Abstract
Plants have evolved a sophisticated mechanism to sense the extracellular sulfur (S) status so that sulfate transport and S assimilation/metabolism can be coordinated. Genetic, biochemical, and molecular studies in Arabidopsis over the past 10 years have started to shed some light on the regulatory mechanism of the S response. Key advances in transcriptional regulation (SLIM1, MYB, and miR395), involvement of hormones (auxin, cytokinin, and abscisic acid) and identification of putative sensors (OASTL and SULTR1;2) are highlighted here. Although our current view of S nutrient sensing and signaling remains fragmented, it is anticipated that through further studies a sensing and signaling network will be revealed in the near future.
Keywords: sulfate, SULTR1;2, transporter, sensor, transceptor
TIGHTLY REGULATED SULFATE UPTAKE IS REQUIRED FOR SULFUR ASSIMILATION AND UTILIZATION
Plants have evolved a biosynthetic pathway to assimilate sulfate (SO42–), a primary source of the essential nutrient sulfur (S), into Cys and Met, which are then used for synthesis of proteins and various S-containing compounds including glucosinolates and glutathione (GSH; Takahashi et al., 2011). SO42– is taken up from the rhizosphere by roots and is subsequently translocated into shoots. Therefore, SO42– transport and assimilation must be tightly coordinated to meet the dynamic demand for S. SO42– uptake and translocation is mediated by transporters (SULTR) with specific gene products performing distinct and also overlapping functions (Gigolashvili and Kopriva, 2014). In Arabidopsis, two members of group 1 (SULTR1;1 and SULTR1;2) are high affinity SO42– transporters and mediate SO42– uptake into roots (Gigolashvili and Kopriva, 2014). Several members of Groups 2 and 3 are likely involved in SO42– translocation from roots to shoots, while Group 4 (SULTR4;1 and SULTR4;2) functions in vacuolar export of SO42– (Takahashi et al., 2011; Gigolashvili and Kopriva, 2014). In response to S deficiency, many of the transporter genes are transcriptionally up-regulated. The two most studied transporters, SULTR1;1 and SULTR1;2, have been shown to act redundantly in controlling SO42– uptake from roots, with SULTR1;2 having a major role (Takahashi et al., 2011; Gigolashvili and Kopriva, 2014). This tightly regulated transport system is critical for plant response and adaptation to the dynamically changing S nutrient environment.
KNOWLEDGE OF S SENSING AND SIGNALING REMAINS FRAGMENTED
To understand the regulatory mechanism of S sensing, transport and signaling, significant efforts have been made and exciting progress is summarized below.
INSIGHTS INTO TRANSCRIPTIONAL CONTROL IN S DEFICIENCY RESPONSE
Several transcriptome profiling studies reported that more than 1500 genes in Arabidopsis are up-or down-regulated by S deficiency (Hirai et al., 2003, 2004; Maruyama-Nakashita et al., 2003, 2006). These studies confirmed up-regulation of SULTR1;2 and other transporter genes, and led to the identification of two novel S-responsive genes, BGLU28 and SDI1, which have received considerable attentions. BGLU28 is the most strongly up-regulated gene in several of the studies and is hypothesized to act by releasing S from glucosinolate, which is potentially a major S storage compound in the vacuole (Maruyama-Nakashita et al., 2003, 2006; Dan et al., 2007). SDI1 is annotated as a protein similar to male sterility family protein MS5 and recent evidence suggests that its expression level can be used as a biosensor of S nutrient status (Howarth et al., 2009). Interestingly, a cis element has been identified called SURE that is necessary for S-deficiency control including transcriptional regulation of BLGU28 (Maruyama-Nakashita et al., 2005). Furthermore, transcriptional regulators have been identified. The SLIM1 mutants lack the ability to up-regulate S-response gene expression including that of SULTR1;2 (Maruyama-Nakashita et al., 2006). Although many of S-responsive genes (including BGLU28 and SDI1) are under SLIM1 control, others (e.g., APR2 and APR3) were not affected, strongly suggesting that although SLIM1 may be a major S-response transcription factor, additional transcriptional regulators are also involved. Consistent with this, several MYB transcription factors, in particular MYB28 and MYB29 which are transcriptionally repressed by S-deficiency, have been shown critical for transcriptional regulation of genes for the biosynthesis of glucosinolate which potentially serves as a critical S storage compound (Yatusevich et al., 2010). Recently, a microRNA gene (miR395) was shown to be important for regulating several target genes involved in S-deficiency response including SULTR2;1/AST68 and APS4 (Kawashima et al., 2009, 2011). Interestingly, miR395 was shown to be controlled by SLIM1 (Kawashima et al., 2011). Taken together, these studies provided an important foundation for understanding the transcriptional events in the nucleus.
