Abstract
The elongation factors of Saccharomyces cerevisiae are extensively methylated, containing a total of ten methyllysine residues. Elongation factor methyltransferases (Efm1, Efm2, Efm3, and Efm4) catalyze at least four of these modifications. Here we report the identification of a new type of protein lysine methyltransferase, Efm5 (Ygr001c), which was initially classified as N6-adenine DNA methyltransferase-like. Efm5 is required for trimethylation of Lys-79 on EF1A. We directly show the loss of this modification in efm5Δ strains by both mass spectrometry and amino acid analysis. Close homologs of Efm5 are found in vertebrates, invertebrates, and plants, although some fungal species apparently lack this enzyme. This suggests possible unique functions of this modification in S. cerevisiae and higher eukaryotes. The misannotation of Efm5 was due to the presence of a DPPF sequence in post-Motif II, typically associated with DNA methylation. Further analysis of this motif and others like it demonstrates a potential consensus sequence for N-methyltransferases.
Keywords: S-adenosylmethionine (AdoMet), protein lysine methylation, elongation factor, methyltransferase, posttranslational modification
1. Introduction
Elongation factor 1A (EF1A) acts as a molecular spell checker during protein translation by ensuring correct codon matches between the incoming amino acylated-tRNA and the mRNA [1]. This highly conserved protein guides charged tRNAs into the ribosomal A-site; a correct codon-anticodon match results in hydrolysis of GTP and release of the amino acyl-tRNA to allow for peptidyl transfer. Intriguingly, Saccharomyces cerevisiae EF1A contains five methylated lysine residues that are near critical areas of the enzyme [2,3]. Monomethyllysine 30 is found in the GTP binding pocket, trimethyllysine 79 is near the nucleotide exchange factor binding pocket and potentially contacts ribosomal components, and dimethyllysine 316, monomethyllysine 390, and the C-terminal lysine alpha-carboxyl methyl ester are located along the tRNA and ribosomal binding interface. Methylation modifications are also abundant in other components of the translational apparatus, including mRNA, tRNA, ribosomal proteins, and release factors [4,5,6,7]. Methyl groups have the potential to block or increase intra- and intermolecular contacts between the translational components [5]. These carefully placed modifications may help EF1A mediate its various interactions with tRNA, other elongation factors, and the ribosome itself.
S. cerevisiae has 86 known and predicted methyltransferases that modify a wide variety of macromolecules including proteins, nucleotides, lipids, and small molecules [18]. Generally, these enzymes fall into three major classes based on their structural folds and binding mechanism of the methyl donor, S-adenosylmethionine (AdoMet): seven-beta-strand (Class I), SET domain, and SPOUT methyltransferases. The majority of SPOUT methyltransferases modify RNA [9,10,11]. The SET domain methyltransferases, such as elongation factor methyltransferase 1 (Efm1), are classical protein lysine modifiers [12,13,14]. Efm4 is an example of a seven-beta-strand methyltransferase, a class of enzymes that have a variety of substrate types [15]. A mass spectrometric screen of knockout strains of predicted protein methyltransferases yielded the identification of these two EF1A methyltransferases [16]. Efm1 and Efm4 monomethylate Lys-30 and dimethylate Lys-316, respectively [16,17]. The remaining EF1A methyltransferases were not found among the 37 SET domain and Class I enzymes tested in the initial screen, indicating that they may be unconventional protein lysine methyltransferases or that more than one enzyme is responsible for catalysis. We therefore broadened our selection of putative methyltransferases to include the minor classification families of methyltransferases [8,15,18].
The protein expressed from ORF YGR001C was initially classified as N6-adenine DNA methyltransferase-like based on primary sequence and predicted secondary structure [8]. This annotation is due to two deviations in the Ygr001c amino acid sequence from that of other Class I methyltransferases. First, Ygr001c lacks the canonical AdoMet binding Motif I sequence. Second, it contains a “DPPF” post-Motif II sequence primarily associated with adenine nucleotide methyltransferases [19,20,21]. However, a similar post-Motif II sequence of “(D/E)XX(Y/F)” has recently been indicated as a hallmark of certain protein lysine methyltransferases [22,23].
Here we show that the protein encoded by YGR001C, which we now refer to as EFM5, is required for the trimethylation of lysine 79 on EF1A. Deletion of EFM5 leads to complete loss of trimethyllysine in 50 kDa polypeptides, specifically on the Lys-79-containing peptide of EF1A. Bioinformatic analysis reveals that Efm5 is not as widely conserved as is its methylation site, possibly indicating a role in only a subset of eukaryotes. Lastly, structural analyses of various (D/E)XX(Y/F) post-Motif II sequences indicate that this motif may generally be associated with N-methylation and structurally nearby active site residues may help select the methyl-accepting molecule.
