Abstract
Metabolic reprogramming is a hallmark of cancer cells. Strap (stress-responsive activator of p300) is a novel TPR motif OB-fold protein that contributes to p53 transcriptional activation. We show here that, in addition to its established transcriptional role, Strap is localised at mitochondria where one of its key interaction partners is ATP synthase. Significantly, the interaction between Strap and ATP synthase downregulates mitochondrial ATP production. Under glucose-limiting conditions, cancer cells are sensitised by mitochondrial Strap to apoptosis, which is rescued by supplementing cells with an extracellular source of ATP. Furthermore, Strap augments the apoptotic effects of mitochondrial p53. These findings define Strap as a dual regulator of cellular reprogramming: first as a nuclear transcription cofactor and second in the direct regulation of mitochondrial respiration.
An established characteristic of tumour cells is their underlying metabolic changes.1 Early observations that tumour cells had persistently high glycolysis even under aerobic conditions led to the suggestion that this underlying metabolic change was fundamental to the process of tumourigenesis.2 In normal cells, ATP production is primarily produced in mitochondria by oxidative phosphorylation, driven by the TCA cycle in the presence of oxygen.3 When energy is needed rapidly or oxygen becomes limiting, cells turn on glycolysis to meet the extra energy demands. In contrast, tumour cells consume large amounts of glucose and maintain high levels of glycolysis even in the presence of oxygen.4
p53 has a critical role in mediating cell cycle arrest and cell death, which occurs via transcription-dependent and -independent mechanisms.5 As a transcription factor, p53 regulates a variety of target genes connected with cell fate (cell cycle arrest, apoptosis, autophagy and senescence, for example) and metabolic reprogramming.5, 6, 7 Further, its ability to function as a transcription factor is influenced by a complex array of posttranslational modifications and cofactors.6, 8 Strap is one such cofactor that regulates p53 transcriptional activity under a variety of cellular conditions.9 Upon DNA damage, for example, it is phosphorylated by the DNA damage signalling ATM and Chk2 kinases, resulting in Strap stabilisation, altered cellular location and enhanced p53 activity.10, 11 An analysis of the three-dimensional structure of Strap highlighted its unusual domain organisation, being composed of tandem TPR motifs together with an OB-fold, with both domains required for it to function in the p53 response.12 The location of phosphorylated residues in Strap suggested a structural model whereby Strap unfolds to become more accessible during the DNA-damage response.12
It has become increasingly evident that p53 is able to modify a variety of metabolic pathways, including glycolysis and oxidative phosphorylation, enabling cells to respond to metabolic stress.13, 14, 15, 16, 17 Further, p53 is essential for cell survival under glucose deprivation and in tumour cells under metabolic stress is able to augment apoptosis.13, 16 Some studies have even suggested that the ability of p53 to act as a tumour suppressor may be influenced by its role in metabolic regulation, rather than its ability to influence apoptosis, senescence or cell cycle arrest.18
In addition to its established transcription-dependent mechanisms, p53 also induces apoptosis in a transcription-independent fashion through its localisation to mitochondria.19 At mitochondria, p53 interacts with and regulates Bcl2 family members, such as Bax and Bak.20, 21, 22 This allows p53 to influence the intrinsic pathway of apoptosis through the release of soluble proteins, like cytochrome c, which then initiate caspase activation in the cytosol.23 The release of cytochrome c occurs as a consequence of compromised integrity of the outer mitochondrial membrane, referred to as mitochondrial outer membrane permeabilisation (MOMP).20, 21 Bax and Bak oligomerise at the mitochondrial membrane influencing MOMP and the subsequent release of pro-apoptotic factors, such as cytochrome c.24
In continuing the analysis of Strap and its role in regulating p53 biology, we have uncovered a new and hitherto unexpected level of control. Remarkably, we have found that Strap is, like p53, localised to mitochondria and identified one of its key interaction partners as mitochondrial ATP synthase. This interaction causes reduced ATP synthesis which, in turn, sensitises cells to mitochondrial-dependent apoptosis. Most significantly, mitochondrial Strap augments apoptosis stimulated by the mitochondrial arm of the p53 response. These results highlight an unanticipated overlap and convergence in the pathways affected by Strap and p53 and provide further support for their interplay in transcription and metabolism.
Results
Strap locates to mitochondria and regulates ATP synthase
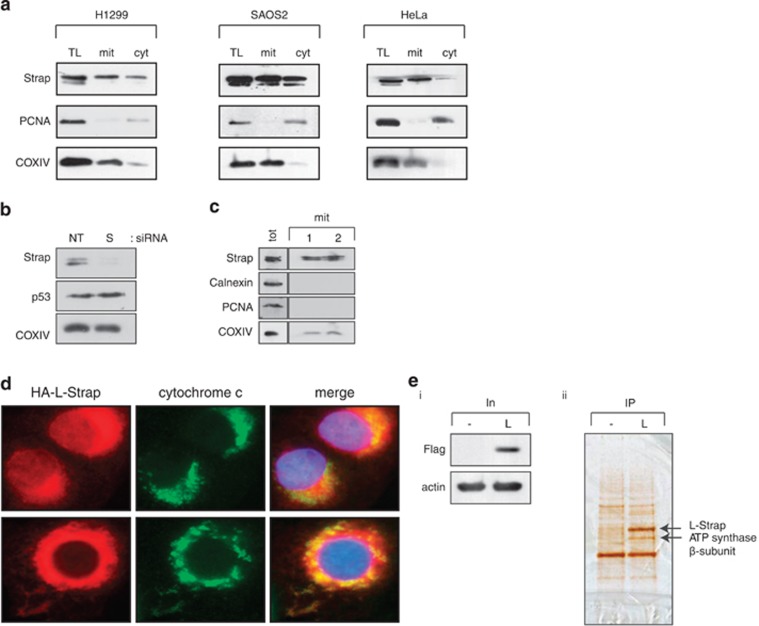
To further explore Strap's subcellular localisation, we prepared fractions from different cell types and measured the level of Strap in total lysate, mitochondria and cytoplasm. Surprisingly, we identified Strap in the mitochondrial fraction (Figure 1a), which was evident when mitochondria were prepared according to different methods and in a variety of cell types (Figures 1a–c and Supplementary Figure S1a).
