Abstract
Background
Species belonging to the genus Aegilops L. are an important source of genetic material for expanding genetic variability of wheat. Ae. triuncialis is an allotetraploid in this genus which was originated from hybridization of Ae. umbellulata and Ae. markgrafii (Greuter) Hammer. Although the Ae. triuncialis karyotype was thoroughly examined by conventional chromosome staining and Giemsa C-banding, it is still poorly characterized using FISH markers. The objective of this study was to test the fluorescence in situ hybridization (FISH) patterns of Ae. triuncialis (2n = 4x = 28, CtCtUtUt) chromosomes using different repetitive sequences and to compare the produced patterns to the chromosomes of its diploid ancestors, with the aim of establishing a generalized Ae. triuncialis idiogram and detection of Aegilops chromosomes in the background of wheat.
Results
The probes pSc119.2-1, pTa535-1, pAs1-1, (CTT)10 and the 45S rDNA clone from wheat (pTa71) were hybridized to chromosomes of Ae. triuncialis and compared with its diploid progenitors (Ae. umbellulata Zhuk., 2n = 2x = 14, UU and Ae. markgrafii (Greuter) Hammer, 2n = 2x = 14, CC) and Ae. cylindrica Host. (2n = 4x = 28, DcDcCcCc), another tetraploid species containing the C-genome. Ae. cylindrica was further analyzed by genomic in situ hybridization (GISH) using C genome probe in order to identify any possible translocation.
Conclusions
In general, FISH patterns of the Ut- and Ct-genome chromosomes of Ae. triuncialis were similar to those of U- and C-genome chromosomes of the diploid progenitor species Ae. umbellulata and Ae. markgrafii respectively, although some differences were observed. Two major 45S rDNA loci were revealed in the short arm of chromosomes A and C, of the Ct genome which correspond to homoeologous groups 1 and 5 respectively. Minor 45S rDNA loci were mapped on the short arm of chromosomes 1Ut and 5Ut. GISH analysis revealed three different non-reciprocal homologous or heterologous translocations between Cc and Dc chromosomes in all studied accessions of Ae. cylindrica.
Electronic supplementary material
The online version of this article (doi:10.1186/s13039-014-0091-6) contains supplementary material, which is available to authorized users.
Keywords: Aegilops triuncialis, Evolution, Wheat–Aegilops triuncialis hybrid, Chromosome marker, Allopolyploidization
Background
Species belonging to the genus Aegilops L. are an important source of genetic material for expanding genetic variability of cultivated bread wheat, Triticum aestivum L. em Thell. (2n = 6x = 42, BBAADD) [1]. The genus Aegilops comprises 11 diploid and 12 allopolyploid species [2] with different types of nuclear and cytoplasmic genomes [3]. Ae. triuncialis is included in the section Aegilops together with diploid Ae. umbellulata and several polyploid species sharing the U-genome [2]. Ae. triuncialis is subdivided into two subspecies, : ssp. triuncialis and ssp. persica, which carry the same type of nuclear genome, but different cytoplasmic genomes. Ae. triuncialis ssp. persica was originated from hybridization of Ae. umbellulata as female parent with Ae. markgrafii (Greuter) Hammer (syn. Ae. caudata L.), whereas ssp. triuncialis arose from a reciprocal cross [4,5].
Many accessions of Ae. triuncialis are tolerant to biotic and abiotic stresses. It has been exploited for a wide range of traits including resistance to pests and diseases [6-14] and may harbor many other, yet unidentified traits for wheat improvement.
Giemsa C-banding technique has been used to characterize the genomes and chromosomes of wheat and Aegilops species [15-17]. In particular C-banding has been employed to examine genetic diversity and to construct the karyotypes of Ae. umbellulata [18], Ae. markgrafii [19,20], Ae. triuncialis [16] and Ae. cylindrica [15,21]. Fluorescence in situ hybridization (FISH) using repetitive sequences as probes is an alternative powerful technique for chromosome characterization.
