Abstract
Bacterium-like particles (BLPs), derived from Lactococcus lactis, offer a self-adjuvanting delivery vehicle for subunit protein vaccines. Proteins can be specifically loaded onto the BLPs via a peptidoglycan anchoring domain (PA). In this study, the tip proteins IpaD, SipD and LcrV belonging to type three secretion systems of Shigella flexneri, Salmonella enterica and Yersinia enterocolitica, respectively, were fused to the PA and loaded onto the BLPs. Herein, we biophysically characterized these nine samples and condensed the spectroscopic results into three-index empirical phase diagrams (EPDs). The EPDs show distinctions between the IpaD/SipD and LcrV subfamilies of tip proteins, based on their physical stability, even upon addition of the PA. Upon attachment to the BLPs, the BLPs become defining moiety in the spectroscopic measurements, leaving the tip proteins to have a subtle yet modulating effect on the structural integrity of the tip proteins-BLPs binding. In summary, this work provides a comprehensive view of physical stability of the tip proteins and tip protein-BLPs and serves as a baseline for screening of excipients to increase the stability of the tip protein-BLPs for future vaccine formulation.
Introduction
Subunit antigens in vaccines require a suitable adjuvant and delivery vehicle to increase vaccine efficacy. Soluble antigens are generally poorly immunogenic, especially when delivered via mucosal routes. We developed a mucosal vaccine technology that is based on non-living particles derived from the food-grade gram-positive bacterium Lactococcus lactis.1 This bacterium is widely used in the food industry and has the Generally Recognized As Safe (GRAS) status of the FDA. The particles derived from L. lactis are referred to as Bacterium-like Particles (BLPs). Previously they were designated GEMs (Gram-positive Enhancer Matrix). BLPs have the same size and shape as living lactococci (approximately 1 µm diameter) and because of a chemical pretreatment with acid, the particles lack intact proteins, DNA, RNA, and lipids.2 The main component of the bacterial cell wall, peptidoglycan, is essentially unaffected and this polymer network preserves BLPs structural integrity. BLPs have an immunostimulating activity and activate the innate immune system through TLR2-mediated signaling.3,4 The immunostimulating activity of BLPs has also been demonstrated in a proof-of-concept Phase I human clinical trial with an intranasal BLP adjuvanted influenza vaccine.5 In addition, BLPs can act as a carrier by loading subunit antigens in trans onto the particle surface. For this purpose, antigens are produced as fusions with a peptidoglycan anchoring (PA) domain in a recombinant expression system. After purification, antigens are loaded onto the surface of BLPs by means of the PA domain which binds non-covalently with high affinity to the peptidoglycan surface of BLPs.6 This vaccine format circumvents the use of recombinant DNA in the carrier, while it preserves the immunostimulating properties of bacterial particles. The efficacy of carrier-based BLP vaccines has been demonstrated in various animal models with vaccines that contain parasitic, viral, or bacterial antigens.3,7–9
Many gram-negative bacterial pathogens possess a type III secretion system (T3SS) that translocates effector proteins into a host eukaryotic cell to manipulate normal host cell functions. The type III secretion apparatus (T3SA) is comprised of a basal body that spans the inner and outer membrane of the pathogen and a needle that protrudes beyond the lipopolysacchride layer. At the T3SA needle tip is a protein complex that is involved in the control of protein secretion.10,11 In Shigella flexneri the tip complex is a pentamer of invasion plasmid antigen D (IpaD).12 IpaD’s closest relative is Salmonella invasion protein D (SipD) which is the T3SA needle tip complex protein for Salmonella enterica.13 For the more distantly related T3SA of Yersinia enterocolitica the needle tip complex is comprised of a pentamer of the low-calcium response protein LcrV.14 Because of the essential role that these tip complex proteins play in the virulence of their respective pathogens, they represent attractive targets for the development of broadly protective enteric vaccines. Indeed, all three have been demonstrated to be protective antigens alone or in combination with other T3SS proteins15–18 (Harrison et al., in preparation). We have previously described the biophysical properties of these three proteins for the purpose of vaccine development, 19–21 however, those data are now updated using more highly purified preparations and improved methods for generating empirical phase diagrams. We have also extended those studies by analyzing the proteins fused with a protein anchor before and after attachment to the surface of BLPs.
The stability of a protein based vaccine is often a major concern associated with formulation development, transport and storage. Thermal and pH alterations are among the primary variables that often compromise stability of the antigenic protein components in a formulation. Thus it becomes a necessity to biophysically characterize and determine the stability of these proteins under different stress conditions of pH and temperature. In this study, the tip proteins were biophysically characterized using circular dichroism (CD) and intrinsic fluorescence spectroscopies as well as static light scattering to assess the structural integrity with regard to secondary structure, tertiary structure and aggregation state of the protein. The acquired data were incorporated into a color map (three-index empirical phase diagram or EPD) that indicated different physical states of the protein using an RGB color scheme. The EPDs provide a comprehensive view of the structural integrity of the proteins and protein-BLPs under pH and temperature stress conditions. The major goal of this work is to examine aspects of the structure which may be helpful for potential formulation development of type III secretion system tip protein-BLPs vaccines. Note that we use here the term “stability” to represent changes in structure or aggregation state, not in a thermodynamic sense due to sure lack or reversibility.
Materials and Methods
Expression and purification of the tip proteins
Expression and purification of the histidine tagged tip proteins IpaD, SipD and LcrV using standard immobilized metal affinity chromatography (IMAC) has been described previously.19,20 Further purification of the tip proteins was achieved using anion exchange chromatography as described.16 The purified proteins were dialyzed into phosphate buffered saline (PBS) and stored at −80 °C.
Expression and purification of the tip proteins-PA
Cloning, expression and purification of LcrV-PA has been described before.3 IpaD-PA protein was prepared as follows: ipaD from S. flexneri was codon-optimized for L. lactis, synthesised and sub-cloned into a L. lactis expression vector pPA224 such that the resulting recombinant gene contains a C-terminal PA and the expressed hybrid protein is secreted into the medium. The resulting plasmid pMUC052 was electroporated into L. lactis PA1001 for expression and secretion of the fusion protein, as previously described.2 The IpaD-PA protein was purified from the clarified culture media using cation exchange chromatography followed by dialysis against PBS.
SipD-PA protein was prepared as follows: sipD from S. enterica was codon-optimized for L. lactis, synthesised and sub-cloned into a L. lactis expression vector pMUC059 such that the resulting recombinant protein contains a C-terminal PA, which is secreted into the medium. The resulting plasmid pMUC108 was electroporated into L. lactis PA1001 for expression and secretion of the fusion protein, as previously described.2 The SipD-PA protein was purified from the clarified culture media by cation exchange chromatography.
