Abstract
The genomes of free-living bacteria frequently exchange genes via lateral gene transfer (LGT), which has played a major role in bacterial evolution. LGT also played a significant role in the acquisition of genes from non-cyanobacterial bacteria to the lineage of “primary” algae and land plants. Small GTPases are widely distributed among prokaryotes and eukaryotes. In this study, we inferred the evolutionary history of organelle-targeted small GTPases in plants. Arabidopsis thaliana contains at least one ortholog in seven subfamilies of OBG-HflX-like and TrmE-Era-EngA-YihA-Septin-like GTPase superfamilies (together referred to as Era-like GTPases). Subcellular localization analysis of all Era-like GTPases in Arabidopsis revealed that all 30 eubacteria-related GTPases are localized to chloroplasts and/or mitochondria, whereas archaea-related DRG and NOG1 are localized to the cytoplasm and nucleus, respectively, suggesting that chloroplast- and mitochondrion-localized GTPases are derived from the ancestral cyanobacterium and α-proteobacterium, respectively, through endosymbiotic gene transfer (EGT). However, phylogenetic analyses revealed that plant organelle GTPase evolution is rather complex. Among the eubacterium-related GTPases, only four localized to chloroplasts (including one dual targeting GTPase) and two localized to mitochondria were derived from cyanobacteria and α-proteobacteria, respectively. Three other chloroplast-targeted GTPases were related to α-proteobacterial proteins, rather than to cyanobacterial GTPases. Furthermore, we found that four other GTPases showed neither cyanobacterial nor α-proteobacterial affiliation. Instead, these GTPases were closely related to clades from other eubacteria, such as Bacteroides (Era1, EngB-1, and EngB-2) and green non-sulfur bacteria (HflX). This study thus provides novel evidence that LGT significantly contributed to the evolution of organelle-targeted Era-like GTPases in plants.
Keywords: endosymbiotic gene transfer, genomic analysis, lateral gene transfer, small GTPase, evolution of organelle
Introduction
Plant cells contain two types of endosymbiotic organelle, chloroplasts and mitochondria, which arose from cyanobacterium and α-proteobacterium-like ancestors, respectively. During the course of plant evolution, many cyanobacterium and α-proteobacterium-derived genes were either lost from the organelles or transferred to the nucleus (endosymbiotic gene transfer: EGT). Thus, extant chloroplasts and mitochondria retain many prokaryotic proteins that are encoded by the nuclear genome, whereas organelle genomes encode a limited number of proteins.
Lateral gene transfer (LGT) refers to the transmission of genetic material between distinct evolutionary lineages, and plays a substantial role in generating the diversity of genes in host cells. It is well known that LGT is an important process in the evolution of prokaryotes, particularly in the evolution of antibiotic resistance (Barlow, 2009). In contrast to prokaryotic cells, LGT between multicellular eukaryotes is generally believed to be rare, due to the barrier of germline in multicellular animals and apical meristem in plants (Andersson, 2005; Bock, 2010). However, several lines of evidence suggest that there were ancient gene transfers from non-cyanobacterial bacteria to the lineage of “primary” algae and land plants. For example, Arabidopsis thaliana has 24 genes of chlamydial origin (Qiu et al., 2013). Furthermore, at least 55 Chlamydiae-derived genes have been identified in algae and plants, most of which are predominantly involved in plastid functions (Moustafa et al., 2008), suggesting an ancient LGT from Chlamydiae to the ancestor of primary photosynthetic eukaryotes (Huang and Gogarten, 2007; Becker et al., 2008; Moustafa et al., 2008; Ball et al., 2013). Moreover, extensive analysis of plastid proteome data revealed that 15% of Arabidopsis plastid proteins are originated through HGT from non-cyanobacterial bacteria, including Proteobacteria and Chlamydiae (Qiu et al., 2013). In addition, five shikimate pathway proteins in chloroplasts have also been obtained by LGT from β/γ-proteobacteria and Rhodopirellula baltica (Richards et al., 2006). It is known that some secondary plastid-containing unicellular algae acquired many chloroplast-targeted proteins through LGT from non-cyanobacterial bacteria (Archibald et al., 2003; Nosenko et al., 2006; Grauvogela and Petersen, 2007; Teicha et al., 2007). Furthermore, recent genome analysis of the moss Physcomitrella patens provided evidence for the impact of LGT on the acquisition of genes involved in several plant specific processes during the evolution of early land plants (Yue et al., 2012). These results suggest that LGT plays a more important role in the evolution of plants than previously thought.
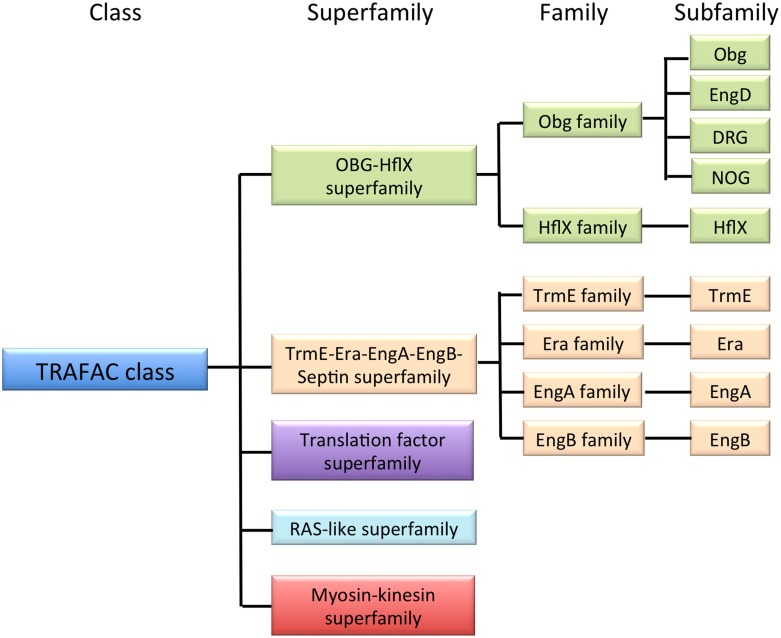
The small GTP-binding proteins (GTPases) are found in all domains of life. They are critical regulators of many aspects of basic cellular processes, including translation, cellular transport and signal transduction. Comprehensive genome sequence analysis has revealed that the TRAFAC (translation factor) class GTPases can be divided into five superfamilies, among which are the OBG-HflX-like and TrmE-Era-EngA-YihA-Septin-like superfamilies (Figure 1). The OBG-HflX superfamily consists of the Obg and HflX families, and the Obg family can be further divided into four subfamilies: Obg, EngD, Drg, and Nog1 (Leipe et al., 2002; Verstraeten et al., 2011). The TrmE-Era-EngA-YihA-Septin superfamily is made up of the TrmE, Era, EngA, EngB families. The OBG-HflX-like and TrmE-Era-EngA-YihA-Septin-like superfamilies (hereafter, together referred to as Era-like GTPases) are represented by Obg and Era, which were identified originally in Bacillus subtilis and Escherichia coli, respectively. Obg proteins are involved in multiple cellular processes, including cell growth (Morimoto et al., 2002), morphological differentiation, DNA replication (Slominska et al., 2002), chromosome partitioning (Kobayashi et al., 2001) and the regulation of protein synthesis and/or ribosome functions (Datta et al., 2004; Sato et al., 2005; Schaefer et al., 2006) in Bacillus subtilis and other eubacteria. Era has also been shown to play an important role in the cell cycle and ribosome assembly (Britton et al., 1998) by binding to 16S rRNA in E. coli (Hang and Zhao, 2003) and to the 30S ribosomal subunit in E. coli and B. subtilis (Morimoto et al., 2002). Other Era-like GTPases are also known to be involved in ribosome maturation and/or RNA modification in eubacteria.
Figure 1.
Classification of GTPases. The TRAFAC class is a member of the P-loop GTPase superclass and is composed of conserved protein superfamilies, as shown. The OBG-HflX-like superfamily and the TrmE-Era-EngA-YihA-septin-like superfamily together contain nine subfamilies (Era-like GTPases).
Among the subfamilies composing the Era-like GTPases, seven are eubacterium-related (Obg, HflX, TrmE, EngD, EngB, Era, and EngA) and conserved from eubacteria to eukaryotes, whereas two are archaea-related (Nog1 and Drg) and conserved in eukaryotes. It is expected that the eubacterium-related GTPase genes were acquired through EGT in eukaryotic cells and are localized to the symbiotic organelles, namely mitochondria and chloroplasts. On the other hand, the archaea-related DRG and NOG1 must have originated from an archaeaic host cell, and likely function in cytoplasm and/or nuclei. However, subcellular localization and functions of the Era-like GTPases remain largely unknown in eukaryotes, except for Obg, Drg, and Nog1. It has been shown that Obg homologs are targeted to mitochondria in yeast (Datta et al., 2005), and to mitochondria and the nucleolus in human cells (Hirano et al., 2006). By contrast, Drg and Nog1 GTPases play important roles in the cytoplasm and mitochondria, respectively, in animal and yeast cells (Mittenhuber, 2001; Park et al., 2001). These results suggest that the Era-like GTPases may be involved in the regulation of organelle functions in eukaryotes.
