Abstract
Since their discovery 20 years ago, miRNAs have attracted much attention from all areas of biology. These short (∼22 nt) non-coding RNA molecules are highly conserved in evolution and are present in nearly all eukaryotes. They have critical roles in virtually every cellular process, particularly determination of cell fate in development and regulation of the cell cycle. Although it has long been known that miRNAs bind to mRNAs to trigger translational repression and degradation, there had been much debate regarding their precise mode of action. It is now believed that translational control is the primary event, only later followed by mRNA destabilisation. This review will discuss the most recent advances in our understanding of the molecular underpinnings of miRNA-mediated repression. Moreover, we highlight the multitude of regulatory mechanisms that modulate miRNA function.
Facts
miRNA-mediated translational repression is a pre-requisite for target mRNA degradation.
RNA helicases are critical to the inhibition of translation initiation by miRNA.
Modifications of RISC components have critical roles in controlling the miRNA pathway.
Competing endogenous RNAs constitute one of the mechanisms through which miRNAs can be removed from target mRNA.
Relief from miRNA-mediated repression has been documented for specific mRNAs in stress conditions and in response to stimuli.
Open Questions
What is the contribution of the Ccr4–NOT complex to translational repression?
What is the precise role of DEAD-box helicases in miRNA mechanism?
Are all miRNA-repressed mRNAs bound by the same complement of proteins or can the extent and timing of the repression be mediated by different complexes?
Is relief of miRNA-mediated repression a widespread phenomenon?
miRNAs function primarily by imperfectly base pairing with the 3′-untranslated region (3′UTR) of target mRNAs to negatively impact their protein output. The dysregulation of miRNA expression in many disease conditions has been thoroughly documented (for details see Croce1). In addition, extensive shortening of mRNA 3′UTRs, which causes loss of miRNA target sites resulting in the post-transcriptional upregulation of critical oncogenes, has been observed in cancer cells (Figure 2d).2, 3 Conversely, lengthening of 3′UTRs and thus an increase in the number of potential miRNA binding sites in mRNAs has been observed during mouse embryonic development.4 These are just some of the indications that miRNAs play a very important role in determining cell fate, and alterations in their regulatory ability can lead to pathological dedifferentiation.
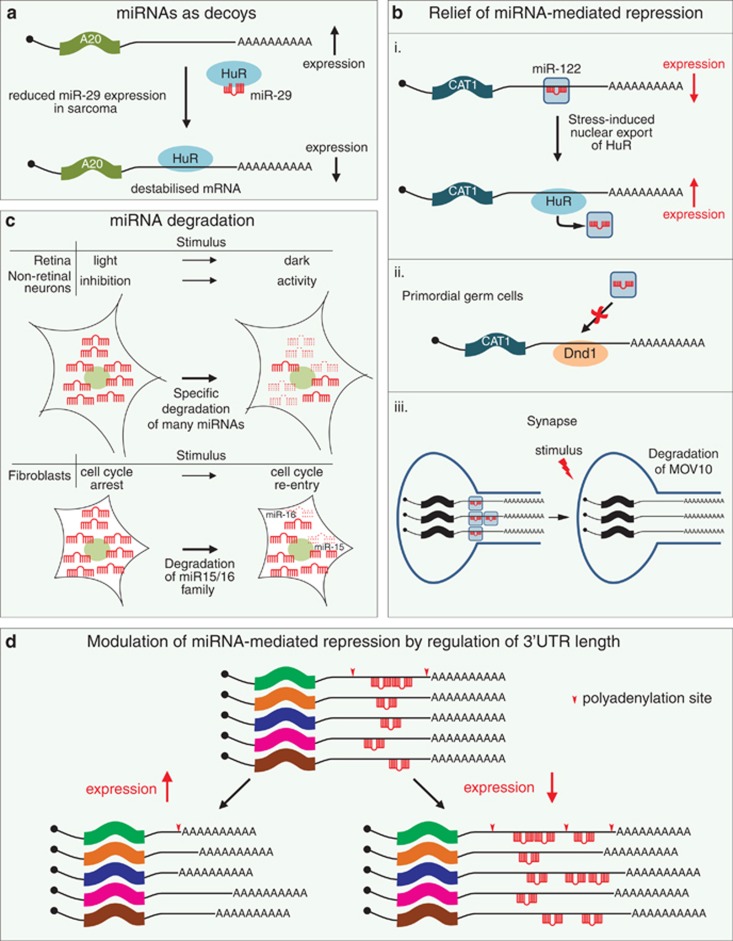
Figure 2.
miRNA-mediated control of gene expression may be regulated in many ways. (a) miRNAs may act as decoys, competing for an RNA-binding protein with an mRNA, an activity beyond the scope of their predominant role in guiding the repressive RISC complex to a target mRNA. For example, miR-29 acts as a decoy for the RNA-binding protein HuR. (b) miRNA-mediated translational repression of an mRNA may be relieved by the preferential binding of a positive-regulator RNA-binding protein. (i) Cellular stress can cause the relief of repression of CAT-1 mRNA by miR-122, which is mediated by the HuR. (ii) Binding of Dnd1 to mRNA in primordial germ cells prevents their association with miRISC, ensuring continued translation. (iii) In the synapse, the stimulus-dependent degradation of a RISC component (MOV10) results in the relief of repression of localised mRNAs. (c) Particular miRNAs may be degraded in upon exposure to specific stimuli, which will lead to the reactivation of mRNAs involved in the cellular response to the stimulus. Selective miRNA degradation has been observed in the retina. Members of the miR-15/16 family are rapidly degraded upon re-entry into the cell cycle. (d) Alternative poly(A) site selection may influence protein expression by removal of miRNA target sites (use of proximal poly(A) site) or increase in their number (use of more distal poly(A) site)
As required sequence complementarity with the target mRNA only spans a fraction of the miRNA's length, each miRNA has hundreds of potential targets, which implicates them in nearly every biological pathway.5 Much effort has in recent years been put into creating a unified model by which miRNAs exert their repressive role on gene expression. Initial work suggested that miRNAs primarily affected the translation of the target mRNA6 and much evidence has accumulated to support this model. Subsequently, it was shown that association of miRNA-bound RNA-induced silencing complex (miRISC) with target mRNAs also resulted in deadenylation-dependent target mRNA decapping and degradation.7, 8, 9, 10, 11, 12, 13, 14 Understanding the relative contribution and temporal characteristics of how translational repression and mRNA destabilisation contribute to miRNA-mediated control of gene expression is central for defining the biological impact of miRNAs as well as for the development of strategies to address pathologies involving their dysregulation.
miRNAs and their Targets
Our current understanding of miRNA target recognition suggests that the miRNA, following extensive processing (for reviews covering miRNA biogenesis, see Tran and Hutvagner,15 Graves and Zeng16 and Finnegan and Pasquinelli17), is bound and presented by the RNA-induced silencing complex (RISC). The miRISC then associates with a target mRNA, usually within its 3′UTR.5, 18 In most cases, there seems to be a requirement for complementarity between the mRNA and bases 2–8 of the miRNA (known as the seed sequence)19 and a certain degree of interaction with the miRNA 3′ end to ensure thermodynamic stability.5 Whereas there are hundreds of potential mRNA targets for each miRNA, based on the presence of seed complementarity and theoretical free energy of interaction, the actual principles of target recognition remain obscure. A recent study suggested that the presence of methylation of the N6 position of adenosine within the mRNA itself might contribute to target selection.20 The identification of true miRNA targets has been the topic of extensive research, and there is emerging evidence that the full impact of some miRNAs upon their targets may only be seen in very specific conditions, such as particular developmental stages,10 stress,21 response to certain environmental queues and extracellular signalling.22 This allows miRNAs to participate in the precise fine-tuning of gene expression, adding an additional level of complexity to the regulation of an mRNA.
