Abstract
MicroRNAs (miRs) are a class of small noncoding RNAs that suppress the expression of protein-coding genes by repressing protein translation. Although the roles that miRs and the miR processing machinery have in regulating epithelial stem cell biology are not fully understood, their fundamental contributions to these processes have been demonstrated over the last few years. The p53-family member p63 is an essential transcription factor for epidermal morphogenesis and homeostasis. p63 functions as a determinant for keratinocyte cell fate and helps to regulate the balance between stemness, differentiation and senescence. An important factor that regulates p63 function is the reciprocal interaction between p63 and miRs. Some miRs control p63 expression, and p63 regulates the miR expression profile in the epidermis. p63 controls miR expression at different levels. It directly regulates the transcription of several miRs and indirectly regulates their processing by regulating the expression of the miR processing components Dicer and DGCR8. In this review, we will discuss the recent findings on the miR–p63 interaction in epidermal biology, particularly focusing on the ΔNp63-dependent regulation of DGCR8 recently described in the ΔNp63−/− mouse. We provide a unified view of the current knowledge and discuss the apparent discrepancies and perspective therapeutic opportunities.
Facts
Epidermal morphogenesis is governed by sets of microRNAs(miRs) with different expression patterns.
Epidermal-conditional deletion of miR processing machinery components, such as DICER, Drosha and DGCR8, has underscored the importance of miRs in epidermal morphogenesis and homeostasis.
p63 is a master regulator of epidermal homeostasis; p63−/− and ΔNp63−/− mice fail to develop a functional epidermis.
p63 directly, via transcriptional regulation, and indirectly, by promoting the transcription of Dicer and DGCR8, controls the epidermal miR expression profile.
The complex network between miRs and p63 regulates the fate of epidermal cells.
Open Questions
Where and when are epidermal/epithelial miRs precisely expressed during epidermal/epithelial morphogenesis, homeostasis and in different diseases?
How do the isoforms ΔNp63 and TAp63 contribute to epidermal homeostasis? And to what extent do miRs contribute to the functioning of the p63 isoforms?
Will the miR expression profiles provide innovative biomarker tools in epidermal cancers?
Will drugs targeting miR expression provide alternative therapeutic approaches in epidermal oncology?
MicroRNAs (miRs) are a class of small noncoding RNA (single-stranded RNAs consisting of 19–22 nucleotides) that suppress the expression of protein-coding genes by repressing protein translation by interacting with the 3′UTR region of messenger RNAs.1 The epidermal-conditional deletion of components of the enzymatic complexes responsible for miR maturation, such as DICER, Drosha and DGCR8, has underscored the importance of miRs in the morphogenesis and adult homeostasis of the epidermis.2, 3, 4, 5
The epidermis is the first barrier of an animal that protects against water loss and the biological and physical stress caused by the external environment. The epidermis consists of a keratinised stratified epithelium (interfollicular epidermis, IFE) that can also include appendages such as hair follicles (HFs) and sebaceous glands (SG). Together, these structures help to protect and thermoregulate the organism. The epidermis originates from a single layer of ectodermal cells during embryonic development. This single layer of ectodermal precursor cells undergoes a stratification process that forms the different layers of the differentiated cells (basal, spinous, granular and cornified layers), which all together function as a barrier. The basal layer consists of highly proliferative cells that express high levels of basal integrins, keratin 5 (K5) and keratin 14 (K14). The mature epidermis undergoes homeostatic regulation as basal cells periodically terminally differentiate and migrate outward in a columnar fashion (Figure 1a). Homeostasis and the repair capability of adult skin are maintained by the epidermal stem cell (SC) compartments in the IFE, HFs and SGs.6 SCs in the HFs are maintained in a specialised microenvironment defined as the bulge (Figure 1b). Bulge SCs are characterised by their slow cycle and the expression of specific markers, such as K15, K19 and Lgr5.7, 8 During the hair cycle process, HFs alternate phases of growth and degeneration. The SCs from the bulge mediate the regeneration phase of the HF cycle by producing transient amplifying cells (TA) in the HF matrix.7 In response to injury, bulge SCs rapidly migrate up to repair the damaged epidermis, and this migration appears to be tightly regulated because it stops once the wound is healed. This is a unique case in which bulge SCs adopt an IFE differentiation programme.9 Under normal conditions, these cells do not contribute to the maintenance of the IFE, which suggests that a separate SC pool in the IFE exists.10 A small unit of SCs in the basal layer of the IFE provides the precursors that replace the terminal differentiated keratinocytes.11 These SCs undergo an asymmetric division that gives rise to a new SC and a committed cell, which undergoes repeated rounds of symmetric division to supply the basal compartment with highly proliferative keratinocytes. In addition to the bulge and IFE SC populations, there is also evidence of epidermal progenitors in SGs. A population of resident SCs in the upper HF that express a high level of MTS24 have been shown to maintain SG homeostasis (Figure 1b).12, 13 Two studies from the laboratories of Fuchs and Millar have identified the specific miRs that are expressed in the developing skin and hair follicles of the mouse.2, 4 Although they used different approaches – Fuchs analysed the E17.5 mouse embryo IFE and HF and Millard analysed fully thick skin of P1 mice – there is a significant overlap in the miR profiles reported. The most significant findings indicate that miR-16, miR-20, miR-21, miR-24 and miR-125b are the most highly expressed miRs in the IFE and HF. miR-203, the miR-200 family (i.e., miR-200a, miR-200b, miR-200c, miR-141 and miR-149) and the miR-19 family (i.e., miR-19b, miR-17–5p and miR-93) appear to be preferentially expressed in the epidermis, whereas the miR-199 family (miR-199a and miR-199b) is only expressed in hair follicles.2, 4
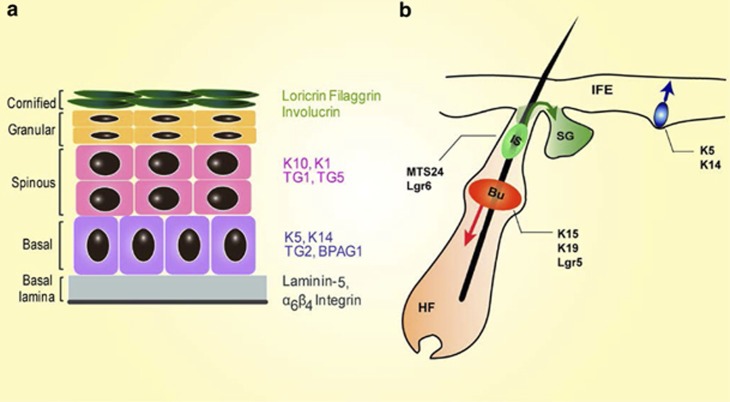
Figure 1.