Repetitive DNA sequences are major components of the plant genome; in some species they can account for up to 90% of the genome size [22]. Dissimilarity of repetitive DNAs may reflect evolutionary distances between species and these repetitive DNA sequences account for the major differences between genomes [23,24]. The chromosomal localization of various repetitive DNA sequences, including single sequence repeats (SSRs) such as (ACG)n and (GAA)n, satellite sequences (pSc119.2, Afa family) and ribosomal genes have been used to identify the chromosomes of wheat and its wild relatives [25-28]. Recently, some new tandemly repeated sequences, such as pTa-535, pTa-713, and pTa-86, were also isolated and tested as FISH probes to identify wheat chromosomes [29]. The hybridization patterns of pSc119.2, Afa family and rDNA probes were described previously for diploid and polyploid Aegilops species [16,30,31]. Some species, like Ae. umbellulata [32], Ae. biuncialis [33], Ae. cylindrica [21] were studied in more detail. These studies showed that combinations of pSc119.2 and Afa probes in most species do not permit the complete identification of all chromosomes, because pSc119.2 probe hybridized mainly to subtelomeric chromosome regions, while the Afa family produces just few signals on chromosomes of the S-genome group, T-, U- and C-genomic species. To solve this problem some authors [26,34,35] suggested to use two or three base-pair synthetic oligo probes as diagnostic markers. These studies demonstrated that the GAA microsatellite is valuable to identify chromosomes of the wheat A- and B-genomes. The labeling patterns generated with this sequence in general corresponded to the Giemsa N-banding patterns of the respective chromosomes thus allowing linking the FISH and Giemsa N- or C-banding analyses of wheat. The GAA repeat was further used to characterize the chromosomes of some Aegilops species including Ae. biuncialis, Ae. comosa and Ae. umbellulata [33].
The Ae. triuncialis karyotype was thoroughly examined by conventional chromosome staining [36] and C-banding [16], however it is still poorly characterized using FISH markers. Although Badaeva et al. described the labeling patterns of pSc119.2, pTa71, pTa794 DNA probes on Ae. triuncialis chromosomes, their correlation with a pattern of Giemsa staining was established only on the basis of chromosome morphology [16]. Morphological similarity of many Ae. triuncialis chromosomes however can impede their correct classification. Hence the main objective of this study was to analyze the FISH patterns of Ae. triuncialis chromosomes using different repetitive sequences and to compare the produced patterns to the chromosomes of its diploid ancestors, with the aim of establishing a generalized Ae. triuncialis idiogram for facilitating the accurate chromosome identification, tracing possible chromosome changes over the course of evolution and detection of Ae. triuncialis chromatin introgressed into wheat. Another tetraploid Aegilops species carrying the C-genome – Ae. cylindrica – was taken for the comparison of the C-genome chromosomes in a different genetic background.
Results and discussion
To develop an informative combination of FISH markers that allows precise identification of all Ae. triuncialis chromosomes, the following combinations of probes were tested: pSc119.2-1 + pTa535-1, pSc119.2-1 + pAs1-1, pSc119.2-1 + (CTT)10 and pTa71.
Examination of two accessions each of diploid Ae. umbellulata and Ae. markgrafii showed only few within-species hybridization pattern polymorphisms, though some minor differences were observed between Ae. markgrafii accessions. All Ae. markgrafii, Ae. umbellulata and Ae. triuncialis accessions analyzed showed similar hybridization patterns with the probe pSc119.2-1, which hybridized predominantly to the subtelomeric regions of one or both arms of all chromosomes. Additional interstitial signals were observed on the long arm of chromosomes 6 and 7 of the U-genome and chromosomes F and G of the C-genome (Figure 1). Thus, these chromosomes can be distinguished using pSc119.2-1 alone. However, the labeling pattern of Ae. triuncialis chromosomes should be treated with care because few minor changes in the position and signal intensity have been recorded for some chromosomes. The most distinct changes were observed for chromosome G, which lost a marker pSc119.2 site in the middle of the long arm, but acquired an increased site in the telomeric region of the same arm (Figure 1).
Figure 1.
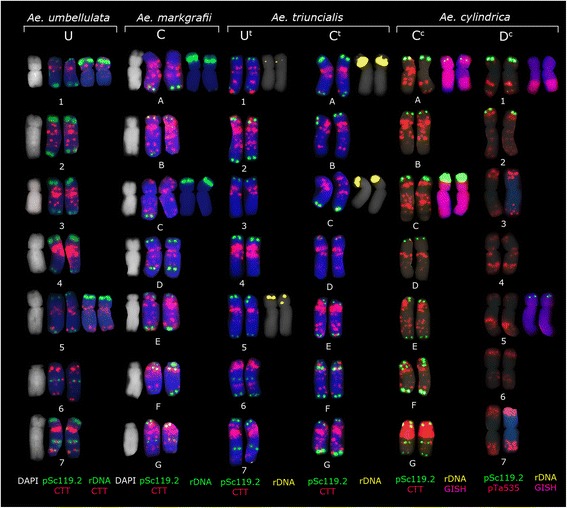
Representative karyotypes of Ae. umbellulata (accession S234), Ae. markgrafii (accession AE1418), Ae. triuncialis (accession S197) and Ae. cylindrica after FISH with different repetitive DNA probes. The yellow signals on Ae. triuncialis chromosomes represent 45S rDNA with the sizes relevant to the intensity of produced signals. The probe combination on each genome chromosomes is presented at the bottom of the figure with the related colour.