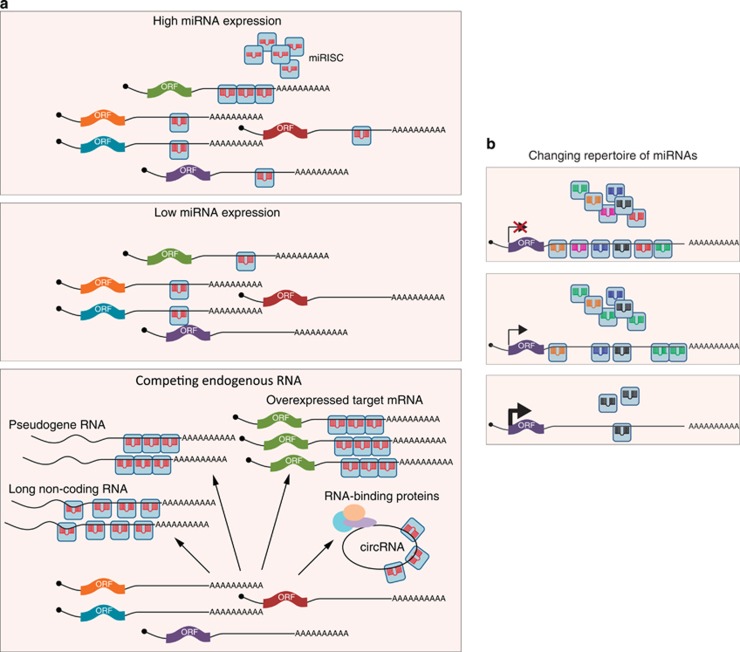
There is some debate as to whether most miRNAs have one primary mRNA target or whether they exert their effect by targeting multiple mRNAs at once (Figure 1b). Indeed, certain miRNAs have been shown to have an impact on mRNAs that participate in a common cellular pathway (for example, see Judson et al.23). Conversely, a single mRNA might be jointly regulated by more than one miRNA (Figure 1b), as in the example of p21/Waf, which is targeted by many miRNAs.24 Some studies have attempted to identify the targets of individual miRNAs within a limited number of cell types or conditions.25, 26 Recently developed methods based on cross-linking and immunoprecipitation (CLIP) of miRISC proteins have allowed the direct identification of mRNAs bound by any miRNA,27, 28, 29 providing us with a broader view of the nature of this regulation. These large data sets suggest the existence of extensive target repertoires for most miRNAs. This, however, does not exclude the possibility that, under certain conditions a single miRNA species may function predominantly as a regulator of one target. The repertoire of mRNAs expressed in a given cell is as important as that of the miRNAs. Whereas some cell types may express only a few targets of a particular miRNA, others may have an abundance of them, resulting in the dilution of the impact of a particular miRNA (Figure 1a, upper panels).30 It is therefore critical to assess the influence of the miRNA in the context of specific cell or tissue types.
Figure 1.
(a) A single mRNA can target hundreds of mRNAs and coordinately regulate them. The extent of regulation of a particular mRNA will depend on the expression levels of the miRNA that targets it as well as competing RNA. (b) One mRNA can have target sites for multiple miRNA. The repertoire of miRNAs in a given cell type, or under certain cellular conditions may result in differential regulation of the same target mRNA
An interesting example of this is seen in a phenomenon called competing endogenous RNA (ceRNA), in which certain RNAs can act to titrate away miRNAs from their target mRNA (Figure 1a, lower panel).31, 32 The HMGA2 transcript has recently been demonstrated to act as a ceRNA.33 It was shown that the overexpression of HMGA2 mRNA promotes lung cancer progression via a mechanism that is independent of its protein-coding region, but rather reliant on the presence of let-7 target sites within its 3′UTR. These are thought to sequester let-7 away from the Tgfbr3 transcript, which as a result is upregulated in metastatic non-small cell lung cancer. Pseudogenes of several cancer-related genes have also been demonstrated to act as miRNA sponges, such as the PTENP1 pseudogene, which normally titrates miRNAs away from the PTEN tumour suppressor mRNA.34 Similarly, long non-coding RNAs (lncRNA) have been implicated in the regulation of miRNA abundance in disease and development. For example, in liver cancer, the HULC lncRNA is upregulated and acts as a sponge for miR-372, which results in a deregulation of transcription.35 Timing of myoblast differentiation has been shown to be dependent on the expression levels of linc-MD1 RNA, which competes for miR-133 binding with transcription factors that regulate muscle differentiation.36
Recently, a new class of RNAs has been added to the repertoire of competing RNAs—circular RNA (circRNA), formed by the covalent linkage between the 5′ and 3′ ends of an exon by the spliceosome.37 Two studies showed how a brain-specific circRNA acts as a sponge for a brain-specific miRNA, miR-7, particularly in the neocortex and hippocampus.38, 39 CircRNA expression is regulated in a tissue- and development stage-dependent manner. CircRNAs are particularly stable as they are not susceptible to the standard RNA degradation pathways, which require linear molecules. The importance of these competing RNAs might manifest itself not only in the sequestration of miRNAs, but also through the direct or indirect binding of RNA-binding proteins.
The complexity of regulatory mechanisms involved in target recognition makes elucidation of the actual impact of individual miRNAs very difficult. Whereas direct molecular identification of miRNA–mRNA interactions is usually employed, comparison of expression profiles of miRNAs and potential target mRNAs in different cell types can help in this process. Normally, an anti-correlation in expression would be expected for a true miRNA–mRNA pair. However, some unexpected relationships can be uncovered, as highlighted in two publications that proposed a novel function for miRNAs as decoys for regulatory RNA-binding proteins.40, 41 The first of these studies showed that miR-328 is bound by hnRNP E2 and sequesters it away from its mRNA targets. Patients with chronic myeloid leukaemia were found to have decreased expression of miR-328, meaning that there is less competition for hnRNP E2. As a result, hnRNP E2 is free to induce the translational inhibition of the key myeloid differentiation factor C/EBPα by binding to the 5′UTR of its mRNA.40 A more recent study showed a similar relationship between HuR and miR-29, which prevents the protein from repressing the mRNA of tumour suppressor A20.41 Future research may uncover involvement of more miRNAs in similar non-canonical mechanisms.
Initially, mature miRNAs were thought to be extremely stable molecules with little evidence for active degradation, which limited the scope of potential mechanisms for relief of miRNA-mediated repression. Subsequent studies have revealed that some miRNAs are readily turned over (Figure 2c).42 Indeed, this is highlighted in the results from the Filipowicz group showing very rapid activity-dependent turnover of certain miRNAs in retinal neurons.43 Such changes may allow quick up- or downregulation of localised miRNA targets required for neuronal activity.44 Another example of miRNA destabilisation is provided by members of the miR-15/16 family, which have been shown to be specifically downregulated during re-entry of cells into the cell cycle from G0 arrest.45 The exonucleases Xrn1 and Xrn2 have been implicated in the regulation of miRNA levels as to have modifications such as uridylation (for reviews see Grosshans and Chatterjee,46 Kai and Pasquinelli47 and Zhang et al.48).
Steps to Repression
During the final steps of biogenesis, miRNAs are bound directly by members of the Argonaute protein family (Ago1–4 in humans), which form part of RISC, that also includes the GW repeat-containing protein GW182 (TNRC6A-C in humans) (for a recent review on the protein–protein interactions of these two proteins, see Pfaff and Meister49 and Wilson and Doudna50). Association of miRISC with an mRNA via base pairing with the miRNA is known to induce both translational repression and mRNA degradation. Whereas the mechanism of translational repression has remained elusive and controversial, the molecular underpinnings of mRNA destabilisation by miRNA are better understood. GW182 interacts not only with Ago proteins, but also with the poly(A)-binding protein (PABP), as well as recruiting the Ccr4–NOT and PAN2–PAN3 complexes, which trigger deadenylation, targeting the mRNA for degradation.51, 52, 53 Recently, it has been shown that in Drosophila miRISC is capable of recruiting the decapping factors Dcp1, HPat and Me31B (DDX6 in humans) to miRNA-targeted mRNAs.54 The relative contribution of translational repression and mRNA degradation and whether these pathways act in sequence or in parallel is still under debate. However, a series of recent publications appears to have brought us closer to some final answers.