EMERGING UNDERSTANDING OF THE ROLE OF PROTEIN PHOSPHORYLATION, DEGRADATION, AND HORMONES IN S DEFICIENCY RESPONSE
SULTR1;2 was shown to be regulated posttranscriptionally (Yoshimoto et al., 2007). The effects of inhibitors of protein kinase and proteasome have indicated that protein phosphorylation (Maruyama-Nakashita et al., 2004a) and degradation (Pootakham et al., 2010) are involved in regulating S transport and S-starvation response in Arabidopsis and Chlamydomonas, respectively. On the other hand, the role of hormones has been increasingly recognized as a key factor in S response. Based on surveys for the impact of several hormones on the S deficiency-activated expression of beta-conglycinin (Ohkama et al., 2002),SULTR1;2 (Maruyama-Nakashita et al., 2004b), and BGLU28 (Dan et al., 2007), it seems that auxin, cytokinin, and abscisic acid (ABA) are involved in negatively regulating S deficiency response. Cytokinin seems to have a broader effect in S response as all of the above three S response genes could be suppressed by exogenous application of this hormone. Furthermore, genetic evidence using a cytokinin receptor mutant cre1 demonstrated the negative regulatory role of cytokine on S uptake (Maruyama-Nakashita et al., 2004b). The negative regulatory role of ABA was first implicated by the observed suppression by S deficiency of an ABA response marker RD29B:GUS and down-regulation of BGLU28 by externally applied ABA (Dan et al., 2007). A role for ABA biosynthesis in S response was recently reported (Cao et al., 2014). Compared to ABA and cytokinin, the role of auxin in S response has received more attentions. Auxin was first implicated as a regulator of S deficiency response by the observed up-regulation of auxin-inducible genes (such as IAA28) and NITs (likely involved in auxin synthesis) under S deficiency (Nikiforova et al., 2003), although S deficiency did not significantly alter auxin level (Kutz et al., 2002). However, evidence obtained from the use of DR5:GUS, an auxin response marker, suggests that S deficiency inhibits auxin accumulation or response (Dan et al., 2007). Such an inhibitory effect of auxin biosynthesis was confirmed recently (Zhao et al., 2014). Furthermore, by applying auxin externally, the S deficiency-activated BGLU28 expression is down-regulated. The role of auxin response regulators such as IAA28 and ARF-2 in controlling expression of S metabolism genes has been implicated using a transgenic approach (Falkenberg et al., 2008), and a definite role of auxin was demonstrated by two genetic studies. An auxin signaling component called AXR1, which is a component of the 26S proteasome, was shown to be involved in the S deficiency response (Dan et al., 2007), in agreement with the subsequently reported role of protein degradation in Chlamydomonas S response (Pootakham et al., 2010). Another S response mutant is allelic to BIG (a calossin-like protein involved in polar auxin transport), indicating a role for auxin transport as well as auxin biosynthesis or response in S signaling (Kasajima et al., 2007).
