Summary
Ionotropic glutamate receptors (iGluRs) mediate the majority of fast excitatory signaling in the nervous system. Despite the profound importance of iGluRs in the nervous system, little is known about the structures and dynamics of intact receptors in distinct functional states. Here we elucidate the structures of the intact GluA2 AMPA receptor in an apo resting/closed state, in an activated/pre-open state bound with the partial agonists and a positive allosteric modulator and in a desensitized/closed state in complex with FW alone. To probe the conformational properties of these states, we carried out double electron-electron resonance experiments on cysteine mutants and cryo-electron microscopy studies. We show how agonist binding modulates the conformation of the ligand binding domain 'layer' of the intact receptors and how, upon desensitization, the receptor undergoes large conformational rearrangements of amino-terminal and ligand-binding domains. We define mechanistic principles by which to understand antagonism, activation and desensitization in AMPA iGluRs.
Introduction
Ionotropic glutamate receptors (iGluRs) harness the chemical potential of glutamate released from presynaptic neurons to drive the opening of a transmembrane, cation-conductive ion channel pore and thus membrane depolarization and the influx of sodium and calcium ions (Traynelis et al., 2010). The major subtypes of iGluRs, AMPA (Boulter et al., 1990) (Keinänen et al., 1990), kainate (Hollmann et al., 1989) and NMDA receptors (Moriyoshi et al., 1991) (Monyer et al., 1992), are related in amino acid sequence yet vary in their pharmacological, physiological and biophysical properties. AMPA receptors are primarily localized to excitatory synapses where they mediate the majority of fast synaptic transmission and participate in synaptic plasticity (Huganir and Nicoll, 2013). Four AMPA receptor subunits – GluA1-4 –assemble to functional homo and heterotetrameric receptor complexes (Keinänen et al., 1990). In native tissues, the AMPA receptor heterotetramers predominate, with calcium-impermeable GluA1/GluA2 and GluA2/GluA3 receptors prevalent at hippocampal synapses (Lu et al., 2009) and calcium-permeable GluA1/GluA4 receptors found in Bergman glia of the cerebellum (Saab et al., 2012). The functional characteristics of AMPA receptors are further diversified by RNA editing, RNA splicing, posttranslational modification by phosphorylation and palmitoylation, and to coassembly with transmembrane AMPA receptor auxiliary proteins (Traynelis et al., 2010).
Hallmarks of AMPA receptors are rapid kinetics of activation, deactivation and desensitization, together with profound reduction of steady-state currents upon prolonged application of agonist (Boulter et al., 1990) (Keinänen et al., 1990). Activation of ion channel gating of AMPA receptors is complex and characterized by multiple sub-conductance states (Rosenmund et al., 1998), differential activation and desensitization by full and partial agonists, such as glutamate and kainate (Jin et al., 2003; Patneau et al., 1993), respectively, and population of progressively larger sub conductance states as a function of increasing agonist concentration (Smith and Howe, 2000). Allosteric modulators, such as aniracetam (Isaacson and Nicoll, 1991) and diazoxide (Yamada and Rothman, 1992), slow receptor deactivation and desensitization and have provided chemical leads to small molecule positive allosteric modulators in clinical trials for treating depression and cognitive impairment (O'Neill et al., 2004). Mutations that slow or block desensitization have profound effects in the context of transgenic animals, proving lethal when homozygous and resulting in severely perturbed animal behavior when heterozygous (Christie et al., 2010). Despite decades of research on AMPA receptors and over 100 structures of the isolated domains, little is known about the molecular mechanisms of agonist activation, of allosteric modulator action and of receptor desensitization.
AMPA receptors harbor a modular architecture composed of an amino terminal domain (ATD), an agonist-recognizing ligand-binding domain (LBD), a transmembrane domain (TMD) that forms the ion channel pore, and intracellular carboxyl-terminal domains (Kumar and Mayer, 2013). Whereas the ATDs and LBDs are organized as dimers-of-dimers (Jin et al., 2009; Kuusinen et al., 1999; Sun et al., 2002), the TMD possesses ~4-fold symmetry, thus giving rise to a symmetry mismatch between the LBDs and TMDs (Sobolevsky et al., 2009). Structural and functional studies show that the LBDs bind agonists and competitive antagonists in the cleft of a bilobed ‘clamshell’, with antagonists stabilizing the cleft in a more ‘open’ conformation and partial and full agonists yielding progressively greater closure of the clamshell cleft (Armstrong and Gouaux, 2000). Under non desensitizing conditions, the LBDs are organized as back-to-back dimers, and perturbations that weaken the dimer interface enhance receptor desensitization whereas mutations and modulators that strengthen the interface block slow desensitization (Sun et al., 2002). Indeed, a site-directed cysteine mutant, S729C, stabilizes the dimer interface in an interface-ruptured, possibly desensitized conformation, thus illustrating how agonist-binding can be decoupled from ion channel gating (Armstrong et al., 2006).
