Abstract
The transcription factor p53 regulates the expression of genes crucial for biological processes such as cell proliferation, metabolism, cell repair, senescence and apoptosis. Activation of p53 also suppresses neoplastic transformations, thereby inhibiting the growth of mutated and/or damaged cells. p53-binding proteins, such as mouse double minute 2 homolog (MDM2), inhibit p53 activation and thus regulate p53-mediated stress responses. Here, we found that long glucocorticoid-induced leucine zipper (L-GILZ), a recently identified isoform of GILZ, activates p53 and that the overexpression of L-GILZ in p53+/+ HCT116 human colorectal carcinoma cells suppresses the growth of xenografts in mice. In the presence of both p53 and MDM2, L-GILZ binds preferentially to MDM2 and interferes with p53/MDM2 complex formation, making p53 available for downstream gene activation. Consistent with this finding, L-GILZ induced p21 and p53 upregulated modulator of apoptosis (PUMA) expression only in p53+/+ cells, while L-GILZ silencing reversed the anti-proliferative activity of dexamethasone as well as expression of p53, p21 and PUMA. Furthermore, L-GILZ stabilizes p53 proteins by decreasing p53 ubiquitination and increasing MDM2 ubiquitination. These findings reveal L-GILZ as a regulator of p53 and a candidate for new therapeutic anti-cancer strategies for tumors associated with p53 deregulation.
By inducing cell cycle arrest, the tumor suppressor gene TP53 repairs damage caused by multiple physiological and pathological stressors.1, 2 The p53 transcription factor is activated by any stressor that threatens the integrity of the genome.3, 4 Once activated, p53 induces the transcription of genes that regulate cell cycle, senescence and apoptosis.1, 5 A critical role for p53 in preventing tumor development is supported by observations that the gene is mutated in 50% of human cancers.6 Therefore, it appears that cancerous cells grow and metastasize by escaping p53 regulation, whereas normal cells maintain low levels of p53 via feedback mechanisms involving partner molecules that monitor p53 expression.7 One critical molecule in the p53 regulation network is mouse double minute 2 homolog (MDM2), an E3 ubiquitin (Ub) ligase that modulates the strength, timing and rapidity of p53 downstream signals and thus enables appropriate biological responses. In addition, p53 upregulates the transcription of MDM2, which in turn downregulates p53 activity through multiple mechanisms.8 By binding to a p53 transactivation domain, MDM2 interferes with p53-mediated transcription, promotes p53 nuclear export and induces Ub-mediated proteasomal p53 degradation.9, 10, 11, 12 This tightly controlled mechanism helps maintain physiological levels of p53 and restores these levels following stress responses.
Glucocorticoids (GCs) exert major anti-inflammatory/immunosuppressive effects and act as anti-tumor agents in the treatment of several hematologic malignancies and solid tumors.13 Most GC-elicited effects result from the transcriptional regulation of GC receptor (GR) target genes.14 Among these genes, GCs increase the transcription of GC-induced leucine zipper (GILZ)15, 16 and its transcriptional variant long (L)-GILZ.17 GILZ mediates several anti-inflammatory and immunomodulatory GC functions.18, 19, 20, 21, 22, 23 GILZ and L-GILZ bind Ras and contribute to cell differentiation and the control of tumorigenesis.24, 25
Crosstalk between p53 and GCs has been described as either antagonistic or synergistic depending on cell type and environmental context.26 Some reports suggest that p53 activation is not necessary for GC-induced apoptotic cell death27 but may instead increase resistance to GC therapy.26 GCs may also rescue cells from p53-induced apoptosis,28 suggesting an antagonistic relationship between GCs and p53. Furthermore, p53 and GRs have been shown to physically interact and form a complex that becomes sequestered within the cytosol, leading to inhibited transcription of both genes.29 The formation of a p53/MDM2/GR triple complex may account for the proteasomal degradation of p53 induced by dexamethasone (DEX);26 however, in other experimental models, p53- and GC-mediated cell cycle arrest involves the upregulation of cyclin-dependent kinase inhibitor p21, and it has been suggested that p53 cooperates in GR-induced p21 upregulation and the consequent inhibition of cell proliferation.30 Moreover, experiments using antisense p53 oligonucleotides show that blocking p53 expression reverses DEX-induced p21 upregulation and inhibition of proliferation.31 Furthermore, p53 was recently found to be involved in GR-mediated repression of nuclear factor-κB transcription.32
Most of these functional interactions between p53 and GCs involve GRs; yet little is known about functional interactions between p53 and GC-regulated proteins. Therefore, we investigated the effect of L-GILZ and GILZ on p53 activation. We observed that L-GILZ, but not GILZ, binds and activates p53. Therefore, our study revealed that L-GILZ is a regulator of p53 activation.
Results
L-GILZ but not GILZ directly binds p53
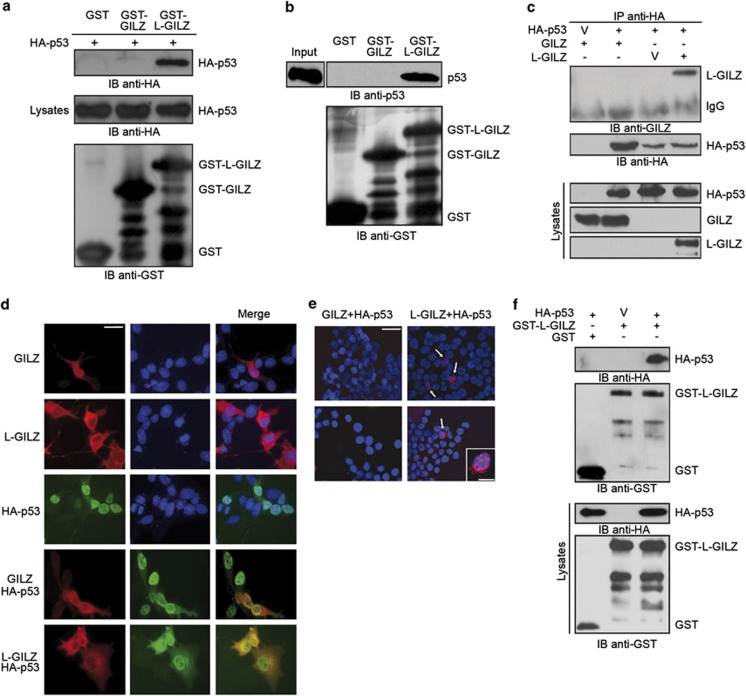
To determine whether GILZ and/or L-GILZ binds p53, we first conducted glutathione S-transferase (GST) pull-down experiments using GST–GILZ, GST–L-GILZ, or GST alone fusion proteins. Immobilized fusion proteins were incubated with cellular lysates from p53−/− HCT116 colon cancer cells transfected with HA-tagged-p53 (HA–p53), and the composition of complexes was examined. We found that GST–L-GILZ, but not GST–GILZ or GST alone bound p53 (Figure 1a). Because p53−/− cell lysates overexpressing p53 may contain other proteins that bridge L-GILZ and p53, we repeated the same experiment using in vitro translated p53. Again, GST–L-GILZ, but not GST–GILZ or GST alone, bound to in vitro translated p53 (Figure 1b), demonstrating a direct p53/L-GILZ interaction.
Figure 1.
