Abstract
Several cellular signaling pathways are regulated by ADP-ribosylation, a posttranslational modification catalyzed by members of the ARTD superfamily. Tankyrases are distinguishable from the rest of this family by their unique domain organization, notably the sterile alpha motif responsible for oligomerization and ankyrin repeats mediating protein-protein interactions. Tankyrases are involved in various cellular functions, such as telomere homeostasis, Wnt/β-catenin signaling, glucose metabolism, and cell cycle progression. In these processes, Tankyrases regulate the interactions and stability of target proteins by poly (ADP-ribosyl)ation. Modified proteins are subsequently recognized by the E3 ubiquitin ligase RNF146, poly-ubiquitinated and predominantly guided to 26S proteasomal degradation. Several small molecule inhibitors have been described for Tankyrases; they compete with the co-substrate NAD+ for binding to the ARTD catalytic domain. The recent, highly potent and selective inhibitors possess several properties of lead compounds and can be used for proof-of-concept studies in cancer and other Tankyrase linked diseases.
Keywords: Cancer, drug design, enzyme, inhibitor, tankyrase, telomere, Wnt/β-catenin signaling.
1. INTRODUCTION
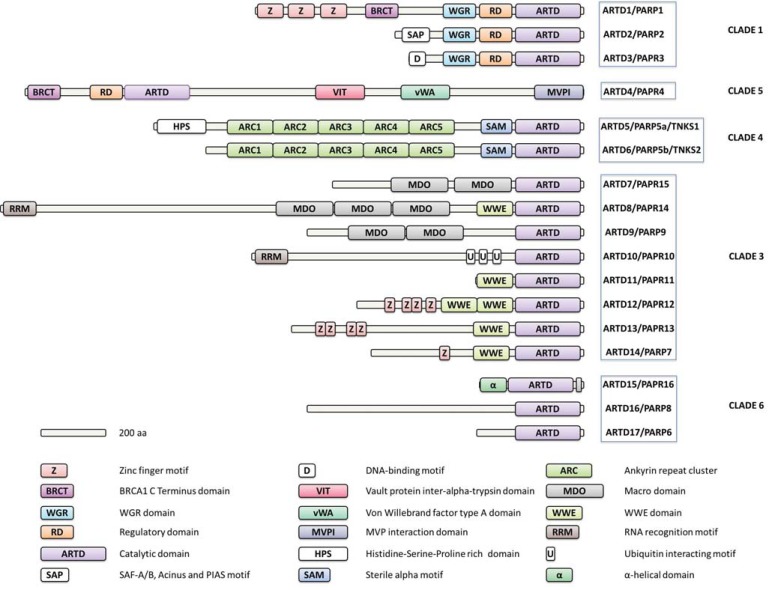
Tankyrases belong to the Diphtheria toxin-like ADP-ribosyltransferase (ARTD) enzyme superfamily (EC 2.4.2.30) that comprises 17 members in humans but has deep evolutionary roots and members of the family are found in lower eukaryotes, plants, bacteria, and archaea [1]. ARTDs, also known as poly(ADP-ribose)polymerases (PARPs), catalyze the transfer of an ADP-ribose from the co-substrate NAD+ on a target protein. This covalent posttranslational modification leads to the attachment of one or several ADP-ribose (ADPr) molecules to the target protein. ARTDs can be classified as polymerases (pARTDs: ARTD1-6), monotransferases (mARTDs: ARTD7, 8, 10-12, 14-17) and supposedly inactive enzymes (ARTD9, 13) based on the amino acid differences at the active site supported by experimental data on some enzymes of this class [1, 2].
Tankyrase 1 (telomeric repeat binding factor 1 (TRF1)-inter-acting ankyrin-related ADP-ribose polymerase; TNKS1/ARTD5/ PARP5a) and Tankyrase 2 (TNKS2/ARTD6/PARP5b) form a distinct subgroup of the polymer forming ARTDs. The two Tankyrases share 82% sequence identity and are distinguished from the rest of the family by a unique domain structure containing several ankyrin repeats and a sterile alpha motif (SAM) (Fig. 1). Ankyrin repeats are a common protein motif that functions as protein interaction modules [3] whereas SAM domain is prone to oligomerization and forms homo- and heterooligomers [4]. In addition to structural similarities, Tankyrases also appear to share significant functional redundancies: While a double knockout of TNKS1 and TNKS2 is embryonically lethal, knockouts of either TNKS1 or TNKS2 in mice bear only mild phenotypes [5-8].
Fig. (1).
Domain organization of human ARTD enzymes. Evolutionary relation according to Citarelli and co-workers [40].
Since their discovery as telomere-associated proteins in human cells [9-11], Tankyrases have been linked to additional cellular functions, such as mitotic progression, glucose metabolism, stress granule formation, Wnt signaling, and possibly proteasome regulation [9, 12-16]. Reflecting the multitude of their target proteins andcellular functions, Tankyrases have been implied in a variety of diseases including cancer, Cherubism, systemic sclerosis, Herpes simplex and Epstein Barr viral infections, and severe obesity [7, 17-24]. Altered levels of TNKS1 and/or TNKS2 expression have, in particular, been reported in lung cancer [25], gastric cancer [26], bladder cancer [27], astroglial brain tumors [28, 29], pancreatic adenocarcinoma [30], breast cancer [31], and colon cancer [32, 33]. While a therapeutic effect of TNKS inhibition has been observed in selected tumor models, it remains to be established in which setting TNKS inhibition may give a reliable therapeutic benefit [15, 23, 34-38].
Within the ARTD superfamily, significant academic and industry effort has been put into developing compounds that inhibit the catalytic domain of ARTD1, due to its role in DNA repair. Currently several ARTD1 inhibitors are tested in clinical trials (reviewed recently by Curtin & Szabo [39]). Tankyrases have more recently emerged as attractive drug targets, and increasing interest has been put into validating Tankyrases as targets and into developing selective TNKS inhibitors. In this review we summarize the molecular basis for Tankyrases as drug targets, recent advances in the field of TNKS inhibitor development, and give future perspectives for TNKS studies, inhibitor development, and potential applications.
2. STRUCTURE OF TANKYRASES
Human Tankyrases are multidomain proteins encompasing 1327 and 1166 residues for TNKS1 and TNKS2, respectively. No structural data are available for the full-length Tankyrases, but some insights have been gained through crystallography of isolated protein domains. At the C-terminus, both Tankyrases have a catalytic ARTD domain. The ARTD domain has been characterized extensively by protein crystallography and its structure has contributed to TNKS inhibitor development [41, 42]. The ARTD domain is highly conserved between TNKS1 and TNKS2 showing 89% overall sequence identity. Structural details of the ARTD domain and similarities, as well as differences, to other ARTD enzymes will be discussed below. A conserved SAM domain is located N-terminal to the ARTD domain (Fig. 1). It is unclear whether the SAM domain influences the structure and activity of the adjacent ARTD domain. However, in analogy to SAM domains in other proteins [43], the tankyrase SAM domain is implicated in the formation of either homo- or heterooligomers [4]. The bulk of the Tankyrases consists of five ankyrin repeat clusters that are implicated in protein-protein interactions [44]. Two parts of the Tankyrase ankyrin repeats (ARC2-ARC3 and ARC4; Fig. 1) and their protein binding interactions with peptides have been studied by protein crystallography, yeast two hybrid systems, and pull down experiments [18, 44, 45]. In addition to the three described conserved domains that are shared by the two Tankyrases, TNKS1 contains a His, Pro, Ser rich (HPS) region at the N-terminus. Notably, a TNKS1a protein lacking the HPS region due to the use of an alternative promoter has been reported [8]. So far the structure and function of the HPS motif are unknown.
2.1. Catalytic Domain of Tankyrases
2.1.1. Function
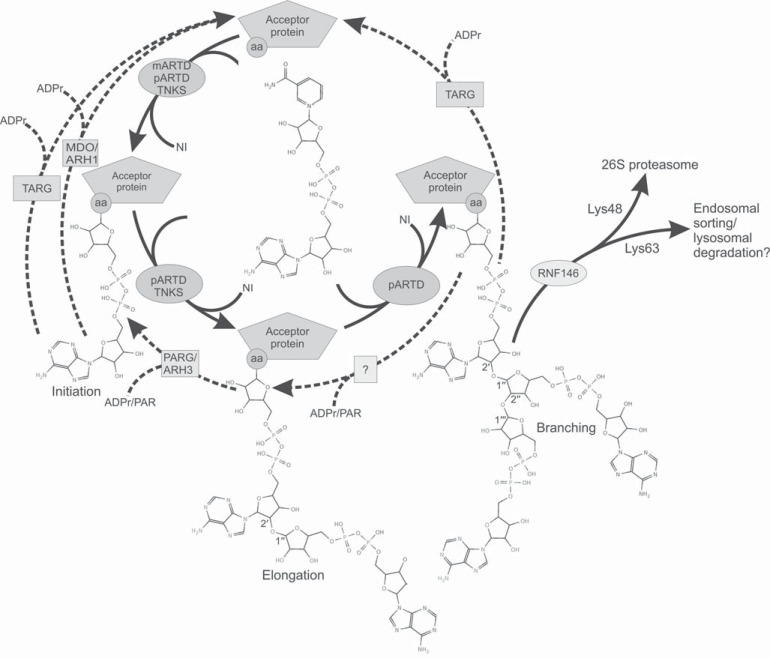
The catalytic domain of ARTDs is responsible for the ADP-ribosyltransferase activity. The domain catalyzes the hydrolysis of NAD+ to ADP-ribose (ADPr) and nicotinamide (Fig. 2). The nicotinamide is released from the binding site while ADPr is transferred to the amino acid side-chain of an acceptor protein in the initiation reaction. Known amino acid acceptors of the human ARTD catalyzed modification are Glu, Asp, and Lys [46]. However, Cys, Arg, Asn, diphthamide, and phospho-serine can also be modified by other ADP-ribosyltransferases [47]. The preferred acceptor amino acids for tankyrase modification have not been mapped in any of the acceptor proteins. The reaction continues with an elongation when the next ADPr unit is added to the existing modification leading to a poly ADP-ribose (PAR) chain. Based on the analogy to ARTD1 this process requires a conserved Glu at the active site (residue 1291 in TNKS1 and 1138 in TNKS2), which stabilizes the oxocarbenium intermediate and activates the ribose hydroxyl of the ADPr bound to the acceptor site for a nucleophilic attack [48, 49]. A branching reaction of the PAR chain has been demonstrated by two-dimensional thin-layer chromatography for ARTD1, but so far not for tankyrase-mediated PARsylation [50]. In vitro, the length on the PAR chain depends on the NAD+ concentration. While the average length of the PAR chain for Tankyrases has been shown to be 20 ADPr units [50], the chain length may increase with augmented NAD+ concentration, reaching 100 units with 100 µM NAD+ concentration in vitro. However, the prevalent form of modification at low NAD+ concentrations (100 nM) may be mono-ADP-ribosylation.
Fig. (2).
Covalent protein modification through ADP-ribosylation as catalyzed by ARTDs and removal of ADP-ribosylation. ADP-ribosylation reactions are shown with solid arrows, while the removal of the modifications is shown as dashed arrows. ARTDs catalyze the ADP-ribosylation onto Asp, Glu, and Lys residues (aa) of the acceptor proteins. PARG/ARH3 hydrolyze the O-glycosidic ribose-ribose 1"-2' bond [51, 52], while MDO1/2 remove the proximal modification [53]. TARG catalyzes the removal of PAR but also a single ADPr modification [54]. The catalytic activity responsible for removing the branching from the PAR chain has not been identified. RNF146 poly-ubiquitinates PARsylated proteins, which may subsequently be targeted for degradation in the 26S proteasome [55]. PARG, poly(ADP-ribose) glycohydrolase; ARH, ADP-ribosylhydrolase; TARG, terminal ADP-ribose protein glycohydrolase, MDO, macro domain; RNF146, E3 ubiquitin-protein ligase RNF146.
Kinetic constants for TNKS automodification (Km 1.5 mM, kcat 0.7 s−1) have been reported to be significantly lower than those measured for ARTD1 and this could affect the outcome of TNKS activities [50, 56]. ARTD1 is allosterically activated due to binding of damaged DNA [57]. An analogous mechanism could exist in Tankyrases via binding of substrate proteins, but so far such a mechanism has not been observed [44, 58, 59]. It is known, however, that the catalytic activity of tankyrase activity and other properties such as protein binding are modulated by posttranslational modifications.
2.1.2. Fold
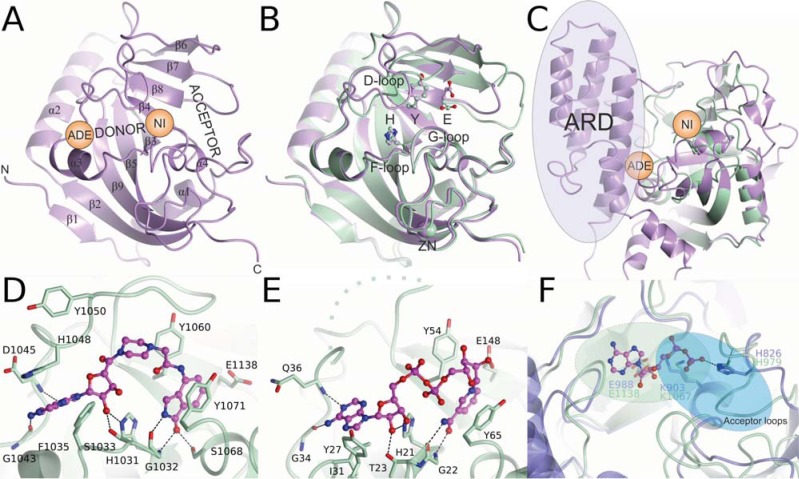
The catalytic domain of Tankyrases consists of two anti-parallel β-sheets surrounded by four α-helices (Fig. 3A). The overall structure of the domain is well-conserved within the ARTD family. However, Tankyrases lack the α-helical regulatory domain (ARD) present in other polymer forming ARTDs adjacent to the catalytic domain (Fig. 1 & 3C). The ARD of ARTD1 is located N-terminally to the catalytic domain and is shown to be involved in the DNA-dependent activation of ARTD1 [57]. A unique feature of the catalytic domain of Tankyrases is the presence of a CHCC-type zinc-finger motif of unknown function (Fig. 3B) [41]. This motif is located 25 Å from the catalytic Glu (1291 in TNKS1 and 1138 in TNKS2) and is unlikely to have a role in the catalytic activity but might play a structural role or may mediate interactions with nucleotides or proteins.
Fig. (3).
