Abstract
The Caco-2 cell line is one of the most important in vitro models for enterocytes, and is used to study drug absorption and disease, including inflammatory bowel disease and cancer. In order to use the model optimally, it is necessary to map its functional entities. In this study, we have generated genome-wide maps of active transcription start sites (TSSs), and active enhancers in Caco-2 cells with or without tumour necrosis factor (TNF)-α stimulation to mimic an inflammatory state. We found 520 promoters that significantly changed their usage level upon TNF-α stimulation; of these, 52% are not annotated. A subset of these has the potential to confer change in protein function due to protein domain exclusion. Moreover, we locate 890 transcribed enhancer candidates, where ∼50% are changing in usage after TNF-α stimulation. These enhancers share motif enrichments with similarly responding gene promoters. As a case example, we characterize an enhancer regulating the laminin-5 γ2-chain (LAMC2) gene by nuclear factor (NF)-κB binding. This report is the first to present comprehensive TSS and enhancer maps over Caco-2 cells, and highlights many novel inflammation-specific promoters and enhancers.
Keywords: alternative promoters, inflammation, non-coding RNAs, transcribed enhancers, transcriptional regulation
1. Introduction
Intestinal epithelial cells (IECs) act as a mucosal barrier between the luminal environment (i.e. microbes and their toxins as well as food-derived antigens) and the internal milieu, and actively contribute to the gut immune system.1 Maintenance of this barrier is crucial for the homeostasis, i.e. the immune system's ability to remain tolerant to antigens. Therefore, dysregulation within the epithelial layer can increase permeability, lead to abnormalities in interactions between IECs and immune cells, and disturb the intestinal immune homeostasis, all of which are linked to the clinical disease course of several intestinal disorders including inflammatory bowel disease (IBD).2 IBD is a chronic inflammation of the intestine and is characterized by an imbalanced production of a wide range of pro-inflammatory mediators including tumour necrosis factor (TNF)-α3. TNF-α is a central pro-inflammatory cytokine in IBD and the use of TNF-inhibitors in the treatment of IBD has been successful.4 The Caco-2 cell line is a widely used model for human IECs, resembling the in vivo differentiation of enterocytes/colonocytes both morphologically and biochemically.5 Because of this, it is an important model for molecular studies focused on gut disease including IBD. At the same time, differentiated Caco-2 cells are widely used to predict the absorption of orally administrated drugs in the pharmaceutical industry.
Despite its wide usage in both applied and basic science, there have been only few comprehensive genome-wide efforts to characterize Caco-2 on the transcriptional level. This is needed, because genome-wide methods have shown that the majority of genes have multiple isoforms, driven by alternative promoter usage,6,7 alternative splicing,8 or alternative transcription termination.9 Usage of alternative isoforms can have drastic functional impact, for instance by loss of functional protein domains.10 Microarray-based methods will fail to pinpoint the exact promoters that are responding to cellular changes, because most genes have alternative promoters.6 Alternative promoters are often tissue- or context-specific6 and can thereby be viewed as independent regulatory platforms in which different cellular contexts can influence the expression of a gene. An example of this is the DLGAP gene which has four alternative promoters, each specific for a different brain tissue.10 Despite its importance, alternative promoter usage is uncharacterized in most medically relevant models.
State-of-the-art methods for the detection of active promoters or transcription start sites (TSSs) are CAGE11 and TSS-seq.12 These methods are genome-wide and based on generating full-length cDNAs from mRNAs, followed by sequencing the first 20–40 nucleotides from their 5′-end. These techniques have been used to chart mammalian genomes,13,14 dissect TSS usage in core promoters, investigate evolutionary conservation and turnover, and for systems biology of developing macrophages (reviewed in refs15,16). The CAGE technique has been thoroughly validated by gene reporter assays, histone marks and other RNA sequencing techniques (e.g. refs6,10,14,17,18).
Surprisingly, the CAGE technique can also identify active enhancers, since the active enhancers are lowly transcribed in a bidirectional pattern.19 Using massive enhancer reporter screens, we have previously shown19 that this approach is more than two times as accurate as non-transcribed enhancer candidates identified by histone modification ChIP-seq or DHS-seq (e.g. refs20–22).
In this study, we present the first genome-wide map of promoters for the Caco-2 cell line, before and after stimulation with pro-inflammatory TNF-α. We show that TNF-α stimulation induces expression from a large number of promoters, where many are novel alternative promoters of well-characterized genes and some represent completely novel TNF-α-specific non-protein-coding transcripts. Similarly, we identify many candidate enhancer regions that respond to TNF-α stimulation and can be linked with similarly responding genes. Finally, we show that many TNF-α-responsive promoters and enhancers are likely regulated by known inflammatory factors like NF-κB.
2. Materials and methods
2.1. Cell culture and stimulation
The human intestinal cell line Caco-2 (American Tissue Type Culture Collection, Rockville, MD, USA) cells were cultivated as monolayers and maintained as previously described.23 For stimulation experiments, 106 cells were seeded in 24-well plates (NUNC Brand, Thermo Fisher, Rochester, NY, USA) and grown to >95% confluence. Cells were then stimulated in medium with or without TNF-α (R&D Systems, Minneapolis, MN, USA) in the presence or absence of tosyl phenylalanyl chloromethyl ketone (TPCK) NF-κB inhibitor; 100 µM; Sigma-Aldrich, St. Louis, MO, USA), FR180204 (ERK inhibitor; 30 µM; Merck Chemicals, Darmstadt, Germany), or vehicle [0.4% dimethyl sulphoxide (DMSO); Sigma-Aldrich] as done previously.24 In experiments involving treatment with inhibitors, cells were exposed to the inhibitors 1 h prior to the addition of TNF-α and subsequently stimulated with TNF-α (10 nM) for 24 h. For CAGE analysis, we used biological triplicates with or without TNF-α (10 nM).
2.2. Protein extraction and immunoblotting
Protein extraction and immunoblotting was done as described in Seidelin et al.,25 using LAMC2 (1 : 333; mouse monoclonal, Millipore, Billerica, MA, USA) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) antibodies (1 : 20,000; mouse monoclonal; Fitzgerald, Concord, MA, USA).
2.3. CAGE library preparation and processing
CAGE libraries were prepared from 5 µg of total RNA purified using the Purelink mini kit (Ambion, Austin, TX, USA) as recommended by the manufacturer. Libraries were prepared and mapped to the hg19 assembly as in ref.19 CAGE tags that mapped close to each other on the same strand were grouped into 52,451 tag clusters (TCs) as in ref.26 The expression level for each library in each TC was normalized to sample size (tags per million mapped tags, TPM). For differential expression assessment, we used EdgeR27 on the TCs having >4TPM and used an FDR threshold of <0.05, comparing TNF-α stimulation and control replicates. This defined a set of TCs that were induced by TNF-α [TNF-α(+)] and a set of TCs that are depleted [TNF-α (−)].
2.4. Gene ontology analysis
TNF-α(+), TNF-α(−) and the set of all TNF-α TCs with TPM > 4 were intersected with RefSeq genes. Genes with more than two TCs were counted only once. DAVID28 was used to identify enriched gene ontology (GO) terms, using default settings. The RefSeq gene symbols were uploaded and converted to DAVID gene symbols. For the TNF-α(+) and TNF-α(−) sets, only Homo sapiens annotations were used. All Caco-2 TCs with TPM > 4 was used as background. Only overrepresentations of GO terms, where P < 0.05 (Benjamini corrected), were reported.