Most interestingly, putative S sensors or sensing components have been reported. Cys homeostasis is tightly controlled by the Cys synthase complex which consists of Ser acetyltransferase (SAT, the enzyme producing the substrate for Cys biosynthesis) and O-acetylserine (thiol) lyase (OASTL, the enzyme producing L-Cys; Yi et al., 2010). Arabidopsis OASTL has three isoforms, OASTL-A1, OASTL-B, and OASTL-C, which are located in the cytosol, plastids, and mitochondria, respectively. OASTL-A1, the most abundant isoform, has been demonstrated in vitro to specifically interact with the STAS domain of SULTR1;2 (Shibagaki and Grossman, 2010). Interestingly, this interaction may be physiologically relevant as demonstrated in a heterologous yeast system. The interaction could enhance OASTL-A1 Cys synthesis activity at the same time it inhibits SULTR1;2 transport activity. This reciprocal activity regulation has led to the proposal that OASTL-A1 is involved in sensing of S status (Shibagaki and Grossman, 2010). OASTL-C has also been reported to act in Cys sensing (Wirtz et al., 2012). The questions remain whether these two differentially localized OASTL members sense Cys or SO42– located in different compartments and how they act to sense S status.
Most recent genetic and physiological evidence obtained from our groups have shown that besides its high affinity transport function, SULTR1;2 has a novel regulatory function (Zhang et al., 2014). Using BGLU28 promoter:GUS as a mutant screening tool, two novel alleles of SULTR1;2 were isolated that exhibit high GUS activity even under sufficient S conditions: sel1-15 (D108N) and sel1-16 (G208D). These two mutations lie in the predicted transmembrane (TM) helices TM11 and TM5. In contrast to all prior studies in which up-regulation of S response genes in sel1 mutants were interpreted as the result of compromised SO42– uptake and consequently lower accumulation of internal SO42– or its metabolites (Shibagaki et al., 2002; Maruyama-Nakashita et al., 2003; El Kassis et al., 2007), we have provided two lines of convincing physiological evidence that support the hypothesis that up-regulation of BGLU28 and three other genes (SULTR4;2, SDI1, and LSU1) could be independent of the compromised SO42– uptake and internal S status of the mutants (Zhang et al., 2014). First, under high concentration of SO42– (10 mM) which did not lead to a difference in internal SO42– concentration and GSH level, sel1-15/16 and a null allele (sel1-18) still had higher gene expression level than their wild-type (WT) backgrounds. Second, treatments with 1 mM Cys or 1 mM GSH in the SO42– deficiency medium (which did not lead to any difference in Cys uptake and/or internal GSH contents between the sel1 alleles and WT) also led to higher gene expression level in sel1-15/16/18. These results strongly suggest that the sel1 seedlings (in particular the expression in roots) grown under sufficient S behave as if they have been treated by certain degrees of S deficiency. In other words, the mutations in SULTR1;2 reduce sensitivity to the S-induced suppression of S response genes. The evidence points toward a novel function for SULTR1;2 in regulating S nutrient response besides its transport function. The possibility that SULTR1;2 acts as an S sensor is discussed in the next section.
CAN SULTR1;2 ACT AS A PUTATIVE PM-LOCALIZED SULFATE TRANSPORTING RECEPTOR?
Dual function transporters, like SULTR1;2 described above, are not unusual. Studies in yeast and animal nutrient transport and sensing have revealed the existence of classic receptors (which are not involved in transport, e.g., G-protein-coupled receptor Gpr1), transceptors (which are either transporting receptors, e.g., Gap1, or non-transporting receptors, e.g., Snf3) and the majority of common transporters (which do not have a sensing function; Thevelein and Voordeckers, 2009). Therefore, transceptors can be considered at the boundary between receptors and transporters. In general, to demonstrate a receptor function for a transporter molecule, genetic or pharmacological evidence is required that shows decoupling of nutrient transport and signaling, i.e., the signaling output is independent of transport.