The emergence of high-throughput methods in the past decade has allowed researchers to address the question of the relative contribution of translational repression and mRNA degradation on a global scale. One of the first studies used a proteomic approach to demonstrate that miRNAs elicit both translational repression and destabilisation of the mRNA.25 A later study utilised ribosome protection assays, a relatively new method that involves immobilisation of translationally active ribosomes on mRNAs followed by isolation and high-throughput sequencing of RNAse-protected fragments.55 The sequencing data were compared with total mRNA levels and proteomics data.56 The analysis led the authors to draw the conclusion that the overwhelming majority of effects involved in the downregulation of expression of miRNA targets came from mRNA degradation, though a later reanalysis of the data revealed a greater degree of translational repression than previously thought.57 Other studies have shown that miRNA-mediated repression is a time-sequenced process. There is now accumulating evidence that translational repression of miRNA targets is the primary event in the mechanism; several groups have demonstrated that repression can be seen without the necessity for mRNA degradation.58, 59, 60, 61, 62, 63, 64, 65 In vitro studies showed that miRNA-mediated repression is a two-step process, the first of which affects translation of the target mRNA.59, 66 These were followed by a series of reports that employed temporal studies to demonstrate that translational repression of miRNA targets precedes deadenylation and degradation in Drosophila, zebrafish and mammalian cells.60, 61, 62 Utilisation of mRNA reporters possessing a poly(A) tail followed by a non-poly(A) sequence, which protects it from deadenylation, proved that translational repression does not require the removal of the poly(A) tail.60, 61, 67, 68 In a recent work from our laboratory, we were able to show that repression is required for subsequent target degradation by determining that a miRNA target mRNA, which is resistant to miRNA-mediated translational repression, does not undergo degradation despite being bound by miRISC.63
Consistent with the notion that not all miRNA-targeted mRNAs are ultimately degraded following translational repression imposed by RISC, several reports have demonstrated that a miRNA-repressed mRNA can be translationally reactivated (see next section for details; Figure 3b).
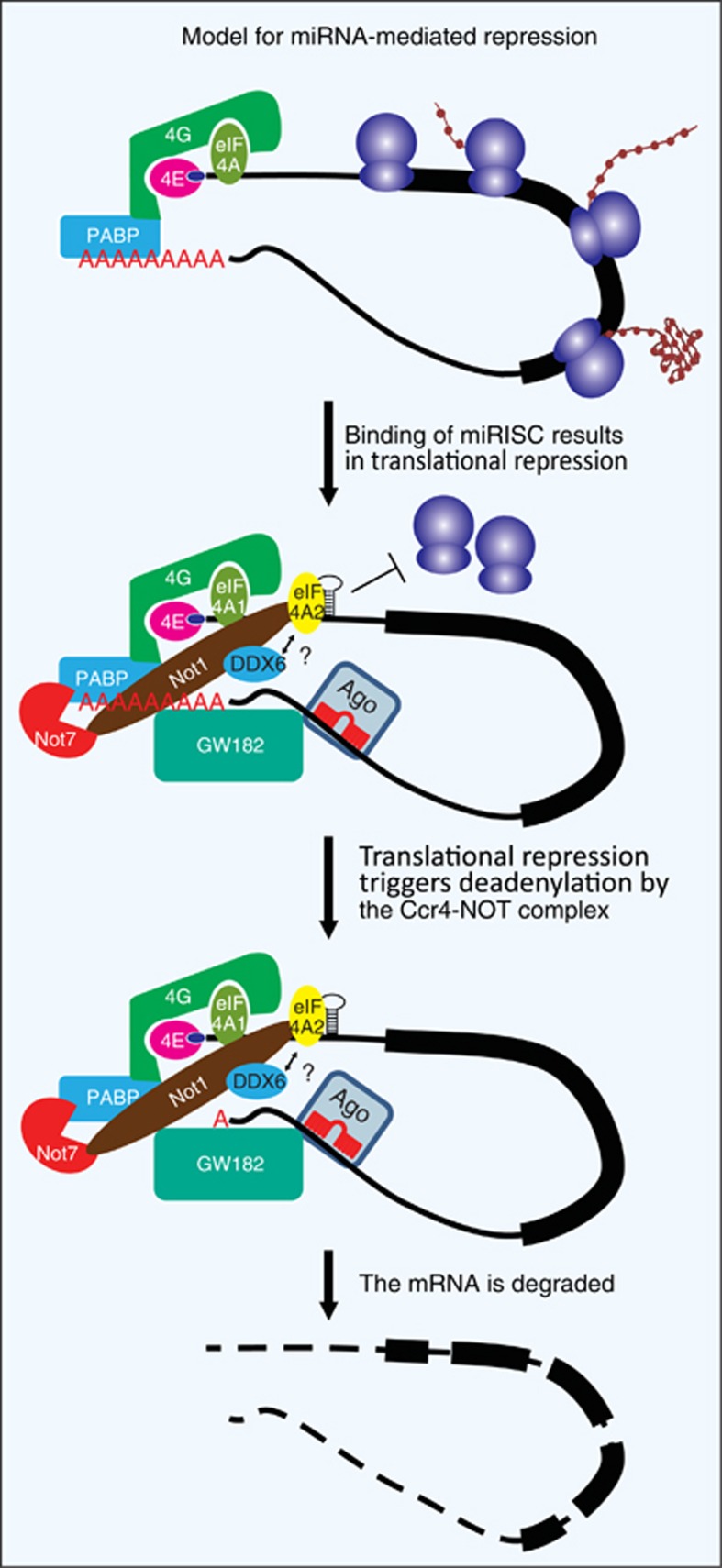
Figure 3.
Working model of miRNA-mediated control of gene expression. The recruitment of RISC-bound miRNA to an mRNA results in translational repression. This is mediated by the DEAD-box helicase eIF4A2, which might be clamping on to the 5′UTR and inhibiting 40S scanning. Another RNA helicase, DDX6, is likely also be involved in this process. Translational repression is followed by deadenylation of the mRNA by Ccr4–NOT deadenylases and subsequent decapping and degradation. The relative contribution of the two DEAD-box RNA helicases to the translational repression, degradation and decapping remains to be determined
Although many suggestions have been made regarding the precise step of translation affected by miRISC, from initiation, through elongation, ribosomal drop-off to polypeptide degradation,69, 70 there is mounting evidence from the work of many groups pointing strongly in favour of an interference with the translation initiation process as being the target of miRNA function (Figure 3).58, 71, 72, 73, 74, 75, 76, 77 Studies comparing the translational activity of cap-dependent and reporter mRNAs containing internal ribosome entry sites (IRES) have been instrumental in shedding light on the onset of miRNA-mediated repression. IRES elements, often present in viral genomes, are sequences that allow for cap-independent initiation of translation. IRES-driven translation has been studied extensively and much is known about the initiation factors required by many of these sequences to drive translation (for review see Thompson78). Initial reports demonstrated that miRNA-mediated repression occurs only on capped mRNAs.71, 75 Humphreys et al.75 showed that a cricket paralysis virus (CrPV) IRES-driven mRNA is completely resistant to miRNA-mediated repression, whereas an mRNA bearing an encephalomyocarditis virus (EMCV) IRES and a poly(A) tail is still partially repressed. Given that the CrPV IRES requires no initiation factors to bind directly to the ribosomal subunits to trigger translation,79 this was a strong indication that translational initiation was affected by miRNAs. Our group extended previous observations by directly comparing cap-dependent miRNA target reporters with EMCV, hepatitis C virus (HCV) and CrPV IRES-bearing ones. Reporter mRNAs bearing the HCV and CrPV IRES were both resistant to repression, whereas an EMCV IRES-containing mRNA showed sustained sensitivity to miRNA. As of these three, the EMCV IRES is the only one to require the presence of eIF4F complex for translational initiation, we concluded that this complex can be directly implicated in translational repression by miRNAs.63
Previous studies had demonstrated that in vitro an excess of eIF4F can cause mRNAs to overcome miRNA-mediated repression in Krebs ascites extract.58 This observation was extended by a study in rabbit reticulocyte lysate (RRL), which established a role in miRNA-mediated repression for the C-terminal domain of eIF4G, which interacts with eIF3 and eIF4A.80 This was also the first demonstration that 43S ribosomal scanning along the 5′UTR was important in the repression process. In agreement with this, a bioinformatic analysis revealed that mRNAs containing miRNA target sites are more likely to have highly structured 5′UTRs.63
Unexpectedly, a detailed siRNA screen for eIF4F components revealed a role for the DEAD-box RNA helicase eIF4A2 in miRNA-mediated repression.63 Previously thought to be a functionally redundant paralog of eIF4A1,81 there is mounting evidence that eIF4A2 has distinct roles in the cell.63, 82 Although earlier in vitro studies had shown both eIF4A1 and eIF4A2 can readily interact with eIF4G as part of the eIF4F complex,81, 83 our in vivo data suggest that eIF4A2 preferentially binds to the CCR4–NOT complex rather than to eIF4G.63 CNOT1, the core component of the Ccr4–NOT complex, has been recently shown to possess a MIF4G domain, a structural domain very similar to the middle domain of eIF4G that harbours the eIF4A binding site.84 The MIF4G domain of CNOT1 resides within the region required for interaction with the deadenylases CNOT7 and CNOT6; however, the interaction interface is located on the opposite side to the putative eIF4A interaction face.85, 86 A third paralog of the eIF4A family—eIF4A3, a core component of the exon-junction complex—rather than unwinding secondary structure, acts as a clamp on RNA.85 It is therefore possible that inhibition of translation initiation by miRISC may occur via enforcement of a similar closed conformation on eIF4A2 by its binding partners and this would inhibit 43S ribosome scanning of the 5′UTR of the target mRNA (Figure 3).