2. Methods and Materials
2.1 In vivo radiolabeling and amino acid analysis
Strains used in this study are listed in Table 1. Yeast culture, radiolabeling, and amino acid analysis were carried out as previously described with some minor changes [22]. After lysates from [3H]AdoMet in vivo labeled wildtype and EFM knockout cells were resolved by SDS-PAGE, the Coomassie-stained protein band running just above the 45 kDa marker was excised and subjected to acid hydrolysis as described [22]. Hydrolysates were loaded onto a cation-exchange column (Beckman AA-15 sulfonated polystyrene resin, 0.9 cm inner diameter by 12 cm height) equilibrated with running buffer (sodium citrate, 0.3 M Na+, pH 3.85) at 55 °C. Amino acids were eluted in the equilibration buffer at 1 mL/min while collecting 1 min fractions at the expected elution position of the methyllysine standards. Standards and [3H]-methylated amino acids detected as previously described [22].
Table 1.
Strains used in this study
| Strain | Genotype | Biological function | Source |
|---|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Wild type | Open Biosystems |
| BY4742 | MATalpha his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 | Wild type | Open Biosystems |
| efm1Δ a | BY4741 background, yhl039wΔ | Elongation factor methyltransferase | Open Biosystems |
| efm1Δ α | BY4742 background, yhl039wΔ | Elongation factor methyltransferase | Open Biosystems |
| efm4Δ a | BY4741 background, yil064wΔ | Elongation factor methyltransferase | Open Biosystems |
| efm4Δ α | BY4742 background, yil064wΔ | Elongation factor methyltransferase | Open Biosystems |
| efm5Δ a | BY4741 background, ygr001cΔ | Putative/Elongation factor methyltransferase | Open Biosystems |
| efm5Δ α | BY4742 background, ygr001cΔ | Putative/Elongation factor methyltransferase | Open Biosystems |
2.2 In-gel trypsin digestion and mass spectrometry
Coomassie-stained gel slices from the 50 kDa region of fractionated polypeptides of unlabeled yeast cell lysates were destained and subjected to in-gel trypsin digestion with sequencing grade trypsin (Promega, V5111) as previously described [22]. Tryptic peptides from the 50 kDa SDS-gel band of wildtype, efm1Δ, efm4Δ and efm5Δ lysates were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) with a Q-Exactive Orbitrap mass spectrometer (Thermo Scientific, Waltham, MA) as previously described [22]. Peptides detected by LC-MS/MS were identified using Proteome Discoverer (Version 1.4; Thermo Scientific, Waltham, MA) coupled with MASCOT (Version 2.4.1; Matrix Science, London, UK). Tryptic peptides were searched against the SwissProt Saccharomyces cerevisiae database (2013; 7,798 sequences) with the following settings: missed cleavages ≤ 1, precursor mass tolerance = 10 ppm, product ion mass tolerance = 0.005 Da, dynamic modifications for carbamidomethyl (C), deamidation (N, Q), oxidation (M), monomethyl (K), dimethyl (K), and trimethyl (K). Peptides identified with methylations at K30, K79, K316 (along with their un-methylated counterparts) were inputted into an inclusion list and wildtype, efm1Δ, efm4Δ, efm5Δ samples reanalyzed by targeted parallel-reaction-monitoring mass spectrometry (PRM-MS) to confirm the methylation status of EFM mutants. Full-scan MS2 data was used to confirm the identity of each observed peptide.
2.3 Bioinformatic alignments and phylogenetic tree construction
Whole protein sequences for EF1A were aligned using Clustal Omega and the region containing Lys-79 was examined. A protein-protein BLAST search was performed using S. cerevisiae Efm5 (UniProt P53200) as the query. Sequences were aligned using MUltiple Sequence Comparison by Log-Expectation (MUSCLE) in MEGA 6 as previously described [22].
2.4 Phyre2 and post-Motif II analysis
The structure of Efm5 was modeled using the Protein Homology/analogY Recognition Engine V2.0 (Phyre2) in one-to-one threading mode with Mtq2 (PDB: 3Q87, chain B) as a template. Coordinates for the model were calculated using the global alignment method with default settings for secondary structure scoring and weight. Structural figures of the catalytic regions of this Phyre2 model as well as crystal structures of related methyltransferases were created using MacPyMol (DeLano Scientific). The crystal structure for VCP-KMT (PDB: 4LG1, chain B) contained two rotamers of Ile-146. As the electron density and difference maps for this structure were unavailable, the rotamer with the most steric hindrance (as observed in PyMol) was removed.