2.5. Enhancer detection, expression quantification and promoter associations
Putative Caco-2 enhancers were identified from CAGE-derived bidirectionally transcribed loci, as discussed in ref.19 In summary, bidirectionally transcribed loci were defined from forward and reverse strand CAGE tag TCs supported by at least two CAGE tags in at least one sample. Only TCs not overlapping antisense TCs were used. We identified divergent (reverse–forward) TC pairs separated by at most 400 bp and merged all such pairs containing the same TC, while at the same time avoiding overlapping forward and reverse strand transcribed regions (prioritization by expression ranking). A centre position was defined for each bidirectional locus as the mid position between the rightmost reverse strand TC and leftmost forward strand TC included in the merged bidirectional pair. Each bidirectional locus was further associated with two 200 bp regions immediately flanking the centre position, one (left) for reverse strand transcription and one (right) for forward strand transcription, in a divergent manner. The merged bidirectional pairs were further required to be bidirectionally transcribed (CAGE tags supporting both windows flanking the centre) in at least one individual sample, and to have a greater aggregate of reverse CAGE tags (over all samples) than forward CAGE tags in the 200 bp region associated with reverse strand transcription, and vice versa. We quantified the expression of bidirectional loci for each strand and 200 bp flanking window in each sample separately by counting the CAGE tags whose 5′ ends were located within these windows. The expression values of both flanking windows were normalized by converting tag counts to TPM. The number of CAGE tags aligned to ChrM was subtracted from the total number of aligned CAGE tags in each library before normalization. The normalized expression values from both windows were used to calculate a sample-set wide directionality score, D, for each enhancer over aggregated normalized reverse, R, and forward, F, strand expression values across all samples; D = (F − R)/(F + R). D ranges between −1 and 1 and specifies the bias in expression to reverse and forward strand, respectively [D = 0 means 50% reverse and 50% forward strand expression, whereas abs(D) close to 1 indicates unidirectional transcription]. Each bidirectional locus was assigned one expression value for each sample by summing the normalized expression of the two flanking windows. Bidirectional loci were further filtered to have low, non-promoter-like, directionality scores [abs(D) <0.8] and to be located distant to TSSs and exons of protein- and multi-exonic non-coding genes. This set was used to extend the set of CAGE-defined enhancers by the functional annotation of mammalian genomes (FANTOM) consortium from a large number of tissues and cells.19 The expression of these enhancers were quantified in each individual Caco-2 CAGE library as described above, and enhancers supported by at least three CAGE tags in any Caco-2 library were considered for further analyses. Enhancers with an absolute log2 expression fold change ≥2 between average TNF-α stimulated and unstimulated samples were considered differentially expressed. Putative enhancer–promoter associations were derived from proximal enhancer–promoter pairs (within 200 kb) with correlated expression (Pearson's r >0.5).
2.6. Motif overrepresentation analysis
Regions −300 to +100 bp around the gene TSS (as in ref.17) were scanned with all JASPAR29 motifs using the Clover motif overrepresentation tool30 with default settings, and all not-significantly regulated TCs as background. When analysing TNF-responsive enhancers, the regions ±300 bp around the mid-positions were used, using non-responsive enhancers as background.
2.7. qPCR validation of promoter usage
Total RNA from Caco-2 cells was extracted using the NucleoSpin columns (Macherey-Nagel, Düren, Germany) following the manufacturer's protocol. Total RNA (1–2 µg) was used to prepare cDNA by SuperScript III reverse transcriptase (Invitrogen, Paisley, UK) and quantitative real-time PCR (qPCR) reactions were performed on a Stratagene Mx3000P thermocycler (Stratagene, La Jolla, CA, USA) using the Maxima SYBR Green qPCR Master Mix (Fisher Scientific, Pittsburgh, PA, USA) according to manufacturer's instructions. All primers were purchased from Integrated DNA Technologies (Leuven, Belgium). Primers were designed to amplify regions immediately downstream of the CAGE signal (Supplementary Table S1). Target gene expression values were normalized to the RPLP0 gene, which was amplified in parallel reactions.
2.8. Transient transfection assays
One day prior to transfection, Caco-2 cells were seeded in 24-well plates at a density of 5 × 104 cells/well and transiently transfected the following day as described previously.31 In stimulation experiments, the medium was changed 24 h after transfection, washed and subsequently stimulated with or without TNF-α (10 nM) for the next 24 h. LAMC2 luciferase reporter constructs were: pGL3-LAMC2 containing the 1.2 kb LAMC2 promoter region upstream of TSS32 and pGL3-LAMC2 + Enhancer containing the 1.2 kb LAMC2 promoter and the enhancer region at chr1:183,149,939–183,150,336 (hg19). The pGL3-LAMC2 + enhancer plasmid was constructed by amplifying the enhancer region by PCR and subcloning it into pGL3-LAMC2 in the Sal1 site of pGL3-plasmid upstream of the LAMC2 promoter using the In-Fusion® HD Cloning kit from Clontech. For assays investigating the effect of overexpression of NF-κB subunits on the LAMC2 transcriptional activity, pCMV4-p65 and pCMV4-p50 (Addgene plasmid number: 21966 and 2196533) or pCMV4-p52 (Addgene plasmid number: 2328934) were used. Plasmids were co-transfected with the LAMC2 reporter constructs. Forty-eight hours post-transfection, cells were harvested and lysed; luciferase and β-galactosidase activities were determined using the Dual Light system (Perkin Elmer, Waltham, MA, USA) according to manufacturer's instructions.
3. Results
3.1. CAGE identifies novel TNF-α-responsive promoters
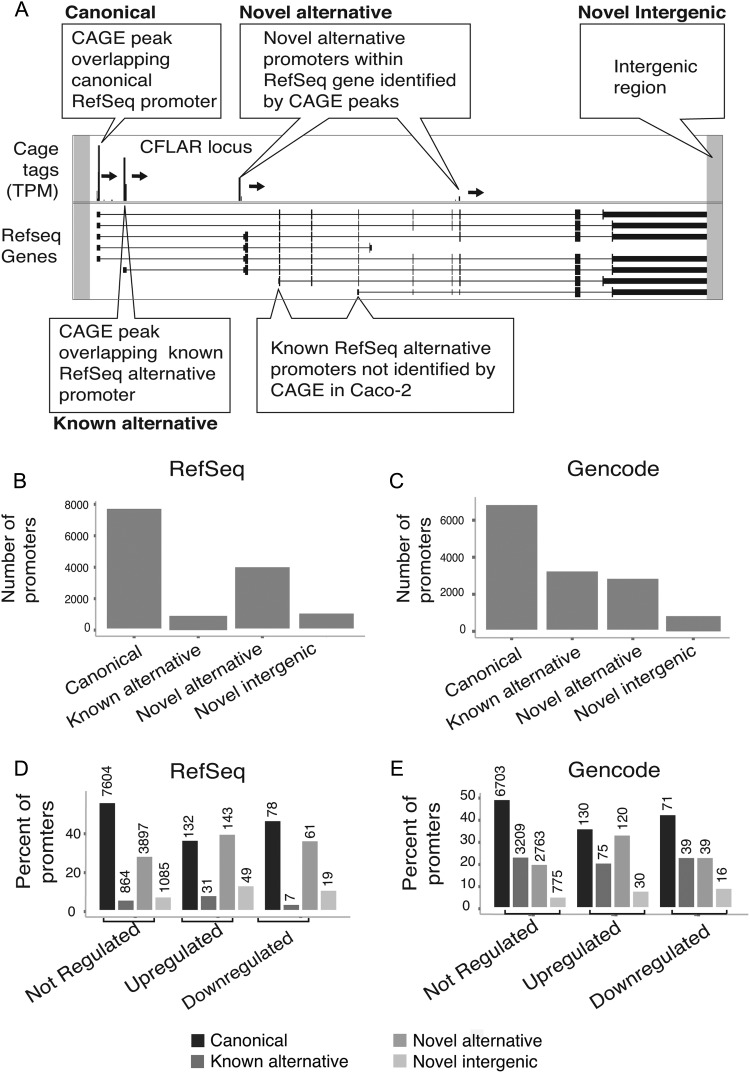
We prepared CAGE libraries from total RNA from Caco-2 cells pre-treated with or without TNF-α in biological triplicates. CAGE tags were mapped to the human genome, and nearby CAGE tags were grouped into 52,451 TCs. As in previous work, we will for simplicity refer to these TCs as candidate ‘promoters’. Using an expression cut-off of four TPM to focus on strong initiation events, 13,970 TCs were retained, of which 92% overlapped an annotated RefSeq gene model. The remaining 8% were labelled novel intergenic promoter candidates. A substantial fraction of genes (23%) had two or more overlapping TCs on the same strand, suggesting alternative initiation events that would not be detected by, for example, microarray techniques. As in the ENSEMBL annotation pipeline35 we defined the canonical TSS as the TSS that is the most upstream in annotated gene models. Of the TCs overlapping RefSeq genes, 61% (7.814 TCs) were within 100 nt from the canonical annotated TSS, while 7% (902 TCs) were within 100 nt of annotated alternative promoters, and the final 32% (4,101) did not overlap any annotated TSS; we labelled these candidate novel alternative promoters (Fig. 1A and B). In order to compare with a more comprehensive, but less conservative, gene annotation, the same TCs were overlapped with the Gencode V19 basic gene annotation.36 While the number of TCs mapping to gene models and canonical TSSs did not change substantially (94 and 53%, respectively) compared with the RefSeq results, a larger fraction of TCs within genes were overlapping Gencode-annotated TSSs (25%), indicating that many of the intragenic TCs correspond to real mRNA TSSs (Fig. 1C).
