Abstract
The identification of molecular subtypes of non-small-cell lung cancer has transformed the clinical management of this disease. This is best exemplified by the clinical success of targeting the EGFR or ALK with tyrosine kinase inhibitors in the front-line setting. Our ability to further improve patient outcomes with biomarker-based targeted therapies will depend on a more comprehensive genetic platform that can rationally interrogate the cancer genome of an individual patient. Novel technologies, including multiplex genotyping and next-generation sequencing are rapidly evolving and will soon challenge the oncologist with a wealth of genetic information for each patient. Although there are many barriers to overcome, the integration of these genetic platforms into clinical care has the potential to transform the management of lung cancer through improved molecular categorization, patient stratification, and drug development, thereby, improving clinical outcomes through personalized lung cancer medicine.
Keywords: multiplex genotyping, next-generation sequencing, non-small-cell lung cancer, targeted therapy
The discovery of genetic alterations that drive tumor progression in subsets of non-small-cell lung cancer (NSCLC) has transformed the clinical management of this disease. In particular, recent therapeutic advances in NSCLC have been established by focusing on unique somatic genetic variations between patients that predict response to targeted therapies. This biomarker-driven paradigm in NSCLC has not only revealed significant interpatient tumor heterogeneity, but also extensive intratumor heterogeneity [1]. While this biological variability poses significant challenges to identify clinically relevant driver genes, our ability to molecularly dissect individual tumor types and classify them based on their genetic profile provides tremendous opportunity to rationally design and therapeutically target these genetic events to improve patient care.
The prototypical example of how genotype-directed, biomarker-driven lung cancer care can dramatically change an entire treatment paradigm is exemplified through the discovery of EGFR mutations in NSCLC that predict tumor responsiveness and improve progression-free survival (PFS) during therapy with EGFR tyrosine kinase inhibitors (TKIs) compared with standard chemotherapy [2]. More recently, the identification of the EML4-ALK gene rearrangement is yet another example of a therapeutic biomarker that has demonstrated tremendous clinical success as a predictive marker of effective ALK inhibitor (crizotinib) treatment [3]. These two clinically validated oncogenic drivers exemplify the unique benefits of genotype-directed targeted therapy and demonstrate the importance of patient stratification to enrich and enhance treatment responses in drug development and clinical trials.
As the list of potential oncogenic drivers in lung cancer continues to grow, our ability to match an individual patient’s tumor profile to a specific targeted therapy hinges upon a comprehensive molecular diagnostic platform that can not only rapidly screen for known targetable genes, but also identify novel actionable drug targets that can be therapeutically exploited to improve clinical outcomes. In this review, we will outline the current genomic landscape of targetable oncogenic driver mutations in lung adenocarcinoma and explore the current and future molecular diagnostics tools to detect and therapeutically guide the treatment of patients with NSCLC.
Current landscape of targetable oncogenic drivers in lung adenocarcinoma
The identification of actionable oncogenic EGFR mutations has transformed the clinical management of NSCLC from a predominantly clinicopathologic to a genotype-directed therapeutic approach. Accompanying this transition, however, is an increasing awareness of the genetic complexity that will ultimately challenge the practicing oncologist to correctly identify and categorize patients into molecular subsets that will drive the clinical decision-making process. Here, we outline the landscape of targetable oncogenic drivers in lung adenocarcinoma, with an emphasis on currently approved targeted agents and novel small molecule inhibitors in clinical development (Table 1).
Table 1.
Targetable oncogenic drivers in non-small-cell lung cancer.
| Gene | Alteration | Frequency (%) | Targeted therapies | Current clinical trials† |
|---|---|---|---|---|
| EGFR | Mutation | 10–15 | Erlotinib, gefitinib, afatinib, CO-1686, AZD9291 | NCT01836341, NCT01542437, NCT01953913, NCT01931306, NCT01526928, NCT01802632 |
| BRAF | Mutation | 3–4 | Dabrafenib, trametinib, dasatinib | NCT01336634, NCT01362296 NCT01514864 |
| PI3KCA | Mutation | 1–3 | BKM120, XL147 | NCT01297452, NCT01570296, NCT01297491, NCT01723800, NCT01390818 |
| HER2 | Mutation | 1–4 | Afatinib, neratinib, dacomitinib | NCT01542437, NCT01827267, NCT01858389, NCT00818441 |
| EML4-ALK, KIF5B-ALK, TFG-ALK | Fusion | 3–5 | Crizotinib, LDK378, CH5424802 | NCT00932451, NCT01639001, NCT01685060, NCT01685138, NCT01828112, NCT01828099, NCT01579994, NCT01871805 |
| ROS1 | Fusion | 1–2 | Crizotinib | NCT01945021 |
| RET | Fusion | 1–2 | Cabozantinib, vandetanib | NCT01639508, NCT01823068 |
Trials can be accessed via the ClinicalTrials.govwebsite.
Targetable genetic mutations
EGFR
The INTACT-1 and INTACT-2 trials were randomized, Phase III trials that evaluated the therapeutic efficacy of the EGFR inhibitor gefinitib plus standard platinum-based chemotherapy in treatment-naive patients with advanced NSCLC [4,5]. These trials demonstrated no significant difference in overall survival, time to progression or response rate, with the addition of gefitinib to standard platinum doublet therapy. Similar results were reported in the erlotinib-based TALENT and TRIBUTE trials, suggesting that, in an unselected population, the addition of an EGFR small molecule inhibitor to systemic chemotherapy provided no significant improvement in clinical outcomes [6,7]. Despite these modest results, astute clinical observation defined a clinical subset of NSCLC patients with dramatic clinical and radiographic responses during therapy with gefitinib or erlotinib. This subpopulation of patients comprised approximately 10–15% of unselected patients and were retrospectively identified to be East Asian, female never-smokers, with a high frequency of adenocarcinoma histology [8]. The clinical responses to EGFR TKIs observed in these patients led to the sequencing of the EGFR gene and subsequent identification of somatic activating mutations in the EGFR kinase domain that drive the malignant phenotype [2]. Both retrospective and prospective genotyping of the EGFR kinase domain in patients with NSCLC have revealed conserved mutations that clinically correlate with EGFR kinase activation and inhibitor sensitivity [9]. These mutations target four exons (18–21) that cluster around the ATP-binding pocket of EGFR and are classified as in-frame deletions in exon 19 that involve the conserved LREA motif (~50% of all EGFR-sensitizing mutations), missense mutations in exon 18 that affect the G719 residue (~5% of all sensitizing mutations) and exon 21 that results in the L858R substitution (~45% of EGFR-sensitizing mutations; Figure 1). Crystallographic studies showed that EGFR-TKI sensitizing mutations destabilize the inactive state of the kinase and shift the equilibrium to favor the active conformation, which imparts decreased ATP affinity and renders the kinase more susceptible to quinazoline-based EGFR inhibitors that selectively compete for the ATP-binding site in the active conformation [10,11]. Moreover, because they destabilize the inactive state of the kinase, EGFR kinase domain mutations lead to increased catalytic activity and oncogenic potential by activating downstream prosurvival pathways [12]. In vitro studies in NSCLC cell lines that harbor EGFR-sensitizing mutations invariably identify hyperactive EGFR signaling outputs through the Raf-MEK-ERK, PI3K-AKT and STAT pathways, which drive the proliferative and anti-apoptotic signals on which these tumor cells are dependent on for survival. This dependency on mutated EGFR is therapeutically exploited by the EGFR TKIs, gefitinib, erlotinib and afatinib, which each interfere and ablate the EGFR-driven prosurvival signals and promote cell death.