3. Results
3.1 YGR001C/EFM5 is required for the presence of trimethyllysine on Lys-79 of EF1A
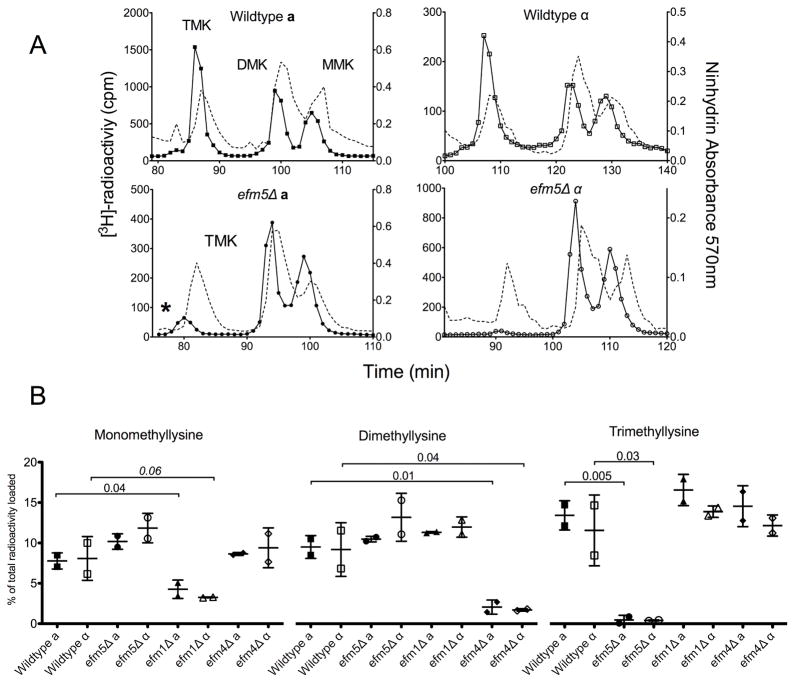
To identify the remaining EF1A methyltransferases, yeast cells from knockout strains of candidate methyltransferases were in vivo radiolabeled with [3H]AdoMet. By supplementing cultures with [3H]AdoMet, substrates methylated during incubation incorporate tritiated methyl groups. After lysate fractionation by SDS-PAGE, the 50 kDa gel slice, corresponding to the molecular weight of EF1A, was excised and acid hydrolyzed. The resulting hydrolysates were loaded onto a high-resolution cation exchange column capable of separating methyl derivatives of the amino acids. Wildtype [3H]-hydrolysates from both mating type backgrounds contained significant levels of [3H]-trimethyllysine, [3H]-dimethyllysine, and [3H]-monomethyllysine (Figure 1A, top panels). Relative quantification of the modified residues demonstrated a radioactivity ratio of MMK:DMK:TMK as 2:2:3, indicating the presence of two monomethyllysines, one dimethyllysine, and one trimethyllysine (Fig. 1B), consistent with previous reports of EF1A methylation [2].
Figure 1. Deletion of EFM5 results in loss of trimethylated lysine in 50 kDa polypeptides.
(A) Radiolabelled methylated lysine derivatives from 50 kDa gel slice hydrolysates were separated. The position of the standards, detected by ninhydrin reactivity, is shown in the dashed line and the methylated lysine peaks are labeled (MMK, DMK, TMK). Due to a tritium isotope effect, the radiolabeled derivatives, shown in the solid lines, elute slightly before the non-labeled standards [32]. Separation of hydrolysates from radiolabeled wildtype and efm5Δ from both mating type backgrounds are shown. Each trace is representative of at least three independent experiments. The asterisk indicates the presence of radioactive 3-methylhistidine. (B) The amount of mono-, di-, and trimethyllysine radioactivity as a percentage of the total radioactivity in the hydrolysate is shown with error bars reflecting the standard deviation. P-values from Student’s t-test are shown. All changes from respective wildtype values were significant with the exception of efm1Δα, indicated by the italicized value, which is likely due to large standard deviation in the wildtype.
To confirm that the radioactive methylated amino acids were derived from EF1A, knockouts of known EF1A methyltransferases were analyzed. Deletion of Efm1 resulted in a 50% reduction in MMK and deletion of Efm4 resulted in almost complete loss of DMK, consistent with their reported activities (Fig. S1 and Fig. 1B) [16]. A complete loss of the TMK radioactive peak was observed only in a deletion strain of YGR001C (Fig 1A, bottom panels). This was seen in both mating type backgrounds, which indicates that this is not due to a secondary mutation in the yeast genome. These results suggest that YGR001C encodes a protein lysine methyltransferase that catalyzes the trimethylation of EF1A. In some experiments, there was residual radioactivity eluting slightly before the trimethyllysine peak (Fig. 1A, asterisk). This peak correlates to 3-methylhistidine and is likely from the closely migrating Rpl3 polypeptide (molecular weight 43.8 kDa) [4,24]. This peak was not present in all runs due to variations in gel slicing that would include or exclude different amounts of Rpl3.
