Abstract
The epithelial lining of the respiratory system originates from a small group of progenitor cells in the ventral foregut endoderm of the early embryo. Research in the last decade has revealed a number of paracrine signaling pathways that are critical for the development of these respiratory progenitors. In the post genomic era the challenge now is to figure out at the genome wide level how these different signaling pathways and their downstream transcription factors interact in a complex “gene regulatory network” (GRN) to orchestrate early lung development. In this prospective we review our growing understanding of the GRN governing lung specification. We discuss key gaps in our knowledge and describe emerging opportunities that will soon provide an unprecedented understanding of lung development and accelerate our ability to apply this knowledge to regenerative medicine.
Keywords: lung development, gene regulatory network, Nkx2.1, respiratory epithelium, specification
Introduction
The billions of epithelial cells lining your respiratory system including ciliated, secretory, neuroendocrine, and the alveolar type 1 and type 2 cells that absorb oxygen and produce surfactant, all originate from a few hundred respiratory progenitor cells located in the ventral foregut endoderm of the early embryo. The first evidence of respiratory lineage specification is the localized expression of the homeodomain transcription factor Nkx2.1 (also known as TTF-1) in these progenitor cells around 28 days of human gestation and at embryonic day (E) 9.0 in the mouse (Figure 1) (Lazzaro et al., 1991). By E10 in the mouse, the ventral Nkx2.1+ epithelial cells evaginate to form the lung buds and trachea, which eventually separate from the dorsal esophagus. As organogenesis proceeds the lung buds grow through a stereotypical branching morphogenesis as different epithelial subtypes are patterned along the proximal-distal axis of the fetal lung (Figure 1B). Finally in perinatal period the distinct epithelial cells types required for breathing and lung function differentiate (Herriges and Morrisey, 2014).
Figure 1. Development of Nkx2.1-expressing respiratory progenitors.
A) Three-dimensional confocal immunostaining of an E9.5 mouse embryo showing the Foxa2-expressing primitive gut tube (yellow) with Nkx2.1 expression (magenta) in the respiratory progenitors and thyroid primordial. The colored image of the gut tube is superimposed on the epifluorescent image of the embryo in gray scale.
B) A lineage diagram showing that the epithelial cells lining the trachea and lungs develops from respiratory progenitors in the ventral foregut of the early embryo. These respiratory progenitors arise from multipotent foregut endoderm cells that also contribute to the digestive system. Segregation of the epiblast into endoderm, mesoderm and ectoderm germ layers occurs during gastrulation. N.E.; neuroendocrine
Over the past decade intensive research has implicated five conserved signaling pathways that mediate paracrine signaling between the foregut endoderm and surrounding mesoderm to control respiratory cell fate specification in the early embryo: the Fibroblast Growth Factor (FGF), Wnt/β-catenin, Bone Morphogenetic Protein (BMP), Retinoic Acid (RA), and Hedgehog (HH) pathways (Figure 2). These same factors have additional roles during fetal lung growth, patterning, morphogenesis and differentiation, and increasing evidence indicates that these pathways are misregulated in lung disease and reactivated during lung repair (Beers and Morrisey, 2011; Whitsett et al., 2011). Knowledge of lung development in animal models has informed our understanding of human congenital malformations such as pulmonary dysplasia and tracheoesophageal fistula, and has led to strategies to differentiate respiratory epithelium-like tissue from human embryonic stem (ES) and induced pluripotent stem (iPS) cells (Wong and Rossant, 2013).
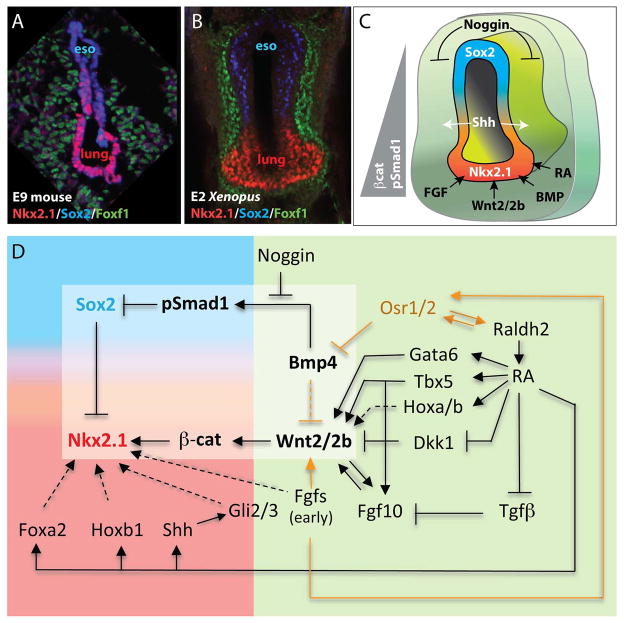
Figure 2. The GRN controlling respiratory specification.
A–B) Confocal immunostaining of a cross section through the foregut a E9 mouse embryo (A) and a two-day-old (E2) Xenopus embryo (B). Nkx2.1+ lung progenitors (red) are located in the ventral foregut, whereas Sox2+ esophagus progenitors (eso) are restricted to the dorsal foregut. The surrounding splanchnic mesenchyme expresses the transcription factor Foxf1 (green).
