Abstract
Study Objectives:
Although HLA-DQB1*06:02 is the strongest predisposing genetic factor for narcolepsy, the effect of this gene must be considered alongside that of its polymorphic partner, DQA1. In this paper, we extend an analysis of the effect of HLA-DQB1 on narcolepsy risk published recently by Tafti et al.
Results:
Imputing allelic variation at the level of HLA-DQA1, we show that this locus also has a considerable effect on disease susceptibility. Our data are also compatible with previous findings in multi-ethnic group data sets showing that allele competition effects within the DQ1 group determine the amount of DQ0602 (the DQA1*01:02/DQB1*06:02 heterodimer), and consequently, the risk of developing narcolepsy. We also found an independent predisposing effect of DQB1*03:01 via a currently unknown mechanism.
Conclusions:
Both DQA1 and DQB1 influence narcolepsy risk.
Citation:
Ollila HM, Fernandez-Vina M, Mignot E. HLA-DQ allele competition in narcolepsy: a comment on Tafti et al. DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. SLEEP 2015;38(1):147–151.
Keywords: narcolepsy, HLA, MHC, cataplexy, hypocretin
It is with great interest that we read the publication of Tafti et al.1 entitled “DQB1 Locus Alone Explains Most of the Risk and Protection in Narcolepsy with Cataplexy in Europe,” one of the largest human leukocyte antigen (HLA) association studies in narcolepsy published to date. The data are fully consistent with previously published studies.2–5 The authors also saw significant replication of known narcolepsy predisposing single nucleotide polymorphisms (SNPs) in HLA and in T cell receptor alpha loci and nominal replication for several other loci (P2RY11, CTSH, and TNFSF4). Although the findings are solid, we do not fully agree with the interpretation that the authors propose in the paper, and would like to offer a few comments and an alternate interpretation.
The study by Tafti and coworkers examines the effect of DQB1 while other HLA loci are not investigated in depth, and thus the title “DQB1 Locus Alone Explains Most of the Risk and Protection in Narcolepsy” does not appropriately define the findings. We believe that it is essential to include alleles of DQA1 in the analysis since the functional DQ molecule requires both DQA1 and DQB1 subunits to be present (see Figure 1 for HLA function). DQA1 variations most likely contribute substantially to susceptibility and protection. Strong linkage dis-equilibrium between DQA1 and DQB1 results from the close physical proximity between these loci (i.e., a few kb apart) and from strong selection pressure to keep alleles that can form functional heterodimer molecules. Isolating the contributory effects of DQB1 and DQA1 alleles is quite challenging, although, as described below, it is not impossible. In all other HLA-DQ associated diseases, both DQα and DQβ have joint effects on disease susceptibility, which is logical as polymorphisms in both genes participate to peptide binding.
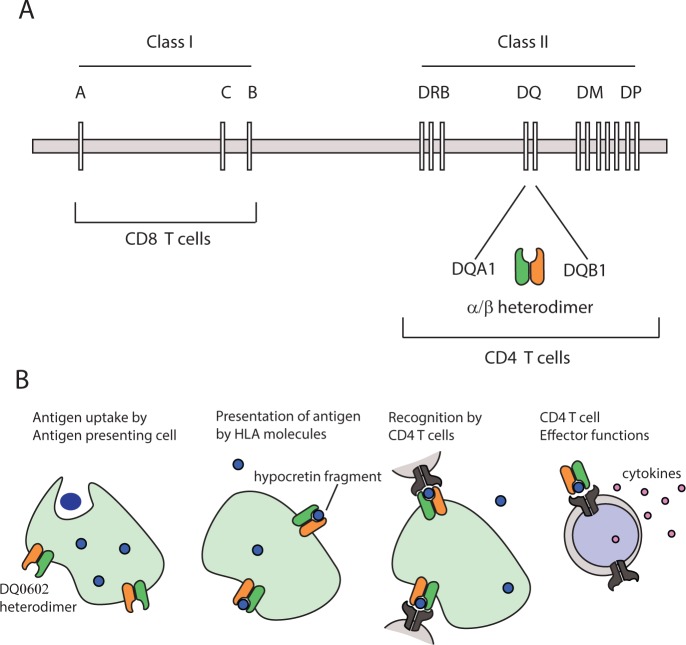
Figure 1.