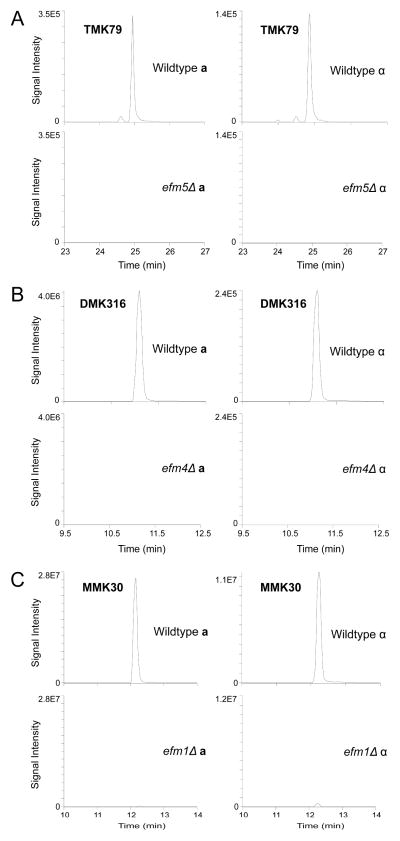
To confirm that Ygr001c acts on the elongation factor, the 50 kDa region on a gel containing non-radioactive lysates was subjected to in-gel trypsin digestion, and the resulting peptides were analyzed by LC-MS/MS. Extracted ion chromatograms demonstrated the presence of all previously reported EF1A side chain lysine methylations (Fig. 2 and Fig. S2) and loss of respective methylations in efm1Δ and efm4Δ strains (Fig. 2B and 2C). Increases in the abundance of corresponding unmethylated peptides were also observed (data not shown). This experiment also demonstrated that Lys-79 is found both in di- and trimethylated states in wildtype cells, consistent with the presence of low levels of [3H]-dimethyllysine in efm4Δ hydrolysates (Fig S1). Importantly, methylated Lys-79 was undetectable in ygr001cΔ (Fig. 2A). We thus designate Ygr001c as Efm5, the fifth known elongation factor methyltransferase in yeast.
Figure 2. Deletion of EFM5 results in loss of trimethyllysine 79 on elongation factor 1A.
The 50 kDa protein bands from wildtype and knockout lysates were excised, in-gel trypsin digested, and analyzed by LC-MS/MS. Extracted ion chromatograms correlating to the EF1A methylated peptides (precursor → product ion transition) from wildtype and each EFM knockout strain are shown. MS/MS data of individual peptides is shown in Supplemental Figure S2. (A) Loss of EF1A trimethylation on peptide GITIDIALWKtrimethylFETPK in efm5Δ was visualized by loss of the 592.010 m/z → 791.466 m/z transition. (B) Loss of EF1A dimethylation on NVSVKdimethylEIR in efm4Δ was visualized by loss of the 324.866 m/z → 573.371 m/z transition. (C) Loss of EF1A monomethylation on peptide STTTGHLIYKmonomethyl in efm1Δ was visualized by loss of the 567.811 m/z → 744.440 m/z transition.
3.2 Evolutionary conservation of Efm5 and the conservation of the EF1A Lys-79 methylation site
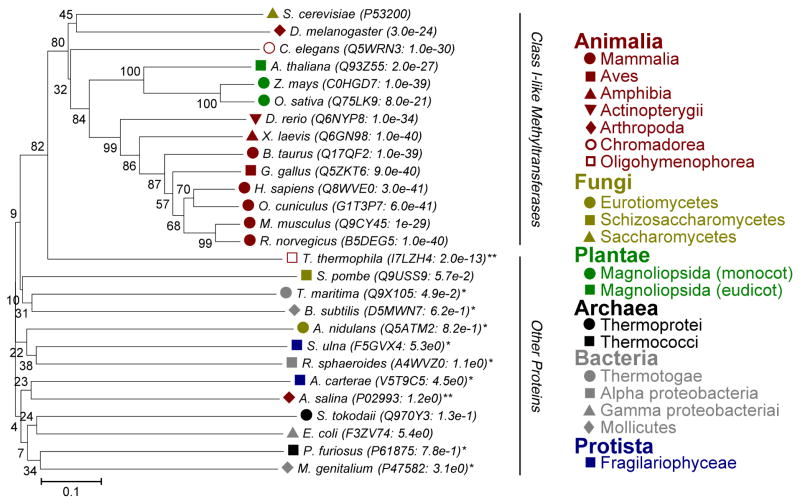
Trimethylation of Lys-79 has been previously confirmed in other eukaryotes, including rabbits and brine shrimp [25]. Alignment of EF1A sequences from every kingdom indicates high conservation of this lysine residue, its flanking aromatic residues, and the surrounding sequence amongst eukaryotes (Fig. S3). In the S. cerevisiae EF1A structure, the W-K-F sequence is located on a small beta-strand; the aromatic residues face the interior of the protein forcing the lysine side chain to face the solvent (PDB: 1F60). The significant similarity of this site suggests EF1A is methylated in higher eukaryotes, including humans. If EF1A from other organisms contain this modification, they should also have a homologous methyltransferase. A BLAST search was thus performed to identify potential homologs of Efm5 and a phylogenetic tree was constructed to visually demonstrate the conservation (Fig. 3). Interestingly, Efm5 is well conserved in higher eukaryotes (vertebrates, invertebrates, and plants), but absent in at least two of S. cerevisiae’s fungal relatives (S. pombe and A. nidulans). This result indicates that TMK79 may not be essential for EF1A’s translational functions but perhaps has more specialized roles in specific organisms. Notably, the brine shrimp A. salina did not appear to have an Efm5 homolog despite the presence of manually confirmed TMK79. However, the genome of this arthropod has not been fully sequenced to date.