The purified proteins were filter sterilized (0.45 µm) and stored at +4 °C at a concentration of 10 mg/mL before binding to BLPs.
Production of Bacterium-Like Particles (BLPs) and binding the tip proteins-PA to BLPs
Production of BLPs and binding of protein-PA fusions to BLPs has been described previously.2,5 Final tip protein-BLPs were stored in PBS at −80 °C at a concentration of 50 mg BLPs/mL. The amounts tip protein bound to BLPs was determined by using Coomassie stained SDS-PAA gels containing a bovine serum albumin (BSA) standard. The tip protein-BLPs batches contained 85 µg of IpaD-PA, 45 µg of SipD-PA and 70 µg of LcrV-PA per mg BLPs.
Sample Preparation
The protein samples were dialyzed into 20 mM citrate phosphate buffer with ionic strength of 0.15 (adjusted with sodium chloride) at a pH ranging from 3 to 8. The protein samples were filtered with 0.45 µm syringe-driven filter units and the concentration was determined using UV absorbance spectroscopy at 280 nm. The final concentration of the proteins was adjusted to 0.15 mg/mL for these studies (Suppl. Table S1).
Each of the tip protein-BLPs (IpaD-BLPs, SipD-BLPs and LcrV-BLPs) samples were diluted to 1 mg BLPs/mL separately. The final solutions contained 85 µg of IpaD-PA, 45 µg of SipD-PA and 70 µg of LcrV-PA per mL. Each sample was centrifuged and the pellet containing tip protein-BLPs was resuspended with citrate-phosphate buffer at the desired pH. This step was repeated three times to stabilize the pH of samples.
Far-UV Circular Dichroism (CD)
The secondary structure of the tip proteins, tip-PA proteins and tip-BLPs was evaluated using a Jasco J-815 CD spectrapolarimeter (Jasco inc., Easton, MD) equipped with a Peltier temperature controller and a 6-position sample holder. CD spectra were acquired from 260 to 200 nm in 0.1 cm pathlength cuvettes at 10 °C. The data were recorded with a scanning speed of 50 nm/min, data integration time of 2 s and 1.0 nm resolution. Thermally induced transitions of tip proteins and tip-PA proteins were acquired by monitoring the CD signal at 222 nm as a function of temperature every 2.5 °C over 10–90 °C employing a temperature ramp rate of 15 °C/h. The transitions for tip protein-BLPs were monitored using CD signal at 225 nm, the wavelength at which the BLPs showed maximum CD signal. Samples were equilibrated to the target temperature (± 0.1 °C) for 5 s prior to each data point measurement. The CD signal for the tip proteins and tip-PA proteins was converted to molar ellipticity. Each sample was measured in triplicate and buffer blank readings were subtracted from the sample reading prior to data analysis.
Intrinsic Trp Fluorescence
IpaD and SipD each possess four Trp while LcrV contains one.19 Thus, intrinsic Trp fluorescence was measured as a function of temperature to monitor changes in the tertiary structure of the proteins. Fluorescence spectra for the samples were acquired using a Photon Technology International (PTI) spectrofluorometer (Birmingham, New Jersey) equipped with a temperature-controlled four position cuvette holder. To selectively excite Trp residues, samples were excited at 295 nm and the emission spectra were collected from 300 to 400 nm with a step size of 1 nm and 0.5 s integration time. The spectra were collected in 1 cm pathlength quartz cuvette every 2.5 °C over 10–85 °C. The samples were equilibrated for 3 min prior to each measurement. The excitation and emission slit width was maintained at 3 nm throughout the study. Peak position wavelengths of the emission spectra were calculated using a mean spectral center of mass (msm) algorithm using Origin 8.6 software. The peak positions obtained using this method are usually shifted 10–12 nm higher than the actual peak positions, but display improved signal/noise ratios. The fluorescence intensity at 330 nm was also monitored as a function of temperature. Each sample was measured in triplicate and scans of buffer solutions were subtracted from the sample readings prior to data analysis.
Static Light Scattering
Static light scattering (SLS) can provide a measure of thermally induced aggregation of proteins.22 These measurements were acquired simultaneously with intrinsic Trp fluorescence measurements using a second detector oriented 180° from the fluorescence photomultiplier tube but still 90° relative to the excitation source. Light scattering intensity at 295 nm was monitored as a function of temperature every 2.5 °C over 10–85 °C. Buffer scans were subtracted from the sample readings prior to data analysis.
Data Visualization Techniques
Three-Index Empirical Phase Diagrams (EPDs)
The thermal stability data acquired from multiple techniques for each protein sample were summarized in the form of three-index empirical phase diagrams to provide comprehensive overviews of the data using a red, green and blue (RGB) color scheme. Detailed description on construction of EPDs is described elsewhere.23 Briefly, structural indices defined as correlation of the degree of structural change within a given range of environmental stress conditions, are calculated for each data set. In this study, the secondary structural index (SI) is calculated from the 222 nm CD molar ellipticity, the tertiary structural index (TI) was calculated from intrinsic Trp fluorescence peak position data and the aggregation index (AI) from the static light scattering data. The values for the structural index for a particular data set are normalized from 0 to 1, in which 1 represents native secondary or tertiary structure while 0 represents extensively conformationally altered secondary or tertiary structure. A value of 0 for AI indicates no aggregation while a value of 1 indicates the maximum extent of aggregation seen. Each of these structural indices was assigned to a color in an RGB color scheme to yield a map that provides a correlation between color and physical state of the protein at a given pH and temperature. In this case, the strongest negative CD signal represents the largest amount of secondary structure and is assigned to red while the weakest signal is assigned to black. For fluorescence peak position data, the native tertiary structure is assigned to green and the most shifted wavelength is black. Aggregation is represented by static light scattering where the minimum signal is assigned to black assuming there is no or minimal aggregation while the highest level of aggregation seen is assigned to blue. The contribution of each RGB component can be seen in the right hand panels next to the associated EPD. The three-index EPDs can then be interpreted as follows: Yellow (i.e. the sum of the “native state” colors) is associated with the native state of a protein where both secondary and tertiary structure are intact and no aggregation has occurred. Black is associated with significantly unfolded states with minimal retention of secondary and tertiary structure and no aggregation. Blue is associated with aggregated states with minimal secondary and tertiary structure and maximal aggregation. Finally, brown is associated with intermediate states of a protein with an intermediate loss of secondary and tertiary structure.