A growing body of evidence suggests that the involvement of another DEAD-box RNA helicase in the translational repression. DDX6 (RCK-p54, Me31B in Drosophila) is a DEAD-box RNA helicase involved in mRNA destabilisation by enhancing decapping and is a core component of P-bodies.87 It has been suggested to form part of the miRISC complex in human cells and its depletion caused partial relief of miRNA-mediated translational repression.88 In Drosophila, it interacts with other decapping enhancers, including HPat (Pat1 in humans), which in turn interacts with the CCR4–NOT complex,89 providing a link between the deadenylation and decapping steps of mRNA degradation. A recent study shows CNOT1-dependent association of HPat with GW182.90 These results suggest that DDX6 may participate in the aggregation of miRISC-associated mRNAs in visible P-bodies, where mRNA deadenylation, decapping as well as storage can occur. Moreover, three groups have recently described that DDX6 interacts with the MIF4G domain of CNOT1.91, 92, 93 Translational repression of reporter mRNAs tethered to either DDX6 or CNOT1 was shown to be dependent on this interaction. Furthermore, the ATPase activity of DDX6 is stimulated by CNOT1 and this activity is important for the function of DDX6 in miRNA-mediated repression.91 It is not yet known whether the DDX6–CNOT1 interaction is critical for translational repression or the subsequent destabilisation of the mRNA.
The relative interplay and relevance of the two helicases, DDX6 and eIF4A2, in miRNA-mediated repression requires further investigation (Figure 3). It remains to be determined whether the association of CNOT1 with a DEAD-box RNA helicase requires additional binding partners, as their functionality has been shown to be dependent on associated proteins.87, 94, 95 The activity of RNA helicases is important in unwinding secondary structure of RNA in vivo.96 Thus, as miRNA-mediated translational repression appears to be dependent on a DEAD-box helicase, the complexity of 5′UTR sequences may have evolved to contribute to the miRNA pathway.63
Translational repression of miRNA-targeted mRNAs is usually followed in short succession by their degradation, which is initiated by the Ago2- and GW182-dependent recruitment of deadenylase complexes.9, 11, 12 There are two such complexes in eukaryotes: the multi-protein, 1 MDa Ccr4–NOT, and the two-subunit Pan2–Pan3 complex (for extensive reviews of their structure and function, see Wahle and Winkler97 and Collart and Panasenko98). The Ccr4–NOT complex comprises two catalytic subunits—Ccr4 (CNOT6 and CNOT6L in mammals) and Caf1 (CNOT7, CNOT8 and Caf1z in mammals). CNOT1 protein is considered to function as a recruitment scaffold for the remaining components of the complex as well as other interacting proteins. Ccr4 is bound to Caf1, which in turn interacts with the MIF4G domain of CNOT1.71, 99 As for the Pan2–Pan3 complex, Pan2 possesses deadenylase activity, whereas it is believed that the complex is canonically recruited to mRNA through Pan3-binding PABP via a PAM2 domain.100
A number of reports have convincingly demonstrated that miRISC recruits the Ccr4–NOT complex to trigger deadenylation and degradation of the mRNA target. Initially shown to cause deadenylation that leads to subsequent decapping,11, 13, 14 this complex has been demonstrated to interact with sequences within the tryptophan-rich C-terminal silencing domain of GW182 (TNRC6 in human).51, 53, 101 The protein responsible for binding to GW182 is the scaffold protein CNOT1, and binding studies have revealed that similarly to Caf1 association, the interaction with GW182 is mediated by the middle domain of the protein.86 New data show that Ccr4–NOT can also bind to the W-rich region of TNRC6 via CNOT9, which in turn is bound to CNOT1, indicating there are multiple interaction sites between RISC and CCr4–NOT, the relative importance of which is not yet fully understood.91, 92
Initial studies suggested that the Pan2–Pan3 complex either has a minor role in miRNA target deadenylation and destabilisation or is active on a subset of mRNAs.14, 52 Its interaction with miRNA targets is facilitated by interaction of Pan3 with the C-terminal domain of GW182.52, 53 Recently, Pan3 was demonstrated to exist as an ATP-binding homodimer, which forms the W-binding pocket required for recruitment to miRNA target mRNAs via GW182.102 It is as yet unclear whether the deadenylase complexes have redundant function or introduce an additional regulatory element to miRNA-mediated repression.
Aside from its activity in deadenylation, components of Ccr4–NOT, namely CNOT7 (Caf1) and CNOT8 (Pop2), have been shown to repress translation in Xenopus oocytes independently of their deadenylase activity, which suggests that they mediate the recruitment of a repressor complex to the mRNA.103 Interestingly, the eIF4F complex has been implicated in their intrinsic repressive function. Similar effects on translation have been reported for dCaf1 and human CNOT1 in S2 cells.53 Repression seen upon tethering of GW182 to an unadeylated reporter mRNA was completely relieved upon NOT1 or Pan3 depletion,52 strengthening the notion that deadenylation is only one of the pathways in which these complexes exert post-transcriptional regulation.
It should be noted that, for certain miRNA target reporter mRNAs, knockdown of NOT1 and PAN3 in Drosophila cells did not result in translational derepression, whereas for others, complete restoration of translation was observed.52, 104 This may indicate that mRNAs may undergo differential regulation following miRISC binding, some being predominantly translationally repressed, whereas others undergoing very rapid decay.62 This would introduce additional levels of post-transcriptional control and allow reactivation of some targets. Future work may reveal sequence determinants that govern such differential effects, and potentially also cell- and tissue-specific regulation.
PABP has been shown to have an important role in the miRNA pathway. It interacts with the TNRC6/GW182 proteins via the silencing domain, which compete with eIF4G for binding.59, 105, 106, 107, 108 The concomitant binding of PABP and the Ccr4–NOT complex105 directs deadenylation of the mRNA.59 Not surprisingly therefore, the poly(A) tail is a contributing cis factor in miRNA-mediated repression and longer poly(A) tails have been shown to confer greater repressive effects on the mRNA.63, 109, 110 Indeed, the role of the poly(A) tail was highlighted in a study describing the recapitulation of miRNA-mediated repression in RRL, where free poly(A) was shown to act in trans to stimulate miRNA-mediated repression in nuclease-treated RRL, possibly by modulating the interaction of PABP with eIF4G.80 Importantly, PABP has been recently demonstrated to dissociate from the poly(A) tails of miRNA-targeted mRNAs in the absence of deadenylation and this effect was triggered by the recruitment of the Ccr4–NOT complex by RISC,111, 112 which could be an additional contributing factor to translational repression as PABP is known to stimulate translation by stabilising the mRNA closed-loop.113, 114
Regulation of miRISC and its Functional Diversity
Modulation of miRNA expression can result in global changes in gene expression. However, rearrangement and modification of miRNP particles and their mRNA component in response to intra- and extracellular queues can also improve the cell's ability for rapid response and adaptation. Below we discuss some examples of how components of the miRNA repression pathway are regulated by post-translational modifications and by accessory proteins (summarised in Table 1).