Figure 1.
Strap is localised at the mitochondria and interacts with ATP synthase. (a) Endogenous Strap from various cell types was detected in total lysate (TL), mitochondrial (mit) and cytoplasmic (cyt) fractions. Cytochrome oxidase IV (COXIV) was used as a mitochondrial marker. PCNA was used as a control for nuclear contamination. The mitochondrial fractions were estimated to be 95% enriched. (b) U2OS cells were transfected with non-targeting (NT) or Strap (S) siRNA for 72 h. Mitochondrial fractions were prepared and immunoblotted with the indicated antibodies. COXIV served as a mitochondrial marker. (c) Endogenous Strap from U2OS cells was detected in mitochondrial (mit) fractions, prepared using different extraction techniques (1 and 2; see Materials and Methods). Nuclear and endoplasmic reticulum material was monitored using PCNA and calnexin antibodies respectively. COXIV served as a mitochondrial marker. Total cell lysate (tot) serves as a comparison. (d) U2OS cells were transfected with L-Strap and immunostained with an anti-HA antibody to detect ectopic Strap. DAPI was used to visualise the nuclei and cytochrome c for mitochondria. The merged imaged is shown (magnification × 600). (e) L-Strap (L) or control (-) transfected U2OS cells (i) were immunoprecipitated (IP) with anti-Flag antibody and the indicated silver-stained protein bands (ii) excised and subjected to tryptic digestion and liquid chromatography tandem mass spectrometry; the position of L-Strap and ATP synthase β-subunit is indicated
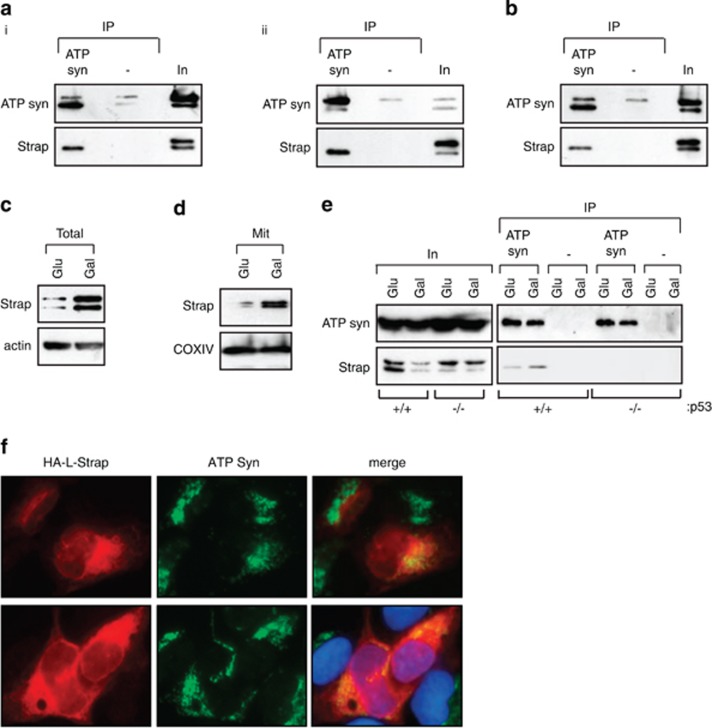
We therefore considered the possibility that Strap may interact with mitochondrial proteins. To facilitate an analysis of Strap's mitochondrial role, we made a derivative of Strap that was exclusively directed to mitochondria using a targeting sequence taken from Bcl2 (referred to as L-Strap); as expected, L-Strap localised to mitochondria (Figure 1d) and lacked any transcription cofactor activity on p53 (Supplementary Figure S1e). We then immunoprecipitated L-Strap from cells, and any bound proteins were subsequently identified by mass spectrometry. The ATP synthase β-subunit was found to be a predominant interacting protein (Figure 1e). The β-subunit is an essential component of ATP synthase, which is a large molecular complex localised at the inner mitochondrial membrane responsible for synthesising ATP by oxidative phosphorylation.3 We verified that the ATP synthase β-subunit interacted with both wild-type and L-Strap by immunoprecipitation (Supplementary Figure S1b), which was consistent with the co-localisation of L-Strap and ATP synthase β-subunit (Figure 2f). Further, the interaction was relevant to endogenous Strap, which also co-immunoprecipitated with ATP synthase β-subunit in different cell types (Figure 2a); the interaction was also detectable in purified mitochondria (Figure 2b).
Figure 2.