Our result confirms previous observations that probe pAs1 is not informative for the identification of U- and C-genome chromosomes [16,30,33], because it generates only few weak signals on only some chromosomes of these genomes. In situ hybridization using the (CTT)10 repeat allowed the identification of all seven chromosomes of Ae. markgrafii and Ae. umbellulata (Figure 1). (CTT)10 hybridization patterns on Ae. triuncialis chromosomes were comparable to those on the ancestral species, however some minor differences were observed in signal distribution and intensity (Figure 1). In particular, we revealed a significantly reduced number of hybridization sites in the long arm of chromosome A, which possessed profound signals in pericentromeric regions of short and long arms and two weak telomeric sites. Another NOR-bearing chromosome – C, carried large signals in proximal region of both arms and a small site in subtelomeric region of the long arm, whereas in the parental species we observed numerous, relatively weak interstitial signals of the same probe distribute over the length of the long arm. Labeling of pericentromeric region of chromosome E was much stronger in Ae. triuncialis than in Ae. caudata karyotype. By contrast, pericentromeric signals on chromosome F were smaller in allotetraploid species as compared to the diploid progenitor.
The (CTT)10 hybridization pattern of Ae. markgrafii and Ae. umbellulata chromosomes was similar with previously reported pattern on Ae. umbellulata chromosomes [33] and basically coincided with the position of previously reported Giemsa C-bands [18,19]. Similarity of the hybridization patterns on Ae. triuncialis chromosomes compared to Ae. markgrafii and Ae. umbellulata indicates that the C- and U-genomes have not underwent the significant structural changes relative to the parental species.
FISH analysis using labeled 45S rDNA showed four pairs of Ae. triuncialis chromosomes with rDNA signals of different size (Figure 1). A suppression of nucleolar organizing regions of the C-genome chromosomes in Ae. triuncialis was suggested earlier by [37] based on Ag-NOR staining and then by [16] based on FISH with two rRNA gene families. Our results also confirm suppression of 45S rRNA gene loci on one of the parental genome of Ae. triuncialis, however, with the use of genomic in situ hybridization (GISH) followed by FISH with pTa71 probe we showed that two major loci are located in the short arm of the C-genome chromosomes A and C, but not on the chromosomes A (1Ct) and 5Ut, as was suggested in [16]. The discrepancies in classification of NOR-bearing chromosome 5 can be due to the fact that in a previous paper the genome affinity of chromosomes was determined by their morphology and therefore should be considered as tentative. Two minor NOR clusters are located in the short arms of chromosomes 1Ut and 5Ut. The signal size decreased in the order A(1Ct) > C(5Ct) > 5Ut > 1Ut (Figure 1). Differences in signal size on chromosomes 1Ut and 5Ut observed in our study compared to the previous one can be due to intraspecific polymorphism as we used different accessions of Ae. triuncialis.
Each of the diploid species Ae. markgrafii and Ae. umbellulata carries two pairs of NOR-bearing chromosomes that were identified as chromosomes A = 1C and C = 5C in Ae. markgrafii and 1U and 5U in Ae. umbellulata, respectively. FISH analysis suggests that in Ae. triuncialis elimination of most of the rRNA gene copies occurred from the Ut genome of Ae. umbellulata (Figure 1). A similar loss of 45S rRNA genes occurred in the A/At genome of Triticum turgidum and T. timopheevii [38] after allopolyploidization.
A mitotic metaphase cell of the F1 hybrid T. aestivum cv ‘Zarin’-Ae. triuncialis (n = 5x = 35, genomically BADUtCt) after FISH using pSc119.2-1 and pTa535-1 as probe combination is shown in Figure 2E. All A-, B- and D-genome chromosomes of wheat can be identified by their characteristic repetitive sequences patterns, arm ratios and chromosome sizes. A 5B:7B translocation was identified in the background ‘Zarin’ (Figure 2E). All Ut- and Ct-genome chromosomes of Ae. triuncialis can be identified, however a combination of pSc119.2-1 and (CTT)10 probes proves to be most useful for the identification of all individual Ae. triuncialis chromosomes in wheat background (Figure 2F) because pTa535-1 probe does produce only few signals on the Ct or Ut chromosomes.
Figure 2.