Figure 1.
CAGE-defined promoters in Caco-2 cells. (A) Schematic illustration showing classification of CAGE-defined promoters based on RefSeq annotation. The upper panel shows the number of CAGE tag clusters (TCs) mapping around an example gene loci (the CFLAR gene) on the plus strand. The lower panel shows the RefSeq gene model exon–intron structure of the gene (blocks are exons, lines are introns). CAGE peaks are commented as belonging to specific classes based on their location. CAGE tags falling outside of gene loci will be classified as ‘novel intergenic’ (grey areas). Box labels indicate names used for respective classes in (B) and (C). RefSeq-annotated alternative promoters that are not hit by CAGE are also indicated. (B) Distribution of Caco-2 TCs (TPM > 4) according to the TC classes defined above using RefSeq annotation. (C) Distribution of Caco-2 TCs (TPM > 4) according to the TC classes defined above using Gencode annotation. (D and E) Per cent of TCs falling into each of the classes from (B) and (C) split up by TNF-α response, using RefSeq (D) or Gencode annotation (E). Bars shaded from black to light grey represent canonical, known alternative, novel intragenic and novel intergenic TCs, respectively. Numbers above bars indicate the number of promoters in respective category.
Next, we used EdgeR27 on the 13.970 TCs and identified 520 TCs with significant expression changes between TNF-α stimulation and control (FDR <0.05); 355 (68%) had a significant increase in expression and 165 TCs had a significant decrease after stimulation (Supplementary Table S2). We subsequently refer to these two categories as TNF-α(+) and TNF-α(−) promoters/TCs.
RefSeq genes overlapping TNF-α(+) promoters were enriched in GO terms involving immune response, defence and cytokine activity (Table 1). Many of the TNF-α(+) genes are well-studied responders to TNF-α stimulation and/or inflammation including the TNFΑIP3,37 PTGS2,38 CFLAR,39 IL-8,40 HDAC941 and SGPP242 genes. The TNF-α(−) genes include genes associated with cell growth, including histone 1 genes. In both TNF-α(+) and TNF-α(−) sets, we observed a remarkably high number of alternative promoters—either novel or overlapping annotated cDNAs starting within known genes; this is particularly true for TNF-α(+) promoters, where 54% are novel: 143 correspond to candidate alternative promoters within known genes, and 49 correspond to novel intergenic promoters (Fig. 1D and E).
Table 1.
A list of the five most overrepresented GO terms in each GO category associated with the genes associated with differentially regulated TCs found by CAGE in Caco-2 in response to the TNF-α stimulation
| Genes linked to up-regulated TCs |
Genes linked to down-regulated TCs |
||||
|---|---|---|---|---|---|
| Term | P-value | Adjusted P | Term | P-value | Adjusted P |
| Biological process | |||||
| Immune response | 1.60E−33 | 2.70E−30 | Nucleosome assembly | 6.90E−12 | 6.20E−09 |
| Defence response | 3.60E−22 | 3.10E−19 | Chromatin assembly | 1.10E−11 | 4.80E−09 |
| Response to wounding | 1.30E−17 | 7.60E−15 | Nucleosome organization | 1.40E−11 | 4.20E−09 |
| Inflammatory response | 7.70E−13 | 3.30E−10 | DNA packaging | 1.50E−11 | 3.30E−09 |
| Locomotory behaviour | 1.20E−12 | 4.10E−10 | Protein-DNA complex assembly | 9.40E−11 | 1.70E−08 |
| Cellular component | |||||
| Extracellular region | 5.50E−19 | 1.10E−16 | Nucleosome | 1.80E−11 | 2.80E−09 |
| Extracellular region part | 2.30E−16 | 2.30E−14 | Preotein-DNA complex | 3.00E−09 | 2.40E−07 |
| Extracellular space | 1.80E−14 | 1.30E−12 | Chromosome | 7.80E−08 | 4.10E−06 |
| Intrinsic to plasma membrane | 1.40E−11 | 7.30E−10 | Chomosomal part | 1.00E−07 | 4.00E−06 |
| Integral to plasma | 4.20E−11 | 1.80E−09 | Chromatin | 9.70E−06 | 3.00E−04 |
| KEGG pathway | |||||
| Cytokine-cytokine receptor interaction | 3.70E−34 | 3.90E−32 | Systemic lupus erythematosus | 9.80E−17 | 6.90E−15 |
| Pathways in cancer | 1.70E−18 | 8.80E−17 | Drug metabolism | 3.10E−04 | 9.70E−03 |
| Small cell lung cancer | 3.20E−13 | 1.10E−11 | Regulation of actin cytoskeleton | 4.70E−04 | 9.60E−03 |
| RIG-I like receptor signalling pathway | 1.30E−12 | 3.40E−11 | Glutathione metabolism | 8.70E−04 | 1.30E−02 |
| Chemokine signalling pathway | 7.10E−12 | 1.50E−10 | Cyanoamino acid metabolism | 3.90E−03 | 4.80E−02 |
| Molecular function | |||||
| Cytokine activity | 9.20E−18 | 4.40E−15 | No GO terms with adjusted P < 0.05 | ||
| Chemokine activity | 8.70E−10 | 2.10E−07 | |||
| Chemokine receptor binding | 1.20E−09 | 1.90E−07 | |||
| Growth factor activity | 1.70E−05 | 2.10E−03 | |||
| Cytokine binding | 2.50E−05 | 2.40E−03 | |||
For details, see Materials and methods.
To verify the findings, we randomly selected 18 TNF-α(+) and 2 TNF-α(−) TCs within different classes: known promoters, novel alternative promoters and novel intergenic promoters. We validated their response to TNF-α by qPCR in quadruplicates and four different TNF-α concentrations. For validating alternative promoters, we used two primer pairs: one pair located immediately downstream of the TC of interest and the other pair just downstream of the most upstream annotated promoter. In the large majority (16 of 20) of cases, the qPCR results validated the CAGE data as well as the response to TNF-α (Fig. 2 and Supplementary Figure S1). We highlight a few examples below; fold changes and P-values refer to qPCR validations at 10 nM TNF-α, two-sided t-tests.
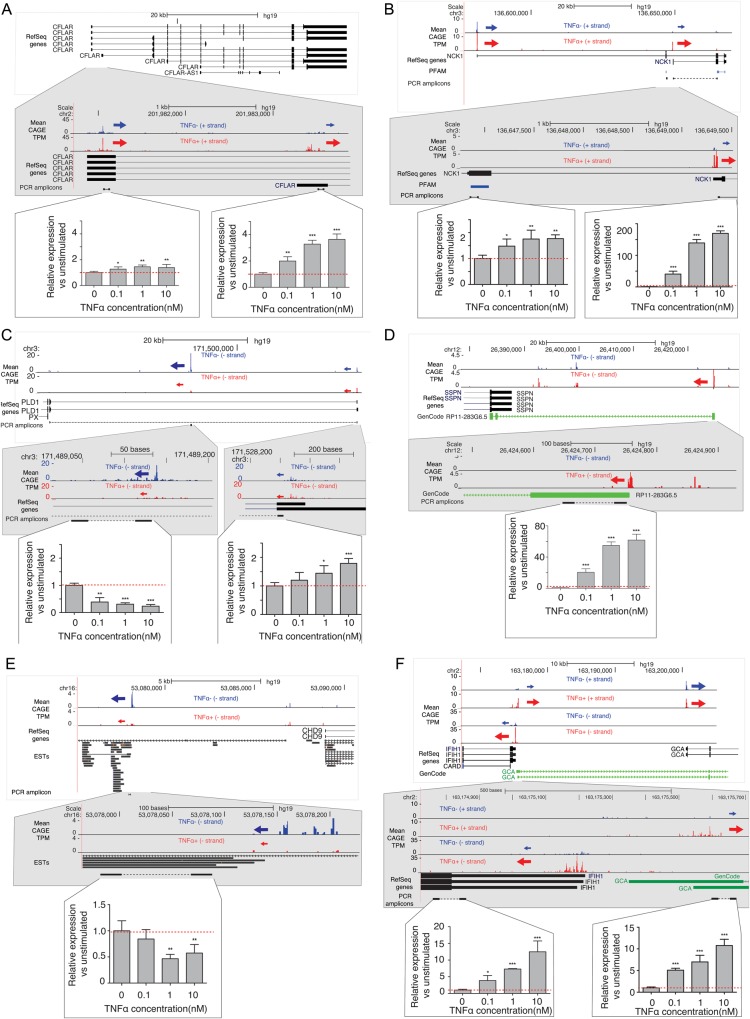
Figure 2.