Figure 3. Efm5 is conserved amongst higher eukaryotes.
Phylogenetic tree of Efm5 homologs. Accession numbers and E-values are indicated for each protein. All homologs were mutual best hits except those indicated by an asterisk (*). Double asterisk (**) indicates the organism has the methylation site but not the enzyme potentially due to incomplete genome sequences. Proteins with high similarity are all predicted methyltransferases; those with low similarities are categorized as other proteins.
3.3 A closer look at post-Motif II residues reveals similarities between N-methyltransferases
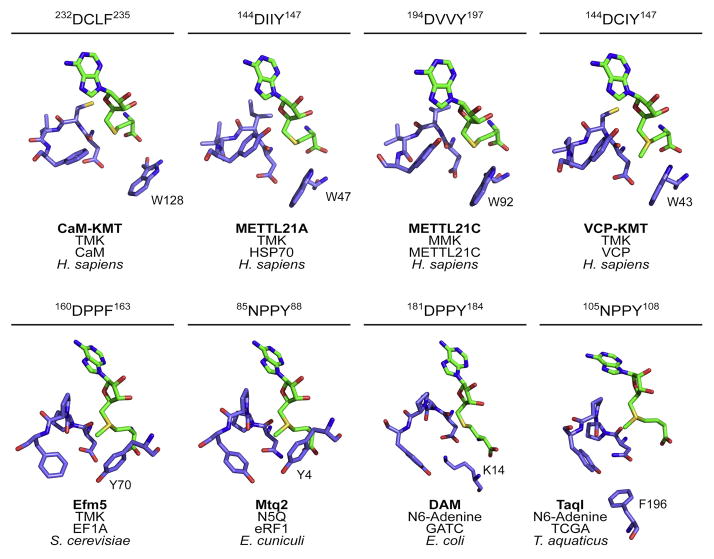
The apparent misannotation of Efm5 as an adenine methyltransferase due to its DPPF post-Motif II raised questions as to how similar active site amino acids could give rise to very different substrate preferences. We compared the structural location and arrangement of post-Motif II residues from multiple lysine methyltransferases, a DNA methyltransferase, two glutamine methyltransferases and the predicted structure of Efm5 (Fig. 4).
Figure 4. Post-Motif II sequences reveal structural similarities of N-methyltransferases.
The post-Motif II region from the crystal structures of CaM-KMT (PDB: 4PWY), METLL21A (PDB: 4LEC), METTL21C (PDB: 4MTL), METTL21D (PDB: 4LG1), Mtq2 (PDB: 3Q87), DAM (PDB: 2G1P), TaqI (PDB: 2ADM) and that of a model of Efm5 (Phyre2) were compared. Above each structure is the post-Motif II sequence and residue numbers. Below each structure is the enzyme name, the type of methylation, the substrate, and the biological species. AdoMet or AdoHcy is shown in green and the enzyme residues are in purple.
CAM-KMT, METTL21A, METTL21C, and METTL21D (VCP-KMT), members of the human Family 16 protein lysine methyltransferases, each contain the DXX(Y/F) post-Motif II where the variable cysteine or aliphatic residues face away from the methyl-accepting substrate site (Fig. 4, top row). Orientation of these residues positions the D and (Y/F) towards the substrate. It has been hypothesized that the aspartate residue helps balance the positive charge on the substrate lysine and the aromatic residue stabilizes the substrate via cation-π interactions [22]. Additionally, each enzyme contains a tryptophan residue, distant in sequence but close in structure, which is relatively planar to the motif tyrosine/phenylalanine. This large aromatic residue could help keep the positive lysine in place for methylation by forming more cation-π interactions on the other side of the lysine and creating a hydrophobic cage [22,26,27].
The fungal protein glutamine and protein lysine methyltransferases Mtq2 and Efm5 have post-Motif II sequences NPPY and DPPF, respectively (Fig. 4). The double proline hinders the rotational freedom of the protein backbone, limiting the relative conformations of the polar and aromatic sidechains found in this motif, maintaining the spatial relationship of the N/D and Y/F residues for substrate recognition. Additionally, these two enzymes lack the tryptophan residue seen in the human lysine methyltransferases but have a tyrosine residue in a similar position. A major difference between Mtq2 and Efm5 is the replacement of asparagine with aspartate. Asparagine would provide the polar contacts to stabilize a substrate glutamine whereas the aspartate would provide a formal negative charge to balance out a formal positive charge on the substrate lysine. This change from Asp to Asn is found in other glutamine methyltransferases such as the E. coli enzyme HemK.