In the case of SULTR1;2, the mutations in TM1 (sel1-15) or TM5 (sel1-16) could abolish both SO42– transport and signaling (as measured by expression of S response genes), but the defect in signaling could be independent of SO42– transport and accumulation (Zhang et al., 2014). Because of this, we propose that SULTR1;2 can function as a putative SO42– transceptor (Figure 1). Although SULTR1;2 cannot be the only S-sensor since the sel1-15/16 mutants show reduced sensitivity to S but does not entirely abolish the S-limitation response, this finding provides a first intriguing insight into S-sensing in plants given its PM location where extracellular SO42– is first in contact with the PM-localized sensors. Note that a dual-affinity nitrate transporter called NRT1.1 has been demonstrated to act as a nitrate sensor (Ho et al., 2009; Bouguyon et al., 2012), and thus using nutrient transporters to sense the external nutrient status may be evolutionally conserved and advantageous to plants. Indeed, a phosphate transceptor (Pho84) has been reported in yeast (Popova et al., 2010). More encouraging is that in yeast SO42– transporters Sul1/2 have also been described as being transceptors (Conrad et al., 2014). To gain further insights into the evolutionarily conserved mechanism of using sulfate transporters as sensors, we performed a sequence alignment using transporters from Arabidopsis, rice, Chlamydomonas, yeast, Drosophila and humans that are most closely related to Arabidopsis SULR1;2. The result (Figure 2) shows that while D108 is only specific to SULTR1 group in Arabidopsis and rice, G208 is highly conserved in all transporters. It will be interesting to determine whether G208 is critical for SO42– transport and signaling in many eukaryotes.
FIGURE 1.
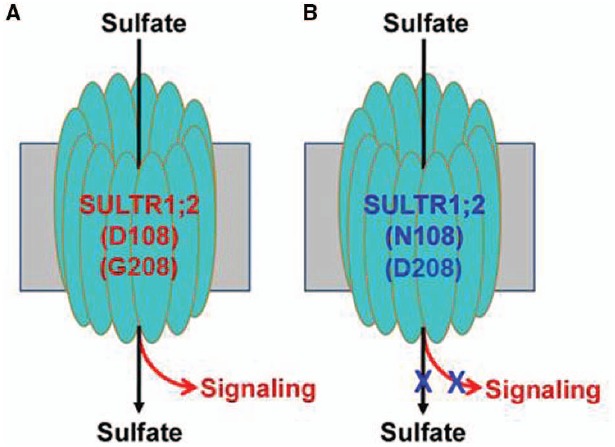
A hypothetical model for the dual function transceptor SULTR1;2. (A) The normal (wild-type) transceptor functions both in SO42– transport and signaling; (B) the transceptor is defective both in transport and signaling due to the mutations of D108N or G208D.
FIGURE 2.
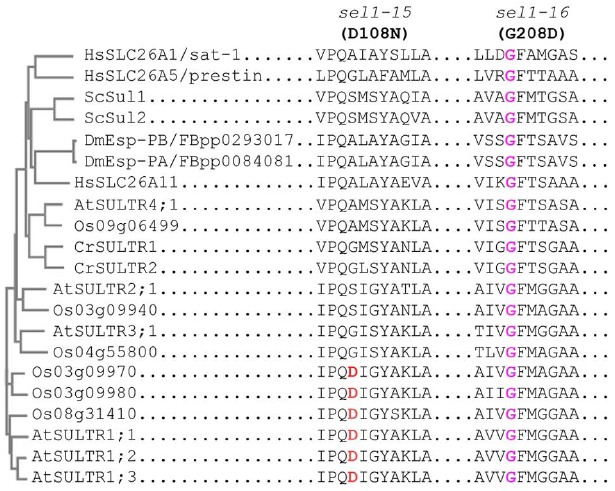
Phylogenetic relationships of SULTR1;2 and its closely related members of transporters in representative eukaryotes. The phylogenetic tree for SULTR1;2 and its closely related transporters, which is constructed using their full-length amino acid sequences, is shown on the left. The amino acid sequence alignment of the motifs surrounding D108 (sel1-15) and G208 (sel1-16) of SULTR1;2 and similar regions for other closely related transporters is shown on the right. At, Arabidopsis thaliana; Cr, Chlamydomonas reinhardtti; Dm, Drosophila melanogaster; Hs, Homo sapiens; Os, Oryza sativa; Sc, Saccharomyces cerevisiae.