Strap interacts with ATP synthase β-subunit. (a) ATP synthase β-subunit from U2OS (i) and HeLa (ii) cells was immunoprecipitated (IP) with anti-ATP synthase β-subunit (ATP syn) or a non-specific (-) antibody and immunoblotted with ATP synthase β-subunit or Strap antibodies as indicated. Input (In) represents 5% of total proteins. (b) Mitochondrial fractions prepared from U2OS cells were immunoprecipitated (IP) with anti-ATP synthase β-subunit or non-specific (-) antibody and immunoblotted with ATP synthase β-subunit or Strap antibodies as indicated. (c and d) Total lysates (c) and mitochondrial (mit) (d) fractions from HCT116 cells grown either in glucose (Glu) or galactose (Gal)-supplemented medium. COXIV and actin served as mitochondrial and total lysate loading controls, respectively. (e) ATP synthase β-subunit was immunoprecipitated (IP) from p53+/+ and p53−/− HCT116 cells grown either in glucose (Glu) or galactose (Gal)-supplemented medium, and immunoblotted with ATP synthase β-subunit or Strap antibodies as indicated. A three-fold increase in the interaction between Strap and ATP synthase β-subunit occurred under galactose-supplemented growth conditions. Input (In) represents 5% of total proteins. (f) U2OS cells were transfected with L-Strap and immunostained with an anti-HA antibody to detect ectopic Strap. Endogenous ATP synthase β-subunit was detected with an anti-ATP synthase β antibody, and DAPI was used to visualise the nuclei
Thus, having established a biochemical interaction between Strap and ATP synthase, we then addressed whether the interaction was influenced by the metabolic status of cells. We therefore measured Strap in cells grown in galactose-supplemented (to promote oxidative phosphorylation) and glucose-supplemented (to promote glycolysis) medium.25, 26 Strap protein levels increased under galactose compared with glucose-supplemented conditions in HCT116 cells (Figure 2c), with a similar pattern apparent in mitochondrial fractions (Figure 2d); moreover, this increase in Strap levels required p53 activity (Supplementary Figure S1g). We then reasoned that the increased protein level might influence Strap binding to the ATP synthase β-subunit and tested this possibility by immunoprecipitating the Strap–ATP synthase complex. An enhanced interaction (three-fold) was apparent between Strap and the β-subunit in cells grown in galactose-supplemented compared with glucose-supplemented medium (Figure 2e).
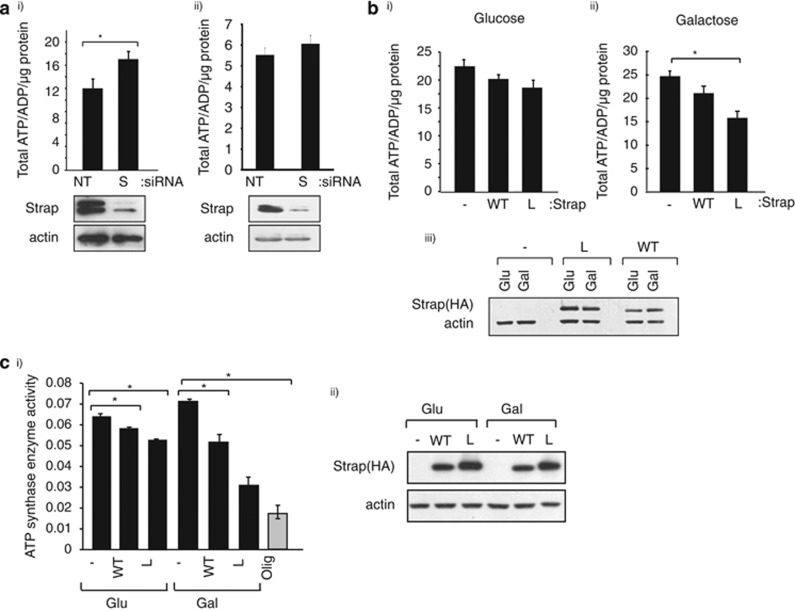
As ATP synthase is the major regulator of cellular ATP,3 we subsequently addressed whether Strap affected cellular ATP levels. Depleting Strap (with siRNA) under normal culture conditions resulted in enhanced cellular ATP levels (Figure 3a), arguing that Strap influenced ATP production in cells growing in normal physiological conditions. Moreover, the effect of Strap on cellular ATP levels was pronounced in p53-expressing compared with mutant p53-expressing cells (Figure 3ai and ii). We then measured ATP levels in cells expressing L-Strap comparing the effect to that observed with wild-type Strap (Figure 3b and Supplementary Figure S1h). The effect of L-Strap on ATP levels was more pronounced in cells grown in galactose compared with glucose-supplemented medium (40% inhibition compared with 10% Figure 3b). Wild-type Strap also caused reduced ATP levels under galactose treatment, but this was, as might be expected, less than observed for L-Strap (15% compared with 40% inhibition; Figure 3bii).
Figure 3.
Strap regulates cellular ATP levels. (a) U2OS (i) or SAOS2 (ii) cells grown in normal culture conditions were transfected with non-targeting (NT) or Strap (S) siRNA for 72 h before cellular ATP was measured. The graph shows the ATP/ADP ratio, and the level of Strap is shown underneath. (b) U2OS-inducible stable cells expressing vector (-), wild-type Strap (WT) or L-Strap (L) induced with doxycycline (1 μg/ml) were grown in glucose (i) or galactose (ii) -supplemented medium, and the cellular ATP level was measured. The level of Strap proteins is shown (iii). *P<0.05, Student's t-test. (c) (i) ATP synthase activity was measured as described in U2OS-inducible stable cells expressing vector (-), wild-type Strap (WT) or L-Strap (L) induced with doxycycline (1 μg/ml), grown in either glucose (Glu) or galactose (Gal)-supplemented medium. Cells were also treated with oligomycin (olig; 2 μM for 30 min) as indicated. The level of Strap is shown (ii). *P<0.05, Student's t-test
We then addressed whether the influence of Strap on ATP levels reflected a direct change in ATP synthase activity using an immune-capture assay that measured cellular ATP synthase activity. Although wild-type and L-Strap had a modest inhibitory effect on ATP synthase activity when harvested from cells cultured in glucose-supplemented medium, the effect was far more significant in cells grown in galactose-supplemented medium (Figure 3c). In fact, L-Strap reduced ATP synthase activity to a level that was very similar to that caused by oligomycin treatment of cells, a small-molecule antagonist which directly inhibits ATP synthase27 (60% compared with 70% Figure 3c). Collectively, therefore the results indicate that Strap downregulates cellular ATP levels and suggest that this is achieved in part through interacting with mitochondrial ATP synthase β-subunit.