(A) The human leukocyte antigen (HLA) genes are located in chromosome 6. The Class I genes (A, B, and C) are recognized by cytotoxic CD8 T-cells whereas the Class II genes that are relevant for narcolepsy are recognized by helper CD4 T-cells. HLA Class II molecules such as HLA DQ are α/β heterodimers encoded by two genes, DQA1 and DQB1. (B) Antigen presenting cells take up the antigen. The antigen is then presented to T cells (CD4 T cells in the case of HLA class II genes) by the HLA molecules. If the antigen-HLA combination is recognized by T cells, an immune reaction takes place and the cells containing the antigen may be destroyed.
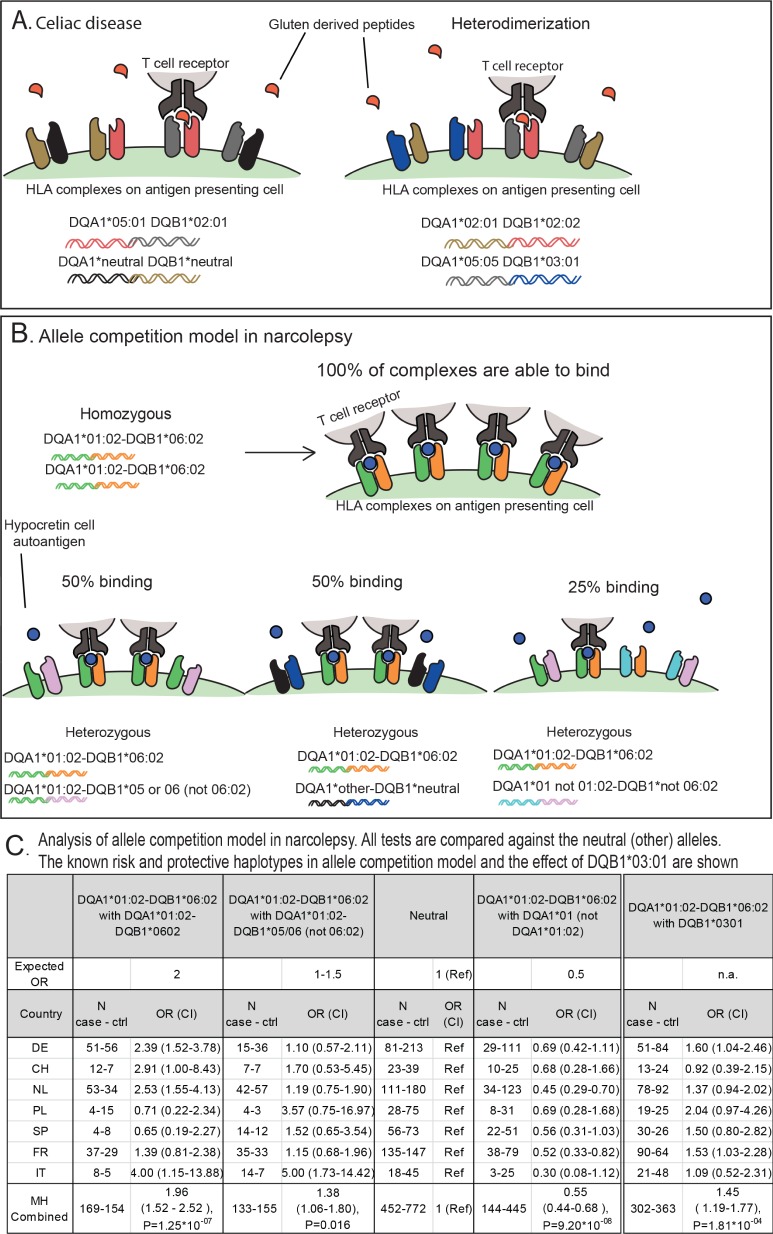
One way to demonstrate that both DQA1 and DQB1 have independent effects in other diseases is through the analysis of specific heterodimer combinations (so-called “trans effects”). As an example (Figure 2), in the classic case of gluten intolerance/celiac disease, almost all cases (70%) share the HLA DQA1*05:01∼DQB1*02:01 haplotype, generating a DQα05:01/DQβ02:01 heterodimer that is encoded in cis in haplotypes bearing DRB1*03:01 (DR3 or DR17 as typed by serologic methods: note that haplotypes are denoted by “∼”, while cis and trans heterodimers are denoted by “/”). In this disease the majority of the patients that do not have the haplotype DQA1*05:01∼DQB1*02:01 carry a specific heterozygous DQ combination, including the DQA1*02:01∼DQB1*02:02 (on the DRB1*07:01 bearing DR7 haplotypes) and DQA1*05:05∼DQB1*03:01 haplotypes in trans. As can be seen, when all 4 DQ allele combinations are expressed, four heterodimers can be formed, including one combination with the DQ α-subunit encoded by DQA1*05:05, which can then pair with the DQ β-subunit DQB1*02:02 from the other haplotype creating a DQα05:05/DQβ02:02 heterodimer. Because the amino acid substitutions that distinguish DQA1*05:01 from DQA1*05:05 and DQB1*02:01 from DQB1*02:02 are located outside of the antigen recognition site, the resulting DQα05:01/DQβ02:01 and DQα05:05/DQβ02:02 heterodimers (generally called DQ2.5 in the literature) are considered to be functionally equivalent. The occurrence of these DQ genotypes may be explained by proposing that genotypes that result in the formation of these specific DQ heterodimers predispose equally to susceptibility for celiac disease. This explains why both combinations pre-dispose