Examples of CAGE-inferred promoters and their response to TNF-α stimulation. Each panel describes a validated gene locus: (A) CFLAR, (B) NCK1, (C) PLD1, (D) the RP11-283G6.5 lncRNA, (E) novel putative lncRNA, (F) IFIH1 and a putative long distal promoter of GCA. Each panel consists of three sub-panels: top, an UCSC browser43 overview of the gene landscape around the promoter(s) of interest; middle, a zoom-in version of the above; bottom, qPCR validations of respective RNA isoforms emanating from the promoters of interests as a function of TNF-α concentration. UCSC browser sub-panels show (when relevant and available) RefSeq genes, expressed sequenced tags (ESTs), GenCode36 annotation and mean normalized CAGE signal per nucleotide on relevant strand over three replicates. Arrows indicate TSS clusters (core promoters) of interest and their direction of transcription. Sizes of arrows correspond to the CAGE signal strength. Locations of qPCR primer pairs are shown also in Supplementary Table S1. Respective examples are commented in detail in the main text. For qPCR bar plots, the vertical axis shows the mean fold change vs. un-stimulated cells (four replicates), error bars indicate standard error of mean. The dotted line indicates a fold change of 1. The statistical significance (t-test) is indicated above the bars: *P < 0.05, **P < 0.01, ***P < 0.001. This figure appears in colour in the online version of DNA Research.
The first example is the CFLAR gene (Fig. 2A), a regulator of apoptosis, known to be up-regulated in IECs from patients with active UC.25 The most upstream promoter is only slightly TNF-α-responsive (1.3-fold increase, P < 0.01), while the alternative promoter is more responsive (3.6-fold, P < 0.001). Similarly, the NCK1 gene, involved in signal transduction, has an alternative promoter with a high up-regulation (170-fold, P < 0.001) at the highest concentration of TNF-α, while the response of the most upstream promoter is less than two-fold (P < 0.01) (Fig. 2B). The alternative promoter is predicted to lead to loss of an SH3 domain, described further below.
The PLD1 gene, a phospoholipase used in signal transduction (Fig. 2C) and utilized by TNF-α to activate the NF-κB and extracellular signal regulated kinase (ERK1/2) during an immune response,44 has two promoters defined by CAGE, where the annotated most upstream promoter is slightly activated by TNF-α (1.8-fold increase P < 0.001), but the novel promoter found ∼30 kb downstream is down-regulated more than 4-fold (P < 0.001) (Fig. 2C). This demonstrates that the genes are not only up-regulated as a consequence of TNF-α stimulation.
We also confirmed that two uncharacterized, long, non-coding RNAs (lncRNAs) found by CAGE are responding to TNF-α stimulation. The first (Fig. 2D), annotated by GENCODE36 as RP11-283G6.5 extends 40 kb downstream to overlap the 3′ UTR of the SSPN gene on the antisense strand, is increased more than 60-fold in response to TNF-α (P < 0.001). Such chains of linked sense–antisense RNAs have been previously described45 and sense–antisense pairs have the potential to influence each other's expression levels.46 The other lncRNA (Fig. 2E) only has support from ENCODE RNA-seq and ESTs, is situated in a large intergenic region and is 50% down-regulated as a response to TNF-α (P < 0.01). Since this lncRNA is functionally uncharacterized, further experiments are needed to deduce the impact of this down-regulation, but we note that there is evidence for other lncRNAs with direct functional roles in inflammation, such as the LEHTE lncRNA which is induced by TNF-α, and binds NF-κB in a negative feedback loop.47
Finally, we validated a TNF-α-responsive bidirectional promoter composed of the annotated IFIH1 gene (involved in interferon response and linked to autoimmune diseases) and a paired novel promoter starting on the opposite strand ∼200 bp upstream of the IFIH1 promoter (both promoters are >12-fold up-regulated, P < 0.001) (Fig. 2F). GENCODE gene annotation36 shows that the novel promoter on the plus strand is likely a distal promoter of the GCA gene, a regulator of adhesion. We note that the canonical promoter of GCA is expressed on similar levels before and after TNF-α. Therefore, we hypothesize that usage of the bidirectional promoter will make GCA partially responsive to TNF-α.
3.2. Loss of protein domain-coding exons due to alternative promoter usage
As discussed above, many of the responding TCs correspond to known or novel alternative promoter candidates. Related to this, 41 RefSeq gene models contained two or more significantly responding TCs. In most of the cases (36), all TCs within a gene were up-regulated, in four cases both TCs were down-regulated and in one case (TMEM150B) one TC was significantly up-regulated while the other TC was significantly down-regulated.
A handful of genes where the use of an alternative promoter leads to exclusion of coding exons have been described previously.48 One example is the promoter usage in the LEF1 gene. This gene contains an N-terminal domain necessary and sufficient to bind β-catenin, a central domain that interacts with co-repressors and a C-terminal DNA-binding domain. The protein has two promoters: promoter 1 produces full-length proteins which recruit β-catenin to Wnt signalling, and promoter 2 produces proteins lacking the β-catenin-binding domain and therefore functions as repressors of Wnt signalling, hence the two isoforms will have opposing effects. An imbalance in the usage of these to promoters is, for example, seen in colon cancer, where promoter 2 is silenced.49
Our CAGE data make it possible to assess the frequency of these types of events in Caco-2 as a response to TNF-α. We found that 75 of the TNF-α(+) promoters are within an annotated coding region. These represent shorter RNA isoforms, which, if translated, would give rise to N-termini-truncated proteins. To identify cases similar to the LEF1 example above, we investigated if any of the intragenic TNF-α(+) promoters were located within or downstream of PFAM50 protein domains mapped to the coding sequence. In total, we found 39 promoters in 33 genes, where usage of the TNF-α(+) promoter would lead to RNAs that exclude one or more protein domains. Five of the promoters were within 100 bp from a known (alternative) RefSeq promoter, where all the gene products are annotated as protein coding. In these cases, it is likely that these TNF-α(+)-responsive promoters are producing proteins with truncated N-termini and loss of functional domains. For non-annotated alternative promoters, further validation is needed to prove that the produced RNAs are translated. We here show four examples of genes where exons containing domains are lost when using the TNF-α(+) promoters (Fig. 3).
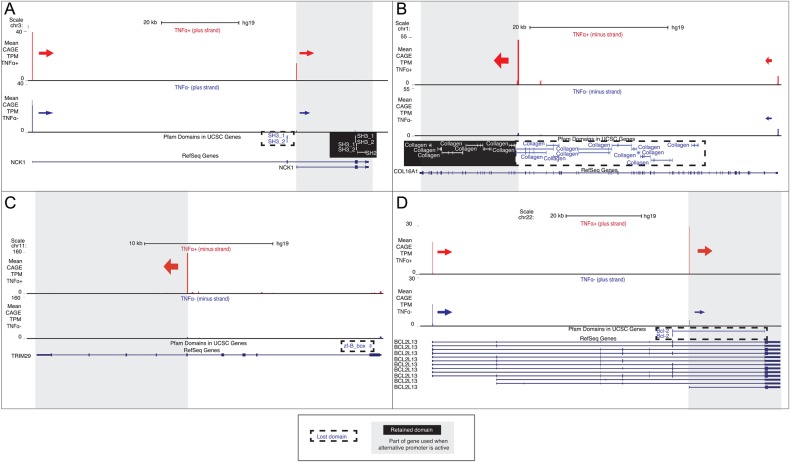
Figure 3.
