Last year three different groups led by Feng Zhang1 at the Broad Institute, George Church2 at Harvard Medical School, Jin-Soo Kim3 at Seoul National University and Jennifer Doudna4 at the University of California Berkeley, have adapted the prokaryotic immunity component, Clustered, Regularly Interspaced, Short Palindromic Repeats (CRISPR), to achieve genome engineering in mammalian cells. The impact of this approach was immediate; within a few months, several scientific reports have implemented or adapted this technique, highlighting its potential power. How effective is this new technology and will it render obsolete classical RNA interference and conventional in vivo mouse knockout strategies? And importantly, will it have an impact on the lives of the non-scientific population, such as by accelerating the generation of genetically modified foodstuffs and opening the possibility to correct and cure human genetic diseases?
Effective genome-editing systems are the Holy Grail for molecular biologists although the methods currently in use appear cumbersome by comparison with the CRISPR technology. CRISPR has only been developed as a result of the continued interest of biologists in elementary organisms, and the identification in prokaryotes of an innovative tool with which to modify the mammalian genome.
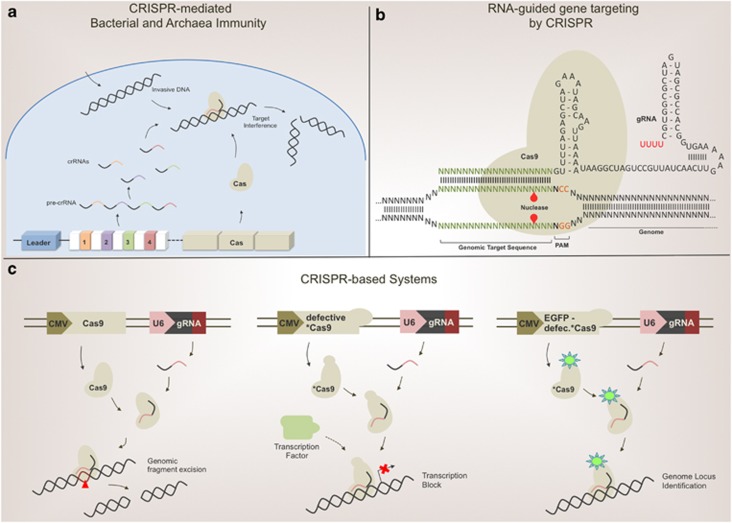
CRISPR is a natural RNA-guided DNA nuclease system used by archaea and bacteria to protect against invasive phage or plasmid DNAs. The CRISPR system is genomically encoded by the prokaryotic chromosome and includes a variable number of short repeats separated by non-repetitive spacer sequences, which are derived from previously encountered foreign DNAs. These spacers can match ‘proto-spacer' sequences of invasive DNAs, functioning as a genomically recorded immunological memory. This region is flanked by a ‘leader' sequence and by cas genes. The cas genes encode the protein component of the interference machinery. In the active machinery, the Cas protein loaded with the mature CRISPR RNA (crRNA) is delivered to the target DNA by double-strand base pair complementarity, and the CRISPR-associated nucleases cleave the invader nucleic acid (Figure 1a).5 The Cas9 protein, a type II CRISPR-associated Cas, exerts an endonuclease function by causing double-strand breaks (DSBs) in DNA. Exploiting the Cas9's activity and designing crRNA (also named guide RNAs, gRNA) to target mammalian genome sequences, Zhang, Church, Kim and Doudna have developed a method to surgically operate on any genomic region. Transfection of mammalian expression vectors containing CRISP components, including a nuclear-localized Cas9 protein, resulted in the disruption of intended target genes in different cell lines. Cas9-dependent DSBs were generated at the designed loci and consequent activation of the mutagenic non-homologous end joining (NHEJ) machinery finalized gene disruption with reasonable efficiency (Figure 1b).
Figure 1.