(a) Schematic diagram of the mouse skin epithelium. Basal layer keratinocytes (violet) adhere to the underlying basement membrane (basal lamina, grey). Basal keratinocytes lose their proliferative ability when they detach from the basement membrane and migrate towards the epidermal surface (spinous layer, orange; granular layer, green; stratum corneum, dark green). An epidermal interfollicular proliferative unit (basal layer) is composed of stem cells and transiently amplifying cells, which divide a few times and then leave the basal layer to execute their differentiation programme. K5, K14, TG2 and BPAG1 are specific markers for the basal layer. Different intermediate differentiation steps occur in the spinous and granular layers. Markers for the spinous layer include K10, K1, TG1 and TG5. Markers for the granular layer – Loricrin, Filaggrin and Involucrin. The cornified layer represents the final phase of terminal differentiation. miR-205 is exclusively and specifically expressed in the basal layer. miR-24, miR-203, miR-574, miR-720 and mir-34a are pro-differentiation miRs expressed in the epidermal suprabasal compartment. (b) Schematic diagram illustrating the following different epidermal SC populations found in the skin epidermis: the IFE SCs (shown in violet), the bulge SCs (Bu in orange), the isthmus SCs (Is in green). IFE SCs specifically express K5 and K14; bulge SCs specifically express K15, K19 and Lgr5; and isthmus SCs specifically express MTS24 and Kgr6. miR-205 is expressed in IFE and bulge SCs, and miR-125b is only expressed in bulge SC. Panel b is a modified version of a previously published illustration.69 K5, cytokeratin-5; K14, cytokeratin-14; K10, TG2, transglutaminase-2; cytokeratin-10; K1, cytokeratin-1; TG1, transglutaminase-1; TG5, transglutaminase-5; Fila, Filaggrin; SC, stem cell; HF, hair follicle; IS, isthmus; SG, sebaceous gland
p63, a member of the p53 family of genes, is thought to be a master regulator in ectodermal/epithelial development. The presence of an additional internal promoter in the gene allows for the expression of a N-truncated isoform, ΔNp63, in addition to the full-length isoform TAp63. TAp63 is the isoform mainly sharing common function with the better-characterized member of the family, p53. TAp63 indeed is able to induce cell cycle arrest and cell death following DNA damage, similarly to p53 and TAp73.14 TAp63 has been additionally involved in metastasis suppression, through different mechanisms including regulation of Sharp-1, Dicer and miRs.15, 16 TAp63 can therefore be considered a tumour suppressor protein.17 ΔNp63 is highly expressed in SCs and the TA cells of the epidermis (basal layers) and other stratified epithelia, and it is progressively downregulated during terminal differentiation.18, 19 Since the generation of two independent p63−/− mouse models, from the laboratories of Mckeon20 and Bradley,21 the role that p63 has in epidermal development has become controversial. Although both of the p63−/− mouse models lack an epidermis and epidermal appendages, have severe limb and craniofacial defects, and die soon after birth, the precise role that p63 has in ectodermal/epithelial biology remains unknown. In contrast with the mouse model generated in the Bradley laboratory, the p63−/− mouse described by Mckeon displays patches of differentiated keratinocytes that express terminal differentiation markers (involucrin, loricrin and filaggrin). These patches were interpreted to be remnants of a developing epidermis that could not be maintained during embryogenesis. This finding suggests that p63 is required for epidermal stem cell maintenance.20 In contrast, the p63−/− mouse model described by Bradley does not appear to express any of the differentiation markers in the epidermis or stratified epithelia. Because the epidermis of this p63−/− mouse model exhibits only a faint and patchy K14 expression pattern and lacks a stratified structure and because epidermal development appears to cease at approximately E9.5, p63 was considered to be a determining factor in epithelial stratification. Another study by Mckeon using epithelial thymus cells, however, demonstrated that p63 does not has a role in proliferation per se but is required to maintain the highly proliferative potential of the SCs of the thymic epithelium. p63 ablation did not affect the incorporation of 5-bromodeoxyuridine in the overall epithelial thymic population, but did affect the amount of apoptosis in the epithelial thymic SC population.22 This study supports the importance of p63 in epithelial SC and distinguishes p63 contribution to proliferative potential versus its contribution in maintaining the stem cell niche or forestalling differentiation. In the debate on interpretation of the failure of p63−/− embryos to develop stratified epithelia, a study by Shalom-Feuerestein et al.23 showed that p63−/− epithelia (either mouse model) failed to develop beyond ectodermal stage and remained a monolayer of nonproliferating cells expressing K8/K18. This suggested the function of p63 as an ectodermal gatekeeper, promoting epidermal commitment during ES cell differentiation. This is in line with the demonstration by Khavari's group24 that somatic epidermal cells in which p63 has been inactivated become K8-positive ectodermal cells.
Recent findings indicate that a complex network and reciprocal interactions between miRs and p63 regulate the fate of epidermal cells. In this review, we discuss the recent findings in this field and try to provide a unified view of how the balance between p63 and miRs regulates epidermal biology.
ΔNp63 is Essential for Epidermal Morphogenesis
To elucidate the precise role that p63 has in epidermal biology, selective knockout models for the most highly expressed p63 isoform in the epithelia, ΔNp63, have been generated. The recently generated ΔNp63-null mouse described by Flores (hereafter indicated as ΔNp63Δ/Δ), however, raised further complexity in the dissection of p63 function in epithelia.25 Lack of ΔNp63 in Flores' model mainly phenocopied the gross morphology of the ΔNp63 mouse epidermis described by Romano et al.26 (hereafter indicated as ΔNp63gfp/gfp), which is partially similar to the phenotypes of both of the p63−/− mouse models that lack all of the p63 isoforms.20, 21 The Flores and Romano mouse models clearly indicate that ΔNp63 is indispensable for normal epidermal development and stratification. The ΔNp63Δ/Δ mice were born at the proper Mendelian ratios but died within hours after birth, which is similar to the p63−/− mice. ΔNp63Δ/Δ mice developed a rudimentary stratified epithelium with nests of basal epidermal cells that appeared in patches in over 20–30% of the embryo (over 5–10% of the embryo). ΔNp63Δ/Δ embryos (E18.5) expressed the basal markers K5 and K14 in a few epidermal patches that were organised in an aberrantly expanded basal layer. In some areas, the suprabasal layer makers K10 and Filaggrin were also detected. These findings suggest that the ΔNp63Δ/Δ epidermis attempts to terminally differentiate but the process is severely defective. The expression pattern of the suprabasal markers resembles the expression patterns observed in the p63−/− Mckeon strain in which some patches of differentiated keratinocytes were identified.20 Interestingly, in contrast with heterozygous ΔNp63+/gfp mice, which appear to be indistinguishable from their wild-type littermates, the heterozygous ΔNp63+/Δ mice described by Flores have a phenotype that is an intermediate between the wild-type and ΔNp63Δ/Δ phenotypes. A morphological analysis of the epidermis revealed that the ΔNp63+/Δ mice have an expanded epidermal basal layer with basaloid cells above the basal epithelium and multiple cell layers with uneven expression patterns of K5, K14, K10, K1 and Filaggrin. The presence of an expanded epidermis suggests that the epidermal cells were hyperproliferative and may have the ability to self-renew. The epidermis of ΔNp63+/Δ mice and the epidermal patches in the ΔNp63Δ/Δ embryos incorporated 5-bromodeoxyuridine at a higher rate, which indicates that they have a hyperproliferative phenotype. Furthermore, serial passages of ΔNp63+/+ and ΔNp63Δ/Δ epidermal cells demonstrated that the ΔNp63 depleted cells retain their proliferation potential in late passages, which is a point at which the wild-type cells have already undergone irreversible cell cycle arrest. Therefore, the deletion of ΔNp63 appears to sustain the proliferation and self-renewal capabilities of these cells. However, contradictory findings have also been reported. By using an ex vivo model of regenerating human epidermis, Khavari lab reported that loss of p63 resulted in severe tissue hypoplasia and inhibition of stratification and differentiation of the keratinocytes, indicating the necessity of p63 for both proliferative and differentiation potential of the epidermis.24 In agreement with Flores, Roop's group instead reported that suprabasal proliferating cells were consistently observed in the skin of an epidermal-specific inducible knockdown mouse model for ΔNp63, once ΔNp63 silencing was induced in newborn mouse epidermis after normal embryonic development. This finding indicates that there is a delay or a defect in the ability of basal keratinocytes to withdraw from the cell cycle.27 This finding also suggests that ΔNp63 and TAp63 may have contrasting roles in epidermal proliferation. The selective inactivation of TAp63 is associated with defective proliferation and the early senescence of dermal and epidermal precursors, which results in premature ageing, skin ulcerations and epidermal cell senescence.28 A balance between the p63 isoforms may be required for proper epidermal homeostasis. Indeed, although the ΔNp63 knockout mouse models have phenotypes that are similar to the full p63−/− mouse model, their phenotypes are not completely identical to the phenotype observed with total p63 ablation.