Genomic data on the Era-like GTPase genes show that plants have a larger number of GTPase genes than do bacteria, yeast, or mammals (Leipe et al., 2002). It is expected that plants acquired additional chloroplast-localized GTPases from cyanobacteria through EGT (McFaddan, 2001). In fact, Bang et al. (2009, 2012) reported that there are two Obg homologs that target to chloroplasts and mitochondria in Arabidopsis. However, very little is known about the intracellular compartmentation and evolution of other Era-like GTPases in plants. To address these questions, we performed comprehensive phylogenetic and subcellular localization analyses of eubacterial Era-like GTPase proteins in Arabidopsis. We found that all 13 eubacteria-related GTPases (of the Obg, HflX, TrmE, EngD, EngB, Era, and EngA subfamilies) were localized to chloroplasts and/or mitochondria in Arabidopsis, whereas archaea-related DRG and NOG1 were localized to the cytoplasm and nuclei, respectively. Unexpectedly, however, EGT likely played a limited role in the evolution of chloroplast and mitochondrial GTPases. There were only three chloroplast GTPases and one dual-targeting GTPase derived from the ancestral cyanobacterium and two mitochondrial GTPases derived from the ancestral α-proteobacterium through EGT. On the other hand, three chloroplast other GTPases were related to α-proteobacterial proteins, but not to cyanobacterial GTPases, suggesting re-compartmentation of mitochondrial GTPases to chloroplasts during plant evolution. Moreover, four Era-like GTPases were closely related to clades from other eubacteria, such as Bacteroides (Era1, EngB-1, and EngB-2) and green non-sulfur bacteria (HflX). These results suggest that LGT from Bacteroides and green non-sulfur bacteria has played a significant role in the evolution of genes for chloroplast- and mitochondria-target GTPases in land plants.
Materials and methods
Phylogenetic analyses and classification
Obg/Era superfamily genes were retrieved from public databases (NCBI, TAIR, and KEGG) by genome screening with the known amino acid sequences of members of each subfamily as queries. Genes that are only detected in the query and potential donor groups will also be identified. Detailed phylogenetic analyses were performed for each of the candidates. Taxonomic distribution of sequence homologs was also investigated.
Multiple protein sequence alignments were performed using the Clustal X program (Jeanmougin et al., 1998) followed by manual refinement. Gaps and ambiguously aligned sites were removed manually. The well-aligned regions were used for the construction of phylogenetic trees. Phylogenetic analyses were performed using the protdist program with JTT amino acids substitution model, and followed by neighbor program in the PHYLIP 3.6 package (Ratief, 2000). The phylogenetic tree was inferred using the neighbor-joining method (Saitou and Nei, 1987) and tested using 100 replications of bootstrap analysis using the seqboot and consense programs in the same package. The data were subsequently visualized as phylogenetic trees using the treeview program (Page, 1996). The names and classifications proposed herein are based on P-loop protein classification (Leipe et al., 2002).
Plant and cell growth conditions
Arabidopsis thaliana ecotype Colombia were germinated and grown on Murashige–Skoog (MS) medium containing 0.8% (w/v) agar and 1% (w/v) sucrose at 22°C with 80–100 μmol m−2 s−1 illumination for a daily 16-h light period. Arabidopsis suspension-culture cells were cultured in MS medium at 23°C with continuous agitation under dark conditions. Onion bulb was purchased from local market.
Molecular cloning and transient expression assays
GFP fusion genes were constructed as follows. First-strand cDNA was synthesized from total RNA prepared from Arabidopsis seedlings using AMV reverse transcriptase (TaKaRa). cDNA was amplified by PCR using KOD-plus-DNA polymerase (TOYOBO) according to the manufacturer's protocol. Transient expression vectors were constructed using the GFP reporter plasmid 35Ω-sGFP(S65T). The PCR fragments containing full length Era-like GTPase genes were ligated in frame into the 35Ω-sGFP(S65T) plasmid. All sets of primers used in this study are listed in Supplemental data 1. Transient expression of the GFP fusion proteins in Arabidopsis protoplasts was performed as previously described (Yanagisawa et al., 2003). Briefly, rosette leaves of 4–6-week-old plants were used for the transient expression experiments. After overnight incubation at 23°C in the dark, GFP signal was observed using a confocal laser scanning microscope (LSM5 PASCAL; Carl Zeiss Inc.) equipped with green HeNe and argon lasers. The assay using Arabidopsis culture cells was performed as previously described (Uemura et al., 2004). Mitochondrial GTPases were transiently expressed in onion epidermal cells by using particle bombardment. 1.5 μg of GFP fusion plasmids coated on 0.6 μm gold particles were bombarded into epidermal sheaths peeled from onion bulbs placed on ½ MS plates. The epidermal cells were stained with MitoTracker Red to label the mitochondria. Expression assays were performed with at least three independent repetitions and mitochondrial signals were confirmed by MitoTracker Red staining.
Isolation of mitochondria from arabidopsis seedlings and immunoblot analysis
Intact mitochondria were isolated from Arabidopsis hydroponic seedling cultures as described previously (Sweetlove et al., 2007). Mitochondria were subsequently separated into membrane and soluble fractions. Immunoblot analyses of the mitochondrial fractions were performed using antibodies against E. coli ObgE and mitochondrial outer membrane marker, voltage-dependent anion-selective channel protein (VDAC).
Results
Era-like GTPase proteins in plants
We conducted genome-wide searches for proteins containing Era-like GTPase signatures to identify all Era-like GTPases in three model plant genomes: Arabidopsis thaliana (dicot), Oryza sativa (monocot), and Cyanidioschyzon merolae (red algae). Arabidopsis was found to have 18 GTPase genes, including members of all nine Era-like GTPase subfamilies (Table 1). Arabidopsis, rice and C. merolae had at least one gene in each of the nine subfamilies, suggesting that plants require similar sets of Obg/Era GTPase genes. Furthermore, humans have the same sets of genes as plants, except for EngA, suggesting that Obg, Drg, NOG1, EngD, HflX, TrmE, Era, and EngB subfamily genes are shared between plants and animals. By contrast, S. cerevisiae lacks HflX, Era, and EngA genes, suggesting that the unicellular fungi Saccharomyces has lost several gene sets during evolution. Drg and Nog1 belong to the Obg family, and were found in two domains of life, archaea and eukaryotes, but not in eubacteria (Suwastika et al., 2014). By contrast, Obg, EngD, HflX, TrmE, Era, EngA, and EngB genes were found in eubacteria and eukaryotes (Table 1). It is likely that the archaea-related genes were derived from a eukaryotic host cell, but eubacteria-related genes from eubacterial ancestors. Both HflX and EngB are also shared among eubacteria, eukaryotes and some archaea.
Table 1.
OBG-Hflx-like Superfamily and TrmE-Era-EngA-YihA-Septin-like Superfamily genes in Arabidopsis genom.