Table 1. Known modifications of RISC proteins and biological effects of accessory TRIM proteins.
| Protein | Modification | Residue | Biological effect | Cell type | Reference |
|---|---|---|---|---|---|
| Ago2 | Phosphorylation | S253 | HEK293 | 123 | |
| T303 | HEK293 | 123 | |||
| T307 | HEK293 | 123 | |||
| S387 | P body localisation; enhancement of miRNA-mediated repression | HEK293, HEK293T, MCF-7, H1299 | 123, 124, 125 | ||
| Y393 | Induced in hypoxia; inhibition of miRNA maturation | HEK293; HTC-1080 | 115, 133 | ||
| Y529 | Reduced P body localisation; impaired miRNA binding | HEK293 | 123 | ||
| Transient loss of miRNA binding by Ago2 following LPS treatment | Macrophage-like cell line RAW 264.7 | 75 | |||
| S798 | 123 | ||||
| Ubiquitination | Specific degradation of Ago2 | HEK293; EC | 130 | ||
| Degradation during T-cell activation | CD4+ T cells | 132 | |||
| Hydroxylation | P700 | Stabilisation of Ago2 and enhancement of miRISC function in hypoxia; increase in miRNA abundance | Human primary pulmonary artery smooth muscle cells (PASMCs), U2OS, HeLa S3 | 134, 135 | |
| Sumoylation | Increase in protein stability | HeLa, primary MEF | 122 | ||
| ADP-ribosylation | Enhancement of stress granule formation; relief of miRNA repression | HeLa | 136 | ||
| TNRC6/GW182 | Phosphorylation | Unknown | Diminished interaction with PABPC, derepression of miRNA targets | NIH3T3, HeLa | 137, 139 |
| Not4p | Phosphorylation | S92, S312, S542, T543 | Modification required for yeast cell stress tolerance | Yeast | 140 |
| Not1p | Phosphorylation | T2002 | Unknown | ||
| Caf1p | Phosphorylation | S39 | Unknown | ||
| PABP | Acetylation | K95, K188, K312, K606 | Potential differential interaction with protein partners | MEF, HeLa | 143 |
| Methylation | E180, E182, D209, E239, K299 | Unknown | |||
| Dimethylation | K312, K361, R493, K606 | Unknown | |||
| Phosphorylation | unknown | Phosphorylation state affects poly(A) binding and interaction with translation initiation factors | Wheat | 144 | |
| MOV10 | Ubiquitination | unknown | Specific activity-dependent destabilisation at synapse | Drosophila, rat | 145, 146, 147, 148 |
| TRIM proteins | Organism | Biological effect | Reference |
|---|---|---|---|
| TRIM proteins and their biological effect on RISC function | |||
| nhl2 | C. elegans | Enhanced repression of certain targets | 127 |
| MeiP26 | Drosophila | Reduction of miRNA levels | |
| TRIM32 | Mouse | Enhanced let-7 activity in neuronal differentiation | 128 |
| TRIM71/lin-41 | Mouse | Degradation of Ago2 in ES cells | 130 |
| This view was challenged in | 104 | ||
| Human | Inhibition of miRNA silencing | 131 | |
Mass spectrometry studies have revealed multiple phosphorylation sites in Ago2 and that phosphorylation at Y529 in the RNA-binding pocket changes the protein's ability to bind small RNA, suggesting a mechanism for the alternate binding and release of miRNAs.115 Phosphorylation of Ago2 by AKT3 and MK2 at S387 has been shown to enhance miRNA-mediated translational repression of endogenous miRNA targets. This is likely due to enhanced interaction of phosphorylated Ago2 with GW182 and P-bodies.116, 117
Two recent studies investigated the role of Ago2 in stress response. One showed that EGFR induces phosphorylation of Y393 of Ago2 in hypoxia, which in turn inhibits miRNA maturation, possibly contributing to tumour cell survival.118 The second used CLIP followed by high-throughput sequencing (CLIP-seq) to detect the increased interaction of Ago2 with many mRNAs and a release of a subset of mRNAs translated following arsenite stress.28 This remodelling of Ago2 mRNPs may well be caused by post-translational modifications. In addition to the above-mentioned phosphorylation events, Ago2 hydroxylation has been shown to stabilise the protein and enhance miRISC function following hypoxia.119, 120 Conversely, ADP-ribosylation of Ago2 upon oxidative stress has been shown to lead to the relief of miRNA-mediated repression.121 It is possible that different stress stimuli result in different responses of the miRNA machinery. Most recently, Ago2 has been shown to be sumoylated at Lys402 negatively affecting its stability.122
Several years ago, TRIM-NHL proteins, which are thought to function as E3 ubiquitin ligases, were demonstrated to interact with the miRNP complex.123 The Caenorhabditis elegans protein nhl2 was shown to enhance miRNA activity, but only for several specific miRNAs.124 The mouse protein TRIM32, which is non-symmetrically distributed in neural progenitor cells and induces their differentiation, co-immunoprecipitates with certain miRNAs, and has been suggested to specifically enhance the activity of let-7 in neuronal differentiation.125 The possibility of asymmetric distribution of miRNAs in association with proteins such as TRIM32 to establish cell fate is an intriguing possibility. Other TRIM-NHL proteins have been implicated in the miRNA pathway in Drosophila.126 Another member of this family, TRIM71—itself a miRNA target, has been shown to be involved in a complex network of interactions with the miRNA pathway (reviewed in Ecsedi and Grosshans127). TRIM71 has been proposed to target Ago2 for proteasomal degradation in mouse embryonic stem cells and neural progenitor cells,128 but this claim was later challenged with the suggestion that it in fact enhances miRNA function through its interaction with miRISC, possibly by exerting additional translational repression itself.101 Moreover, although TRIM71 was also shown to be inactive in neural progenitor cells as a factor mediating Ago2 ubiquitination, it does affect AKT signalling,129 which may in turn affect Ago2 function, as discussed above. More recently, Ago2 was shown to be regulated via ubiquitination in T-cell activation, resulting in global downregulation of miRNAs in these cells.130
The core RISC protein TNRC6 (GW182) was originally identified as an autoimmune antigen and was shown to be a phosphoprotein localising to cytoplasmic foci.131 It is still unclear which of the many putative phosphorylation sites within this large protein are actively modified. TNRC6 is known to interact with the C-terminal MLLE domain of PABPC via its PAM2 motif.132 This intrinsically disordered region surrounding the TNRC6C PAM2 motif contains many potential phosphorylation sites. Phosphatase treatment of TNRC6C results in an increased interaction with PABPC.133 It was therefore proposed that, in light of the recent demonstration that PABPC enhances miRISC binding to target mRNA in the early stages of translational repression,111 differential phosphorylation may be responsible for stronger interactions between the two proteins at the initial phase, and weaker binding at the onset of deadenylation. More research is required to understand the roles of phosphorylation of this important protein.
Little is known about the regulation of Ccr4–NOT complex function despite its central importance in mRNA metabolism. Several accessory proteins have already been identified, although how and when they affect the function of the complex remains unknown.97 Some components of the yeast Ccr4–NOT complex, namely Not4p, Not1p and Caf1p, have been shown to be phosphoproteins,134 but nothing is yet known about their higher eukaryote counterparts.