Figure 1. Schematic representation of EGF receptor mutations associated with tyrosine kinase inhibitor sensitivity.
EGFR: EGF receptor; TKI: Tyrosine kinase inhibitor; TM: Transmembrane domain.
The molecular and structural mechanisms that underlie the EGFR TKI response has had a rapid and profound clinical impact in the way we select and treat patients with EGFR-mutant NSCLC. Specifically, the exquisite dependence on EGFR signaling in this subset of NSCLC provides a uniquely susceptible therapeutic target that leads to dramatic tumor regression and durable clinical responses when treated with EGFR TKIs. Recent randomized Phase III clinical trials utilizing upfront genotype-directed patient stratification to identify mutant EGFR in patients with NSCLC has unequivocally demonstrated an improvement in PFS with the reversible EGFR TKIs, gefinitib and erlotinib when compared with standard platinum doublet chemotherapy. This data emphasizes the importance of identifying molecular subsets of NSCLC in the front-line setting. Specifically, among the patients who harbor an EGFR TKI-sensitizing mutation and receive first-line TKI therapy, there was a significant improvement in PFS observed when gefitinib was compared with carboplatin and paclitaxel (10.8 vs 5.4 months; hazard ratio [HR]: 0.30; 95% CI: 0.22–0.41; p < 0.001) or cisplatin and docetaxel (9.2 vs 6.3 months; HR: 0.49; 95% CI: 0.34–0.71; p < 0.001) [13–16]. Further evidence of the superiority of genotype-directed therapeutic selection was recently demonstrated in a Phase III, randomized trial comparing afatinib, a potent irreversible inhibitor of EGFR, with standard platinum doublet chemotherapy. In the LUX-Lung 3 trial, afatinib demonstrated prolonged PFS when compared with cisplatin and pemetrexed (11.1 vs 6.9 months; HR: 0.58; 95% CI: 0.43–0.78; p < 0.001) in patients with advanced lung adenocarcinoma whose tumors harbored an activating mutation in EGFR [17].
The overall superior efficacy of EGFR TKIs in the front-line setting in patients whose tumors harbor activating EGFR mutations has validated the importance of comprehensively profiling an individual patient’s tumor to genetically classify and rapidly direct the selection of molecularly targeted therapies.
BRAF mutations
The discovery that activating mutations in EGFR predict responsiveness to EGFR TKIs and clinically improve PFS with decreased overall toxicity compared with standard chemotherapy led to large collaborative efforts to comprehensively identify and catalog all somatic mutations in histopathologically validated lung adenocarcinoma tumor specimens [18]. These large genomic studies have identified multiple candidate genes that are mutated at increased frequency in lung adenocarcinoma, many of which are currently being validated clinically as potential therapeutic targets. Considerable interest has been generated by these sequencing efforts because some of these genes represent known driver mutations in other cancers and are thus associated with increased sensitivity to clinically approved targeted therapies. Specifically, BRAF, PIK3CA and HER2 all occur at low frequencies (<5%) in lung adenocarcinoma; however, they serve as actionable molecular targets in melanoma, glioblastoma and breast cancer, respectively [19–21].
BRAF is a serine/threonine kinase that lies upstream of MEK and ERK in the RAF-MEK-ERK signaling cascade, a key molecular pathway that regulates cell growth and proliferation [22]. Activating somatic mutations in BRAF constitutively activate MEK and ERK and leads to oncogenic cell proliferation [23]. In approximately 50% of melanoma cases, the BRAF Val-600Glu (V600E) mutation drives tumorigenesis and is effectively targeted with the selective BRAF and MEK inhibitors, vemurafenib and dabrafenib (BRAF inhibitors), and trametinib (MEK inhibitor). BRAF mutations have also been identified in 3–4% of patients with NSCLC [24,25]. Unlike melanoma, however, where 80–90% of all BRAF mutant tumors harbor the V600E activating mutation, the BRAF mutational landscape in NSCLC consists of 40–50% of non-V600E mutations that can either active or inactivate the kinase. While the pharmacologic responses to BRAF inhibitors in patients with non-V600E BRAF mutations is an area of active investigation, a recent Phase II study evaluating the therapeutic efficacy of dasatinib in 34 patients with advanced NSCLC identified one patient with a dramatic durable response [26]. Interestingly, through comprehensive tumor profiling, this patient was found to harbor a BRAFY472C kinase-inactivating mutation that drove sensitivity to dasatinib [27]. The complex intragenic mutational diversity of BRAF in NSCLC has important implications on clinical trial design. Specifically, clinical investigators recognize that, in order to establish BRAF as a valid therapeutic target in NSCLC, distinct therapeutic strategies will be required to treat patients with BRAF V600E, non-V600E and inactivating mutations. Thus, current clinical trials in BRAF-mutant NSCLC are prospectively stratifying patients to receive the BRAF inhibitor, dabrafenib for patients with V600E mutations; the MEK inhibitor, trametinib for patients with non-V600E BRAF mutations; and dasatinib, a multitargeted TKI for patients with inactivating or uncharacterized BRAF mutations (Table 1).