C) A diagram of the foregut showing the spatial arrangement of paracrine factors regulating respiratory specification and D–V patterning of the foregut. Differential Wnt and BMP signaling results in a gradient of β-catenin and pSmad1 activity in the epithelium, highest in the Nkx2.1+ ventral foregut and lowest in the Sox2+ dorsal foregut.
D) A diagram of the evolutionarily conserved GRN controlling specification of the respiratory progenitors. This model represents gene interactions data from the published literature combining results from mouse, chicken and Xenopus embryos. Orange lines indicate Xenopus data, which remain to be tested in other species. Dashed lines indicate relationships that are only weekly supported; mostly inferred from in vitro data. The epithelial compartment is indicated in the blue-red gradient, whereas gene expressed in the mesenchymal compartment is in the green area. The light box highlights the core Wnt/BMP regulatory cassette immediately upstream of foregut patterning and specification of the respiratory progenitors.
Despite tremendous progress we are just beginning to understand how these pathways interact in a “gene regulatory network” (GRN) to orchestrate respiratory system development. In this prospective we discuss what GRNs are and review our current understanding of the evolutionarily conserved GRN governing respiratory specification highlighting some important unresolved questions. Finally we describe how high throughput DNA sequencing, new genome editing techniques, and improved computational methods together with rapidly emerging stem cell differentiation approaches and alternative animal models will soon allow us to interrogate the GRN controlling lung development at an unprecedented genome-wide scale and with single cell mechanistic depth.
Gene Regulatory Networks: what are they and what are they good for?
A “gene regulatory network” is essentially a wiring diagram that represents how signaling pathways, transcription factors, and their target genes interact to orchestrate complex biological processes such as organ development (Figure 2D) (Bolouri, 2008; Peter et al., 2012). However GRNs are not static like the electric wiring diagram of your house, but rather reflect the temporally and spatially changing microenvironment during organogenesis. At first glance GRN diagrams can appear overwhelming in their complexity, but embedded within the web of interactions are simpler “sub-circuits” or “regulatory cassettes” such as feedback loops (that limit the activity of a pathway) or combinatorial reinforcing motifs (that lock a previously plastic cell state into a robust set gene expression pattern). The interconnectivity of the different sub-circuits describes the underlying regulatory logic and provides a systems-level view of how the GRN controls biological processes, which is not possible with single gene studies.
Various software programs such as Cytoscape and Biotapestry can process a large body of experimental data and generate a visualization of a GRN. Underpinning the GRN is the input data, the basic discovery of individual genes and their role in the system. Of critical importance are mechanistic experiments that define the epistatic relationship between genes and distinguish direct from indirect interactions to ultimately elucidate the DNA cis-regulatory modules or enhancers that transcription factors interact with to control gene expression. Importantly a GRN model is not just a visualization tool; if it is populated with sufficient data, it can have predictive power (Bolouri, 2008; Peter et al., 2012). Computational modeling of GRNs can predict how gene mutations or changes in timing of gene expression might impact subsequent development. For example GRNs describing endocrine pancreas development, have informed our understanding of type 1 diabetes, and influenced protocols to differentiate beta-cells in vitro (Arda et al., 2013).
Our understanding of the GRN governing respiratory specification is still in its infancy and is largely based on single gene studies in mice with limited analysis of downstream targets. However, increasingly sophisticated mouse genetics and the use of alternative models more amenable to experimental manipulations, such as Xenopus and ES/iPS cell cultures, are rapidly advancing our understanding of how signaling pathways and transcription factors are integrated into a respiratory specification GRN.
The GRN controlling lung specification: knowns and unknowns
The evolution of the vertebrate lung was an essential adaptation to terrestrial air breathing and it has become clear that a conserved genetic program controls lung specification in frogs, chickens, rodents, and humans. While it remains to be determined exactly how much of the molecular details are conserved between species, a comparative approach has proven to be useful as each model system has different experimental advantages. In the following section we describe what is known about the GRN governing lung specification (Figure 2D), comparing studies from different animal models and highlighting key outstanding questions.
Endoderm formation and anterior-posterior pattering
Conserved in all vertebrates, endoderm formation and early patterning begins at gastrulation (E6.5–7.5 in mice) when the secreted TGFβ family ligand Nodal induces definitive endoderm tissue (Zorn and Wells, 2009). As the naïve endoderm forms a primitive gut tube (E7.5–8.5 in mice), it is patterned along the anterior-posterior axis into broad foregut and hindgut domains (Figure 1B) by mesodermal FGF, Wnt, and BMP signals, which promote Cdx2+ hindgut fate and repress Sox2+ foregut fate (Zorn and Wells, 2009). By E8.5 the future respiratory progenitors, along with cells that give rise to the esophagus, thyroid, liver, stomach, and pancreas (Figure 1B), are located in the Sox2+ ventral foregut domain adjacent to the developing cardiac mesoderm. Specification of the respiratory progenitors is defined by the induction of Nkx2.1 expression and the down-regulation of Sox2 in a subset of these ventral foregut cells around E9–9.5 (Figure 2). It is important to point out that Nkx2.1 is also expressed in the developing thyroid (Lazzaro et al., 1991) and although Nkx2.1−/− mutant mice have pulmonary dysplasia and tracheoesophageal fistula (Minoo et al., 1999), lung tissue is still present indicating that other transcription factors in addition to Nkx2.1 must also be required for respiratory fate. Transcription profiling in early mouse foregut has begun to shed light on the combinatorial transcription factor code of different lineages (Fagman et al., 2011; Millien et al., 2008), but those that uniquely define the respiratory epithelium are still unknown.