Strap augments mitochondrial-dependent apoptosis
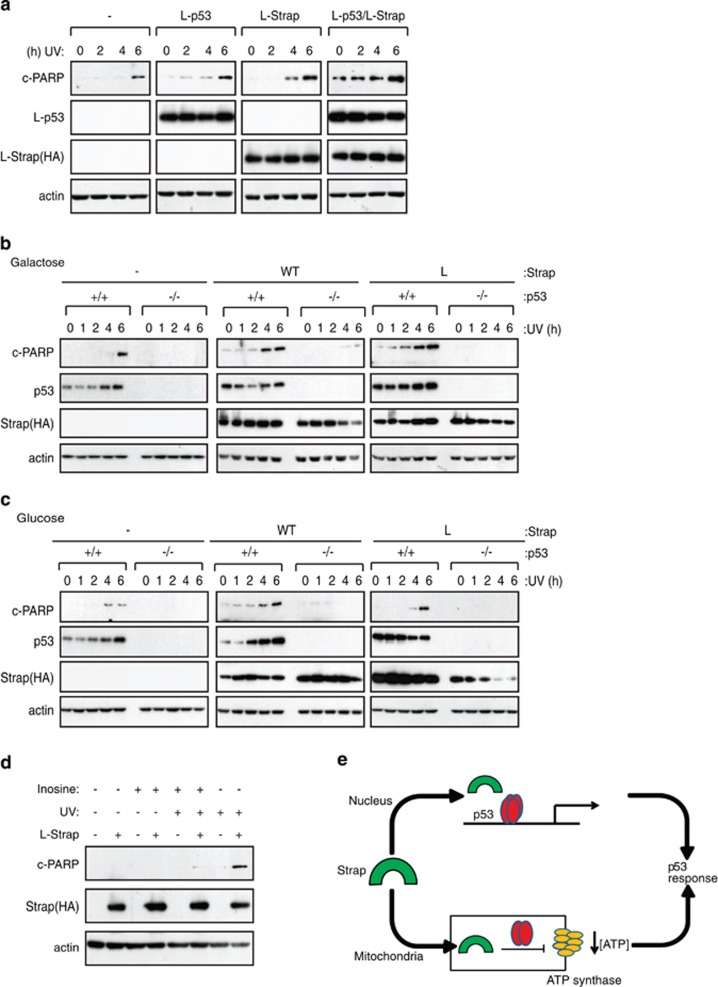
Altered levels of cellular ATP have been implicated in regulating apoptosis.28 We therefore explored the functional consequence of mitochondrial localised Strap by addressing whether L-Strap could sensitise cells to apoptosis and further if this occurred in an ATP-dependent fashion. At the same time, given the dependence of the Strap-ATP synthase interaction on p53 (Figure 2e and Supplementary Figure S1g) and the interaction between Strap and p53 in mitochondria (Supplementary Figure S1c), it was of interest to investigate whether Strap could influence the mitochondrial activity of p53. For this, we expressed L-Strap and monitored the level of apoptosis and observed that L-Strap was able to cause a modest increase in PARP cleavage (Figure 4a).
Figure 4.
Strap augments apoptosis in the presence of p53. (a) SAOS2 cells were transfected (1 μg) with either L-Strap or L-p53 alone or together with vector control (-) as indicated. Cells were treated with ultraviolet (UV) light (50 J/m2) and harvested at the indicated time points. L-Strap was detected with an anti-HA antibody, and L-p53 was detected using an anti-p53 antibody. Cleaved (c) PARP was used as the apoptotic marker while actin served as the loading control. (b and c) p53+/+ and p53 −/− HCT116 cells transfected (1 μg) with either vector control (-), wild-type (WT) or L-Strap (L) were grown in galactose (b) or glucose (c) -supplemented medium as indicated. Cells were treated with UV light (50 J/m2) and harvested at the indicated time points. WT and L-Strap were detected with an anti-HA antibody. Cleaved (c) PARP was used as the apoptotic marker while actin served as the loading control. (d) U2OS stable cells induced with doxycycline (1 μg/ml) grown in galactose-supplemented medium expressing vector (-) or L-Strap (+) were treated with either inosine (20 mM, 6 h) and/or UV light (50 J/m2, 3 h) as indicated. Strap was detected with an anti-HA antibody. Cleaved (c) PARP was used as the apoptotic marker, and actin served as the loading control. (e) Model describing the role of Strap in the p53 response, where Strap (green) augments nuclear transcriptional activation by p53 (red), while inhibiting ATP production in mitochondria by downregulating ATP synthase (yellow) activity and stimulating apoptosis
L-p53 is a p53 derivative that is expressed in mitochondria (Supplementary Figure S1d;22) and lacks any nuclear transcription activity (Supplementary Figure S1f). When L-p53 was co-expressed with L-Strap, there was a marked increase in the level of cleaved PARP in both normal and UV light-treated cells compared with each one when expressed alone (Figure 4a), consistent with an interaction between the proteins. The influence of p53 on the apoptotic activity of L-Strap was explored further in p53+/+ and p53−/− HCT116 cells grown in the presence of galactose or glucose. In p53+/+cells, the expression of L-Strap caused enhanced PARP cleavage in galactose-supplemented cells, which was less apparent in cells grown in glucose-supplemented medium (Figures 4b and c; compare PARP cleavage at 1 and 4 h, respectively). This contrasted with the effect of wild-type Strap, which had an equivalent effect on PARP cleavage regardless of the culture conditions (Figures 4b and c). Further, the effect was not apparent in p53−/− cells, where Strap had minimal influence on PARP cleavage (Figures 4b and c), thus establishing the dependency on p53 activity for mitochondrically localised Strap to mediate apoptosis. Notably, similar results were evident in U2OS cells (expressing wild-type p53), where L-Strap expression resulted in increased levels of apoptotic (sub-G1 DNA content) cells under galactose but not under glucose culture conditions, in contrast to SAOS2 (p53-defective) cells where L-Strap failed to influence apoptosis (Supplementary Figures S2a and b). Significantly, Strap was required for cytochrome c release from mitochondria, an effect which was similarly dependent upon p53 (Supplementary Figure S1c). Moreover, the ability of L-Strap to influence apoptosis and the dependence on p53 activity occurred in the absence of activating p53 target genes, because in conditions where L-Strap induced apoptosis, it failed to activate a variety of p53 target genes connected with metabolism (e.g., GLUT4, TIGAR, SCO2, GLS2) contrasting with wild-type Strap (Supplementary Figure S2d). These results show that mitochondrial Strap stimulates apoptosis and further that the ability of Strap to augment mitochondrial-dependent apoptosis requires p53 activity but occurs independently of transcriptional activity.