Radar Charts and Chernoff Face Diagrams
Multivariable data obtained from biophysical measurements could also be visualized through the use of radar charts and Chernoff faces, which serve as alternative data visualization techniques to the EPDs. These techniques map stability of the protein through image shapes rather than through colors. For example, in radar charts experimental values are related to the length of spikes of equiangular polygons, while facial expressions were used to depict physical state of the proteins under different pH-temperature conditions. These diagrams are presented in the supplementary material (Suppl. Fig. S12–S17). A detailed description of these techniques can be found elsewhere.23
The Clustering Analysis
The three-index EPDs are displayed with dotted boundary that indicate regions with similar conformational states (i.e. similar colors). The boundary is first calculated using the k-Means clustering algorithm.24 The three indices used to make EPDs are supplied as an input to the algorithm with a range of values (2–10) as a number of clusters to be classified. Among these results, one is chosen that matches well with the interpretation of raw data. Because the k-Means clustering algorithm is stochastic and converges to a local optimum, even the chosen clustering result is not optimal. Based on the interpretation of experimental data, the clustering boundary can be manually edited afterwards.
Results
Biophysical characterization of the tip proteins IpaD, SipD and LcrV
Although we have previously published characterization of T3SS tip proteins,19–21 the purification schemes have been improved 16 and the mathematical methods used to construct EPDs has been optimized, as described above.23 Therefore, to compare the changes in the tip proteins attributable to the PA moiety and the BLPs, CD signals, fluorescence peak position and static light scattering data were collected as a function of temperature and pH to derive new three index EPDs. The far-UV CD spectra at 10 °C showed double minima at 208 nm and 222 nm for IpaD, SipD and LcrV at every pH tested (Suppl. Fig. S1A–S3A respectively), indicating the expected significant amount of α-helical structure content. To obtain the secondary structure index, the molar ellipticity values at 222 nm as a function of temperature at pH 3–8 for all three tip proteins were obtained and demonstrated a pH dependent thermo-stability for each protein (Fig. 1A–C, red diagrams at right, and Suppl. Fig. S1B–S3B). For all three proteins, secondary structure under neutral buffer conditions (pH 6–8, right half of red diagrams) was maintained until ~50 °C where a gradual decrease in secondary structure began with a substantial loss of structure being seen by 75 °C. The gradual loss of secondary structure of IpaD and SipD occurred at lower temperatures as the pH was lowered with the degree of secondary structure loss by 70 °C being much greater at these pH values (pH 3–5). While this trend was also seen at pH 4 and 5 for LcrV, it appeared to retain most of its secondary structure at pH 3 regardless of temperature. When the tertiary structural index was examined (Fig. 1A–C, green diagrams at right, and Suppl. Fig. S1C–S3C), a similar pattern is seen in which the loss of tertiary structure is greater in all proteins in acidic conditions relative to neutral conditions. Nevertheless, the initial disruption of the tertiary structures of IpaD and SipD in neutral buffers occurs earlier than seen in the secondary structural measurements with this phenomenon being more pronounced in SipD. Under acidic conditions the loss of tertiary structure increased with decreasing pH, as was seen in secondary structure results. Interestingly, the tertiary structure of LcrV in the neutral pH range is quite stable with a striking decrease in thermal stability as the pH is lowered. It is noteworthy that some of this apparent stability could be attributed to the method of calculating the diagram in which a longer wavelength associated with a red shift is assigned black while LcrV undergoes a blue shift before it undergoes a red shift in Trp fluorescence. Light scattering was measured to obtain the aggregation index (Fig. 1A–C, blue diagrams at right, and Suppl. Suppl. Fig. S1E–S3E). As the pH was decreased, an increase in aggregation was visualized with each protein exhibiting the least amount of light scattering at neutral pH. Examination of the tertiary structure and aggregation state of LcrV at pH 3 indicated that the thermal destabilization begins to occur at 40 °C in contrast to secondary structural measurements. It may be worth noting that LcrV is significantly different in primary structure relative to IpaD and SipD and contains only a single tryptophan residue.
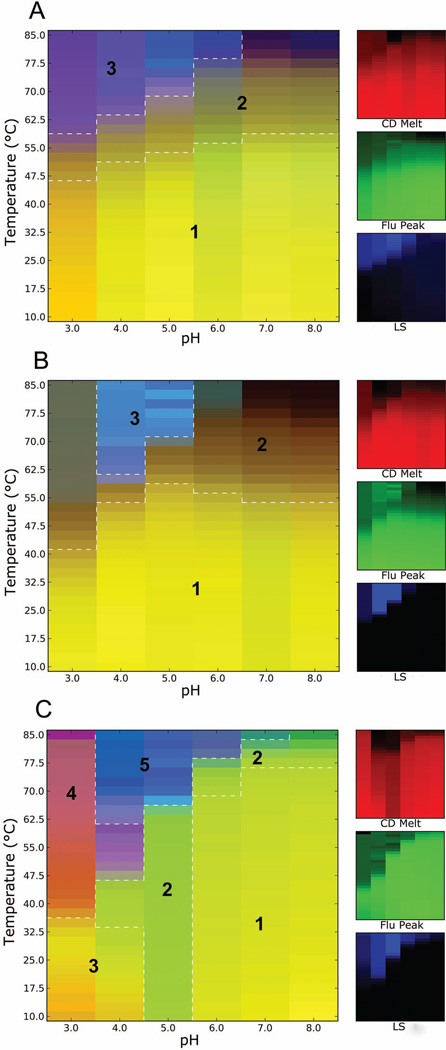
Figure 1.
Three-index empirical phase diagrams (EPDs) for IpaD (A), SipD (B) and LcrV (C), representing the conformational stability of the tip proteins as a function of pH and temperature. The red, green and blue panels at the right define individual component indices for secondary structure (CD), tertiary structure (fluorescence peak position) and aggregation behavior (LS), respectively.