PI3K
PI3Ks are lipid kinases that regulate pathways important for cellular proliferation, adhesion, motility and survival [28]. In 2004, somatic mutations in PIK3CA, the gene that encodes the p110α catalytic subunit of PI3K were identified in approximately 30% of colon and gastric cancer, and glioblastomas, with a much lower frequency in lung cancer (4%) [28]. Two subsequent studies found that 1.6% (11/691) and 3.4% (8/235) of NSCLC tissue specimens harbor mutations in the PIK3CA gene, with the majority of mutations clustering into two distinct hotspots, the helical (E542 and E545) and kinase (H1047) domains of PI3K [29,30]. PIK3CA mutations in lung cancer cell lines are associated with increased enzymatic activity and oncogenic potential, which is mediated, in part, through the AKT/mTOR pathway [31]. Furthermore, in vivo mouse models have demonstrated that isolated expression of mutant PIK3CA results in lung adenocarcinomas and treatment with a dual pan-PI3K and mTOR inhibitor led to marked tumor regression, suggesting that these cancers are in part, dependent on oncogenic PI3K [32].
While the PI3K/AKT/mTOR pathway remains an attractive therapeutic target in NSCLC, the clinical efficacy of targeting the key regulators of this pathway has been hampered by an inability to prospectively identify specific patient cohorts who are likely to respond to PI3K inhibition. This is likely due, in large part, to the recent finding that approximately 70% of PIK3CA mutations in lung adenocarcinoma coexist with validated oncogenic driver mutations, including EGFR, KRAS and ALK rearrangements [33]. Thus, the future success of PI3K pathway inhibition will depend on the prospective identification of these overlapping mutations through comprehensive molecular profiling of patient’s tumors that will genotypically direct combination therapies in clinical trials. To this end, PI3K, AKT and dual PI3K-mTOR inhibitors are currently in early-phase clinical trials, both as mono- and combination therapy with standard chemotherapy protocols [34].
HER2
HER2 (or ERBB2) is a membrane-bound tyrosine kinase in the ERBB family of receptors. In contrast to EGFR, ERBB3 and ERBB4, HER2 has no known ligand but instead heterodimerizes with these receptors upon ligand binding to mediate downstream signaling [35]. HER2 amplification occurs in approximately 30% of breast cancer, where it is associated with improved survival outcomes since the advent of HER2-targeted therapies, such as traztuzumab. While HER2 overexpression is observed in 13–20% of NSCLC tumors, the predictive association of HER expression with response to HER2-targeted agents has not been demonstrated [36]. Importantly, these studies were performed in NSCLC patients unselected for HER2 mutational status and thus were not designed to detect a clinical benefit in patients with genomic activation of HER2, which has been identified in a subset of NSCLC patients. In particular, genomic sequencing of NSCLC tumor specimens identified in-frame insertions involving exon 20, which result in constitutive kinase activation in a ligand-independent manner [37]. Clinically, these mutations are associated with never smoking status and are found in 1.2–4.2% of resected NSCLC tumor specimens, with enrichment in adenocarcinoma histology [38]. In a recent retrospective study dedicated to HER2-positive (insertion exon 20) NSCLC patients (n = 65), the clinical efficacy of HER2-directed targeted therapies, including trastuzumab and afatinib, were associated with a disease control rate of 93% in the trastuzumab-based cohort (n = 15) and 100% in the afatinib group (n = 3) [39]. These data suggest a potential therapeutic benefit for HER2-targeted agents in this subpopulation and have prompted further investigation and ongoing clinical development of multiple TKIs, including the irreversible dual HER2 and EGFR TKIs, afatinib, neratinib and dacomitinib [37]. A recent exploratory Phase II trial of single agent afatinib evaluated five patients with mutations in the HER2 kinase domain and objective responses were observed in two of the five evaluable patients, with clinical toxicity noted in the remaining three patients [40]. In a prospective, Phase II study evaluating dacomitinib in HER2-amplified and -mutant NSCLC, early response data from 16 patients identified two with partial clinical responses, both harboring HER2 mutations [41]. Larger trials are needed to define the clinical activity and efficacy of targeting HER2-dependent NSCLC.
Targetable genetic rearrangements
ALK rearrangements
ALK is a highly conserved receptor tyrosine kinase that was first identified as a fusion protein in anaplastic large-cell lymphoma [42]. Since this time, multiple additional ALK chimeric variants have been identified in several types of cancer, including neuroblastoma, inflammatory myofibroblastic tumor and NSCLC [43]. In 2007, the identification of an inversion within the short arm of chromosome 2, which results in the fusion of the EML4 gene with the ALK gene, was identified in a patient with NSCLC [44]. The resultant EML4-ALK translocation encodes for a cytoplasmic chimeric protein with constitutive kinase activity [45]. Since this initial discovery, multiple variants of EML-ALK have been identified in NSCLC, all of which encode for the cytoplasmic kinase domain of ALK but contain different EML4 truncations [46]. Regardless of the precise EML4 breakpoint, all identified EML4-ALK fusion proteins to date contain a constitutively active kinase with transforming capabilities in cell culture, largely functioning through the MAPK, AKT and JAK/STAT pathways [42]. In transgenic mouse models, lung-specific expression of EML4-ALK results in the development of lung adenocarcinomas, which regress upon treatment with a TKI of ALK [47]. This preclinical data suggested that ALK rearrangements are oncogenic drivers that are susceptible to targeted therapies in a subset of lung adenocarcinomas.
EML4-ALK rearrangements are present in approximately 3–5% of patients with lung adenocarcinoma, with a much lower frequency observed for the other known ALK fusion partners including KIF5B and TFG [45]. Patients that harbor EML4-ALK fusions share many of the clinical hallmarks, including younger age of onset (median 50 years of age at diagnosis) and light or never-smoker status, as observed in some other genetically defined lung cancers, including EGFR-mutant NSCLC [48]. EML4-ALK rearrangements and EGFR mutations are mutually exclusive with the exception of rare cases and thus, detailed molecular characterization to isolate these populations becomes paramount in genotype-directed clinical trial design. To this end, in order to directly evaluate the effect of ALK inhibition in patients with EML4-ALK rearrangements, a multicenter, single-arm study of crizotinib was conducted in 2010, which has already demonstrated impressive clinical efficacy, with an objective response rate (ORR) of 61% and median duration of response of 47 weeks [49]. These findings served as the foundation for accelerated approval of crizotinib by the US FDA in 2011 for the treatment of advanced ALK-positive NSCLC. More recently, a randomized, multicenter, Phase III study of crizotinib versus standard of care chemotherapy in ALK-positive NSCLC patients who had previously received platinum doublet therapy demonstrated a median PFS of 7.7 versus 3.0 months in the crizotinib and chemotherapy (pemetrexed or docetaxel) groups, respectively. Consistent with the PFS results, ORR greatly favored the crizotinib group (65%) over pemetrexed (29%) or docetaxel (7%) [3]. In order to evaluate the role of crizotinib in the front-line setting, a randomized controlled trial of crizotinib versus standard platinum-based chemotherapy in patients with advanced ALK-positive NSCLC is currently being conducted.