The relationship between Strap and its ability to downregulate ATP synthase activity, with the reduced level of cellular ATP and increase in mitochondrial-dependent apoptosis, prompted us to examine whether the level of cellular ATP was responsible for the enhanced apoptosis. We addressed this idea by testing whether we could rescue cells from apoptosis by supplying an extracellular source of ATP. Thus we induced mitochondrial-dependent apoptosis with L-Strap and UV light treatment and supplemented the medium with inosine, to provide an extracellular source of ATP.29 Apoptosis was apparent upon expressing L-Strap and treating with UV light (Figure 4d). Most significantly, in inosine-supplemented medium apoptosis was less evident, reflected as a reduction in the level of cleaved PARP in cells expressing L-Strap (Figure 4d). These results argue that the reduced level of cellular ATP caused by targeting Strap to mitochondria is responsible for the heightened level of apoptosis.
Discussion
We have described a novel mitochondrial role for cofactor Strap in reprogramming cellular metabolism through its ability to influence oxidative phosphorylation. This is achieved by means of an interaction between Strap and ATP synthase, enabling Strap to downregulate ATP synthase activity and lower the level of cellular ATP. Significantly, a reduced level of cellular ATP, as a consequence of mitochondrially localised Strap, sensitised cells to apoptosis. This effect could be rescued by supplying an extracellular source of ATP. Our results thus connect Strap with mitochondria and suggest that it acts on ATP synthesis to influence the pool of available cellular ATP (Figure 4e).
Mitochondrial ATP production is the main source of energy for intracellular pathways.3 A large body of evidence connects the level of intracellular ATP with apoptosis; thus defective ATP production or excessive ATP consumption can lead to reduced activity of ATP-dependent ion pumps with subsequent apoptosis.30 Moreover, cellular stress can often result in ATP depletion and manipulating cell metabolism also impacts on cell fate.31 During apoptosis, a sufficient supply of ATP is necessary to favour apoptosome formation and trigger caspase activation,32 and the severity of reduction in cellular ATP often determines the mode of programmed cell death.32 A decrease in intracellular ATP can initiate cytochrome c release from the inner mitochondrial membrane and trigger apoptosis;33 in this respect, overexpression of L-Strap resulted in a decrease in the ATP/ADP ratio and was able to sensitise the cells to p53-dependent apoptosis.
Studies have suggested a role for mitochondrial p53 in influencing MOMP during the early stages of p53-mediated apoptosis, which precedes its nuclear translocation during the DNA-damage response.34, 35 Our data also support this model and suggest that similar to p53, Strap, in the early stages of the damage response, can be found at the mitochondria where it may influence mitochondrial MOMP and cytochrome c release. At the later stages of damage, both Strap and p53 take on a more nuclear localisation (Figure 4e) where Strap can function as a nuclear p53 co-factor.9, 10, 11 Although our study supports a role of Strap in mitochondrial-dependent apoptosis, we cannot rule out the possibility that Strap, as a general stress-responsive p53 co-factor, may have a role in other p53 functions, such as p53-dependent necrosis.36 It is also interesting to note that alterations in ATP levels would likely influence AMPK (AMP-activated protein kinase), which monitors cellular energy status. AMPK has been shown to phosphorylate and stabilise mitochondrial p53 leading to Bak oligomerisation and apoptosis.37 It is possible that Strap-mediated alterations in ATP levels could, in turn, impact on AMPK activity and this may feed-forward into p53 activity during metabolic reconditioning and would be of interest to evaluate in future studies.
Strap is a previously described p53 cofactor that undergoes nuclear accumulation upon DNA damage.10 Its nuclear localisation is dependent on DNA-damage-activated protein kinases, mediated by both ATM and Chk2 kinases.10, 11 The results described here reveal an unexpected level of Strap control, through its localisation to mitochondria, interaction with ATP synthase and its subsequent influence on ATP production. Our results thus define a dual role for Strap that integrates metabolic reprogramming mediated by the key enzyme ATP synthase with the p53 response.