FGF mediated foregut pattering
In vitro studies using mouse foregut explants suggest that FGF ligands produced by the cardiac mesoderm around E8.0–9.0 play an important role in segregating lung, liver, or pancreas lineages from a common pool of foregut cells in a concentration-dependent fashion with the highest levels of FGF promoting Nkx2.1 expression and lung fate (Figure 2D) (Serls et al., 2005). Pharmacological inhibition in Xenopus embryos confirmed that FGF signaling via both the MAP kinase and AKT pathways are required for respiratory specification in vivo, and suggested that it is the prolonged duration of FGF signaling that promotes lung fate over liver and pancreas (Shifley et al., 2012; Wang et al., 2011). The precise FGF ligands and receptors (FGFRs) that mediate this are unknown but multiple ligands are expressed in the mesoderm surrounding the presumptive lung including Fgf1, 2, 3,7,9 and 10, and it is likely that some combination of these act redundantly. Fgf10−/− and FGFR2b−/− mutant mice both exhibit lung atresia but a rudimentary trachea is present consistent with Fgf10-FgfR2b signaling acting after initial respiratory specification to regulate fetal lung bud growth (Min et al., 1998; Sekine et al., 1999).
Gain-of-function studies in both chick (Sakiyama et al., 2003) and Xenopus (Shifley et al., 2012) indicate that increased FGF signaling is sufficient to expand the Nkx2.1 domain, however it is unclear whether FGFs act directly on the epithelium. Another question is: how do FGFs exert distinct roles at different times in lung development, repressing foregut fate at E7.5–8.5, then promoting Nkx2.1+ respiratory induction between E8–9 and subsequently regulating lung bud growth and morphogenesis after E9.5. Signaling by FGFR tyrosine kinases often impacts gene expression by modulating the activity of ETS transcription factors and although several of these are expressed in the early lung, their role in specification of respiratory progenitors is unknown (Herriges and Morrisey, 2014).
Wnt/β-catenin induction of lung fate
Wnt2 and Wnt2b ligands expressed in the mouse splanchnic mesenchyme surrounding the ventral foregut at E8.5–E10.5 are redundantly required for respiratory specification (Figure. 2C,D). Compound Wnt2−/−; Wnt2b−/− mutants, or embryos with endoderm-specific deletion of the canonical Wnt effector β-catenin, fail to express Nkx2.1 and exhibit pulmonary agenesis (Goss et al., 2009; Harris-Johnson et al., 2009). In Xenopus redundant Wnt2 and Wnt2b signaling via β-catenin is also required for respiratory specification (Rankin et al., 2012). Moreover ectopic activation of β-catenin in the endoderm is sufficient to expand Nkx2.1 and Sftpc expression in both mouse and Xenopus. Intriguingly, the ectopic expression was limited indicating that only a subset of the foregut is competent to respond, but the molecular basis of this is unclear. Experiments in Xenopus and human thyroid carcinoma cells in vitro suggest that β-catenin, in a complex with TCF transcription factors, can directly activate Nkx2.1 transcription, although this remains to be validated by Chromatin immunoprecipitation (ChIP) analysis. Indeed the DNA cis-regulatory elements and transcription factors controlling Nkx2.1 transcription in foregut have not been characterized in any species and is a major gap in our understanding of the GRN controlling lung specification.
The available evidence suggests that foregut patterning by FGFs (E8–9 in mice) acts earlier than Wnt2/2b (E8.5–9.5). In Xenopus early FGF signaling is required for wnt2b expression (Figure 2D) and experimental activation of β-catenin signaling rescued lung specification in embryos where FGF signaling was blocked (Rankin et al., 2012), suggesting that Wnt acts downstream of FGF. Consistent with this interpretation, transgenic Fgf10 overexpression could not rescue Nkx2.1 transcription in mouse foregut explants in which, Wnt signaling was experimentally inhibited by the Wnt-antagonist Dkk1 (Volckaert et al., 2013). However there may not be a simple linear relationship of FGF
 Wnt because Wnt2/2b double mutants fail to express Fgf10 in the mesenchyme around the developing lung (Goss et al., 2009). This and other data suggest that Wnts and Fgfs promote each other’s expression in a feed forward loop (Figure 2D), to maintain lung fate and coordinate growth and differentiation of both the epithelium and lung mesenchyme (Herriges and Morrisey, 2014; Yin et al., 2008), although the precise mechanisms remain to be determined.
Wnt because Wnt2/2b double mutants fail to express Fgf10 in the mesenchyme around the developing lung (Goss et al., 2009). This and other data suggest that Wnts and Fgfs promote each other’s expression in a feed forward loop (Figure 2D), to maintain lung fate and coordinate growth and differentiation of both the epithelium and lung mesenchyme (Herriges and Morrisey, 2014; Yin et al., 2008), although the precise mechanisms remain to be determined.