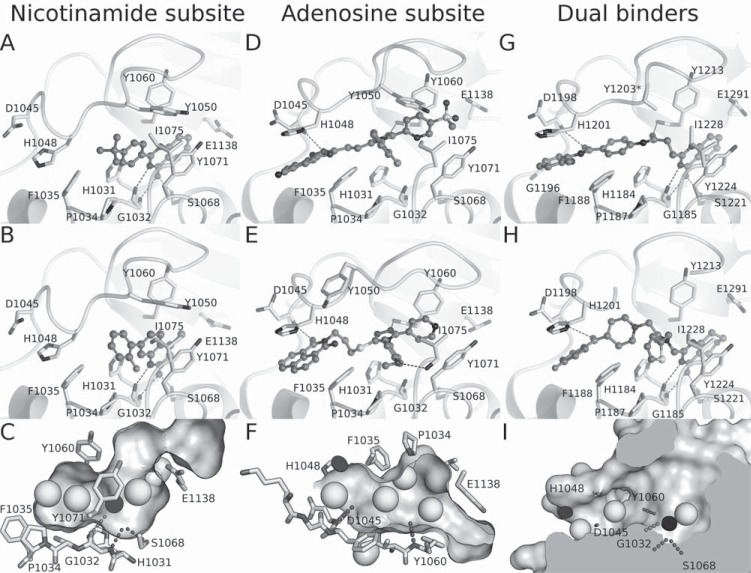
Structure and catalytic sites of Tankyrases. A) The donor and acceptor NAD+ binding sites of TNKS1 (PDB ID 2RF5). The nicotinamide (NI) and adenosine (ADE) subsites are labeled. N-terminus marks the approximate position of the SAM domain which is connected to the catalytic domain with a linker of 18 residues. B) Superposition of TNKS1 (purple) and TNKS2 (aquamarine) (PDB ID 3KR7) showing the HYE conserved triad and the zinc binding site. C) Superposition of TNKS2 and ARTD1 (purple) (PDB ID 3GJW). The regulatory domain (ARD) of ARTD1 is missing in Tankyrases. D) Binding of EB-47 to tankyrase 2 (PDB ID 4BJ9). E) Binding of NAD+ to Diphtheria toxin (PDB ID 1TOX). The disordered D-loop is shown as a dashed line. F) Differences of the acceptor sites of ARTD1 (PDB ID 1A26) and TNKS2 (PDB ID 4HYF). The ADP moiety of an NAD+ analog bound to the ARTD1 is shown. For branching reaction ADP should rotate 180 degrees (from green to blue area), which is blocked in TNKS by acceptor loops.
2.1.3. Catalytic Site
The catalytic domain of ARTDs consists of a donor site, which binds and hydrolyses NAD+, and an acceptor site, which accommodates the target protein to be modified or a PAR chain to be elongated (Fig. 3A). No crystal structures of any ARTD in complex with NAD+ have been determined hampering the analysis of the catalytic mechanism. Based on the Diphtheria toxin (a bacterial ADP-ribosyltransferase)-NAD+ complex (PDB ID: 1TOX) [60] the donor site can be divided into two parts, namely the nicotinamide and adenosine subsites. The catalytic domain includes three central amino acids (the conserved HYX triad) that are situated near the nicotinamide subsite, where the hydrolysis of the NAD+ occurs. These residues are His1184, Tyr1213, Glu1291 for TNKS1, and His1031, Tyr1060, Glu1138 for TNKS2 (Fig. 3B). The conserved triad of the active ARTDs always contains His and Tyr while the third amino acid varies. A Glu in the triad (HYE) is found in all pARTDs, while variant triads HYI, HYL, and HYY have presumably only mono-transferase activity [2] (Fig. 2). This is also supported by the observation that a Glu-to-Gln mutation converts ARTD1 to a mARTD [61].
In extension of the studies on Diphtheria toxin and other ARTDs, the crystal structure of TNKS2 in complex with nicotinamide validated the binding of a nicotinamide moiety of NAD+ to the subsite [62]. Crystallographic evidence of NAD+ binding to ARTDs was also acquired through a crystal structure of TNKS2 in complex with a NAD+ mimic inhibitor, EB-47 [63]. The isoindolinone moiety, a nicotinamide isostere of EB-47, binds to the nicotinamide subsite in a similar fashion as nicotinamide (Fig. 3D). The nicotinamide isostere extends to the adenosine subsite and the adenosine moiety of the inhibitor overlaps with the adenosine of NAD+ in Diphtheria toxin (Fig. 3E). In the tankyrase crystal, the adenosine moiety of EB-47 is rotated by 180 degrees in comparison to the NAD+ in Diphtheria toxin structure.
The donor NAD+ binding site in Tankyrases is rather flexible and it has a closed conformation in the crystal structures in the absence of ligand binding. This is due to a D-loop lining the donor site, which closes the NAD+ binding groove. The D-loop opens up upon binding of NAD+ or bulky inhibitors and it is often poorly resolved in reported crystal structures. Although identical in sequence, the D-loop assumes rather different conformations in the apo-structures of TNKS1 and TNKS2 (Fig. 3B).
The acceptor site in the ARTD domain is adjacent to the nicotinamide subsite allowing the transfer of an ADPr to a target protein in the initiation reaction, or to a growing PAR during chain elongation. In the initiation reaction, when ADPr is transferred to an acceptor amino acid side chain (Fig. 2), NAD+ is bound to the donor site, while the target protein occupies the acceptor site. After initiation the next NAD+ binds to the donor site while the growing PAR chain is situated in the acceptor site with its 2′ ribose hydroxyl activated by the catalytic Glu. There is no direct crystallographic evidence of the ADPr binding to the acceptor site. However, a fragment of a NAD+ analog containing the adenosine diphosphate (ADP) part has been observed in the acceptor site of ARTD1 [49] (Fig. 3F). The residues positioning the ADP moiety in the acceptor site and allowing the growing PAR chain to bind to the acceptor site for further elongation are conserved between pARTDs. However, in mARTDs, these residues are not conserved implying that these proteins are not capable of binding ADP-ribose but only the substrate protein. The branching reaction, which occurs at the 1′′ hydroxyl instead of the 2′ hydroxyl of the ADPr, has not been observed in Tankyrases. For positioning of the 1′′ hydroxyl for the catalysis of the branching, the polymer would have to rotate 180 degrees in relation to the elongation reaction. With the exception of Tankyrases, the acceptor site groove in pARTDs is wide enough to accommodate the polymer in both directions (Fig. 3F). The more constricted character of the tankyrase acceptor site is due to acceptor site loops closing a part of the site (Fig. 3F) and may explain the observed lack of TNKS catalyzed branching reaction.
2.2. SAM Domain
The catalytic domain of both Tankyrases is preceded by a SAM domain. SAM domains are all α-helical domains of approximately 70 residues. They have been identified in several proteins where they mediate protein-protein interactions and also form homo- and hetero-oligomers with themselves and other SAM domains, respectively. SAM domains have also been found to bind DNA, RNA, and lipids [64-66].
The SAM domain of TNKS1 has been reported to oligomerize to form large (>30 molecule) complexes [4]. Also the full-length TNKS1 polymerizes through SAM domain and TNKS1 and TNKS2 are capable of forming hetero-oligomers in vivo although it remains to be seen whether that reflects physiological conditions [58]. The polymerization of Tankyrases is reversible, and the dissociation of the molecules is followed by autoPARsylation [4]. Tankyrase oligomers interact with and PARsylate TRF1 suggesting that SAM domain is required for optimal catalytic activity [4]. The SAM domain is located close to the catalytic domain in primary structure with 18 residue linkers implying that it may affect the donor NAD+ binding site (Fig. 3A). We have observed that the SAM domain affects the potency of certain inhibitors against TNKS2, when compared to the catalytic domain alone [67, 68]. This suggests that the SAM domain interacts with the catalytic domain, or possibly affects the catalytic properties through dimerization or multimerization.
2.3. Ankyrin Repeats
Ankyrin repeats, a common protein motif functioning as protein-protein interaction modules, constitute an extensive part of Tankyrases (Fig. 1). The ankyrin repeats of Tankyrases can be divided into five domains called ankyrin repeat clusters (ARCs) and each of these ARCs consist of five stacked ankyrin repeats [18]. The binding properties of the ARCs have mostly been studied using peptides. ARCs 1, 2, 4, and 5 bind substrate peptides of various proteins with a similar binding mode. The tankyrase-binding motif (TBM) of the substrate proteins usually contains a consensus sequence RXXPDG, which appears to be the same for the four ARCs [44, 58, 69, 70]. The motif was recently extended by Guettler and co-workers. The Arg and Gly at positions 1 and 6 are the most critical for the binding, while positions 2 and 3 can be almost any amino acid except Phe. Position 4 favors small, hydrophobic residues and position 5 has - predominantly but not exclusively - Asp. Finally, a wide range of amino acids except Pro are found at position 7 and a preference for acidic residues has been observed at position 8 [18].
The target protein Axin [15, 55, 71] has been shown to bind to tankyrase in a bivalent binding mode with its N-terminal end [45] involving the Axin residues 19-30 (motif 1) and residues 60-71 (motif 2). The tankyrase binding sites in Axin are close to the binding site of the structural protein Adenomatous polyposis coli (APC). APC frequently interacts with Axin and both form the core structural proteins of the β-catenin destruction complex (DC) that regulates canonical Wnt signaling, but Axin may also interact with other protein complexes [45, 72-74].
Curiously, TNKS ARC3, whose substrate binding surface is poorly conserved, does not bind tankyrase substrate peptides [18, 70]. The exact function of ARC3 remains unknown, although it could interact with unidentified substrates or participate in the binding of full-length proteins.
Individual ARCs have micromolar affinities for substrate peptides but the presence of more than one TBM in a protein increases the avidity [18, 45]. The binding of a substrate protein by ARCs is required for subsequent PARsylation, but may not be sufficient for the modification as certain substrates are not modified upon binding (Table 1). Verified binding partners link Tankyrases to various signaling events, metabolism, and when interrupted, to diseases such as Cherubism (Table 1). Based on sequence conservation, over 100 proteins potentially interacting with Tankyrases have been predicted [18]. Interestingly, although the binding partners are often PARsylated by Tankyrases, some binding partners can inhibit the PARsylation activity [75, 76].
Table 1.
Tankyrase binding partners.
| Protein | PARsylated | Reference |
|---|---|---|
| SH3 domain-binding protein 2 (3BP2) | Yes | [19] |
| Axis inhibition protein 1/2 (AXIN1/2) | Yes | [15] |
| Breakpoint cluster region protein (BCR) | Yes | [18] |
| Golgin-45 (BLZF1) | Yes | [77, 78] |
| Voltage-dependent L-type calcium channel subunit alpha-1S (CACNA1S) | N.D. | [79] |
| Cancer susceptibility candidate gene 3 protein (CASC3) | N.D. | [77] |
| Disrupted in schizophrenia 1 protein (Disc1) | Yes | [18] |
| Epstein-Barr nuclear antigen 1 (EBNA1) | Yes | [21, 80] |
| Cadherin family member 14 (Fat4) | Yes | [18] |
| Formin-binding protein 17 (FBP17) | N.D. | [81] |
| Glucose transporter type 4, insulin-responsive (GLUT4) | Yes | [82] |
| Growth factor receptor-bound protein 14 (GRB14) | N.D. | [12] |
| Homeobox protein Hox-B2 (HOXB2) | N.D. | [79] |
| Insulin-responsive aminopeptidase (IRAP) | Yes | [59] |
| Induced myeloid leukemia cell differentiation protein, short/long isoform (MCL-1L/MCL-1S) | No | [83] |
| BRISC and BRCA1-A complex member 1 (MERIT40) | Yes | [18] |
| NUMA1 protein (NUMA1) | Yes | [13] |
| Protein phosphatase 1 regulatory subunit 12C (PPP1R12C) | N.D. | [79] |
| DNA repair and recombination protein RAD54 (RAD54) | No | [18] |
| E3 ubiquitin-protein ligase RNF146 (RNF146) | N.D. | [55, 77] |
| Striatin | Yes | [18] |
| Telomeric repeat-binding factor 1 (TRF1) | Yes | [9] |
| 182 kDa tankyrase-1-binding protein (TAB182) | Yes | [44] |
| Tax1-binding protein 1 (TXBP151) | N.D. | [79] |
| Ubiquitin carboxyl-terminal hydrolase 25 (USP25) | N.D. | [79] |
N.D., not determined.
3. POSTTRANSLATIONAL MODIFICATIONS AND TANKYRASE ACTIVITY
Tankyrases catalyze a posttranslational modification of target proteins, but also itself through automodification. Only very recently we have gained some understanding of how this large modification controls the stability of proteins and of protein complexes. PARsylation by Tankyrases appears to be tightly coupled to ubitiquitination and proteasomal degradation of proteins. It is also reversible through the catalytic activity of several enzymes attenuating the effect of Tankyrases on substrate proteins. Furthermore, the activity and interactions of Tankyrases is controlled under certain conditions through phosphorylation and hydroxylation, although little is known about the effects of these modifications on the activity of Tankyrases.
3.1. PARsylation and Ubiquitination
The functions of Tankyrases are mediated by an interplay between the formation of protein/protein complexes through its ankyrin (and SAM) domains, and the disassembly of protein complexes through PARsylation, followed by protein degradation. Thus, a central protein modification controlling TNKS protein partnering and turn-over is the PARsylation reaction itself. The PAR chain, which is negatively charged, serves as a recognition site for the E3 ubiquitin-protein ligase RNF146, which interacts with iso-ADP-ribose moieties [55, 77]. Poly-ubiquitination of tankyrase target proteins by RNF146 is frequently followed by degradation in the 26S proteasome. It has been pointed out by Citarelli and co-workers that PARsylation and ubiquitination may both compete for Lys residues at target proteins [40], although the amino acid residues that are ADP-ribosylated by Tankyrases have not been identified. A further functional link between ADP-ribosylation and ubiquitination is the presence of a structurally related WWE domains in both E3 ubiquitin ligases such as RNF146 (but also the E3 ligases Deltex1, Deltex2, Deltex4, HUWE1, and ULF) and in some mARTDs (mARTD8, 11, 12, 13, and 14) (Fig. 1) but not in pARTDs [40, 84]. WWE domains of mARTDs such as ARTD11 recognizes the terminal ADPr, including the ones created by mARTDs, while WWE domain of the E3 ubiquitin ligases, such as RNF146, recognizes the iso-ADP-ribose moiety that is present only in PAR chains [85]. Hence, MARsylation through the WWE domain may induce the recruitment of further mARTDs as in the case of ARTD10 and ARTD8 [86]. In contrast, PARsylation may trigger an interaction with E3 ubiquitin ligases followed by poly-ubiquitination and ubiquitin-Lys residues have been identified in both TNKS1 and TNKS2. Ubiquitination itself can come in various forms and has not been studied thoroughly in the context of TNKS mediated PARsylation. Poly-ubiquitinated protein substrates conjugated by Lys48-linked ubiquitin chains (Lys48 of ubiquitin) are routed to the 26S proteasome. It has been shown in early studies that a chain of at least four Lys48 linked ubiquitins is required to cause destruction of a protein via the 26 proteasome [87-89]. Lys48 linked poly-ubiquitination has been coupled to the ubiquitination of Axin, RNF146, and tankyrase [15, 55, 77]. Other ubiquitin links involving any Lys (Lys6, Lys11, Lys27, Lys29, Lys33, or Lys63) or the amino-terminal Met (Met1) of the ubiquitin monomer are also possible, but may trigger alternative biological responses including trafficking and endosomal sorting or lysosomal degradation [87]. RNF146, when overexpressed in HEK293 cells, also induces Lys63 chains on TNKS2 [55]. It is hitherto unclear what biological response, if any, a Lys63 ubiquitin link in TNKS2 would cause.