Materials and Methods
Cell lines, plasmids and expression vectors
U2OS, HeLa, SAOS2, H1299, HCT116 (p53+/+ and p53−/−) cells were maintained at 37 °C with 5% CO2. Strap and L-Strap stable inducible cell lines were created in U2OS cells using the TET-On gene expression system (Clontech, Saint-Germain-en-Laye, France). Cells were seeded in DMEM containing 5% foetal calf serum in the presence of antibiotics and were allowed to grow for 24–48 h. Cells were then washed twice with PBS and received glucose- or galactose-supplemented media for 24 h. Glucose-supplemented media consisted of 25 mM glucose and 2 mM glutamine supplemented with antibiotics, and the galactose-supplemented media consisted of glucose-free media with 10 mM galactose, 5 mM HEPES and 2 mM glutamine supplemented with antibiotics.26 The following plasmids have previously been described: pcDNA3 HA-WT Strap,9, 10 pCMV-L-p53 was a generous gift from Prof Ute Moll22 and p3xFlag-CMV-7.1 expression vector (Sigma, Dorset, UK). L-Strap was prepared by fusing the Bcl2 mitochondrial targeting signal (DFSWLSLKTLLSLALVGACITLGAYGHK) onto the C-terminal region of Strap in HA-WT Strap.9, 10
Antibodies
The following antibodies were used: Flag peptide M2 (Sigma), HA11 (Covance, Cambridge, UK), calnexin (Millipore, Nottingham, UK), cleaved PARP and cytochrome oxidase IV (Cell Signaling Technology, Leiden, The Netherlands), β-actin (Sigma), ATP synthase β-subunit (BD Pharmingen, Oxford, UK), PCNA, p53 and cytochrome c (Santa Cruz, Heidelberg, Germany) and Strap.11
Transfection
Cells were transfected using Genejuice (Novagen, Nottingham, UK), with the indicated plasmids. Empty pcDNA3.1 vector was used to equalise amounts of DNA transfected when necessary. For siRNA, cells were plated and transfected for 72 h using oligofectamine (Invitrogen, Paisley, UK) following the manufacturer's guidelines. A mixture of three siRNAs11, 38 were used at a final concentration of 90 nM (Dharmacon, Lafayette, CO, USA).
Immunoprecipitation
Cells were harvested in TNN lysis buffer (150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 5 mM EDTA, 0.5% NP40, 50 mM NaF, β-mercaptoethanol and 0.2 mM sodium orthovanadate) and protease inhibitor cocktail (leupeptin (0.5 μg/ml), pepstatin (0.5 μg/ml), aprotinin (0.5 μg/ml)). After 20-min incubation on ice, samples were spun at 16 000 × g for 10 min. The cell extract was incubated with Flag agarose beads (Sigma) for 2 h at 4 °C. The beads were washed using lysis buffer three times. The beads were eluted using 500 μg of FLAG peptide in 25 mM Tris-HCl pH 6.5 at 1000 r.p.m. for 1 h. The samples were denatured and run on a SDS-PAGE gel and silver stained.
In-gel tryptic digestion and mass spectrometry
Following SDS-PAGE and silver staining, bands were excised and subjected to tryptic digestion as described.39 Briefly, the excised bands were washed and subjected to a series of dehydration, reduction and rehydration. The gel pieces were dehydrated in 200 μl of acetonitrile before being reduced with 30 μl of dithiothreitol and 30 μl of iodoacetamide. This was followed by rehydration of the gel pieces in 200 μl of 50 mM ammonium bicarbonate. The bands were digested with 30 μl of trypsin, and the samples were extracted twice using extraction buffer (50% acetonitrile: 45% water: 5% formic acid). The samples were dried using a vacuum centrifuge and reconstituted with 20 μl of 97.9% water, 2% acetonitrile, 0.1% formic acid prior to mass spectrometry analysis. The individual tryptic peptides were analysed by LC-MS/MS. MS/MS spectra were analysed using the Mascot Software package (Version 2.2, Matrixscience, London, UK).
Immunostaining
Cells were seeded onto 13-mm glass coverslips and fixed with 3.7% formaldehyde (or formaldehyde followed by ice-cold methanol for ATP synthase β-subunit detection). Permeabilisation of the cells was performed for 5 min with 0.5% Triton X-100 in PBS followed by incubation with primary antibody for an hour at room temperature. Coverslips were washed with 0.025% Tween in PBS extensively before adding secondary antibody. Coverslips were mounted on microscope slides using Vectashield containing DAPI (4,6-diamino-2-phenylindole).
Mitochondrial fractionation
Mitochondrial fractions from cells seeded in 10-cm dishes were isolated using commercially available mitochondrial isolation kits, which used a reagent-based procedure (Pierce Biotechnology, Rockford, IL, USA) or Dounce homogenisation (Sigma-Aldrich, Nottingham, UK) to disrupt cells. Both kits used differential centrifugation to isolate intact mitochondria, and purification of mitochondria was performed as recommended by the manufacturer. Mitochondria purified from MCF-7 cells by sucrose gradient was a kind gift from Professor A Storey (University of Oxford) and commercially prepared pure mitochondria from bovine heart were purchased from MitoSciences (Eugene, OR, USA).
Cellular ATP measurement
The total ATP levels were measured as described previously40 with slight modifications. Cells were seeded in 10-cm dishes. In all, 50–75% confluent cells were harvested by removing the media, washing with PBS and adding 200 μl of 5% tricholoroacetic acid. The cells were centrifuged at 16 000 × g for 5 min at 4 °C. The supernatant was removed to a fresh tube and immediately frozen at −80 °C. The ATP was neutralised for analysis by extracting the defrosted samples with an equal volume (100 μl) of freon (1,1,2 trichlorotrifluorethane) and trioctylamine. The aqueous upper layer was removed, and 5–10 μl was injected for HPLC analysis as described previously.40 ATP was identified and quantitated against commercially available ATP.