to celiac disease, something that has also been shown by the similar affinity of these DQ molecules to bind the causal gluten epitopes.6 Similar trans heterozygous effects have been proposed to result in susceptibility to type I diabetes, although in this disease, the situation is much more complex, as multiple factors encoded in DRB1 and DQ loci appear to determine susceptibility and protective effects.7 It is worth noting that not all DQA1 alleles can pair with all DQB1 alleles and form heterodimers, as was elegantly demonstrated by Kwok and coworkers.8,9 The DQA1 and DQB1 alleles that are able to effectively pair can be roughly classified in mutually exclusive groups: the DQ1 group (includes DQA1*01 alleles that can pair with alleles of the DQB1*05 and DQB1*06 groups) and the nonDQ1 group (including alleles of DQA1*02, DQA1*03, DQA1*04, DQA1*06 groups that can pair effectively with DQB1*02, DQB1*03, DQB1*04 alleles). The DQA1 and DQB1 alleles of different groups appear to have divergent evolutionary origins and sequences.10,11
Figure 2.
(A) In celiac disease the majority of patients carry two predisposing alleles: DQA1*05 together with DQB1*02. The predisposing HLA genes do not have to be on the same chromosome but instead can also be on different chromosomes (trans). Heterodimerization of the alleles causes the risk alleles to form a HLA-heterodimer that can detect gluten-derived peptides, thus increasing the risk for celiac disease. (B,C) In narcolepsy, the allele competition model predicts decreasing DQ0602 availability in various genotype combinations. Similarly to celiac disease, the HLA-molecules can heterodimerize and affect the risk of narcolepsy. The DQ0602 homozygotes have the highest risk since they can present hypocretin autoantigen in all DQ0602 complexes. In contrast, those that carry one copy of DQ0602 and have DQA1*01 (not 01:02) at the other chromosome will be protected: DQA1*01 (not 01:02) can heterodimerize with DQB1*06:02 thus competing with DQA1*01:02 binding and reducing the amount of functional, hypocretin-presenting DQ0602. Carrying DQA1*01:02 at both chromosomes confers a modest increase in risk as DQB1*06:02 will be able to always bind with DQA1*01:02. DQB1*03:01 increases risk for narcolepsy which is not explained by the amount of available DQ0602. Data obtained from Tafti et al., 2014, and courtesy of the authors.