3.2. De-PARsylation
Both MARsylation and PARsylation can be reversed by a number of enzymes, although studies have predominantly been carried out in the context of ARTD1 and need to be verified in the context of TNKS mediated PARsylation. Terminal ADP-ribose protein glucohydrolase (TARG/C6orf130) can remove the PAR chain en bloc at its ester bond with the target protein [54] (Fig. 2). The proteins poly(ADP-ribose) glycohydrolase (PARG) as well as the ADP-ribosyl hydrolase 3 (ARH-3) target poly ADP (1´´-2´) ribose-ribose bonds [90, 91] that are found within PAR chains (Fig. 2). PARG can have exoglycosidic activity in which the chain is reduced to single ADPr moieties. This activity appears to be the predominant form of PARG activity. Alternatively, PARG can also have endoglycosidic activity that causes release of larger PAR fragments [92]. It has been suggested that the relation between exoglycosidic and endoglycosidic PAR hydrolysis depends on the PAR/PARG ratio whereby endoglycosidic hydrolysis is seen when cells show excess PAR production as observed during cellular insults [92]. In contrast to TARG, PARG cannot cleave the ester bond linking the proximal ADPr unit directly to proteins [52] and thus is not capable of reversing MARsylations [92]. This task is taken by members of the Macro Domain (MDO) protein family [53, 54, 92, 93] that catalyze the removal of a single ADPr moiety (Fig. 2). This removal is dependent on the ADPr-amino acid both with the substrate protein. The proteins MDO1, MDO2 and TARG catalyze the hydrolysis of an Asp- or Glu-ADP ribose linkage [92]. The ADP-ribosyl hydrolase 1 (ARH-1) removes an Arg-ADP ribose linkage [94], whereas an enzyme that resolves Lys-ADP ribose linkages remains unaccounted for [95]. A link between de-MARsylation and tumorigenicity has been established for ARH-1. Arh-/- mice show a higher incidence of spontaneous lymphomas and adenocarcinomas with increased metastasis [96].
3.3. Phosphorylation and Hydroxylation of Tankyrases
The binding of TNKS to protein partners, as well as its catalytic activity can be attenuated by protein modifications. During mitosis, TNKS1 is phosphorylated by Polo-like kinase-1 (Plk1) and this modification leads to increased PARsylation activity of TNKS1 in cells due to an increased stability of the enzyme. Inhibition of Plk1 mediated phosphorylation impairs TNKS function in mitotic spindles and telomere homeostasis [97]. The identified phosphorylation sites are located in ARC5, but one (Thr1128 in TNKS1) is located near the acceptor site of the catalytic domain. Furthermore, mitotic phosphorylation of TNKS on Ser978, Thr982, Ser987, and Ser991 has been associated with Glycogen synthase kinase 3-β (GSK3β). GSK3β localizes to spindle poles similar to TNKS and its inhibition can lead to mitotic arrest. GSK3β mediated phosphorylation of TNKS did not alter autoPARsylation in vitro, but it has been speculated that GSK3β phosphorylation alters substrate binding [98]. Phosphorylation of Tankyrases has also been reported in the context of IRAP (insulin-responsive amino peptidase) and GLUT4 storage vesicles in 3T3-L1 adipocytes [98]. Tankyrase binds to IRAP and regulates GLUT4 storage vesicle release to the membrane. Tankyrase has been shown to be phosphorylated by mitogen-activated protein kinase (MAPK) in vivo as a result of insulin and growth-factor stimulation. This phosphorylation enhances PARsylation activity of TNKS in vitro but does not affect GLUT4 targeting [59]. The release of GLUT4 vesicle to the plasma membrane has been connected to Protein Kinase B (Akt) dependent phosphorylation in a process that involves a TNKS, Axin, and the Kinesin-like protein KIF3A. While insulin stimulates the TNKS, Axin, and KIF3A complex formation and membrane release of storage vesicles through inhibition of TNKS mediated PARsylation, Akt inhibition abrogates insulin mediated complex stabilization. It remains unclear whether TNKS is directly phosphorylated by Akt [82].
In a further posttranslational modification, ankyrin repeats, at least in TNKS2, are hydroxylated on several Asn and His residues by hypoxia-inducible factor 1-alpha inhibitor (FIH) [99, 100]. The role of this modification remains unclear, although it has been proposed that hydroxylation of the ankyrin domains could affect the interactions between TNKS and target proteins. Hydroxylation could also modulate hypoxic response by regulating the amount of FIH that is bound to ankyrin repeat domains and thus its availability to hydroxylate Hypoxia-inducible factor 1-alpha (HIF1α) [99, 101].
4. CELLULAR ROLES OF TANKYRASES
Tankyrases are segregated to multiple cellular locations. In human cells, TNKS has been found at telomeres [9, 11], at nuclear pores [102], at the Golgi complex [59, 82, 103], in the cytoplasm [18], at the cell membrane [104] and at the spindle poles [97, 103, 105-108]. The dynamic sub-cellular localizations of Tankyrases as well as their posttranslational protein modifications will influence protein/protein interactions. While TNKS1 and TNKS2 have a substantial functional overlap, subtle differences in their functionality, sub-cellular localization, and protein-protein interactions may exist, although they have not been thoroughly explored. The implications of substrate protein binding and PARsylation have been studied only in a few proteins (Table 1).
4.1. Interplay of Tankyrase and Axin in Wnt/β-catenin Signaling
Even for better studied TNKS target proteins such as the structural proteins Axin1/2, the field is far from understanding all intricate biological and biochemical consequences of TNKS interactions with its target protein. In the context of Wnt/β-catenin signaling, Axin is found at three sub-cellular localizations: the Wnt signalosome, the β-catenin destruction complex (DC) and the nucleus [109]. So far, TNKS/Axin interactions have only been mapped at the DC [15, 71, 110]. In addition to its role in Wnt/β-catenin signaling, Axin has been linked to transforming growth factor beta (TGF-β) signaling and stress-activated protein kinase (SAPK) signaling [111]. Moreover, Axin has been mapped to the mitotic spindle during mitosis [112] and to exocytic GLUT4-containing vesicles [82, 113].
In the Wnt/β-catenin signaling pathway, the DC is a gatekeeper that regulates β-catenin turnover. Axin is the rate limiting structural protein in the DC, where it forms a multiprotein complex with the structural protein APC, the priming kinase CK1α (casein kinase I isoform α), and the Ser/Thr kinase GSK3β [114]. The DC phosphorylates β-catenin, leading to its 26S proteasomal degradation [45, 72-74]. Alternatively, upon active Wnt signaling, Axin can associate with the Wnt signalosome whereby it binds the structural protein disheveled, recruits the kinases CK1α and GSK3β and forms a multiprotein complex around the transmembrane proteins LRP5/6 (low-density lipoprotein receptor-related protein 6) and frizzled [115]. As Axin is a non-abundant protein, its binding to the Wnt signalosome reduces the ability of the formation of a functional DC [74]. Furthermore, Axin has been described as a molecular chaperon that can shuttle β-catenin from the nucleus to the cytoplasm [109, 116]. Axin is enriched in the nuclei of cells from diverse cancer cell lines and tissues, pointing towards problems in the export of Axin from the nucleus in such cells [117, 118].
The current model predicts that in the presence of a functional DC, active TNKS reduces the stability of the DC by increasing the turnover of Axin. This leads to reduced β-catenin degradation and thus increased levels of Wnt signaling. TNKS PARsylates itself and the structural protein Axin [15]. Next the E3 ubiquitin ligase RNF146 binds to the PAR tails on both proteins through its WWE domain [119]. Subsequently, all three proteins are poly-ubiquitinated by RNF146 and undergo proteolysis in the 26S proteasome [55, 77, 104, 119]. TNKS inhibition in this scenario leads to increased DC stability and reduced Wnt/β-catenin signaling [15, 23, 71, 120]. TNKS- and RNF146-dependent ubiquitination of Axin may be opposed by the ubiquitin specific protease 34 (USP34) [121]. USP34 reverts the poly-ubiquitination of Axin, thereby stabilizing it and reducing, similar to TNKS inhibition, β-catenin signaling. A second ubiquitin-specific protease, USP25, has been linked to TNKS2. However, the function of this interaction remains unknown [18, 79].
Recently it was shown that the effect of TNKS inhibition on Wnt/β-catenin signaling through the DC may be rendered ineffective in colorectal cancer cells after prolonged Wnt stimulation. Chronic Wnt stimulation can induce the nuclear accumulation of LEF1 and B9L that shield β-catenin from binding to cytoplasmic Axin and thus from the regulation by TNKS [110]. Also a nuclear accumulation of Axin can attenuate the role of TNKS on Wnt/β-catenin signaling and examples have been presented that nuclear Axin no longer acts as an inhibitor but may act as a positive regulator of Wnt/β-catenin signaling [121, 122]. In colon carcinoma cell lines that show activation of Wnt/β-catenin signaling downstream of the DC, Axin and USP34 were found to positively regulate β-catenin-dependent transcription [121] whereby USP34 supported the nuclear accumulation of Axin [121].
The emerging picture is that TNKS and Axin act as a strongly context-dependent master regulators of Wnt/β-catenin signaling. This could have significant biological consequences. In mouse, a mutation that renders Axin2 protein more stable (Axin2canp) leads, as predicted, to decreased Wnt/β-catenin signaling in most tissues. However, in the late primitive streak stage, the mutation leads to a phenotype that is consistent with increased Wnt/β-catenin signaling and associated with the formation of an ectopic second tail [122]. A similar phenotype could be reproduced with the TNKS inhibitor IWR-1 [122].
The multiple roles of Axin may help explain the differential response of tumors to the TNKS inhibitor G007-LK as observed by Lau and co-workers in a broad array of colorectal cancer (CRC) cells harboring mutations in either APC or β-catenin [23]. In the APC mutant CRC cell lines Colo320DM, SW403, HCT-15, DLD-1, SW480, and SW620, a significant cytoplasmic stabilization of Axin2 was observed upon TNKS inhibition, accompanied by a reduction of nuclear active β-catenin. In the CRC cell lines Colo205 and HT-29, cytoplasmic Axin stabilization was seen but without an apparent effect on nuclear β-catenin. In the β-catenin mutant cell line LS174T, Axin stabilization was observed but nuclear β-catenin was increased. Understanding the engagement of TNKS in CRC cells is important as about 80% of all colon cancers show functionally relevant mutations in Wnt/β-catenin signaling pathway components comprising either inactivating truncations of the APC protein [123], loss-of-function mutations in Axin [124], or activating mutations in β-catenin [125].
In addition to colon carcinomas, which are seen as classic Wnt dependent tumors, a correlation between TNKS and β-catenin was reported in a variety of further tumors. In 51 patient astrocytomas a significant association was claimed between TNKS1 up-regulation, a positive β-catenin immunostaining, and the pathological grade [29]. Also in a subset of castration resistant prostate cancer cell lines an over-expression of β-catenin was observed by immunohistochemistry, and in the androgen-independent LNCaP-AI cell line the TNKS inhibitor XAV939 reduced growth [126]. Furthermore, a correlation between components of the Wnt/β-catenin signaling pathway and TNKS was found in cyclin E-expressing murine transgenic Non-small-cell lung carcinoma (NSCLC) models and a panel of 12 human lung cancer cell lines. In a subset of these NSCLC models TNKS1 and TNKS2 siRNA treatment, as well as XAV939 and IWR-1 treatment, increased cellular Axin1 and TNKS levels. In particular, XAV939 and IWR-1 treatments of the human lung cancer cell lines A549, Hop62, and H522 decreased proliferation of each of these lines, although high doses were needed [25]. Bao and co-workers showed in multiple breast cancer cell lines that TNKS inhibition by XAV939 or siRNA-mediated abrogation of TNKS expression increased Axin1 and Axin2 protein levels and attenuated Wnt-induced transcriptional responses but lead to growth reduction only in the serum deprived basal-like triple-negative breast cancer cell line MDA-MB-231 [36].
An interesting observation that was made by Tenbaum and co-workers is that PI3K or Akt inhibitor treatment leads to the nuclear accumulation of the transcription factor FOXO3a and induced metastasis in patient derived primary colon carcinoma that showed high nuclear β-catenin. This effect could be reversed by the TNKS inhibitor XAV939 [127]. Another interesting inhibitor connection has been established in NSCLC where it was shown that the Wnt/β-catenin pathway contributes to the maintenance of NSCLC cells during EGFR inhibition, while inhibition of TNKS by either XAV939 or IWR-1 in cell lines H322C (only XAV939 sensitive) and HCC4006 significantly increased the efficacy of EGFR inhibitors on growth inhibition and colony formation [37].
4.2. Tankyrase and GLUT4 Vesicles
TNKS and Axin interact not only in the context of Wnt/β-catenin signaling. In 3T3-L1 adipocytes, an Axin/KIF1a/TNKS complex has been implicated in an insulin dependent transport of GLUT4 vesicles from the Golgi apparatus to the cell surface. At the cell surface, the GLUT4 protein contributes to glucose transport into cells. GLUT4 exocytosis is dependent on the inhibition of PARsylation by TNKS through insulin induced Akt signaling [82]. In the absence of insulin-Akt signaling, Axin is PARsylated and the tertiary complex disintegrates, preventing GLUT4 from being passed to the cell surface [82]. How Akt signaling affects the tertiary complex has not been mapped. Insulin also modulates GLUT4 recycling from endosomes during insulin exposure [128]. In accordance with a role of TNKS in GLUT4 exocytosis, TNKS knock-out mice show altered insulin action and glucose metabolism and reduced epididymal white fat pads [7].
In human body, GLUT4 is expressed in select somatic cells such as muscle cells and adipocytes. Tumor cells exhibit elevated levels of glucose metabolism and a key rate-limiting step in glucose utilization is the transport of glucose across the plasma membrane. It has been shown that multiple myeloma cell lines exhibit dependence on GLUT4 [129]. However, a comprehensive analysis of the involvement of GLUT transporters and in particular GLUT4 in tumors is still lacking.
TNKS interacts also with IRAP, which co-localizes with GLUT4 in exocytotic vesicles [59]. IRAP has been described as the principal trimming aminopeptidase in endosomes and phagosomes, involved in MHC-I ligands presentation, but it is unclear whether TNKS would affect MHC-I cross presentation [130]. Insulin has a regulatory role in the IRAP/TNKS complex, and a stoichiometric phosphorylation of TNKS by MAPK has been shown upon insulin stimulation [59].
4.3. Tankyrases at Spindles and Centrosomes
The described locations and interactions of TNKS and Axin change during the M phase when TNKS re-localize to the spindles and centrosomes where the assembly and function of mitotic spindles require TNKS mediated PARsylation of spindle components [98]. Axin2 is also found in spindles. Furthermore, GSK3β, which is frequently associated with Axin, is recruited to the spindles and phosphorylates TNKS1 during mitosis [98]. It is not known how the GSK3β mediated phosphorylation affects the activity and binding properties of TNKS during mitosis but it has been speculated that it may affect TNKS interaction with NuMA [98]. Interestingly, another ARTD, ARTD3, also interacts with NuMA but the differential roles of both ARTD3 and TNKS has not been determined in the context of mitosis [13, 131, 132]. Nevertheless, both TNKS1 and ARTD3 are functionally required for mitotic progression and are co-precipitated with NuMA in COS-1 cells [11, 132]. PARsylation of TNKS1 and NuMA were found to be strongly enhanced in the presence of functional ARTD3 [132], and it has been speculated that their malfunction may lead to aneuploid cancer cell survival [133]. PARsylation of NuMA by TNKS1 is controlled through phosphorylation by Plk1, which colocalizes with TNKS1. Phosphorylation increases TNKS1 stability and TNKS1 catalyzed PARsylation [97]. Centrosome clustering that is regulated by NuMA is an important process in cell cycle progression, and centrosomes are frequently altered in tumors [134].