ATP synthase assay
ATP synthase activity from total cell lysates was measured by a kinetic colorimetric assay using the ATP synthase Specific Activity Microplate Assay kit (Abcam plc, Cambridge, UK) according to the manufacturer's instructions. Briefly, cells were detergent-extracted, and the ATP synthase was immune-captured within the 96-well plates. The ATP synthase activity was measured as the rate of ATP being hydrolysed to ADP. The production of ADP is coupled to the oxidation of NADH to NAD+, which was measured by absorbance at 340 nm using a POLARstar Omega multimode plate reader (Tecan, Reading, UK). The ATP synthase activity was calculated by expressing the ATP synthase activity relative to the quantity of ATP synthase captured in each well.
RT-PCR
RNA was isolated from cells using the TriZol reagent according to the manufacturer's instructions. One milligram of RNA was reverse transcribed using moloney murine leukaemia virus reverse transcriptase (MMLV) and random hexamers. PCR was performed using PAQ5000 DNA polymerase using standard conditions. Sequences for primers: p53: 5′ cccaagcaatggatgatttga, 3′ ggcattctgggagcttcatct; Bax: 5′ gatgattgccgccgtgga, 3′ cccaaccaccctggtcttg; p21: 5′ ggcagaccagcatgacagatt, 3′ gcggattagggcttcctctt; GLUT4: 5′ cttcatcattggcatgggttt, 3′ aggaccgcaaatagaaggaaga; TIGAR: 5′ ctgactgaaactcgctaagg, 3′ cagaactagcagaggagaga; SCO2: 5′ gcagcaaaagcgaacagaa, 3′ gtgatccagcaggtggaagt; GLS2: 5′ tgcctatagtggcgatgtctca, 3′ gttccatatccatggctgacaa.
Reporter assays
U2OS cells were seeded into 6-cm dishes and transfected with the appropriate plasmid DNA along with 100 ng β-galactosidase plasmid to monitor transfection efficiency. Cells were harvested 48 h posttransfection and lysed with 125 μl of reporter lysis buffer (Promega, Southampton, UK) and snap frozen and thawed at 37 °C. Luciferase activity was measured and normalised to β-galactosidase activity.
FACS analysis
Cells were seeded into 6-cm dishes and treated as appropriate before harvesting. Growth media were collected and adherent cells were lifted by adding 2 ml of trypsin per dish. The cells were pelleted (1000 r.p.m.) for 5 min at 4 °C and washed once with PBS. The cells were fixed overnight in ice-cold 70% ethanol/PBS (v/v). Fixed cells were washed once with PBS and stained in 4% (v/v) propidium iodide in the presence of 125 U/ml RNAse A. Stained cells were analysed by using flow cytometry (Accuri C6, BD Bioscience, Oxford, UK).
Acknowledgments
We thank the MRC, CRUK (Programme Grant C300/A13058), Rosetrees Trust and Eurocan Platform for supporting this work. SM was supported by a Graduate Scholarship from the Malaysian Ministry of Higher Education.
Author contributions
SM designed and performed the majority of the experiments and data analysis. MRS measured the cellular ATP levels. JM and BK performed the mass spectrometry. ASC guided the research, performed experiments, assisted in data analysis and wrote the paper. NLT conceived the project, directed the research and wrote the paper.
Glossary
- AMPK
AMP-activated protein kinase
- ADP
adenosine diphosphate
- ATM
ataxia telangiectasia mutated
- ATP
adenosine triphosphate
- Bcl2
B-cell lymphoma 2
- Chk2
checkpoint kinase 2
- COXIV
cytochrome c oxidase IV subunit
- DAPI
4′,6-diamidino-2-phenylindole
- GLS2
glutaminase 2
- GLUT4
glucose transporter type 4
- MOMP
mitochondrial outer membrane permeabilisation
- NT
non-targeting; OB, oligonucleotide/oligosaccharide-binding fold
- PARP
Poly [ADP-ribose] polymerase
- PCNA
proliferating cell nuclear antigen
- SCO2
SCO2 cytochrome c oxidase assembly protein
- Strap
stress-responsive activator of p300
- TCA
tricarboxylic acid cycle
- TIGAR
TP53-inducible glycolysis and apoptosis regulator
- TPR
tetratricopeptide repeat
- UV
ultraviolet
- WT
wild type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
Supplementary Material
References
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Coutts AS, La Thangue N. The p53 response during DNA damage: impact of transcriptional cofactors. Biochem Soc Symp. 2006;73:181–189. doi: 10.1042/bss0730181. [DOI] [PubMed] [Google Scholar]
- Wang SJ, Gu W. To be, or not to be: functional dilemma of p53 metabolic regulation. Curr Opin Oncol. 2014;26:78–85. doi: 10.1097/CCO.0000000000000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray-Zmijewski FSE, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- Demonacos C, Krstic-Demonacos M, La Thangue NBA. TPR motif cofactor contributes to p300 activity in the p53 response. Mol Cell. 2001;8:71–84. doi: 10.1016/s1097-2765(01)00277-5. [DOI] [PubMed] [Google Scholar]
- Demonacos C, Krstic-Demonacos M, Smith L, Xu D, O'Connor DP, Jansson M, et al. A new effector pathway links ATM kinase with the DNA damage response. Nat Cell Biol. 2004;6:968–976. doi: 10.1038/ncb1170. [DOI] [PubMed] [Google Scholar]