It remains unclear how plants use SULTR1;2 to sense external SO42– status and adopt a high or low affinity transport system in response to dynamic S environment. However, studies from the yeast amino acid transceptor Gap1 or Arabidopsis nitrate transceptor NRT1.1 may provide some hints for the SULTR1;2-mediated sensing mechanism. In NRT1.1-mediated nitrate sensing and signaling, auxin transport and NRT1.1 phosphorylation have been shown to be critical (Ho et al., 2009; Bouguyon et al., 2012). In yeast, Gap1 uses the same sites for amino acid binding/transport and signaling (Van Zeebroeck et al., 2009; Conrad et al., 2014). Once amino acid is bound to Gap1, it triggers a conformational change in Gap1 that subsequently allows the amino acid be transported into the cytoplasm and in the same time a signaling cascade is activated. If the amino acid status is perceived to be sufficient, Gap1 undergoes a rapid endocytic process that removes it from the PM and sorts it for degradation.
FUTURE PROSPECT
Exciting findings in the past 10 years have led to the identification of several components from the PM to the cytoplasm and to the nucleus that are involved in S sensing, transport and downstream response. Several outstanding questions remained to be answered. What is the SULTR1;2 topology and does SULTR1;2 have separate sensing and transport domains? Can SULTR1;2 interact with OASTL in vivo (if so, which OASTL isoform?) and exert the effect of S sensing in planta? How does SULTR1;2 link to various signaling intermediates acting at the PM, the cytoplasm or the nucleus? Are there additional partners that may form a larger SULTR1;2-based S sensing complex? If such complex cannot account for all S responses, what other sensors are involved? Further, what are the roles of these sensing components in local and systemic S signaling (Hubberten et al., 2012)? Although our current view of S sensing and signaling remains fragmented, further studies into these questions will allow us to piece together individual components and ultimately construct the SULTR1;2-mediated S sensing and signaling pathway or network.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
ACKNOWLEDGMENTS
Research in our groups was initially funded in part by an NIH grant 3S06GM008225-20S1 (to Zhi-Liang Zheng) and a USDA HATCH grant (to Thomas Leustek) and is currently by a collaborative NSF grant (IOS-1121521 to Thomas Leustek and IOS-1121551 to Zhi-Liang Zheng). We apologize for not being able to cite many original or review articles due to the space limitation. We appreciate the reviewers comments.
REFERENCES
- Bouguyon E., Gojon A., Nacry P. (2012). Nitrate sensing and signaling in plants. Semin. Cell Dev. Biol. 23, 648–654. 10.1016/j.semcdb.2012.01.004 [DOI] [PubMed] [Google Scholar]
- Cao M. J., Wang Z., Zhao Q., Mao J. L., Speiser A., Wirtz M., et al. (2014). Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana. Plant J. 77, 604–615. 10.1111/tpj.12407 [DOI] [PubMed] [Google Scholar]
- Conrad M., Schothorst J., Kankipati H. N., Van Zeebroeck G., Rubio-Texeira M., Thevelein J. M. (2014). Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 38, 254–299. 10.1111/1574-6976.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan H., Yang G., Zheng Z. L. (2007). A negative regulatory role for auxin in sulphate deficiency response in Arabidopsis thaliana. Plant Mol. Biol. 63, 221–235. 10.1007/s11103-006-9084-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kassis E., Cathala N., Rouached H., Fourcroy P., Berthomieu P., Terry N., et al. (2007). Characterization of a selenate-resistant Arabidopsis mutant. Root growth as a potential target for selenate toxicity. Plant Physiol. 143, 1231–1241. 