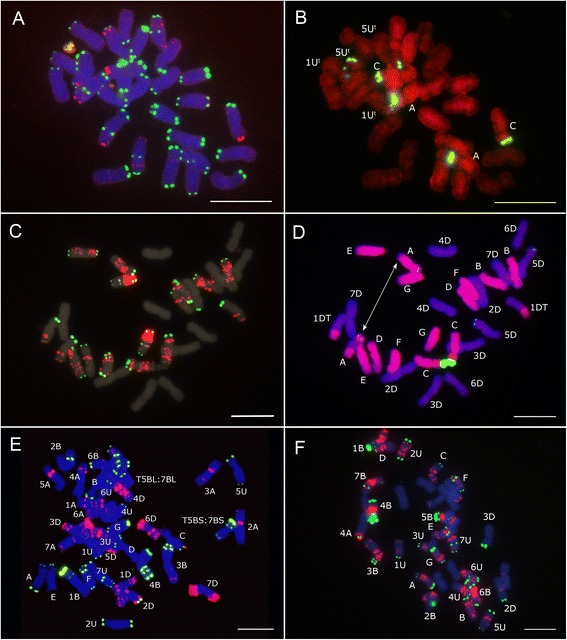
FISH and GISH patterns of Ae. triuncialis , Ae. cylindrica and F 1 hybrids derived from wheat and Ae. triuncialis crosses. A: FISH signals from oligonucleotide probes pAS1-1 (FAM 5′ labeled), pSc119.2-1 (Tamra labelled) on the Ae. triuncialis (accession S197); B: Re-probing on the same preparation using Ae. markgrafii genomic DNA (red signals) and pTa71 clone (green signals); C: FISH signals from oligonucleotide probes pSc119.2-1 (FAM 5′ labeled), pTa535-1 (Tamra labelled) and (CTT)10 (Cy3 labelled) on the Ae. cylindrica ecotype. Just signals of (CTT)10 (mainly on Cc chromosomes) and pSc119.2-1 are presented in this picture. Using Texas Red filter, pTa535-1 signal was observed and identified Dc chromosomes which are presented in Figure 1; D: Re-probing on the same preparation using Ae. markgrafii genomic DNA (red signals) and pTa71 clone (green signals) which shows unbalanced translocation on chromosome pair 1D and 3C. The translocation on 3C is heterologous; E: FISH pattern of repetitive oligonucleotide probes pSc119.2-1 (FAM 5′ labelled), pTa535-1 (Tamra labelled) on mitotic metaphase chromosomes of a derived F1 hybrid T. aestivum cv ‘Zarin’- Ae. triuncialis (accession S101); F: FISH pattern of repetitive oligonucleotide probes pSc119.2-1 (FAM 5′ labelled) and (CTT)10 (Cy3 labelled) on mitotic metaphase chromosomes of a derived F1 hybrid T. aestivum cv ‘Pishgam’- Ae. triuncialis (accession S101). Bar = 10 μm.
So far, the homoeologous relationship of only three chromosomes of Ae. markgrafii has been established. Analysis of the chromosome substitution lines A (1C), C (5C) [19] and B (2C) [39] indicated that the chromosomes A, B and C should be assigned to homoeologous groups 1, 2 and 5 of wheat, respectively. The Ae. markgrafii chromosomes - D, E, F, and G showed homoeology to more than one group, based on isozyme [40] and restriction fragment length polymorphisms (RFLP) [20,41] analyses. Structural rearrangements of the U-genome chromosomes of Ae. umbellulata have been earlier deduced based on comparative chromosome mapping [42]. That is why both Ae. markgrafii and Ae. umbellulata have highly asymmetrical karyotypes, compared to the more symmetrical karyotypes of the other diploid species of this genus [36].
The C-genome is also present in another tetraploid Aegilops species with a different genome constitution, Ae. cylindrica (DcDcCcCc) [43]. Ae. triuncialis and Ae. cylindrica is suggested to evolve rather recently with only few modifications of the parental genomes [44]. To find possible changes of the C-genome due to polyploidization, we included Ae. cylindrica into the analysis of the C-genome evolution. The pattern produced by the (CTT)10 repeat on Cc-genome chromosomes of Ae. cylindrica was more similar to that in the ancestral species Ae. markgrafii than the Ct-genome chromosomes of Ae. triuncialis. However, three different non-reciprocal homologous or heterologous translocations between Cc and Dc- genome chromosomes where observed in all the studied accessions of this species. Thus, Ae. cylindrica accession S376 carried an intergenomic translocation (Additional file 1: Figure S1) and the Ae. cylindrica ecotype consisted of genotypes with two different translocations (Figure 2D and Additional file 1: Figure S1). Reciprocal translocations between Cc and Dc chromosomes have been reported in this species previously [21]. Sequential FISH and GISH showed that the breakpoints were mainly located near the (CTT)10 hybridization sites. Similar relationship between intergenomic translocation breakpoints and SSR-rich chromosomal regions in the allopolyploid species has been reported by other authors [33] which suggests that SSR DNA sequences might facilitate the formation of chromosomal rearrangements. Frequent incidence of reciprocal and non-reciprocal translocations can be an indicative of an extensive speciation process in this relatively new allopolyploid species. A more recent origin of Ae. cylindrica than Ae. triuncialis can also be presumed based on the higher similarity of the Cc-genome chromosomes with the C-genome chromosomes of diploid Ae. markgrafii as compared to the Ct-genome of Ae. triuncialis. We could hardly discriminate the Ct and Ut genome chromosomes of Ae. triuncialis (accession S197) by GISH using Ae. markgrafii as probe (Figure 2A and B). This can be due to very close evolutionary relationships of these genomes which share many repetitive sequences [44] thus precluding their discrimination. No intergenomic translocations was detected in the accessions of Ae. triuncialis used.