Examples of loss of exons coding for protein domains due to alternative promoter usage. UCSC browser overview of the genes of interest: (A) NCK1, (B) COL16A1, (C) TRIM29, (D) BCL2L13 similar to Fig. 2. From top; (i) bar graph of normalized CAGE tag counts (TPM) from TNF-α(+) and TNF-α(−). Only tags on the relevant strand are shown; (ii) Pfam domains mapped to UCSC genes and (iii) RefSeq genes. Arrows indicate TSS clusters (core promoters) of interest and their direction of transcription. Sizes of arrows correspond to the CAGE signal strength. Grey regions indicate the part of the gene that is retained by usage of alternative promoters of interest. Lost, or partially lost, domains upstream of this region with dashed lines, while retained domains have a black background. Gene examples are commented in detail in the main text. This figure appears in colour in the online version of DNA Research.
The NCK1 gene, described and validated above (Fig. 2B), have two promoters; the upstream P1 at the canonical promoter and P2, which is an alternative TSS and preferentially used after TNF-α treatment. When using P2, one of the SH3_1 domains and one of the SH3_2 domains are lost (Fig. 3A). SH3 and SH2 domains are important for protein–protein interaction, and the alternative promoter usage could potentially lead to an up-regulation of Ras activity in the cell.51
A more extreme example of loss of multiple domains is COL16A1 (qPCR validation showed in Supplementary Figure S1G). This gene encodes the alpha chain of type XVI collagen and maintains the integrity of the extracellular matrix. COL16A1 contains several collagen domains and usage of the alternative downstream promoter decreases the number of collagen repeats from 18 to 10 (Fig. 3B). This exemplifies a trend in the dataset: if a domain is lost there is often (7 of 33 genes with alternative promoters analysed) another instance of the domain still present in the resulting transcript.
In some cases, usage of an alternative downstream promoter leaves a transcript without predicted domains. This is the case for the zinc finger protein TRIM29 (Fig. 3C). In the unstimulated state little transcription is observed from this gene, but after stimulation we observe high expression from an alternative promoter situated approximately in the middle of the gene. Using this promoter leads to loss of a B-box-type zinc finger domain, often involved in ubiquitinylation, and it is unclear whether the remaining transcript has any function on protein level (Fig. 3C).
Another example of a gene where no downstream domains are kept is the apoptotic regulator BCL2L13 (Fig. 3D). Most of the expression of this gene comes from two distinct promoters, P1 at the annotated 5′ end of the gene, and P2 mapping to an annotated start site, which corresponds to an RNA including only the last exon of the gene. In the unstimulated state P1 is primarily used. When stimulated the usage is switched: P2 now shows the highest expression. When P2 is used, the PFAM protein domain Bcl-2 is lost (Fig. 3D). The shorter transcript, produced from P2, has been described previously:52 overexpression of the transcript resulted in higher activation of caspase-3 and apoptosis than the full-length variant. A necessary caveat for these examples is that whereas CAGE data might reveal whether a protein-coding domains is lost due to alternative promoter usage, it cannot prove whether a certain alternative promoter produces a transcript that is translated into a protein, in particular in cases where the alternative promoter is not previously annotated and linked to sequenced full-length RNAs.
3.3. Identification of enhancer candidates responding to TNF-α
We have previously shown that bidirectional CAGE TCs situated at most 200 nt from each other can identify enhancer RNA.19 The method has been extensively validated with >100 in vitro reporter gene assays and shown to be a more accurate prediction method for enhancer activity than untranscribed hypersensitive sites or enhancer candidates predicted by chromatin immunoprecipitation.19 We have also shown that expression correlations between the enhancer activity and that of nearby promoters can identify physical interactions. With the same approach as in ref.,19 using a combination of the CAGE data from this study supplemented by CAGE data from a wide range of tissues,14,19 we found 890 Caco-2-expressed candidate enhancers, where 62% overlap Caco-2 DNase hypersensitive sites20 (Supplementary Table S3). Of these, 443 (49.7%) responded 4-fold or more to stimulation by TNF-α compared with control and 222 (of the 443) were up-regulated as a response to TNF-α stimulation. Within the list of 443 TNF-α-responsive enhancers, we found 37 unique enhancer–promoter pairs with a maximal distance of 200 kb, where the enhancer and the promoter expression are highly correlated before and after stimulation, suggesting regulatory interaction. An interesting example of such pairings is the enhancer 60 kb downstream of the UBR4 gene,53 linked by co-expression to a novel alternative promoter within the UBR4 locus. We subsequently validated the promoter expression change by qPCR (Fig. 4A). Other examples include the TNF-α-induced candidate enhancer region located within the first intron of the TNFSF10 gene—a member of the TNF ligand superfamily, whose annotated promoter is also highly induced by TNF-α (Fig. 4B), and an enhancer candidate 2 kb upstream of the ANXA13 gene (known to increase in expression during epithelial cell differentiation54) where both the gene promoter and the enhancer expression is decreased as a response to TNF-α (Fig. 4C).
Figure 4.
Examples of CAGE-inferred TNF-α responsive enhancers and their potential targets. Each panel consists of sub-panels: top, an UCSC browser43 overview of the gene landscape around the enhancer and promoter of interest; bottom, zoom-in version(s) of the above. (A) An enhancer is predicted downstream of the UBR4 gene identified by bidirectional CAGE tag pairs (left panel shows a zoom-in). It has support from multiple data types from the ENCODE project,21 including ChIP-seq for transcription factors, and histone marks typical of enhancers, as well as Caco-2-specific DNase sensitivity site peaks. CAGE data are shown as in Fig. 2. The enhancer is induced by TNF-α, and is predicted to interact with a CAGE-defined alternative promoter within the gene (middle panel), which is also induced by TNF-α (verified by qPCR as in Fig. 2). Conversely, the annotated promoter is not responding highly to TNF-α (right panel). (B) A TNF-α-induced enhancer is predicted within the first intron of the TNFSF10 gene, and predicted to interact with the annotated TSS, which also is highly TNF-α-induced. The enhancer has support by ENCODE histone marks and multiple transcription factor ChIP peaks. (C) An enhancer, which is only used in non-induced cells, is predicted 2 kb upstream of the annotated TSS of the ANXA13 gene, which also has much higher expression in the non-induced state. The enhancer has support by multiple ENCODE transcription factor ChIP peaks. See main text for further discussion. This figure appears in colour in the online version of DNA Research.
3.4. Regulation of LAMC2 TNF-α response: an example of CAGE-enabled targeted analysis
Computational prediction of transcription factor-binding sites in promoter regions identified by CAGE has proved to be a powerful approach for the identification of key transcription factors in a given system.6,10,16,17 Indeed, NF-κB-binding sites, and binding sites of other known inflammatory transcription factors such as SPIB (Table 2), are significantly overrepresented in TNF-α(+) promoters, which fits with previous work.56
Table 2.
Clover analysis of overrepresented transcription factor-binding sites in the −300 to +100 bp region covering CAGE TCs found in differentially expressed promoters of Caco-2, in response to TNF-α stimulation
| JASPAR motif | Clover score | Clover P-value | Promoter response |
|---|---|---|---|
| RELA | 245 | <0.001 | TNF-α(+) |
| NF-kappaB | 214 | <0.001 | TNF-α(+) |
| REL | 199 | <0.001 | TNF-α(+) |
| IRF1 | 198 | <0.001 | TNF-α(+) |
| SPIB | 142 | 0.006 | TNF-α(+) |
| IRF2 | 135 | <0.001 | TNF-α(+) |
| dl_2 | 131 | <0.001 | TNF-α(+) |
| MZF1_1-4 | 131 | <0.001 | TNF-α(+) |
| ACE2 | 103 | <0.001 | TNF-α(+) |
| AP1 | 95.4 | <0.001 | TNF-α(+) |
| NFIC | 41.7 | <0.001 | TNF-α(−) |
| HAP4 | 38.8 | 0.002 | TNF-α(−) |
| HAP3 | 37.5 | 0.007 | TNF-α(−) |
| ACE2 | 32.3 | 0.001 | TNF-α(−) |
| HNF4A | 24.8 | 0.001 | TNF-α(−) |
| nub | 23.7 | 0.002 | TNF-α(−) |
| SOK2 | 22.5 | <0.001 | TNF-α(−) |
| FOXA1 | 21.6 | 0.001 | TNF-α(−) |
| fkh | 20.5 | 0.002 | TNF-α(−) |
| sna | 14 | 0.009 | TNF-α(−) |
P-values are calculated by Clover.55 Motif names are taken from the JASPAR database.