BMPs regulate dorsal-ventral patterning
Work from both mouse and Xenopus indicates that differential BMP signaling patterns the foregut along the dorsal-ventral (D–V) axis and cooperates with Wnt2/2b to specify respiratory fate. BMP ligands (Bmp2, Bmp4, and Bmp7) produced in the cardiac and splanchnic mesenchyme signal to the adjacent ventral foregut epithelium to phosphorylate and activate the transcriptional co-activators Smad1/5/8 (pSmad1). Endoderm-specific deletion of BMP type I receptor genes Bmpr1a and Bmpr1b (Bmpr1a/b), which prevents the endoderm from responding to mesenchymal BMP ligands, caused an intriguing phenotype of tracheal agenesis but ectopic bronchi development (Domyan et al., 2011). The Bmpr1a/b mutant tracheal epithelium failed to maintain Nkx2.1 and exhibited expanded Sox2, yet Nkx2.1 was still expressed in the lung buds. This differential requirement for BMP suggests that not all Nkx2.1+ progenitors are equivalent and that segregation of the trachea and lung lineages occurs early in development. Moreover, BMP signaling was required for the ectopic Nkx2.1 caused by hyper-activation of β-catenin, suggesting that the endoderm needs pSmad1 activity in order to respond to Wnt signaling (Domyan et al., 2011). BMP appears to act primarily by repressing expression of the transcription factor Sox2 in the ventral epithelium, which would otherwise inhibit Nkx2.1 transcription (Domyan et al., 2011), although it is unclear whether Sox2 acts directly on Nkx2.1 enhancers. It is interesting that during respiratory specification Sox2 and Nkx2.1 acquire a mutually exclusive expression pattern, whereas just a day and a half later (E11–11.5), Sox2 and Nkx2.1 are co-expressed in the upper airway epithelium (Que et al., 2009). How this dynamic expression is regulated is still poorly understood.
In normal development ventrally-produced BMPs are counterbalanced by the BMP-antagonist Noggin secreted from the notochord, resulting in a graded level of pSmad1 along the D–V axis of the foregut epithelium: highest in the Nkx2.1+ ventral foregut and lowest in the Sox2+ dorsal epithelium (Figure 2C). This distribution of BMP activity is essential for correct separation of the trachea and esophagus. Bmp4−/− mutant mice exhibit tracheal atresia, whereas Noggin−/− mutant mice display esophageal atresia and tracheoesophageal fistula (EA/TEF), with the Noggin−/− phenotype being alleviated in Bmp4+/− or Bmp7−/− mutant backgrounds (Li et al., 2008; Que et al., 2006). This foregut patterning into dorsal Sox2+ esophagus and ventral repression of Sox2 by BMP being required for Wnt-mediated Nkx2.1 induction appears to be a key regulatory cassette conserved from frogs to humans (Figure 2D).
Recent experiments revealed that in Xenopus BMPs not only have a “pro-lung” activity in the epithelium, but BMPs also exert an “anti-lung” activity in the mesenchyme. In Xenopus the zinc finger transcriptional repressors Osr1 and Osr2 (Osr1/2) act redundantly to spatially regulate mesenchymal bmp4 expression, and BMP signaling in turn acts within the mesenchyme to restrict the wnt2/2b expression domain (Rankin et al., 2012). In Osr1/2-depleted embryos bmp4 expression is expanded and there is excessive pSmad1 both in the epithelium and mesenchyme resulting in a down regulation of both sox2 and wnt2/2b; the end result is a failure to specify the Nkx2.1+ respiratory lineage and lung agenesis. This highlights an important feedback loop that regulates BMP activity in both the epithelium and mesenchyme to coordinate epithelial competence with the mesenchymal inducing signals (Figure 2D). It remains to be determined if mammalian Osr1/2 have a similar function, although mouse Osr1 is expressed at the right time and place. It is also unclear how pSmad1 activity restricts wnt2/2b, as the DNA cis-regulatory elements controlling their expression in the foregut mesenchyme have not been characterized in any species.
Retinoic acid
A number of genetic and pharmacological studies in mice and Xenopus have shown that RA signaling is important to maintain early Nkx2.1 expression and promote lung bud outgrowth, although it is still unclear whether RA is absolutely required for initial Nkx2.1 transcription (Chen et al., 2010; Chen et al., 2007; Wang et al., 2011; Wang et al., 2006). Two key functions of RA are: 1) to inhibit activation of the TGFβ-pSmad2 pathway which restricts lung development by repressing Fgf10 expression (Chen et al., 2007) and 2) to inhibit expression of Dkk1, a Wnt/β-catenin antagonist (Chen et al., 2010). Thus, RA creates a “pro-respiratory” field by allowing FGF10 and Wnt signaling. Experiments in Xenopus suggest that the RA
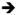 FGF/Wnt module may be integrated with the Osr -| BMP regulatory cassette (Figure 2D) because in frogs Osr1/2 are required for expression of Raldh2, the rate-limiting enzyme for RA production (Rankin et al., 2012).
FGF/Wnt module may be integrated with the Osr -| BMP regulatory cassette (Figure 2D) because in frogs Osr1/2 are required for expression of Raldh2, the rate-limiting enzyme for RA production (Rankin et al., 2012).
RA probably acts in part by regulating expression of Hox, Tbx, and Gata transcription factors in the mesenchyme. Several RA-regulated Hoxa/b genes are expressed in the foregut mesenchyme (Packer et al., 2000) and Hoxa5 mutant mice have hypoplastic lungs (Aubin et al., 1997). RA is also required for robust levels of Tbx5 and Gata4/5/6 in the cardiac and splanchnic mesenchyme (Ryckebusch et al., 2008). Tbx5 (along with Tbx4) positively regulate Wnt2b and Fgf10 expression in chick and mice (Arora et al., 2012; Sakiyama et al., 2003), whereas Gata6 and Wnt2 promote each other’s expression in a positive feed forward loop (Tian et al., 2010). This raises the possibility that an RA
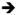 Hox/Tbx5/Gata6
Hox/Tbx5/Gata6
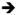 Wnt2/2b sub-circuit (Figure 2D) may control the lung inducing signals in the mesenchyme.