ΔNp63 Depletion Induces Multipotency in Epidermal Cells
A striking phenomenon observed in Flores' mouse model was that, owing to their self-renewal capacity, the ΔNp63Δ/Δ epidermal cells expressed higher levels of markers for embryonic stem cells and fibroblasts induced to become pluripotent. The ablation of ΔNp63 induced epidermal cells to become multipotent.25 These multipotent cells demonstrated features and expression profiles that resembled the those observed in induced pluripotent stem cells obtained from fibroblasts using Yamanaka factors (miPSYam).29 Epidermal ΔNp63Δ/Δ cells expressed Nanog, Oct4, Sox2 and SSEA-1 (Figure 2). All of the epidermal ΔNp63Δ/Δ cells, however, did not express the pluripotency markers at the same level. This high variation in expression levels suggests that a specific in vitro clone selection process may have occurred and raises additional questions. What additional factors are these cells acquiring to be selected as highly pluripotent cells? Are these selected clones derived from epidermal stem cells or are they stochastically selected from the entire epidermal population?
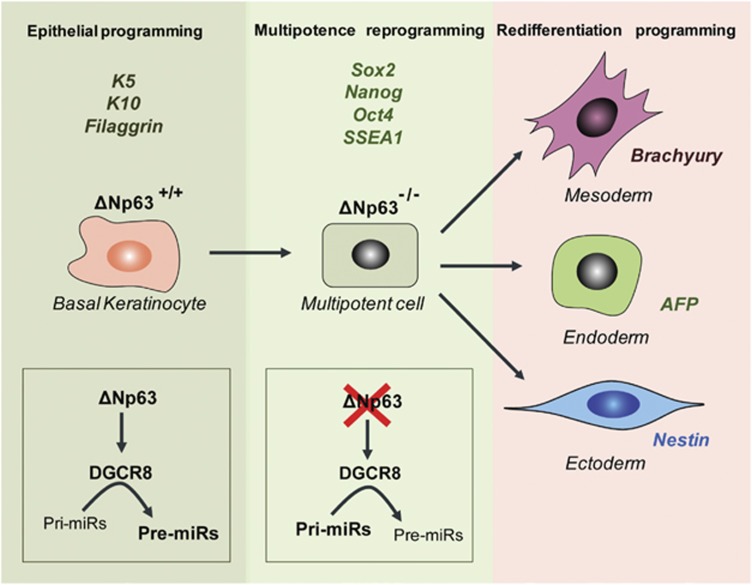
Figure 2.
ΔNp63 downregulation promotes epidermal cells to become multipotent. Epithelial programming is characterised by the expression of specific markers, such as K5, K14 and filaggrin. The basal keratinocyte epithelial programme is maintained by an active ΔNp63/DGCR8 axis, which ensures a correct miR expression profile. Loss of ΔNp63 results in DGCR8 downregulation, which impairs normal miR maturation. These events result in the cells becoming multipotent and expressing the multipotent stem cell marker Sox2, Nanog, Oct4 and SSEA1. Specific environmental conditions allow these cells to re-differentiate into different tissue types, such as mesoderm, endoderm or ectoderm. Each re-differentiation programme is characterised by the expression of specific markers (Brachyury, AFP and Nestin)
miR biogenesis has been shown to be critical for maintaining pluripotency.30 miRs help to silence the ability of ES cells to self-renew, and this process normally occurs with the induction of differentiation.3, 4, 31 Experiments using knockout ES cells have shown that DGCR8, the RNA-binding protein that assists Drosha in miR processing, is essential for downregulating the expression of pluripotency markers and promoting ES cell colony differentiation.31 ΔNp63 transcriptionally regulates DGCR8. ΔNp63Δ/Δ epidermal cells express a lower level of DGCR8 compared with wt cells. Furthermore, it has also been shown that ΔNp63 can directly bind to and transactivate a p53-like responsive element in intron 1 of the DGCR8 gene. A global miR expression profile demonstrated that the expression levels of miRs previously described as crucial for cell reprogramming, such as miR-369,30 are downregulated in ΔNp63Δ/Δ epidermal cells. The overall miR signature revealed that the ΔNp63Δ/Δ epidermal cell line partially resembled the miPSYam and mES cell expression profiles, and the ectopic expression of DGCR8 in these cells produced a signature that was very similar to the signature observed in wt keratinocytes. Furthermore, the expression profile of ΔNp63Δ/Δ did not reveal a global downregulation in miR biogenesis, but specific changes in the expression pattern that included the downregulation of some miRs and the upregulation of others.25 This finding suggests that in addition to reducing DGCR8 expression and the consequential downregulation in miR biogenesis, additional mechanisms may also be responsible for the ΔNp63Δ/Δ epidermal cell phenotype.
It has also been shown that ΔNp63Δ/Δ epidermal cells have the ability to differentiate into multiple cell types in vitro and in vivo. Culturing these cells in specific epidermal or neuroectodermal medium promotes terminal differentiation, which results in the expression of specific differentiation markers for two different cell lineages. An additional requirement for terminal differentiation, however, was the ectopic expression of DGCR8 in ΔNp63Δ/Δ cells. This requirement was confirmed in vivo in a teratoma model and by injecting ΔNp63Δ/Δ cells in mouse blastocysts. ΔNp63Δ/Δ epidermal cells that expressed DGCR8 were able to differentiate into skin, muscle, liver and brain cells in vivo (Figure 2). E18.5 chimera embryos generated from a GFP-expressing ΔNp63Δ/Δ epidermal cell line showed that the ΔNp63Δ/Δ cells contributed to 70–80% of the mouse embryo. The GFP-positive cells were also found to express keratin 5 in the skin, villin in the intestinal tract, AFP in the liver and nestin in the brain.25 These chimera embryos, however, demonstrated severe abnormalities in the expression patterns of epithelia markers, and they died shortly after birth because of skin defects. Contrasting findings on the involvement of p63 in stem cell reprogramming have been reported by the other groups. ΔNp63 appeared to be essential for the reprogramming of MEFs by OSK (Oct4, Sox2 and Klf4). Moll32 reported that loss of p63 affects reprogramming through several mechanisms, such as reducing the expression of mesenchymal–epithelial transition and pluripotency genes, hypoproliferation and loss of the most reprogrammable cell populations. The depletion of p63, however, did not affect the maintenance of the iPSCs derived from MEFs, which indicates that it is only important in the reprogramming process. Another study suggests that p63 loss in mouse ES cells still allows the generation of K8/K18 epithelial cells, but impairs the full commitment to K5/K14 positive epidermal cells.33 Clearly, the work by Flores adds another level of complexity to the functional role that p63 has in epidermal biology and stem cell maintenance. Although many questions remain in this field, this study alludes to an additional strategy to achieve the fascinating possibility of extracting epidermal cells from patient skin biopsies and reprogramming the keratinocytes into multipotent stem cells for various therapeutic strategies.