| OBG-HflX-like SUPERFAMILY | ||||||||
|---|---|---|---|---|---|---|---|---|
| Fam. | Sub fam. | A. thaliana | Name | O. zativa | C. merolae | S. sereviciae | H. sapien | E.coli |
| Obg | Obg | At1g07615 | Obg A-1 | Os03g58540 | CMG146C | YHR168W | hsa26164 | JW3150 |
| At5g18570 | Obg A-2 | Os07g47300 | hsa85865 | |||||
| Os11g47800 | ||||||||
| EngD/YyaF/YchF | At1g30580 | EngD-1 | Os08g019930 | CME188C | YRR025C | hsa29789 | JW1194 | |
| At1g56050 | EngD-2 | CMT184C | YHL014C | |||||
| Ygr210 | YGR210C | |||||||
| Drg | At4g39520 | Drg1-1 | Os07g43470 | CMG124C | YAL036C | hsa151457 | ||
| At1g17470 | Drg1-2 | Os05g28940 | CMN324C | YGR173W | hsa1819 | |||
| At1g72660 | Drg1-3 | hsa4733 | ||||||
| Nog | At1g50920 | Nog1-1 | Os07g01920 | CMB146C | YPL093W | hsa23560 | ||
| At1g10300 | Nog1-2 | Os06g09570 | ||||||
| Hflx | At5g57960 | Hflx | Os03g51820 | CMT373C | hsa8225 | JW4131 | ||
| Os11g38020 | ||||||||
| TrmE-Era-EngA-YihA-Septin-like SUPERFAMILY | |||||||
|---|---|---|---|---|---|---|---|
| Fam. | A. thaliana | Name | O. zativa | C. merolae | S. sereviciae | H. sapien | E. coli |
| TrmE/ThdF | At1g78010 | TrmE | Os08g31460 | CMK223C | YMR023C | hsa84705 | JW3684 |
| CMV025C | |||||||
| FeoB | JW3372 | ||||||
| EngB/YihA | At2g22870 | EngB-1 | Os03g23250 | CMQ232C | YDR336W | hsa29083 | JW5930 |
| At5g11480 | EngB-2 | Os01g73220 | |||||
| At5g58370 | EngB-3 | Os03g81640 | |||||
| Era | At5g66470 | Era-1 | Os05g49220 | CMN201C | hsa26284 | JW2550 | |
| At1g30960 | Era-2 | ||||||
| EngA/YfgK | At3g12080 | EngA-1 | Os01g12540 | CMC059C | JW5403 | ||
| At5g39960 | EngA-2 | Os11g41910 | |||||
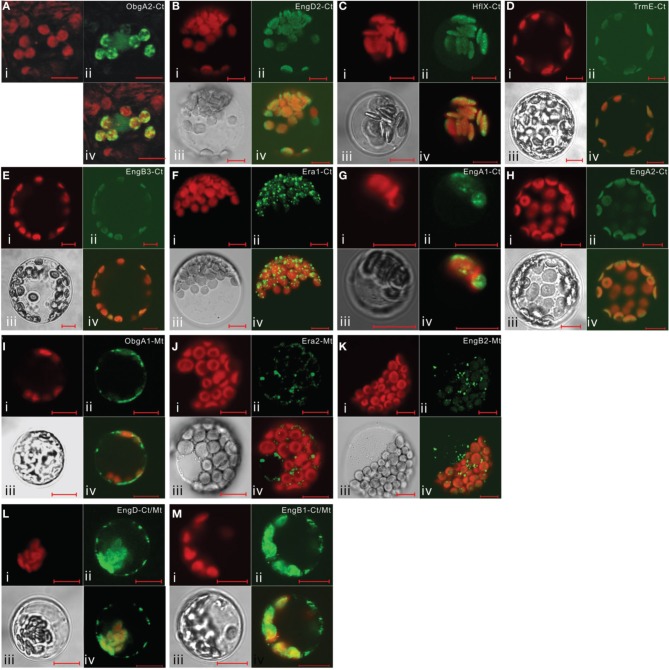
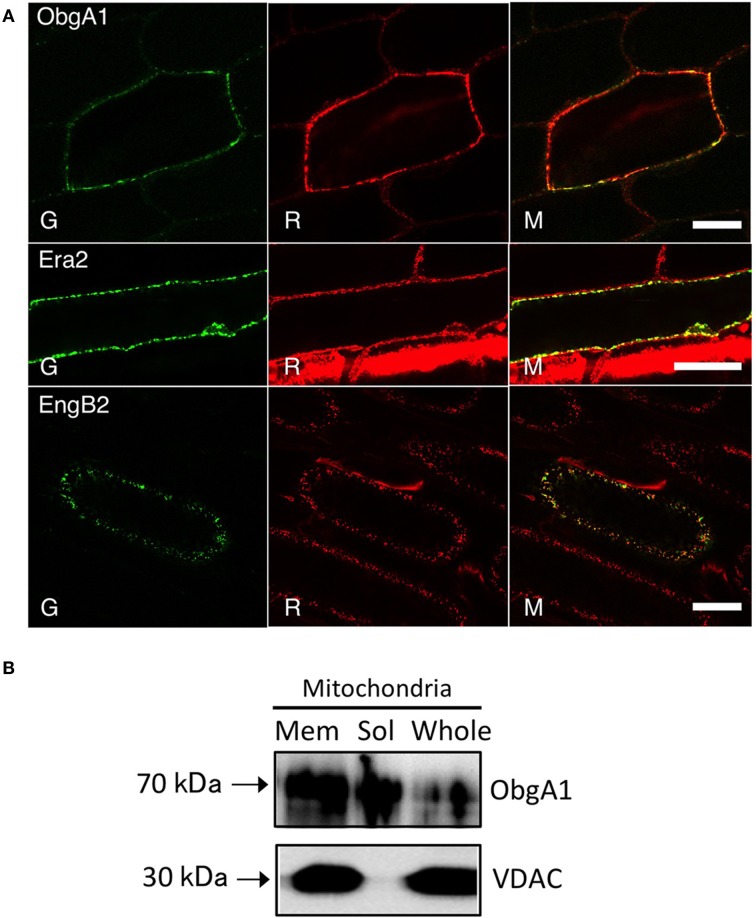
It is noteworthy that vacsular plants have a larger number of Era-like GTPase genes (18 genes in Arabidopsis and 17 genes in rice) compared to human (11 genes) and yeast (9 genes) (Table 1). Although the human genome contains a single gene of each Era-like GTPase subfamily except for the Obg and Drg subfamilies, plant Era-like GTPase subfamilies contain multiple genes. It is predicted that multiple Era-like GTPase proteins are targeted to different cellular compartments, such as chloroplasts, mitochondria and nuclei (Table 2). However, the subcellular localization of most Obg/Era superfamily proteins has not been determined in plants, except for chloroplastic and mitochondrial Obg proteins (Bang et al., 2009). In this study, we examined subcellular localization of Era-like GTPases in Arabidopsis using in vivo analysis of GFP-tagged proteins. C-terminal GFP fusions were transiently expressed in Arabidopsis protoplasts or cultured cells under the transcriptional control of the cauliflower mosaic virus 35S promoter. As predicted, all eubacterium-related GTPases were localized in chloroplasts and/or mitochondria, but not other organelles nor cytoplasm (Figure 2). We identified eight proteins that were targeted exclusively to chloroplasts (Figures 2A–H) and two dual-targeting proteins transported into both chloroplasts and mitochondria (Figures 2L,M). Interestingly, each family/subfamily contained at least one chloroplast protein, suggesting that eubacteria-related Era-like GTPases play an important role in chloroplasts (Table 2). On the other hand, only three mitochondrion-specific proteins (ObgA1, Era2 and EngB2) were identified (Figures 2I–K). The colocalization of the GFP fluorescence with the red fluorescence of the MitoTracker dye confirms the mitochondrial targeting of these respective GFP fusions in onion epidermal cells (Figure 3A). Mitochondrial localization of ObgA1 was further confirmed by western blotting analysis of mitochondrial fractions isolated from Arabidopsis seedlings. Anti-ObgE antibody specifically detected ObgA1 in both membrane and soluble fractions of mitochondria (Figure 3B). By contrast, all Drg GTPases were localized to the cytoplasm in Arabidopsis (Suwastika et al., 2014), whereas NOG1 homologs were localized to the nucleus (Figure 4).
Table 2.
Subcellular localization of Obg-TrmE GTPases in Arabidobsis.
| Subfamily | Gene name | AGI number | *TargetP/Wolf PSORT | Proteome | **GFP |
|---|---|---|---|---|---|
| Obg | Obg A1 | At1g07615 | Mit 6/Chl 4, Mit 4 | n.d | Mit |
| Subfamily | Obg A2 | At5g18570 | Mit 9/Chl 8 | Chl | Chl |
| EngD | EngD-1 | At1g30580 | Other 5/Cysk 8 | Cyto | Chl/Mit |
| EngD-2 | At1g56050 | Chl 3/Chl 10 | Chl | Chl | |
| Drg*** | Drg1-1 | At4g39520 | Other 8/Cyto 7 | Cyto/PM | Cyto |
| Drg1-2 | At1g17470 | Other 8/Cyto 7 | n.d | Cyto | |
| Drg1-3 | At1g72660 | Other 8/Cyto 7 | n.d | Cyto/Nucl | |
| Nog | Nog1-1 | At1g50920 | Other 8/Cyto 8 | PM | Nucl |
| Nog1-2 | At1g10300 | Other 8/Chl 6 | n.d | Nucl | |
| Hflx | Hflx | At5g57960 | Chl 8/Chl 7 | Chl | Chl |
| TrmE | TrmE | At1g78010 | Chl 5, Mit 3/Chl 9 | Chl | Chl |
| Era | Era 1 | At5g66470 | Chl 8/Mit, Chl 3 | Chl | Chl |
| Era 2 | At1g30960 | Mit 9/Chl 5, Mit 5 | n.d | Mit | |
| EngA | EngA-1 | At3g12080 | Chl 8/Mit 5, Chl 4 | Chl | Chl |
| EngA-2 | At5g39960 | Mit 7/Chl 6, Mit 3 | n.d | Chl | |
| EngB | EngB-1 | At2g22870 | Mit 7/Chl 7 | n.d | Chl/Mit |
| EngB-2 | At5g11480 | Chl 7/Chl 8 | n.d | Chl/Mit | |
| EngB-3 | At5g58370 | Mit 5/Chl 4, nucl 3 | n.d | Chl |
Figure 2.
Subcellular localization of eubacterium-related Era-like GTPases in Arabidopsis. Transient expression of GFP-fusion proteins in Arabidopsis protoplasts: (A–H) chloroplast targeting of ObgA2, EngD2, Hflx, TrmE, EngB3, Era1, EngA1, and EngA2 proteins. (I–K) mitochondrial targeting of ObgA1, Era2, and EngB2 proteins. (L,M) dual targeting of EngD1 and EngB1 to mitochondria and chloroplasts. (i) Chlorophyll auto-fluorescence, (ii) GFP fluorescence, (iii) DIC image, (iv) merged image of (i), (ii), and (iii). Scale bars are 10 μm.
Figure 3.