The DNA methyltransferases, DAM and TaqI, contain the classical (D/N)PP(Y/F) motif associated with adenine modifiers, although there is more variability in the terminal residues of the motif [19]. TaqI contains a post-Motif II asparagine while DAM has an aspartate. As with Mtq2, a formal negative charge would not be required for stabilizing an adenine and therefore asparagine should suffice in the first position. Interestingly, the biggest deviation of DAM from the lysine modifiers is the absence of the sequentially distant aromatic residue. In its place is a lysine residue that would present a formal positive charge near the active site. TaqI, on the other hand, has a nearby phenylalanine. These nearby residues could have more involvement with recognition of the specific DNA sequence than that of the substrate nitrogen atom.
The importance of the proline-proline motif has been widely studied in the DNA methyltransferases, likely adding to the association of this motif with DNA methylation. The proline backbone of the NPPY motif in TaqI participates in substrate stabilization directly through hydrogen bonding between its backbone carbonyl and substrate adenine [28]. It also confers rigidity in the backbone of this motif, helping to position the motif tyrosine for face-to-face π-stacking interactions with the same adenine. P172A and P172T mutations of phage T4 N6-adenine specific DNA methyltransferase resulted in a 5 to 20-fold increase in Km for AdoMet binding and a 2 to 4-fold decrease in Kcat [20]. This indicates that the rigidity of the (D/N)PP(Y/F) motif may be important for DNA methylation. This requirement seems less important in some of the Class I protein lysine methyltransferases, which feature a conserved smaller hydrophobic residues instead of prolines.
Taken together, these analyses indicate that post-Motif II residues may dictate the atom methylated by N-methyltransferases. Variations of the motif residues in conjunction with nearby active site residues can direct enzyme preference towards adenine, glutamine, or lysine.
Discussion
We have identified a protein encoded by the open reading frame YGR001C that is required for the trimethylation of EF1A. We now designate this protein as elongation factor methyltransferase 5. Deletion of EFM5 led to complete loss of trimethyllysine in EF1A. Mass spectrometry revealed that trimethylated Lys-79 residue on EF1A is undetectable in the absence of Efm5.
The previous annotation of Efm5 as N-6 adenine DNA methyltransferase-like was based on its amino acid similarity to DNA modifying enzymes containing similar post-Motif II residues. This annotation has now been propagated for its homologs in other organisms. Our finding here that Efm5 is a protein lysine methyltransferase adds another example of the difficulty predicting substrate specificity from sequence alone. SPOUT methyltransferases were considered to exclusively have RNA substrates until one member (Sfm1) was shown to generate an omega-monomethylarginine residue on ribosomal protein Rps3 [29]. Additionally, the Mtq1 and Mtq2 protein glutamine methyltransferases were also originally annotated as DNA adenine methyltransferases. Interestingly, Mtq1 and Mtq2 contain a similar post-Motif II (NPPY) as Efm5 (DPPF). In both cases, the unusual proline-proline sequence likely contributed to their misannotation. There may be additional methyltransferases with unusual post-Motif II sequences with novel substrate specificity.
Closer examination of post-Motif II-containing methyltransferases and their respective modifications reveals commonalities that could be linked to N-methylation (Fig. 4). Comparison of adenine, glutamine, and lysine methyltransferase sequences and structures demonstrated the presence of a (D/N)XX(Y/F) motif surrounding the catalytic center. We previously predicted this motif to be associated with lysine methylation. Now that it is clear this motif extends beyond protein lysine methyltransferases, we have amended our prediction to associate this with recognition of nitrogen atoms regardless of macromolecule type. The (D/N)PP(Y/F) motif associated with DNA methylation is found in prokaryotes and eukaryotes while the (D/N)XX(Y/F) motif described above seems restricted to eukaryotes alone. This may suggest an evolutionary lineage where eukaryotes took advantage of mutations of the double proline to repurpose the enzymes for protein modification.
The remaining EF1A methyltransferase that monomethylates Lys-390 is still unaccounted for and the functional relevance of elongation factor methylation remains largely unexplored. Recent work on the elongation factor 2 methyltransferases, Efm2 and Efm3, revealed that these methylations are important for proper protein synthesis [22,30,31]. Investigation into EF1A methylation could reveal importance to translational function. Alternatively, EF1A has been implicated in various non-canonical functions such as nuclear export of tRNAs, actin binding, and detection of misfolded proteins [1]. Since EF1A methylation is not completely conserved in all eukaryotes, it is possible that these modifications assist in these accessory functions.
Supplementary Material
Highlights.