miRs-p63 Loops in Epidermal Differentiation
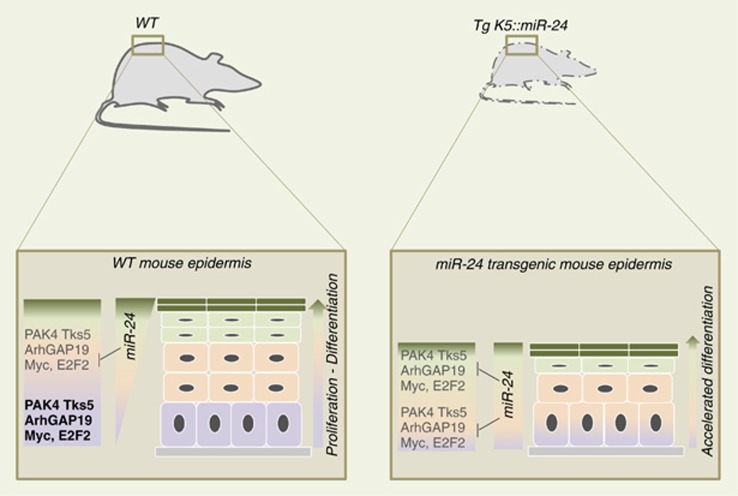
Disruption of miR processing in the skin, by the conditional deletion of Dicer, results in abnormalities in epidermal stratification and suggests that miRs have important roles in keratinocyte differentiation. Several recent studies further highlight the importance of miR-p63 loops in regulating epidermal stratification (Table 1). miR-203 was the first miR implicated in driving epidermal differentiation. miR-203 is rapidly upregulated when primary keratinocytes exit the cell cycle and are induced to differentiate.34, 35 In the mouse skin, miR-203 expression is only detectable in the suprabasal layers and the differentiated sheets of mature hair follicles.35 MiR-203 targets the 3′UTR of p63 and directly represses ΔNp63 expression in the suprabasal layers. The ectopic expression of miR-203 in the basal layer of mouse skin resulted in perinatal lethality, which was associated with a lack of epidermal basal progenitors and the detection of sporadic ΔNp63-expressing cells.35 These findings strongly implicate that p63 is an essential regulator of stem cell maintenance in stratified epithelial tissues. It has been shown that miR-203 is transcriptionally activated in the differentiating daughter cells resulting from the asymmetric cell division of interfollicular progenitor cells. miR-203 rapidly promotes exiting the cell cycle within 6 h and abolishes the self-renewal ability of the progenitor cells. However, only the co-suppression of the miR-203 individual targets (including p63, Skp2 and Msi2) is sufficient for promoting exiting the cell cycle and inhibiting long-term proliferation.36 This finding demonstrates that not all of the effects of miR-203 are the result of p63-dependent repression. miR-203 defines a molecular boundary between proliferative basal progenitors and terminally differentiating suprabasal cells, which ensures the proper identification of neighbouring layers. A novel autoregulatory feedback loop involving iASPP/p63/miRs has been implicated in maintaining homeostasis of the stratified epithelia.37 iASPP belongs to the family of apoptosis-stimulating proteins of p53 (ASPP), which regulates p53-family mediated apoptosis. iASPP is mostly expressed in epithelial tissues and preferentially binds to p63. ΔNp63 and iASPP are coexpressed in basal epidermal cells at E15.5 and reciprocally regulate each other. ΔNp63 transcriptionally controls the expression of iASPP while iASPP represses the expression of two miRs, miR-574 and miR-720, that translationally repress ΔNp63 expression (Figure 3). iASPP−/− keratinocytes are not as proliferative and prematurely differentiate, and these phenotypes are partially rescued by inducing the expression of miR-574 and miR-720. However, iASPP depletion does not completely photocopy p63 depletion. The downregulated expression pattern of adhesion molecules observed in p63-depleted cells (reduced expression of integrin β1, β2 and α3) is not precisely reproduced in iASPP-depleted cells (reduced expression of integrin β1 and increased expression of integrin α3). Another difference between the two phenotypes is the premature differentiation of iASPP−/− keratinocytes, which does not occur with ΔNp6325, 26 or full p63 depletion.24 Other miRs have also been implicated in regulating the proliferation of epidermal progenitor cells and maintaining the stem cell population (Figure 4). Recently, miR-24 has been identified as a critical regulator of keratinocyte differentiation that functions by regulating the rearrangement of the actin cytoskeleton. Similar to miR-203, miR-24 is expressed in differentiated compartments of the epidermis. The ectopic expression of miR-24 in the basal layer of the mouse skin results in the premature expression of differentiation markers, and an early exit from the cell cycle. A few days after birth, miR-24 transgenic mice (K14::miR-24 Tg) are significantly smaller compared with their wt littermates and die, most likely from malnutrition. Although the general architecture appeared normal, their skin was thinner and several basaloid cells were positive for the spinous layer marker K10, which is indicative of accelerated differentiation. It has been shown that miR-24 targets actin cytoskeleton modulators, including PAK4, Tsk5 and ArhGAP19, that promote a switch from a basal to a suprabasal actin cytoskeleton architecture, which promotes keratinocyte differentiation (Figure 5).38 Furthermore, the miR-24 transgenic mice HF bulge has a severely depleted SC compartment.39 The balance between stem cell renewal and differentiation is also tightly regulated by extrinsic signals from the extracellular matrix and the arrangement of the actin cytoskeleton. SC differentiation required the concomitant regulation of the serum response factor activity and a dense cortical F-actin cytoskeleton. Thus, actin cytoskeleton-mediated shape-induced differentiation has been shown to be a determining factor in epidermal cell fate.40
Table 1. MiRs involved in epidermal homeostasis.
| miR | Function | Target/s | Ref |
|---|---|---|---|
| miR-203 | Keratinocyte proliferation/differentiation | p63 Skp2 Msi2 | 34, 35, 36 |
| miR-574 miR-720 | Keratinocyte proliferation/differentiation | p63 | 37 |
| miR-24 | Keratinocyte differentiation/actin cytoskeleton | PAK4 Tsk5 ArhGAP19 Tcf3 | 38, 39 |
| miR-205 | Epidermal Stem cells | 41 | |
| miR-180 miR-130a miR-181a miR-181b miR-138 | Keratinocyte senescence | p63 SIRT1 | 46, 50 |
| miR-191 | Keratinocyte senescence | Satb1 | 49 |
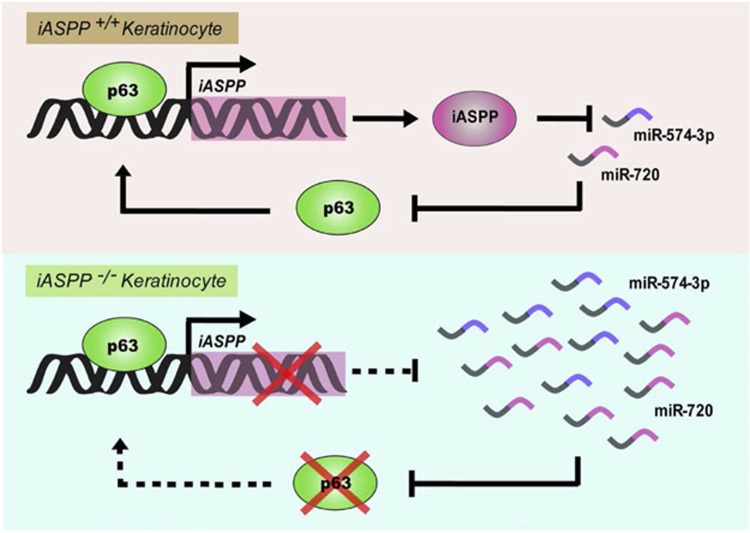
Figure 3.
