentiation. Blood. 2007;109(8):3253–9. doi: 10.1182/blood-2006-09-048249. [DOI] [PubMed] [Google Scholar]
- 51.Miyauchi Y, Ninomiya K, Miyamoto H, Sakamoto A, Iwasaki R, Hoshi H, et al. The Blimp1-Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J Exp Med. 2010;207(4):751–62. doi: 10.1084/jem.20091957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishikawa K, Nakashima T, Hayashi M, Fukunaga T, Kato S, Kodama T, et al. Blimp1-mediated repression of negative regulators is required for osteoclast differentiation. Proc Natl Acad Sci U S A. 2010;107(7):3117–22. doi: 10.1073/pnas.0912779107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yarilina A, Xu K, Chen J, Ivashkiv LB. TNF activates calcium-nuclear factor of activated T cells (NFAT)c1 signaling pathways in human macrophages. Proc Natl Acad Sci U S A. 2011;108(4):1573–8. doi: 10.1073/pnas.1010030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chihara K, Kimura Y, Honjoh C, Yamauchi S, Takeuchi K, Sada K. Tyrosine phosphorylation of 3BP2 is indispensable for the interaction with VAV3 in chicken DT40 cells. Exp Cell Res. 2014;322(1):99–107. doi: 10.1016/j.yexcr.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 55.Mocsai A, Ruland J, Tybulewicz VL. The SYK tyrosine kinase: a crucial player in diverse biological functions. Nat Rev Immunol. 2010;10(6):387–402. doi: 10.1038/nri2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.GuezGuez A, Prod'homme V, Mouska X, Baudot A, Blin-Wakkach C, Rottapel R, et al. 3BP2 adapter protein is required for receptor activator of NFkappaB ligand (RANKL)-induced osteoclast differentiation of RAW264.7 cells. J Biol Chem. 2010;285(27):20952–63. doi: 10.1074/jbc.M109.091124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu B, Yu SF, Li TJ. Multinucleated giant cells in various forms of giant cell containing lesions of the jaws express features of osteoclasts. J Oral Pathol Med. 2003;32(6):367–75. doi: 10.1034/j.1600-0714.2003.00126.x. [DOI] [PubMed] [Google Scholar]
- 58.Lee JY, Jung YS, Kim SA, Lee SH, Ahn SG, Yoon JH. Investigation of the SH3BP2 gene mutation in cherubism. Acta Med Okayama. 2008;62(3):209–12. doi: 10.18926/AMO/30977. [DOI] [PubMed] [Google Scholar]
- 59.Yoshitaka T, Ishida S, Mukai T, Kittaka M, Reichenberger E, Ueki Y. Etanercept Administration to Neonatal SH3BP2 Knock-In Cherubism Mice Prevents TNF-α-induced Inflammation and Bone Loss. J Bone Miner Res. 2014;29(5):1170–1182. doi: 10.1002/jbmr.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamashita T, Yao Z, Li F, Zhang Q, Badell IR, Schwarz EM, et al. NF-kappaB p50 and p52 regulate receptor activator of NF-kappaB ligand (RANKL) and tumor necrosis factor-induced osteoclast precursor differentiation by activating c-Fos and NFATc1. J Biol Chem. 2007;282(25):18245–53. doi: 10.1074/jbc.M610701200. [DOI] [PubMed] [Google Scholar]
- 61.Lee SE, Chung WJ, Kwak HB, Chung CH, Kwack KB, Lee ZH, et al. Tumor necrosis factor-alpha supports the survival of osteoclasts through the activation of Akt and ERK. J Biol Chem. 2001;276(52):49343–9. doi: 10.1074/jbc.M103642200. [DOI] [PubMed] [Google Scholar]
- 62.Mocsai A, Humphrey MB, Van Ziffle JA, Hu Y, Burghardt A, Spusta SC, et al. The immunomodulatory adapter proteins DAP12 and Fc receptor gamma-chain (FcRgamma) regulate development of functional osteoclasts through the Syk tyrosine kinase. Proc Natl Acad Sci U S A. 2004;101(16):6158–63. doi: 10.1073/pnas.0401602101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Faccio R, Zou W, Colaianni G, Teitelbaum SL, Ross FP. High dose M-CSF partially rescues the Dap12−/− osteoclast phenotype. J Cell Biochem. 2003;90(5):871–83. doi: 10.1002/jcb.10694. [DOI] [PubMed] [Google Scholar]
- 64.Mao D, Epple H, Uthgenannt B, Novack DV, Faccio R. PLCgamma2 regulates osteoclastogenesis via its interaction with ITAM proteins and GAB2. J Clin Invest. 2006;116(11):2869–79. doi: 10.1172/JCI28775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takada Y, Aggarwal BB. TNF activates Syk protein tyrosine kinase leading to TNF-induced MAPK activation, NF-kappaB activation, and apoptosis. J Immunol. 2004;173(2):1066–77. doi: 10.4049/jimmunol.173.2.1066. [DOI] [PubMed] [Google Scholar]