We recently extended the trans heterodimerization concept to narcolepsy, proposing a more general model called the “allele competition effect,” which we believe explains the protective effects of various HLA combinations in narcolepsy.2,3 One particular aspect of narcolepsy is that almost all patients with the disease carry DQA1*01:02∼DQB1*06:02 versus approximately 25% of controls. Crystal structure of this heterodimer, often simply called DQ0602, predicts the importance of both DQA1*01:02 and DQB1*06:02 for peptide binding.12 Because DQA1*01:02 is always present together with DQB1*06:02, it is impossible to test with available data whether DQB1*06:02 alone would predispose to narcolepsy. It is however possible to test the fact that DQA1*01:02 alone does not predispose to narcolepsy, as DQA1*01:02 is also commonly found in haplotypes bearing DQB1*05:02, DQB1*06:04 or DQB1*06:09, and those combinations are never found in narcolepsy without DQB1*06:02. Our allele competition hypothesis predicts that DQB1*06:02 alone would not predispose to narcolepsy. This will likely be testable only once we have very large number of cases and controls that have rare combinations of DQB1*06:02 without DQA1*01:02. These combinations do exist at very low frequency in controls, notably in African Americans.
In the meantime, it is however possible to test the involvement of DQA1 in the context of specific heterozygote genotypes by taking into account the other DQB1 alleles that pair with DQA1. For example, as reported in Tafti et al.1 and in several prior studies, two frequent alleles “DQB1*06:03 and DQB1*05:01” are protective in the presence of DQA1*01:02∼DQB1*06:02. Importantly, those two DQB1 alleles are almost always associated with DQA1*01:03 and DQA1*01:01 or DQA1*01:05, respectively, so it is impossible to claim that “DQB1 explains most of the protective effects alone.” As a case in point, in Japanese, DQB1*06:01 is also very protective.13 As DQB1*06:01 associates strongly with DQA1*01:03 in this ethnicity, the most logical explanation would be that DQA1*01:03 mediates the protective effect of DQB1*06:01 and DQB1*06:03 across all ethnic groups. Similarly, the fact that DQB1*05:01 is protective in this study and prior study could also indicate protective effects of DQA1*01:01 or DQA1*01:05.
Our allele competition model2,3 suggests that both DQB1 and DQA1 are involved in predisposition and protection, and that relative risk is proportional to the amount of the resulting DQα01:02/DQβ06:02 heterodimer expressed on antigen presenting cells. We hypothesized that the more of the heterodimer that is present, the greater its chance to bind auto-antigens and thus trigger autoimmunity. This model predicts that homo-zygotes are at the highest risk (2 doses of DQα01:02/DQβ06:02). The second highest risk is in heterozygous genotype combinations that include DQα01:02/DQβ06:02 with other DQα/DQβ pairs of “neutral” alleles that cannot heterodimerize with either DQα01:02 or DQβ06:02 (1 dose of DQα01:02/DQβ06:02). In contrast, the genotype combinations of DQA1*01:02∼DQB1*06:02 with genotypes of the DQ1 group (DQA1*01, DQB1*05 or DQB1*06) that are not DQA1*01:02 or DQB1*06:02 would be protective as they create additional DQ heterodimers that cannot bind the putative pathogenic peptides, reducing the availability of DQα01:02/DQβ06:02 heterodimers. The model also proposes that DQA1*01:02∼DQB1*06:02 heterozygous combinations that carry DQA1*01:02∼DQB1*05:02 or DQA1*01:02∼DQB1*06:04 or DQA1*01:02∼DQB1*06:09 in the second haplotype would present an intermediate risk, having two doses of DQA1*01:02 but only one dose of DQB1*06:02. As pointed in our prior publications,2 only the trans effects of DQB1*03:01 are not in line with this model, with recent studies suggesting a strong effect on disease onset (unlike with the effects of DQB1*06:02 dosage). DQB1*03:01 allele may thus affect disease predisposition differently, perhaps via effects on shaping the TCR repertoire.