Three-Index Empirical Phase Diagrams (EPDs) for IpaD, SipD and LcrV
Clustering analysis for the EPD of IpaD indicated the presence of three predominant regions (Fig. 1A, multi-colored panel). Region 1 in yellow at pH 3 to 8 up to 45–55 °C represented the native state of the protein as indicated by CD melts (red), fluorescence peak position (green) and aggregation (blue). Region 2 (at pH 3–8, from 45 up to 85 °C) represented a structurally altered physical state of IpaD, primarily reflecting a loss of secondary and tertiary structure. Region 3 (at pH 3–6, above 60 °C) represented an aggregated state of the protein as observed by LS (blue component) with the accompanying loss of some protein structure. Thus, the EPD clearly shows minimal protein disruption at lower temperatures regardless of pH, while a pH dependent increase in the onset of the aggregated state can be seen from pH 3 to 6. The EPD for SipD also depicted three regions. Like IpaD, region 1 represented the native state of the protein and was quite broad with greater thermostability seen around neutral pH environments (Fig. 1B). As thermal stress was applied to SipD, the altered physical state with minimal aggregation accounted for most of the rest of the diagram seen as region 2. At higher temperatures at pH 3, a gray color was formed illustrating some loss of SipD secondary and tertiary structure along with aggregation (presumabily a significantly unfolded state) which is in contrast to pH 7 and 8 where less loss of secondary structure is seen and a brown color is created. Blue colored region 3 was dominated by the LS signal, reflecting the significant aggregation observed at pH 4 and 5. In contrast, the EPD for LcrV could be divided into 5 distinct regions (Fig. 1C). Region 1 constituted the native conformation of the protein. Unlike IpaD and SipD, region 1 for LcrV spanned pH 6 to 8 up to 75 °C while region 2 occupied pH 5 up to 65 °C and at pH 7 and 8 at higher temperatures, representing a partially unfolded state of the protein. The partially unfolded state of LcrV at pH 5 may be explained by low secondary structure content as indicated by the low CD signal at pH 5. Region 3 at pH 3 and 4 can be distinguished from region 1 for LcrV by comparing fluorescence intensities at the lower pH values to the intensities under neutral pH conditions. The intensity counts were lower at pH 3 and 4, while at pH 4 the intensity data possessed a transition occurring at a comparatively low temperature (~30 °C). This region depicted an altered conformation of LcrV under low pH conditions. Region 4 then represented an unfolded and aggregated state of the protein due to acidic pH, whereas region 5 represented completely aggregated protein primarily at pH 4 and 5 above 62 °C as indicated by the SLS data. Comparing all three EPDs, LcrV illustrated a completely different pattern of stability than did IpaD and SipD. This was signified by lower stability of LcrV under acidic pH conditions. In general, maximum stability for all three tip proteins was observed near neutral pH conditions up to 55 °C.
Biophysical characterization of the proteins IpaD-PA, SipD-PA and LcrV-PA
Far-UV CD spectra for IpaD-PA, SipD-PA and LcrV-PA show an intense minima around 207–208 nm with presence of a weak shoulder centered around 222 nm. This suggests presence of significant amount of 310-helix, and alpha helical structure with perhaps some cross-beta structure.25 The ratio of the intensity at 222 nm over 207 nm (R = [θ]222nm /[θ]207nm) provides a criterion for distinguishing alpha and 310-helix. The R value is closer to unity for an α-helix, and a value between 0.4 to 1 suggests a mixed population of α-helix and 310-helix 26,27. The R values calculated were ~0.75 for IpaD PA, ~0.63 for SipD-PA and ~0.85 for LcrV-PA (Suppl. Fig. S4A, S5A and S6A respectively). This implies the conservation of secondary structure upon the production of the fusion tip-PA proteins, although the molar ellipticity was less intense for each fusion than it was for the respective tip proteins alone. Different stability patterns, however, were noted for the three tip-PA proteins when compared to the tip proteins themselves (Fig. 2A–C, red diagrams, and Suppl. Fig. –S6B). IpaD-PA exhibited a lower initial molar ellipticity and thus presumably less secondary structure at pH 7 and 8, which only modestly decreases with increasing thermal stress (Fig. 2A, red diagram). At the lower four pH values, only a modest loss of secondary structure was detected as temperature was increased. Little loss of secondary structure in SipD-PA was detected as thermal stress was increased except for pH 5 and 6 where a complete loss of secondary structure is seen at high temperatures (Fig. 2B, red diagram, Suppl. Fig. S2B). In contrast to IpaD and SipD, the fusion of PA to LcrV had a significant impact on LcrV structure that was manifested by the formation of insoluble aggregates during dialysis at pH 3 and 4. As a result, no spectroscopic data could be collected on LcrV-PA at low pH. Little change in secondary structure was detected in LcrV under neutral pH conditions (Fig. 2C, red diagram, Suppl. Fig. S3B) while at pH 5 a modest decrease in secondary structure integrity was detected at lower temperatures with a complete loss of secondary structure seen above 70 °C. As with the secondary structure analyses, tertiary structural measurements indicated destabilization upon the fusion of PA to IpaD at higher pH with a significant loss of tertiary structure at higher temperatures (Fig. 2A, green diagram and Suppl. Fig. S4C). Regardless of pH, a distinct transition occurs at ~45–50 °C. At pH 3 the loss is more significant as the temperature increases. A modest loss of tertiary structure in SipD-PA was seen as thermal stress was applied regardless of pH (Fig. 2B, green diagram and Suppl. Fig. S5C). A more severe loss of tertiary structure was seen above 50 °C at pH 3 and a complete loss is seen at pH 5 and 6 at high temperatures as was seen in secondary structural analysis. LcrV-PA exhibited a dual transition during tertiary structure upon thermally induced unfolding at neutral pH (Suppl. Fig. S6C). At pH 5, a complete loss of structure was detected above ~50 °C. Static light scattering data for the IpaD-PA revealed little aggregation at pH 3, 7 and 8 with complete aggregation occurring at higher temperatures at pH 4–6 (Fig. 2A, blue diagram and Suppl. Fig. S4E). SipD-PA began to aggregate at ~42–45 °C independent of the pH (Fig. 2B, blue diagram and Suppl. Fig. S5E), whereas, LcrV-PA was found to resist thermally induced aggregation at pH 6–8 with complete aggregation induced by thermal stress at ~47 °C at pH 5 (Fig. 2C, blue diagram and Suppl. Fig. S6E).