- Adams CJ, Graham AL, Jansson M, Coutts AS, Edelmann M, Smith L, et al. ATM and Chk2 kinase target the p53 cofactor Strap. EMBO Rep. 2008;9:1222–1229. doi: 10.1038/embor.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams CJ, Pike AC, Maniam S, Sharpe TD, Coutts AS, Knapp S, et al. The p53 cofactor Strap exhibits an unexpected TPR motif and oligonucleotide-binding (OB)-fold structure. Proc Natl Acad Sci USA. 2012;109:3778–3783. doi: 10.1073/pnas.1113731109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Thorens B, Mueckler M. Glucose transporters in the 21st century. Am J Physiol Endocrinol Metab. 2010;298:E141–E145. doi: 10.1152/ajpendo.00712.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. Metabolic regulation by p53 family members. Cell Metab. 2013;18:617–633. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, et al. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi L, Morselli E, Kepp O, Vitale I, Pinti M, Kroemer G. Mitochondrial liaisons of p53. Antioxid Redox Signal. 2011;15:1691–1714. doi: 10.1089/ars.2010.3504. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, et al. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- Leu JI, Dumont P, Hafey M, Murphy ME, George DL. Mitochondrial p53 activates Bak and causes disruption of a Bak-Mcl1 complex. Nat Cell Biol. 2004;6:443–450. doi: 10.1038/ncb1123. [DOI] [PubMed] [Google Scholar]
- Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, et al. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Green DR, Kroemer G. The pathophysiology of mitochondrial cell death. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- Westphal D, Dewson G, Czabotar PE, Kluck RM. Molecular biology of Bax and Bak activation and action. Biochim Biophys Acta. 2011;1813:521–531. doi: 10.1016/j.bbamcr.2010.12.019. [DOI] [PubMed] [Google Scholar]
- Rossignol R, Gilkerson R, Aggeler R, Yamagata K, Remington SJ, Capaldi RA. Energy substrate modulates mitochondrial structure and oxidative capacity in cancer cells. Cancer Res. 2004;64:985–993. doi: 10.1158/0008-5472.can-03-1101. [DOI] [PubMed] [Google Scholar]
- Marroquin LD, Hynes J, Dykens JA, Jamieson JD, Will Y. Circumventing the Crabtree effect: replacing media glucose with galactose increases susceptibility of HepG2 cells to mitochondrial toxicants. Toxicol Sci. 2007;97:539–547. doi: 10.1093/toxsci/kfm052. [DOI] [PubMed] [Google Scholar]
- Salomon AR, Voehringer DW, Herzenberg LA, Khosla C. Understanding and exploiting the mechanistic basis for selectivity of polyketide inhibitors of F(0)F(1)-ATPase. Proc Natl Acad Sci USA. 2000;97:14766–14771. doi: 10.1073/pnas.97.26.14766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M, Single B, Castoldi AF, Kuhnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J Exp Med. 1997;185:1481–1486. doi: 10.1084/jem.185.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hresko RC, Heimberg H, Chi MM, Mueckler M. Glucosamine-induced insulin resistance in 3T3-L1 adipocytes is caused by depletion of intracellular ATP. J Biol Chem. 1998;273:20658–20668. doi: 10.1074/jbc.273.32.20658. [DOI] [PubMed] [Google Scholar]
- Skulachev VP. Bioenergetic aspects of apoptosis, necrosis and mitoptosis. Apoptosis. 2006;11:473–485. doi: 10.1007/s10495-006-5881-9. [DOI] [PubMed] [Google Scholar]
- Altman BJ, Rathmell JC. Metabolic stress in autophagy and cell death pathways. Cold Spring Harb Perspect Biol. 2012;4:a008763. doi: 10.1101/cshperspect.a008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Arago M, Formentini L, Cuezva JM. Mitochondria-mediated energy adaption in cancer: the H(+)-ATP synthase-geared switch of metabolism in human tumors. Antioxid Redox Signal. 2012;19:285–298. doi: 10.1089/ars.2012.4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izyumov DS, Avetisyan AV, Pletjushkina OY, Sakharov DV, Wirtz KW, Chernyak BV, et al. "Wages of fear": transient threefold decrease in intracellular ATP level imposes apoptosis. Biochim Biophys Acta. 2004;1658:141–147. doi: 10.1016/j.bbabio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Sorrentino G, Mioni M, Giorgi C, Ruggeri N, Pinton P, Moll U, et al. The prolyl-isomerase Pin1 activates the mitochondrial death program of p53. Cell Death Differ. 2013;20:198–208. doi: 10.1038/cdd.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erster S, Mihara M, Kim RH, Petrenko O, Moll UM. In vivo mitochondrial p53 translocation triggers a rapid first wave of cell death in response to DNA damage that can precede p53 target gene activation. Mol Cell Biol. 2004;24:6728–6741. doi: 10.1128/MCB.24.15.6728-6741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Sesaki H, Qi X. Drp1 stabilizes p53 on the mitochondria to trigger necrosis under oxidative stress conditions, in vitro and in vivo. Biochem J. 2014;461:137–146. doi: 10.1042/BJ20131438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminen AI, Eskelinen VM, Haikala HM, Tervonen TA, Yan Y, Partanen JI, et al. Myc-induced AMPK-phospho p53 pathway activates Bak to sensitize mitochondrial apoptosis. Proc Natl Acad Sci USA. 2013;110:E1839–E1848. doi: 10.1073/pnas.1208530110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies L, Paraskevopoulou E, Sadeq M, Symeou C, Pantelidou C, Demonacos C, et al. Regulation of glucocorticoid receptor activity by a stress responsive transcriptional cofactor. Mol Endocrinol. 2011;25:58–71. doi: 10.1210/me.2010-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- Moss J, Tinline-Purvis H, Walker CA, Folkes LK, Stratford MR, Hayles J, et al. Break-induced ATR and Ddb1-Cul4(Cdt)(2) ubiquitin ligase-dependent nucleotide synthesis promotes homologous recombination repair in fission yeast. Genes Dev. 2010;24:2705–2716. doi: 10.1101/gad.1970810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.