The preclinical development of second-generation ALK inhibitors with increased potency and selectivity toward ALK has translated rapidly into clinical success. Of these next-generation ALK inhibitors, LDK378 demonstrated significant antitumor activity in an EML4-ALK xenograft model and has since been evaluated in a multicenter, Phase I study, where preliminary results have demonstrated an ORR of 70% in ALK-positive treatment-naive NSCLC and 73% in ALK-positive patients who clinically acquired resistance to crizotinib [50]. Given these results, LDK378 was designated as a ‘breakthrough therapy’ by the FDA in March 2013 for treatment of both crizotinib-naive and crizotinib-resistant ALK-positive NSCLC. As a result, two Phase III randomized trials are currently underway to evaluate the clinical efficacy of LDK378 in the up-front setting when compared with standard platinum doublet therapy in patients with ALK-positive NSCLC (Table 1).
ROS1 rearrangements
The initial identification of a ROS1 rearrangement was demonstrated in a human glioblastoma cell line, in which a deletion on chromosome 6 resulted in a ROS1-FIG gene fusion [51]. The transforming properties of the resultant ROS1-FIG chimeric protein has since been demonstrated in a glioblastoma transgenic mouse model [52]. Although ROS1 rearrangements have been identified in a diverse set of human cancers (glioblastoma, angiosarcoma, gastric, colon and ovarian cancers), its potential as a therapeutic target has been unexplored due to its relatively low frequency in these tumors. More recently, ROS1 fusions have been identified in a NSCLC tumor specimen (CD74-ROS1) and cell line (SLC34A2-ROS1) through a large phosphoproteomic screen [53]. Additionally, novel fusion partners for ROS1 including EZR have demonstrated oncogenic properties in transgenic mouse models [54]. With these discoveries, a renewed interest in ROS1 fusions has led to several studies focused on the prevalence of ROS1 rearrangements in NSCLC. The largest and most comprehensive study was a retrospective analysis that screened 1073 NSCLC patient samples using a break-apart FISH probe, which identified 1.7% with ROS1 fusions [55]. The clinical characteristics of NSCLC patients with ROS1 rearrangements are similar to those that define the ALK fusion-positive lung cancers, with a predominance of never-smokers, younger age (median 49 years of age at diagnosis) of onset and adenocarcinoma histology [51]. The clinical efficacy of targeting ROS1-driven NSCLCs is actively being explored with the adventitious finding that crizotinib, which is now approved for the treatment of ALK-positive NSCLC, is also a potent ROS1 inhibitor [56]. Moreover, based on preclinical biochemical data demonstrating decreased ROS1 phosphorylation in response to crizotinib treatment and the rapid, near complete resolution of a ROS1-driven tumor in a patient with NSCLC, the clinical development of ROS1 inhibitors including crizotinib could greatly and rapidly benefit this subgroup of NSCLC patients. The recent results of a genotype-directed Phase I trial of crizotinib in patients with advanced NSCLC harboring a ROS1 gene rearrangement closely parallels the clinical success for ALK-positive NSCLC, with an ORR of 57% and a disease control rate at 8 weeks of 79% [57]. While the early clinical effects of crizotinib in patients with ROS1 fusions is encouraging, further clinical validation is warranted to clarify the specific target of crizotinib, given its ability to potently inhibit multiple receptor kinases including MET and ALK, both of which are validated oncogenic drivers in lung cancer. To meet this need, Phase II trials assessing the efficacy of crizotinib in ROS1-positive patients are prospectively screening tumors for ALK rearrangements and retrospective histological analysis should be performed to assess the status of MET in these patients.
RET rearrangements
RET is a well-studied receptor tyrosine kinase that has oncogenic properties in several human cancers. Both familial and sporadic gain-of-function mutations have been identified in multiple endocrine neoplasia type 2 and medullary thyroid cancers, respectively [58]. RET gene fusions have also been observed in up to 50% of sporadic papillary thyroid cancer cases and, most recently, in 1.9% of lung adenocarcinomas [59–61]. The most common RET fusion in lung adenocarcinoma involves an inversion in chromosome 10 that leads to the fusion of the KIF5B and RET genes, resulting in constitutive RET kinase activity [62]. Patients that harbor KIF5B-RET fusions are young, light to never smokers with a predominance of adenocarcinoma histology [58]. Precedence for RET oncogenic dependence has already been demonstrated in thyroid cancers, in which two FDA-approved RET inhibitors, vandetanib (ZD6474) and cabozantinib (XL184), are in clinical use [62]. The therapeutic potential of these inhibitors in patients with RET fusion-positive NSCLC is currently being addressed and the preliminary antitumor effects in the first three patients treated with cabozantinib are encouraging, demonstrating either partial responses (n = 2) or stable disease (n = 1) [63]. While early, this small cohort provides clinical validation that targeting the RET oncogene is a promising new therapeutic approach in this molecular subset of NSCLC.
Genotyping & whole-genome profiling to guide clinical decision-making
While the genomic landscape of targetable oncogenic drivers in lung adenocarcinoma has exponentially increased within the past few years, we are only beginning to understand the complexities that make each individual tumor unique. As we continue to catalog the genetic alterations that define NSCLC subtypes, our ability to rapidly identify and treat patients based on their mutational profile will require a more high-throughput diagnostic platform to enhance speed and sensitivity. Here we outline the current clinical state of molecular diagnostics in lung cancer medicine and provide a glimpse of next-generation sequencing (NGS) technologies that can offer rapid detection of standard-of-care mutations, while generating additional information through genome-wide analysis that may lead to rationally designed alternative therapies (Table 2).
Table 2.
Comparison of genotyping platforms.
| Method | Coverage | Specimen | Turnaround time (days) | Cost (US$) |
|---|---|---|---|---|
| Single gene analysis | Single gene (RT-PCR, FISH) | ~600 ng FFPE tumor DNA | 10 | 200–800 |
| Multiplex genotyping [64] | >50 hotspot mutation sites in 8–14 genes (SNaPshot, Applied Biosystems). >200 hotspot mutations in 19 genes (Sequenom) | ~500 ng FFPE tumor DNA | 14–21 | 1500–2000 |
| NGS | >500 genes. Whole exon, copy number, rearrangements, methylation, transcriptome | 100–3000 ng FFPE tumor DNA | 21 | 3500–5000 |
FFPE: Formalin-fixed paraffin-embedded; NGS: Next-generation sequencing; RT-PCR: Real-time PCR.