Yeast YGR001C was predicted as an adenine-specific DNA methyltransferase
Deletion of YGR001C results in loss of trimethyllysine-79 on elongation factor 1A
We now designate this protein elongation factor methyltransferase 5 (Efm5)
Efm5 is widely conserved in higher eukaryotes
Efm5 has a unique post-Motif II sequence common to N-methyltransferases
Acknowledgments
This work was supported, in whole or in part, by National Institutes of Health Grants GM026020 (to S.G.C.) and GM007185, a Ruth L Kirschstein National Research Service Award (to M.C.D.). K.J.T. was supported by a UCLA Department of Chemistry and Biochemistry Alumni Undergraduate Summer Research Fellowship, a UCLA College Honors Summer Research Fellowship and a MacDowell Senior Undergraduate Research Scholarship.
GLOSSARY
- EFM
elongation factor methyltransferase
- AdoMet
S-adenosylmethionine
- [3H]AdoMet
S-adenosyl-L-[methyl-3H]-methionine
- MMK
monomethyllysine
- DMK
dimethyllysine
- TMK
trimethyllysine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mateyak MK, Kinzy TG. eEF1A: thinking outside the ribosome. J Biol Chem. 2010;285:21209–21213. doi: 10.1074/jbc.R110.113795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavallius J, Zoll W, Chakraburtty K, Merrick WC. Characterization of yeast EF-1 alpha: non-conservation of post-translational modifications. Biochim Biophys Acta. 1993;1163:75–80. doi: 10.1016/0167-4838(93)90281-u. [DOI] [PubMed] [Google Scholar]
- 3.Zobel-Thropp P, Yang MC, Machado L, Clarke S. A novel post-translational modification of yeast elongation factor 1A. Methylesterification at the C terminus. J Biol Chem. 2000;275:37150–37158. doi: 10.1074/jbc.M001005200. [DOI] [PubMed] [Google Scholar]
- 4.Al-Hadid Q, Roy K, Munroe W, Dzialo MC, Chanfreau GF, Clarke SG. Histidine Methylation of Yeast Ribosomal Protein Rpl3p Is Required for Proper 60S Subunit Assembly. Mol Cell Biol. 2014;34:2903–2916. doi: 10.1128/MCB.01634-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke SG. Protein methylation at the surface and buried deep: thinking outside the histone box. Trends Biochem Sci. 2013;38:243–252. doi: 10.1016/j.tibs.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graille M, Figaro S, Kervestin S, Buckingham RH, Liger D, Heurgue-Hamard V. Methylation of class I translation termination factors: structural and functional aspects. Biochimie. 2012;94:1533–1543. doi: 10.1016/j.biochi.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Polevoda B, Sherman F. Methylation of proteins involved in translation. Mol Microbiol. 2007;65:590–606. doi: 10.1111/j.1365-2958.2007.05831.x. [DOI] [PubMed] [Google Scholar]
- 8.Petrossian TC, Clarke SG. Multiple Motif Scanning to identify methyltransferases from the yeast proteome. Mol Cell Proteomics. 2009;8:1516–1526. doi: 10.1074/mcp.M900025-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szczepinska T, Kutner J, Kopczynski M, Pawlowski K, Dziembowski A, Kudlicki A, Ginalski K, Rowicka M. Probabilistic approach to predicting substrate specificity of methyltransferases. PLoS Comput Biol. 2014;10:e1003514. doi: 10.1371/journal.pcbi.1003514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shao Z, Yan W, Peng J, Zuo X, Zou Y, Li F, Gong D, Ma R, Wu J, Shi Y, Zhang Z, Teng M, Li X, Gong Q. Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate. Nucleic Acids Res. 2014;42:509–525. doi: 10.1093/nar/gkt869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas SR, Keller CA, Szyk A, Cannon JR, Laronde-Leblanc NA. Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic Acids Res. 2011;39:2445–2457. doi: 10.1093/nar/gkq1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Rizzo PA, Trievel RC. Substrate and product specificities of SET domain methyltransferases. Epigenetics. 2011;6:1059–1067. doi: 10.4161/epi.6.9.16069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang Y, Levy D, Horton JR, Peng J, Zhang X, Gozani O, Cheng X. Structural basis of SETD6-mediated regulation of the NF-kB network via methyl-lysine signaling. Nucleic Acids Res. 2011;39:6380–6389. doi: 10.1093/nar/gkr256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wlodarski T, Kutner J, Towpik J, Knizewski L, Rychlewski L, Kudlicki A, Rowicka M, Dziembowski A, Ginalski K. Comprehensive structural and substrate specificity classification of the Saccharomyces cerevisiae methyltransferome. PLoS One. 2011;6:e23168. doi: 10.1371/journal.pone.0023168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lipson RS, Webb KJ, Clarke SG. Two novel methyltransferases acting upon eukaryotic elongation factor 1A in Saccharomyces cerevisiae. Arch Biochem Biophys. 2010;500:137–143. doi: 10.1016/j.abb.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couttas TA, Raftery MJ, Padula MP, Herbert BR, Wilkins MR. Methylation of translation-associated proteins in Saccharomyces cerevisiae: Identification of methylated lysines and their methyltransferases. Proteomics. 2012;12:960–972. doi: 10.1002/pmic.201100570. [DOI] [PubMed] [Google Scholar]