PABP may also modulate miRNA activity. The extent of miRNA-mediated repression has been shown to be dependent on levels of the protein.110, 135 Paip2 is a negative regulator of PABP function, and acts by competing with eIF4G for interaction with PABP.136, 137 In accordance with the effects of PABP overexpression, increased levels of Paip2 were shown to enhance miRNA-mediated repression.110 PABP also undergoes a number of post-translational modifications,138, 139 which may in the future be shown to influence its effects in the miRNA repression pathway.
Cellular mechanisms that counteract miRNA function are only beginning to emerge, but understanding them will no doubt reveal the complexity of regulatory mechanisms these small RNA molecules are involved in. Localisation of mRNA, miRNAs and translational machinery in neurons has received much attention as rapid spatially restricted protein production at the synapse can have an important role in regulation of neuronal plasticity (for review of miRNA-mediated repression at the synapse, see Schratt44). Stimulus-dependent relief of repression of miRNA targets has been demonstrated for local synaptic translation.140, 141 There is evidence of activity-dependent destabilisation of the RISC component MOV10 (Armitage in Drosophila) at the synapse, an event that leads to release of certain dendritically localised mRNAs from RISC and their subsequent upregulation (Figure 2b).141, 142, 143, 144, 145 Stress conditions were the focus of two elegant studies from the Filipowicz lab that demonstrated how another mRNA-binding protein, HuR, can cause the relief of miRNA-mediated repression of the CAT-1 mRNA by evicting miRISC (Figure 2b).72, 146 Whether modifications of the components of RISC are involved in this mechanism is unknown, although HuR does not appear to interact directly with the complex. More recently, a transient relief of miRNA-mediated repression was shown to be involved in the early stages of macrophage activation upon exposure to lipopolysaccharide.99 This effect is mediated by the temporary increase in Ago2 phosphorylation, which decreases its association with miRNAs, thereby releasing pro-inflammatory target mRNAs to initiate immune response. In primordial germ cells, the RNA-binding protein Deadend (Dnd1) was shown to protect a subset of miR-430 targets from repression by occluding the target site.147 DAZL, which is known to promote translation in oocytes and embryos,148 was also shown to antagonise miR-430 on its mRNA targets in primordial germ cells.149
The multifunctional RNA-binding protein PTB has been demonstrated to also be a regulator of miRNA function in the control of cell differentiation.150 Strikingly, depletion of PTB resulted in transdifferentiation of fibroblasts into neurons. This effect is primarily due to PTB normally competing with miRNAs that repress negative regulators of neuronal differentiation. Interestingly, the study shows that for a subset of mRNAs PTB actually enhances miRISC binding by modifying the structure of the 3′UTR. Time will tell whether this multifunctional protein has set a paradigm for the coordinated regulation of gene expression by RNA-binding proteins and miRNAs.
Conclusions
Many high-profile publications of recent years have highlighted the importance of miRNAs as critical factors contributing to the complexity and specificity in the control of mRNA translation and turnover. The diverse networks through which miRNAs exert their biological functions in development, differentiation and disease are only beginning to be elucidated. The effect of miRNA gene knockout is often not obvious in the phenotype under normal conditions,151, 152, 153, 154, 155 only to become apparent in certain cellular contexts or under stress conditions, highlighting the importance of miRNA in the ‘micromanagement' of gene expression. More questions remain about the specific protein interactions that are involved in the inhibition of translational initiation by miRNAs. However, a better understanding of the mechanism of miRNA-mediated control of gene expression opens up the possibility of manipulating the pathway in therapeutic, diagnostic and preventive interventions.
Nonetheless, practical applications targeting miRNA function are already beginning to emerge. Indeed, clinical trials using a miR-122 antagonist as a treatment for chronic HCV infection have shown very promising results.156 miR-122 acts upon HCV in an unusual manner, targeting its 5′UTR to promote viral replication.157 Interestingly, miR-10a has also been implicated in targeting the 5′UTR sequences of endogenous mRNAs. Unlike the canonical function of miRNAs in inhibition of gene expression, miR-10a was shown to upregulate translation of 5′-terminal oligopyrimidine mRNAs by binding to their 5′UTR.158 Several reports have also suggested that cellular conditions may impart activating capability to miRNAs targeting sites in the 3′UTR, such as the upregulation of TNFα translation by miR-369-3p in quiescent cells.159 Understanding the scope and mechanism of this unusual activity requires further studies. These examples underscore the breadth of regulatory activities of these small but powerful RNAs.
Notes Added in Proof
A new publication by Zhang et al. has shown that specific upregulation of translation is mediated by the binding of Ago2 in complex with miRNAs to mitochrondrially encoded mRNAs during muscle differentiation. The ceRNA theory has recently been challenged by a study in mice that showed that for high-abundance miRNAs, like miR-122, achieving the required concentration of a physiologically relevant ceRNA is unrealistic: Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell 2014; 54: 766–776.
Glossary
- 3′UTR
3′-untranslated region
- 5′UTR
5′-untranslated region
- Ago1–4
argonaute 1–4
- AKT
protein kinase B
- ATP
adenosine triphosphate
- ATPase
ATP hydrolase
- C/EBPα
CCAAT-enhancer-binding protein alpha
- ceRNA
competing endogenous RNA
- circRNA
circular RNA
- CLIP
cross-linking and immunoprecipitation
- CrPV
cricket paralysis virus
- eIF4
eukaryotic translation initiation factor 4
- EMCV
encephalomyocarditis virus
- HCV
hepatitis C virus
- hnRNP
heterogenous nuclear ribonucleoprotein
- HULC
highly upregulated in liver cancer non-coding RNA
- HuR
human antigen R protein
- IRES
internal ribosome entry site
- lncRNA
long non-coding RNA
- MIF4G
middle domain of eukaryotic initiation factor 4G
- miRISC
miRNA-bound RNA-induced silencing complex
- miRNA
microRNA
- MK2
MAP kinase-activated protein kinase 2.
- mRNA
messenger RNA
- PABP
poly(A)-binding protein
- PTEN
phosphatase and tensin homolog
- PTENP1
phosphatase and tensin homolog pseudogene 1
- RISC
RNA-induced silencing complex
- RRL
rabbit reticulocyte lysate
- S2 cells
Schneider 2 cells
- Tgfbr3
transforming growth factor beta receptor III
- TNRC6A-C
trinucleotide repeat containing 6 A-C (also known as GW182)
- Xrn1/2
exoribonuclease 1/2
The authors declare no conflict of interest.