Conclusion
In conclusion, FISH patterns of the Ut- and Ct-genome chromosomes of Ae. triuncialis using different repetitive were similar to those of U- and C-genome chromosomes of the diploid progenitor species Ae. umbellulata and Ae. markgrafii respectively, although some differences were observed. In situ hybridization using the (CTT)10 repeat allowed the identification of all chromosomes of Ae. triuncialis and its diploid ancestors Ae. markgrafii and Ae. umbellulata. Ae. triuncialis chromosomes could also be identified in the background of bread wheat. GISH analysis revealed different non-reciprocal homologous or heterologous translocations between Cc and Dc chromosomes in the studied accessions of Ae. cylindrica.
Methods
Plant materials
Two accessions of Ae. markgrafii (AE1418, AE1082), two accessions of Ae. umbellulata (S147, S234), three accessions of Ae. triuncialis (S101, S146, S197), and one accession (S376) and one ecotype (collected in the Kurdistan province, Iran) of Ae. cylindrica, were examined. The Ae. markgrafii accessions were obtained from the germplasm collections of the IPK, Germany and other accessions are maintained in the Research Institute of Forests and Rangelands (RIFR) of Iran. T. aestivum cultivars ‘Zarin’ and ‘Pishgam’ were crossed with Ae. triuncialis (accession S101) and the resulted F1 seeds were used for identification of Ae. triuncialis chromosomes in the background of wheat.
DNA probes
The 5′ with 6-carboxyfluorescein (6-FAM) or 6-carboxytetramethylrhodamine (Tamra) end-labelled oligo probes oligo-pAs1-1 (Tamra-5′-CCT TTC TGA CTT CAT TTG TTA TTT TTC ATG CAT TTA CTA ATT ATT TTG AGC TAT AAG AC-3′), oligo-pSc119.2-1 (6-FAM-5′-CCG TTT TGT GGA CTA TTA CTC ACC GCT TTG GGG TCC CAT AGC TAT-3′) and oligo-pTa535-1 (Tamra-5′-AAA AAC TTG ACG CAC GTC ACG TAC AAA TTG GAC AAA CTC TTT CGG AGT ATC AGG GTT TC-3′) [29,45] were synthesized by MWG (Germany). The oligo (CTT)10 probe was directly labelled with 5/6 sulforhodamine 101PEG3_azide by click chemistry (Baseclick). The plasmid pTA71 containing the 45S rDNA repeat from wheat was directly labelled by nick translation and used to detect the NORs.
FISH and GISH
Root tips were pretreated with ice cold water for 24 h and were then fixed in ethanol-glacial acetic acid (3:1). Mitotic chromosome spreads were prepared using dropping technique. FISH was carried out according to [33]. After documentation of the FISH signals, some of the preparations were rehybridized to detect the 45S rDNA sites or the parental genomes by genomic in situ hybridization (GISH).
GISH was carried out after FISH on the same preparations. Therefore, total Ae. markgrafii genomic DNA was labelled with Atto-550 11-dUTP by nick translation, and used as a probe to detect the C-genome chromosomes of Ae. triuncialis and Ae. cylindrica. Unlabelled, fragmented wheat DNA was used as blocking DNA at 60 times the quantity.
Acknowledgments
We wish to thank Katrin Kumke for the excellent technical support and advices in ISH.
Funding
This work was supported by a fellowship within the 3 month Program of the German Academic Exchange Service (DAAD) and the DFG grant HO 1779/20-1.
Additional file
A GISH using labelled Ae. markgrafii genomic DNA on a mitotic metaphase cell of the Ae. cylindrica ecotype showing unbalanced intergenomic translocations on two pairs of chromosomes. The red labelling is the GISH with the C genome, and the green is the rDNA loci. This ecotype consisted of genotypes with two different translocations. The other one is shown in Figure 2D; B: Ae. cylindrica accession S376 showed an unbalanced intergenomic translocation on one pair of chromosomes. Bar = 10 μm.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GM: Conception and design of the study; providing of genetic material; acquisition of data; analysis and interpretation of data; manuscript writing. AH: Supervision; Provision of study material; analysis and interpretation of data; critical discussion and manuscript corrections. EDB: Supervision; Analysis and interpretation of data; Manuscript corrections. All authors read and approved the final manuscript.