Wnt2/2b sub-circuit (Figure 2D) may control the lung inducing signals in the mesenchyme.
RA may also have a direct role in the endoderm, as Raldh2−/− mutant mouse embryos display reduced Shh and Foxa2 levels in the epithelium (Wang et al., 2006). The regulation of Foxa2 is particularly interesting because Fox transcription factors are “pioneering” DNA-binding proteins which regulate the state of chromatin and modulate the ability of target genes to respond to inductive signals (Zaret and Carroll, 2011). In some contexts Foxa genes are direct transcriptional targets of RA and both Foxa1 and Foxa2 can activate a Nkx2.1 minimal promoter construct in tissue culture (Ikeda et al., 1996). The combined deletion of Foxa1 and Foxa2 in the fetal mouse lung epithelium leads to severe differentiation defects (Wan et al., 2005), although it is unclear whether they are required earlier for initial activation of Nkx2.1 transcription. Hoxb1 is another candidate RA target expressed in the foregut epithelium, and Hoxb proteins can also activate Nkx2.1 transcription in vitro (Guazzi et al., 1994). Thus in addition to regulating inductive signals in the mesenchyme, RA may also act in the epithelium to regulate the competence to respond to those signals.
Hedgehog
There are 3 Hedgehog family members in vertebrates, Sonic hedgehog (Shh), Indian hedgehog, and Desert hedgehog all of which modulate target gene expression through Gli transcription factors (Robbins et al., 2012). Shh is expressed in the foregut epithelium of mouse, chick, and Xenopus embryos where it is thought to act on the adjacent mesenchyme through paracrine signaling. Consistent with epithelial to mesenchymal signaling, Shh−/− mutant mice have defects in pulmonary vasculature (Peng et al., 2013), as well as displaying an abnormal trachea and hypoplastic lungs (Litingtung et al., 1998); this phenotype is similar to that seen in human patients with HH pathway mutations (Spilde et al., 2003). Since Shh mutants still make respiratory tissue, if HH signaling is required for respiratory specification then there must be functional redundancy with other ligands. Consistent with this possibility, Gli2−/−; Gli3−/− compound mutant mouse embryos exhibit respiratory agenesis (Motoyama et al., 1998), however the early molecular phenotype in these mutants has not been analyzed, so it is unclear if HH/Gli signaling is required for respiratory specification or rather for lung bud outgrowth.
Using developmental paradigms to generate lung from pluripotent stem cells
A major motivation for studying embryonic development is to apply the knowledge to differentiate pluripotent stem cells (ES or iPS cells) into therapeutically useful tissue. For a while progress in making respiratory tissue lagged behind successes in other endodermal tissues such as liver, pancreas, and intestine, in part because the early development of those organs was better understood. However in the last three years major advances in our understanding of lung development have enabled a number of groups to generate respiratory-like tissue from both mouse and human ES and iPS cells by treating cultures with different cocktails of FGF, Wnt, BMP, RA, and HH agonists or inhibitors (Huang et al., 2014; Longmire et al., 2012; Mou et al., 2012; Wong and Rossant, 2013). The best results to date closely mimic development and generated robust populations of Nkx2.1+ progenitor cells from human ES cultures that can be further differentiated into respiratory-like tissue containing all of the major airway lineages (Huang et al., 2014).
Interestingly, there is considerable variation in the combination, dose, and timing of factors used by different groups, resulting in variable efficiency in differentiated respiratory epithelium. The fact that different protocols generate what appear to be equivalent respiratory progenitors begs the question of whether the Nkx2.1+ cells produced in different protocols are really the same? It is possible that some protocols only activate a part of the lung specification GRN and as a result the cells are impaired in their ability to further differentiate. Another limitation with the current approaches is that the co-differentiating mesenchyme cells (which are invariably present) are poorly characterized and probably produce paracrine signals that impact the cultures. A deeper understanding of the GRN controlling lung specification will no doubt accelerate further progress.
Unanswered Questions and emerging opportunities
It has become clear that in the post genomic era, the elucidation of GRNs will be necessary to fully understand the inherent complexity of organogenesis, stem cell differentiation, disease, and regeneration. Over the next few years a convergence of new enabling technologies, alternative models, and high throughput analysis will allow us to address important outstanding questions and more completely define the GRN controlling lung development.
First, we need a more complete understanding of the transcriptional signature that defines respiratory progenitors and the epithelial lineages that they develop into. The overreliance on just a few markers like Nkx2.1 has left us a relatively shallow view. Recognizing the importance of this issue the National Heart, Lung and Blood Institute has called for a high resolution, genome wide molecular atlas of lung development (RFA-HL-14-008). With new technologies that enable robust RNA-seq transcription profiling of single cells (Jaitin et al., 2014) and increasingly sophisticated transgenic tools in mice allowing cell-specific conditional gene targeting and lineage tracing, we will gain critical insight into progenitor cell heterogeneity and how transcription profiles progressively change as lineages become restricted.