- 18.Petrossian T, Clarke S. Bioinformatic Identification of Novel Methyltransferases. Epigenomics. 2009;1:163–175. doi: 10.2217/epi.09.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guschlbauer W. The DNA and S-adenosylmethionine-binding regions of EcoDam and related methyltransferases. Gene. 1988;74:211–214. doi: 10.1016/0378-1119(88)90289-2. [DOI] [PubMed] [Google Scholar]
- 20.Kossykh VG, Schlagman SL, Hattman S. Conserved sequence motif DPPY in region IV of the phage T4 Dam DNA-[N-adenine]-methyltransferase is important for S-adenosyl-L-methionine binding. Nucleic Acids Res. 1993;21:3563–3566. doi: 10.1093/nar/21.15.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas CB, Scavetta RD, Gumport RI, Churchill ME. Structures of liganded and unliganded RsrI N6-adenine DNA methyltransferase: a distinct orientation for active cofactor binding. J Biol Chem. 2003;278:26094–26101. doi: 10.1074/jbc.M303751200. [DOI] [PubMed] [Google Scholar]
- 22.Dzialo MC, Travaglini KJ, Shen S, Roy K, Chanfreau GF, Loo JA, Clarke SG. Translational Roles of Elongation Factor 2 Protein Lysine Methylation. J Biol Chem. 2014 doi: 10.1074/jbc.M114.605527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kernstock S, Davydova E, Jakobsson M, Moen A, Pettersen S, Maelandsmo GM, Egge-Jacobsen W, Falnes PO. Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat Commun. 2012;3:1038. doi: 10.1038/ncomms2041. [DOI] [PubMed] [Google Scholar]
- 24.Webb KJ, Zurita-Lopez CI, Al-Hadid Q, Laganowsky A, Young BD, Lipson RS, Souda P, Faull KF, Whitelegge JP, Clarke SG. A novel 3-methylhistidine modification of yeast ribosomal protein Rpl3 is dependent upon the YIL110W methyltransferase. J Biol Chem. 2010;285:37598–37606. doi: 10.1074/jbc.M110.170787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amons R, Muranova TA, Rykunova AI, Eliseikina IA, Sedelnikova SE. The complete primary structure of ribosomal protein L1 from Thermus thermophilus. J Protein Chem. 1993;12:725–734. doi: 10.1007/BF01024930. [DOI] [PubMed] [Google Scholar]
- 26.van Ingen H, van Schaik FM, Wienk H, Ballering J, Rehmann H, Dechesne AC, Kruijzer JA, Liskamp RM, Timmers HT, Boelens R. Structural insight into the recognition of the H3K4me3 mark by the TFIID subunit TAF3. Structure. 2008;16:1245–1256. doi: 10.1016/j.str.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 27.Roy S, Musselman CA, Kachirskaia I, Hayashi R, Glass KC, Nix JC, Gozani O, Appella E, Kutateladze TG. Structural insight into p53 recognition by the 53BP1 tandem Tudor domain. J Mol Biol. 2010;398:489–496. doi: 10.1016/j.jmb.2010.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goedecke K, Pignot M, Goody RS, Scheidig AJ, Weinhold E. Structure of the N6-adenine DNA methyltransferase M.TaqI in complex with DNA and a cofactor analog. Nat Struct Biol. 2001;8:121–125. doi: 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- 29.Young BD, Weiss DI, Zurita-Lopez CI, Webb KJ, Clarke SG, McBride AE. Identification of methylated proteins in the yeast small ribosomal subunit: a role for SPOUT methyltransferases in protein arginine methylation. Biochemistry. 2012;51:5091–5104. doi: 10.1021/bi300186g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang L, Hamey JJ, Hart-Smith G, Erce MA, Wilkins MR. Elongation factor methyltransferase 3 - A novel eukaryotic lysine methyltransferase. Biochem Biophys Res Commun. 2014;451:229–234. doi: 10.1016/j.bbrc.2014.07.110. [DOI] [PubMed] [Google Scholar]
- 31.Davydova E, Ho AY, Malecki J, Moen A, Enserink JM, Jakobsson ME, Loenarz C, Falnes PO. Identification and characterization of a novel evolutionarily conserved lysine-specific methyltransferase targeting eukaryotic translation elongation factor 2 (eEF2) J Biol Chem. 2014 doi: 10.1074/jbc.M114.601658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG. Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming omega-NG-monomethylated arginine residues. J Biol Chem. 2012;287:7859–7870. doi: 10.1074/jbc.M111.336271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.