L-GILZ interacts with p53. (a) Cell lysates of HA–p53-transfected p53−/− HCT116 cells were incubated with GST, GST–GILZ or GST–L-GILZ attached to glutathione–sepharose beads. Proteins bound to the resin were eluted, resolved by SDS-PAGE, and subjected to Western blot using anti-HA antibody. Immunoblot (IB) of immobilized bait proteins that were incubated in the absence of target protein shows equal amounts of GST-tagged proteins. IB of lysates shows comparable p53 transfection levels across groups. (b) The in vitro transcription/translation product of full-length wild-type p53 was incubated with GST, GST–GILZ or GST–L-GILZ attached to glutathione–sepharose beads. Proteins bound to the resin were eluted and analyzed by Western blot using anti-p53 antibody. ‘Input' indicates 10% volume of the in vitro translated product used in the pull-down assay. IB of GST-tagged proteins was performed to show that equal amounts of immobilized bait proteins were used. (c) p53−/− HCT116 cells were co-transfected with pcDNA3-GILZ or pcDNA3-L-GILZ together with HA–p53 vector. Immunoprecipitation (IP) was performed with anti-HA antibody, and immunoreactive proteins were revealed with anti-GILZ or anti-HA antibodies. V, vector. Whole-cell lysates were loaded to control for plasmid expression (lower panels). (d) Microscopic analysis of GILZ/L-GILZ (red) and p53 (green) localization in GILZ-, L-GILZ-, HA–p53-, GILZ/ HA–p53-, or L-GILZ/ HA–p53-transfected p53−/− HCT116 cells. DAPI (blue) was used as a nuclear counterstain. Right panels show merged images. Scale bar represents 30 μm. (e) In situ PLA in p53−/− HCT116 cells transfected with HA–p53 together with GILZ or L-GILZ. Cells were fixed and processed by incubating with a goat anti-GILZ and mouse anti-HA antibody mixture, followed by an anti-goat PLA-plus probe for GILZ and L-GILZ and anti-mouse PLA-minus probe for HA. Representative merged images from two experiments with the PLA (red) and DAPI (blue) channels are shown. No signals were observed in non-transfected or single-transfected cells (not shown). Positive cells are indicated by arrows. Scale bar represents 30 μm. The insert shows a single cell at 100 × magnification. Scale bar represents 10 μm in the insert. (f) H1299 cells were co-transfected with GST–L-GILZ or GST (in pEBG) together with either HA–p53 or HA–empty vector. GST–L-GILZ and GST were purified by glutathione–sepharose resin. Co-purified p53 was detected by anti-HA antibody, and purified GST–L-GILZ and GST were detected by anti-GST antibody. Lysates were analyzed by Western blot using anti-GST or anti-HA antibodies
We next examined this interaction in vivo. The p53−/− HCT116 cells were transiently co-transfected with HA–p53 vector together with an expression vector carrying either L-GILZ or GILZ. Immunoblot analysis of HA–p53 immunoprecipitates indicated the presence of L-GILZ but not GILZ (Figure 1c). Consistently, fluorescent microscopy revealed extensive co-localization of p53 and L-GILZ but not of p53 and GILZ (Figure 1d) in p53−/− cells overexpressing L-GILZ/HA–p53 or GILZ/HA–p53, respectively. In situ proximity ligation assay (PLA), an antibody-based method for the selective and highly sensitive detection of protein interactions,33 confirmed these observations. The signal, in the form of red spots, revealed that p53 interacts with L-GILZ but not GILZ (Figure 1e). This L-GILZ/p53 binding was also observed in p53-deficient H1299 cells transfected with these same vectors (Figure 1f).
L-GILZ binds p53 at its N-terminal domain
We next investigated the domain that mediates p53 binding to L-GILZ. GST–L-GILZ or GST alone (in pEBG vector) together with wild-type (full-length) or mutant (deleted) forms of p5334 (Figure 2a) were transfected into p53−/− HCT116 cells. We found that L-GILZ bound full-length p53, mutant 1 p53 (amino acids 1–355, deletion of the regulatory domain) and mutant 2 p53 (amino acids 1–298, deletion of the regulatory and oligomerization domains). By contrast, L-GILZ did not bind mutant 3 p53 (amino acids 94–298, deletion of regulatory, oligomerization and transactivation domains as well as part of the proline-rich region; Figure 2b). In situ PLA confirmed that L-GILZ interacts with full-length p53 and mutant 1 and 2 p53 but not with mutant 3 p53 (Figure 2c). Together, these results demonstrate that the regulatory N-terminal domain (amino acids 1–93) of p53 is necessary for L-GILZ/p53 binding. As MDM2 also binds p53 at its N-terminal domain,8 we next addressed the possibility that L-GILZ disturbs the interaction between p53 and MDM2.
Figure 2.
Mapping the L-GILZ-binding domains of p53. (a) Schematic illustration of full-length and mutant p53 constructs. TA, transactivation domain; PR, proline-rich region; DBD, DNA-binding domain; OD, oligomerization domain; RD, regulatory domain. (b) p53−/− HCT116 cells were co-transfected with GST–L-GILZ or GST (in pEBG) together with full-length or mutant p53 constructs. GST–L-GILZ and GST were purified by glutathione–sepharose resin. Co-purified full-length or mutant p53 was detected with an anti-p53 antibody (upper left), and purified GST–L-GILZ or GST was detected by anti-GST antibody (lower left). Lysates were analyzed by Western blot using anti-p53 (upper right) or anti-GST (lower right) antibodies to control for plasmid expression. (c) Complex formation was measured by in situ PLA in p53−/− HCT116 cells transiently transfected with L-GILZ together with full-length or mutant p53. Primary antibodies were goat anti-GILZ and rabbit anti-p53. Secondary antibodies were anti-goat PLA-plus probe for L-GILZ and anti-rabbit PLA-minus probe for p53. In situ PLA is indicated by red signals from rolling cycle amplification products. Positive cells are indicated by arrows. Scale bar represents 30 μm. Images are representative of three experiments with consistent results
L-GILZ interacts with MDM2 and disturbs p53/MDM2 complexes
First, we performed co-immunoprecipitation (co-IP) experiments to determine whether L-GILZ binds MDM2 using p53−/− HCT116 cells transfected with GST–L-GILZ, GST–GILZ or empty vector together with pCMV–MDM2. We found that lysates contained immunoreactive MDM2 when cells were co-transfected with GST–L-GILZ and pCMV–MDM2 but not when cells were transfected with GST–GILZ or empty vector along with pCMV–MDM2 (Figure 3a). The binding of L-GILZ and p53 was confirmed using in situ PLA (Figure 3b). Moreover, both the p53/L-GILZ and MDM2/L-GILZ interactions were also observed in p53/MDM2 double-knockout mouse embryo fibroblasts (MEFs) transfected with either p53 or MDM2 vectors together with L-GILZ (Figure 3c).
Figure 3.
L-GILZ interacts with MDM2 and disturbs p53/MDM2 complexes. (a) p53−/− HCT116 cells were co-transfected with GST alone or with GST–L-GILZ or GST–GILZ together with pCMV–MDM2 expression vector. After 24 h, GST-tagged proteins were purified by adsorption to glutathione–sepharose beads. In Western blots, co-purified proteins were revealed with anti-MDM2 antibody, and purified GST/GST–L-GILZ/GST–GILZ was detected with anti-GST antibody. Whole-cell lysates were loaded to control for plasmid expression (lower panel). (b) In situ PLA was used to confirm the interaction between L-GILZ or GILZ and p53. p53−/− HCT116 cells transiently transfected with L-GILZ or GILZ and pCMV–MDM2 expression vectors were treated with a mixture of goat anti-GILZ and mouse anti-MDM2 antibodies and then with anti-goat PLA-plus probe and anti-mouse PLA-minus probe. The signal (red spots) was analyzed by fluorescence microscopy. Positive cells are indicated by arrows. Scale bar represents 30 μm. (c) p53/MDM2 double-knockout MEFs were co-transfected with GST–L-GILZ or GST (in pEBG) together with either HA–p53 or pCMV–MDM2 expression vectors. GST–L-GILZ and GST were purified by glutathione–sepharose resin. Co-purified p53 and MDM2 were detected by anti-HA and anti-MDM2 antibodies, and purified GST–L-GILZ or GST was detected by anti-GST antibody. Lysates were analyzed by Western blot using anti-GST, anti-HA and anti-MDM2 antibodies. (d) Schematic illustration of full-length and mutant MDM2 constructs. p53BD, p53-binding domain; NLS, nuclear localization sequence; Acidic, acidic domain; Zn, Zn finger. (e) The in vitro transcription and translation products of full-length (FL) MDM2 or the deleted MDM2 mutants 4, 8 and 9 (M) were incubated with GST or GST–L-GILZ immobilized on glutathione–sepharose beads. Equal amounts of GST-tagged protein were used (not shown). The proteins bound to the resin were eluted, resolved by SDS-PAGE, and detected using anti-MDM2 antibodies. Input was 10% of the volume of the transcription and translation products used in the pull-down assays. The proteins were resolved by SDS-PAGE and revealed by anti-MDM2 antibodies. (f) p53−/− HCT116 cells were co-transfected with GST–L-GILZ or GST alone together with pCMV–MDM2 and/or HA–p53 expression vectors. Approximately 48 h after transfection, cells were treated with MG132 (25 μM) for 5 h. Protein complexes were purified by adsorption to glutathione–sepharose beads and analyzed via Western blot with anti-MDM2, anti-HA, or anti-GST antibodies. Whole-cell lysates were loaded to control for transfection levels (lower panels). V, vector. (g) Lysates from p53−/− HCT116 cells transiently transfected with the indicated expression vectors and then treated with MG132 (25 μM) for 5 h were immunoprecipitated with anti-HA antibodies, and immunocomplexes were analyzed by Western blot with anti-HA, anti-Myc or anti-MDM2 antibodies. Whole-cell lysates were loaded to control for transfection levels (lower panels). Images are representative of three experiments with consistent results
To identify the MDM2 domain that binds L-GILZ, we performed in vitro GST-pull-down experiments using GST–L-GILZ as a bait with in vitro translated MDM2 or deleted forms of MDM2, namely mutant 4 (6–200 aa), mutant 8 (150–490 aa) and mutant 9 (1–290 aa; Figure 3d). L-GILZ bound full-length, mutant 4 and mutant 9 MDM2, but not mutant 8 MDM2 that lacks the N-terminal region including the p53-binding domain35 (Figure 3e).