Mitochondrial localization of ObgA1, Era2, and EngB2 proteins. (A) Confocal images of ObgA1-GFP, Era2-GFP, and EngB2-GFP fusion proteins transiently expressed in onion epidermal cells. All proteins were targeted to mitochondria as confirmed by mitotracker staining. G, GFP fluorescence; R, mitotracker Red; M, merged image. Scale bars are 10 μm. (B) Western blot analysis of ObgA1 in mitochondria fractions. The Arabidopsis mitochondria whole lysates (whole) were fractionated into membrane (Mem) and matrix (Sol) fractions. Fractions were resolved on a 10% SDS-PAGE and detected with the anti-ObgE and anti-VDAC (mitochondrial outer membrane marker) antibodies. Ten micrograms protein were loaded.
Figure 4.
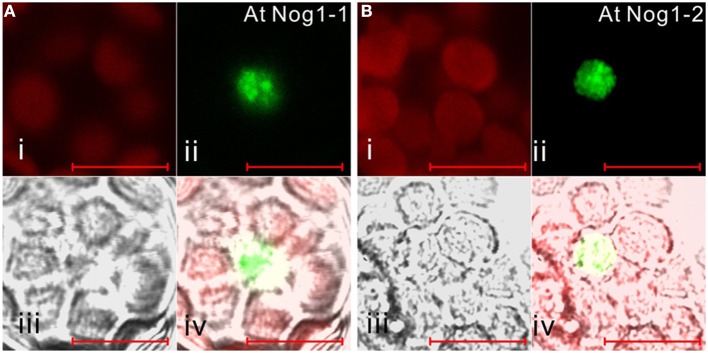
Subcellular localization of archaea-related Nog1 in Arabidopsis. GFP-fusion Nog1-1 (A) and Nog1-2 (B) proteins are transiently expressed in Arabidopsis protoplasts. (i) Chlorophyll auto-fluorescence, (ii) GFP fluorescence, (iii) DIC image, (iv) merged image of (i), (ii), and (iii). Scale bars are 10 μm.
Chloroplast-targeted Obg and TrmE are of cyanobacterial origin
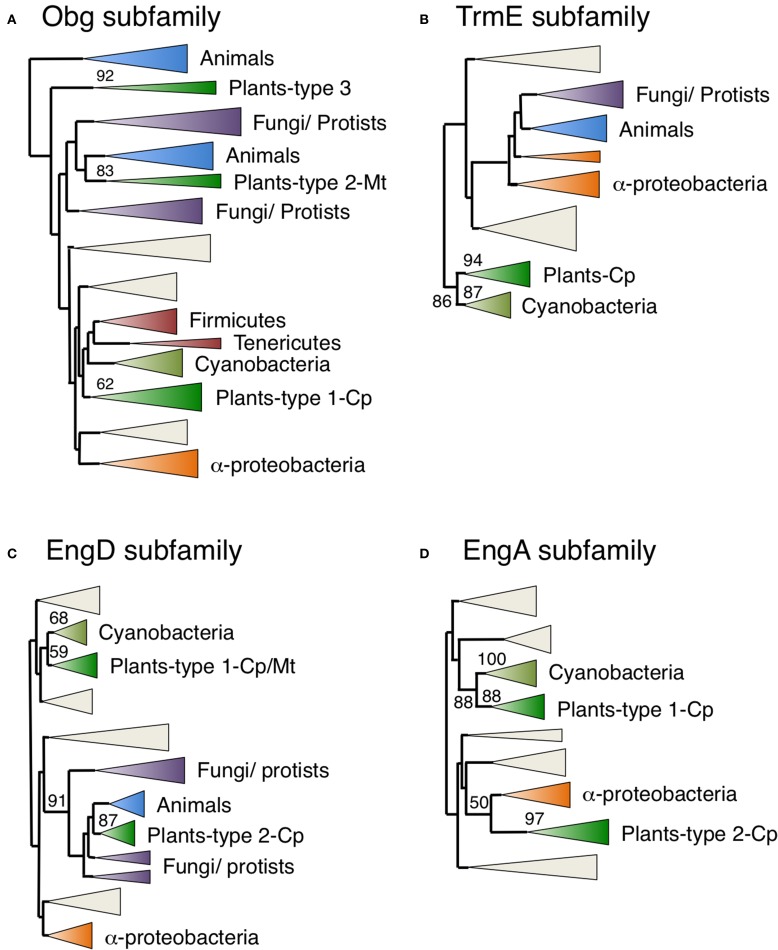
Obg and TrmE genes are found in eubacteria, animals, fungi and plants (Table 1). Several lines of evidence imply that Obg GTPases function in ribosome maturation in eubacteria (Sato et al., 2005), mitochondria of yeast (Datta et al., 2005) and human nuclei (Hirano et al., 2006). Figure 5, Figure S1 portray a NJ tree of Obg homologs, demonstrating that plant Obg homologs formed three distinct monophyletic clusters (types 1–3) with robust support of 62, 83, and 92%, respectively. Arabidopsis had two Obg homologs, ObgA1 (At1g07615) and ObgA2 (At5g18570). ObgA2 (ObgC/Obg target to chloroplast) in the type 1 cluster has been shown to be localized to chloroplasts (Bang et al., 2009; Figure 2A). GFP-tagged ObgA2 appeared in small dot-like structures in chloroplasts, suggesting that ObgA2 is associated with chloroplast nucleoids. The type 1 plant Obg homologs were closely related to cyanobacterial homologs, suggesting that they have cyanobacterial endosymbiotic ancestry.
Figure 5.
Phylogenetic tree of Obg, TrmE, EngD, and EngA subfamily proteins. Comprehensive comparison of Obg (A), TrmE (B), EngD (C), and EngA (D) subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 185 (Obg), 152 (TrmE), 182 (EngD), and 147 (EngA) proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates. The horizontal length of the triangles is equivalent to the average branch length. Green triangles, plant clade; light green triangles, cyanobacterial clade; blue triangles, animal clade; purple triangles, fungus-protist clade; orange triangles, α-proteobacteria clade. The original phylogenetic trees are shown in Figures S1–S4.
By contrast, the type 2 plant Obg homologs were closely related to animal and fungal homologs. The human Obg homolog ObgH1 is localized to mitochondria in HeLa cells (Hirano et al., 2006). Similarly, we showed that Arabidopsis ObgA1 (a type 2 Obg) was also exclusively localized in mitochondria (Figures 2I, 3A,B). However, it should be noted that there was not a close relationship between type 2 plant Obg and α-proteobacterial Obg. The chloroplast and cyanobacterium-like Obg proteins have a TGS domain in the C-terminal region, whereas mitochondrial Obg proteins lack the TGS domain. The TGS domain is known to be involved in stress responses in eubacteria. Therefore, chloroplast Obg GTPases might have specific a role in plant stress responses.
The type 3 plant Obg proteins were related to another animal Obg homologs, represented by ObgH2, which is localized in nucleus (Hirano et al., 2006). Plants including green algae, moss and some vacsular plants have one type 3 Obg homolog, whereas Arabidopsis lacks the type 3 Obg. The subcellular localization of type 3 plant Obg homologs remains to be examined.
Finally, it is noteworthy that C. merolae retained the Type 1 chloroplast Obg homolog, but lacked the type 2 and type 3 mitochondrial and nuclear Obg homologs. It is conceivable that type 1 Obg or other Obg-related proteins might take over the function of mitochondria Obg in C. merolae.
On the other hand, green plant TrmE proteins formed a single monophyletic group that was closely related to a cyanobacterial clade with a strong bootstrap value (87%) (Figure 5, Figure S2), supporting their cyanobacterial endosymbiotic ancestry. In fact, Arabidopsis TrmE protein was targeted exclusively to chloroplasts. E. coli TrmE is involved in the modification of uridine bases at the first anticodon of tRNA. Therefore, plant TrmE might have a role in tRNA modification in chloroplasts. It should be noted that animal and fungal proteins form distinct clades that are unrelated to plant proteins, but are grouped with α-proteobacterial genes. The TrmE protein is known to be targeted to mitochondria in yeast (Decoster et al., 1993; Colby et al., 1998), suggesting that mitochondrial TrmE was derived from α-proteobacteria. Interestingly C. merolae has two animal-related TrmE genes but not the cyanobacterium-related chloroplast genes. It is likely that C. merolae has lost the cyanobacterium-derived TrmE gene, while green plants have lost the animal-type mitochondrial TrmE during evolution. It is possible that other mitochondrion-localized GTPases have taken over the function of mitochondrial TrmE in green plants.
Chloroplast-targeted EngD and EngA are of α-proteobacterial origin
EngD and EngA encode GTP-dependent nucleic acid binding protein (Tomar et al., 2011) and 50S ribosome associated protein (Bharat et al., 2006), respectively. Both plant EngD and EngA homologs formed two monophyletic clusters. The type 1 plant clusters grouped with cyanobacterial clusters with 68% support for EngD1 (Figure 5, Figure S3) and 88% for EngA1 (Figure 5, Figure S4). On the other hand, the type 2 EngD2 and EngA2 proteins formed monophyletic clusters with 87 and 97% support, respectively, and were closely related to animal/fungal and/or α-proteobacterial genes. C. merolae also had two EngD proteins that were divided into type 1 and type 2 groups, and one EngA related to the type 1 group. These results suggest that type 1 EngD and EngA proteins were derived from cyanobacterial endosymbiotic ancestors, whereas type 2 EngA proteins were derived from the α-proteobacterial endosymbiont via EGT. The type 1 cyanobacterium-related EngD1 was localized in both chloroplasts and mitochondria (dual targeting; Figure 2L), whereas EngA1 was localized exclusively to chloroplasts (Figure 2G). Interestingly, the type 2 α-proteobacteria-related EngD2 (Figure 2B) and EngA2 GTPases (Figure 2H) were also exclusively targeted to chloroplasts. These findings support the idea that chloroplasts acquired additional type 2 EngD2 and EngA2 GTPases through re-compartmentation of α-proteobacterium-related GTPases from mitochondria.