Direct single gene analysis
Randomized controlled trials have demonstrated that NSCLC patients with wild-type EGFR respond better to systemic chemotherapy [13,14]. Therefore, in most clinical circumstances, EGFR TKI therapy should not be administered as first-line therapy without documented evidence of a sensitizing EGFR mutation. Based on these clinical findings, coupled with the fact that patients with stage IV NSCLC have a median survival of approximately 16 weeks if left untreated, evidence-based guidelines have been established that mandate the turnaround time for EGFR molecular testing to be no longer than 10 days [65]. To meet this need, the majority of Clinical Laboratory Improvement Act-certified laboratories are now implementing FDA-approved mutational detection assays to identify single gene alterations. These detection methods are variable, but are all based on sequencing, amplification or FISH analysis of the mutant EGFR or KRAS allele or ALK rearrangements, respectively. The current guidelines established by three professional organizations (College of American Pathologists, Association for Molecular Pathology and the International Association for the Study of Lung Cancer) do not favor one testing platform over the other, but do recommend that all methods be able to detect mutations in clinical specimens with at least 50% cancer cell content [65].
Multiplex genotyping
Given the relatively small number of actionable targets in NSCLC, single gene-based molecular tests for EGFR, KRAS and EML4-ALK continue to be the standard of care. As we continue to clinically validate the growing list of potential driver mutations in lung cancer, however, we will need to develop a more comprehensive, cost-effective tumor genotyping protocol to rapidly screen patients for all actionable therapeutic targets. Multiplex genotyping, a PCR-based assay designed to simultaneously detect the expression of known mutational hotspots in multiple target genes in a single reaction, is currently being employed throughout the cancer research community and being validated in clinical trials [66]. The most well known of these platforms are the SNaPShot (Applied Biosystems) and Sequenom (Seqeunom), the former of which is currently being validated in a large prospective trial by the Lung Cancer Mutational Consortium. In order to assess the clinical feasibility and applicability of a multiplex genotyping assay, the Lung Cancer Mutational Consortium utilized the SNaPShot platform to detect alterations in ten genes (KRAS, EGFR, HER2, BRAF, PIK3CA, AKT1, MEK1, NRAS mutations, ALK rearrangements and MET amplifications) and subsequently employed rationally based therapeutic approaches based on an individual’s molecular subtype. To date, of the 1007 patients enrolled in the study, 64% of tumors harbored an actionable genetic driver mutation, and 28% of these patients were either treated with a clinically approved targeted therapy or matched to a clinical trial based on their genotype. The preliminary results from this trial suggest that multiplex genomic methodologies can enhance mutational detection and lead to increased median survival rates when patients are appropriately matched to a targeted therapy or clinical trial based on their tumor genetic profile [67]. While promising, multiplex genotyping has two major pitfalls. The first is the turnaround time, which is currently 2–3 weeks given the labor-intensive protocol involved, and the second is the limited ability to discover new drug targets given the predetermined genetic hotspots that are utilized in these assays. In order to implement these multiplex genotyping platforms into widespread clinical practice, we will first need to rigorously validate and confirm that these approaches can improve clinical outcomes in patients with NSCLC. Furthermore, as the mutational spectrum continues to evolve and new actionable drivers are identified, we will need to rapidly addend the predefined mutational hotspots profiled in these assays.
Next-generation sequencing
NGS technologies offer a rapid and comprehensive approach to genome-wide analysis of an individual genome, transcriptome or epigenome in a cost-effective manner [1]. The increased sensitivity, speed, and lower cost make NGS an attractive clinical platform that will likely replace or, at the very minimum, augment existing mutational detection methodologies. Unlike current single gene or multiplex genotyping, NGS provides unbiased detection of point mutations, copy number variations and complex structural rearrangements, ultimately leading to a more complete survey of a patient’s cancer genome. Importantly, while this global assessment holds unprecedented potential to personalize lung cancer medicine, the analytical complexity inherent in whole-genome sequencing is a challenge that must be overcome to ascertain clinically relevant and actionable biomarkers. Thus, in order to effectively implement NGS into clinical practice, we must first solve the issues that accompany comprehensive analysis, including intratumor heterogeneity, quality of tumor tissue, and the ethical issues surrounding the use of whole-genome sequencing data as a diagnostic tool. To begin to meet these challenges, a recent pilot study evaluating the clinical applicability of NGS in various tumor types, the Michigan Oncology Sequencing Project, identified on average 150 genomic alterations within each tumor specimen analyzed, reinforcing the genetic complexity that will ultimately require rigorous biological and clinical validation to distinguish between actionable driver mutations from passenger events [68]. The results of this study highlight both the biological and clinical challenges that emerge as a result of intratumor genetic heterogeneity. Specifically, we are now beginning to recognize that the clinical utility of predictive biomarkers can be influenced by low frequency subclones that evade detection by conventional molecular diagnostic approaches and that emerge during therapy as a dominant drug-resistant clone. Clinically, this manifests as mixed tumor responses between primary and metastatic sites or through the development of acquired drug resistance [69,70]. In order to gain a deeper understanding of the molecular complexities that drive intratumor heterogeneity, we will need to obtain serial biopsy specimens from patients to fully interrogate the real-time genomic changes that arise during an individual patient’s treatment course. The evolutionary dynamics observed both within individual tumor biopsies and between separate biopsies of the same tumor in response to the selective pressures of systemic therapy can ultimately provide a clinically relevant roadmap to guide rational drug selection.
In addition to distinct clonal populations, stromal and immune cell contamination poses a considerable challenge for large-scale genome and transcriptome analysis that can bias the molecular profile and potentially lead to erroneous prognostic and therapeutic outcomes. Bioinformatics and computational software that have been extensively reviewed elsewhere are now being employed to help navigate through mixed cell populations and estimate tumor purity [71]. As these tools evolve, detailed histopathologic assessment through tumor microdissection can be used in parallel to minimize tissue contamination.
If we are able to overcome the technical hurdles that currently impede our ability to implement NGS into clinical practice, we will then be faced with the ethical implications that accompany such a comprehensive genomic analysis. In particular, while current NGS platforms are more accurate than traditional Sanger sequencing methods, the massive quantity of data produced translates into a significant number of false-positive reads or variants of unknown significance or variants of significance in other diseases [72]. For these reasons, informed consent is imperative and patients that elect to pursue NGS for a disease-specific diagnosis must be properly educated about the potential of this technology to reveal information about their risk for alternative, unrelated disease states. While not currently implemented in routine clinical practice, the development of clinical protocols to delineate genetic information that relates specifically to a patient’s presenting disease or potential risk for an unrelated disease will need to be explicitly reviewed and individualized to each patient.