4.4. Tankyrases and Telomeres
Tankyrases have been associated with telomeres. Based on work in human HeLa cells, TNKS has initially been suggested to be involved in telomere homeostasis. Recently TNKS has shown to been linked to the regulation of telomere cohesion during mitosis [9, 11, 97]. Telomeres consist of repetitive TTAGGG nucleotide sequences that protect the chromatid ends and prevent fusion to other chromosomes during mitosis. Telomere repeats associate with a six-protein complex termed shelterin that includes TRF1, TRF2, the TRF1 interacting protein 2 (TIN2), the protection of telomeres 1 protein (POT1), the TIN2 and POT1 interacting protein TPP1, and the transcriptional repressor/activator protein RAP1 [135]. Through its ankyrin binding sites [18], TNKS associates with TRF1 and PARsylation of TRF1 by TNKS has been proposed to open the shelterin complex [9, 10]. While it is not clear whether the TNKS/TRF1 interactions occur during interphase - tankyrase lacks a nuclear localization signal [102] - TNKS has shown to be necessary for the resolution of sister chromatid association at mitosis. A knock down of TNKS1 leads to a mitotic arrest with unresolved sister-chromatid cohesion resulting in prolonged anaphase, and abnormal chromosome distribution and spindle morphology [11, 13, 136, 137].
The TNKS1/TRF1 interaction can be enhanced by Plk1 mediated phosphorylation of TNKS, increasing TNKS1 stability at the telomeres [97]. The Plk1 mediated phosphorylation appears to be a central regulatory process as TNKS that contains mutated Plk1 phosphorylation sites shows a sharply reduced presence at the telomeres and spindle poles [97]. As a consequence, Plk1 inhibitor-treated HeLa cells show abnormal telomeric fusions [97]. TNKS function can also be altered by the Fanconi anemia protein FANCD2, which has recently been demonstrated to inhibit TNKS1 mediated PARsylation at telomeres in human transformed fibroblasts [76]. Conversely, FANCD2 deficiency leads to increased TRF1 PARsylation [76]. Somatic inactivation of the Fanconi anemia (FA)/BRCA pathway by mutation or epigenetic silencing has been observed in several different types of sporadic cancers [138].
Recently, a further process through which TNKS impacts telomeres was revealed. In human tumors, telomeres are often maintained at a constant length by a telomerase dependent process [139] and telomerase is activated in the majority of advanced human carcinoma [140]. However, some tumors utilize a telomerase independent mechanism to maintain telomere length. This process is based on telomere-sister chromatid exchange (T-SCE) through recombination, a process that is commonly repressed by the shelterin complex. Removal of TNKS1 in human lymphoblast cells has been shown to significantly elevate the recombination within telomeres, leading to increased levels of T-SCE. This process is accompanied by a proteasome mediated degradation of DNA-PKc (the catalytic subunit DNA-dependent protein kinase), a component of the non-homologous end joining (NHEJ) pathway [141]. One consequence of this interaction was that reduced levels of TNKS1 resulted in increased sensitivity to ionizing radiation suggestive of a role for TNKS1 in DNA repair [141].
While TNKS has been shown to be up-regulated in selected tumors and correlates in some but not all tumors with poor prognosis [9, 12, 25-27, 29-34, 142, 143], TNKS expression in the context of telomerase activity has only been addressed in a few studies on tumor samples. Gao and co-workers reported up-regulation of TNKS in gastric cancerous tissue and statistically correlate increased TNKS expression with increased telomerase activity and tumor stage, although it is unclear whether telomerase activity is functionally linked to TNKS levels [26]. In astroglial tumors, telomerase activity significantly correlates with the WHO grade of the analyzed tissue, but only in anaplastic astrocytomas a concomitant up-regulation of TNKS1 has been observed [28]. Clearly possible links between TNKS expression and telomerase activity await further scrutiny.
Nevertheless, a possible therapeutic role of TNKS inhibitors in the context of telomeres, either as single agent or in combination with telomerase inhibitors has been proposed [144]. Recently, inhibition of TNKS1 was shown to play a synergistic role with telomerase inhibition in the gastric cancer cell line SGC-7901 [35]. Furthermore, apoptosis was promoted when antisense oligonucleotides towards TNKS were combined with antisense oligonucleotides for telomerase in the human lung adenocarcinoma cells A549. The observed effect was linked to the cell leukemia-1 (MCL-1) protein which can directly interact with TNKS [83, 145].
Curiously, while the TNKS/TRF1 interaction has been observed in human cells, it does not appear to be of relevance in mice where TRF1 lacks the Tankyrase binding motif (TBM) [8], nor in Drosophila, where telomeres are structured differently and consist of transposons in place of telomeric repeats [146].
4.5. Tankyrases in Stress Granule Assembly
Stress granules (SG) can form in the cytoplasm as a response to oxidative, osmotic, hypoxic, thermal, viral, and genotoxic stress [147]. SGs contain mRNA and RNA binding proteins but also TNKS1, PARG99, PARG102, and mARTDs 12, 13, 14, and 15 have been shown to co-localize in the SGs. Interestingly, PARsylation of SG associated proteins appears to enable the formation of SGs and Argonaute 2 (Ago2) (miRNA binding protein), G3BP1 (RNA decay factor), TIA-1 (translational suppressor), and PABP (poly(A)-binding protein) have been found PARsylated in SGs. Conversely, PARGs cause the disassembly of SGs [148]. It has been proposed that the suppression of miRNA through Ago2, which is bound in SGs, may contribute to dynamic stress responses in cancer cells [133, 149]. Interestingly, in the chondrosarcoma cell line U2OS, TNKS inhibitor reduced growth and induced differentiation accompanied by an increase of let-7 miRNA [150].
5. TANKYRASE INHIBITORS
The first tankyrase-selective inhibitors were discovered through Wnt-responsive luciferase reporter assay screening [15, 151]. The inhibitor of Wnt Response (IWR) compounds, which belong to a class of endo bridged phthalimides connected to a quinolinyl group with a benzoyl linker, were found to inhibit Wnt signaling by promoting the degradation of β-catenin, which was accredited to an increased stability of the DC [151]. Similarly, a compound named XAV939 has been shown to block the accumulation of β-catenin by increasing the activity of the DC. Subsequently, both XAV939 and IWR-1 were found to act through TNKS inhibition [15] and XAV939 also inhibited the proliferation of β-catenin dependent cancer cells [15].
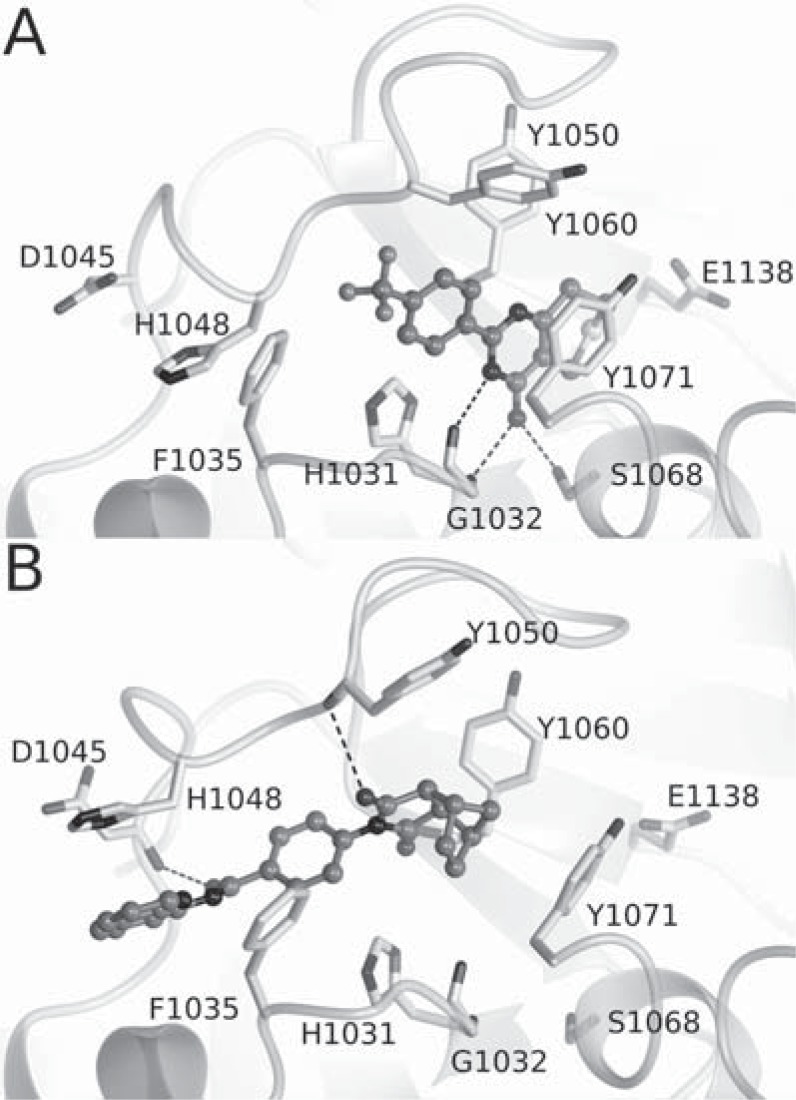
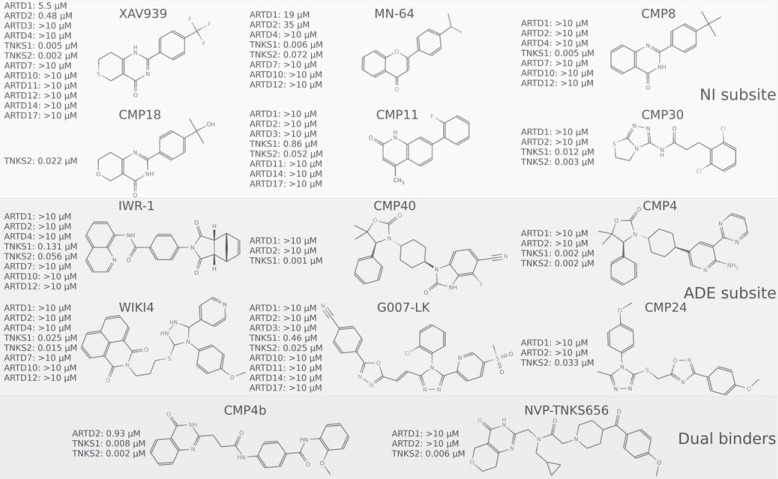
The crystal structures of XAV939 in complex with TNKS1 and TNKS2 revealed that the compound binds to the nicotinamide subsite in the catalytic domain (Fig. 4A) [42, 152]. This was the known conserved binding site of all the ARTD inhibitors studied to that date. XAV939 also shared the typical interactions of other ARTD inhibitors binding to the pocket; the hydrogen bonds with Gly and Ser (Gly1032 and Ser1068 in TNKS2) by the carboxamide group of XAV939 and a π-π stacking interaction with a Tyr at the active site (Tyr1071 in TNKS2) (Fig. 4A). Despite being the first reported TNKS-specific inhibitor, XAV939 inhibits also other ARTDs, especially ARTD2 with reported IC50 values between 110 - 480 nM [15, 68] (Fig. 5).
Fig. (4).
The binding of canonical TNKS inhibitors to the donor NAD+ binding site. A) Binding of XAV939 to TNKS2 nicotinamide subsite (PDB ID 3KR8). B) Binding of IWR-1 to the adenosine subsite of TNKS2 (PDB ID 3UA9).
Fig. (5).
In vitro selectivity profile of selected TNKS inhibitors. No inhibitors have so far been profiled using activity assays with ARTD8, ARTD9, ARTD13, ARTD15, or ARTD16. NI subsite, nicotinamide subsite; ADE subsite, adenosine subsite. IC50 values have been gathered from the literature [15, 23, 68, 157, 159-168].
In contrast, IWR-1 and IWR-2 do not contain the characteristic nicotinamide motif of the most ARTD inhibitors. The crystal structures of the compounds in complex with Tankyrases revealed a novel binding mode to the adenosine sub-site of the donor NAD+ binding groove in the catalytic pocket [62, 153] (Fig. 4B). In TNKS2, IWR-1 forms hydrogen bonds with the backbone amides Asp1045 and Tyr1050 of the D-loop and backbone amide of Tyr1060. The quinoline moiety of the compound stacks between an α-helix and His1048 from the D-loop and the norbornyl binds in the middle of a tyrosine triangle formed by Tyr1050, Tyr1060, and Tyr1071.
In recent years TNKS inhibitor development has increased substantially as Tankyrases show initial promise as drug targets against various conditions, including selected tumors. In the following sections we will summarize the latest advances and perspectives of TNKS inhibitor development.
5.1. Targeting the Nicotinamide Subsite
Most of the ARTD and TNKS inhibitors target the nicotinamide subsite and this was also the sole binding site of ARTD inhibitors since 1970s until very recently. An exception to this was Iniparib/BSI-201, which progressed to clinical trials as ARTD1 inhibitor, but later turned out not to inhibit ARTDs [154, 155]. The nicotinamide site inhibitors usually contain a nicotinamide mimic, which anchors the inhibitors at the bottom of the pocket (Fig. 4A). Small inhibitors mimicking nicotinamide, such as TIQ-A and phenanthridinone function as general ARTD inhibitors lacking isoenzyme specificity [63, 156]. The extension of the scaffold towards the opening of the subsite allows the introduction of interactions leading to TNKS selectivity (Fig. 5). The nicotinamide subsite in Tankyrases is more hydrophobic and restricted compared to other structurally characterized ARTDs [156, 157]. The hydrophobicity of the site is mainly due to Pro1034 and Phe1035 situated in the F-loop lining the site, and Ile1075 in the G-loop, which points toward the binding pocket (Fig. 3B, 6A). This region is poorly conserved in ARTDs and especially Pro1034 and Phe1035 are often replaced by hydrophilic residues. In addition, the extension of the G-loop containing Ile1075 is unique to Tankyrases.
Fig. (6).
The binding of selected inhibitors to TNKS1/TNKS2. Inhibitors binding to the nicotinamide subsite: A) CMP8 (PDB ID 4BUD), B) CMP11 (PDB ID 4IUE), and C) the general phramacophore of the inhibitors binding to this site. Hydrogen bond interactions of the compounds are shown in dashed lines. Hydrophobic features are shown as spheres and π-π stacking features are black discs. Inhibitors binding mainly to the adenosine subsite are shown in D) G007- LK (PDB ID 4HYF) and E) WIKI4 (PDB ID 4BFP), together with F) showing the common pharmacophoric features at this site. Compounds spanning both sites G) CMP4b (PDB ID 4I9I) and H) NVP-TNKS656-analog (PDB ID 4LI8) are shown as well as I) the combined pharmacophoric features of the dual binders.