Data provided in Table 4 of Tafti et al.1 are in line with our model. As mentioned above, DQB1*06:03 and DQB1*05:01, two DQ1 alleles also associated with DQA1*01 that are non-DQA1*01:02, are strongly protective. Similarly, DQB1*05:03 (an allele that associates with DQA1*01:04) is also protective, while DQB1*05:02 and DQB1*06:04, associated with DQA1*01:02, have less protective effects. The only outlier for this model is DQB1*06:09, a rare allele that is associated with DQA1*01:02 and is slightly more protective than predicted by our allele competition model, an observation that could result from chance alone considering the numerous alleles studied and the small sample size for this allele (27 controls and 4 with narcolepsy). A prior study did not find such a strong effect of DQB1*06:09.4 Alternatively, differences in affinities of DQB1*06:09 with DQA1*01:02 may be involved, as protective effects should vary slightly, dependent on specific DQα/ DQβ allelic heterodimeric affinities. It should be noted that the DQB1*06:09 allele is more common in Jewish and Middle Eastern populations; therefore, it cannot be ruled out that the findings for DQB1*06:09 may have resulted from different ethnic stratification of patients and controls.
To test the hypothesis of allele competition in this particular dataset more formally, we used the raw data kindly provided by the authors to impute DQA1*01 genotypes in the reported sample based on the known tight linkage disequilibrium between the DQA1 and DQB1 loci.14 We ranked effects of the various allelic combinations as predicted by the allele competition model (Figure 1). Similar to prior findings, we found that DQB1*06:02 homozygotes are approximately at 2× increased risk versus “neutral” DQB1*06:02 heterozygotes, and that DQB1*06:02+DQB1*03:01 heterozygotes are also at increased risk, although less than in our recent Chinese study, most probably because onset in the Chinese sample is much younger, in line with a recent study of a strong effect of DQB1*03:01 on earlier disease onset.15 Finally, as expected DQA1*01:02∼DQB1*06:02 + DQA1*01:02∼DQB1*05 or 06 (non DQB1*06:02) were intermediate, due to double dosage of DQα01:02 but not DQβ06:02 while DQB1*06:02 + DQA1*01 (non-01:02)-DQB1*05 or 06 (non DQB1*06:02) were the most protective. We conclude that the dataset of Tafti1 is very much in line with prior studies2–5 and our proposed allele competition model.2
We also would like to caution against the overinterpretation of a 98% DQB1*06:02 association in narcolepsy, and the relative risk of 251, in the absence of verified low CSF hypo-cretin-1 levels in all patients. Indeed, it is impossible not to consider that clinicians commonly used HLA typing to confirm diagnosis, thus an ascertainment bias surely inflates this value. Similarly, high relative risks were reported for DR2 in the past16 (almost equivalent to DQB1*06:02 in Caucasians) to be reduced in samples with unbiased, clinical diagnosis.17
In conclusion, we are enthusiastic about the findings of the study by Tafti and coworkers1 since this is the largest study that includes DQB1 typing performed to date, including over a thousand patients and matched controls. The results are entirely consistent with prior findings. Minor differences include slightly stronger protective effects of DQA1*01:03∼DQB1*06:03 and DQA1*01:02∼DQB1*06:09 in comparison to other Caucasian samples, an effect that can be attributed to chance, small biases in patient and control populations samples, or small sample sizes for these haplotypes.
DISCLOSURE STATEMENT
This was not an industry supported study. This study has been supported by Sigrid Juselius Foundation, Orion Farmos Research Foundation, Päivikki and Sakari Sohlberg Foundation for H.M.O. The authors have indicated no financial conflicts of interest.