Contributor Information
Ghader Mirzaghaderi, Email: gh.mirzaghaderi@uok.ac.ir.
Andreas Houben, Email: houben@ipk-gatersleben.de.
Ekaterina D Badaeva, Email: katerinabadaeva@gmail.com.
References
- 1.Schneider A, Molnár I, Molnár-Láng M. Utilisation of Aegilops (goatgrass) species to widen the genetic diversity of cultivated wheat. Euphytica. 2008;163:1–19. doi: 10.1007/s10681-007-9624-y. [DOI] [Google Scholar]
- 2.Van Slageren MW. Wild Wheats: a monograph of Aegilops L. and Amblyopyrum (Jaub. et Spach) Eig (Poaceae) Wageningen Agricultural University, Wageningen and ICARDA, Aleppo, Syria: Wageningen; 1994. [Google Scholar]
- 3.Kimber G, Tsunewaki K. Genome symbols and plasma types in the wheat group. In: Miller TE, Koebner RMD, editors. Proceedings of the 7th International Wheat Genetics Symposium, 13–19 July 1988. Cambridge, England: Institute of Plant Science Research, Cambridge Laboratory, Trumpington, England; 1996. pp. 1209–1210. [Google Scholar]
- 4.Murai K, Tsunewaki K. Molecular basis of genetic diversity among cytoplasms of Triticum and Aegilops species. IV. CtDNA variation in Ae. triuncialis. Heredity. 1986;57:335–339. doi: 10.1038/hdy.1986.132. [DOI] [Google Scholar]
- 5.Vanichanon A, Blake N, Sherman J, Talbert L. Multiple origins of allopolyploid Aegilops triuncialis. Theor Appl Genet. 2003;106:804–810. doi: 10.1007/s00122-002-1092-0. [DOI] [PubMed] [Google Scholar]
- 6.Aghaee-Sarbarzeh M, Ferrahi M, Singh S, Singh H, Friebe B, Gill B, Dhaliwal H. PhI-induced transfer of leaf and stripe rust-resistance genes from Aegilops triuncialis and Ae. geniculata to bread wheat. Euphytica. 2002;127:377–382. doi: 10.1023/A:1020334821122. [DOI] [Google Scholar]
- 7.Baalbaki R, Hajj-Hassan N, Zurayk R. Aegilops species from semiarid areas of Lebanon: Variation in quantitative attributes under water stress. Crop Sci. 2006;46:799–806. doi: 10.2135/cropsci2005.0120. [DOI] [Google Scholar]
- 8.El Bouhssini M, Benlhabib O, Nachit M, Houari A, Bentika A, Nsarellah N, Lhaloui S. Identification in Aegilops species of resistant sources to Hessian fly (Diptera: Cecidomyiidae) in Morocco. Genet Resour Crop Evol. 1998;45:343–345. doi: 10.1023/A:1008675029389. [DOI] [Google Scholar]
- 9.Ghazvini H, Hiebert CW, Zegeye T, Fetch T. Inheritance of stem rust resistance derived from Aegilops triuncialis in wheat line Tr129. Can J Plant Sci. 2012;92:1037–1041. doi: 10.4141/cjps2011-248. [DOI] [Google Scholar]
- 10.Makkouk KM, Comeau A, Ghulam W. Resistance to barley yellow dwarf luteovirus in Aegilops species. Can J Plant Sci. 1994;74:631–634. doi: 10.4141/cjps94-113. [DOI] [Google Scholar]
- 11.Martin-Sanchez J, Gomez-Colmenarejo M, Del Moral J, Sin E, Montes M, Gonzalez-Belinchon C, Lopez-Brana I, Delibes A. A new Hessian fly resistance gene (H30) transferred from the wild grass Aegilops triuncialis to hexaploid wheat. Theor Appl Genet. 2003;106:1248–1255. doi: 10.1007/s00122-002-1182-z. [DOI] [PubMed] [Google Scholar]
- 12.Romero MD, Montes MJ, Sin E, Lopez-Braña I, Duce A, Martín-Sanchez JA, Andrés MF, Delibes A. A cereal cyst nematode (Heterodera avenae Woll.) resistance gene transferred from Aegilops triuncialis to hexaploid wheat. Theor Appl Genet. 1998;96:1135–1140. doi: 10.1007/s001220050849. [DOI] [Google Scholar]
- 13.Singh H, Dhaliwal H. Intraspecific genetic diversity for resistance to wheat rusts in wild Triticum and Aegilops species. Wheat Information Service. 2000;90:21–30. [Google Scholar]
- 14.Wang S, Yin L, Tanaka H, Tanaka K, Tsujimoto H. Wheat-Aegilops chromosome addition lines showing high iron and zinc contents in grains. Breed Sci. 2011;61:189–195. doi: 10.1270/jsbbs.61.189. [DOI] [Google Scholar]