Two XAV939-like, TNKS-specific inhibitor scaffolds, namely flavones and 2-phenyl-3, 4-dihydroquinazolin-4-ones bind to the nicotinamide subsite and extend towards the opening of the site [68, 157] (Fig. 6A). The 2-phenyl-3, 4-dihydroquinazolin-4-one derivatives have also been independently discovered and preliminary characterized by others [156, 158, 159]. These scaffolds take advantage of the TNKS-specific features of the binding site. Accordingly, hydrophobic substituents at the para position of the scaffolds highly increase the potency of the inhibitors and the profiling of inhibitors against several ARTDs revealed that hydrophobicity of the substituents is important for compound selectivity (Fig. 5). These studies have demonstrated that it is possible to design highly selective TNKS inhibitors binding to the nicotinamide subsite despite the high conservation of the site among ARTDs. The best inhibitors from these studies (MN-64 and CMP8) (Fig. 5) had nanomolar potencies against Tankyrases and displayed under 100 nM activities in a cell-based β-catenin reporter assay.
Other selective compounds have also been discovered that make use of the hydrophobic character of the binding site. One interesting set of compounds was identified through fragment-based ligand design. Expansion of a hit fragment resulted in a series of potent inhibitors that, despite not exactly mimicking nicotinamide, still shared the interactions found in XAV939 (Fig. 6B) [160]. Compound 11 (CMP11) of the series was demonstrated to be highly selective over several ARTDs but had considerably lower potency towards TNKS1 than for TNKS2 (Fig. 5).
Many optimized TNKS inhibitors of various scaffolds, contain hydrophobic moieties near the F- and G-loops. Recently Novartis opted a lipophilic efficiency (LipE) driven approach to optimize compounds from hit identification to lead optimization stage and this strategy lead to the identification of several new compounds binding to the nicotinamide subsite [159, 161]. Interestingly, despite the differences of the scaffolds, all the compounds anchor to the nicotinamide pocket with similar interactions as seen with XAV939. LipE driven optimization of an aminotriazole based high throughput screening (HTS) hit resulted in a compound series that had several favorable properties for a lead compound. Several compounds in the series exhibited Axin2 stabilization indicating cellular inhibition of Tankyrases. Compound 30 (CMP30) of the series demonstrated submicromolar Axin2 stabilization with low nanomolar in vitro potencies against Tankyrases (Fig. 5). It was also demonstrated to be selective over ARTD1/2 and against other off targets on a panel of 120 Novartis safety targets. However, the most potent compounds of the series had low solubility that might have led to somewhat inconsistent cellular activities of the compounds [161].
An optimization of XAV939 by Novartis was driven on the hypothesis that by reducing lipophilicity, the low selectivity, low microsomal stability, and low solubility of the compound could be improved. This optimization led to compound 18 (CMP18) with the trifluorophenyl group of XAV939 replaced by a hydrophobic 2-phenyl-2-propanol group. CMP18 displayed high potency (IC50 TNKS2: 22 nM) (Fig. 5), improved solubility over XAV939, and good microsomal stability. The compound also possessed good pharmacokinetic properties with good overall exposure, bioavailability, and low clearance with indications of enterohepatic recirculation. The selectivity of the compound to Tankyrases was not profiled but a close analog showed a high selectivity over ARTD1 [159].
Taken together, various compounds binding to the nicotinamide subsite have displayed low nanomolar potencies, high selectivity towards Tankyrases, and in some cases good bioavailability and moderate clearance both in vitro and in vivo. This binding site is highly conserved in ARTDs and the compounds have rather typical ARTD inhibitor pharmacophoric features (Fig. 6C). Despite this, many of the compounds are more selective TNKS inhibitors than XAV939, and should be better suited for studies where selective TNKS inhibition is required. Especially the hydrophobic interactions outside the most conserved nicotinamide binding pocket and interactions with hydrophobic G-loop improve TNKS specificity of the inhibitors targeting this site (Fig. 6C) [68, 157].
5.2. Targeting the Adenosine Subsite
The adenosine subsite was only recently identified as a potential target for TNKS inhibitors [62]. So far inhibitors binding exclusively to the adenosine subsite have not been described for other ARTDs, providing exciting opportunities for achieving high selectivity with these compounds. Indeed, inhibitors targeting this subsite have been shown to have remarkable selectivity towards Tankyrases (Fig. 5).
The identification of a novel TNKS inhibitor from HTS, JW74 [120], lead to independent efforts by two groups to optimize the compound properties [164, 168]. The JW74 derivative G007-LK [164] is a potent TNKS inhibitor with an excellent selectivity over several other isoenzymes (Fig. 5), no inhibition of tested kinases, phosphatases, and GPCRs, and a good activity in vitro. The compound binds to the adenosine site utilizing similar interactions as IWR-1 (Fig. 6D) but also extends towards the hydrophobic pocket lined by the F- and G-loops, making extensive van der Waals interactions with the region and giving a possible rationale for the high selectivity of G007-LK. G007-LK showed a Super Top Flash (STF) IC50 of 50 nM in selected colon cancer cell lines, accompanied by an inhibition of cell cycle progression, reduction of colony formation, and induction of differentiation. Moreover, G007-LK was found to be stable in human liver microsomes and exhibited excellent i.p. and p.o. pharmacokinetics in mice [164]. In xenografts, G007-LK inhibited tumor growth in a dose dependent manner in the colon cancer cell lines COLO-320DM and SW403. In contrast, no growth inhibition was observed in the colon cancer cell lines HCT-15 or DLD-1 [23].
The optimization of the JW74 chemotype by Novartis led to compound 24 (CMP24) (Fig. 5). Essentially, the compound has similar binding mode and interactions as G007-LK. CMP24 had good selectivity over ARTD1/2, 20-fold higher potency (Fig. 5) when compared to JW74, and exhibited similar level of Axin stabilization, and higher level of Wnt signaling inhibition compared to JW74. The microsomal stability of this compound series was very low, and several potential metabolic soft spots were identified from the compounds but unfortunately they were all required for effective TNKS inhibition. The compounds also had a rapid in vivo clearance probably due to the low microsomal stability.
Another compound, WIKI4, was recently discovered with HTS as a novel TNKS inhibitor. The compound was found to regulate β-catenin levels in several cancer cell lines and also in human embryonic stem cells [163]. Structural characterization revealed that WIKI4 binds to the adenosine subsite of TNKS2 [162] (Fig. 6E). In addition to the typical hydrogen bonds to the backbone amides of Asp1045 and Tyr1060, WIKI4 extends towards the hydrophobic pocket similar to CMP24 and G007-LK but also forms a hydrogen bond with the backbone amide of Ile1075 in the pocket. WIKI4 showed high potency and selectivity towards Tankyrases but has not yet been reported to be developed beyond the hit stage.
A structure-based design starting from IWR-1 to improve the potency and pharmacokinetic properties led to the discovery of compounds with highly improved potencies and pharmacokinetic properties [165]. The compounds show similar binding modes as IWR-1 and IWR-2 and the top lead compounds were selective over ARTD1/2 (Fig. 5). Earlier compounds in the series suffered from moderate cellular potencies and poor oral exposure but further optimization led to improvements, such as with compound 40 (CMP40), which had significantly improved cellular potencies and when dosed orally in mice, had unbound exposure levels over their cellular IC50 values. This compound series was further optimized by combining features from a dual binder acquired from HTS [167]. The optimized compound (CMP4) added a hydrogen bond to a glycine (Gly1196 in TNKS1). The compound had high potency towards Tankyrases and exhibited excellent cellular potency in STF assay (IC50 12 nM). The compound was also highly selective over ARTD1/2 (Fig. 5), did not inhibit 13 GPCRs, hERG, and BSEP, and exhibited only weak inhibitory activity when profiled against a panel of kinases. Furthermore CMP4 showed no genotoxicity in microAmes test. Pharmacokinetic studies in rats showed that the compound had moderate clearance and volume of distribution with an elimination half-life of 3.6 h upon intravenous administration. When dosed orally in rats, CMP4 exhibited moderate plasma exposure and bioavailability. In pharmacodynamic assay in DLD-1 human tumor xenograft mice CMP4 promoted accumulation of Axin and showed Wnt/β-catenin signaling inhibition in STF assay and the same dosing regime was well tolerated in naïve athymic nude mice for eight days.
The compounds binding to the adenosine subsite all share certain pharmacophoric features, although they were discovered using different strategies. They all have two conserved hydrogen bond acceptors, π-π stacking with a histidine of the D-loop and various hydrophobic regions (Fig. 6F). Small variations and extensions to the general pharmacophore exist and typically one or two hydrophobic interactions are shared between the TNKS selective adenosine site and nicotinamide site binders (Fig. 6C, F).
5.3. Dual Binders
Using a substructure search with IWR-2 pharmacophore as a model, Bregman and co-workers [166] recently identified a long quinazolinone compound with low nanomolar potency against TNKS1 (CMP4b) (Fig. 5). The crystal structure of the compound in complex with TNKS1 revealed that instead of binding to the adenosine subsite, the compound occupied both adenosine and nicotinamide subsites (Fig. 6G). The interactions made by the compound with the nicotinamide subsite are similar to what have been seen with nicotinamide binders. In the adenosine sub-site, the compound makes conventional hydrogen bonds with Tyr1213 and Asp1198 (Tyr1060 and Asp1045 in TNKS2) (Fig. 6G).
Shortly thereafter, another dual binder was reported by Novartis. NVP-TNKS656 was shown to be a highly potent TNKS inhibitor (Fig. 5) with good pharmacokinetic properties [159]. This dual binder shares the common interactions in both sub-sites and also extends towards the hydrophobic pocket between the F- and G-loops (Fig. 6H). In mice the compound had low clearance after intravenous administration and good exposure and moderate bioavailability after oral administration. Mice allografted with mammary adenocarcinomas showed good plasma and tumor exposures of NVP-TNKS656 and the tumors exhibited both Axin1 stabilization and reduction in the Axin2 mRNA levels.
Dual site binders are not unique to Tankyrases. Also ARTD1 inhibitors binding to both sub-sites have been described. Olaparib, an ARTD1 inhibitor currently in phase III clinical trials [169], utilizes both sub-sites. Olaparib is a moderately weak TNKS inhibitor (TNKS1 IC50 = 1.5 µM) but shares similar hydrogen bonding pattern as the long quinazolinone (CMP4b). The low potency of Olaparib towards TNKS1 can be explained by unfavorable interactions of the compound with the TNKS binding pocket, especially by the interaction of the fluorine of the fluorophenyl group with the D-loop [62].
EB-47 is an ARTD1 inhibitor (IC50 45nM) designed as an NAD+ mimic [170] and it was recently verified to be a dual binder [63]. The compound is also a potent TNKS inhibitor (IC50 410 nM and 45 nM, for TNKS1 and TNKS2, respectively). As an NAD+ mimic, the binding of EB-47 possibly resembles that of NAD+ (Fig. 3D, E). In TNKS2 it makes the conventional interactions at the nicotinamide subsite. In the adenosine subsite, the ribose hydroxyl of the compound interacts with His1031 and Ser1033, which are well-conserved in the ARTD family but not utilized by other inhibitors. The adenosine moiety forms a hydrogen bond with the backbone amide of Asp1045 and main chain carbonyl of Gly1043, which are also found in adenosine binders and other dual binders. Also other ARTD1 dual binders have been described [171, 172] but their selectivity towards Tankyrases has not been profiled.
5.4. Future Perspectives in Inhibitor Development
Clearly there is a need for validation of the selectivity of both current and future TNKS inhibitors. To be useful as tools in proof-of-concept in vivo studies inhibitors have to be selective for Tankyrases over other ARTD isoenzymes. Although high specificity is important for the analysis of the biological response, it may not be clinically the most beneficial property of a drug candidate [173]. In particular in a cancer context, inhibition of several ARTD enzymes might be a more efficient strategy. Indeed, Rucaparib, an initially optimized ARTD1 inhibitor which is currently in phase II studies was found to be a very potent TNKS inhibitor with 25 nM and 14 nM IC50 for TNKS1 and TNKS2, respectively [63]. Therefore, some of the observed beneficial effects in clinical studies might be due to inhibition of Tankyrases. Also for a number of other ARTD inhibitors, possible inhibitory effects on Tankyrase have been tested. Veliparib is a poor tankyrase inhibitor (IC50 > 10 µM), Olaparib is inhibiting Tankyrases at high dose (IC50 1.5 µM), and Rucaparib is an efficient tankyrase inhibitor [63]. Unfortunately, specificity data are not routinely available for new compounds, making a clear link between the reported effects and a particular ARTD, or multiple ARTDs difficult. The same holds true for TNKS inhibitors. XAV939 is often used in studies that are aimed to reveal Tankyrase functions, even though it is also a potent ARTD2 inhibitor (Fig. 5). Careful analysis of inhibition profiles within the ARTD family are needed as a prerequisite to evaluate the results of studies done with ARTD inhibitors.
Currently all the TNKS inhibitors are targeting the donor NAD+ binding site. Two other druggable sites exist on Tankyrases, namely the acceptor site and the ankyrin repeats. There are several reasons why these sites have been overlooked in the inhibitor design. Ankyrin repeat inhibitors are not found in in vitro enzyme activity screening where automodification is used as a signal. Protein-protein interactions are also difficult to target with inhibitors and compounds binding to these interfaces are not commonly found in HTS. Several projects start the inhibitor development from the existing pharmacophores binding to nicotinamide or adenosine subsites. However, ankyrin repeats and the acceptor sites offer novel avenues for selectively targeting Tankyrases as ankyrin repeats are not present in other ARTDs (although they are present in several other proteins) and acceptor sites are poorly conserved among ARTDs.
Some of the described TNKS inhibitors have up to ten fold selectivity between the two TNKS isoforms indicating that it could be possible to develop isoform selective inhibitors. However, as Tankyrases have overlapping roles, an inhibitor may need to target both isoforms to cause a measurable cellular response in most contexts. There are many unanswered questions in the roles of Tankyrases in various processes as highlighted in the number of binding partners (Table 1) and therefore it may be that under certain conditions one of the isoforms would be the preferred target. As the donor binding sites are identical in the amino acid sequence, the differences seen in the compound potencies are likely due to different dynamics of the catalytic domain imparted by sequence divergence outside the active site (Fig. 5) [41]. Apo crystal structures are also different mainly due to the changes in the D-loop (Fig. 3B) and further studies are needed to fully understand what causes these differences in the catalytic domains.
6. CONCLUSION
Tankyrases are challenging drug targets. The rules at which they engage in their functions in individual cells, healthy or diseased, are still only partially understood, as is their systemic impact.