As our data suggest that the L-GILZ-binding domain of p53 and MDM2 includes the regions responsible for p53/MDM2 interaction, we characterized the molecular interactions among the three proteins. As expected, GST–L-GILZ bound MDM2 and p53 in lysates from p53−/− HCT116 cells transiently co-transfected with MDM2 and p53, respectively; however, when cells were simultaneously transfected with all three plasmids, GST–L-GILZ interacted with MDM2 but not with p53 (Figure 3f). These experiments demonstrate that L-GILZ mainly binds MDM2 in the presence of simultaneous overexpression of p53 and MDM2, suggesting that L-GILZ has a greater affinity for MDM2 than for p53.
Next, we tested whether L-GILZ disturbs p53/MDM2 complexes and thereby affects p53 activation. As expected, p53 bound MDM2 in lysates from cells co-transfected with HA–p53 and MDM2, but the simultaneous presence of L-GILZ prevented the formation of this complex (Figure 3g).
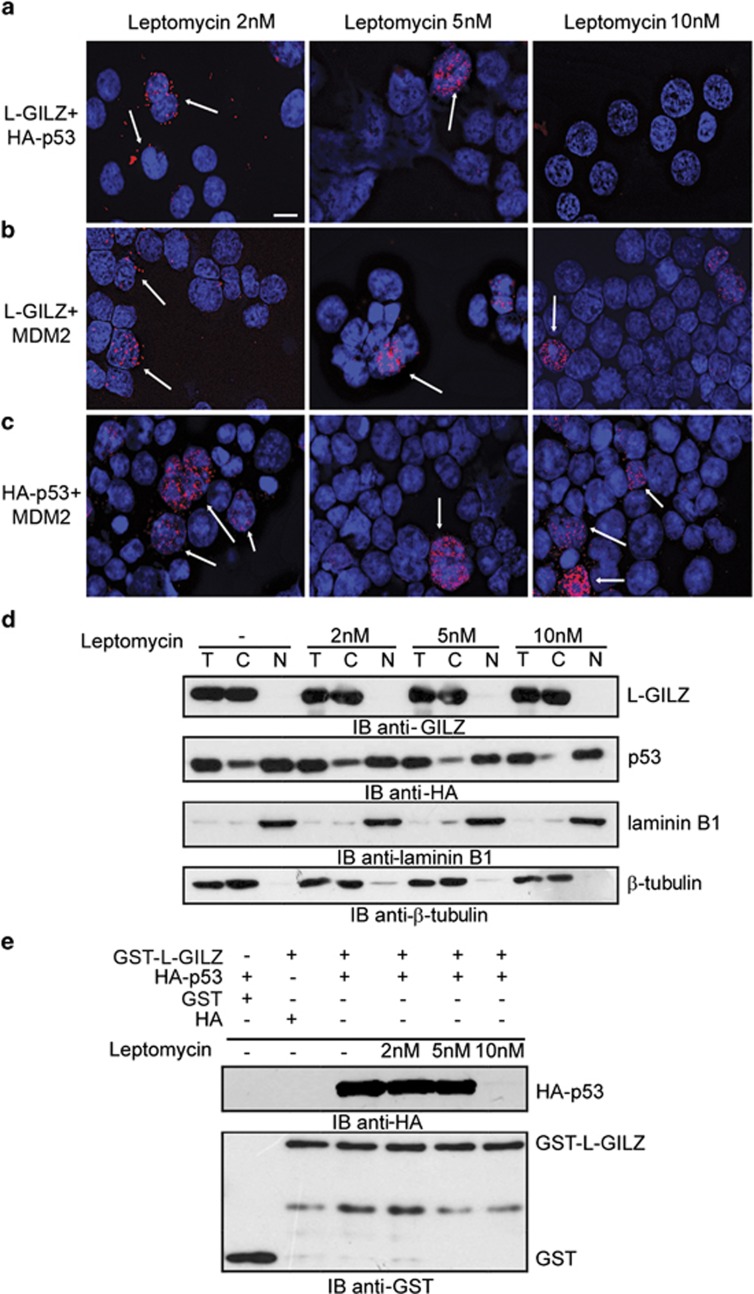
As nuclear–cytoplasmic shuttling is crucial for p53 function, we investigated whether the interaction among L-GILZ, p53 and MDM2 is affected by molecular shuttle inhibition. p53−/− HCT116 cells were transfected with L-GILZ together with p53 or MDM2 in the presence of increasing amounts of leptomycin, which reduces nuclear export by inhibiting the receptor chromosome region maintenance 1.36 In situ PLA revealed that the L-GILZ/p53 interaction was reduced by 10 nM leptomycin (Figure 4a), whereas no reduction of L-GILZ/MDM2 or p53/MDM2 co-localization was observed (Figures 4b and c). To further explore this observation, we separated cytoplasmic and nuclear proteins from GST–L-GILZ/Ha–p53-co-transfected p53−/− HCT116 cells that were treated with leptomycin to examine subcellular distributions of p53 and L-GILZ proteins. Western blot analysis of total, cytoplasmic and nuclear proteins of transfected cells showed cytoplasmic localization for GST–L-GILZ, while HA–p53 was mainly detected in the nucleus and to a lesser extent, in the cytoplasm; however, the highest dose of leptomycin inhibited the cytoplasmic localization of p53 but had no effect on L-GILZ localization (Figure 4d). As a consequence, the co-IP of cytoplasmic p53 and L-GILZ was inhibited by 10 nM leptomycin (Figure 4e). Therefore the binding of p53 and L-GILZ depends on p53 nuclear export.
Figure 4.