REFERENCES
- 1.Tafti M, Hor H, Dauvilliers Y, et al. DQB1 locus alone explains most of the risk and protection in narcolepsy with cataplexy in Europe. Sleep. 2014;37:19–25. doi: 10.5665/sleep.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han F, Lin L, Li J, et al. HLA-DQ association and allele competition in Chinese narcolepsy. Tissue Antigens. 2012;80:328–35. doi: 10.1111/j.1399-0039.2012.01948.x. [DOI] [PubMed] [Google Scholar]
- 3.Hong SC, Lin L, Lo B, et al. DQB1*0301 and DQB1*0601 modulate narcolepsy susceptibility in Koreans. Hum Immunol. 2007;68:59–68. doi: 10.1016/j.humimm.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 4.Mignot E, Lin L, Rogers W, et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet. 2001;68:686–99. doi: 10.1086/318799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roh EY, Park MH, Park H, et al. Association of HLA-DR and -DQ genes with narcolepsy in Koreans: comparison with two control groups, randomly selected subjects and DRB1*1501-DQB1*0602--positive subjects. Hum Immunol. 2006;67:749–55. doi: 10.1016/j.humimm.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 6.Sollid LM, Qiao SW, Anderson RP, Gianfrani C, Koning F. Nomenclature and listing of celiac disease relevant gluten T-cell epitopes restricted by HLA-DQ molecules. Immunogenetics. 2012;64:455–60. doi: 10.1007/s00251-012-0599-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koeleman BP, Lie BA, Undlien DE, et al. Genotype effects and epistasis in type 1 diabetes and HLA-DQ trans dimer associations with disease. Genes Immun. 2004;5:381–8. doi: 10.1038/sj.gene.6364106. [DOI] [PubMed] [Google Scholar]
- 8.Kwok WW, Kovats S, Thurtle P, Nepom GT. HLA-DQ allelic polymorphisms constrain patterns of class II heterodimer formation. J Immunol. 1993;150:2263–72. [PubMed] [Google Scholar]
- 9.Temme S, Zacharias M, Neumann J, et al. A novel family of human leukocyte antigen class II receptors may have its origin in archaic human species. J Biol Chem. 2014;289:639–53. doi: 10.1074/jbc.M113.515767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gyllensten UB, Erlich HA. Evolution of HLA class-II polymorphism in primates: the DQA locus. Immunol Res. 1990;9:223–33. doi: 10.1007/BF02918181. [DOI] [PubMed] [Google Scholar]
- 11.Otting N, de Groot NG, Doxiadis GG, Bontrop RE. Extensive Mhc-DQB variation in humans and non-human primate species. Immunogenetics. 2002;54:230–9. doi: 10.1007/s00251-002-0461-9. [DOI] [PubMed] [Google Scholar]
- 12.Siebold C, Hansen BE, Wyer JR, et al. Crystal structure of HLA-DQ0602 that protects against type 1 diabetes and confers strong susceptibility to narcolepsy. Proc Natl Acad Sci U S A. 2004;101:1999–2004. doi: 10.1073/pnas.0308458100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hohjoh H, Terada N, Honda Y, Juji T, Tokunaga K. Negative association of the HLA-DRB1*1502-DQB1*0601 haplotype with human narcolepsy. Immunogenetics. 2001;52:299–301. doi: 10.1007/s002510000269. [DOI] [PubMed] [Google Scholar]
- 14.Voorter CE, Lee KW, Smillie D, Tilanus MG, van den Berg-Loonen EM. Sequence-based typing of HLA-DQA1: comprehensive approach showed molecular heterogeneity. Tissue Antigens. 2007;69(Suppl 1):76–81. doi: 10.1111/j.1399-0039.2006.761_1.x. [DOI] [PubMed] [Google Scholar]
- 15.Han F, Faraco J, Dong XS, et al. Genome wide analysis of narcolepsy in China implicates novel immune loci and reveals changes in association prior to versus after the 2009 H1N1 influenza pandemic. PLoS Genetics. 2013;9:e1003880. doi: 10.1371/journal.pgen.1003880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Juji T, Satake M, Honda Y, Doi Y. HLA antigens in Japanese patients with narcolepsy. All the patients were DR2 positive. Tissue Antigens. 1984;24:316–9. doi: 10.1111/j.1399-0039.1984.tb02144.x. [DOI] [PubMed] [Google Scholar]
- 17.Rogers AE, Meehan J, Guilleminault C, Grumet FC, Mignot E. HLA DR15 (DR2) and DQB1*0602 typing studies in 188 narcoleptic patients with cataplexy. Neurology. 1997;48:1550–6. doi: 10.1212/wnl.48.6.1550. [DOI] [PubMed] [Google Scholar]