To be useful as tools for in vitro and in vivo proof-of-concept studies, TNKS inhibitors have to be selective for Tankyrases over other ARTD isoenzymes and recent advances in the design of TNKS inhibitors have provided such specific tools. These tools are now used to dissect the benefits and pitfalls of specific TNKS inhibition in a variety of disease conditions including cancer, but also systemic sclerosis, metabolic disease, virus infections, and others. In some conditions, such as cancer, questions on the therapeutic window have to be addressed as the TNKS inhibitor doses required for achieving single agent efficacy may approach doses that lead to toxicity. While a selected sub-set of cancer cell lines responds to TNKS inhibition in a way that establishes a predictive link to known TNKS dependent biomarkers such as β-catenin in Wnt/β-catenin signaling, other cancer cell lines respond in a manner that is not understood, demanding further biological characterization and additional predictive biomarkers. Moreover, TNKS inhibition appears to be strongly context dependent and the biological consequence of TNKS inhibition will be influenced by the activity of other cellular pathways, cellular NAD+ levels, hypoxia, and other metabolic alterations; conditions that require further attention in the context of cancer cells.
If a single agent regime will not be sufficient in the cancer arena, the effect of other pathways inhibitors may be potentiated by TNKS inhibition as has been shown with TNKS and PI3K or Akt inhibitors and TNKS and EGFR inhibitors. It remains to be seen whether selective TNKS inhibition can be established as a clinically beneficial property for a drug candidate in the cancer arena. Simultaneous inhibition of several ARTD enzymes, as demonstrated for the ARTD1 inhibitors, might be an attractive strategy. Despite the pitfalls and open questions on the therapeutic benefits and applications of TNKS specific inhibitors, the recent flush of novel TNKS inhibitors indicates a substantial interest both in the pharmaceutical industry, and in academic research.
ACKNOWLEDGEMENTS
We like to thank the members of our laboratories and many colleagues for excellent discussions. In particular, we want to thank Nai-Wen Chi (UCSF) and Mike Costa (Genentech) for critical reading of the manuscript. The work was funded by Biocenter Oulu (LL), the Academy of Finland (TH; grant No. 266922), by the Research Council of Norway and by the Oslo University Hospital (SK).
CONFLICT OF INTEREST
The author SK declares competing financial interest.
LIST OF ABBREVIATIONS
- ADE
= Adenosine
- ADP
= Adenosine diphosphate
- ADPr
= ADP-ribose
- Ago2
= Argonaute 2
- APC
= Adenomatous polyposis coli
- ARC
= Ankyrin repeat cluster
- ARD
= α-helical regulatory domain
- ARH
= ADP-ribosylhydrolase
- ARTD
= ADP-ribosyl transferase with Diphtheria toxin homology
- BRCA
= Breast cancer susceptibility protein
- CDKα
= Casein kinase I isoform α
- DC
= β-catenin destruction complex
- DNA-PKc
= Catalytic subunit DNA-dependent protein kinase
- EGFR
= Epidermal growth factor receptor
- FANCD2
= Fanconi anemia protein 2
- FIH
= Hypoxia-inducible factor 1-alpha inhibitor
- G3BP1
= Ras GTPase-activating protein-binding protein 1
- GSK3β
= Glycogen synthase kinase 3-β
- HTS
= High throughput screening
- IRAP
= Insulin-responsive aminopeptidase
- LRP6
= Low-density lipoprotein receptor-related protein 6
- MAR
= Mono(ADP-ribose)
- mARTD
= ARTD with mono ADP-ribosylation activity
- MAPK
= Mitogen-activated protein kinase
- MDO
= Macro domain
- NHEJ
= Non-homologous end joining
- NI
= Nicotinamide
- NSCLC
= Non-small-cell lung carcinoma
- PARG
= poly(ADP-ribose) glycohydrolase
- pARTD
= polymer forming ARTD
- PAR
= poly(ADP-ribose)
- PARP
= poly(ADP-ribose)polymerase
- Plk1
= polo-like kinase-1
- POT1
= Protection of telomeres 1 protein
- RAP1
= Transcriptional repressor/activator protein
- SAM
= Steril alpha motif
- SG
= Stress granule
- STF
= Super Top Flash
- USP
= Ubiquitin specific protease
- TARG
= Terminal ADP-ribose protein glycohydrolase
- TBM
= Tankyrase binding motif
- TGF-β
= Transforming growth factor beta
- TIN
= TRF1 interacting protein
- TPP1
= TIN2 and POT1 interacting protein
- TRF
= Telomeric repeat-binding factor
- UPS
= Ubiquitin-proteasome system
REFERENCES
- 1.Otto H, Reche PA, Bazan F, Dittmar K, Haag F, Koch-Nolte F. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics. 2005;6: 139. doi: 10.1186/1471-2164-6-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleine H, Poreba E, Lesniewicz K , et al. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32: 57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Mahajan A, Tsai M-D. Ankyrin repeat a unique motif mediating protein-protein interactions. Biochemistry. 2006;45: 15168–78. doi: 10.1021/bi062188q. [DOI] [PubMed] [Google Scholar]
- 4.De Rycker M, Price CM. Tankyrase polymerization is controlled by its sterile alpha motif and poly(ADP-ribose) polymerase domains. Mol Cell Biol. 2004;24: 9802–12. doi: 10.1128/MCB.24.22.9802-9812.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsiao SJ, Poitras MF, Cook BD, Liu Y, Smith S. Tankyrase 2 poly(ADP-ribose) polymerase domain-deleted mice exhibit growth defects but have normal telomere length and capping. Mol Cell Biol. 2006;26: 2044–54. doi: 10.1128/MCB.26.6.2044-2054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang YJ, Nguyen M-L, Gurunathan S , et al. Generation and characterization of telomere length maintenance in tankyrase 2-deficient mice. Mol Cell Biol. 2006;26: 2037–43. doi: 10.1128/MCB.26.6.2037-2043.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeh T-YJ, Beiswenger KK, Li P , et al. Hypermetabolism, hyperphagia, and reduced adiposity in tankyrase-deficient mice. Diabetes. 2009;58: 2476–85. doi: 10.2337/db08-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang YJ, Hsiao SJ, Yver D , et al. Tankyrase 1 and tankyrase 2 are essential but redundant for mouse embryonic development. PloS One. 2008;3: e2639. doi: 10.1371/journal.pone.0002639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith S, Giriat I, Schmitt A, de Lange T. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres. Science. 1998;282: 1484–7. doi: 10.1126/science.282.5393.1484. [DOI] [PubMed] [Google Scholar]
- 10.Smith S, de Lange T. Tankyrase promotes telomere elongation in human cells. Curr Biol. 2000;10: 1299–302. doi: 10.1016/s0960-9822(00)00752-1. [DOI] [PubMed] [Google Scholar]
- 11.Kim MK, Smith S. Persistent telomere cohesion triggers a prolonged anaphase. Mol Biol Cell. 2014;25: 30–40. doi: 10.1091/mbc.E13-08-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyons RJ, Deane R, Lynch DK , et al. Identification of a novel human tankyrase through its interaction with the adaptor protein Grb14. J Biol Chem. 2001;276: 17172–80. doi: 10.1074/jbc.M009756200. [DOI] [PubMed] [Google Scholar]
- 13.Chang W, Dynek JN, Smith S. NuMA is a major acceptor of poly(ADP-ribosyl)ation by tankyrase 1 in mitosis. Biochem J. 2005;391: 177–84. doi: 10.1042/BJ20050885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho-Park PF, Steller H. Proteasome regulation by ADP-ribosylation. Cell. 2013;153: 614–27. doi: 10.1016/j.cell.2013.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang S-MA, Mishina YM, Liu S , et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461: 614–20. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 16.Leung AKL, Todorova T, Ando Y, Chang P. Poly(ADP-ribose) regulates post-transcriptional gene regulation in the cytoplasm. RNA Biol. 2012;9: 542–8. doi: 10.4161/rna.19899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scherag A, Dina C, Hinney A , et al. Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and german study groups. PLoS Genet. 2010;6: e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guettler S, LaRose J, Petsalaki E , et al. Structural basis and sequence rules for substrate recognition by Tankyrase explain the basis for cherubism disease. Cell. 2011;147: 1340–54. doi: 10.1016/j.cell.2011.10.046. [DOI] [PubMed] [Google Scholar]
- 19.Levaot N, Voytyuk O, Dimitriou I , et al. Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell. 2011;147: 1324–39. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Yamauchi Y, Kamakura M , et al. Herpes simplex virus requires poly(ADP-ribose) polymerase activity for efficient replication and induces extracellular signal-related kinase-dependent phosphorylation and ICP0-dependent nuclear localization of tankyrase 1. J Virol. 2012;86: 492–503. doi: 10.1128/JVI.05897-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng Z, Atanasiu C, Zhao K , et al. Inhibition of Epstein-Barr virus OriP function by tankyrase, a telomere-associated poly-ADP ribose polymerase that binds and modifies EBNA1. J Virol. 2005;79: 4640–50. doi: 10.1128/JVI.79.8.4640-4650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Distler A, Deloch L, Huang J , et al. Inactivation of tankyrases reduces experimental fibrosis by inhibiting canonical Wnt signalling. Ann Rheum Dis. 2013;72: 1575–80. doi: 10.1136/annrheumdis-2012-202275. [DOI] [PubMed] [Google Scholar]
- 23.Lau T, Chan E, Callow M , et al. A novel tankyrase small-molecule inhibitor suppresses APC mutation-driven colorectal tumor growth. Cancer Res. 2013;73: 3132–44. doi: 10.1158/0008-5472.CAN-12-4562. [DOI] [PubMed] [Google Scholar]
- 24.Sangu N, Shimosato T, Inoda H , et al. Novel nucleotide mutation leading to a recurrent amino acid alteration in SH3BP2 in a patient with cherubism. Congenit Anom. 2013;53: 166–9. doi: 10.1111/cga.12013. [DOI] [PubMed] [Google Scholar]
- 25.Busch AM, Johnson KC, Stan RV , et al. Evidence for tankyrases as antineoplastic targets in lung cancer. BMC Cancer. 2013;13: 211. doi: 10.1186/1471-2407-13-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao J, Zhang J, Long Y, Tian Y, Lu X. Expression of tankyrase 1 in gastric cancer and its correlation with telomerase activity. Pathol Oncol Res. 2011;17: 685–90. doi: 10.1007/s12253-011-9369-8. [DOI] [PubMed] [Google Scholar]
- 27.Gelmini S, Quattrone S, Malentacchi F , et al. Tankyrase-1 mRNA expression in bladder cancer and paired urine sediment preliminary experience. Clin Chem Lab Med. 2007;45: 862–6. doi: 10.1515/CCLM.2007.133. [DOI] [PubMed] [Google Scholar]
- 28.La Torre D, Conti A, Aguennouz MH , et al. Telomere length modulation in human astroglial brain tumors. PloS One. 2013;8: e64296. doi: 10.1371/journal.pone.0064296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang B, Wang J, Fang J , et al. Expression of TNKS1 is correlated with pathologic grade and Wnt/κ-catenin pathway in human astrocytomas. J Clin Neurosci. 2012;19: 139–43. doi: 10.1016/j.jocn.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 30.Zhao F, Vermeer B, Lehmann U , et al. Identification of a novel murine pancreatic tumour antigen, which elicits antibody responses in patients with pancreatic carcinoma. Immunology. 2009;128: 134–40. doi: 10.1111/j.1365-2567.2009.03090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gelmini S, Poggesi M, Distante V , et al. Tankyrase, a positive regulator of telomere elongation, is over expressed in human breast cancer. Cancer Lett. 2004;216: 81–7. doi: 10.1016/j.canlet.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Gelmini S, Poggesi M, Pinzani P , et al. Distribution of Tankyrase-1 mRNA expression in colon cancer and its prospective correlation with progression stage. Oncol Rep. 2006;16: 1261–6. [PubMed] [Google Scholar]
- 33.Shebzukhov YV, Lavrik IN, Karbach J , et al. Human tankyrases are aberrantly expressed in colon tumors and contain multiple epitopes that induce humoral and cellular immune responses in cancer patients. Cancer Immunol Immunother. 2008;57: 871–81. doi: 10.1007/s00262-007-0423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCabe N, Cerone MA, Ohishi T, Seimiya H, Lord CJ, Ashworth A. Targeting Tankyrase 1 as a therapeutic strategy for BRCA-associated cancer. Oncogene. 2009;28: 1465–70. doi: 10.1038/onc.2008.483. [DOI] [PubMed] [Google Scholar]
- 35.Zhang H, Yang M-H, Zhao J-J , et al. Inhibition of tankyrase 1 in human gastric cancer cells enhances telomere shortening by telomerase inhibitors. Oncol Rep. 2010;24: 1059–65. doi: 10.3892/or.2010.1059. [DOI] [PubMed] [Google Scholar]
- 36.Bao R, Christova T, Song S, Angers S, Yan X, Attisano L. Inhibition of tankyrases induces Axin stabilization and blocks Wnt signalling in breast cancer cells. PloS One. 2012;7: e48670. doi: 10.1371/journal.pone.0048670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casás-Selves M, Kim J, Zhang Z , et al. Tankyrase and the canonical Wnt pathway protect lung cancer cells from EGFR inhibition. Cancer Res. 2012;72: 4154–64. doi: 10.1158/0008-5472.CAN-11-2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osherovich L. Targeting tankyrase SciBX Sci-Bus. Exch [Internet] 2013 [cited 2013 Nov 26] 6. Available from http //www.nature.com/scibx/journal/v6/n15/full/scibx.2013.353.html.