Finally, in order to bring this technology closer to clinical mainstream, we will need to optimize the NGS platform to be as efficient as the standard single gene analysis. By scaling down the sequencing coverage to identify specific cancer-associated genes, expense and turnaround time can be reduced, ultimately leading to a more clinically applicable and feasible diagnostic tool that can guide therapeutic decision-making.
Personalizing NSCLC treatment through genotype-directed targeted therapy
The identification of targetable oncogenic drivers that define molecular subsets in NSCLC has transformed the clinical management of this disease. Initial clinical design based on large, unselected populations failed to show a significant benefit in clinical outcomes in patients with NSCLC treated with targeted therapies. Conversely, when patients are prospectively selected based on their molecular profile and rationally directed to an appropriate targeted therapy, unprecedented results emerge as evidenced by EGFR and EML4-ALK in NSCLC. The remarkable efficacy of these molecularly targeted therapies in selected patient populations has led to a significant surge in the number of genotype-directed clinical trials, heralding the beginning of personalized medicine in lung cancer.
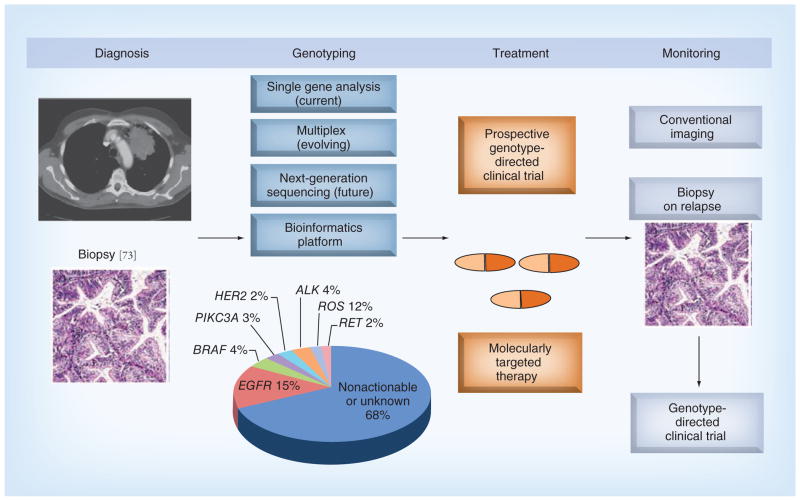
While this brings great optimism, there are still many hurdles that we must overcome in order to systematically tailor drugs to each patient’s tumor (Figure 2). We will first need to empower and educate our patients about the importance of routine molecular testing to guide therapeutic decision-making. The need for patient participation in clinical trials to assess the predictive and prognostic value of novel biomarkers and to establish the efficacy of molecularly targeted therapies is paramount. Patients, clinicians and translational scientists need to collaborate effectively to ensure routine biopsy specimens are obtained at the time of diagnosis and relapse to maximize therapeutic efficacy. Second, we will need to enhance our bioinformatics and computational biology platforms to optimize our current high throughput genotyping assays. While NGS technologies in cancer patients is providing an unprecedented amount of genomic information, the ability of clinical oncologists to rapidly digest the massive amounts of sequencing data and translate these unique blueprints into patient care is a significant barrier that could limit its full potential to improve clinical care. To this end, we will likely need to narrow the spectrum of coverage of NGS to include the most biologically relevant cancer-associated genes. This will increase speed and sensitivity while decreasing overall costs, at least until NGS technologies improve to enable democratization of the assays, analysis and interpretation. Third, as we continue to refine these state-of-the-art genomic sequencing technologies, we will need to understand the biological relevance of the multiple genomic alterations both as isolated entities and within the context of an evolving tumor over time. This will allow us to more effectively translate this information into biomarker-driven clinical trials to improve patient outcomes and personalize lung cancer therapy.
Figure 2.
Personalizing non-small-cell lung cancer treatment through genotype-directed therapy.
Conclusion
The identification of targetable oncogenic drivers in NSCLC has led to a new understanding of the genetic complexity of these tumors. These advancements provide the foundation to uniquely characterize molecular subsets of lung cancer and allow for the development of rationally designed clinical trials to evaluate novel molecularly targeted therapeutics. Despite early success with biomarker-driven lung cancer trials, however, our ability to personalize lung cancer medicine will depend on the parallel development of genomic technologies to identify and monitor in real-time the evolution of an individual patient’s cancer genome. To meet this need, many oncologists are implementing multiplex genotyping platforms of actionable mutational hotspots into clinical practice. As the list of actionable targets continues to grow, however, we will need to transition to a more rapid, sensitive and cost-effective genomic approach. NGS technology offers a comprehensive genome-wide survey that holds great promise for the advancement of genotype-directed lung cancer care. Our ability to effectively utilize this technology in the clinic will depend on the development of a bioinformatics platform that can rapidly digest and reveal the most biologically relevant alterations to ultimately guide clinical decision-making.
Future perspective
Genotype-directed clinical trial design has transformed the clinical management of NSCLC. Our ability to systematically categorize NSCLC by molecular subtype optimizes patient selection and enables rational selection of patients for the most appropriate molecularly targeted therapy. While there are still many obstacles to overcome before genome-wide technologies become clinical mainstay, the continued generation of cancer genome data coupled with rapid targeted therapy development will catalyze continued advances in personalized lung cancer medicine.
Executive summary.
Background
The discovery of genetic alterations that drive tumor progression in subsets of non-small-cell lung cancer (NSCLC) has transformed the clinical management of this disease.
Molecular characterization of NSCLC tumors has revealed a new layer of genetic complexity.
Advancements in genotyping technologies are needed to guide molecularly targeted therapies and clinical trial design.
Targetable oncogenic drivers in lung adenocarcinoma
Biomarker-driven lung cancer medicine is the new paradigm.
Targeting mutant EGFR or the EML4-ALK rearrangement with tyrosine kinase inhibitors in the front-line setting has translated into improved patient outcomes.
The identification of novel oncogenic drivers in lung adenocarcinoma, including mutations in BRAF, PIK3CA, HER2, and fusions in ROS1 and RET are currently being validated as therapeutic targets in clinical trials.
Genotyping platforms to guide clinical decision-making
The most widely used genotyping platform in clinical lung cancer medicine relies on single gene analysis of EGFR mutations, ALK rearrangements and KRAS mutations.
Multiplex genotyping is currently being implemented in select academic centers to guide biomarker-driven patient stratification.
Next-generation sequencing has led to an unprecedented understanding of the lung cancer genome.