iASPP regulates the proliferation/differentiation switch in keratinocytes by regulating a miRs/p63 axis. Under normal conditions, p63 regulates iASPP expression, which represses the expression of miR-574-3p and miR-720. miR-574-3p and miR-720 repress p63 expression, providing a feedback loop, which maintains the proliferation/differentiation balance. iASPP knockout keratinocytes demonstrate defective differentiation programmes that are associated with an upregulation in the expression of these miRs and the inhibition of p6312
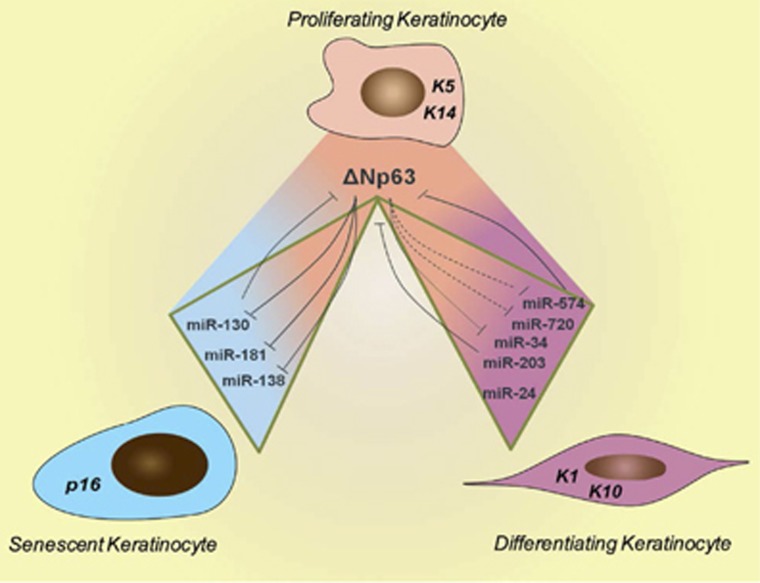
Figure 4.
Senescence and differentiation of keratinocytes are regulated by a complex ΔNp63/miRs network. ΔNp63 is highly expressed in proliferating keratinocytes (K5 and K14 positive). Upon differentiation or the initiation of the senescence programme, ΔNp63 expression is progressively downregulated. During differentiation the miR-574, miR-720, miR-34 and miR-203 expression levels are progressively upregulated. In this context, miR-574, miR-720 and miR-203 directly target ΔNp63, repressing its expression, and ΔNp63 maintains the proliferative ability of the cell by repressing the expression of miR-574, miR-720 and mir-34a. During senescence, the miR-130, miR-181 and miR-138 expression levels are upregulated. miR-130/ΔNp63 reciprocally repress the expression of each other. ΔNp63 also represses the expression of the pro-senescence miRs miR-181 and miR-138. The fine balance of this complex network regulates the keratinocyte fate
Figure 5.
miR-24 promotes keratinocyte differentiation by repressing pro-proliferation factors and actin cytoskeleton modulators. In wt mice, miR-24 expression is confined to the suprabasal layers, where it represses the expression of Myc and E2F2, which determine when the cell exits the cell cycle, and PAK4, ArhGAP19 and Tks5, which promote actin cytoskeleton rearrangement. miR-24-dependent cell cycle exit and actin cytoskeleton rearrangement promote the proliferation/differentiation switch. In Tg K5::miR-24 mice, the ectopic expression of miR-24 in basal keratinocytes promotes early cell cycle exit and actin cytoskeleton rearrangement, which accelerates differentiation. The premature differentiation is confirmed by the expression of suprabasal markers in the basal layer. Tg K5::miR-24 mice demonstrate a severe body weight loss and have a thinner epidermis. Although the normal epidermal architecture is maintained, the epidermal compartments have a reduced number of cell layers38
In addition to these pro-differentiation miRs, miR-205 has been shown to have a pivotal role in the expansion of the SC population. The genetic deletion of miR-205 results in neonatal lethality and severely compromised epidermal and hair follicle growth. miR-205−/− epidermal SCs prematurely exit the cell cycle and undergo transition towards quiescence.41 An expression profile analysis of miR-205−/− HF–SCs revealed a downregulation in the PI(3)K/Akt pathway activity, which stresses the importance of this pathway in sustaining the SC cell cycle. Therefore, miR-205 has a role in the expansion of the SC population, and its downregulation results in the transition to quiescence and towards differentiation.
miR-p63 Loops in Epidermal Senescence
Epidermal tissue needs to be continuously renewed throughout life, and keratinocyte stem cells contribute to normal tissue homeostasis and regeneration in response to injury or stress.42 There is a decline in the regenerative potential of the tissue that occurs with age that may be due to the result of the intrinsic ageing of the stem cells overtime. The accumulation of senescent cells over time most likely reduces the tissue regenerative capability and contributes to the physiological ageing of the tissue.43, 44 In vitro, senescence can result from multiple types of cellular stress, such as overproduction of ROS, oncogene activation, telomere shortening and by simply terminating the replicative potential, which is known as replicative senescence. In tissues with a high turnover rate, such as the epidermis, replicative senescence has a crucial role in regulating time-dependent changes, and several studies have demonstrated that miRs are intimately involved in these processes. p63 has a crucial role in counteracting ageing/senescence in vitro and in vivo. Furthermore, p63 and p63-isoform specific depletions induce premature ageing in different mice models.28, 45 An interesting correlation between the p63 expression levels and cellular senescence and/or ageing has also been demonstrated in human keratinocytes.46, 47 Most notably, a genomic-wide miR profiling study has shown that several miRs (miR-138, miR-181a, miR-181b, miR-191 and miR130b) are significantly upregulated in senescence human keratinocytes compared with proliferating keratinocytes. Of these identified miRs, miR-130a, miR-181a, miR-181b and miR-138 target the 3′UTRs of p63 and SIRT1, which are the molecules that are expressed in the basal layer and are strongly associated with keratinocyte proliferation and the maintenance of epidermal progenitor cells. The importance of miR-181a, miR181, miR-138 and miR-130a and their targets, p63 and SIRT1, has also been studied in skin ageing in vivo. A study evaluating groups of healthy young (<10 years old) and older (>60 years old) human subjects showed that the senescence-associated miRs were significantly upregulated in aged skin, which paralleled a significant downregulation in their targets p63 and SIRT1.46, 47 The importance of p63 in regulating the epidermal differentiation/proliferation programme is eloquently illustrated in two recent papers that demonstrate that p63 transcriptionally controls DNA- and chromatin-remodelling factors (Brg1 and Satb148, 49). In mouse epidermal progenitor cells, p63 directly regulates the transcription of the ATP-dependent chromatin remodeller Brg1 and the genome organiser Satb1. Both factors bind to distinct domains within the epidermal differentiation complex, which is required for epidermal morphogenesis. Interestingly, the senescence-associated miR-191 targets the Satb1 3′UTR and links miRs to the remodelling of higher-order chromatin structures.48, 50
miRs and p63 in Cancer
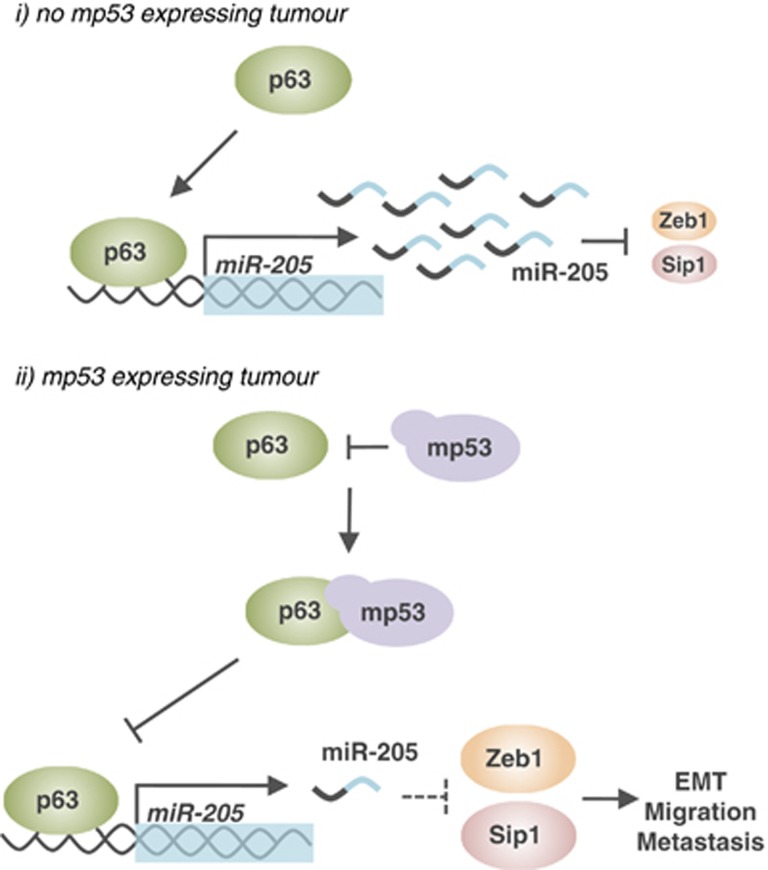