- 39.Curtin NJ, Szabo C. Therapeutic applications of PARP inhibitors anticancer therapy and beyond. Mol Aspects Med. 2013;34: 1217–56. doi: 10.1016/j.mam.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Citarelli M, Teotia S, Lamb RS. Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol Biol. 2010;10: 308. doi: 10.1186/1471-2148-10-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lehtiö L, Collins R, van den Berg S , et al. Zinc binding catalytic domain of human tankyrase 1. J Mol Biol. 2008;379: 136–45. doi: 10.1016/j.jmb.2008.03.058. [DOI] [PubMed] [Google Scholar]
- 42.Karlberg T, Markova N, Johansson I , et al. Structural basis for the interaction between tankyrase-2 and a potent Wnt-signaling inhibitor. J Med Chem. 2010;53: 5352–5. doi: 10.1021/jm100249w. [DOI] [PubMed] [Google Scholar]
- 43.Meruelo AD, Bowie JU. Identifying polymer-forming SAM domains. Proteins. 2009;74: 1–5. doi: 10.1002/prot.22232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seimiya H, Smith S. The telomeric poly(ADP-ribose) polymerase, tankyrase 1, contains multiple binding sites for telomeric repeat binding factor 1 (TRF1):and a novel acceptor, 182-kDa tankyrase-binding protein (TAB182). J Biol Chem. 2002;277: 14116–26. doi: 10.1074/jbc.M112266200. [DOI] [PubMed] [Google Scholar]
- 45.Morrone S, Cheng Z, Moon RT, Cong F, Xu W. Crystal structure of a Tankyrase-Axin complex and its implications for Axin turnover and Tankyrase substrate recruitment. Proc Natl Acad Sci USA. 2012;109: 1500–5. doi: 10.1073/pnas.1116618109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37: 3723–38. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hottiger MO, Hassa PO, Lüscher B, Schüler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35: 208–19. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 48.Ruf A, Mennissier de Murcia J, de Murcia G, Schulz GE. Structure of the catalytic fragment of poly(AD-ribose) polymerase from chicken. Proc Natl Acad Sci USA. 1996;93: 7481–5. doi: 10.1073/pnas.93.15.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruf A, Rolli V, de Murcia G, Schulz GE. The mechanism of the elongation and branching reaction of poly(ADP-ribose) polymerase as derived from crystal structures and mutagenesis. J Mol Biol. 1998;278: 57–65. doi: 10.1006/jmbi.1998.1673. [DOI] [PubMed] [Google Scholar]
- 50.Rippmann JF, Damm K, Schnapp A. Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. J Mol Biol. 2002;323: 217–24. doi: 10.1016/s0022-2836(02)00946-4. [DOI] [PubMed] [Google Scholar]
- 51.Mueller-Dieckmann C, Kernstock S, Lisurek M , et al. The structure of human ADP-ribosylhydrolase 3 (ARH3):provides insights into the reversibility of protein ADP-ribosylation. Proc Natl Acad Sci USA. 2006;103: 15026–31. doi: 10.1073/pnas.0606762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Slade D, Dunstan MS, Barkauskaite E , et al. The structure and catalytic mechanism of a poly(ADP-ribose) glycohydrolase. Nature. 2011;477: 616–20. doi: 10.1038/nature10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosenthal F, Feijs KLH, Frugier E , et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat Struct Mol Biol. 2013;20: 502–7. doi: 10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- 54.Sharifi R, Morra R, Appel CD , et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32: 1225–37. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Callow MG, Tran H, Phu L , et al. Ubiquitin ligase RNF146 regulates tankyrase and Axin to promote Wnt signaling. PloS One. 2011;6: e22595. doi: 10.1371/journal.pone.0022595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langelier M-F, Ruhl DD, Planck JL, Kraus WL, Pascal JM. The Zn3 domain of human poly(ADP-ribose) polymerase-1 (PARP-1):functions in both DNA-dependent poly(ADP-ribose) synthesis activity and chromatin compaction. J Biol Chem. 2010;285: 18877–87. doi: 10.1074/jbc.M110.105668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langelier M-F, Planck JL, Roy S, Pascal JM. Structural basis for DNA damage-dependent poly(ADP-ribosyl)ation by human PARP-1. Science. 2012;336: 728–32. doi: 10.1126/science.1216338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sbodio JI, Lodish HF, Chi N-W. Tankyrase-2 oligomerizes with tankyrase-1 and binds to both TRF1 (telomere-repeat-binding factor 1):and IRAP (insulin-responsive aminopeptidase). Biochem J. 2002;361: 451–9. doi: 10.1042/0264-6021:3610451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chi NW, Lodish HF. Tankyrase is a golgi-associated mitogen-activated protein kinase substrate that interacts with IRAP in GLUT4 vesicles. J Biol Chem. 2000;275: 38437–44. doi: 10.1074/jbc.M007635200. [DOI] [PubMed] [Google Scholar]
- 60.Bell CE, Eisenberg D. Crystal structure of diphtheria toxin bound to nicotinamide adenine dinucleotide. Biochemistry. 1996;35: 1137–49. doi: 10.1021/bi9520848. [DOI] [PubMed] [Google Scholar]
- 61.Marsischky GT, Wilson BA, Collier RJ. Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation.Evidence for active site similarities to the ADP-ribosylating toxins. J Biol Chem. 1995;270: 3247–54. doi: 10.1074/jbc.270.7.3247. [DOI] [PubMed] [Google Scholar]
- 62.Narwal M, Venkannagari H, Lehtiö L. Structural basis of selective inhibition of human tankyrases. J Med Chem. 2012;55: 1360–7. doi: 10.1021/jm201510p. [DOI] [PubMed] [Google Scholar]
- 63.Haikarainen T, Narwal M, Joensuu P, Lehtiö L. Evaluation and Structural Basis for the Inhibition of Tankyrases by PARP Inhibitors. ACS Med Chem Lett. 2014;5: 18–22. doi: 10.1021/ml400292s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H, Fung K-L, Jin D-Y , et al. Solution structures, dynamics, and lipid-binding of the sterile alpha-motif domain of the deleted in liver cancer 2. Proteins. 2007;67: 1154–66. doi: 10.1002/prot.21361. [DOI] [PubMed] [Google Scholar]
- 65.Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FH-T. Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nat Struct Mol Biol. 2006;13: 160–7. doi: 10.1038/nsmb1038. [DOI] [PubMed] [Google Scholar]
- 66.Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat Struct Biol. 2003;10: 614–21. doi: 10.1038/nsb956. [DOI] [PubMed] [Google Scholar]
- 67.Narwal M, Haikarainen T, Fallarero A, Vuorela PM, Lehtiö L. Screening and structural analysis of flavones inhibiting tankyrases. J Med Chem. 2013;56: 3507–17. doi: 10.1021/jm3018783. [DOI] [PubMed] [Google Scholar]
- 68.Narwal M, Koivunen J, Haikarainen T , et al. Discovery of Tankyrase Inhibiting Flavones with Increased Potency and Isoenzyme Selectivity. J Med Chem. 2013;56: 7880–9. doi: 10.1021/jm401463y. [DOI] [PubMed] [Google Scholar]
- 69.De Rycker M, Venkatesan RN, Wei C, Price CM. Vertebrate tankyrase domain structure and sterile alpha motif (SAM)-mediated multimerization. Biochem J. 2003;372: 87–96. doi: 10.1042/BJ20021450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seimiya H, Muramatsu Y, Smith S, Tsuruo T. Functional subdomain in the ankyrin domain of tankyrase 1 required for poly(ADP-ribosyl)ation of TRF1 and telomere elongation. Mol Cell Biol. 2004;24: 1944–55. doi: 10.1128/MCB.24.5.1944-1955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waaler J, Machon O, Tumova L , et al. A novel tankyrase inhibitor decreases canonical Wnt signaling in colon carcinoma cells and reduces tumor growth in conditional APC mutant mice. Cancer Res. 2012;72: 2822–32. doi: 10.1158/0008-5472.CAN-11-3336. [DOI] [PubMed] [Google Scholar]
- 72.Stamos JL, Weis WI. The κ-catenin destruction complex. Cold Spring Harb. Perspect Biol. 2013;5: a007898. doi: 10.1101/cshperspect.a007898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voronkov A, Krauss S. Wnt/beta-catenin signaling and small molecule inhibitors. Curr Pharm Des. 2013;19: 634–64. doi: 10.2174/138161213804581837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li VSW, Ng SS, Boersema PJ , et al. Wnt signaling through inhibition of κ-catenin degradation in an intact Axin1 complex. Cell. 2012;149: 1245–56. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 75.Bisht KK, Dudognon C, Chang WG, Sokol ES, Ramirez A, Smith S. GDP-mannose-4, 6-dehydratase is a cytosolic partner of tankyrase 1 that inhibits its poly(ADP-ribose) polymerase activity. Mol Cell Biol. 2012;32: 3044–53. doi: 10.1128/MCB.00258-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lyakhovich A, Ramirez MJ, Castellanos A , et al. Fanconi anemia protein FANCD2 inhibits TRF1 polyADP-ribosylation through tankyrase1-dependent manner. Genome Integr. 2011;2: 4. doi: 10.1186/2041-9414-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Y, Liu S, Mickanin C , et al. RNF146 is a poly(ADP-ribose)-directed E3 ligase that regulates axin degradation and Wnt signalling. Nat Cell Biol. 2011;13: 623–9. doi: 10.1038/ncb2222. [DOI] [PubMed] [Google Scholar]
- 78.Short B, Preisinger C, Körner R, Kopajtich R, Byron O, Barr FA. A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J Cell Biol. 2001;155: 877–83. doi: 10.1083/jcb.200108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sbodio JI, Chi N-W. Identification of a tankyrase-binding motif shared by IRAP, TAB182, and human TRF1 but not mouse TRF1.NuMA contains this RXXPDG motif and is a novel tankyrase partner. J Biol Chem. 2002;277: 31887–92. doi: 10.1074/jbc.M203916200. [DOI] [PubMed] [Google Scholar]
- 80.Deng Z, Lezina L, Chen C-J, Shtivelband S, So W, Lieberman PM. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol Cell. 2002;9: 493–503. doi: 10.1016/s1097-2765(02)00476-8. [DOI] [PubMed] [Google Scholar]
- 81.Fuchs U, Rehkamp GF, Slany R, Follo M, Borkhardt A. The formin-binding protein 17, FBP17, binds via a TNKS binding motif to tankyrase, a protein involved in telomere maintenance. FEBS Lett. 2003;554: 10–6. doi: 10.1016/s0014-5793(03)01063-9. [DOI] [PubMed] [Google Scholar]
- 82.Guo H-L, Zhang C, Liu Q , et al. The Axin/TNKS complex interacts with KIF3A and is required for insulin-stimulated GLUT4 translocation. Cell Res. 2012;22: 1246–57. doi: 10.1038/cr.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bae J, Donigian Jr, Hsueh AJW. Tankyrase 1 interacts with Mcl-1 proteins and inhibits their regulation of apoptosis. J Biol Chem. 2003;278: 5195–204. doi: 10.1074/jbc.M201988200. [DOI] [PubMed] [Google Scholar]
- 84.Aravind L. The WWE domain a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci. 2001;26: 273–5. doi: 10.1016/s0968-0004(01)01787-x. [DOI] [PubMed] [Google Scholar]
- 85.He F, Tsuda K, Takahashi M , et al. Structural insight into the interaction of ADP-ribose with the PARP WWE domains. FEBS Lett. 2012;586: 3858–64. doi: 10.1016/j.febslet.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 86.Forst AH, Karlberg T, Herzog N , et al. Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure. 2013;21: 462–75. doi: 10.1016/j.str.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 87.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12: 295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chau V, Tobias JW, Bachmair A , et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243: 1576–83. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 89.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000;19: 94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feng X, Koh DW. Roles of poly(ADP-ribose) glycohydrolase in DNA damage and apoptosis. Int Rev Cell Mol Biol. 2013;304: 227–81. doi: 10.1016/B978-0-12-407696-9.00005-1. [DOI] [PubMed] [Google Scholar]
- 91.Dunstan MS, Barkauskaite E, Lafite P , et al. Structure and mechanism of a canonical poly(ADP-ribose) glycohydrolase. Nat Commun. 2012;3: 878. doi: 10.1038/ncomms1889. [DOI] [PubMed] [Google Scholar]
- 92.Barkauskaite E, Brassington A, Tan ES , et al. Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nat Commun. 2013;4: 2164. doi: 10.1038/ncomms3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jankevicius G, Hassler M, Golia B , et al. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat Struct Mol Biol. 2013;20: 508–14. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moss J, Stanley SJ, Nightingale MS , et al. Molecular and immunological characterization of ADP-ribosylarginine hydrolases. J Biol Chem. 1992;267: 10481–8. [PubMed] [Google Scholar]
- 95.Steffen JD, Pascal JM. New players to the field of ADP-ribosylation make the final cut. EMBO J. 2013;32: 1205–7. doi: 10.1038/emboj.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kato J, Zhu J, Liu C , et al. ADP-ribosylarginine hydrolase regulates cell proliferation and tumorigenesis. Cancer Res. 2011;71: 5327–35. doi: 10.1158/0008-5472.CAN-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ha G-H, Kim H-S, Go H , et al. Tankyrase-1 function at telomeres and during mitosis is regulated by Polo-like kinase-1-mediated phosphorylation. Cell Death Differ. 2012;19: 321–32. doi: 10.1038/cdd.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yeh T-YJ, Sbodio JI, Chi N-W. Mitotic phosphorylation of tankyrase, a PARP that promotes spindle assembly, by GSK3. Biochem Biophys Res Commun. 2006;350: 574–9. doi: 10.1016/j.bbrc.2006.09.080. [DOI] [PubMed] [Google Scholar]
- 99.Yang M, Chowdhury R, Ge W , et al. Factor-inhibiting hypoxia-inducible factor (FIH) catalyses the post-translational hydroxylation of histidinyl residues within ankyrin repeat domains. FEBS J. 2011;278: 1086–97. doi: 10.1111/j.1742-4658.2011.08022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cockman ME, Webb JD, Kramer HB, Kessler BM, Ratcliffe PJ. Proteomics-based identification of novel factor inhibiting hypoxia-inducible factor (FIH) substrates indicates widespread asparaginyl hydroxylation of ankyrin repeat domain-containing proteins. Mol Cell Proteomics. 2009;8: 535–46. doi: 10.1074/mcp.M800340-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Coleman ML, McDonough MA, Hewitson KS , et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem. 2007;282: 24027–38. doi: 10.1074/jbc.M704102200. [DOI] [PubMed] [Google Scholar]
- 102.Smith S, de Lange T. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci. 1999;112(Pt 21): 3649–56. doi: 10.1242/jcs.112.21.3649. [DOI] [PubMed] [Google Scholar]
- 103.Ozaki Y, Matsui H, Asou H , et al. Poly-ADP ribosylation of Miki by tankyrase-1 promotes centrosome maturation. Mol Cell. 2012;47: 694–706. doi: 10.1016/j.molcel.2012.06.033. [DOI] [PubMed] [Google Scholar]