Personalizing NSCLC treatment through genotype-directed targeted therapy
Biomarker-driven clinical trial design will become increasingly complex due to the growing number of novel oncogenic drivers in NSCLC.
The continued success of genotype-directed molecularly targeted therapy will depend on the optimization and implementation of a more rapid and comprehensive genotyping platform.
Clinical application of next-generation sequencing will require the parallel development of computational methodologies that can guide clinicians to the most appropriate, biologically relevant targeted therapy or clinical trial.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
TG Bivona is a consultant of the Cancer Therapeutics Innovation Group. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• • of considerable interest
- 1.Li T, Kung HJ, Mack PC, et al. Genotyping and genomic profiling of non-small cell lung cancer: implications for current and future therapies. J Clin Oncol. 2013;31(8):1039–1049. doi: 10.1200/JCO.2012.45.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2• •.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. Discovery of EGFR mutations in non-small-cell lung cancer (NSCLC) tumor samples that confer sensitivity to the tyrosine kinase inhibitor, gefitinib. [DOI] [PubMed] [Google Scholar]
- 3.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368(25):2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 4.Giaccone G, Herbst RS, Manegold C, et al. Gefintinib in combination with gemcitabine and cisplatin in advanced non-small cell lung cancer: a Phase III trial – INTACT 1. J Clin Oncol. 2004;22(5):777–784. doi: 10.1200/JCO.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Herbst RS, Giaccone G, Schiller JH, et al. Geftinitib in combination with paclitaxel and carboplatin in advanced non-small-cell lung cancer: a Phase III trial – INTACT 2. J Clin Oncol. 2004;22(5):785–794. doi: 10.1200/JCO.2004.07.215. [DOI] [PubMed] [Google Scholar]
- 6.Gatzemeier U, Pluzanska A, Szczesna A, et al. Results of a Phase III trial of erlotinib (OSI-774) combined with cisplatin and gemcitabine chemotherapy in advanced non-small cell lung cancer (NSCLC) J Clin Oncol. 2004;22:619S. [Google Scholar]
- 7.Herbst RS, Prager D, Hermann R, et al. TRIBUTE: a Phase III trial of erlotinib hydrochloride (OSI-774) combined with carboplatin and paclitaxel chemotherapy in advance non-small cell lung cancer. J Clin Oncol. 2005;23(25):5892–5899. doi: 10.1200/JCO.2005.02.840. [DOI] [PubMed] [Google Scholar]
- 8• •.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. doi: 10.1126/science.1099314. Establishes the clinical correlates of EGFR mutations in NSCLC (Asian ethnicity, females and never-smoker status) [DOI] [PubMed] [Google Scholar]
- 9.Bronte G, Rizzo S, Paglia LL, et al. Driver mutations and differential sensitivity to targeted therapies: a new approach to the treatment of lung adenocarcinoma. Cancer Treat Rev. 2010;36(3):S21–S29. doi: 10.1016/S0305-7372(10)70016-5. [DOI] [PubMed] [Google Scholar]
- 10.Kumar A, Petri ET, Halmos B, et al. Structure and clinical relevance of the epidermal growth factor receptor in human cancer. J Clin Oncol. 2008;26(10):1742–1750. doi: 10.1200/JCO.2007.12.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang X, Gureasko J, Shen K, et al. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125(6):1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 12.Sharma S, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutation in lung cancer. Nat Rev Cancer. 2007;7(3):169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 13.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 14.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised Phase 3 trial. Lancet Oncol. 2010;11(2):121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 15• •.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362(25):2380–2388. doi: 10.1056/NEJMoa0909530. Establishes the clinical efficacy of genotype-directed targeted therapy in the front-line setting. [DOI] [PubMed] [Google Scholar]
- 16.Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multi-centre, open-label, randomised, Phase 3 study. Lancet Oncol. 2011;12(8):735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 17.Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31(27):3327–3334. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 18.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen PY, Lee EQ, Reardon DA. Current clinical development of PI3K pathway inhibitors in glioblastoma. Neuro Oncol. 2012;14(7):819–829. doi: 10.1093/neuonc/nos117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 22.Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 23.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 24.Naoki K, Chen TH, Richards WG, et al. Missense mutations of the BRAF gene in human lung adenocarcinoma. Cancer Res. 2002;62(23):7001–7003. [PubMed] [Google Scholar]
- 25.Brose MS, Volpe P, Feldman M, et al. BRAF and RAS mutations in human lung cancer and melanoma. Cancer Res. 2002;62(23):6997–7000. [PubMed] [Google Scholar]
- 26.Johnson FM, Bekele BN, Fong L, et al. Phase II study of dasatinib in patients with advanced non-small cell lung cancer. J Clin Oncol. 2010;28(30):4609–4615. doi: 10.1200/JCO.2010.30.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen B, Peng S, Tang X, et al. Kinase-impaired BRAF mutations in lung cancer confer sensitivity to dasatinib. Sci Transl Med. 2012;4(136):136ra70. doi: 10.1126/scitranslmed.3003513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 29.Kawano O, Sasaki H, Endo K, et al. PIK3CA mutation status in Japanese lung caner patients. Lung Cancer. 2006;54(2):209–215. doi: 10.1016/j.lungcan.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68(17):6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci USA. 2006;103(5):1475–1479. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engelman JA, Chen L, Tan X, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008;14(12):1351–1356. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heavey S, O’Bryne KJ, Gately K. Strategies for co-targeting the PI3K/AKT/mTOR pathway in NSCLC. Cancer Treat Rev. 2013;40(3):445–456. doi: 10.1016/j.ctrv.2013.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Oxnard GR, Binder A, Janne PA, et al. New targetable oncogenes in non-small cell lung cancer. J Clin Oncol. 2013;31(8):1097–1104. doi: 10.1200/JCO.2012.42.9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garrido-Castro AC, Felip E. HER2 driven non-small cell lung cancer (NSCLC): potential therapeutic approaches. Transl Lung Cancer Res. 2013;2(2):122–127. doi: 10.3978/j.issn.2218-6751.2013.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clamon G, Herndon J, Kern J, et al. Lack of trastuzumab activity in nonsmall cell lung carcinoma with overexpression of erb-B2: 39810 – a Phase II trial of Cancer and Leukemia Group B. Cancer. 2005;103(8):1670–1675. doi: 10.1002/cncr.20950. [DOI] [PubMed] [Google Scholar]
- 37.Shigematsu M, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65(5):1642–1646. doi: 10.1158/0008-5472.CAN-04-4235. [DOI] [PubMed] [Google Scholar]