We were encouraged that the same transcription factor-binding sites were also overrepresented in the CAGE-defined enhancers activated by TNF-α (Table 3). These sites were not overrepresented in TNF-α(−) promoters and enhancers, which instead had an overrepresentation of hepatocyte nuclear factor motifs (HNF-4α and HNF-3α) (Tables 2 and 3). Since HNF-4α is important for the differentiation of the IECs during embryonic development, the overrepresentation of HNF-4α sites in the non-stimulated promoters and enhancers could indicate that unstimulated cells are differentiating towards an IEC-like state57 while differentiation is repressed in stimulated cells.
Table 3.
Clover analysis of overrepresented transcription factor-binding sites in the −300 to +300 bp region around the centre of CAGE-defined enhancers active in Caco-2, in response to TNF-α stimulation
| JASPAR motif | Clover score | Clover P-value | Enhancer response |
|---|---|---|---|
| SPIB | 114 | 0.008 | TNF-α(+) |
| AP1 | 87.3 | <0.001 | TNF-α(+) |
| RUNX1 | 57.1 | <0.001 | TNF-α(+) |
| NFE2L2 | 53.3 | <0.001 | TNF-α(+) |
| ARG81 | 53.2 | 0.001 | TNF-α(+) |
| HCM1 | 49.5 | 0.002 | TNF-α(+) |
| FOXI1 | 32.8 | <0.001 | TNF-α(+) |
| RELA | 28.4 | <0.001 | TNF-α(+) |
| NF-kappaB | 23.4 | <0.001 | TNF-α(+) |
| MET32 | 21.9 | 0.003 | TNF-α(+) |
| HNF4A | 103 | <0.001 | TNF-α(−) |
| EWSR1-FLI1 | 17.3 | <0.001 | TNF-α(−) |
| Gata1 | 6.82 | 0.003 | TNF-α(−) |
| NDT80 | 5.24 | 0.006 | TNF-α(−) |
| CG15696 | 1.74 | 0.007 | TNF-α(−) |
| En | 0.827 | 0.009 | TNF-α(−) |
| slou | 0.742 | 0.004 | TNF-α(−) |
| Hmx | 0.513 | 0.001 | TNF-α(−) |
| Unc-4 | 0.473 | 0.002 | TNF-α(−) |
| tup | −0.18 | 0.002 | TNF-α(−) |
P-values are calculated by Clover.55
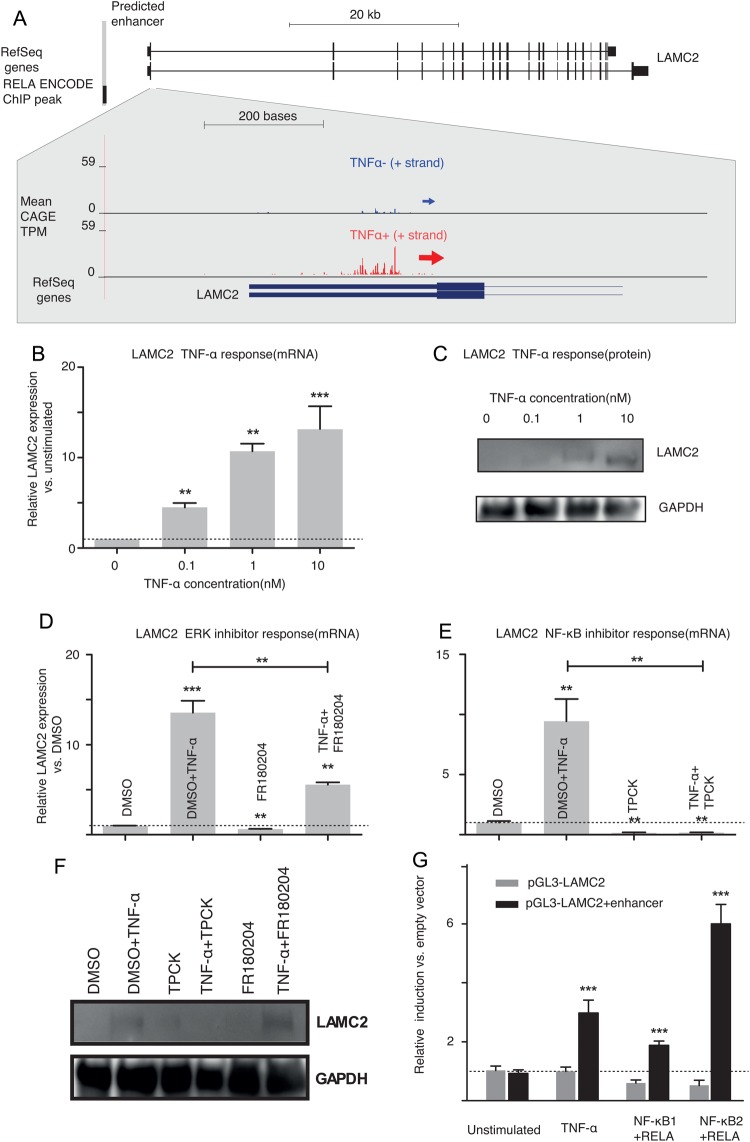
As a case example, we focused on the LAMC2 gene (Fig. 5A) whose main promoter is >8-fold up-regulated in TNF-α-stimulated Caco-2 cells as measured by CAGE. LAMC2 has previously been shown to be induced by TNF-α in other cell lines.58 Indeed, we validated that TNF-α stimulation (P < 0.05, t-test) increased the expression of LAMC2 mRNA and protein levels in a dose-dependent manner with a maximal effect observed at 10 nM (Fig. 5B and C). TNF-α can initiate pro-inflammatory signalling by activating the NF-κB pathway and the mitogen-activated protein kinase (MAPK) pathway. In mammals there are three major constituents of the MAPK superfamily: ERKs (ERK1/2 or p42/p44), c-Jun N-terminal kinases (JNK1/2/3) and the p38 MAPK family.56 The NF-κB family of transcription factors consists of five mammalian members [NFKB1(p50), NFKB2(p52), RELA(p65) and RELB]59,60 that can form either homodimers or heterodimers. Therefore, the role of these signalling pathways in TNF-α-mediated up-regulation of endogenous LAMC2 expression in Caco-2 cells was investigated. Interestingly, we noticed a ∼400 nt long candidate enhancer region (chr1: 183,149,939–183,150,336, hg19 assembly) upstream of the LAMC2 TSS, which is predicted to have a single NF-κB-binding site and is also bound by RELA as indicated by ENCODE ChIP-seq21 data (Fig. 5A). Consistent with the hypothesis of NF-κB-mediated regulation, the NF-κB inhibitor TPCK effectively diminished the effect of TNF-α on LAMC2 mRNA and protein levels (Fig. 5E and F). Moreover, it has previously been demonstrated that an active ERK pathway is involved in the regulation of LAMC2 in squamous cell carcinoma.61 However, inhibition of ERK1/2 with FR180204 had only a minor effect on the TNF-α-induced LAMC2 mRNA expression (Fig. 5D) and almost no effect on the LAMC2 protein levels (Fig. 5F) in our experiments. Since these findings indicate a transcriptional up-regulation by TNF-α and NF-κB, we measured the effect of TNF-α on the activity of an LAMC2 promoter construct32 with and without the predicted enhancer; the enhancerless construct did not respond to TNF-α, but the enhancer–promoter construct gave a 3-fold increase in reporter signal (P < 0.001 (t-test)) (Fig. 5G). Validating the causal role of NF-κB, co-transfection of LAMC2 reporter constructs with RELA in combination with NFKB1 or NFKB2-expression plasmids could significantly (P < 0.001 (t-test)) up-regulate the enhancer–promoter construct but not the enhancerless construct. This effect was highest in Caco-2 cells overexpressing both RELA and NFKB2 subunits (P < 0.001) (Fig. 5G).
Figure 5.