Leptomycin inhibits L-GILZ/p53 but not L-GILZ/MDM2 co-localization. p53−/− HCT116 cells transiently transfected with (a) L-GILZ/HA–p53, (b) L-GILZ/MDM2, or (c) HA–p53/MDM2 were treated with scalar concentrations of leptomycin and analyzed by in situ PLA. Positive cells are indicated by arrows. Scale bar represents 30 μm. (d) p53−/− HCT116 cells transiently transfected with GST–L-GILZ/HA–p53 were treated for 3 h with the indicated concentrations of leptomycin. Total lysates (T) as well as cytosolic (C) and nuclear (N) proteins were analyzed via Western blot using anti-GILZ, anti-HA, anti-laminin B1 (for nuclear protein detection) and anti-tubulin (for cytoplasmic protein detection) antibodies. (e) GST–L-GILZ and GST from the cytoplasmic fraction of GST–L-GILZ/HA–p53-transfected- p53−/− HCT116 cells (experiment shown in d) were purified by glutathione–sepharose resin. Western blot analysis with anti-HA antibody revealed the co-purified proteins, and anti-GST antibody was used to reveal the purified GST/GST–L-GILZ
L-GILZ activates the p53 signaling pathway and inhibits tumor cell growth
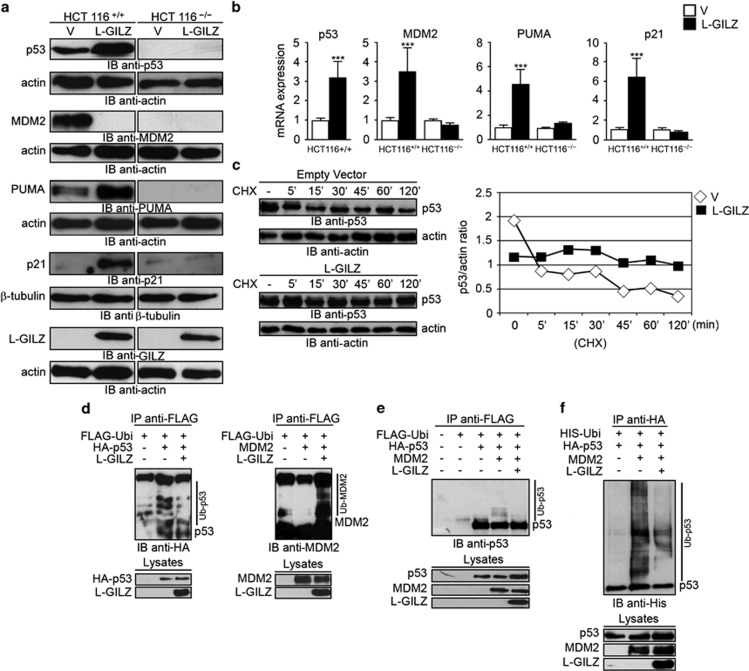
To verify whether L-GILZ induces p53 activation, we compared the expression of MDM2, p53, and its main target genes p21 and p53 upregulated modulator of apoptosis (PUMA) in p53+/+ and p53−/− HCT116 cells overexpressing L-GILZ. We observed increased expression of p53, p21 and PUMA and reduced expression of MDM2 in p53+/+ cells expressing L-GILZ but not in p53+/+ cells transfected with empty vector or in p53−/− cells (Figure 5a). Similar results were obtained by mRNA analysis using real-time PCR, although MDM2 expression was increased (Figure 5b). This apparent discrepancy may be explained by the different times for analysis of protein and mRNA (48 and 24 h after transfection, respectively). In addition, MDM2 expression is regulated at the transcriptional level by p53; thus, not surprisingly, its mRNA is upregulated following p53 activation. Moreover, we found that MDM2 mRNA was downregulated when p53 protein was low (data not shown), and this result is in agreement with a negative autoregulatory feedback loop, wherein p53 stimulates MDM2 synthesis, which in turn shuts off p53 activity.37
Figure 5.
L-GILZ activates p53 and modulates p53/MDM2 ubiquitination. (a) Lysates from pcDNA3-L-GILZ- or empty vector-transfected p53+/+ and p53−/− HCT116 cells were analyzed by Western blot 48 h after transfection using anti-p53, anti-MDM2, anti-PUMA, anti-p21, anti-L-GILZ, anti-actin and anti-tubulin antibodies. (b) mRNAs from pcDNA3-L-GILZ- or empty vector-transfected p53+/+ and p53−/− HCT116 cells were analyzed by real-time PCR for p53, MDM2, PUMA and p21 expression 24 h after transfection. For clarity, expression levels of empty vector-transfected p53+/+ and p53−/− HCT116 cells were set to 1. ‘***' indicates the direct comparison of L-GILZ- versus empty vector-transfected cells. (c) Stability of p53 proteins was assessed in L-GILZ- or empty vector-transfected p53+/+ HCT116 cells treated for the indicated times with cycloheximide (30 μg/ml). Cell lysates were analyzed by Western blot with anti-p53 antibody. Quantitative densitometry analysis is shown. (d) HEK293 cells were transfected with the indicated expression vectors. Approximately 48 h after transfection, cells were treated with MG132 (25 μM) for 5 h. Cell lysates were immunoprecipitated with anti-Flag antibody, and ubiquitinated p53 and MDM2 were revealed by Western blot with anti-HA and anti-MDM2 antibodies, respectively. Transfection levels are shown in the lower panel. Of note, the smearing pattern of MDM2 blinds Ub-free MDM2 fraction. (e) H1299 cells were transfected with Flag–ubiquitin along with the indicated plasmids. Cell were treated with MG132 (25 μM) for 5 h before harvest. Cell lysates were immunoprecipitated with anti-Flag antibody followed by Western blot with anti-p53 (FL-393). (f) H1299 cells were transfected with His–ubiquitin along with the indicated plasmids. Cells were treated with MG132 for 5 h before harvest. Cell lysates were immunoprecipitated with anti-HA antibody followed by Western blot with anti-His antibody
Protein stability is the main mechanism by which p53 activity is controlled. MDM2 maintains low levels of p53 through polyubiquitination and proteasomal degradation.38 Therefore, we reasoned that L-GILZ may affect the ubiquitination machinery. To test this hypothesis, we first evaluated p53 stability in L-GILZ- or empty vector-transfected p53+/+ HCT116 cells after cycloheximide treatment and found that L-GILZ stabilized and enhanced endogenous p53 protein levels (Figure 5c). To determine whether L-GILZ interferes with p53 ubiquitination, HEK293 cells were transfected with FLAG–Ub, HA–p53 or MDM2, and L-GILZ and treated with the proteasome inhibitor MG132. L-GILZ overexpression decreased p53 and increased MDM2 ubiquitination (Figure 5d), suggesting that L-GILZ stabilizes and activates p53 proteins by affecting p53/MDM2 ubiquitination. Furthermore, in p53-null H1299 cells, increased ubiquitination of ectopic p53 occurred in the presence of MDM2, and this effect was inhibited by L-GILZ (Figures 5e and f). Finally, we examined whether L-GILZ plays a role in the control of cell proliferation and tumor cell growth. First, proliferation of p53−/− or p53+/+ HCT116 cells expressing L-GILZ or empty vector was evaluated using thymidine uptake assays. The results indicate that L-GILZ reduced the proliferation of p53+/+ but not p53−/− HCT116 cells (Figure 6a). Second, using the transfected cells described above, we performed a colony formation assay. L-GILZ reduced the number of colonies in p53+/+ HCT116 but not in p53−/− HCT116 cells (Figure 6b). Furthermore, we examined whether L-GILZ affects tumor cell growth by subcutaneously injecting p53−/− or p53+/+ HCT116 cells expressing L-GILZ or empty vector into nude mice (NU) and observing tumor growth for 40 days. Tumors were readily observed in mice injected with empty vector-transfected p53+/+ cells (four out of five mice had tumors at day 16), whereas tumors appeared later and were significantly smaller in mice injected with L-GILZ-transfected p53+/+ cells (one out of five mice with tumors at day 16). By contrast, L-GILZ did not affect tumor growth in mice injected with p53−/− cells. After excision, images were taken of representative tumors, and all tumors were weighed. We found a significant difference in average tumor weight between mice injected with L-GILZ-transfected versus empty vector-transfected p53+/+ cells (Figure 6c), but no significant difference was observed for mice injected with p53−/− cells, suggesting that the anti-tumor activity of L-GILZ depends on p53 activation.
Figure 6.
L-GILZ inhibits p53-dependent proliferation and tumor growth. p53+/+ or p53−/− HCT116 cells (0.25 × 106) were transfected with L-GILZ or empty vector, and 24 h after transfection, these cells were (a) seeded in triplicate in 96-well plates for [3H]-thymidine assays (*L-GILZ-transfected versus empty vector) or (b) plated in 10-cm dishes in triplicate for clonogenic assays (***L-GILZ-transfected versus empty vector) as described in Materials and Methods. Number of colonies represents the mean±S.D. of three independent experiments. Representative plates for each group are shown. (c) p53+/+ or p53−/− HCT116 cells (5 × 106) transfected with L-GILZ or empty vector were subcutaneously injected into the right flank of nude mice. Tumor volume (n=5 per group) was determined in vivo at the indicated time points using an external caliper. Mice were killed 40 days after injection, and tumors were excised, imaged and weighed. *L-GILZ- versus empty vector-transfected cells
L-GILZ silencing reverts DEX-induced anti-proliferative effects and downregulates expression of p53, p21 and PUMA
The biological relevance of these findings was evaluated by testing whether a similar interaction occurs between endogenous proteins under basal conditions and whether DEX, which upregulates GILZ,15 also increases L-GILZ expression. Indeed, IP/Western blot analysis revealed that p53 immunoprecipitates from MCF-7 cells that simultaneously expresses p5339 and L-GILZ (for hL-GILZ, NCBI Reference Sequence: NM198057) contained immunoreactive L-GILZ and that DEX treatment increased L-GILZ expression in cell lysates and in p53 immunoprecipitates (Figures 7a and b). Similar results were obtained using the PLA technique (Figure 7c). Indeed, a significant increase in red interaction spots was observed following DEX treatment. Next, we determine whether L-GILZ is responsible for the DEX-induced anti-proliferative effects. We treated MCF-7 cells with L-GILZ-specific siRNA prior to DEX treatment. As expected, DEX treatment significantly reduced the number of cells in S phase, while L-GILZ silencing (Figure 7e, left panel) blocked the anti-proliferative effect of DEX (Figure 7d). Using real-time PCR, we measured the expression of p53, p21 and PUMA in DEX-treated MCF-7 cells in the presence of either scramble or L-GILZ-specific siRNA. Silencing of L-GILZ reduced mRNA expression of p53, p21 and PUMA (Figure 7e), further supporting our conclusion that L-GILZ modulates p53 activity.