- 38.Stephens P, Hunter C, Bignell G, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431(7008):525–526. doi: 10.1038/431525b. [DOI] [PubMed] [Google Scholar]
- 39.Mazieres J, Peters S, Lepage B, et al. Lung cancer that harbors a HER2 mutation: epideiologic characteristics and therapeutic perspectives. J Clin Oncol. 2012;31(16):1997–2003. doi: 10.1200/JCO.2012.45.6095. [DOI] [PubMed] [Google Scholar]
- 40.De Grève J, Teugels E, Geers C, et al. Clinical activity of afatinib (BIBW 2992) in patients with lung adenocarcinoma with mutations in the kinase domain of HER2/neu. Lung Cancer. 2012;76(1):123–127. doi: 10.1016/j.lungcan.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Jänne PA, Kris MG, Goldberg Z, et al. Dacomitinib (PF-00299804), an irreversible pan-HER tyrosine kinase inhibitor, for first-line treatment of EGFR-mutant or HER2-mutant or -amplified lung cancers. Ann Oncol. 2012;23:1228O. [Google Scholar]
- 42.Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8(1):11–23. doi: 10.1038/nrc2291. [DOI] [PubMed] [Google Scholar]
- 43.Pulford K, Lamant L, Espinos E, et al. The emerging normal and disease-related roles of anaplastic lymphoma kinase. Cell Mol Life Sci. 2004;61(23):2939–2953. doi: 10.1007/s00018-004-4275-9. [DOI] [PubMed] [Google Scholar]
- 44.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 45.Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer. 2013;13(10):685–700. doi: 10.1038/nrc3580. [DOI] [PubMed] [Google Scholar]
- 46.Choi YL, Takeuchi K, Soda M, et al. Identification of novel isoforms of the EML4-ALK transforming gene in non-small cell lung cancer. Cancer Res. 2008;68(13):4971–4976. doi: 10.1158/0008-5472.CAN-07-6158. [DOI] [PubMed] [Google Scholar]
- 47.Soda M, Takada S, Takuchi K, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci USA. 2008;105(50):19893–19897. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw AT, Yeap BY, Solomon BJ, et al. Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. Lancet Oncol. 2011;12(11):1004–1012. doi: 10.1016/S1470-2045(11)70232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49• •.Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363(18):1693–1703. doi: 10.1056/NEJMoa1006448. The impressive results from this study led to the accelerated US FDA approval of crizotinib in patients with ALK-positive NSCLC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaw A, Mehra R, Kim DW, et al. Clinical activity of the ALK inhibitor LDK378 in advanced, ALK-positive NSCLC. J Clin Oncol. 2013;31(Suppl):Abstract 8010. [Google Scholar]
- 51.Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res. 2013;19(15):4040–4045. doi: 10.1158/1078-0432.CCR-12-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Charest A, Wilker EW, McLaughlin ME, et al. ROS fusion tyrosine kinases activates a SH2 domain containing phosphatase-2/phosphatidylinositol 3-kinase/mammalian target of rapamycin signaling axis to form glioblastoma in mice. Cancer Res. 2008;68(15):3389–3395. doi: 10.1158/0008-5472.CAN-06-1193. [DOI] [PubMed] [Google Scholar]
- 53.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 54.Takeuchi K, Soda M, Togashi Y, et al. Ret, Ros1 and ALK fusions in lung cancer. Nat Med. 2012;18(3):378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 55.Bergethon K, Shaw AT, Ignatius SH, et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol. 2012;30(8):863–870. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lovly CM, Heuckmann JM, de Stanchina E, et al. Insights into ALK-driven cancers revealed through development of novel ALK tyrosine kinase inhibitors. Cancer Res. 2011;71(14):4920–4931. doi: 10.1158/0008-5472.CAN-10-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ou SH, Camidge DR, Engelman J, et al. Clinical activity of crizotinib in patients with advanced non-small cell lung cancer (NSCLC) harboring ROS1 gene rearrangement. Ann Oncol. 2012;23(Suppl 9):ix389–ix399. [Google Scholar]
- 58.Kohno T, Tsuta K, Tsuchihara K, et al. RET fusion gene: translation to personalized lung cancer therapy. Cancer Sci. 2013;104(11):1396–1400. doi: 10.1111/cas.12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeuchi K, Soda M, Togashi Y, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18(3):378–381. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 60.Lipson D, Capelletti M, Yelensky R, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18(3):382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18(3):375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Borrello MG, Ardini E, Locati LD, et al. RET inhibition, implications for cancer therapy. Expert Opin Ther Targets. 2013;17(4):403–419. doi: 10.1517/14728222.2013.758715. [DOI] [PubMed] [Google Scholar]
- 63.Drilon A, Wang L, Hasanovic A, et al. Response to cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013;3(6):630–635. doi: 10.1158/2159-8290.CD-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Su Z, Dias-Santagata D, Duke M, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn. 2011;13(1):74–84. doi: 10.1016/j.jmoldx.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol. 2013;8(7):823–859. doi: 10.1097/JTO.0b013e318290868f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pikor LA, Ramnarine VR, Lam WL, et al. Genetic alterations defining NSCLC subtypes and their therapeutic implications. Lung Cancer. 2013;82(2):179–189. doi: 10.1016/j.lungcan.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 67.Johnson BE, Kris MG, Berry LD, et al. A multicenter effort to identify driver mutations and employ targeted therapy in patients with lung adenocarcinomas: the Lung Cancer Mutation Consortium (LCMC) J Clin Oncol. 2013;31(Suppl):Abstract 8019. [Google Scholar]
- 68• •.Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3(111):111ra121. doi: 10.1126/scitranslmed.3003161. Exposes the unique challenges that hinder the implementation of whole-genome sequencing platforms into clinical practice. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sequest LV, Waitman BA, Dias-Santagata D, et al. Genotypic and histologic evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bedard PL, Hansen AR, Ratain MJ, et al. Tumor heterogeneity in the clinic. Nature. 2013;501(7467):355–364. doi: 10.1038/nature12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yadav VK, De S. An assessment of computational methods for estimating purity and clonality using genomic data derived from heterogenous tumor tissue samples. Brief Bioinform. 2014 doi: 10.1093/bib/bbu002. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ulahannan D, Kovac MB, Mulholland PJ, et al. Technical and implementation issues in using next-generation sequencing of cancer in clinical practice. Br J Cancer. 2013;109(4):827–835. doi: 10.1038/bjc.2013.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He L, Long LR, Antani S, et al. Histology image analysis for carcinoma detection and grading. Comput Methods Programs Biomed. 2012;107(3):538–556. doi: 10.1016/j.cmpb.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]