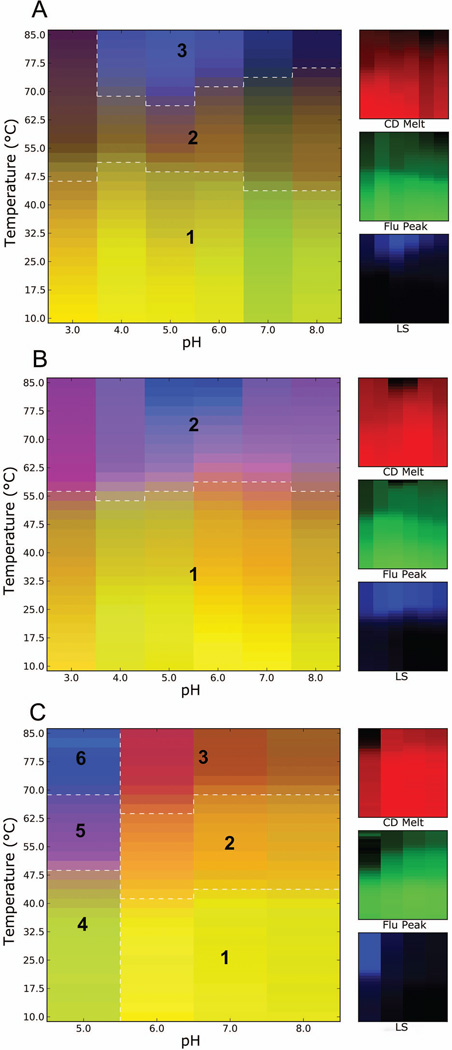
Figure 2.
Three-index empirical phase diagrams (EPDs) for IpaD-PA (A), SipD-PA (B) and LcrV-PA (C), representing the conformational stability of the proteins as a function of pH and temperature. The red, green and blue panels at the right define individual component indices for secondary structure (CD), tertiary structure (fluorescence peak position) and aggregation behavior (LS), respectively.
Three-Index Empirical Phase Diagrams (EPDs) for IpaD-PA, SipD-PA and LcrV-PA
The EPD for IpaD-PA was differentiated into three regions based on clustering analysis (Fig. 2A). Region 1 (yellow) is seen from 10 °C to 47–50 °C at every pH tested and represents the native conformation of IpaD-PA. In agreement with the three indices, the EPD indicated increased thermal stability at low pH than at higher pH values. Region 2 (above 50 °C, pH 3–8) indicates a structurally altered physical state of the protein, probably undergoing a transition from native to at least partially unfolded states. At pH 7 and 8, region 1 showed a greenish tint since the molar ellipticity was low under these pH conditions. This was not, however, identified as a distinct region in the cluster analysis. Aggregation and permanently unfolded states of the protein were seen as the blue colored region 3 towards higher temperatures. The EPD for SipD-PA displayed two distinct regions (Fig. 2B) with the native state (region 1) extending up to 55 °C across pH 3 to 8. The pH conditions 6 and 7 gave rise to the most stable forms of SipD-PA up to about 60 °C. Region 2 was mainly comprised of a structurally altered state of the protein. No transitional region was clearly visible, although it might consist of a narrow boundary between regions 1 and 2. The EPD for LcrV-PA defines the native state of the protein (region 1) as that encompassed by the near neutral pH environment of pH 6–8 up to 40 °C (Fig. 2C). Regions 2 (at pH 6–8, 40–70 °C) and 3 (at pH 6–8, above 65 °C) are assumed to represent molten globular and partially unfolded states, respectively. At pH 5 (region 4, up to 48 °C) LcrV-PA structure is conformationally altered which is manifested by decreased secondary and tertiary structure compared to higher pH. Regions 5 (between 48 and 68 °C) and 6 (above 68 °C) represent aggregated and extensively denatured state of the protein. Comparison of the EPDs for the three tip-PA proteins again demonstrates that LcrV-PA behaves differently than does IpaD-PA and SipD-PA under different environmental stresses. This is particularly true at acidic pH where it either completely aggregates to form insoluble material or quickly aggregates under thermal stress. The PA seems to have only a marginal impact on IpaD and SipD behavior.
Biophysical characterization of IpaD-BLPs, SipD-BLPs and LcrV-BLPs
The minima in the far-UV CD spectra at 10 °C for the tip protein-BLP complexes (lpaD-BLPs, SipD-BLPs and LcrV-BLPs) were shifted to ~225 nm. The minimum at 208 nm, however, was totally missing for each protein-BLPs complex (Suppl. Fig. S7A–S9A). As a general observation, the 225 nm minimum was more intense under neutral pH conditions for all three tip protein-BLPs. The majority of the CD signal appeared to arise from BLPs as suggested by the spectra derived from BLPs itself (Suppl. Fig. S10A). This is similar to the spectra seen for the live attenuated vaccine Salmonella Typhi Ty21a.28 When the IpaD- and SipD-BLPs were transferred into pH 3 buffer, degradation of the tip protein occurred (data not shown). Therefore, biophysical data at this pH were not included in the three indices for generation of a final EPD. Thermal stress on the tip protein-BLPs forced a loss of secondary structure as seen by a decrease in intensity at pH ranging from 4 to 6 (Fig. 3A–C, red diagram). In contrast, the signal remained almost unchanged at pH 7 and 8 for the tip protein-BLPs (Suppl. Fig. S7A, S8A and S9A). Because the signal was dominated by the BLPs, it is likely that the neutral pH environment helps to maintain structural integrity of the BLPs anchored with the tip proteins. The intrinsic Trp fluorescence peak position for IpaD-BLPs at pH 7 and 8 showed a decrease in tertiary structure as the temperature increased indicating partial unfolding of the protein (Suppl. Fig. S7C and D). Conversely, at pH 4, 5 and 6, little change in tertiary structure was detected. The tertiary structure of the SipD-BLPs exhibited spontaneous unfolding regardless of the pH. (Suppl. Fig. S8C and D). For LcrV-BLPs, tertiary structure was disrupted as thermal stress was applied in a pH dependent manner with greater stability observed at higher pH. (Suppl. Fig. S9C and D). Light scattering data provided further insight into the structural integrity of the tip proteins anchored to the BLPs (Suppl. Fig. S7E, S8E and S9E). Initially high light scattering intensities decreased upon applying thermal stress, indicating settling of BLPs in the cuvette. This is probably a result of protein aggregation at higher temperatures. The light scattering data suggested near neutral pH conditions to be suitable for maintaining the structural integrity and absence of aggregation for the tip protein-BLPs.