ΔNp63 lacking the transactivation domain TA can function as a dominant negative protein of TAp63 and TAp73 and inhibit their function in cell cycle arrest and apoptosis.51 Mutations in p63 are extremely rare in human cancers. Most head and neck squamous cell carcinomas, and other squamous cell epithelial malignancies and non-small-cell lung cancer not only retain p63 expression, but are also often overexpressed and occasionally amplified.52, 53, 54 An in vivo mouse model showed that established squamous cell carcinomas (SCCs) are dependent on p63; the acute genetic ablation of p63 rapidly induced apoptosis and tumour regression.55 The p63-targeting miR miR-203 has an important role as a tumour suppressor in different cancer types, including chronic myelogenous leukaemia. In chronic myelogenous leukaemia, miR-203 represses the expression of the oncoprotein c-Abl, which is often involved in the genomic translocation of the Philadelphia chromosome (BCR-ABL).56 miR-203 also acts as a metastasis suppressor in epithelial cancers. miR-203 is sufficient to induce a mesenchymal to epithelial transition with inhibition of cell proliferation, migration and invasiveness in brain and bone metastatic prostate cell lines.57 Another miR that is also involved in metastasis is miR-205. In prostate cancer, miR-205 has been shown to affect p63 metastasis suppressor function through direct transcriptional regulation. miR-205 is essential for the inhibitory effects of p63 on markers of epithelial–mesenchymal transition (EMT), such as ZEB1 and vimentin. p53 mutants inhibit the expression of p63 and miR-205, which promotes cell migration and metastasis. Clinical data have shown that metastasising human prostate cancer cells demonstrate a loss of one or both components of the p63/miR-205 axis (Figure 6).58 p63 has also been shown to regulate the expression of miR-155, thus promoting tumour suppression and metastasis suppression in epithelial cancer cell lines.59 miR-34a is also involved in a wide interplay with the p53 family. miR-34a is a well-established target and mediator of p53 function with a similar context-dependent involvement in growth arrest, senescence, and apoptosis.60 p63 transcriptionally represses the expression of miR-34a, which reduces the proliferation of primary keratinocytes. The deletion of p63 in human keratinocytes increased the miR-34a and miR-34c expression levels and inhibited the expression of cell cycle regulators (cyclin D1 and Cdk4) with a concomitant G1-phase arrest.61 miR-34 also promotes the differentiation of different cell types, including neurons62, 63 and keratinocytes.64 Consistent with this function, miR-34a is frequently repressed in SCCs. Reversing genomic DNA methylation restores miR-34a expression and promotes the differentiation of malignant keratinocytes.64 Among the oncogenic miRs miR-21 is one of the most commonly overexpressed miRs in carcinomas, including prostate, breast, pancreas, colon, head and neck.65 Knockout of miR-21 in mice revealed that this miR has a direct role in promoting epidermal tumourigenesis. miR-21-null mice demonstrated a significant reduction in skin papilloma formation in chemical-induced carcinogenesis experiments. The upregulation of Spry1, PTEN and Pdcd4 was observed in miR-21−/− keratinocytes, which is associated with reduced activation of the major downstream effectors of RAS signalling, which is fundamental for skin carcinogenesis initiation.66 miR-21 also targets Grhl3, a transcription factor that functions as a tumour suppressor in epidermal SCC. miR-21 activates a proto-oncogenic network that represses Grhl3 expression and results in a loss of PTEN expression, which is a direct GRHL3 target. miR-21 activation results in an aggressive SCC induced by PTEN repression and the consequent activation of PI3K/AKT/mTOR signalling.67 A recent study has illustrated the importance of K15-positive epidermal SCs (bulge cells) and miRs in the tumourigenesis and metastasisation of epidermal cancer. A mouse model of SCC, which was generated by targeting two frequent genetic mutations found in human SCCs, oncogene KrasG12D activation and Smad4 deletion in mouse K15–expressing SCs, revealed a fundamental function of miR-9 in regulating the expansion of metastatic cancer stem cells. miR-9 is expressed in metastatic human SSCs and activates the invasive ability of SCC cells repressing α-catenin expression.68 Regulating the expression of these miRs (Table 2) in tumour cells should be considered for use in novel therapeutic strategies in epidermal cancers.
Figure 6.
p63/miR-205 axis inhibits EMT, migration and metastasis. (i) When the p63/miR-205 axis is functional, p63 transcriptionally promotes the expression of miR-205, which negatively regulates the expression levels of the pro-EMT factors Zeb1 and Sip1. These events inhibit EMT, migration and metastasis of cancer cells. (ii) When mutant p53 (mp53) is expressed, it negatively regulates p63 activity. This expression impairs the p63/miR-205 axis, which results in Zeb1 and Sip1 accumulation and increased EMT, migration and metastasis
Table 2. MiRs involved in epidermal cancer.
| miR | Expression | Function | Target/s | Ref |
|---|---|---|---|---|
| miR-21 | Up | Cell cycle progression | PTEN, PCDC4, GRHL3 | 66, 67 |
| miR-34a | Down | Cell cycle progression | Cyc D1, Cdk4, Sirt6 | 61, 64 |
| miR-203 | Down | Cell cycle progression | p63, cJun | 70 |
| miR-9 | Up | Metastasis | α-Catenin | 68 |
| Let-7 | Down | Cell cycle progression | KRas | 71 |
| miR-124 | Down | Cell cycle progression | ERK2 | 72 |
| miR-214 | Down | Cell cycle progression | ERK1 | 72 |
Concluding Remarks
p63 acts as a central hub in epidermal biology. Modulating p63 activity alters the balance between keratinocyte stemness, differentiation and senescence. Several miRs are crucially involved in this network and act as either downstream or upstream regulators of p63 function. Furthermore, the transcriptional regulation of the miR processor complex (DGCR8 by ΔNp63 and Dicer by TAp63) by p63 further illustrates the importance and complexity of the p63-miR network. Characterisation of isoform-specific p63 knockout mice has not definitively solidified our understanding of the roles of the various p63 isoforms in these biological processes. The recent generation of ΔNp63 selective knockouts has instead illustrated the complexity of this network. For example, TAp63 has only a minor role in epidermal development, but contributes to the maintenance of adult skin stem cells and senescence. It appears that both p63 isoforms must be precisely regulated for epidermal homeostasis. How are the functions of these two isoforms integrated? Do their functions partially overlap or counteract each other or do they function in parallel processes?