(a) CRISPR loci are included in the prokaryotic chromosome. A variable number of repeats are separated by spacer sequences, which derive from previously encountered nucleic acids of invasive microorganisms or ancestors. The cas gene, encoding the protein component of CRISPR system, flanks this region. Transcription and processing of precursor crRNA produces mature crRNAs. The protein product of the cas gene is loaded with the crRNA, generating the effector complex. CRISPR pathway activation results in DSBs at matching proto-spacer sequences included within invasive DNAs. This bacterial immunity response overcomes phage infection or exogenous DNA invasion. (b) CRISPR/Cas9-catalyzed cleavage of target genomic DNA in cells. Schematic representation of target DNA and chimeric gRNA. The proto-spacer adjacent motif (PAM) is an NGG sequence required for Cas9 recognition. Red triangles indicate cleavage sites. Cas9 opens the DNA duplex and cleaves both strands upon recognition of a target sequence by the gRNA, but only if the correct PAM is present at the 3′ end. (c) Schematic representation of CRISPR-based systems in action. Left panel, RNA-guided targeting of genes in mammalian cells requires expression of Cas9 protein, modified with a nuclear localization sequence, and a gRNA targeting the genomic locus of interest. The Cas9–gRNA effector complex identifies the targeting locus and excises the sequence from the genome. The central panel describes the CRISPRi system. A defective Cas9 (*Cas9) lacks endonuclease activity. When coexpressed with a specific gRNA, *Cas9 recognizes the locus in genomic regulatory regions, specifically interfering with transcriptional elongation, RNA polymerase binding or transcription factor binding. The CRISPRi system does not alter genomic sequences, but only interferes with regulatory elements. The right panel provides an overview of CRISPR imaging. Imaging of genomic elements in living cells is possible by gRNA-directed defective EGFP-fused Cas9. Sequence-specific enrichment of fluorescence signals can be measured by microscopy techniques. CMV, cytomegalovirus promoter; U6, U6-polymerase III promoter
What about specificity and off-targets effects? One of the major limitations of the widely used genome-editing technologies based on transcription activator-like effector nucleases (TALENs) and zinc finger nucleases (ZNFs) is their off-target effects.6, 7 Homologous recombination minimizes off-target effects of CRISPR. A mutant form of Cas9 (a nickase endonuclease), generating only DNA single-strand breaks, yielded similar homologous recombination but a lower NHEJ rate, thus reducing the NHEJ-dependent insertions, deletions and chromosomal rearrangements. Most of the potential of CRISPR indeed lies in the flexibility of the Cas9 protein. On the other hand, the possibility to design gRNAs for every genomic locus boosts the potential of this technology. With a catalytically active Cas9 protein, the CRISPR approach can generate gene disruption, whereas a defective Cas9 (dCas9) can either enable or disable the expression of a defined gene. Coupling dCas9 with a transcriptional activator or repressor has made possible the manipulation of specific gene transcription without altering genomic sequences.8 A system, called CRISPRi, employs a dCas9 lacking endonuclease activity that, when coexpressed with a gRNA, generates a DNA recognition complex that can specifically interfere with transcriptional elongation, RNA polymerase binding or transcription factor binding (Figure 1c).9 However, the genomic-editing power of CRISPR goes beyond gene disruption. The cellular homologous recombination machinery can replace the DNA locus with an engineered donor DNA fragment, containing a region with homology to the DSB site. This method enables the opportunity to integrate external DNA sequences into the genome. In addition, by adapting CRISPR as an imaging tool it acquires the potential to significantly improve the capacity to study genomic loci in live human cells. Thus, using an EGFP-tagged endonuclease-deficient Cas9 protein and an optimized sgRNA, the Huang group reported effective imaging of repetitive elements in telomeres and coding genes in living cells (Figure 1c).10 Now, the inventors of CRISPR-Cas9, Zhang group, have adapted their method in a genome-wide knockout screen of human cells. Using lentiCRISPR, they have targeted 18 080 genes in melanoma and embryonic stem cells, and cell with a library of 64 751 unique gRNAs.11, 12 However, as for all the possible applications, being a particular new technology the limitations of CRISPR usage in this approach are still under careful assessment.
Heritable gene targeting in the mouse and rat has also been achieved by the CRIPSPR system. The generation of a double-gene knockout rat was obtained with a single microinjection, and the germline-transmission efficiency was reasonably high in both mice and rats.13 In plants, CRISPR has been shown to effectively inactivate multiple genes in rice and wheat.14 This application in plants has the potential, for example, to confer resistance to parasitic organisms.
CRISPR allows the possibility to knockout genes in cells, to easily alter simultaneously multiple genes to study their interactions, and to create animal models much more quickly than before. The opportunity to precisely and selectively manipulate genomes is potentially opening a new era in genetic engineering. The cost- and time-efficiency of this technology compared with previous methods should boost its diffusion not only throughout basic science, but also in agriculture and medicine. Indeed, if high-fidelity target recognition is confirmed, the biggest challenge ahead for CRISPR is to correct human disease-associated mutations, finally providing an effective gene therapy strategy for human genetic diseases.
References
- Cong L, et al. Science. 2013. pp. 819–823. [DOI] [PMC free article] [PubMed]
- Mali P, et al. Science. 2013. pp. 823–826. [DOI] [PMC free article] [PubMed]
- Cho SW, et al. Nat Biotechnol. 2013. pp. 230–232. [DOI] [PubMed]
- Jinek M, et al. Elife. 2013. p. e00471. [DOI] [PMC free article] [PubMed]
- Wiedenheft B, Sternberg SH, Doudna JA. Nature. 2012. pp. 331–338. [DOI] [PubMed]
- Mussolino CM, et al. Nucleic Acids Res. 2011. pp. 9283–9293. [DOI] [PMC free article] [PubMed]
- Kim E, et al. Genome Res. 2012. pp. 1327–1333. [DOI] [PMC free article] [PubMed]
- Gilbert LA, et al. Cell. 2013. pp. 442–451. [DOI] [PMC free article] [PubMed]
- Qi LS, et al. Cell. 2013. pp. 1173–1183. [DOI] [PMC free article] [PubMed]
- Chen B, et al. Cell. 2013. pp. 1479–1491. [DOI] [PMC free article] [PubMed]
- Wang T, et al. Science. 2014. pp. 80–84. [DOI] [PMC free article] [PubMed]
- Shalem O, et al. Science. 2014. pp. 84–87. [DOI] [PMC free article] [PubMed]
- Li D, et al. Nat Biotechnol. 2013. pp. 681–683. [DOI] [PubMed]
- Shan Q, et al. Nat Biotechnol. 2013. pp. 686–688. [DOI] [PubMed]