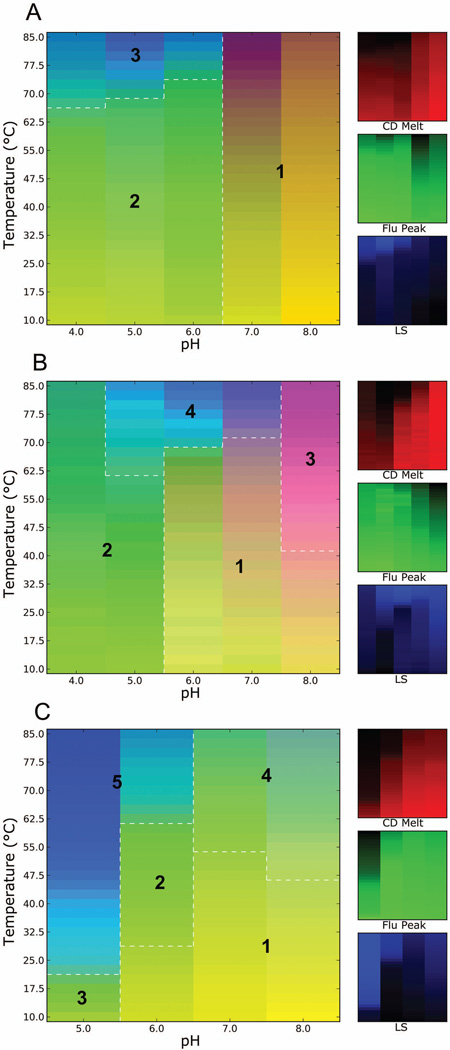
Figure 3.
Three-index empirical phase diagrams (EPDs) for IpaD-BLPs (A), SipD-BLPs (B) and LcrV-BLPs (C), representing the conformational stability of the tip proteins attached to the BLPs as a function of pH and temperature. The red, green and blue panels at the right define individual component indices for secondary structure (CD), tertiary structure (fluorescence peak position) and aggregation behavior (LS), respectively.
Three-Index Empirical Phase Diagrams (EPDs) for IpaD-BLPs, SipD-BLPs and LcrV-BLPs
Unlike the EPDs associated with the tip proteins and tip protein-PAs, the EPDs of the tip protein-BLPs are much more complex. The EPD for IpaD-BLPs represent region 1 at pH 7 and 8 as the most stable state (Fig 3A). This was characterized by the absence of any sharp transitions in the CD and fluorescence data. A comparatively less stable region 2 at pH 4, 5 and 6 up to 65 °C was defined by a lower CD signal and revealed transitions at higher temperatures. Region 3 at pH 4–6 present at higher temperatures (above 65–70 °C) involved protein aggregation including clumping and settling of BLPs. Region 1 of the EPD for SipD-BLPs defined the most stable conditions for these complexes which maintained integrity up to ~65 °C at pH 6 and 7 (Fig. 3B). This region also appeared at pH 8 but extended only up to ~40 °C, thus indicating comparatively less stability at this pH. Region 2 at pH 4 and 5 was classified as an unstable region because of the low CD signal. Region 3 at higher temperatures at pH 8 is consistent with an altered conformation as observed by transitions in the fluorescence data and with aggregation. Region 4 (spanning pH 4–6 at high temperature) included settling of BLPs caused by protein aggregation. This was confirmed by a decrease in the light scattering intensity. The most stable region for LcrV-BLPs spanned pH 7–8 in the EPD (region 1) with maximum stability at pH 7 up to ~50 °C (Fig. 3C). Region 2 at pH 6 below 60 °C and region 3 at pH 5 up to 20 °C represent partially unfolded regions since the secondary structure index begins to darken. Region 4 at pH 7 and 8 above 55 °C defined an altered physical state and also corresponded to settling of BLPs. Region 5 at pH 5 and 6 represented unfolding and aggregation of the LcrV-BLPs.
Discussion
The tip proteins from type three secretion systems of S. flexneri (IpaD), S. Typhimurium (SipD) and Y. enterocolitica (LcrV) have been identified as protective antigens against infection by Shigella spp., 16–18 S. Typhimurium (Harrison, in preparation) and Yersinia spp.15 As a mechanism to increase immune stimulation, these proteins were genetically fused to a peptidoglycan anchor domain (PA) and then bound to BLPs produced from L lactis. While antigen discovery is the initial step in vaccine development, a vaccine can fail if not properly formulated due to instability of the antigen resulting from chemical modification, conformational alteration and/or aggregation. Unfortunately, assessment of antigen stability and subsequent formulation is often not adequately addressed during the early stages of vaccine development. We therefore used multiple spectroscopic and light scattering techniques to assess the structural stability of the proteins alone, fused to PA, and attached to BLPs under a range of pH and temperature stresses. The large amount of resultant data were then consolidated to generate three index EPDs which facilitate visual interpretation and comparison of antigen stability that can then be used to design excipient screens and the formulation of a final vaccine. We previously generated EPDs for these tip proteins. We now, however, apply an improved EPD format by employing the three-index EPD and increasing the level of protein purity. It was thus necessary to create new EPDs for comparison with those generated from the tip protein-PA and tip protein-BLPs.
The T3SS tip protein family is structurally characterized by a dumbbell-like shape where the handle is an anti-parallel coiled-coil that supports overall structural integrity.19,29 In contrast, the size and the shape of the N- and C-termini vary between subfamilies and provide pathogen-specific functions.29 The N-terminal domain of the IpaD family is thermo-labile, which results in two thermal transitions 20,29,30 that can be seen within the three regions of the EPDs for IpaD and SipD. Both proteins exhibit native structural features with no aggregation at all pH values with pH-dependent thermal boundaries at 45–55 °C. Structurally altered states appear at all pH values above this boundary and an aggregated state is seen at high temperatures at low pH. In contrast, LcrV is from a different subfamily and has a distinct EPD. The LcrV native structure is seen at neutral pH and it is retained at higher temperatures than IpaD and SipD. At acidic pH, however, LcrV loses its structural integrity. Based on fluorescence intensity, region 3 (Fig. 1C) represents less stable protein than region 1 since the starting intensity at 10 °C at pH 3 and 4 is reduced compared to higher pH. Nevertheless, all three proteins maintain their native state at neutral pH under low to moderate thermal stress.
The addition of PA to the C-terminus of each tip protein had a negative impact on maintenance of their native structure during thermal stress. Unlike SipD-PA, IpaD-PA was slightly less stable under neutral pH conditions and both IpaD-PA and SipD-PA were vulnerable to aggregation as the temperature was raised above 50 °C. Close examination of data from IpaD-PA and SipD-PA reveals that the CD spectra, thermal melts and fluorescence peak positions differ from IpaD and SipD alone. In fact, they seem to have taken on many of the characteristics of the PA domain (Suppl. Fig. S11). First, the double transitions seen in IpaD and SipD are missing. Second, the initial fluorescence peak position of the PA fusion proteins was ~8 nm higher than that of the tip proteins alone. This may be because the PA domain by itself contains five Trp residues. Then, as thermal stress is applied, a completely different melt is seen with a large blue shift in the middle of the melt rather than the continuous red shift observed with the tip proteins alone. This gives a completely different tertiary index diagram. Striking changes in the behavior of LcrV were observed upon addition of the PA domain. LcrV-PA formed insoluble aggregates during dialysis at pH 3 and 4 and the protein was sensitive to thermal stress at pH 5. Furthermore, the structural stability of LcrV-PA was significantly compromised at pH 6, 7 and 8 relative to LcrV alone.