TNF-α regulates LAMC2 transcriptional activity by an NF-κB-bound enhancer. (A) An overview of the LAMC2 gene and its predicted enhancer location upstream of the LAMC2 TSS. The RELA ChIP peak overlapping the predicted enhancer is taken from the ENCODE UCSC ChIP track.21 Genome browser zoom-in shows CAGE data around the annotated TSS as shown in Fig. 2. (B) Dose-dependent increase of LAMC2 mRNA expression in cells treated with increasing concentrations of TNF-α (0, 0.1, 1 or 10 nM). (C) Western blot analysis of LAMC2 protein levels in TNF-α-treated cells and GAPDH used as an internal loading control. The shown blot represents three independent experiments. The effect of (D) FR180204, an ERK inhibitor, and (E) TPCK, an NF-κB inhibitor, on the TNF-α-mediated up-regulation of LAMC2 mRNA expression and (F) LAMC2 and GAPDH protein levels. Cells were pre-treated with inhibitors or with vehicle DMSO (0.4%) for 1 h and then incubated with TNF-α (10 nM) for 24 h. (G) The effect of TNF-α and overexpression of NF-κB subunits on the LAMC2 promoter activity with and without the enhancer shown in (A). Cells were transiently transfected with the human LAMC2 promoter (pGL3-LAMC2) (grey bars) or LAMC2 promoter/enhancer construct (pGL3-LAMC2 + enhancer) (black bars) and were unstimulated, stimulated with TNF-α for 24 h (black bars), co-transfected with plasmids overexpressing NFKB1(p50) and RELA NF-κB subunits, or co-transfected with plasmids overexpressing NFKB2 and RELA NF-κB subunits. Asterisks indicate levels of significance: *P < 0.05, **P < 0.01, ***P < 0.001 (t-test): error bars indicate the standard error of the mean. This figure appears in colour in the online version of DNA Research.
4. Discussion
Here, we have presented the first TSS map of the Caco-2 cell line, and the changes in TSS usage as a response to TNF-α. We show that almost half of the TNF-α-activated promoters are novel, underscoring the importance of alternative and novel promoters in particular and the merit of experimental methods not based on annotated genes. Several of these have the potential to produce gene products that loose coding regions for important domains. Regardless of whether the candidate promoters were known, overlapped known genes or represented novel transcripts, qPCR-based experiments could validate the large majority of the chosen promoters as TNF-α responsive promoters (negative or positive).
CAGE has previously been used to locate active enhancer regions and their putative gene targets,19 and its application to Caco-2 cells highlights a large number of enhancer candidates, where many are TNF-α-responsive. Interestingly, we observed that the same type of transcription factor-binding sites were overrepresented in both promoters and enhancer regions that were activated as a response to TNF-α. Related to this, a case study guided by the CAGE technique identified an NF-κB-responsive enhancer region regulating the LAMC2 gene, which explains its responsiveness to TNF-α.62 LAMC2 has previously been shown to be a useful biomarker in the diagnosis of the invasiveness of cervical adenocarcinoma63 and in colorectal cancer,64 but was here shown to be a potential inflammatory marker as well.
We have here presented comprehensive maps of the locations and activity of promoters and enhancers in the Caco-2 cell line. Importantly, many of the responding promoters are uncharacterized and comprise either novel alternative promoters (often to key inflammatory genes) or TSSs of uncharacterized RNAs that in all likelihood are non-coding, which could not have been detected by array-based methods. Therefore, our data are a rich resource for empowering detailed, gene-centred studies in Caco-2.
5. Availability
The CAGE data are available at the NCBI GEO database (accession GSE54074).
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Authors’ contributions
M.B. made the CAGE libraries. M.B., M.C. and J.B. made the qPCR validations. M.C., K.D., J.O. and J.T.T. made the experiments on LAMC2. M.C., J.T.T., J.T.B., J.B.S. and O.H.N. prepared Caco-2 cells. B.L., M.B., I.H., R.A., M.V. and A.S .made the computational analysis. M.B., M.C., B.L., and A.S. wrote the paper with input from all authors. J.T.T., M.B., M.C. and A.S. designed the project.
Funding
This work was supported by grants from the Novo Nordisk Foundation, the Lundbeck Foundation, the Foundation of Aase and Ejnar Danielsen, Harboe Foundation, the Foundation of Frode V. Nyegaard and Wife, the Commemorative Foundation of Family Erichsen and the Foundation of Director Emil C. Hertz and wife Inger Hertz. The CAGE libraries were sequenced at the National High-throughput DNA Sequencing Centre, Copenhagen. Funding to pay the Open Access publication charges for this article was provided by the Lundbeck Foundation.
Supplementary Material
References
- 1.Dahan S., Roth-Walter F., Arnaboldi P., Agarwal S., Mayer L. Epithelia: lymphocyte interactions in the gut. Immunol. Rev. 2007;215:243–53. doi: 10.1111/j.1600-065X.2006.00484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koch S., Nusrat A. The life and death of epithelia during inflammation: lessons learned from the gut. Annu. Rev. Pathol. 2012;7:35–60. doi: 10.1146/annurev-pathol-011811-120905. [DOI] [PubMed] [Google Scholar]
- 3.Ordás I., Mould D.R., Feagan B.G., Sandborn W.J. Anti-TNF monoclonal antibodies in inflammatory bowel disease: pharmacokinetics-based dosing paradigms. Clin. Pharmacol. Ther. 2012;91:635–46. doi: 10.1038/clpt.2011.328. [DOI] [PubMed] [Google Scholar]
- 4.Nielsen O.H., Ainsworth M.A. Tumor necrosis factor inhibitors for inflammatory bowel disease. N. Engl. J. Med. 2013;369:754–62. doi: 10.1056/NEJMct1209614. [DOI] [PubMed] [Google Scholar]
- 5.Hidalgo I.J., Raub T.J., Borchardt R.T. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology. 1989;96:736–49. [PubMed] [Google Scholar]
- 6.Carninci P., Sandelin A., Lenhard B., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–35. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 7.Pal S., Gupta R., Kim H., et al. Alternative transcription exceeds alternative splicing in generating the transcriptome diversity of cerebellar development. Genome Res. 2011;21:1260–72. doi: 10.1101/gr.120535.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kornblihtt A.R., Schor I.E., Alló M., Dujardin G., Petrillo E., Muñoz M.J. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol. 2013;14:153–65. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- 9.Sandberg R., Neilson J.R., Sarma A., Sharp P.A., Burge C.B. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320:1643–7. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valen E., Pascarella G., Chalk A., et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2008;19:255–65. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kodzius R., Kojima M., Nishiyori H., et al. CAGE: cap analysis of gene expression. Nat. Methods. 2006;3:211–22. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita R., Sathira N.P., Kanai A., et al. Genome-wide characterization of transcriptional start sites in humans by integrative transcriptome analysis. Genome Res. 2011;21:775–89. doi: 10.1101/gr.110254.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The FANTOM Consortium. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 14.The FANTOM Consortium and the RIKEN PMI and CLST(DGT) A promoter level mammalian expression atlas. Nature. 2014;507:462–70. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandelin A., Carninci P., Lenhard B., Ponjavic J., Hayashizaki Y., Hume D.A. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–36. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 16.Lenhard B., Sandelin A., Carninci P. Metazoan promoters: emerging characteristics and insights into transcriptional regulation. Nat. Rev. Genet. 2012;13:233–45. doi: 10.1038/nrg3163. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki H., Forrest A.R.R., van Nimwegen E., et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat. Genet. 2009;41:553–62. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawaji H., Lizio M., Itoh M., et al. Comparison of CAGE and RNA-seq transcriptome profiling using clonally amplified and single-molecule next-generation sequencing. Genome Res. 2014;24:708–17. doi: 10.1101/gr.156232.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., et al. The FANTOM Consortium. An atlas of active enhancers across the human body. Nature. 2014;507:455–61. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thurman R.E., Rynes E., Humbert R., et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The ENCODE Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ernst J., Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010;28:817–25. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coskun M., Boyd M., Olsen J., Troelsen J.T. Control of intestinal promoter activity of the cellular migratory regulator gene ELMO3 by CDX2 and SP1. J. Cell. Biochem. 2010;109:1118–28. doi: 10.1002/jcb.22490. [DOI] [PubMed] [Google Scholar]