Footnotes
Edited by RA Knight
References
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lembo A, Di Cunto F, Provero P. Shortening of 3′UTRs correlates with poor prognosis in breast and lung cancer. PLoS One. 2012;7:e31129. doi: 10.1371/journal.pone.0031129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA. 2009;106:7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. Rna. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Zheng D, Xia Z, Shyu AB. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran N, Hutvagner G. Biogenesis and the regulation of the maturation of miRNAs. Essays Biochem. 2013;54:17–28. doi: 10.1042/bse0540017. [DOI] [PubMed] [Google Scholar]
- Graves P, Zeng Y. Biogenesis of mammalian microRNAs: a global view. Genomics Proteomics Bioinformatics. 2012;10:239–245. doi: 10.1016/j.gpb.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EF, Pasquinelli AE. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol. 2013;48:51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475–488. doi: 10.1038/nrm3611. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Avraham R, Yarden Y. Regulation of signalling by microRNAs. Biochem Soc Trans. 2012;40:26–30. doi: 10.1042/BST20110623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RL, Greve TS, Parchem RJ, Blelloch R. MicroRNA-based discovery of barriers to dedifferentiation of fibroblasts to pluripotent stem cells. Nat Struct Mol Biol. 2013;20:1227–1235. doi: 10.1038/nsmb.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, et al. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3′ untranslated region. Oncogene. 2010;29:2302–2308. doi: 10.1038/onc.2010.34. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov FV, Hannon GJ. Remodeling of Ago2-mRNA interactions upon cellular stress reflects miRNA complementarity and correlates with altered translation rates. Genes Dev. 2013;27:1624–1632. doi: 10.1101/gad.215939.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H. Redefining microRNA targets. Curr Biol. 2009;19:870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language. Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver AL, Subramanian S. Competing endogenous RNA database. Bioinformation. 2012;8:731–733. doi: 10.6026/97320630008731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Armenteros-Monterroso E, East P, Chakravorty P, Matthews N, Winslow MM, et al. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature. 2014;505:212–217. doi: 10.1038/nature12785. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Preiss T. Circular RNAs: splicing's enigma variations. Embo J. 2013;32:923–925. doi: 10.1038/emboj.2013.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- Eiring AM, Harb JG, Neviani P, Garton C, Oaks JJ, Spizzo R, et al. miR-328 functions as an RNA decoy to modulate hnRNP E2 regulation of mRNA translation in leukemic blasts. Cell. 2010;140:652–665. doi: 10.1016/j.cell.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkhi MY, Iwenofu OH, Bakkar N, Ladner KJ, Chandler DS, Houghton PJ, et al. miR-29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR. Sci Signal. 2013;6:ra63. doi: 10.1126/scisignal.2004177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger S, Grosshans H. MicroRNA turnover: when, how, and why. Trends Biochem Sci. 2012;37:436–446. doi: 10.1016/j.tibs.2012.07.002. [DOI] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- Schratt G. microRNAs at the synapse. Nat Rev Neurosci. 2009;10:842–849. doi: 10.1038/nrn2763. [DOI] [PubMed] [Google Scholar]
- Rissland OS, Hong SJ, Bartel DP. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol Cell. 2011;43:993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H, Chatterjee S. MicroRNases and the Regulated Degradation of Mature Animal miRNAs. Adv Exp Med Biol. 2011;700:140–155. doi: 10.1007/978-1-4419-7823-3_12. [DOI] [PubMed] [Google Scholar]
- Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol. 2010;17:5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Qin Y-W, Brewer G, Jing Q. MicroRNA degradation and turnover: regulating the regulators. Wiley Interdiscip Rev RNA. 2012;3:593–600. doi: 10.1002/wrna.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff J, Meister G. Argonaute and GW182 proteins: an effective alliance in gene silencing. Biochem Soc Trans. 2013;41:855–860. doi: 10.1042/BST20130047. [DOI] [PubMed] [Google Scholar]
- Wilson RC, Doudna JA. Molecular mechanisms of RNA interference. Annu Rev Biophys. 2013;42:217–239. doi: 10.1146/annurev-biophys-083012-130404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, et al. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol. 2011;18:1211–1217. doi: 10.1038/nsmb.2149. [DOI] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell. 2011;44:120–133. doi: 10.1016/j.molcel.2011.09.007. [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, et al. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat StructMol Biol. 2011;18:1218–1226. doi: 10.1038/nsmb.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara T, Zekri L, Braun JE, Izaurralde E. miRISC recruits decapping factors to miRNA targets to enhance their degradation. Nucleic Acids Res. 2013;41:8692–8705. doi: 10.1093/nar/gkt619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O, Nadon R. Re-analysis of genome wide data on mammalian microRNA-mediated suppression of gene expression. Translation. 2013;1:e24557. doi: 10.4161/trla.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–240. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–237. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–723. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, et al. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–85. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- Cannell IG, Kong YW, Johnston SJ, Chen ML, Collins HM, Dobbyn HC, et al. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc Natl Acad Sci USA. 2010;107:5375–5380. doi: 10.1073/pnas.0910015107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferland-McCollough D, Fernandez-Twinn DS, Cannell IG, David H, Warner M, Vaag AA, et al. Programming of adipose tissue miR-483-3p and GDF-3 expression by maternal diet in type 2 diabetes. Cell Death Differ. 2012;19:1003–1012. doi: 10.1038/cdd.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdanowicz A, Thermann R, Kowalska J, Jemielity J, Duncan K, Preiss T, et al. Drosophila miR2 primarily targets the m7GpppN cap structure for translational repression. Mol Cell. 2009;35:881–888. doi: 10.1016/j.molcel.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Mishima Y, Fukao A, Kishimoto T, Sakamoto H, Fujiwara T, Inoue K. Translational inhibition by deadenylation-independent mechanisms is central to microRNA-mediated silencing in zebrafish. Proc Natl Acad Sci USA. 2012;109:1104–1109. doi: 10.1073/pnas.1113350109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, Tomari Y. PABP is not essential for microRNA-mediated translational repression and deadenylation in vitro. Embo J. 2011;30:4998–5009. doi: 10.1038/emboj.2011.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression. Biochem Soc Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nat Struct Mol Biol. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kawamata T, Tomari Y. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol Cell. 2009;34:58–67. doi: 10.1016/j.molcel.2009.02.010. [DOI] [PubMed] [Google Scholar]
- Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG, Hamilton TL, et al. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci USA. 2008;105:8866–8871. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SR. Tricks an IRES uses to enslave ribosomes. Trends Microbiol. 2012;20:558–566. doi: 10.1016/j.tim.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci EP, Limousin T, Soto-Rifo R, Allison R, Poyry T, Decimo D, et al. Activation of a microRNA response in trans reveals a new role for poly(A) in translational repression. Nucleic Acids Res. 2011;39:5215–5231. doi: 10.1093/nar/gkr086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder-Hill J, Pause A, Sonenberg N, Merrick WC. The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF-4A. J Biol Chem. 1993;268:5566–5573. [PubMed] [Google Scholar]
- Galicia-Vazquez G, Cencic R, Robert F, Agenor AQ, Pelletier J. A cellular response linking eIF4AI activity to eIF4AII transcription. RNA. 2012;18:1373–1384. doi: 10.1261/rna.033209.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Belsham GJ, Proud CG. Eukaryotic initiation factors 4A (eIF4A) and 4G (eIF4G) mutually interact in a 1:1 ratio in vivo. J Biol Chem. 2001;276:29111–29115. doi: 10.1074/jbc.C100284200. [DOI] [PubMed] [Google Scholar]
- Schutz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, et al. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basquin J, Roudko VV, Rode M, Basquin C, Seraphin B, Conti E. Architecture of the nuclease module of the yeast Ccr4-not complex: the Not1-Caf1-Ccr4 interaction. Mol Cell. 2012;48:207–218. doi: 10.1016/j.molcel.2012.08.014. [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Kuzuoglu-Ozturk D, Braun JE, Eulalio A, Wohlbold L, Izaurralde E. The interactions of GW182 proteins with PABP and deadenylases are required for both translational repression and degradation of miRNA targets. Nucleic Acids Res. 2013;41:978–994. doi: 10.1093/nar/gks1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E. HPat provides a link between deadenylation and decapping in metazoa. J Cell Biol. 2010;189:289–302. doi: 10.1083/jcb.200910141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barisic-Jager E, Krecioch I, Hosiner S, Antic S, Dorner S. HPat a decapping activator interacting with the miRNA effector complex. PLoS One. 2013;8:e71860. doi: 10.1371/journal.pone.0071860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H, Basquin J, Ozgur S, Czarnocki-Cieciura M, Bonneau F, Aartse A, et al. Structural and biochemical insights to the role of the CCR4-NOT complex and DDX6 ATPase in microRNA repression. Mol Cell. 2014;54:751–765. doi: 10.1016/j.molcel.2014.03.036. [DOI] [PubMed] [Google Scholar]