Only five pH conditions could be examined for IpaD-BLPs and SipD-BLPs since these tip proteins degraded when transferred to pH 3. While it is unclear why these proteins become physically unstable, it is consistent with the instability seen for all the tip proteins at low pH when they are fused to the PA before or after binding BLPs. In general, many of the spectroscopic characteristics of the tip protein-BLPs can be attributed to the BLP itself which is not completely devoid of protein content. This is most notable in the CD spectra where the signals for all tip protein-BLPs were dominated by the BLPs with the presence of only a single minimum at ~225 nm. At neutral pH, a strong signal minimum ranged from 222 to 225 nm. At lower pH, the minimum shifted to 225–227 nm with reduced intensity. The BLPs alone tend to follow this same pattern at pH 7 and 8, which is distinct from what is seen at pH 4 to 6. While this phenomenon can generally describe the tip protein-BLP complexes, protein-specific traits that are unique to each tip protein-BLP can also be seen in the EPDs.
The EPD for IpaD-BLP clearly exhibits a stability boundary between regions 1 and 2 with pH 7 and 8 samples exhibiting higher stability than at lower pH and pH 8 exhibiting the highest stability (Fig. 3). At pH 4–6 the secondary structure index initially shows less intensity with a modest decrease until higher temperatures are reached where aggregation occurs. This stability boundary can be attributed to IpaD rather than the BLPs because a less distinct boundary can be detected between pH 6 and 7 in the IpaD and IpaD-PA EPDs. Regions 1 and 2 in the SipD-BLPs EPD were differentiated based on reduced secondary structure content at low pH. The subtle changes within region 1 are associated with the LS intensity data, which for the pH 6 to 8 range begin at about the same level but at pH 4 and 5 start at a higher intensity. Thus, region 1 in the EPD represents the most stable region for SipD-BLPs at lower temperatures from pH 6 to 8. LcrV-BLPs reflected the pH dependent stability pattern attributable to LcrV-PA and, like IpaD-BLPs, had greatest overall stability at pH 7 and 8, but with greater sensitivity to thermal stress than IpaD-BLPs and SipD-BLPs. This again suggested that LcrV has a biophysical character that is different from IpaD and SipD.
The EPDs of the three tip proteins suggest that the two subfamilies represented here have distinct characteristics before and after fusion to PA with the anchor having a more moderate effect on the properties of LcrV. Perhaps the most significant finding of this study is the protein specific stability conferred upon anchoring of the tip proteins to the BLPs along with the ability to differentiate the three tip proteins’ behavior. Nevertheless, the BLP moiety was the major contributor to all three indices leaving the protein moiety to represent the more subtle yet defining contributor. These EPDs will now be used to direct excipient screening. For IpaD-BLPs, which show minimal aggregation, excipients will be screened to promote structural stability at pH 7 and 8 as thermal stress is applied. Similarly, the SipD-BLPs and LcrV-BLPs exhibit aggregation tendencies at low thermal stress that must be addressed by addition of stabilizers (excipients). Excipients will be needed to raise the thermal boundaries of the EPD, thereby increasing the window in which these candidate vaccines are stable. It is worth noting that a large amount of spectral data was collected here under thermal and pH stress and then consolidated into an EPD format. These modified EPDs and their associated indices provide a new and convenient platform for analyzing and stabilizing a growing number of new biopharmaceuticals.
Supplementary Material
Acknowledgements
This work was supported, in part, by grants the NIH R01 AI089519) to W.L.P. We thank the members of the Picking laboratory Philip Adam, Olivia Arizmendi and Kelly Harrison for critical discussions and review of the manuscript. We also thank Kirk Pendleton and Daniel Picking for assistance in protein expression and purification.
References
- 1.Leenhouts K. Mimopath-Based Vaccine Delivery. In: Singh M, editor. Novel Immune Potentiators and Delivery Technologies for Next Generation Vaccines. New York: Springer; 2013. pp. 245–265. [Google Scholar]
- 2.van Roosmalen ML, Kanninga R, El Khattabi M, Neef J, Audouy S, Bosma T, Kuipers A, Post E, Steen A, Kok J, Buist G, Kuipers OP, Robillard G, Leenhouts K. Mucosal vaccine delivery of antigens tightly bound to an adjuvant particle made from food-grade bacteria. Methods. 2006;38(2):144–149. doi: 10.1016/j.ymeth.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Ramirez K, Ditamo Y, Rodriguez L, Picking WL, van Roosmalen ML, Leenhouts K, Pasetti MF. Neonatal mucosal immunization with a non-living, non-genetically modified Lactococcus lactis vaccine carrier induces systemic and local Th1-type immunity and protects against lethal bacterial infection. Mucosal immunology. 2010;3(2):159–171. doi: 10.1038/mi.2009.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keijzer CMT, Voorn P, de Haan A, Haijema BJ, Leenhouts K, van Roosmalen ML, van Eden W, Broere F. Inactivated influenza vaccine adjuvanted with bacterium-like particles induce systemic and mucosal influenza A virus specific T-cell and B-cell responses after nasal administration in a TLR2 dependent fashion. Vaccine. 2013 doi: 10.1016/j.vaccine.2014.02.019. in press. [DOI] [PubMed] [Google Scholar]