We also need a better understanding of the functional relationships between the FGF, Wnt, BMP, RA, and HH pathways and how dynamic changes in combinatorial signaling during lung development can activate distinct genetic programs at different times. The rapid functional genomics possible in externally developing Xenopus embryos will continue to be useful in this regard as the epistatic relationships between multiple genes can be easily tested in a single experiment. The differentiation of pluripotent stem cells, in addition to being an important end goal, will also provide a powerful tool to biochemically examine signaling cross-talk in human lung development.
Furthermore, it will be essential to determine how this combinatorial signaling is integrated on DNA cis-regulatory elements at the whole genome level. Although many transcription factors have been implicated in regulating Nkx2.1 transcription in vitro, whether they control Nkx2.1 expression in respiratory progenitors in vivo is unknown. We need to define enhancers and characterize the in vivo DNA-binding of transcription factors in both the foregut epithelium and mesenchyme during respiratory specification. It will also be important to determine how epigenetic regulation and non-coding RNAs impact lung specification. For example the epigenetic modification of chromatin by histone deacetylase (HDAC) regulates Bmp4 and Sox2 expression in fetal airway progenitors (Wang et al., 2013), whereas microRNAs regulate Gata6 to influence the balance between lung progenitor proliferation and differentiation (Tian et al., 2011). The impact of similar mechanisms on early lung specification remains to be elucidated.
The ENCODE project and a growing number of follow-up studies have begun to reveal how dynamic modulation of the chromatin landscape is critical for enhancer activity and gene expression (Consortium et al., 2012; Kieffer-Kwon et al., 2013). However most of these studies to date have focused on cell lines or adult tissues, which provide the ample biological material need for ChIP-seq type analyses. The limited tissue available from the early mouse embryos has hampered similar analyses of organogenesis, however the abundant material available from Xenopus embryos and ES/iPS cells cultures should help resolve this bottleneck. For example ChIP-Seq studies have revealed genome-wide dynamic changes in histone modifications and transcription factor binding during endoderm formation and endocrine pancreas development in Xenopus embryos and human stem cell cultures (Kim et al., 2011; Xie et al., 2013). Similar approaches need to be applied to studies of lung development. Importantly putative enhancers controlling gene expression in the early foregut will need to be validated in vivo. New genome editing technologies using CRISPR/Cas9 or TALENS will enable the rapid mutation of candidate enhancers and the introduction of transgenic reporter constructs, at any location in the genome of frogs, mice and human stem cells (Burgess, 2013; Kieffer-Kwon et al., 2013).
Finally advanced computational methods are being developed that will enable integration all of these diverse data types, from single gene mutation studies to whole genome epigenetic data into GRN models that have probabilistic predictive power (Mitra et al., 2013; Peter et al., 2012). This will enable investigators to obtain an unprecedented view of the complex gene regulatory network controlling respiratory specification and accelerate our ability to harness this knowledge for disease modeling and regenerative medicine.
Acknowledgments
Lung research in the Zorn lab is supported by NIH grant HL114898 to AMZ. We are grateful to Lu Han and Virgilio Ponferrada for mouse embryo images, to Lu Han and Marcin Wlizla for comments on the manuscript, and members of the Zorn, Wells, Whitsett, and Shannon labs for helpful discussions.
References
- Arda HE, Benitez CM, Kim SK. Gene regulatory networks governing pancreas development. Dev Cell. 2013;25:5–13. doi: 10.1016/j.devcel.2013.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Metzger RJ, Papaioannou VE. Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system. PLoS Genet. 2012;8:e1002866. doi: 10.1371/journal.pgen.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin J, Lemieux M, Tremblay M, Berard J, Jeannotte L. Early postnatal lethality in Hoxa-5 mutant mice is attributable to respiratory tract defects. Dev Biol. 1997;192:432–445. doi: 10.1006/dbio.1997.8746. [DOI] [PubMed] [Google Scholar]
- Beers MF, Morrisey EE. The three R’s of lung health and disease: repair, remodeling, and regeneration. J Clin Invest. 2011;121:2065–2073. doi: 10.1172/JCI45961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri H. Computational modeling of gene regulatory networks: a primer. Imperial College Press; London: 2008. [Google Scholar]
- Burgess DJ. Technology: a CRISPR genome-editing tool. Nat Rev Genet. 2013;14:80. doi: 10.1038/nrg3409. [DOI] [PubMed] [Google Scholar]
- Chen F, Cao Y, Qian J, Shao F, Niederreither K, Cardoso WV. A retinoic acid-dependent network in the foregut controls formation of the mouse lung primordium. J Clin Invest. 2010;120:2040–2048. doi: 10.1172/JCI40253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Desai TJ, Qian J, Niederreither K, Lu J, Cardoso WV. Inhibition of Tgf beta signaling by endogenous retinoic acid is essential for primary lung bud induction. Development. 2007;134:2969–2979. doi: 10.1242/dev.006221. [DOI] [PubMed] [Google Scholar]