- 15.Badaeva ED, Amosova AV, Muravenko OV, Samatadze TE, Chikida NN, Zelenin AV, Friebe B, Gill BS. Genome differentiation in Aegilops. 3. Evolution of the D-genome cluster. Plant Syst Evol. 2002;231:163–190. doi: 10.1007/s006060200018. [DOI] [Google Scholar]
- 16.Badaeva ED, Amosova AV, Samatadze TE, Zoshchuk SA, Shostak NG, Chikida NN, Zelenin AV, Raupp WJ, Friebe B, Gill BS. Genome differentiation in Aegilops. 4. Evolution of the U-genome cluster. Plant Syst Evol. 2004;246:45–76. doi: 10.1007/s00606-003-0072-4. [DOI] [Google Scholar]
- 17.Friebe B, Gill BS. Chromosome banding and genome analysis in diploid and cultivated polyploid wheats. In: Jauhar PP, editor. Methods in Genome Analysis in Plants: Their Merits and Pitfals. New York, London: Tokyo Boca Ration: CRC Press; 1996. pp. 39–60. [Google Scholar]
- 18.Friebe B, Jiang J, Tuleen N, Gill BS. Standard karyotype of Triticum umbellulatum and the characterization of derived chromosome addition and translocation lines in common wheat. Theor Appl Genet. 1995;90:150–156. doi: 10.1007/BF00221010. [DOI] [PubMed] [Google Scholar]
- 19.Friebe B, Schubert V, Bluthner WD, Hammer K. C-banding pattern and polymorphism of Aegilops caudata and chromosomal constitutions of the amphiploid T. aestivum - Ae. caudata and six derived chromosome addition lines. Theor Appl Genet. 1992;83:589–596. doi: 10.1007/BF00226902. [DOI] [PubMed] [Google Scholar]
- 20.Schubert V. Untersuchungen an Triticum aestivum-Aegilops markgrafii Kreuzungen und die Nutzung hochrepetitiver DNA Sequenzen in der squash dot Technik. Halle-Wittenberg: Thesis, Martin-Luther University; 1989. [Google Scholar]
- 21.Linc G, Friebe BR, Kynast RG, Molnar-Lang M, Köszegi B, Sutka J, Gill BS. Molecular cytogenetic analysis of Aegilops cylindrica Host. Genome. 1999;42:497–503. doi: 10.1139/gen-42-3-497. [DOI] [PubMed] [Google Scholar]
- 22.Flavell RB, Bennett MD, Smith JB, Smith DB. Genome size and the proportion of repeated sequence DNA in plants. Biochem Genet. 1974;12:257–269. doi: 10.1007/BF00485947. [DOI] [PubMed] [Google Scholar]
- 23.Mehrotra S, Goyal V. Repetitive sequences in plant nuclear DNA: Types, distribution, evolution and function. Genomics, Proteomics & Bioinformatics. 2014;12:164–171. doi: 10.1016/j.gpb.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dvořák J, Zhang HB. Reconstruction of the phylogeny of the genus Triticum from variation in repeated nucleotide sequences. Theor Appl Genet. 1992;84:419–429. doi: 10.1007/BF00229502. [DOI] [PubMed] [Google Scholar]
- 25.Sepsi A, Molnár I, Szalay D, Molnár-Láng M. Characterization of a leaf rust-resistant wheat–Thinopyrum ponticum partial amphiploid BE-1, using sequential multicolor GISH and FISH. Theor Appl Genet. 2008;116:825–834. doi: 10.1007/s00122-008-0716-4. [DOI] [PubMed] [Google Scholar]
- 26.Pedersen C, Langridge P. Identification of the entire chromosome complement of bread wheat by two-colour FISH. Genome. 1997;40:589–593. doi: 10.1139/g97-077. [DOI] [PubMed] [Google Scholar]
- 27.Rayburn AL, Gill BS. Molecular analysis of the D-genome of the Triticeae. Theor Appl Genet. 1987;73:385–388. doi: 10.1007/BF00262505. [DOI] [PubMed] [Google Scholar]
- 28.Mukai Y. Multicolor fluorescence in situ hybridization: a new strategy for genome analysis. In: Jauhar PP, editor. Methods in Genome Analysis in Plants: Their Merits and Piffals. NY, London, Tokyo: Boca Ration: CRC Press; 1996. pp. 181–194. [Google Scholar]
- 29.Komuro S, Endo R, Shikata K, Kato A. Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome. 2013;56:131–137. doi: 10.1139/gen-2013-0003. [DOI] [PubMed] [Google Scholar]