- 66.Ainsua-Enrich E, Alvarez-Errico D, Gilfillan AM, Picado C, Sayos J, Rivera J, et al. The adaptor 3BP2 is required for early and late events in FcepsilonRI signaling in human mast cells. J Immunol. 2012;189(6):2727–34. doi: 10.4049/jimmunol.1200380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Negishi-Koga T, Takayanagi H. Ca2+-NFATc1 signaling is an essential axis of osteoclast differentiation. Immunol Rev. 2009;231(1):241–56. doi: 10.1111/j.1600-065X.2009.00821.x. [DOI] [PubMed] [Google Scholar]
- 68.Lietman SA, Prescott NL, Hicks DG, Westra WH, Levine MA. SH3BP2 is rarely mutated in exon 9 in giant cell lesions outside cherubism. Clin Orthop Relat Res. 2007;459:22–7. doi: 10.1097/BLO.0b013e31804b4131. [DOI] [PubMed] [Google Scholar]
- 69.Watanabe D, Hashimoto S, Ishiai M, Matsushita M, Baba Y, Kishimoto T, et al. Four tyrosine residues in phospholipase C-gamma 2, identified as Btk-dependent phosphorylation sites, are required for B cell antigen receptor-coupled calcium signaling. J Biol Chem. 2001;276(42):38595–601. doi: 10.1074/jbc.M103675200. [DOI] [PubMed] [Google Scholar]
- 70.Zhang YH, Heulsmann A, Tondravi MM, Mukherjee A, Abu-Amer Y. Tumor necrosis factor-alpha (TNF) stimulates RANKL-induced osteoclastogenesis via coupling of TNF type 1 receptor and RANK signaling pathways. J Biol Chem. 2001;276(1):563–8. doi: 10.1074/jbc.M008198200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Fig. 1. Osteoclast-associated gene expression in TNF-α-stimulated Sh3bp2+/+ and Sh3bp2KI/+ bone marrow cells. BMMs from Sh3bp2+/+ and Sh3bp2KI/+ mice were stimulated with TNF-α (100 ng/ml) for 96 hours. RNA samples were collected at the indicated time points and subjected to qPCR analysis. Gene expression levels relative to Hprt were calculated and normalized to the expression of Sh3bp2+/+ at 0 hour. The data are representative of three independent experiments. Data are presented as mean ± SD. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+. * P < 0.05.
Supplemental Fig. 2. Nuclear translocation of transcription factors during TNF-α-induced TRAP+ MNC formation in Sh3bp2+/+ and Sh3bp2KI/+ BMMs. BMMs from Sh3bp2+/+ and Sh3bp2KI/+ mice were stimulated with TNF-α (100 ng/ml). Nuclear and cytoplasmic protein samples were collected at indicated time points and subjected to Western blot analysis for indicated proteins. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+.
Supplemental Fig. 3. Effect of the inhibition of NF-κB- and c-Fos-mediated signaling pathways on TNF-α-induced TRAP+ MNC formation. (A, B) TRAP staining images and quantitation of TRAP+ MNCs. BMMs from Sh3bp2+/+ and Sh3bp2KI/+ mice were stimulated with TNF-α (100ng/ml) in the presence of wild-type and mutant forms of NEMO-binding domain (NBD) peptides and vehicle (DMSO) for 96 hours. (C, D) TRAP staining images and quantitation of TRAP+ MNCs. Sh3bp2KI/+ mice were crossed with c-Fos-deficient (c-fos−/−) mice. Fetal liver cells from c-Fos-deficient Sh3bp2KI/+ mice (Sh3bp2KI/+/c-fos−/−) were stimulated with TNF-α (100 ng/ml) for 96 hours. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+. Data are presented as mean ± SD. *P < 0.05.
Supplemental Fig. 4. Intracellular signaling pathways during TNF-α-induced TRAP+ MNC formation. (A, B) BMMs from Sh3bp2+/+ and Sh3bp2KI/+ mice were stimulated with TNF-α (100 ng/ml). Protein samples were collected at indicated time points and subjected to Western blot analysis for indicated proteins. Expression levels were examined in (A) short-term culture and (B) long-term culture. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+.
Supplemental Fig. 5. Rankl and Opg mRNA expression in TNF-α-injected calvarial tissues. (A–C) Ten-week-old male Sh3bp2+/+ and Sh3bp2KI/+ mice were subcutaneously injected with either PBS or TNF-α (1.5 μg/mouse/day) over calvariae for 5 days. Mice were euthanized 24 hours after last injection. Gene expression of Rankl and Opg and their ratio. RNA samples were isolated from mouse calvarial tissues and subjected to qPCR analysis (n = 3/group). Gene expression levels relative to Hprt were calculated and normalized to the expression levels of PBS-injected Sh3bp2+/+ control. Data are presented as mean ± SD. +/+: Sh3bp2+/+, KI/+: Sh3bp2KI/+. * P < 0.05, NS: not significant.