Figure 7.
Silencing of L-GILZ decreases DEX anti-proliferative activity and the expression of p53, p21 and PUMA in MCF-7 cells. (a) Lysates from MCF-7 cells that were treated with DEX (10−7 M) for 24 h or left untreated were (b) immunoprecipitated with anti-p53 antibodies, and co-immunoprecipitated L-GILZ was detected by Western blot using anti-GILZ antibody. (c) Co-localization of endogenous L-GILZ and p53 proteins in MCF-7 cells that were treated with DEX (10−7M) for 24 h or left untreated was detected by in situ PLA. Images are representative of three experiments with consistent results. Graph shows RCP values normalized to untreated control (100%), representing the mean of three separate experiments. **DEX-treated versus control cells. (d) MCF-7 cells were transfected with L-GILZ (siL-GILZ) or scramble-L-GILZ (siCtrl) siRNAs 24 h before DEX treatment (10−7M) for 48 h. Cell cycle analysis according to PI staining was performed. A representative experiment is shown (DNA content is on x axis, and number of nuclei is on y axis). (e) L-GILZ, p53, p21 and PUMA mRNA levels were determined by real-time PCR in untransfected (Ctrl), siCtrl and siL-GILZ MCF-7 cells treated 24 h after silencing with DEX (10−7 M) for 3 h. Fold increase is relative to the respective untreated control. ‘***' indicates the direct comparison of L-GILZ versus scramble-L-GILZ siRNAs
Discussion
In the present study, we demonstrated that L-GILZ interacts with p53 and MDM2, leading to the activation of p53 and the inhibition of tumor cell growth.
The ability of L-GILZ to influence the activation of p53 depends on a finely regulated interaction between p53 and MDM2.38 MDM2 stimulates the nuclear export of p53 and targets p53 for proteasomal degradation.9, 40 Upon genotoxic cellular stress, p53 becomes stabilized by post-translational modifications,41 is released from MDM2, and then migrates to the nucleus and binds the promoter region of target genes.38, 42 Although other Ub ligases are also involved in the control of p53 stability,43 MDM2 is the most important Ub ligase that promotes p53 monoubiquitination and nuclear export as well as polyubiquitination and degradation.44, 45 Although MDM2 can be degraded via autoubiquitination,46 other binding partners may function as ligases and contribute to MDM2 degradation or otherwise affect its function.42 For example, p19ARF binds and inhibits MDM2, thereby stabilizing p53.47 Other molecules may bind both p53 and MDM2 as dimers or ternary complexes, such as the Ub-specific protease USP7 (herpesvirus-associated Ub-specific protease), which binds and deubiquitinates MDM2 and p53,48 or Ub carboxyl-terminal hydrolase L1, which deubiquitinates p53 and p19ARF and ubiquitinates MDM2,49 thereby stabilizing p53 and promoting signaling. TGF-β-stimulated clone 22 (TSC-22)50 and Numb51 bind p53 and MDM2 and inhibit MDM2-driven ubiquitination of p53, resulting in p53-dependent apoptosis and inhibition of cell proliferation. Other proteins bind p53 only. For instance, USP10 interacts with and deubiquitinates p53, thereby regulating its localization and stability.52
Here, we found that L-GILZ bound to both p53 and MDM2.The binding of L-GILZ to p53 occurred at the N-terminal domain of p53 (Figure 2), identified by x-ray crystallography as the site responsible for the inhibition of p53 transactivation.53 Furthermore, L-GILZ bound to MDM2 via its N-terminal domain, which is the domain that mediates the MDM2 interaction with p5335 (Figure 3d). Thus, our results suggest that L-GILZ binds p53 in the same region where MDM2 binding occurs and binds MDM2 in the same region where p53 binding occurs, thus inhibiting the p53/MDM2 interaction (Figure 3g). In MEFs co- transfected with L-GILZ, p53, and/or MDM2, dimeric conformation of L-GILZ/p53 and L-GILZ/MDM2 were detected (Figure 3c), although no ternary complexes involving L-GILZ, p53 and MDM2 were observed. This observation is not surprising, however, given that the L-GILZ-binding domain for p53 and MDM2 partially overlaps the regions involved in the p53–MDM2 interaction, resulting in inhibition of p53/MDM2 binding. Notably, formation of a transient trimeric L-GILZ/p53/MDM2 complex within particular pathophysiological states cannot be excluded.
As a consequence of these interactions, p53 proteins are stabilized and MDM2 expression is reduced. By binding to p53 and MDM2, L-GILZ behaves in a manner similar to Numb and TSC-22;50, 51 however, unlike TSC-22, which does not interfere with the formation of p53/MDM2 complexes,50 L-GILZ prevents p53/MDM2 complex formation, similar to Numb.51
Using overexpression models, we also showed the cytoplasmic co-localization of p53 and L-GILZ, whereas ectopic expression of L-GILZ or p53 alone was associated with cytoplasmic and mostly nuclear localization, respectively. Furthermore, blocking nuclear export by leptomycin prevented binding between L-GILZ and p53 but not between L-GILZ and MDM2 or p53 and MDM2 (Figure 4). These findings suggest that molecular interactions involving L-GILZ occur in the cytoplasm and are determined by the pool of p53 that is located in the cytoplasm and ready for MDM2-mediated proteasomal degradation. The ubiquitination of p53 induces nuclear export,43 and whether this exported p53 can migrate back to the nucleus remains unknown. Nevertheless, we demonstrated that the binding of L-GILZ to p53 and MDM2 in the cytoplasm reduced ubiquitination and induced activation of p53 while also increasing MDM2 degradation. In general, deubiquitination of p53 occurs in the nucleus, but, like USP10,52 L-GILZ may induce the deubiquitination of p53 that has been exported to the cytoplasm. Therefore, we propose the following simplest mechanism: L-GILZ, which disrupts the cytoplasmic association between MDM2 and p53, inhibits the MDM2-mediated proteasome degradation of p53 (Figures 5d–f), thus stabilizing and activating p53. The transcriptional activation of p53 and p53 downstream genes, p21 and PUMA (Figure 5b), is then a consequence of L-GILZ-induced post-transcriptional stabilization of p53, which, when activated, functions as transcription factor to regulate downstream target genes as well as its own expression either directly or indirectly.54, 55 Of note, 24 h after transfection, MDM2 mRNA was increased, whereas the MDM2 protein was dramatically decreased 48 h after transfection (Figures 5a and b). This result suggests that the effect of L-GILZ on MDM2 expression is the result of a balance between L-GILZ activity at the post-transcriptional level on MDM2 ubiquitination, resulting in activation of p53 and the effects of activated p53 protein, which increases MDM2 transcription. Despite these interesting findings, we do not know the exact mechanism through which L-GILZ reduces p53 and increases MDM2 ubiquitination nor why L-GILZ preferentially binds to MDM2 but also forms dimers with both p53 and MDM2. We assume that the different interactions between L-GILZ and its binding partners may serve to maintain low levels of p53 or to activate p53. These interactions may involve different pools of p53 and/or MDM2 and depend on the L-GILZ, p53 and MDM2 expression levels. For example, L-GILZ may bind to p53 under stress conditions to protect p53 from MDM2-mediated targeting for proteasomal degradation. In contrast, MDM2 upregulation, which is necessary for the restoration of physiological conditions after the stress response, may favor L-GILZ/MDM2 association, and L-GILZ may contribute to regulation of MDM2 proteins levels. This hypothesis, however, remains to be tested empirically.