The involvement of miRs in these phenomena stresses the complexity of this process. A single miR can simultaneously affect dozens of target genes, which can result in a wide change in the expression profile. The molecular dissection of these signalling pathways is fundamental for fully elucidating these processes. This aspect is particularly relevant for epidermal diseases. Alternative and innovative therapeutic strategies for skin cancer may be developed by targeting miR expression. Answering these and other questions will provide the future research in this field with important challenges.
Acknowledgments
This study was supported by the Medical Research Council, UK, by MIUR, MinSan/IDI-IRCCS (RF73, RF57) and by the Italian Association for Cancer Research (AIRC) investigator grants awarded to GM.
Glossary
- miR
microRNA
- IFE
interfollicular epidermis
- HFs
hair follicles
- SG
sebaceous glands
- SC
stem cell
- PAK
p21-activated kinases
- IPSC
induced pluripotent stem cell
- MEF
mouse embryonic fibroblast
- ECM
extracellular matrix
- K
keratin
Footnotes
Edited by RA Knight
References
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, et al. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci USA. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, O'Carroll D, Pasolli HA, Zhang Z, Dietrich FS, Tarakhovsky A, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- Teta M, Choi YS, Okegbe T, Wong G, Tam OH, Chong MM, et al. Inducible deletion of epidermal Dicer and Drosha reveals multiple functions for miRNAs in postnatal skin. Development. 2012;139:1405–1416. doi: 10.1242/dev.070920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, et al. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- Jaks V, Barker N, Kasper M, van Es JH, Snippert HJ, Clevers H, et al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–1299. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- Ito M, Liu Y, Yang Z, Nguyen J, Liang F, Morris RJ, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- Levy V, Lindon C, Harfe BD, Morgan BA. Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev Cell. 2005;9:855–861. doi: 10.1016/j.devcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Ghazizadeh S, Taichman LB. Organization of stem cells and their progeny in human epidermis. J Invest Dermatol. 2005;124:367–372. doi: 10.1111/j.0022-202X.2004.23599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhof JG, Braun KM, Giangreco A, van Pelt C, Kawamoto H, Boyd RL, et al. The cell-surface marker MTS24 identifies a novel population of follicular keratinocytes with characteristics of progenitor cells. Development. 2006;133:3027–3037. doi: 10.1242/dev.02443. [DOI] [PubMed] [Google Scholar]
- Jensen UB, Yan X, Triel C, Woo SH, Christensen R, Owens DM. A distinct population of clonogenic and multipotent murine follicular keratinocytes residing in the upper isthmus. J Cell Sci. 2008;121:609–617. doi: 10.1242/jcs.025502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- Adorno M, Cordenonsi M, Montagner M, Dupont S, Wong C, Hann B, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- Guo X, Keyes WM, Papazoglu C, Zuber J, Li W, Lowe SW, et al. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009;11:1451–1457. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Chakravarti D, Flores ER. p63 steps into the limelight: crucial roles in the suppression of tumorigenesis and metastasis. Nat Rev Cancer. 2013;13:136–143. doi: 10.1038/nrc3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanbokhoven H, Melino G, Candi E, Declercq W. p63, a story of mice and men. J Invest Dermatol. 2011;131:1196–1207. doi: 10.1038/jid.2011.84. [DOI] [PubMed] [Google Scholar]
- Lena AM, Cipollone R, Amelio I, Catani MV, Ramadan S, Browne G, et al. Skn-1a/Oct-11 and DeltaNp63alpha exert antagonizing effects on human keratin expression. Biochem Biophys Res Commun. 2010;401:568–573. doi: 10.1016/j.bbrc.2010.09.102. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronson RT, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–718. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- Shalom-Feuerstein R, Lena AM, Zhou H, De La Forest Divonne S, Van Bokhoven H, Candi E, et al. DeltaNp63 is an ectodermal gatekeeper of epidermal morphogenesis. Cell Death Differ. 2011;18:887–896. doi: 10.1038/cdd.2010.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong AB, Kretz M, Ridky TW, Kimmel R, Khavari PA. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 2006;20:3185–3197. doi: 10.1101/gad.1463206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarti D, Su X, Cho MS, Bui NH, Coarfa C, Venkatanarayan A, et al. Induced multipotency in adult keratinocytes through down-regulation of DeltaNp63 or DGCR8. Proc Natl Acad Sci USA. 2014. [DOI] [PMC free article] [PubMed]
- Romano RA, Smalley K, Magraw C, Serna VA, Kurita T, Raghavan S, et al. DeltaNp63 knockout mice reveal its indispensable role as a master regulator of epithelial development and differentiation. Development. 2012;139:772–782. doi: 10.1242/dev.071191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster MI, Dai D, Marinari B, Sano Y, Costanzo A, Karin M, et al. p63 induces key target genes required for epidermal morphogenesis. Proc Natl Acad Sci USA. 2007;104:3255–3260. doi: 10.1073/pnas.0611376104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Paris M, Gi YJ, Tsai KY, Cho MS, Lin YL, et al. TAp63 prevents premature aging by promoting adult stem cell maintenance. Cell Stem Cell. 2009;5:64–75. doi: 10.1016/j.stem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova EM, Petrenko O, Nemajerova A, Romano RA, Sinha S, Moll UM. DeltaNp63 regulates select routes of reprogramming via multiple mechanisms. Cell Death Differ. 2013;20:1698–1708. doi: 10.1038/cdd.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medawar A, Virolle T, Rostagno P, de la Forest-Divonne S, Gambaro K, Rouleau M, et al. DeltaNp63 is essential for epidermal commitment of embryonic stem cells. PLoS One. 2008;3:e3441. doi: 10.1371/journal.pone.0003441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, Aberdam D, Knight RA, Melino G, et al. miR-203 represses 'stemness' by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing 'stemness'. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SJ, Zhang Z, Feng D, Flagg M, O'Loughlin E, Wang D, et al. Rapid and widespread suppression of self-renewal by microRNA-203 during epidermal differentiation. Development. 2013;140:1882–1891. doi: 10.1242/dev.089649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikh A, Matin RN, Senatore V, Hufbauer M, Lavery D, Raimondi C, et al. iASPP/p63 autoregulatory feedback loop is required for the homeostasis of stratified epithelia. EMBO J. 2011;30:4261–4273. doi: 10.1038/emboj.2011.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I, Lena AM, Viticchie G, Shalom-Feuerstein R, Terrinoni A, Dinsdale D, et al. miR-24 triggers epidermal differentiation by controlling actin adhesion and cell migration. J Cell Biol. 2012;199:347–363. doi: 10.1083/jcb.201203134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amelio I, Lena AM, Bonanno E, Melino G, Candi E. miR-24 affects hair follicle morphogenesis targeting Tcf-3. Cell Death Dis. 2013;4:e922. doi: 10.1038/cddis.2013.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JT, Gautrot JE, Trappmann B, Tan DW, Donati G, Huck WT, et al. Actin and serum response factor transduce physical cues from the microenvironment to regulate epidermal stem cell fate decisions. Nat Cell Biol. 2010;12:711–718. doi: 10.1038/ncb2074. [DOI] [PubMed] [Google Scholar]