Chloroplast-localized HflX might be derived from green non-sulfur bacteria through lateral gene transfer
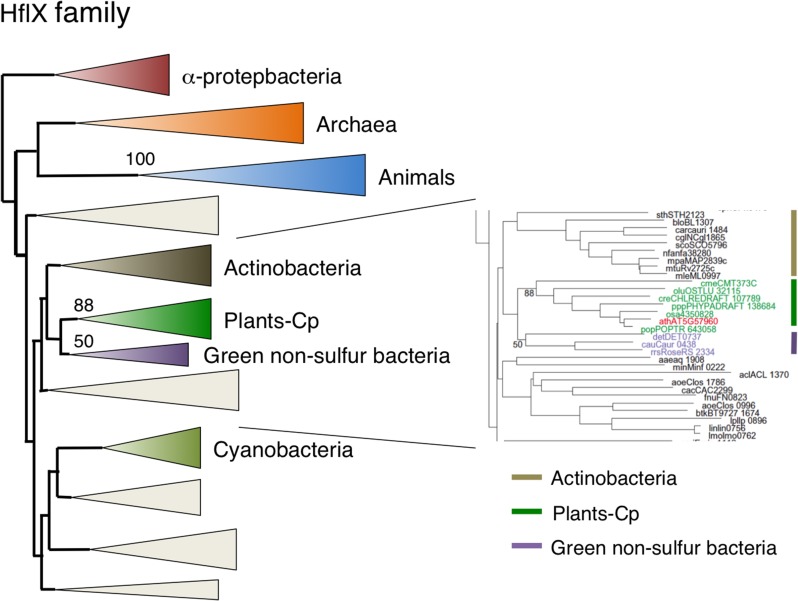
HflX genes are widely conserved among eubacteria, eukaryotes, and some archaea. It was demonstrated recently that Chlamydophila HflX is associated with the 50S ribosome, suggesting a possible role in ribosome maturation and translational regulation (Polkinghorne et al., 2008). Animal HflX homologs formed a monophyletic group with 100% bootstrap support, and were closely related to the archaeal clade (Figure 6, Figure S5), suggesting that animal HflX genes were derived from archaeal ancestors. By contrast, plants lack archaea-like genes. Arabidopsis had a single HflX homolog that was exclusively localized in chloroplasts (Figure 2C). Phylogenetic analysis revealed that plant HflX homologs form a single monophyletic group with strong bootstrap support (88%). Unexpectedly, however, the plant HflX clade was not related to the cyanobacterial or animal clades, but instead was closely related to the green non-sulfur bacteria group. It is conceivable that the plant HflX genes were derived from green non-sulfur bacteria through LGT. The plant clade included the protein from the primitive red algae C. merolae, suggesting that the gene transfer occurred at a very early stage in plant evolution before the red algae lineage and green plant lineage diverged.
Figure 6.
Phylogenetic tree of HflX subfamily proteins. Comprehensive comparison of HflX subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 153 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates. The horizontal length of the triangles is equivalent to the average branch length. Green triangle, plant clade; light green triangle, cyanobacterial clade; blue triangle, animal clade; orange triangle, α-proteobacteria clade; dark blue triangle, archaea clade; yellow triangle, green non-sulfur clade. The original phylogenetic tree is shown in Figure S5.
Chloroplast-localized Era1 is derived from green sulfur bacteria or bacteriodes, but not cyanobacteria
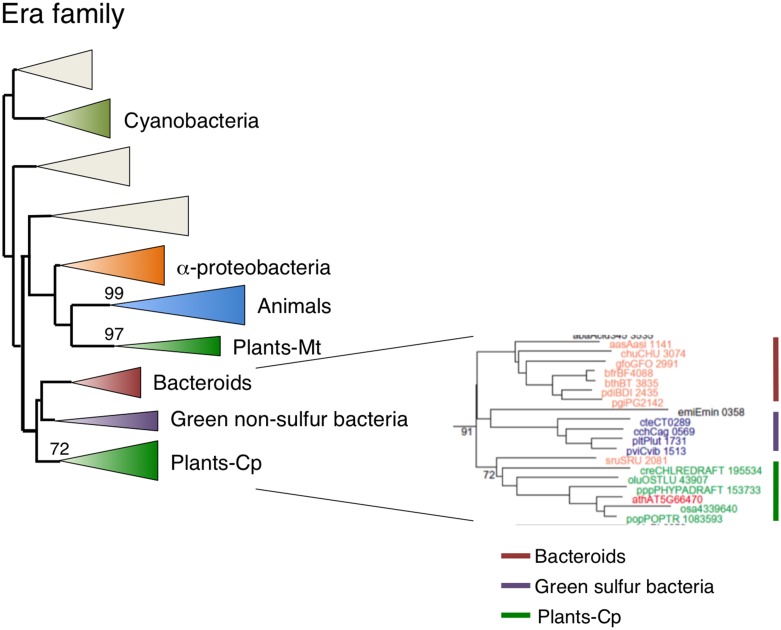
As a homolog of RAS, Era is an extremely important GTPase in E. coli. It has been suggested that Era is directly associated with the 30S ribosomal subunits (Sayed et al., 1999). Human Era (ERAL1) is involved in the regulation of apoptosis (Akiyama et al., 2001). Arabidopsis had two Era homologs: type 1 Era-1 was targeted to chloroplasts (Figure 2F) and type 2 Era2 was a mitochondrial protein (Figures 2J, 3A). GFP-tagged Era1 appeared in small dot-like structures that were observed throughout chloroplasts, suggesting that Era1 is associated with chloroplast nucleoids. Plant Era2 homologs formed a monophyletic group with robust support of 97% and grouped with clusters of animal and α-proteobacteria (Figure 7, Figure S6), suggesting that mitochondrial Era genes were derived from the symbiotic α-proteobacterial ancestors. By contrast, type 1 Era homologs formed a distinct monophyletic group (91%) with Bacteriodes and Green sulfur bacteria clusters. In particular, Salinibacter rubber (Bacteroidetes) was placed at the base of the plant lineage. Cyanobacterial Era homologs formed a separate monophyletic group and were not related to either type 1 or type 2 plant Era clusters. This lineage-specific bacterial affiliation of chloroplast-targeted Era implies that there was LGT from Bacteriodes/Green sulfur bacteria to the plant ancestor. Type 2 mitochondrial Era was conserved in the primitive red alga C. merolae, but the type 1 chloroplast Era was not.
Figure 7.
Phylogenetic tree of Era subfamily proteins. Comprehensive comparison of Era subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 141 genes. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates. The horizontal length of the triangles is equivalent to the average branch length. Green triangle, plant clade; light green triangle, cyanobacterial clade; blue triangle, animal clade; orange triangle, α-proteobacteria clade; yellow triangle, green non-sulfur clade; red triangle, bacteroides clade. The original phylogenetic tree is shown in Figure S6.
Dual-targeting EngB is derived from bacteroides via lateral gene transfer
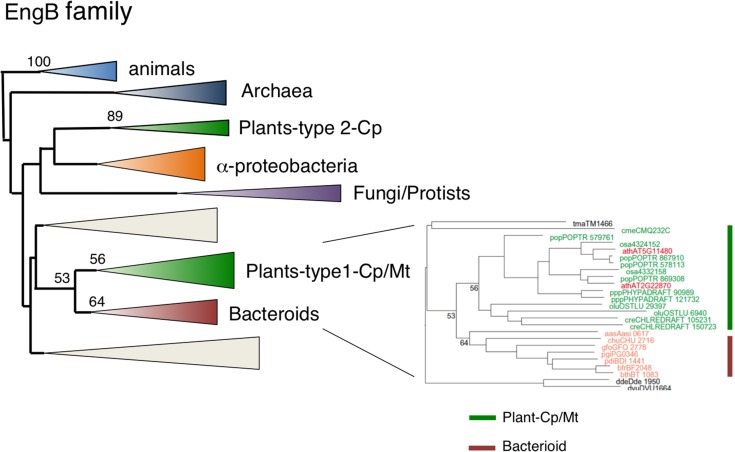
EngB (YihA) has been characterized as an essential gene of unknown function in both E. coli and B. subtilis (Arigoni et al., 1998; Dassain et al., 1999). Arabidopsis encodes three EngB proteins: EngB1 was dual targeted to chloroplasts and mitochondria (Figure 2M), whereas EngB3 was localized exclusively in chloroplasts (Figure 2E). By contrast, EngB2 was localized exclusively to mitochondria (Figures 2K, 3A). Phylogenetic analysis revealed that plant EngB proteins formed two distinct monophyletic clusters: type 1 and type 2 clusters with 56 and 89% support, respectively (Figure 8, Figure S7). The type 1 cluster, including dual-targeting EngB1 and mitochondrial EngB2, was grouped with the Bacteroides clade, suggesting an LGT origin of type 1 genes from Bacteroides. On the other hand, the type 2 cluster, containing chloroplast-targeting EngB3, was grouped with a cluster from α-proteobacteria. Fungi and protist genes were closely related to this clade, but animal genes formed a distinct cluster (100%) that was related to the archaeal cluster, suggesting that type 2 genes were derived from α-proteobacteria. It is expected that α-proteobacteria-related fungal and protist EngB GTPases are localized to mitochondria. Animals probably have lost the type 2 EngB genes although fungi, protists and plants retain them. Type 1 EngB was conserved in C. merolae, but the type 2 EngB was not. These results suggest that the mitochondrion-derived EngB3 has changed its target from mitochondria to chloroplasts.