- Consortium EP, Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domyan ET, Ferretti E, Throckmorton K, Mishina Y, Nicolis SK, Sun X. Signaling through BMP receptors promotes respiratory identity in the foregut via repression of Sox2. Development. 2011;138:971–981. doi: 10.1242/dev.053694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagman H, Amendola E, Parrillo L, Zoppoli P, Marotta P, Scarfo M, De Luca P, de Carvalho DP, Ceccarelli M, De Felice M, Di Lauro R. Gene expression profiling at early organogenesis reveals both common and diverse mechanisms in foregut patterning. Dev Biol. 2011;359:163–175. doi: 10.1016/j.ydbio.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Yamaguchi TP, Morrisey EE. Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev Cell. 2009;17:290–298. doi: 10.1016/j.devcel.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guazzi S, Lonigro R, Pintonello L, Boncinelli E, Di Lauro R, Mavilio F. The thyroid transcription factor-1 gene is a candidate target for regulation by Hox proteins. EMBO J. 1994;13:3339–3347. doi: 10.1002/j.1460-2075.1994.tb06636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Johnson KS, Domyan ET, Vezina CM, Sun X. beta-Catenin promotes respiratory progenitor identity in mouse foregut. Proc Natl Acad Sci U S A. 2009;106:16287–16292. doi: 10.1073/pnas.0902274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriges M, Morrisey EE. Lung development: orchestrating the generation and regeneration of a complex organ. Development. 2014;141:502–513. doi: 10.1242/dev.098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SX, Islam MN, O’Neill J, Hu Z, Yang YG, Chen YW, Mumau M, Green MD, Vunjak-Novakovic G, Bhattacharya J, Snoeck HW. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat Biotechnol. 2014;32:84–91. doi: 10.1038/nbt.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Shaw-White JR, Wert SE, Whitsett JA. Hepatocyte nuclear factor 3 activates transcription of thyroid transcription factor 1 in respiratory epithelial cells. Mol Cell Biol. 1996;16:3626–3636. doi: 10.1128/mcb.16.7.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grontved L, Vian L, Nelson S, Zare H, Hakim O, Reyon D, Yamane A, Nakahashi H, Kovalchuk AL, Zou J, Joung JK, Sartorelli V, Wei CL, Ruan X, Hager GL, Ruan Y, Casellas R. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell. 2013;155:1507–1520. doi: 10.1016/j.cell.2013.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Yoon SJ, Chuong E, Oyolu C, Wills AE, Gupta R, Baker J. Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev Biol. 2011;357:492–504. doi: 10.1016/j.ydbio.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Lazzaro D, Price M, de Felice M, Di Lauro R. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113:1093–1104. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- Li Y, Gordon J, Manley NR, Litingtung Y, Chiang C. Bmp4 is required for tracheal formation: a novel mouse model for tracheal agenesis. Developmental biology. 2008;322:145–155. doi: 10.1016/j.ydbio.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y, Lei L, Westphal H, Chiang C. Sonic hedgehog is essential to foregut development. Nat Genet. 1998;20:58–61. doi: 10.1038/1717. [DOI] [PubMed] [Google Scholar]
- Longmire TA, Ikonomou L, Hawkins F, Christodoulou C, Cao Y, Jean JC, Kwok LW, Mou H, Rajagopal J, Shen SS, Dowton AA, Serra M, Weiss DJ, Green MD, Snoeck HW, Ramirez MI, Kotton DN. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell. 2012;10:398–411. doi: 10.1016/j.stem.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millien G, Beane J, Lenburg M, Tsao PN, Lu J, Spira A, Ramirez MI. Characterization of the mid-foregut transcriptome identifies genes regulated during lung bud induction. Gene Expr Patterns. 2008;8:124–139. doi: 10.1016/j.modgep.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes & development. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoo P, Su G, Drum H, Bringas P, Kimura S. Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Developmental Biology. 1999;209:60–71. doi: 10.1006/dbio.1999.9234. [DOI] [PubMed] [Google Scholar]
- Mitra K, Carvunis AR, Ramesh SK, Ideker T. Integrative approaches for finding modular structure in biological networks. Nat Rev Genet. 2013;14:719–732. doi: 10.1038/nrg3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama J, Liu J, Mo R, Ding Q, Post M, Hui CC. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus [see comments] Nat Genet. 1998;20:54–57. doi: 10.1038/1711. [DOI] [PubMed] [Google Scholar]
- Mou H, Zhao R, Sherwood R, Ahfeldt T, Lapey A, Wain J, Sicilian L, Izvolsky K, Musunuru K, Cowan C, Rajagopal J. Generation of multipotent lung and airway progenitors from mouse ESCs and patient-specific cystic fibrosis iPSCs. Cell Stem Cell. 2012;10:385–397. doi: 10.1016/j.stem.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AI, Mailutha KG, Ambrozewicz LA, Wolgemuth DJ. Regulation of the Hoxa4 and Hoxa5 genes in the embryonic mouse lung by retinoic acid and TGFbeta1: implications for lung development and patterning. Dev Dyn. 2000;217:62–74. doi: 10.1002/(SICI)1097-0177(200001)217:1<62::AID-DVDY6>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Peng T, Tian Y, Boogerd CJ, Lu MM, Kadzik RS, Stewart KM, Evans SM, Morrisey EE. Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature. 2013;500:589–592. doi: 10.1038/nature12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Faure E, Davidson EH. Predictive computation of genomic logic processing functions in embryonic development. Proc Natl Acad Sci U S A. 2012;109:16434–16442. doi: 10.1073/pnas.1207852109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J, Choi M, Ziel JW, Klingensmith J, Hogan BL. Morphogenesis of the trachea and esophagus: current players and new roles for noggin and Bmps. Differentiation. 2006;74:422–437. doi: 10.1111/j.1432-0436.2006.00096.x. [DOI] [PubMed] [Google Scholar]