- Wang D, Zhang Z, O'Loughlin E, Wang L, Fan X, Lai EC, et al. MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat Cell Biol. 2013;15:1153–1163. doi: 10.1038/ncb2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G. The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol. 2005;6:328–340. doi: 10.1038/nrm1619. [DOI] [PubMed] [Google Scholar]
- Satyanarayana A, Rudolph KL. p16 and ARF: activation of teenage proteins in old age. J Clin Invest. 2004;114:1237–1240. doi: 10.1172/JCI23437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Keyes WM, Wu Y, Vogel H, Guo X, Lowe SW, Mills AA. p63 deficiency activates a programme of cellular senescence and leads to accelerated aging. Genes Dev. 2005;19:1986–1999. doi: 10.1101/gad.342305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivetti di Val Cervo P, Lena AM, Nicoloso M, Rossi S, Mancini M, Zhou H, et al. p63-microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci USA. 2012;109:1133–1138. doi: 10.1073/pnas.1112257109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordisco S, Maurelli R, Bondanza S, Stefanini M, Zambruno G, Guerra L, et al. Bmi-1 reduction plays a key role in physiological and premature aging of primary human keratinocytes. J Invest Dermatol. 2010;130:1048–1062. doi: 10.1038/jid.2009.355. [DOI] [PubMed] [Google Scholar]
- Fessing MY, Mardaryev AN, Gdula MR, Sharov AA, Sharova TY, Rapisarda V, et al. p63 regulates Satb1 to control tissue-specific chromatin remodeling during development of the epidermis. J Cell Biol. 2011;194:825–839. doi: 10.1083/jcb.201101148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardaryev AN, Gdula MR, Yarker JL, Emelianov VN, Poterlowicz K, Sharov AA, et al. p63 and Brg1 control developmentally regulated higher-order chromatin remodelling at the epidermal differentiation complex locus in epidermal progenitor cells. Development. 2014;141:101–111. doi: 10.1242/dev.103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lena AM, Mancini M, Rivetti di Val Cervo P, Saintigny G, Mahe C, Melino G, et al. MicroRNA-191 triggers keratinocytes senescence by SATB1 and CDK6 downregulation. Biochem Biophys Res Commun. 2012;423:509–514. doi: 10.1016/j.bbrc.2012.05.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- Massion PP, Taflan PM, Jamshedur Rahman SM, Yildiz P, Shyr Y, Edgerton ME, et al. Significance of p63 amplification and overexpression in lung cancer development and prognosis. Cancer Res. 2003;63:7113–7121. [PubMed] [Google Scholar]
- DeYoung MP, Johannessen CM, Leong CO, Faquin W, Rocco JW, Ellisen LW. Tumor-specific p73 up-regulation mediates p63 dependence in squamous cell carcinoma. Cancer Res. 2006;66:9362–9368. doi: 10.1158/0008-5472.CAN-06-1619. [DOI] [PubMed] [Google Scholar]
- Hibi K, Trink B, Patturajan M, Westra WH, Caballero OL, Hill DE, et al. AIS is an oncogene amplified in squamous cell carcinoma. Proc Natl Acad Sci USA. 2000;97:5462–5467. doi: 10.1073/pnas.97.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey MR, Wilson C, Ory B, Rothenberg SM, Faquin W, Mills AA, et al. FGFR2 signaling underlies p63 oncogenic function in squamous cell carcinoma. J Clin Invest. 2013;123:3525–3538. doi: 10.1172/JCI68899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC, et al. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Viticchie G, Lena AM, Latina A, Formosa A, Gregersen LH, Lund AH, et al. MiR-203 controls proliferation, migration and invasive potential of prostate cancer cell lines. Cell Cycle. 2011;10:1121–1131. doi: 10.4161/cc.10.7.15180. [DOI] [PubMed] [Google Scholar]
- Tucci P, Agostini M, Grespi F, Markert EK, Terrinoni A, Vousden KH, et al. Loss of p63 and its microRNA-205 target results in enhanced cell migration and metastasis in prostate cancer. Proc Natl Acad Sci USA. 2012;109:15312–15317. doi: 10.1073/pnas.1110977109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiske S, Ho K, Noll JE, Neilsen PM, Callen DF, Suetani RJ. TAp63 regulates oncogenic miR-155 to mediate migration and tumour growth. Oncotarget. 2013;4:1894–1903. doi: 10.18632/oncotarget.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Antonini D, Russo MT, De Rosa L, Gorrese M, Del Vecchio L, Missero C. Transcriptional repression of miR-34 family contributes to p63-mediated cell cycle progression in epidermal cells. J Invest Dermatol. 2010;130:1249–1257. doi: 10.1038/jid.2009.438. [DOI] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Steinert JR, Shalom-Feuerstein R, Rouleau M, Aberdam D, et al. microRNA-34a regulates neurite outgrowth, spinal morphology, and function. Proc Natl Acad Sci USA. 2011;108:21099–21104. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M, Tucci P, Killick R, Candi E, Sayan BS, Rivetti di Val Cervo P, et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci USA. 2011;108:21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort K, Brooks Y, Ostano P, Cario-Andre M, Calpini V, Guinea-Viniegra J, et al. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J. 2013;32:2248–2263. doi: 10.1038/emboj.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Galasso M, Costinean S, Tagliavini L, Gamberoni G, Drusco A, et al. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Kumar M, Choudhury SN, Becker Buscaglia LE, Barker JR, Kanakamedala K, et al. Loss of the miR-21 allele elevates the expression of its target genes and reduces tumorigenesis. Proc Natl Acad Sci USA. 2011;108:10144–10149. doi: 10.1073/pnas.1103735108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darido C, Georgy SR, Wilanowski T, Dworkin S, Auden A, Zhao Q, et al. Targeting of the tumor suppressor GRHL3 by a miR-21-dependent proto-oncogenic network results in PTEN loss and tumorigenesis. Cancer Cell. 2011;20:635–648. doi: 10.1016/j.ccr.2011.10.014. [DOI] [PubMed] [Google Scholar]
- White RA, Neiman JM, Reddi A, Han G, Birlea S, Mitra D, et al. Epithelial stem cell mutations that promote squamous cell carcinoma metastasis. J Clin Invest. 2013;123:4390–4404. doi: 10.1172/JCI65856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B, Blanpain C. Mechanisms regulating epidermal stem cells. EMBO J. 2012;31:2067–2075. doi: 10.1038/emboj.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E, Loven J, Xu N, Meisgen F, Wei T, Brodin P, et al. MicroRNA-203 functions as a tumor suppressor in basal cell carcinoma. Oncogenesis. 2012;1:e3. doi: 10.1038/oncsis.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffelfinger C, Ouyang Z, Engberg A, Leffell DJ, Hanlon AM, Gordon PB, et al. Correlation of global microRNA expression with basal cell carcinoma subtype. G3 (Bethesda) 2012;2:279–286. doi: 10.1534/g3.111.001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Jinnin M, Etoh T, Kobayashi Y, Shimozono N, Fukushima S, et al. Down-regulation of miR-124/-214 in cutaneous squamous cell carcinoma mediates abnormal cell proliferation via the induction of ERK. J Mol Med (Berl) 2013;91:69–81. doi: 10.1007/s00109-012-0935-7. [DOI] [PubMed] [Google Scholar]