10.1104/pp.106.091462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenberg B., Witt I., Zanor M. I., Steinhauser D., Mueller-Roeber B., Hesse H., et al. (2008). Transcription factors relevant to auxin signalling coordinate broad-spectrum metabolic shifts including sulphur metabolism. J. Exp. Bot. 59, 2831–2846. 10.1093/jxb/ern144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili T., Kopriva S. (2014). Transporters in plant sulfur metabolism. Front. Plant Sci. 5:442. 10.3389/fpls.2014.00442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M. Y., Fujiwara T., Awazuhara M., Kimura T., Noji M., Saito K. (2003). Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J. 33, 651–663. 10.1046/j.1365-313X.2003.01658.x [DOI] [PubMed] [Google Scholar]
- Hirai M. Y., Yano M., Goodenowe D. B., Kanaya S., Kimura T., Awazuhara M., et al. (2004). Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101, 10205–10210. 10.1073/pnas.0403218101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C. H., Lin S. H., Hu H. C., Tsay Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. 10.1016/j.cell.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Howarth J. R., Parmar S., Barraclough P. B., Hawkesford M. J. (2009). A sulphur deficiency-induced gene, sdi1, involved in the utilization of stored sulphate pools under sulphur-limiting conditions has potential as a diagnostic indicator of sulphur nutritional status. Plant Biotechnol. J. 7, 200–209. 10.1111/j.1467-7652.2008.00391.x [DOI] [PubMed] [Google Scholar]
- Hubberten H. M., Drozd A., Tran B. V., Hesse H., Hoefgen R. (2012). Local and systemic regulation of sulfur homeostasis in roots of Arabidopsis thaliana. Plant J. 72, 625–635. 10.1111/j.1365-313X.2012.05105.x [DOI] [PubMed] [Google Scholar]
- Kasajima I., Ohkama-Ohtsu N., Ide Y., Hayashi H., Yoneyama T., Suzuki Y., et al. (2007). The BIG gene is involved in regulation of sulfur deficiency-response genes in Arabidopsis thaliana. Physiol. Plant. 129, 351–363 10.1111/j.1399-3054.2006.00814.x [DOI] [Google Scholar]
- Kawashima C. G., Matthewman C. A., Huang S., Lee B. R., Yoshimoto N., Koprivova A., et al. (2011). Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis. Plant J. 66, 863–876. 10.1111/j.1365-313X.2011.04547.x [DOI] [PubMed] [Google Scholar]
- Kawashima C. G., Yoshimoto N., Maruyama-Nakashita A., Tsuchiya Y. N., Saito K., Takahashi H., et al. (2009). Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 57, 313–321. 10.1111/j.1365-313X.2008.03690.x [DOI] [PubMed] [Google Scholar]
- Kutz A., Muller A., Hennig P., Kaiser W. M., Piotrowski M., Weiler E. W. (2002). A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 30, 95–106. 10.1046/j.1365-313X.2002.01271.x [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Inoue E., Watanabe-Takahashi A., Yamaya T., Takahashi H. (2003). Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiol. 132, 597–605. 10.1104/pp.102.019802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Tohge T., Saito K., Takahashi H. (2006). Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell 18, 3235–3251. 10.1105/tpc.106.046458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Watanabe-Takahashi A., Inoue E., Yamaya T., Takahashi H. (2005). Identification of a novel cis-acting element conferring sulfur deficiency response in Arabidopsis roots. Plant J. 42, 305–314. 10.1111/j.1365-313X.2005.02363.x [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Watanabe-Takahashi A., Yamaya T., Takahashi H. (2004a). Induction of SULTR1;1 sulfate transporter in Arabidopsis roots involves protein phosphorylation/dephosphorylation circuit for transcriptional regulation. Plant Cell Physiol. 45, 340–345. 10.1093/pcp/pch029 [DOI] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Yamaya T., Takahashi H. (2004b). A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 38, 779–789. 10.1111/j.1365-313X.2004.02079.x [DOI] [PubMed] [Google Scholar]