- Que J, Luo X, Schwartz RJ, Hogan BL. Multiple roles for Sox2 in the developing and adult mouse trachea. Development. 2009;136:1899–1907. doi: 10.1242/dev.034629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Gallas AL, Neto A, Gomez-Skarmeta JL, Zorn AM. Suppression of Bmp4 signaling by the zinc-finger repressors Osr1 and Osr2 is required for Wnt/beta-catenin-mediated lung specification in Xenopus. Development. 2012;139:3010–3020. doi: 10.1242/dev.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins DJ, Fei DL, Riobo NA. The Hedgehog signal transduction network. Sci Signal. 2012;5:re6. doi: 10.1126/scisignal.2002906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckebusch L, Wang Z, Bertrand N, Lin SC, Chi X, Schwartz R, Zaffran S, Niederreither K. Retinoic acid deficiency alters second heart field formation. Proc Natl Acad Sci U S A. 2008;105:2913–2918. doi: 10.1073/pnas.0712344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakiyama J, Yamagishi A, Kuroiwa A. Tbx4-Fgf10 system controls lung bud formation during chicken embryonic development. Development. 2003;130:1225–1234. doi: 10.1242/dev.00345. [DOI] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. Fgf10 is essential for limb and lung formation. Nature genetics. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- Serls AE, Doherty S, Parvatiyar P, Wells JM, Deutsch GH. Different thresholds of fibroblast growth factors pattern the ventral foregut into liver and lung. Development. 2005;132:35–47. doi: 10.1242/dev.01570. [DOI] [PubMed] [Google Scholar]
- Shifley ET, Kenny AP, Rankin SA, Zorn AM. Prolonged FGF signaling is necessary for lung and liver induction in Xenopus. BMC Dev Biol. 2012;12:27. doi: 10.1186/1471-213X-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilde T, Bhatia A, Ostlie D, Marosky J, Holcomb G, 3rd, Snyder C, Gittes G. A role for sonic hedgehog signaling in the pathogenesis of human tracheoesophageal fistula. J Pediatr Surg. 2003;38:465–468. doi: 10.1053/jpsu.2003.50080. [DOI] [PubMed] [Google Scholar]
- Tian Y, Yuan L, Goss AM, Wang T, Yang J, Lepore JJ, Zhou D, Schwartz RJ, Patel V, Cohen ED, Morrisey EE. Characterization and in vivo pharmacological rescue of a Wnt2-Gata6 pathway required for cardiac inflow tract development. Developmental cell. 2010;18:275–287. doi: 10.1016/j.devcel.2010.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Zhang Y, Hurd L, Hannenhalli S, Liu F, Lu MM, Morrisey EE. Regulation of lung endoderm progenitor cell behavior by miR302/367. Development. 2011;138:1235–1245. doi: 10.1242/dev.061762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T, Campbell A, Dill E, Li C, Minoo P, De Langhe S. Localized Fgf10 expression is not required for lung branching morphogenesis but prevents differentiation of epithelial progenitors. Development. 2013;140:3731–3742. doi: 10.1242/dev.096560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Dingle S, Xu Y, Besnard V, Kaestner KH, Ang SL, Wert S, Stahlman MT, Whitsett JA. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem. 2005;280:13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- Wang JH, Deimling SJ, D’Alessandro NE, Zhao L, Possmayer F, Drysdale TA. Retinoic acid is a key regulatory switch determining the difference between lung and thyroid fates in Xenopus laevis. BMC developmental biology. 2011;11:75. doi: 10.1186/1471-213X-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Tian Y, Morley MP, Lu MM, Demayo FJ, Olson EN, Morrisey EE. Development and regeneration of Sox2+ endoderm progenitors are regulated by a Hdac1/2-Bmp4/Rb1 regulatory pathway. Dev Cell. 2013;24:345–358. doi: 10.1016/j.devcel.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Dolle P, Cardoso WV, Niederreither K. Retinoic acid regulates morphogenesis and patterning of posterior foregut derivatives. Dev Biol. 2006;297:433–445. doi: 10.1016/j.ydbio.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Whitsett JA, Haitchi HM, Maeda Y. Intersections between pulmonary development and disease. Am J Respir Crit Care Med. 2011;184:401–406. doi: 10.1164/rccm.201103-0495PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AP, Rossant J. Generation of Lung Epithelium from Pluripotent Stem Cells. Current pathobiology reports. 2013;1:137–145. doi: 10.1007/s40139-013-0016-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, Everett LJ, Lim HW, Patel NA, Schug J, Kroon E, Kelly OG, Wang A, D’Amour KA, Robins AJ, Won KJ, Kaestner KH, Sander M. Dynamic chromatin remodeling mediated by polycomb proteins orchestrates pancreatic differentiation of human embryonic stem cells. Cell Stem Cell. 2013;12:224–237. doi: 10.1016/j.stem.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, White AC, Huh SH, Hilton MJ, Kanazawa H, Long F, Ornitz DM. An FGF-WNT gene regulatory network controls lung mesenchyme development. Dev Biol. 2008;319:426–436. doi: 10.1016/j.ydbio.2008.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, Wells JM. Vertebrate endoderm development and organ formation. Annual review of cell and developmental biology. 2009;25:221–251. doi: 10.1146/annurev.cellbio.042308.113344. [DOI] [PMC free article] [PubMed] [Google Scholar]