Apart from the modulation of p53 ubiquitination, other mechanisms also contribute to p53 stabilization and activation. For example, following cell damage, phosphorylated ataxia-telangiectasia mutated serine/threonine kinase associated with p53 and/or MDM2 may inhibit p53 binding to MDM2 or destabilize MDM2, respectively.56 Here, we propose that the stabilization and activation of p53 also involves L-GILZ; however, in spite of experimental evidence suggesting less binding of MDM2 to p53 in the presence of L-GILZ, we cannot exclude other potential mechanisms of L-GILZ modulation of the p53 pathway, which deserve further investigation.
A consequence of L-GILZ-induced stability of p53 proteins is activation of anti-proliferative and apoptotic pathways, which involve increased expression of p21 and PUMA. This outcome is dependent on the effect of L-GILZ on p53, as p21 and PUMA expression were not detected in p53−/− cells. Also, L-GILZ-induced inhibition of cell proliferation and tumor growth was found only in tumors expressing p53. Furthermore, the involvement of L-GILZ in modulation of p53-dependent proliferative mechanisms is supported by our experimental observation that in the MCF-7 cell line, L-GILZ silencing reversed the anti-proliferative effect of DEX, decreasing the expression of p53 and the p53-dependent genes p21 and PUMA.
One emerging question is whether the redundancy of GILZ isoforms is crucial for cell function or whether isoforms provide specific functions or function in specific cell types. GILZ has been studied thoroughly,18 while the study of L-GILZ has just begun; however, increasing evidence suggests that the two isoforms may exert different functions. Specifically, previous studies indicate that L-GILZ has an exclusive role in the regulation of spermatogenesis,25 and the present study suggests that L-GILZ may have an exclusive role in the control of p53 activation. Also, as deregulation of the p53 signal is one means by which tumors escape death or the homeostatic control of proliferation, our discovery that L-GILZ inhibits tumor growth through the activation of p53 supports our future research to determine the expression of L-GILZ in human tumors, especially those known to involve p53 abnormalities. If a role of L-GILZ in human tumors is established, L-GILZ may be considered a tumor suppressor gene, and thus this work may ultimately lead to new approaches for L-GILZ-targeted cancer therapy.
Materials and Methods
Cell lines and animals
HCT116 human colorectal carcinoma parental cells (p53+/+) and p53-deficient cells (p53−/−),57 generously provided by Dr. Bert Vogelstein (Johns Hopkins University, Baltimore, MD, USA), were maintained in McCoy's 5A medium with 10% fetal bovine serum (FBS) supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin at 37 °C. MCF-7 (breast cancer cell line, p53 wild type), HEK293 (human embryonic kidney) and H1299 (non-small cell lung carcinoma, p53-null) cells were grown in recommended medium containing 10% heat-inactivated FBS, 100 U/ml penicillin and 100 μg/ml streptomycin as previously described.39, 50 MEFs from double null (p53/MDM2) embryos, which were a generous gift from Pier Giuseppe Pelicci (European Institute of Oncology, Milan, Italy), were cultured in Dulbecco's modified Eagle's medium supplemented with 10% FBS.
Male NU (NU/J; Harlan, Udine, Italy), aged 8–10 weeks, were housed in a pathogen-free room. Mice had free access to commercial rodent feed and water and were allowed to acclimatize for 2 weeks prior to treatment. Mice were weighed weekly. Animal care was in compliance with regulations in Italy (D.M. 116192), Europe (O.J. of E.C. L 358/1 12/18/1986) and the United States (Animal Welfare Assurance No A5594-01, Department of Health and Human Services).
For mouse xenograft studies, 5 × 106 p53+/+ or p53−/− HCT116 cells transfected with L-GILZ or empty vector were subcutaneously injected into NU. Tumor growth was monitored using electronic digital calipers. Tumor volume (mm3) was calculated as (length × width2)/2.58 Mice were killed 40 days after treatment, and tumors were excised and weighed.
Plasmid construction and cell transfection
Full-length cDNA encoding MDM2, GILZ and L-GILZ was inserted into pcDNA3 (Invitrogen, Life Technologies, Paisley, UK) or into pcDNA3.1/Myc-His for Myc-L-GILZ epitope tagging as previously described.59 GST-tagged vectors containing L-GILZ were generated by subcloning into the BamHI and NotI sites of the mammalian expression vector pEBG, which was kindly provided by Walter Kolch (the Beatson Institute for Cancer Research, Glasgow, UK). The pcDNA3-HA–p53 and Flag–Ub constructs were a generous gift from Gerry Melino (the Medical Research Council, Leicester University, Leicester, UK); His-tagged Ub construct was provided by Dirk Bohmann (the University of Rochester Medical Center, Rochester, NY, USA); pCMV–MDM2 was provided by Giuseppe Servillo (the Perugia University, Perugia, Italy); p53-truncated mutants were provided by Giannino Del Sal (the Laboratorio Nazionale, Trieste, Italy); and MDM2 deletion mutants in pBS-KS vector were kindly provided by Arnold Levine (the Rutgers Cancer Institute of New Jersey, New Brunswick, NJ, USA). Cells were transfected using jetPEI transfection reagent (Polyplus, Illkirch, France) according to the manufacturer's instructions.
Nuclear and cytoplasmic fractionation was conducted using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer's protocol.
co-IP and Western blot
Approximately 24–48 h after transfection, whole-cell extracts were prepared and IP was performed in IP buffer.20 Antigen–antibody complexes were precipitated with protein A or G bound to sepharose beads (Sigma-Aldrich, Seelze, Germany).
For analysis of complex formation, cells were co-transfected with either pEBG-L-GILZ (eukaryotic GST–L-GILZ), pEBG empty vector, or pEBG-GILZ. Cell extracts were incubated with glutathione–sepharose beads in buffer containing 1 M tris(hydroxymethyl)aminomethane (Tris)-HCl (pH 7.6), 5 M NaCl, 1% NP-40, 10% glycerol, 1 M MgCl2 and 10 μg/ml leupeptin and aprotinin. GST-fusion and associated proteins were eluted after boiling and subjected to Western blot with the indicated antibodies.
Extracted or immunoprecipitated proteins were separated using SDS-PAGE and assessed by Western blotting. Primary antibodies were anti-GILZ (eBioscience, San Diego, CA, USA), anti-p53 (DO-1 and polyclonal FL-393, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-MDM2 (SMP14, Santa Cruz Biotechnology), anti-p21 (Cell Signaling, Danvers, MA, USA) and anti-PUMA (Cell Signaling). We also used antibodies specific for Flag (Sigma-Aldrich), Myc (Invitrogen, Life Technologies), HA, GST or HIS (Cell Signaling) epitopes. Anti-β-tubulin or anti-actin (Sigma-Aldrich) and anti-lamin B1 (Abcam, Cambridge, UK) antibodies were used as controls. Secondary antibodies were labeled with horseradish peroxidase (Pierce, Thermo Scientific). Antigen–antibody complexes were revealed by enhanced chemiluminescence following the manufacturer's instructions (Millipore, Billerica, MA, USA).
Western blot films were scanned, and band signal intensities were determined using ImageJ software (developed by NIH, Bethesda, MD, USA). Expression levels were normalized to β-tubulin, actin or lamin B1 expression. In vivo ubiquitination assays were performed as previously described.52 All IP experiments were controlled with appropriate pre-immune serum (not shown) and considered valid only if similar results were obtained in three repeated experiments.
In vitro protein-binding assays
For pull-down assays, the fusion proteins GST–GILZ or GST–L-GILZ were loaded on glutathione–sepharose beads as previously described24 and incubated with lysates from p53−/− HCT116 cells transfected with HA–p53. Beads were washed extensively, suspended in sample buffer, and analyzed by SDS-PAGE and Western blotting with anti-HA antibodies. For direct complex analysis, wild-type p53, wild-type MDM2 and MDM2-deleted mutants (4, 8 and 9) were translated in vitro using the rabbit reticulocyte-coupled in vitro transcription–translation system under control of the T7 promoter, or T3 promoter for mutant 9, according to the manufacturer's instructions (Promega, Madison, WI, USA). Translated proteins were diluted with binding buffer (25 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid at pH 7.5, 10% glycerol, 50 mM NaCl, 0.05% NP-40, and 1 mM DTT) and pre-cleared with glutathione–sepharose beads for 45 min at 4 °C. GST–GILZ and/or GST–L-GILZ was incubated with translated proteins for 18 h at 4 °C. Beads were subsequently washed five times with binding buffer. Bound proteins were analyzed by SDS-PAGE and revealed by Western blotting with anti-p53 or -MDM2 antibodies.