Figure 8.
Phylogenetic tree of EngB family proteins. Comprehensive comparison of EngB subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 143 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates. The horizontal length of the triangles is equivalent to the average branch length. Green triangle, plant clade; light green triangle, cyanobacterial clade; blue triangle, animal clade; purple triangle, fungus-protist clade; orange triangle, α-proteobacteria clade; yellow triangle, green non-sulfur clade; red triangle, Bacteroides clade. The original phylogenetic tree is shown in Figure S7.
Archaea-related Drg and Nog1 target to the cytoplasm and nucleus, respectively
Eubacteria possess two Obg family proteins, Obg and EngD, which are also conserved in plants and animals. By contrast, archaea encode two other Obg-related proteins, Drg and Nog1. In addition to eubacterium-like Obg and EngD GTPases, all eukaryotes possess Drg and Nog1, suggesting their distinct roles in eukaryotic cells. It has been shown that Drg GTPases are associated with translating ribosomes in the cytoplasm in S. cerevisiae (Li and Trueb, 2000). On the other hand, NOG1 is critical for biogenesis of the 60S ribosomal subunit in the nucleus (Jensen et al., 2003). Arabidopsis encodes three Drg (Drg1–Drg3) and two Nog1 (Nog1-1, Nog1-2) homologs. Subcellular localization analyses using GFP fusion proteins revealed that all Drg GTPases are localized to the cytoplasm in Arabidopsis (Suwastika et al., 2014), whereas NOG1 homologs were localized to the nucleus (Figure 4). Phylogenetic analyses of Drg and Nog1 proteins revealed that both Drg and Nog1 proteins formed a distinct monophyletic cluster with 97% and 100% support, respectively (Figures S8, S9). Plant Drg and Nog1 were related to archaeal Drg and Nog1 proteins. These results suggest that Obg-related Drg and Nog1 GTPases were derived from archaeal GTPases and have acquired specific functions in the cytoplasm and nucleus, respectively, during evolution.
Discussion
Chloroplasts are descended from an ancient endosymbiotic cyanobacterium. Consequently, it has been thought that nuclear genes encoding chloroplast proteins are mainly derived from the endosymbiotic cyanobacterium. Indeed, it is estimated that 14–18% of nuclear-encoded proteins are cyanobacterial in origin (Martin et al., 2002; Deusch et al., 2008). However, chloroplast proteins are not only encoded by cyanobacterium-derived genes, but also by a considerable number of non-cyanobacterial genes. Chloroplasts have recruited significant number of eukaryotic proteins from host cells. Thus, chloroplasts possess unique prokaryotic-eukaryotic hybrid systems in several cellular processes, including transcription (Baumgartner et al., 1993; Yagi and Shiina, 2011), translation and metabolic pathways (Martin and Schnarrenberger, 1997; Reyes-Prieto and Bhattacharya, 2007; Reyes-Prieto and Moustafa, 2012). In total, more than 600 non-cyanobacterial-host-derived proteins contribute to the chloroplast proteome, which includes ~3000 proteins (Abdallah et al., 2000). It has been suggested that Chlamydia genomes encode a large number of plant-related genes (Brinkman et al., 2002). Moreover, previous study identified 31 genes highly related to those from Chlamydiae in green algae and plants, and 20 Chlamydiae-related genes shared by red and green algae (Moustafa et al., 2008). Another study identified 39 proteins of chlamydial origin in photosynthetic eukaryotes (Becker et al., 2008). Chlamydiae are obligate intracellular pathogens/symbionts in many eukaryotes, although not in plants. It is presumed that Chlamydiae temporarily established an endosymbiosis with ancestral plant cells containing chloroplasts and transferred a number of genes into the host cell (Becker et al., 2008; Moustafa et al., 2008). Some evidence suggests that the LGT of chlamydial genes occurred before the divergence of the Glaucoplantae, Rhodoplantae and Viridiplantae (Becker et al., 2008). In addition, several lines of evidence suggest that there were LGTs among other eubacteria and plants. It has been reported that the gene for chloroplast-localized rRNA adenine dimethyltransferase (rAD) was acquired by LGT from Bacteroides/chlorobi in the rhodophyte lineages, whereas rAD genes of chlorophytes/land plants are derived from Chlamydiae genes (Park et al., 2009). Genes for plastid-localized shikimate pathway proteins are derived from prokaryotic sources, including a proteobacterium related to the γ/β group and an α-proteobacterium (Waller et al., 2006). Furthermore, it has been suggested that some enzymes encoded in the host nuclear genome were mistargeted into the plastid during the evolution of plastids (Reyes-Prieto and Moustafa, 2012). In this study, we found that three chloroplast GTPases (EngD2, EngA2, and EngB3) are likely derived from α-proteobacterium-like ancestors, suggesting re-compartmentation of mitochondrial GTPases.
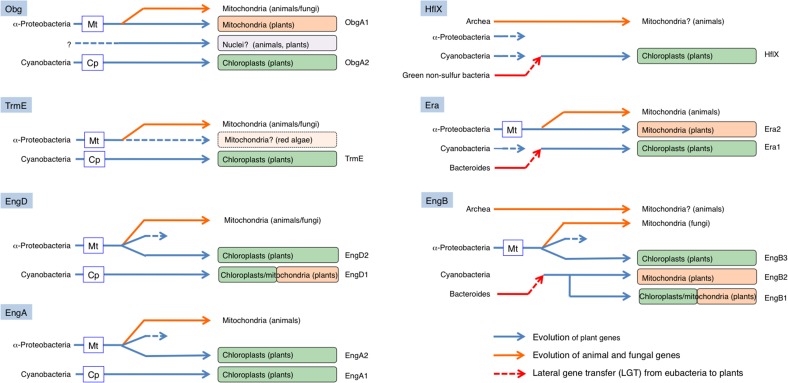
We also identified two novel LGT events among eubacteria and plants. Figure 9 shows a summary of possible evolutionary models including LGT for the Era-like GTPase subfamily genes. First, we found that three genes (chloroplast-targeting Era1, chloroplast/mitochondrion dual-targeting EngB1 and mitochondrion-targeting EngB2) were acquired from Bacteroides through LGT. In these cases, cyanobacterium-derived homologs were likely replaced by novel genes and have disappeared during evolution. In contrast to the situation for rAD, Bacteroides-related Era1, EngB1, and EngB2 genes were also found in red algae C. merolae, suggesting that the LGT event occurred before the divergence of the Glaucoplantae, Rhodoplantae, and Viridiplantae. Secondly, we found that LGT from green non-sulfur bacteria to plants provided a novel type of chloroplast-localized HflX in plants. This is the first evidence of LGT from non-oxygen producing photosynthetic eubacteria to plants. It remains unclear whether green non-sulfur bacterium-derived HflX confers any functional advantage in chloroplasts compared to the cyanobacterium-related gene. Taken together, our work demonstrates that LGT from eubacteria to plants occurred more frequently than previously thought. It is plausible that eubacterial genes provided novel functions in chloroplasts and that they played a crucial role in plant evolution.
Figure 9.