- 24.Coskun M., Olsen A.K., Bzorek M., et al. Involvement of CDX2 in the cross talk between TNF-α and Wnt signaling pathway in the colon cancer cell line Caco-2. Carcinogenesis. 2014;35:1185–92. doi: 10.1093/carcin/bgu037. [DOI] [PubMed] [Google Scholar]
- 25.Seidelin J.B., Coskun M., Vainer B., Riis L., Soendergaard C., Nielsen O.H. ERK controls epithelial cell death receptor signalling and cellular FLICE-like inhibitory protein (c-FLIP) in ulcerative colitis. J. Mol. Med. 2013;91:839–49. doi: 10.1007/s00109-013-1003-7. [DOI] [PubMed] [Google Scholar]
- 26.Ntini E., Järvelin A.I., Bornholdt J., et al. Polyadenylation site-induced decay of upstream transcripts enforces promoter directionality. Nat. Struct. Mol. Biol. 2013;20:923–8. doi: 10.1038/nsmb.2640. [DOI] [PubMed] [Google Scholar]
- 27.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 29.Portales-Casamar E., Thongjuea S., Kwon A.T., et al. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2009;38:D105–10. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frith M.C., Valen E., Krogh A., Hayashizaki Y., Carninci P., Sandelin A. A code for transcription initiation in mammalian genomes. Genome Res. 2007;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coskun M., Troelsen J.T., Nielsen O.H. The role of CDX2 in intestinal homeostasis and inflammation. Biochim. Biophys. Acta Mol. Basis Dis. 2011;18,12:283–9. doi: 10.1016/j.bbadis.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Olsen J., Lefebvre O., Fritsch C., et al. Involvement of activator protein 1 complexes in the epithelium-specific activation of the laminin γ2-chain gene promoter by hepatocyte growth factor (scatter factor) Biochem. J. 2000;347:407–17. doi: 10.1042/0264-6021:3470407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersson S., Davis D.L., Dahlbäck H., Jörnvall H., Russell D.W. Cloning, structure, and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J. Biol. Chem. 1989;264:8222–9. [PubMed] [Google Scholar]
- 34.Béraud C., Sun S.C., Ganchi P., Ballard D.W., Greene W.C. Human T-cell leukemia virus type I Tax associates with and is negatively regulated by the NF-kappa B2 p100 gene product: implications for viral latency. Mol. Cell. Biol. 1994;14:1374–82. doi: 10.1128/mcb.14.2.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hubbard T.J.P., Aken B.L., Ayling S., et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–7. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derrien T., Johnson R., Bussotti G., et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma A., Malynn B.A. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat. Rev. Immunol. 2012;12:774–85. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singer I.I., Kawka D.W., Schloemann S., Tessner T., Riehl T., Stenson W.F. Cyclooxygenase 2 is induced in colonic epithelial cells in inflammatory bowel disease. Gastroenterology. 1998;115:297–306. doi: 10.1016/s0016-5085(98)70196-9. [DOI] [PubMed] [Google Scholar]
- 39.Micheau O., Lens S., Gaide O., Alevizopoulos K., Tschopp J. NF-κB signals Induce the expression of c-FLIP. Mol. Cell Biol. 2001;21:5299–305. doi: 10.1128/MCB.21.16.5299-5305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin H.S., Zhao Z., Satsu H., Totsuka M., Shimizu M. Synergistic effect of tumor necrosis factor-alpha and hydrogen peroxide on the induction of IL-8 production in human intestinal Caco-2 cells. Inflammation. 2010;34:440–7. doi: 10.1007/s10753-010-9251-y. [DOI] [PubMed] [Google Scholar]
- 41.Poelkens F., Lammers G., Pardoel E.M., Tack C.J., Hopman M.T.E. Upregulation of skeletal muscle inflammatory genes links inflammation with insulin resistance in women with the metabolic syndrome. Exp. Physiol. 2013;98:1485–94. doi: 10.1113/expphysiol.2013.072710. [DOI] [PubMed] [Google Scholar]
- 42.Mechtcheriakova D., Wlachos A., Sobanov J., et al. Sphingosine 1-phosphate phosphatase 2 is induced during inflammatory responses. Cell. Signal. 2007;19:748–60. doi: 10.1016/j.cellsig.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 43.Rhead B., Karolchik D., Kuhn R.M., et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2009;38:D613–9. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sethu S., Pushparaj P.N., Melendez A.J. Phospholipase D1 mediates TNFα-induced inflammation in a murine model of TNFα-induced peritonitis. PLoS One. 2010;5:e10506. doi: 10.1371/journal.pone.0010506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engström P.G., Suzuki H., Ninomiya N., et al. Complex loci in human and mouse genomes. PLoS Genet. 2006;2:e47. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katayama S., Tomaru Y., Kasukawa T., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–6. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 47.Rapicavoli N.A., Qu K., Zhang J., Mikhail M., Laberge R.-M., Chang H.Y. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. Elife. 2012;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davuluri R.V., Suzuki Y., Sugano S., Plass C., Huang T.H.M. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–77. doi: 10.1016/j.tig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 49.Li T.W.-H., Ting J.-H.T., Yokoyama N.N., Bernstein A., van de Wetering M., Waterman M.L. Wnt activation and alternative promoter repression of LEF1 in colon cancer. Mol. Cell. Biol. 2006;26:5284–99. doi: 10.1128/MCB.00105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Finn R.D., Bateman A., Clements J., et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ger M., Zitkus Z., Valius M. Adaptor protein Nck1 interacts with p120 Ras GTPase-activating protein and regulates its activity. Cell. Signal. 2011;23:1651–8. doi: 10.1016/j.cellsig.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 52.Kataoka T., Holler N., Micheau O., et al. Bcl-rambo, a novel Bcl-2 homologue that induces apoptosis via its unique C-terminal extension. J. Biol. Chem. 2001;276:19548–54. doi: 10.1074/jbc.M010520200. [DOI] [PubMed] [Google Scholar]
- 53.Nakatani Y., Koneshi H., Vassilev A., et al. p600, a unique protein required for membrane morphogenesis and cell survival. Proc. Natl Acad. Sci. USA. 2005;102:15093–8. doi: 10.1073/pnas.0507458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wice B.M., Gordon J.I. A strategy for isolation of cDNAs encoding proteins affecting human intestinal epithelial cell growth and differentiation: characterization of a novel gut-specific N-myristoylated annexin. J. Cell. Biol. 1992;116:405–22. doi: 10.1083/jcb.116.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frith M.C. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–81. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coskun M., Olsen J., Seidelin J.B., Nielsen O.H. MAP kinases in inflammatory bowel disease. Clin. Chimic. Acta. 2011;412:513–20. doi: 10.1016/j.cca.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 57.Babeu J.-P., Darsigny M., Lussier C.R., Boudreau F. Hepatocyte nuclear factor 4α contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;297:G124–34. doi: 10.1152/ajpgi.90690.2008. [DOI] [PubMed] [Google Scholar]
- 58.Francoeur C., Escaffit F., Vachon P.H., Beaulieu J.-F. Proinflammatory cytokines TNF-α and IFN-γ alter laminin expression under an apoptosis-independent mechanism in human intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2004;287:G592–8. doi: 10.1152/ajpgi.00535.2003. [DOI] [PubMed] [Google Scholar]
- 59.Ghosh S, Michael J., May A., Kopp E.B. NF-κB and REL PROTEINS: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 2003;16:225–60. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 60.Hoffmann A., Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol. Rev. 2006;210:171–86. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- 61.Degen M., Natarajan E., Barron P., Widlund H.R., Rheinwald J.G. MAPK/ERK-dependent translation factor hyperactivation and dysregulated laminin γ2 expression in oral dysplasia and squamous cell carcinoma. Am. J. Pathol. 2012;180:2462–78. doi: 10.1016/j.ajpath.2012.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zboralski D., Warscheid B., Klein-Scory S., et al. Uncoupled responses of Smad4-deficient cancer cells to TNFalpha result in secretion of monomeric laminin-gamma2. Mol. Cancer. 2010;9:65. doi: 10.1186/1476-4598-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Imura J., Uchida Y., Nomoto K., et al. Laminin-5 is a biomarker of invasiveness in cervical adenocarcinoma. Diagn. Pathol. 2012;7:105. doi: 10.1186/1746-1596-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kevans D., Wang L.M., Sheahan K., et al. Epithelial-Mesenchymal Transition (EMT) protein expression in a cohort of stage II colorectal cancer patients with characterized tumor budding and mismatch repair protein status. Int. J. Surg. Pathol. 2011;19:751–60. doi: 10.1177/1066896911414566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.