Indirect immunofluorescence and microscope analysis
HA–p53/GILZ- or HA–p53/L-GILZ-transfected p53−/− HCT116 cells were adhered directly onto glass slides. Cells were permeabilized by incubation in methanol for 20 min at −20 °C. After washing three times in phosphate-buffered saline (PBS) and blocking in PBS containing 3% bovine serum albumin and 1% glycine, cells were incubated for 1 h at room temperature with anti-HA and anti-GILZ antibodies. Cells were then incubated for 1 h with secondary anti-mouse-FITC-conjugated or anti-rabbit-Texas red-conjugated antibodies in blocking buffer containing 1 ng/ml of 4,6 diamidino-2-phenylindole (DAPI). After washing, slides were mounted with cover glass and analyzed using a Zeiss Axioplan fluorescence microscope (Zeiss, Oberkochen, Germany) equipped with a Spot-2 cooled camera (SPOT Imaging Solution, Sterling Heights, MI, USA).
In situ PLA
In situ PLA detection was performed using DUOLINK II In Situ kit components (OLINK Bioscience, Uppsala, Sweden) according to the manufacturer's instructions. Briefly, p53−/− HCT116 cells transfected with the indicated expression vectors were prepared as cytospin slides, fixed with 4% paraformaldehyde for 20 min at room temperature, and blocked in PBS containing 1% bovine serum albumin, 0.1% Triton X-100 and 10% horse serum. Cells were then incubated with primary goat anti-GILZ antibody (recognizing L-GILZ and GILZ) together with either mouse anti-HA (recognizing HA–p53), mouse anti-MDM2 or rabbit p53 (recognizing wild-type and mutant p53) antibody in blocking buffer at 4 °C overnight. After washing slides with appropriate buffers, secondary PLA probes (anti-goat PLA-plus probe for GILZ and L-GILZ, anti-mouse PLA-minus probe for HA–p53, anti-rabbit PLA-minus or plus probe for wild-type and mutant p53 and anti-mouse PLA-minus probe for MDM2) were added, and slides were incubated for 1 h at 37 °C. After washing, the amplification reaction was performed for 100 min at 37 °C. Slides were then washed, air dried and mounted with Olink mounting media containing DAPI. A technical negative control (not shown) was performed with only minus probes. Some groups with ectopic plasmid expression were treated for 3 h with leptomycin (Sigma-Aldrich) prior to in situ PLA.
Detection of interaction signals was performed using a Zeiss Axioplan fluorescence microscope equipped with a Spot-2 cooled camera. The absolute numbers of detected rolling cycle amplification products (RCP, red) visualized as bright fluorescent signals were measured by ImageJ software in 5–10 fields/slide. All RCP signals in the image were divided by the blue (DAPI) signal.
Clonogenic, [3H]-thymidine uptake and propidium iodide (PI) assays
HCT116 p53+/+ and HCT116 p53−/− cells were transfected with either pcDNA3 or L-GILZ. Approximately 24 h after transfection, a total of 1000 cells were plated in 10-cm dish plates in triplicate. The plates were incubated at 37 °C for 10–12 days and stained with 0.5% (w/v) crystal violet. The cell culture dishes were photographed to obtain digital images. The colonies (range 0.002–0.04 in2) were counted using the ImageJ software. The experiment was performed at least three times using triplicate cultures. For [3H]-thymidine uptake, transfected cells were seeded in triplicate in 96-well plates in the presence of [3H]-thymidine (2 μCi/well; Amersham Biosciences, Piscataway, NJ, USA), and the uptake assays were performed as previously described.24 Cell cycle profiles were analyzed by flow cytometry to determine DNA content of cell nuclei stained with PI, as previously described.24
RNA interference
The post-transcriptional silencing of the L-GILZ gene was induced by dsRNA interference using the Trilencer-27 siRNA knockdown duplexes commercial kit (Origene, Rockville, MD, USA) according the manufacturer's instructions. Briefly, MCF-7 cells were transfected with one of the three L-GILZ siRNAs (SR301284, Origene) or with the scrambled control siRNA (SR30004) in the presence of siTran transfection reagent (Origene) in a culture medium for 24 h. After 24 h, cells were treated with DEX (10−7 M) for another 3 or 48 h for real-time PCR and FACS analysis, respectively.
Real-time PCR
The RNA of cells was isolated using the Trizol reagent (Invitrogen, Life Technologies), and conversion of total RNA to cDNA was performed with the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. All real-time PCR reactions were performed using the ABI 7300 Real-Time Cycler (Applied Biosystems, Life Technologies), and amplification was achieved using the TaqMan Gene Expression Assay (Applied Biosystems, Life Technologies). Master mix, primers, and probes for amplification of the L-GILZ, p53, MDM2, p21, PUMA and HPRT-1 genes were all purchased from Applied Biosystems. All experiments were carried out in triplicate, and the ΔΔCt method was used to determine expression of the genes of interest.
Statistical analysis
Each experiment was performed at least three times. Results of representative experiments are shown in the figures. Due to the non-normal distribution of data, non-parametric tests (Kruskall–Wallis analysis of variance) were used for statistical evaluation. Individual group means were compared using the Student's unpaired t-test. Differences were considered statistically significant according to the following criteria: *P<0.05; **P<0.01; ***P<0.001.
Acknowledgments
This study was supported by a grant from the Associazione Italiana per la Ricerca sul Cancro (AIRC), Milan, Italy (IG 10677) to CR.
Glossary
- MDM2
mouse double minute 2 homolog
- GC
glucocorticoid
- GR
glucocorticoid receptor
- GILZ
glucocorticoid-induced leucine zipper
- L-GILZ
long glucocorticoid-induced leucine zipper
- TSC
TGF-β-stimulated clone
- DEX
dexamethasone
- PLA
proximity ligation assay
- MEFs
mouse embryo fibroblasts
- PUMA
p53 upregulated modulator of apoptosis
- ARF
ADP ribosylation factors
- USP
ubiquitin-specific protease
- IB
immunoblot
- NU
nude mice
- IP
immunoprecipitation
- GST
glutathione S-transferase
- Ub
ubiquitin
The authors declare no conflict of interest.
