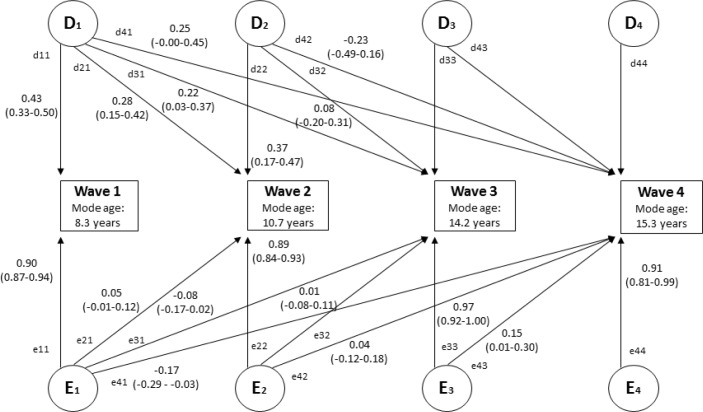
Figure 1.
Multivariate Cholesky decomposition with parameter estimates (95% confidence interval) from best-fitting DE model. Figure is shown for one twin only. D = nonadditive genetic influence; E = nonshared environmental influence. Figure displays unsquared parameter estimates for significant paths. The parameter estimates can be squared to indicate relative proportions of variance (%). The extent to which genetic influences account for the phenotypic correlations between variables can be calculated as follows: (d11*d21)/r(wave 1 and wave 2); (d11*d31)/r(wave 1 and wave 3); (d11*d41)/r(wave 1 and wave 4); (d21*d31) + (d22*d32)/r(wave 2 and wave 3); (d21*d41) + (d22*d42)/r(wave 2 and wave 4); (d31*d41) + (d32*d42) + (d33*d43)/r(wave 3 and wave 4). The same principles apply for calculating the relative proportions of variance accounted for by non-shared environmental influences.