- Chen Y, Boland A, Kuzuoglu-Ozturk D, Bawankar P, Loh B, Chang CT, et al. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell. 2014;54:737–750. doi: 10.1016/j.molcel.2014.03.034. [DOI] [PubMed] [Google Scholar]
- Rouya C, Siddiqui N, Morita M, Duchaine TF, Fabian MR, Sonenberg N.Human DDX6 effects miRNA-mediated gene silencing via direct binding to CNOT1 RNA 2014. e-pub ahead of print 17 July 2014. [DOI] [PMC free article] [PubMed]
- Lu WT, Wilczynska A, Smith E, Bushell M. The diverse roles of the eIF4A family: you are the company you keep. Biochem Soc Trans. 2014;42:166–172. doi: 10.1042/BST20130161. [DOI] [PubMed] [Google Scholar]
- Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol Cell. 2009;33:661–668. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–705. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Winkler GS. RNA decay machines: deadenylation by the Ccr4-not and Pan2-Pan3 complexes. Biochim Biophys Acta. 2013;1829:561–570. doi: 10.1016/j.bbagrm.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Collart MA, Panasenko OO. The Ccr4—not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- Mazumder A, Bose M, Chakraborty A, Chakrabarti S, Bhattacharyya SN. A transient reversal of miRNA-mediated repression controls macrophage activation. EMBO Rep. 2013;14:1008–1016. doi: 10.1038/embor.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui N, Mangus DA, Chang TC, Palermino JM, Shyu AB, Gehring K. Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein. J Biol Chem. 2007;282:25067–25075. doi: 10.1074/jbc.M701256200. [DOI] [PubMed] [Google Scholar]
- Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat Commun. 2012;3:923. doi: 10.1038/ncomms1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie M, Boland A, Huntzinger E, Weichenrieder O, Izaurralde E. Structure of the PAN3 pseudokinase reveals the basis for interactions with the PAN2 deadenylase and the GW182 proteins. Mol Cell. 2013;51:360–373. doi: 10.1016/j.molcel.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Cooke A, Prigge A, Wickens M. Translational repression by deadenylases. J Biol Chem. 2010;285:28506–28513. doi: 10.1074/jbc.M110.150763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Izaurralde E. The role of GW182 proteins in miRNA-mediated gene silencing. Adv Exp Med Biol. 2013;768:147–163. doi: 10.1007/978-1-4614-5107-5_9. [DOI] [PubMed] [Google Scholar]
- Zekri L, Huntzinger E, Heimstadt S, Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol Cell Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. Embo J. 2010;29:4146–4160. doi: 10.1038/emboj.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol. 2010;17:238–240. doi: 10.1038/nsmb.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Safaee N, Rosenauer A, Gehring K. Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein. J Biol Chem. 2010;285:13599–13606. doi: 10.1074/jbc.M109.089540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Walters RW, Bradrick SS, Gromeier M. Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA. 2010;16:239–250. doi: 10.1261/rna.1795410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti F, Kaiser C, Zdanowicz-Specht A, Hentze MW. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat Struct Mol Biol. 2012;19:603–608. doi: 10.1038/nsmb.2309. [DOI] [PubMed] [Google Scholar]
- Zekri L, Kuzuoglu-Ozturk D, Izaurralde E. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. Embo J. 2013;32:1052–1065. doi: 10.1038/emboj.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Imataka H, Sonenberg N. Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr Biol. 2000;10:1147–1150. doi: 10.1016/s0960-9822(00)00701-6. [DOI] [PubMed] [Google Scholar]
- Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. Embo J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel S, Wang Y, Lenobel R, Korner R, Hsiao HH, Urlaub H, et al. Phosphorylation of human Argonaute proteins affects small RNA binding. Nucleic Acids Res. 2011;39:2330–2343. doi: 10.1093/nar/gkq1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horman SR, Janas MM, Litterst C, Wang B, MacRae IJ, Sever MJ, et al. Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Mol Cell. 2013;50:356–367. doi: 10.1016/j.molcel.2013.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem J. 2008;413:429–436. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, So J, Davis-Dusenbery BN, Qi HH, Bloch DB, Shi Y, et al. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol Cell Biol. 2011;31:4760–4774. doi: 10.1128/MCB.05776-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, et al. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–424. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42:489–499. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, Lapaquette P, Andrieux A, Faure G, Dejean A. Sumoylation of human argonaute 2 at lysine-402 regulates its stability. PLoS One. 2014;9:e102957. doi: 10.1371/journal.pone.0102957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn FG, Cuevas E, Franzoni E, Rybak A. MiRNA need a TRIM regulation of miRNA activity by Trim-NHL proteins. Adv Exp Med Biol. 2010;700:85–105. [PubMed] [Google Scholar]
- Hammell CM, Lubin I, Boag PR, Blackwell TK, Ambros V. nhl-2 Modulates microRNA activity in Caenorhabditis elegans. Cell. 2009;136:926–938. doi: 10.1016/j.cell.2009.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn JC, Berezikov E, Knoblich JA. The TRIM-NHL protein TRIM32 activates microRNAs and prevents self-renewal in mouse neural progenitors. Cell. 2009;136:913–925. doi: 10.1016/j.cell.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, et al. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454:241–245. doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecsedi M, Grosshans H. LIN-41/TRIM71: emancipation of a miRNA target. Genes Dev. 2013;27:581–589. doi: 10.1101/gad.207266.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, et al. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol. 2009;11:1411–1420. doi: 10.1038/ncb1987. [DOI] [PubMed] [Google Scholar]
- Chen J, Lai F, Niswander L. The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev. 2012;26:803–815. doi: 10.1101/gad.187641.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, et al. T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med. 2013;210:417–432. doi: 10.1084/jem.20111717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Coyle SM, Doudna JA. Coupled 5′ nucleotide recognition and processivity in Xrn1-mediated mRNA decay. Mol Cell. 2011;41:600–608. doi: 10.1016/j.molcel.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KL, Chadee AB, Chen CY, Zhang Y, Shyu AB. Phosphorylation at intrinsically disordered regions of PAM2 motif-containing proteins modulates their interactions with PABPC1 and influences mRNA fate. RNA. 2013;19:295–305. doi: 10.1261/rna.037317.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau NC, Mulder KW, Brenkman AB, Mohammed S, van den Broek NJ, Heck AJ, et al. Phosphorylation of Not4p functions parallel to BUR2 to regulate resistance to cellular stresses in Saccharomyces cerevisiae. PLoS One. 2010;5:e9864. doi: 10.1371/journal.pone.0009864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni B, Gray NK. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief Funct Genomic Proteomic. 2004;3:125–141. doi: 10.1093/bfgp/3.2.125. [DOI] [PubMed] [Google Scholar]
- Khaleghpour K, Kahvejian A, De Crescenzo G, Roy G, Svitkin YV, Imataka H, et al. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol. 2001;21:5200–5213. doi: 10.1128/MCB.21.15.5200-5213.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim MM, Svitkin YV, Kahvejian A, De Crescenzo G, Costa-Mattioli M, Sonenberg N. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc Natl Acad Sci USA. 2006;103:9494–9499. doi: 10.1073/pnas.0603701103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook M, McCracken L, Reddington JP, Lu ZL, Morrice NA, Gray NK. The multifunctional poly(A)-binding protein (PABP) 1 is subject to extensive dynamic post-translational modification, which molecular modelling suggests plays an important role in co-ordinating its activities. Biochem J. 2012;441:803–812. doi: 10.1042/BJ20111474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le H, Browning KS, Gallie DR. The phosphorylation state of poly(A)-binding protein specifies its binding to poly(A) RNA and its interaction with eukaryotic initiation factor (eIF) 4F, eIFiso4F, and eIF4B. J Biol Chem. 2000;275:17452–17462. doi: 10.1074/jbc.M001186200. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell. 2006;124:191–205. doi: 10.1016/j.cell.2005.12.017. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116:817–829. doi: 10.1016/s0092-8674(04)00250-8. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, et al. RISC assembly defects in the Drosophila RNAi mutant armitage. Cell. 2004;116:831–841. doi: 10.1016/s0092-8674(04)00218-1. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, et al. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic Acids Res. 2012;40:5088–5100. doi: 10.1093/nar/gks148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Collier B, Gorgoni B, Loveridge C, Cooke HJ, Gray NK. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. Embo J. 2005;24:2656–2666. doi: 10.1038/sj.emboj.7600738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One. 2009;4:e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MV, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi J, van den Bosch MW, Palygin O, Mistry RB, McKenzie C, Macdonald A, et al. miR-132/212 knockout mice reveal roles for these miRNAs in regulating cortical synaptic transmission and plasticity. PLoS One. 2013;8:e62509. doi: 10.1371/journal.pone.0062509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Jeker LT, Carver-Moore K, Oh A, Liu HJ, Cameron R, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum Mol Genet. 2010;19:R169–R175. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–373. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]