Footnotes
Edited by M Oren
References
- Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- Giaccia AJ, Kastan MB. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Van Dyke T. p53 and tumor suppression. N Engl J Med. 2007;356:79–81. doi: 10.1056/NEJMcibr066301. [DOI] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Marine JC, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17:93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- Boyd SD, Tsai KY, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids—new mechanisms for old drugs. N Engl J Med. 2005;353:1711–1723. doi: 10.1056/NEJMra050541. [DOI] [PubMed] [Google Scholar]
- Kadmiel M, Cidlowski JA. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol Sci. 2013;34:518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Adamio F, Zollo O, Moraca R, Ayroldi E, Bruscoli S, Bartoli A, et al. A new dexamethasone-induced gene of the leucine zipper family protects T lymphocytes from TCR/CD3-activated cell death. Immunity. 1997;7:803–812. doi: 10.1016/s1074-7613(00)80398-2. [DOI] [PubMed] [Google Scholar]
- Cannarile L, Zollo O, D'Adamio F, Ayroldi E, Marchetti C, Tabilio A, et al. Cloning, chromosomal assignment and tissue distribution of human GILZ, a glucocorticoid hormone-induced gene. Cell Death Differ. 2001;8:201–203. doi: 10.1038/sj.cdd.4400798. [DOI] [PubMed] [Google Scholar]
- Bruscoli S, Donato V, Velardi E, Di Sante M, Migliorati G, Donato R, et al. Glucocorticoid-induced leucine zipper (GILZ) and long GILZ inhibit myogenic differentiation and mediate anti-myogenic effects of glucocorticoids. J Biol Chem. 2010;285:10385–10396. doi: 10.1074/jbc.M109.070136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroldi E, Riccardi C. Glucocorticoid-induced leucine zipper (GILZ): a new important mediator of glucocorticoid action. FASEB J. 2009;23:3649–3658. doi: 10.1096/fj.09-134684. [DOI] [PubMed] [Google Scholar]
- Beaulieu E, Morand EF. Role of GILZ in immune regulation, glucocorticoid actions and rheumatoid arthritis. Nat Rev Rheumatol. 2011;7:340–348. doi: 10.1038/nrrheum.2011.59. [DOI] [PubMed] [Google Scholar]
- Ayroldi E, Migliorati G, Bruscoli S, Marchetti C, Zollo O, Cannarile L, et al. Modulation of T-cell activation by the glucocorticoid-induced leucine zipper factor via inhibition of nuclear factor kappaB. Blood. 2001;98:743–753. doi: 10.1182/blood.v98.3.743. [DOI] [PubMed] [Google Scholar]
- Cannarile L, Fallarino F, Agostini M, Cuzzocrea S, Mazzon E, Vacca C, et al. Increased GILZ expression in transgenic mice up-regulates Th-2 lymphokines. Blood. 2006;107:1039–1047. doi: 10.1182/blood-2005-05-2183. [DOI] [PubMed] [Google Scholar]
- Cannarile L, Cuzzocrea S, Santucci L, Agostini M, Mazzon E, Esposito E, et al. Glucocorticoid-induced leucine zipper is protective in Th1-mediated models of colitis. Gastroenterology. 2009;136:530–541. doi: 10.1053/j.gastro.2008.09.024. [DOI] [PubMed] [Google Scholar]
- Pinheiro I, Dejager L, Petta I, Vandevyver S, Puimege L, Mahieu T, et al. LPS resistance of SPRET/Ei mice is mediated by Gilz, encoded by the Tsc22d3 gene on the X chromosome. EMBO Mol Med. 2013;5:456–470. doi: 10.1002/emmm.201201683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroldi E, Zollo O, Bastianelli A, Marchetti C, Agostini M, Di Virgilio R, et al. GILZ mediates the antiproliferative activity of glucocorticoids by negative regulation of Ras signaling. J Clin Investig. 2007;117:1605–1615. doi: 10.1172/JCI30724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruscoli S, Velardi E, Di Sante M, Bereshchenko O, Venanzi A, Coppo M, et al. Long glucocorticoid-induced leucine zipper (L-GILZ) protein interacts with ras protein pathway and contributes to spermatogenesis control. J Biol Chem. 2012;287:1242–1251. doi: 10.1074/jbc.M111.316372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Wasylyk B. Physiological and pathological consequences of the interactions of the p53 tumor suppressor with the glucocorticoid, androgen, and estrogen receptors. Ann NY Acad Sci. 2004;1024:54–71. doi: 10.1196/annals.1321.005. [DOI] [PubMed] [Google Scholar]
- Clarke AR, Purdie CA, Harrison DJ, Morris RG, Bird CC, Hooper ML, et al. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–852. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- Sasson R, Tajima K, Amsterdam A. Glucocorticoids protect against apoptosis induced by serum deprivation, cyclic adenosine 3',5'-monophosphate and p53 activation in immortalized human granulosa cells: involvement of Bcl-2. Endocrinology. 2001;142:802–811. doi: 10.1210/endo.142.2.7942. [DOI] [PubMed] [Google Scholar]
- Yu C, Yap N, Chen D, Cheng S. Modulation of hormone-dependent transcriptional activity of the glucocorticoid receptor by the tumor suppressor p53. Cancer Lett. 1997;116:191–196. doi: 10.1016/s0304-3835(97)00186-9. [DOI] [PubMed] [Google Scholar]
- Crochemore C, Michaelidis TM, Fischer D, Loeffler JP, Almeida OF. Enhancement of p53 activity and inhibition of neural cell proliferation by glucocorticoid receptor activation. FASEB J. 2002;16:761–770. doi: 10.1096/fj.01-0577com. [DOI] [PubMed] [Google Scholar]
- Urban G, Golden T, Aragon IV, Cowsert L, Cooper SR, Dean NM, et al. Identification of a functional link for the p53 tumor suppressor protein in dexamethasone-induced growth suppression. J Biol Chem. 2003;278:9747–9753. doi: 10.1074/jbc.M210993200. [DOI] [PubMed] [Google Scholar]
- Murphy SH, Suzuki K, Downes M, Welch GL, De Jesus P, Miraglia LJ, et al. Tumor suppressor protein (p)53, is a regulator of NF-kappaB repression by the glucocorticoid receptor. Proc Natl Acad Sci USA. 2011;108:17117–17122. doi: 10.1073/pnas.1114420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, et al. Direct observation of individual endogenous protein complexes In situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Sandy P, Gostissa M, Fogal V, Cecco LD, Szalay K, Rooney RJ, et al. p53 is involved in the p120E4F-mediated growth arrest. Oncogene. 2000;19:188–199. doi: 10.1038/sj.onc.1203250. [DOI] [PubMed] [Google Scholar]
- Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, et al. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112–9117. doi: 10.1073/pnas.96.16.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren M. Decision making by p53: life, death and cancer. Cell Death Differ. 2003;10:431–442. doi: 10.1038/sj.cdd.4401183. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Geng QQ, Dong DF, Chen NZ, Wu YY, Li EX, Wang J, et al. Induction of p53 expression and apoptosis by a recombinant dual-target MDM2/MDMX inhibitory protein in wildtype p53 breast cancer cells. Int J Oncol. 2013;43:1935–1942. doi: 10.3892/ijo.2013.2138. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W. SnapShot: p53 posttranslational modifications. Cell. 2008;133:930–930 e931. doi: 10.1016/j.cell.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahav G, Rosenfeld N, Sigal A, Geva-Zatorsky N, Levine AJ, Elowitz MB, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17:86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman DA, Levine AJ. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- Lai Z, Yang T, Kim YB, Sielecki TM, Diamond MA, Strack P, et al. Differentiation of Hdm2-mediated p53 ubiquitination and Hdm2 autoubiquitination activity by small molecular weight inhibitors. Proc Natl Acad Sci USA. 2002;99:14734–14739. doi: 10.1073/pnas.212428599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Li M, Hu M, Shi Y, Gu W. The p53—Mdm2—HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene. 2007;26:7262–7266. doi: 10.1038/sj.onc.1210531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Tao Q, Jin H, van Hasselt A, Poon FF, Wang X, et al. The tumor suppressor UCHL1 forms a complex with p53/MDM2/ARF to promote p53 signaling and is frequently silenced in nasopharyngeal carcinoma. Clin Cancer Res. 2010;16:2949–2958. doi: 10.1158/1078-0432.CCR-09-3178. [DOI] [PubMed] [Google Scholar]
- Yoon CH, Rho SB, Kim ST, Kho S, Park J, Jang IS, et al. Crucial role of TSC-22 in preventing the proteasomal degradation of p53 in cervical cancer. PloS One. 2012;7:e42006. doi: 10.1371/journal.pone.0042006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaluca IN, Tosoni D, Nuciforo P, Senic-Matuglia F, Galimberti V, Viale G, et al. NUMB controls p53 tumour suppressor activity. Nature. 2008;451:76–80. doi: 10.1038/nature06412. [DOI] [PubMed] [Google Scholar]
- Yuan J, Luo K, Zhang L, Cheville JC, Lou Z. USP10 regulates p53 localization and stability by deubiquitinating p53. Cell. 2010;140:384–396. doi: 10.1016/j.cell.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- Wang S, El-Deiry WS. p73 or p53 directly regulates human p53 transcription to maintain cell cycle checkpoints. Cancer Res. 2006;66:6982–6989. doi: 10.1158/0008-5472.CAN-06-0511. [DOI] [PubMed] [Google Scholar]
- Deffie A, Wu H, Reinke V, Lozano G. The tumor suppressor p53 regulates its own transcription. Mol Cell Biol. 1993;13:3415–3423. doi: 10.1128/mcb.13.6.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci USA. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Investig. 1999;104:263–269. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zeng SX, Zhang Y, Zhang Y, Ding D, Ye Q, et al. A small molecule Inauhzin inhibits SIRT1 activity and suppresses tumour growth through activation of p53. EMBO Mol Med. 2012;4:298–312. doi: 10.1002/emmm.201100211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayroldi E, Zollo O, Macchiarulo A, Di Marco B, Marchetti C, Riccardi C. Glucocorticoid-induced leucine zipper inhibits the Raf-extracellular signal-regulated kinase pathway by binding to Raf-1. Mol Cell Biol. 2002;22:7929–7941. doi: 10.1128/MCB.22.22.7929-7941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]