- 104.Yeh T-YJ, Meyer TN, Schwesinger C, Tsun Z-Y, Lee RM, Chi N-W. Tankyrase recruitment to the lateral membrane in polarized epithelial cells regulation by cell-cell contact and protein poly(ADP-ribosyl)ation. Biochem J. 2006;399: 415–25. doi: 10.1042/BJ20060713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hsiao SJ, Smith S. Tankyrase function at telomeres, spindle poles, and beyond. Biochimie. 2008;90: 83–92. doi: 10.1016/j.biochi.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 106.Chang P, Coughlin M, Mitchison TJ. Interaction between Poly(ADP-ribose) and NuMA contributes to mitotic spindle pole assembly. Mol Biol Cell. 2009;20: 4575–85. doi: 10.1091/mbc.E09-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim MK, Dudognon C, Smith S. Tankyrase 1 regulates centrosome function by controlling CPAP stability. EMBO Rep. 2012;13: 724–32. doi: 10.1038/embor.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lan J, Zhu Y, Xu L , et al. The 68 kDa telomeric repeat binding factor 1 (TRF1):associated protein (TAP68):interacts with and recruits TRF1 to the spindle pole during mitosis. J Biol Chem. 2014 doi: 10.1074/jbc.M113.526244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cong F, Varmus H. Nuclear-cytoplasmic shuttling of Axin regulates subcellular localization of beta-catenin. Proc Natl Acad Sci USA. 2004;101: 2882–7. doi: 10.1073/pnas.0307344101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.De la Roche M, Ibrahim AEK, Mieszczanek J, Bienz M. LEF1 and B9L shield κ-catenin from inactivation by Axin desensitizing colorectal cancer cells to tankyrase inhibitors. Cancer Res. 2014;74: 1495–505. doi: 10.1158/0008-5472.CAN-13-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Parveen N, Hussain MU, Pandith AA, Mudassar S. Diversity of axin in signaling pathways and its relation to colorectal cancer. Med Oncol Northwood Lond Engl. 2011;28(Suppl 1 ): S259–267. doi: 10.1007/s12032-010-9722-x. [DOI] [PubMed] [Google Scholar]
- 112.Kim S-M, Choi E-J, Song K-J , et al. Axin localizes to mitotic spindles and centrosomes in mitotic cells. Exp Cell Res. 2009;315: 943–54. doi: 10.1016/j.yexcr.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 113.Yeh T-YJ, Sbodio JI, Tsun Z-Y, Luo B, Chi N-W. Insulin-stimulated exocytosis of GLUT4 is enhanced by IRAP and its partner tankyrase. Biochem J. 2007;402: 279–90. doi: 10.1042/BJ20060793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Tacchelly-Benites O, Wang Z, Yang E, Lee E, Ahmed Y. Toggling a conformational switch in Wnt/κ-catenin signaling regulation of Axin phosphorylation.The phosphorylation state of Axin controls its scaffold function in two Wnt pathway protein complexes. Bioessays. 2013;35: 1063–70. doi: 10.1002/bies.201300101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ring L, Neth P, Weber C, Steffens S, Faussner A. κ-Catenin-dependent pathway activation by both promiscuous “canonical” WNT3a-, and specific “noncanonical” WNT4- and WNT5a-FZD receptor combinations with strong differences in LRP5 and LRP6 dependency. Cell Signal. 2014;26: 260–7. doi: 10.1016/j.cellsig.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 116.Wiechens N, Heinle K, Englmeier L, Schohl A, Fagotto F. Nucleo-cytoplasmic shuttling of Axin, a negative regulator of the Wnt-beta-catenin Pathway. J Biol Chem. 2004;279: 5263–7. doi: 10.1074/jbc.M307253200. [DOI] [PubMed] [Google Scholar]
- 117.Anderson CB, Neufeld KL, White RL. Subcellular distribution of Wnt pathway proteins in normal and neoplastic colon. Proc Natl Acad Sci USA. 2002;99: 8683–8. doi: 10.1073/pnas.122235399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Salahshor S, Woodgett Jr. The links between axin and carcinogenesis. J Clin Pathol. 2005;58: 225–36. doi: 10.1136/jcp.2003.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Z, Michaud GA, Cheng Z , et al. Recognition of the iso-ADP-ribose moiety in poly(ADP-ribose) by WWE domains suggests a general mechanism for poly(ADP-ribosyl)ation-dependent ubiquitination. Genes Dev. 2012;26: 235–40. doi: 10.1101/gad.182618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Waaler J, Machon O, von Kries JP , et al. Novel synthetic antagonists of canonical Wnt signaling inhibit colorectal cancer cell growth. Cancer Res. 2011;71: 197–205. doi: 10.1158/0008-5472.CAN-10-1282. [DOI] [PubMed] [Google Scholar]
- 121.Lui TTH, Lacroix C, Ahmed SM , et al. The ubiquitin-specific protease USP34 regulates axin stability and Wnt/κ-catenin signaling. Mol Cell Biol. 2011;31: 2053–65. doi: 10.1128/MCB.01094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Qian L, Mahaffey JP, Alcorn HL, Anderson KV. Tissue-specific roles of Axin2 in the inhibition and activation of Wnt signaling in the mouse embryo. Proc Natl Acad Sci USA. 2011;108: 8692–7. doi: 10.1073/pnas.1100328108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Powell SM, Zilz N, Beazer-Barclay Y , et al. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359: 235–7. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 124.Jin L-H, Shao Q-J, Luo W, Ye Z-Y, Li Q, Lin S-C. Detection of point mutations of the Axin1 gene in colorectal cancers. Int. J. Cancer J Int Cancer. 2003;107: 696–9. doi: 10.1002/ijc.11435. [DOI] [PubMed] [Google Scholar]
- 125.Morin PJ, Sparks AB, Korinek V , et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275: 1787–90. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 126.Rajan P, Sudbery IM, Villasevil MEM , et al. Next-generation Sequencing of Advanced Prostate Cancer Treated with Androgen-deprivation Therapy. Eur Urol. 2013 doi: 10.1016/j.eururo.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tenbaum SP, Ordóñez-Morán P, Puig I , et al. κ-catenin confers resistance to PI3K and AKT inhibitors and subverts FOXO3a to promote metastasis in colon cancer. Nat Med. 2012;18: 892–901. doi: 10.1038/nm.2772. [DOI] [PubMed] [Google Scholar]
- 128.Bogan JS. Regulation of glucose transporter translocation in health and diabetes. Annu Rev Biochem. 2012;81: 507–32. doi: 10.1146/annurev-biochem-060109-094246. [DOI] [PubMed] [Google Scholar]
- 129.McBrayer SK, Cheng JC, Singhal S, Krett NL, Rosen ST, Shanmugam M. Multiple myeloma exhibits novel dependence on GLUT4, GLUT8, and GLUT11: implications for glucose transporter-directed therapy. Blood. 2012;119: 4686–97. doi: 10.1182/blood-2011-09-377846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saveanu L, van Endert P. The role of insulin-regulated aminopeptidase in MHC class I antigen presentation. Front Immunol. 2012;3: 57. doi: 10.3389/fimmu.2012.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7: 1133–9. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 132.Boehler C, Gauthier LR, Mortusewicz O , et al. Poly(ADP-ribose) polymerase 3 (PARP3), a newcomer in cellular response to DNA damage and mitotic progression. Proc Natl Acad Sci USA. 2011;108: 2783–8. doi: 10.1073/pnas.1016574108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Masutani M, Fujimori H. Poly(ADP-ribosyl)ation in carcinogenesis. Mol Aspects Med. 2013;34: 1202–16. doi: 10.1016/j.mam.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 134.Korzeniewski N, Hohenfellner M, Duensing S. The centrosome as potential target for cancer therapy and prevention. Expert Opin Ther Targets. 2013;17: 43–52. doi: 10.1517/14728222.2013.731396. [DOI] [PubMed] [Google Scholar]
- 135.De Lange T. Shelterin the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19: 2100–10. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 136.Dynek JN, Smith S. Resolution of sister telomere association is required for progression through mitosis. Science. 2004;304: 97–100. doi: 10.1126/science.1094754. [DOI] [PubMed] [Google Scholar]
- 137.Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S. Protein requirements for sister telomere association in human cells. EMBO J. 2007;26: 4867–78. doi: 10.1038/sj.emboj.7601903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mathew CG. Fanconi anaemia genes and susceptibility to cancer. Oncogene. 2006;25: 5875–84. doi: 10.1038/sj.onc.1209878. [DOI] [PubMed] [Google Scholar]
- 139.Counter CM, Avilion AA, LeFeuvre CE , et al. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 1992;11: 1921–9. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim NW, Piatyszek MA, Prowse KR , et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266: 2011–5. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 141.Dregalla RC, Zhou J, Idate RR, Battaglia CLR, Liber HL, Bailey SM. Regulatory roles of tankyrase 1 at telomeres and in DNA repair suppression of T-SCE and stabilization of DNA-PKcs. Aging. 2010;2: 691–708. doi: 10.18632/aging.100210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Matsutani N, Yokozaki H, Tahara E , et al. Expression of telomeric repeat binding factor 1 and 2 and TRF1-interacting nuclear protein 2 in human gastric carcinomas. Int J Oncol. 2001;19: 507–12. [PubMed] [Google Scholar]
- 143.Shervington A, Patel R, Lu C , et al. Telomerase subunits expression variation between biopsy samples and cell lines derived from malignant glioma. Brain Res. 2007;1134: 45–52. doi: 10.1016/j.brainres.2006.11.093. [DOI] [PubMed] [Google Scholar]
- 144.Seimiya H. The telomeric PARP, tankyrases, as targets for cancer therapy. Br J Cancer. 2006;94: 341–5. doi: 10.1038/sj.bjc.6602951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lu H, Lei Z, Lu Z , et al. Silencing tankyrase and telomerase promotes A549 human lung adenocarcinoma cell apoptosis and inhibits proliferation. Oncol Rep. 2013;30: 1745–52. doi: 10.3892/or.2013.2665. [DOI] [PubMed] [Google Scholar]
- 146.Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75: 1083–93. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 147.Ohn T, Anderson P. The role of posttranslational modifications in the assembly of stress granules. Wiley Interdiscip Rev RNA. 2010;1: 486–93. doi: 10.1002/wrna.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ji Y, Tulin AV. Post-transcriptional regulation by poly(ADP-ribosyl)ation of the RNA-binding proteins. Int J Mol Sci. 2013;14: 16168–83. doi: 10.3390/ijms140816168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Leung AKL, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P. Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell. 2011;42: 489–99. doi: 10.1016/j.molcel.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wessel Stratford E, Daffinrud J, Munthe E , et al. The tankyrase-specific inhibitor JW74 affects cell cycle progression and induces apoptosis and differentiation in osteosarcoma cell lines. Cancer Med. 2014;3: 36–46. doi: 10.1002/cam4.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen B, Dodge ME, Tang W , et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5: 100–7. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kirby CA, Cheung A, Fazal A, Shultz MD, Stams T. Structure of human tankyrase 1 in complex with small-molecule inhibitors PJ34 and XAV939. Acta Crystallograph Sect F Struct Biol Cryst Commun. 2012;68: 115–8. doi: 10.1107/S1744309111051219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gunaydin H, Gu Y, Huang X. Novel binding mode of a potent and selective tankyrase inhibitor. PloS One. 2012;7: e33740. doi: 10.1371/journal.pone.0033740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chuang H-C, Kapuriya N, Kulp SK, Chen C-S, Shapiro CL. Differential anti-proliferative activities of poly(ADP-ribose) polymerase (PARP) inhibitors in triple-negative breast cancer cells. Breast Cancer Res Treat. 2012;134: 649–59. doi: 10.1007/s10549-012-2106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Patel AG, De Lorenzo SB, Flatten KS, Poirier GG, Kaufmann SH. Failure of iniparib to inhibit poly(ADP-Ribose) polymerase in vitro. Clin Cancer Res. 2012;18: 1655–62. doi: 10.1158/1078-0432.CCR-11-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wahlberg E, Karlberg T, Kouznetsova E , et al. Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors. Nat Biotechnol. 2012;30: 283–8. doi: 10.1038/nbt.2121. [DOI] [PubMed] [Google Scholar]
- 157.Haikarainen T, Koivunen J, Narwal M , et al. para-Substituted 2-Phenyl-3, 4-dihydroquinazolin-4-ones As Potent and Selective Tankyrase Inhibitors. ChemMedChem. 2013;8: 1978–85. doi: 10.1002/cmdc.201300337. [DOI] [PubMed] [Google Scholar]
- 158.Nathubhai A, Wood PJ, Lloyd MD, Thompson AS, Threadgill MD. Design and Discovery of 2-Arylquinazolin-4-ones as Potent and Selective Inhibitors of Tankyrases. ACS Med Chem Lett. 2013;4: 1173–7. doi: 10.1021/ml400260b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Shultz MD, Cheung AK, Kirby CA , et al. Identification of NVP-TNKS656: The Use of Structure-Efficiency Relationships To Generate a Highly Potent, Selective, and Orally Active Tankyrase Inhibitor. J Med Chem. 2013;56: 6495–511. doi: 10.1021/jm400807n. [DOI] [PubMed] [Google Scholar]
- 160.Larsson EA, Jansson A, Ng FM , et al. Fragment-based ligand design of novel potent inhibitors of tankyrases. J Med Chem. 2013;56: 4497–508. doi: 10.1021/jm400211f. [DOI] [PubMed] [Google Scholar]
- 161.Shultz MD, Majumdar D, Chin DN , et al. Structure-efficiency relationship of [1, 2, 4]triazol-3-ylamines as novel nicotinamide isosteres that inhibit tankyrases. J Med Chem. 2013;56: 7049–59. doi: 10.1021/jm400826j. [DOI] [PubMed] [Google Scholar]
- 162.Haikarainen T, Venkannagari H, Narwal M , et al. Structural basis and selectivity of tankyrase inhibition by a Wnt signaling inhibitor WIKI4. PloS One. 2013;8: e65404. doi: 10.1371/journal.pone.0065404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.James RG, Davidson KC, Bosch KA , et al. WIKI4, a novel inhibitor of tankyrase and Wnt/ß-catenin signaling. PloS One. 2012;7: e50457. doi: 10.1371/journal.pone.0050457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Voronkov A, Holsworth DD, Waaler J , et al. Structural basis and SAR for G007-LK, a lead stage 1, 2, 4-triazole based specific tankyrase 1/2 inhibitor. J Med Chem. 2013;56: 3012–23. doi: 10.1021/jm4000566. [DOI] [PubMed] [Google Scholar]
- 165.Bregman H, Chakka N, Guzman-Perez A , et al. Discovery of novel, induced-pocket binding oxazolidinones as potent, selective, and orally bioavailable tankyrase inhibitors. J Med Chem. 2013;56: 4320–42. doi: 10.1021/jm4000038. [DOI] [PubMed] [Google Scholar]
- 166.Bregman H, Gunaydin H, Gu Y , et al. Discovery of a class of novel tankyrase inhibitors that bind to both the nicotinamide pocket and the induced pocket. J Med Chem. 2013;56: 1341–5. doi: 10.1021/jm301607v. [DOI] [PubMed] [Google Scholar]
- 167.Huang H, Guzman-Perez A, Acquaviva L , et al. Structure-Based Design of 2-Aminopyridine Oxazolidinones as Potent and Selective Tankyrase Inhibitors. ACS Med Chem Lett. 2013;4: 1218–23. doi: 10.1021/ml4003315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shultz MD, Kirby CA, Stams T , et al. [1, 2, 4]triazol-3-ylsulfanylmethyl)-3-phenyl-[1, 2, 4]oxadiazoles antagonists of the Wnt pathway that inhibit tankyrases 1 and 2 via novel adenosine pocket binding. J Med Chem. 2012;55: 1127–36. doi: 10.1021/jm2011222. [DOI] [PubMed] [Google Scholar]
- 169.Olaparib Enters Phase III Clinical Testing. Cancer Discov. 2013;3 [Google Scholar]
- 170.Jagtap PG, Southan GJ, Baloglu E , et al. The discovery and synthesis of novel adenosine substituted 2, 3-dihydro-1H-isoindol-1-ones potent inhibitors of poly(ADP-ribose) polymerase-1 (PARP-1). Bioorg Med Chem Lett. 2004;14: 81–5. doi: 10.1016/j.bmcl.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 171.Gangloff AR, Brown J, de Jong R , et al. Discovery of novel benzo[b][1, 4]oxazin-3(4H)-ones as poly(ADP-ribose)polymerase inhibitors. Bioorg Med Chem Lett. 2013;23: 4501–5. doi: 10.1016/j.bmcl.2013.06.055. [DOI] [PubMed] [Google Scholar]
- 172.Ye N, Chen C-H, Chen T , et al. Design, synthesis, and biological evaluation of a series of benzo[de][1, 7]naphthyridin-7(8H)-ones bearing a functionalized longer chain appendage as novel PARP1 inhibitors. J Med Chem. 2013;56: 2885–903. doi: 10.1021/jm301825t. [DOI] [PubMed] [Google Scholar]
- 173.Liscio P, Camaioni E, Carotti A, Pellicciari R, Macchiarulo A. From Polypharmacology to Target Specificity The Case of PARP Inhibitors. Curr Top Med Chem. 2013;13: 2939–54. doi: 10.2174/15680266113136660209. [DOI] [PubMed] [Google Scholar]