Schematic model of the evolution of eubacteria-type plant Era-like genes. Chloroplasts in land plants likely have acquired four GTPases from cyanobacteria by endosymbiotic gene transfer (EGT): ObgA2, TrmE, EngD1 (dual targeting), and EngA1. In addition, mitochondria have also acquired two GTPases from α-proteobacteria through EGT; ObgA1, and Era2. On the other hand, chloroplasts have acquired three α-proteobacterium-related GTPases, probably through re-compartmentation of mitochondrial proteins: EngD2, EngA2, and EngB3. Moreover, chloroplasts have acquired one GTPase from Green non-sulfur bacteria (HflX) and two GTPases from Bacteroides [Era1 and EngB-2 (dual-targeting)]. Similarly, mitochondria also have acquired one GTPase from Bacteroides (EngB2).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Y. Ishizaki for helpful discussion and C. Wada (Kyoto University) for providing anti-ObgE antibody. This study was supported by JSPS and MEXT Grants-in-Aid for Scientific Research to Takashi Shiina (24657036, 25291065, 25120723) and Kunio Takeyasu, COE Research from MEXT to Kunio Takeyasu, Grants of NEDO to Takashi Shiina, and Grants from the BK21 plus program of the Ministry of Education, Science and Technology of Korea (for Jeong Dong Bahk). I. Nengah Suwastika was a recipient of an International Student Fellowship from MEXT. This work was also partially supported by JSPS-DGHE international grant between Japan and Indonesia to I. Nengah Suwastika and Takashi Shiina, research Grand from DGHE-Indonesian Gov. (for I. Nengah Suwastika), and a grant from the Mitsubishi Foundation to Takashi Shiina.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2014.00678/abstract
Phylogenetic tree of Obg subfamily proteins. Comprehensive comparison of Obg subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 185 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of TrmE subfamily proteins. Comprehensive comparison of TrmE subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 152 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of EngD subfamily proteins. Comprehensive comparison of EngD subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 182 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of EngA subfamily proteins. Comprehensive comparison of EngA subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 147 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of HflX subfamily proteins. Comprehensive comparison of HflX subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 153 genes. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of Era subfamily proteins. Comprehensive comparison of Era subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 141 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of EngB subfamily proteins. Comprehensive comparison of EngB subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 143 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of Drg subfamily proteins. Comprehensive comparison of Drg subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 185 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of Nog subfamily proteins. Comprehensive comparison of Nog1 subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 185 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
References
- Abdallah F., Salamini F., Leister D. (2000). A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 5, 141–142. 10.1016/S1360-1385(00)01574-0 [DOI] [PubMed] [Google Scholar]
- Akiyama T., Gohda J., Shibata S., Nomura Y., Azuma S., Ohmori Y., et al. (2001). Mammalian homologue of E. coli ras-like GTPase (ERA) is a possible apoptosis regulator with RNA binding activity. Gene Cell 6, 987–1001. 10.1046/j.1365-2443.2001.00480.x [DOI] [PubMed] [Google Scholar]
- Andersson J. O. (2005). Lateral gene transfer in eukaryotes. Cell. Mol. Life Sci. 62, 1182–1197. 10.1007/s00018-005-4539-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald J. M., Rogers M. B., Toop M., Ishida K.-I., Keeling P. J. (2003). Lateral gene transfer and the evolution of plastid-targeted proteins in the secondary plastid-containing alga Bigelowiella natans. Proc. Natl. Acad. Sci. U.S.A. 100, 7678–7683. 10.1073/pnas.1230951100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigoni F., Talabot F., Peitsch M., Edgerton M. D., Meldrum E., Allet E., et al. (1998). A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16, 851–856. 10.1038/nbt0998-851 [DOI] [PubMed] [Google Scholar]
- Ball S. G., Subtil A., Bhattacharya D., Moustafa A., Weber A. P., Gehre L., et al. (2013). Metabolic effectors secreted by bacterial pathogens: essential facilitators of plastid endosymbiosis? Plant Cell 25, 7–21. 10.1105/tpc.112.101329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang W. Y., Chen J., Jeong I. S., Kim S. W., Kim C. W., Jung H. S., et al. (2012). Functional characterization of ObgC in ribosome biogenesis during chloroplast development. Plant J. 71, 122–134. 10.1111/j.1365-313X.2012.04976.x [DOI] [PubMed] [Google Scholar]
- Bang W. Y., Hata A., Im C. H., Ishizaki Y., Suwastika I. N., Jeong I. S., et al. (2009). Chloroplast ribosome-associated AtObgC is crucial for embryogenesis at the early stage. Plant Mol. Biol. 71, 379–390 10.1007/s11103-009-9529-3 [DOI] [PubMed] [Google Scholar]
- Barlow M. (2009). What antimicrobial resistance has taught us about horizontal gene transfer. Methods Mol. Biol. 532, 397–411. 10.1007/978-1-60327-853-9_23 [DOI] [PubMed] [Google Scholar]
- Baumgartner B. J., Rapp J. C., Mullet J. E. (1993). Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development. Plant Physiol. 101, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B., Hoef-Emden K., Melkonian M. (2008). Chlamydial genes shed light on the evolution of photoautotrophic eukaryotes. BMC Evol. Biol. 8:203. 10.1186/1471-2148-8-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharat A., Jiang M., Sullivan S. M., Maddock J. R., Brown E. D. (2006). Cooperative and critical roles for both G domains in the GTPase activity and cellular function of ribosome-associated Escherichia coli EngA. J. Bacteriol. 188, 7992–7996. 10.1128/JB.00959-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock R. (2010). The give-and-take of DNA: horizontal gene transfer in plants. Trends Plant Sci. 15, 11–22. 10.1016/j.tplants.2009.10.001 [DOI] [PubMed] [Google Scholar]
- Brinkman F. S. L., Blanchard J. L., Cherkasov A., Av-Gay Y., Brunham R. C., Fernandez R. C., et al. (2002). Evidence that plant-like genes in Chlamydia species reflect an ancestral relationship between Chlamydiaceae, cyanobacteria, and the chloroplast. Genome Res. 12, 1159–1167. 10.1101/gr.341802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton R. A., Powell B. S., Dasgupta S., Sun Q., Margolin W., Lupski J. R., et al. (1998). Cell cycle arrest in Era GTPase mutants: a potential growth rate-regulated checkpoint in Escherichia coli. Mol. Microbiol. 27, 739–750. 10.1046/j.1365-2958.1998.00719.x [DOI] [PubMed] [Google Scholar]
- Colby G., Wu M., Tzagoloff A. (1998). MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J. Biol. Chem. 273, 27945–27952. 10.1074/jbc.273.43.27945 [DOI] [PubMed] [Google Scholar]
- Dassain M., Leroy A., Colosetti L., Carolé S., Bouché J. P. (1999). A new essential gene of the ‘minimal genome’ affecting cell division. Biochimie 81, 889–895. 10.1016/S0300-9084(99)00207-2 [DOI] [PubMed] [Google Scholar]
- Datta K., Fuentes J. L., Maddock J. R. (2005). The Yeast GTPase Mtg2p is required for mitochondrial translation and partially suppresses an rRNA methyltransferase mutant, mm2. Mol. Biol. Cell 16, 954–963. 10.1091/mbc.E04-07-0622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta K., Skidmore J. M., Pu K., Maddock J. R. (2004). The Caulobacter crescentus GTPase CgtAC is required for progression through the cell cycle and for maintaining 50S ribosomal sublevels. Mol. Microbiol. 54, 1379–1392. 10.1111/j.1365-2958.2004.04354.x [DOI] [PubMed] [Google Scholar]
- Decoster E., Vassal A., Faye G. (1993). MSS1, a nuclear-encoded mitochondrial GTPase involved in the expression of COX1 subunit of cytochrome c oxidase. J. Mol. Biol. 232, 79–88. 10.1006/jmbi.1993.1371 [DOI] [PubMed] [Google Scholar]
- Deusch E. W., Lam H., Aebersold R. (2008). PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434. 10.1038/embor.2008.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grauvogela C., Petersen J. (2007). Isoprenoid biosynthesis authenticates the classification of the green alga Mesostigma viride as an ancient streptophyte. Gene 396, 125–133. 10.1016/j.gene.2007.02.020 [DOI] [PubMed] [Google Scholar]
- Hang J. Q., Zhao G. (2003). Characterization of the 16S rRNA- and membrane-binding domains of Streptococcus pneumoniae Era GTPase structural and functional implications. Eur. J. Biochem. 270, 4164–4172. 10.1046/j.1432-1033.2003.03813.x [DOI] [PubMed] [Google Scholar]
- Hirano Y., Ohniwa R. L., Wada C., Yoshimura S., Takeyasu K. (2006). Human small G proteins, Obg H1 and Obg H2 participate in the maintenance of mitochondria and nucleolar architectures. Genes Cells 11, 1295–1304. 10.1111/j.1365-2443.2006.01017.x [DOI] [PubMed] [Google Scholar]
- Huang J., Gogarten J. P. (2007). Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 8:R99. 10.1186/gb-2007-8-6-r99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanmougin F., Thompson J. D., Gouy M., Higgins D. G., Gibson T. J. (1998). Multiple sequence alignment with CLUSTAL X. Trends Biochem. Sci. 23, 403–405. 10.1016/S0968-0004(98)01285-7 [DOI] [PubMed] [Google Scholar]
- Jensen B. C., Wang Q., Kifer C. T., Parsons M. (2003). The NOG1 GTP-binding protein is required for biogenesis of the 60 s ribosomal subunit. J. Biol. Chem. 278, 32204–32211. 10.1074/jbc.M304198200 [DOI] [PubMed] [Google Scholar]