- 30.Badaeva ED, Friebe B, Gill BS. Genome differentiation in Aegilops. 1. Distribution of highly repetitive DNA sequences on chromosomes of diploid species. Genome. 1996;39:293–306. doi: 10.1139/g96-040. [DOI] [PubMed] [Google Scholar]
- 31.Schneider A, Linc G, Molnár I, Molnár-Láng M. Molecular cytogenetic characterization of Aegilops biuncialis and its use for the identification of 5 derived wheat – Aegilops biuncialis disomic addition lines. Genome. 2005;48:1070–1082. doi: 10.1139/g05-062. [DOI] [PubMed] [Google Scholar]
- 32.Castilho A, Heslop-Harrison JS. Physical mapping of 5S and 18S-25S rDNA and repetitive DNA sequences in Aegilops umbellulata. Genome. 1995;38:91–96. doi: 10.1139/g95-011. [DOI] [PubMed] [Google Scholar]
- 33.Molnár I, Cifuentes M, Schneider A, Benavente E, Molnár-Láng M. Association between simple sequence repeat-rich chromosome regions and intergenomic translocation breakpoints in natural populations of allopolyploid wild wheats. Ann Bot. 2011;107:65–76. doi: 10.1093/aob/mcq215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cuadrado A, Cardoso M, Jouve N. Increasing the physical markers of wheat chromosomes using SSRs as FISH probes. Genome. 2008;51:809–815. doi: 10.1139/G08-065. [DOI] [PubMed] [Google Scholar]
- 35.Cuadrado A, Schwarzacher T, Jouve N. Identification of different chromatin classes in wheat using in situ hybridization with simple sequence repeat oligonucleotides. Theor Appl Genet. 2000;101:711–717. doi: 10.1007/s001220051535. [DOI] [Google Scholar]
- 36.Chennaveeraiah MS. Karyomorphologic and cytotaxonomic studies in Aegilops. Acta Horti Gotoburgensis. 1960;23:85–186. [Google Scholar]
- 37.Cermeno MC, Orellana J, Santos JL, Lacadena JR. Nucleolar activity and competition (amphiplasty) in the genus Aegilops. Heredity. 1984;53:603–611. doi: 10.1038/hdy.1984.118. [DOI] [Google Scholar]
- 38.Jiang J, Gill B. New 18S · 26S ribosomal RNA gene loci: chromosomal landmarks for the evolution of polyploid wheats. Chromosoma. 1994;103:179–185. doi: 10.1007/BF00368010. [DOI] [PubMed] [Google Scholar]
- 39.Endo TR. Allocation of a gametocidal chromosome of Aegilops cylindrica to wheat homoeologous group 2. Genes Genetic Systems. 1996;71:243–246. doi: 10.1266/ggs.71.243. [DOI] [Google Scholar]
- 40.Schmidt J-C, Schubert V, Blüthner W-D. Use of isozymes to characterize Triticum aestivum–Aegilops markgrafii addition lines. Biochem Physiol Pflanz. 1993;188:385–392. doi: 10.1016/S0015-3796(11)80141-1. [DOI] [Google Scholar]
- 41.Peil A, Korzun V, Schubert V, Schumann E, Weber WE, Röder MS. The application of wheat microsatellites to identify disomic Triticum aestivum-Aegilops markgrafii addition lines. Theor Appl Genet. 1998;96:138–146. doi: 10.1007/s001220050720. [DOI] [Google Scholar]
- 42.Zhang HB, Jia J, Gale MD, Devos KM. Relationships between the chromosomes of Aegilops umbellulata and wheat. Theor Appl Genet. 1998;96:69–75. doi: 10.1007/s001220050710. [DOI] [Google Scholar]
- 43.Kihara H. Genomanalyse bei Triticum und Aegilops. II. Aegilotricum und Aegilops cylindrica. Cytologia (Tokyo) 1931;2:106–156. doi: 10.1508/cytologia.2.106. [DOI] [Google Scholar]
- 44.Dubkovsky J, Dvořák J. Genome origin of Triticum cylindricum, Triticum triunciale, and Triticum ventricosum (Poaceae) inferred from variation in repeated nucleotide sequences: a methodological study. Am J Bot. 1994;81:1327–1335. doi: 10.2307/2445408. [DOI] [Google Scholar]
- 45.Tang Z, Yang Z, Fu S. Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J Appl Genetics. 2014;55:313–318. doi: 10.1007/s13353-014-0215-z. [DOI] [PubMed] [Google Scholar]