- Nikiforova V., Freitag J., Kempa S., Adamik M., Hesse H., Hoefgen R. (2003). Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J. 33, 633–650. 10.1046/j.1365-313X.2003.01657.x [DOI] [PubMed] [Google Scholar]
- Ohkama N., Takei K., Sakakibara H., Hayashi H., Yoneyama T., Fujiwara T. (2002). Regulation of sulfur-responsive gene expression by exogenously applied cytokinins in Arabidopsis thaliana. Plant Cell Physiol. 43, 1493–1501. 10.1093/pcp/pcf183 [DOI] [PubMed] [Google Scholar]
- Pootakham W., Gonzalez-Ballester D., Grossman A. R. (2010). Identification and regulation of plasma membrane sulfate transporters in Chlamydomonas. Plant Physiol. 153, 1653–1668. 10.1104/pp.110.157875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova Y., Thayumanavan P., Lonati E., Agrochao M., Thevelein J. M. (2010). Transport and signaling through the phosphate-binding site of the yeast Pho84 phosphate transceptor. Proc. Natl. Acad. Sci. U.S.A. 107, 2890–2895. 10.1073/pnas.0906546107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibagaki N., Grossman A. R. (2010). Binding of cysteine synthase to the STAS domain of sulfate transporter and its regulatory consequences. J. Biol. Chem. 285, 25094–25102. 10.1074/jbc.M110.126888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibagaki N., Rose A., Mcdermott J. P., Fujiwara T., Hayashi H., Yoneyama T., et al. (2002). Selenate-resistant mutants of Arabidopsis thaliana identify Sultr1;2, a sulfate transporter required for efficient transport of sulfate into roots. Plant J. 29, 475–486. 10.1046/j.0960-7412.2001.01232.x [DOI] [PubMed] [Google Scholar]
- Takahashi H., Kopriva S., Giordano M., Saito K., Hell R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184. 10.1146/annurev-arplant-042110-103921 [DOI] [PubMed] [Google Scholar]
- Thevelein J. M., Voordeckers K. (2009). Functioning and evolutionary significance of nutrient transceptors. Mol. Biol. Evol. 26, 2407–2414. 10.1093/molbev/msp168 [DOI] [PubMed] [Google Scholar]
- Van Zeebroeck G., Bonini B. M., Versele M., Thevelein J. M. (2009). Transport and signaling via the amino acid binding site of the yeast Gap1 amino acid transceptor. Nat. Chem. Biol. 5, 45–52. 10.1038/nchembio.132 [DOI] [PubMed] [Google Scholar]
- Wirtz M., Beard K. F., Lee C. P., Boltz A., Schwarzlander M., Fuchs C., et al. (2012). Mitochondrial cysteine synthase complex regulates O-acetylserine biosynthesis in plants. J. Biol. Chem. 287, 27941–27947. 10.1074/jbc.M112.372656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatusevich R., Mugford S. G., Matthewman C., Gigolashvili T., Frerigmann H., Delaney S., et al. (2010). Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana. Plant J. 62, 1–11. 10.1111/j.1365-313X.2009.04118.x [DOI] [PubMed] [Google Scholar]
- Yi H., Galant A., Ravilious G. E., Preuss M. L., Jez J. M. (2010). Sensing sulfur conditions: simple to complex protein regulatory mechanisms in plant thiol metabolism. Mol. Plant 3, 269–279. 10.1093/mp/ssp112 [DOI] [PubMed] [Google Scholar]
- Yoshimoto N., Inoue E., Watanabe-Takahashi A., Saito K., Takahashi H. (2007). Posttranscriptional regulation of high-affinity sulfate transporters in Arabidopsis by sulfur nutrition. Plant Physiol. 145, 378–388. 10.1104/pp.107.105742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Pasini R., Dan H., Joshi N., Zhao Y., Leustek T., et al. (2014). Aberrant gene expression in the Arabidopsis SULTR1;2 mutants suggests a possible regulatory role for this sulfate transporter in response to sulfur nutrient status. Plant J. 77, 185–197. 10.1111/tpj.12376 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Wu Y., Gao L., Ma J., Li C. Y., Xiang C. B. (2014). Sulfur nutrient availability regulates root elongation by affecting root indole-3-acetic acid levels and the stem cell niche. J. Integr. Plant Biol. [Epub ahead of print]. 10.1111/jipb.12217 [DOI] [PubMed] [Google Scholar]