- 5.Van Braeckel-Budimir N, Haijema BJ, Leenhouts K. Bacterium-like particles for efficient immune stimulation of existing vaccines and new subunit vaccines in mucosal applications. Frontiers in immunology. 2013;4:282. doi: 10.3389/fimmu.2013.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosma T, Kanninga R, Neef J, Audouy SA, van Roosmalen ML, Steen A, Buist G, Kok J, Kuipers OP, Robillard G, Leenhouts K. Novel surface display system for proteins on non-genetically modified gram-positive bacteria. Applied and environmental microbiology. 2006;72(1):880–889. doi: 10.1128/AEM.72.1.880-889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Audouy SA, van Selm S, van Roosmalen ML, Post E, Kanninga R, Neef J, Estevao S, Nieuwenhuis EE, Adrian PV, Leenhouts K, Hermans PW. Development of lactococcal GEM-based pneumococcal vaccines. Vaccine. 2007;25(13):2497–2506. doi: 10.1016/j.vaccine.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 8.Nganou-Makamdop K, van Roosmalen ML, Audouy SA, van Gemert GJ, Leenhouts K, Hermsen CC, Sauerwein RW. Bacterium-like particles as multi-epitope delivery platform for Plasmodium berghei circumsporozoite protein induce complete protection against malaria in mice. Malaria journal. 2012;11:50. doi: 10.1186/1475-2875-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rigter A, Widjaja I, Versantvoort H, Coenjaerts FE, van Roosmalen M, Leenhouts K, Rottier PJ, Haijema BJ, de Haan CA. A protective and safe intranasal RSV vaccine based on a recombinant prefusion-like form of the F protein bound to bacterium-like particles. PLoS One. 2013;8(8):e71072. doi: 10.1371/journal.pone.0071072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schroeder GN, Hilbi H. Molecular pathogenesis of Shigella spp.: controlling host cell signaling, invasion, and death by type III secretion. Clin Microbiol Rev. 2008;21(1):134–156. doi: 10.1128/CMR.00032-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galan JE, Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444(7119):567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 12.Epler CR, Dickenson NE, Bullitt E, Picking WL. Ultrastructural Analysis of IpaD at the Tip of the Nascent MxiH Type III Secretion Apparatus of Shigella flexneri. J Mol Biol. 2012;420(1–2):29–39. doi: 10.1016/j.jmb.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lara-Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun. 2009;77(7):2635–2642. doi: 10.1128/IAI.00077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller CA, Broz P, Muller SA, Ringler P, Erne-Brand F, Sorg I, Kuhn M, Engel A, Cornelis GR. The V-antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science. 2005;310(5748):674–676. doi: 10.1126/science.1118476. [DOI] [PubMed] [Google Scholar]
- 15.Burrows TW. An antigen determining virulence in Pasteurella pestis . Nature. 1956;177(4505):426–427. doi: 10.1038/177426b0. [DOI] [PubMed] [Google Scholar]
- 16.Martinez-Becerra FJ, Kissmann JM, Diaz-McNair J, Choudhari SP, Quick AM, Mellado-Sanchez G, Clements JD, Pasetti MF, Picking WL. Broadly protective Shigella vaccine based on type III secretion apparatus proteins. Infection and immunity. 2012;80(3):1222–1231. doi: 10.1128/IAI.06174-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Becerra FJ, Scobey M, Harrison K, Choudhari SP, Quick AM, Joshi SB, Middaugh CR, Picking WL. Parenteral immunization with IpaB/IpaD protects mice against lethal pulmonary infection by Shigella. Vaccine. 2013;31(24):2667–2672. doi: 10.1016/j.vaccine.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Becerra FJCX, Dickenson NE, Choudhari SP, Harrison K, Clements JD, Picking WD, Van De Verg LL, Walker RI, Picking WL. Characterization of a novel fusion protein of IpaB and IpaD of Shigella and its potential as a pan-Shigella vaccine. Infection and Immunity. 2013;81(12) doi: 10.1128/IAI.00859-13. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espina M, Ausar SF, Middaugh CR, Baxter MA, Picking WD, Picking WL. Conformational stability and differential structural analysis of LcrV, PcrV, BipD, and SipD from type III secretion systems. Protein Sci. 2007;16(4):704–714. doi: 10.1110/ps.062645007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espina M, Ausar SF, Middaugh CR, Picking WD, Picking WL. Spectroscopic and Calorimetric Analyses of Invasion Plasmid Antigen D (IpaD) from Shigella flexneri Reveal the Presence of Two Structural Domains. Biochemistry. 2006;45(30):9219–9227. doi: 10.1021/bi060625v. [DOI] [PubMed] [Google Scholar]
- 21.Markham AP, Birket SE, Picking WD, Picking WL, Middaugh CR. pH sensitivity of type III secretion system tip proteins. Proteins. 2008;71(4):1830–1842. doi: 10.1002/prot.21864. [DOI] [PubMed] [Google Scholar]
- 22.Tsunashima Y, Moro K, Chu B, Liu TY. Characterization of group C meningococcal polysaccharide by light-scattering spectroscopy. III. Determination of molecular weight, radius of gyration, and translational diffusional coefficient. Biopolymers. 1978;17(2):251–265. doi: 10.1002/bip.1978.360170202. [DOI] [PubMed] [Google Scholar]
- 23.Kim JH, Iyer V, Joshi SB, Volkin DB, Middaugh CR. Improved data visualization techniques for analyzing macromolecule structural changes. Protein science : a publication of the Protein Society. 2012;21(10):1540–1553. doi: 10.1002/pro.2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacQueen JB. Some Methods for classification and Analysis of Multivariate Observations. Proceedings of 5th Berkeley Symposium on Mathematical Statistics and Probability. 1967;1:281–297. [Google Scholar]
- 25.Manning MC, Woody RW. Theoretical CD studies of polypeptide helices: examination of important electronic and geometric factors. Biopolymers. 1991;31(5):569–586. doi: 10.1002/bip.360310511. [DOI] [PubMed] [Google Scholar]
- 26.Lakshminarayanan R, Fan D, Du C, Moradian-Oldak J. The role of secondary structure in the entropically driven amelogenin self-assembly. Biophys J. 2007;93(10):3664–3674. doi: 10.1529/biophysj.107.113936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen HH, Imhof D, Kronen M, Grafe U, Reissmann S. Circular dichroism studies of ampullosporin-A analogues. Journal of peptide science : an official publication of the European Peptide Society. 2003;9(11–12):714–728. doi: 10.1002/psc.459. [DOI] [PubMed] [Google Scholar]
- 28.Zeng Y, Fan H, Chiueh G, Pham B, Martin R, Lechuga-Ballesteros D, Truong VL, Joshi SB, Middaugh CR. Towards development of stable formulations of a live attenuated bacterial vaccine: a preformulation study facilitated by a biophysical approach. Human vaccines. 2009;5(5):322–331. doi: 10.4161/hv.5.5.7559. [DOI] [PubMed] [Google Scholar]
- 29.Johnson S, Roversi P, Espina M, Olive A, Deane JE, Birket S, Field T, Picking WD, Blocker AJ, Galyov EE, Picking WL, Lea SM. Self-chaperoning of the Type III Secretion System Needle Tip Proteins IpaD and BipD. J Biol Chem. 2007;282(6):4035–4044. doi: 10.1074/jbc.M607945200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chatterjee S, Zhong D, Nordhues BA, Battaile KP, Lovell S, De Guzman RN. The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci. 2011;20(1):75–86. doi: 10.1002/pro.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.