- Kobayashi G., Moriya S., Wada C. (2001). Deficiency of essential GTP binding protein ObgE in Escherichia coli inhibits chromosome partition. Mol. Microbiol. 41, 1037–1051. 10.1046/j.1365-2958.2001.02574.x [DOI] [PubMed] [Google Scholar]
- Leipe D. D., Wolf Y. I., Koonin E. V., Aravind L. (2002). Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 317, 41–72. 10.1006/jmbi.2001.5378 [DOI] [PubMed] [Google Scholar]
- Li B., Trueb B. (2000). DRG represents a family of two closely related GTP-binding proteins. Biochim. Biophys. Acta 149, 196–204. 10.1016/S0167-4781(00)00025-7 [DOI] [PubMed] [Google Scholar]
- Martin W., Rujan T., Richly E., Hansen A., Cornelsen S., Lins T., et al. (2002). Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 99, 12246–12251. 10.1073/pnas.182432999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Schnarrenberger C. (1997). The evolution of the Calvin cycle from prokaryotic to eukaryotic chromosomes: a case study of functional redundancy in ancient pathways through endosymbiosis. Curr. Gen. 32, 1–18. 10.1007/s002940050241 [DOI] [PubMed] [Google Scholar]
- McFaddan G. I. (2001). Primary and secondary endosymbiosis and the origin of plastids. J. Phycol. 37, 951–959 10.1046/j.1529-8817.2001.01126.x [DOI] [Google Scholar]
- Mittenhuber G. (2001). Comparative genomics of prokaryotic GTP-binding proteins (the Era, Obg, EngA, ThdF (TrmE), YchF and YihA families) and their relationship to eukaryotic GTP-binding proteins (the DRG, ARF, RAB, RAN, RAS and RHO families). J. Mol. Microbiol. Biotechnol. 3, 21–35. [PubMed] [Google Scholar]
- Morimoto T., Loh P. C., Hirai T., Asai K., Kobayashi K., Moriya S., et al. (2002). Six GTP binding proteins of the Era/Obg family are essential for cell growth in Bacillus subtilis. Microbiology 148, 3539–3552. [DOI] [PubMed] [Google Scholar]
- Moustafa A., Reyes-Prieto A., Bhattacharya D. (2008). Chlamydiae has contributed at least 55 genes to plantae with predominantly plastid functions. PLoS ONE 3:e2205. 10.1371/journal.pone.0002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosenko T., Lidie K. L., Van Dolah F. M., Lindquist E., Cheng J. F., Bhattacharya D. (2006). Chimeric plastid proteome in the Florida “red tide” dinoflagellate Karenia brevis. Mol. Biol. Evol. 23, 2026–2038. 10.1093/molbev/msl074 [DOI] [PubMed] [Google Scholar]
- Page R. D. (1996). TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Park A. K., Kim H., Jin H. J. (2009). Comprehensive phylogenetic analysis of evolutionarily conserved rRNA adenine dimethyltransferase suggests diverse bacterial contributions to the nucleus-encoded plastid proteome. Mol. Phylogenet. Evol. 50, 282–289. 10.1016/j.ympev.2008.10.020 [DOI] [PubMed] [Google Scholar]
- Park J. H., Jensen B. C., Kifer C. T., Parsons M. (2001). A novel nucleolar G-protein conserved in eukaryotes. J. Cell Sci. 114, 173–185. [DOI] [PubMed] [Google Scholar]
- Polkinghorne A., Ziegler U., González-Hernández Y., Pospischil A., Timms P., Vaughan L. (2008). Chlamydophila pneumoniae HflX belongs to an uncharacterized family of conserved GTPases and associates with the Escherichia coli 50S large ribosomal subunit. Microbiology 154, 3537–3546. 10.1099/mic.0.2008/022137-0 [DOI] [PubMed] [Google Scholar]
- Qiu H., Price D. C., Weber A. P., Facchinelli F., Yoon H. S., Bhattacharya D. (2013). Assessing the bacterial contribution to the plastid proteome. Trends Plant Sci. 18, 680–687. 10.1016/j.tplants.2013.09.007 [DOI] [PubMed] [Google Scholar]
- Ratief J. D. (2000). Phylogenetic analysis using PHYLIP. Methods Mol. Biol. 132, 243–258. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A., Bhattacharya D. (2007). Phylogeny of Calvin cycle enzymes supports Plantae monophyly. Mol. Phylogenet. Evol. 45, 384–391. 10.1016/j.ympev.2007.02.026 [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A., Moustafa A. (2012). Plastid-localized amino acid biosynthetic pathways of Plantae are predominantly composed of non-cyanobacterial enzymes. Sci. Rep. 2, 1–12. 10.1038/srep00955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards T. A., Dacks J. B., Campbell S. A., Blanchard J. L., Foster P. G., McLeod R., et al. (2006). Evolutionary origins of the eukaryotic shikimate pathway: gene fusions, horizontal gene transfer, and endosymbiotic replacements. Eukaryot. Cell 5, 1517–1531. 10.1128/EC.00106-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. [DOI] [PubMed] [Google Scholar]
- Sato A., Kobayashi G., Hayashi H., Yoshida H., Wada A., Maeda M., et al. (2005). The GTP binding protein homolog ObgE is involved in ribosomal maturation. Genes Cells 10, 393–408. 10.1111/j.1365-2443.2005.00851.x [DOI] [PubMed] [Google Scholar]
- Sayed A., Matsuyama S., Inouye M. (1999). Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem. Biophys. Res. Commun. 264, 51–54. 10.1006/bbrc.1999.1471 [DOI] [PubMed] [Google Scholar]
- Schaefer L., Uicker W. C., Wicker-Planquart C., Foucher A.-E., Jault J.-M., Britton R. A. (2006). Multiple GTPases participate in the assembly of the large ribosomal subunit in Bacillus subtilis. J. Bacteriol. 188, 8252–8258. 10.1128/JB.01213-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slominska M., Konopa G., Wegrzyn G., Czyz A. (2002). Impaired chromosome partitioning and chynchronization of DNA replication initiation in an insertional mutant in the Vibrio harveyi cgtA gene coding for common GTP-binding protein. Biochem. J. 362, 579–584. 10.1042/0264-6021:3620579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwastika I. N., Ohniwa R. L., Takeyasu K., Shiina T. (2014). Plant Drg Proteins are Cytoplasmic Small GTPase-ObgHomologue. Proc. Environ. Sci. 20, 357–364 10.1016/j.proenv.2014.03.045 [DOI] [Google Scholar]
- Sweetlove L. J., Taylor N. L., Leaver C. J. (2007). Isolation of intact, functional mitochondria from the model plant Arabidopsis thaliana. Methods Mol. Biol. 372, 125–136. 10.1007/978-1-59745-365-3_9 [DOI] [PubMed] [Google Scholar]
- Teicha R., Zaunerb S., Baurainc D., Brinkmannc H., Petersena J. (2007). Origin and distribution of Calvin cycle fructose and sedoheptulose bisphosphatases in plantae and complex algae: a single secondary origin of complex red plastids and subsequent propagation via tertiary endosymbioses. Protist 158, 263–276. 10.1016/j.protis.2006.12.004 [DOI] [PubMed] [Google Scholar]
- Tomar S. K., Kumar P., Prakash B. (2011). Deciphering the catalytic machinery in a universally conserved ribosome binding ATPase YchF. Biochem. Biophys. Res. Commun. 408, 459–464. 10.1016/j.bbrc.2011.04.052 [DOI] [PubMed] [Google Scholar]
- Uemura T., Ueda T., Ohniwa R. L., Nakano A., Takeyasu K., Sato M. H. (2004). Systemic analysis of SNARE molecules in Arabidopsis: dissection of the post-golgi network in plant cells. Cell Struct. Funct. 29, 49–65. 10.1247/csf.29.49 [DOI] [PubMed] [Google Scholar]
- Verstraeten N., Fauvart M., Versées W., Michiels J. (2011). The universally conserved prokaryotic GTPases. Microbiol. Mol. Biol. Rev. 75, 507–542. 10.1128/MMBR.00009-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller R. F., Slamovits C. H., Keeling P. J. (2006). Lateral gene transfer of a multigene region from cyanobacteria to dinoflagellatesresulting in a novel plastid-targeted fusion protein. Mol. Biol. Evol. 23, 1437–1443. 10.1093/molbev/msl008 [DOI] [PubMed] [Google Scholar]
- Yagi Y., Shiina T. (2011). Evolutionary aspects of plastid proteins involved in transcription the transcription of a tiny genome is mediated by a complicated machinery. Transcription 3, 290–294. 10.4161/trns.21810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S., Yoo S. D., Sheen J. (2003). Deferential regulation of EIN3 stability by glucose and ethylene signaling in plants. Nature 425, 521–525. 10.1038/nature01984 [DOI] [PubMed] [Google Scholar]
- Yue J., Hu X., Sun H., Yang Y., Huang J. (2012). Widespread impact of horizontal gene transfer on plant colonization of land. Nat. Commun. 3:1152. 10.1038/ncomms2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of Obg subfamily proteins. Comprehensive comparison of Obg subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 185 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of TrmE subfamily proteins. Comprehensive comparison of TrmE subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 152 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of EngD subfamily proteins. Comprehensive comparison of EngD subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 182 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of EngA subfamily proteins. Comprehensive comparison of EngA subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 147 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of HflX subfamily proteins. Comprehensive comparison of HflX subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 153 genes. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of Era subfamily proteins. Comprehensive comparison of Era subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 141 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of EngB subfamily proteins. Comprehensive comparison of EngB subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 143 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of Drg subfamily proteins. Comprehensive comparison of Drg subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 185 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.
Phylogenetic tree of Nog subfamily proteins. Comprehensive comparison of Nog1 subfamily proteins in eukaryotes, eubacteria and archaea. Sequences were aligned using Clustal X based on 185 proteins. The tree was inferred using the neighbor-joining method with JTT model. Numbers at the nodes indicate bootstrap values obtained for 100 replicates.