Results and Discussion
To understand the structural changes underlying AMPA receptor gating, we crystallized an intact homotetrameric AMPA (GluA2) receptor in an apo/resting state, an agonist-bound/activated state and agonist-bound/desensitized state. Because AMPA receptors desensitize rapidly and profoundly, crystallization in the presence of full or partial agonists should yield structures representing a desensitized receptor conformation. To trap the receptor in an agonist-bound, activated state, we employed agonists in combination with a high affinity positive allosteric modulator (R,R)-2b which block desensitizations (Kaae et al., 2007). Finally, crystallization under ligand-free conditions was carried out to determine a structure of the full-length GluA2 receptor in an apo/resting state.
To optimize diffraction quality we screened single amino acid substitutions in the TMD region for enhanced thermostability (Hattori et al., 2012). Five or ten of the most thermostabilizing mutations were combined into crystallization constructs 5M and 10M, respectively (see extended experimental procedures). To further improve crystallization of the 10M construct, a five amino acid stretch in the M1M2 loop was deleted (Figure S1A), yielding construct 10Mdel. The resulting constructs 5M and 10Mdel exhibit significantly higher melting temperatures compared to construct GluA2cryst (Sobolevsky et al., 2009) previously used for crystallization of the antagonist-bound receptor (41° C and 51° C versus 35° C, see Figure S1B). Importantly, radio ligand binding experiments and two-electrode voltage clamp (TEVC) recordings confirmed that binding of full and partial agonists and gating properties, such as ion channel gating, desensitization and allosteric modulation, are maintained for these constructs (Figure S2).
Here we report X-ray structures at resolutions of 3.5–4.2 Å for the full-length receptor in the apo state, in complex with the partial agonist kainate (KA), and two structures with positive allosteric modulator (R,R)-2b and the partial agonists fluorowilliardiine (FW) or KA, respectively. We obtained two different crystal forms (A and B) for the KA +(R,R)-2b condition and will refer to form A unless otherwise noted, because form A has similar cell dimensions and ATD arrangements to the apo, KA alone and FW+(R,R)-2b structures. Furthermore, we determined the domain arrangement from a low resolution X-ray structure in complex with FW. Except for the low-resolution FW-bound structure, all new structures belong to the orthorhombic space group P212121, with similar cell dimensions (see Table 1).
Table 1.
Crystallographic data collection and refinement statistics
| Apo formA (4U2P) |
KA+(R,R)- 2b_formA (4U1W) |
KA+(R,R)- 2b_formB (4U1X) |
FW+(R,R)-2b formA (4U1Y) |
KA formA (4U2Q) |
FW | |
|---|---|---|---|---|---|---|
| Construct | 5M | 5M | 10M | 10Mdel | 5M | 5M |
| Data collection | ALS 5.0.2 | ALS 5.0.2 | ALS 5.0.2 | ALS 5.0.2 | ALS 5.0.2 | APS24ID-C |
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P21 |
| Cell dimensions a, b, c (Å) |
107.8, 149.1, 352.8 |
104.5, 151.1, 332.5 |
96.5, 160.7, 338.9 |
105.2, 151.4, 330.5 |
104.9, 148.7, 337.1 |
150.9, 114.7, 158.7 |
| Cell angles α, β, γ (°) | 90,90,90 | 90,90,90 | 90,90,90 | 90,90,90 | 90,90,90 | 90,98.13,90 |
| Resolution (Å)a | 49.00–3.71 (3.84–3.71) |
83.21–3.31 (3.73–3.61) |
73.38– 3.20(3.97–3.82) |
64.99–3.70 (4.42–4.27) |
83.08–4.05 (4.19–4.05) |
49.91–7.94 (8.16–7.94) |
| Completeness | 96.6 (99.5) | 98.4 (99.6) | 91.6(91.3) | 99.1(99.6) | 80.8 (83.3) | 91.7 (92.8) |
| Multiplicity | 3.4(3.5) | 4.1 (4.2) | 2.9(2.7) | 4.12(4.00) | 4.9 (4.9) | 3.1 (2.5) |
| I/σI | 7.77 (1.82) | 11.1(2.3) | 5.4(1.9) | 6.93(1.97) | 6.45 (1.92) | 7.8 (1.85) |
| Rmerge (%) | 9.9 (78.9) | 8.5 (61.5) | 17.1(70.4) | 11.6(75.2) | 11.2 (81.4) | 6.9 (43.2) |
| Anisotropy (Å: a*/b*/c*)b |
3.2, 3.4, 4.1 | 3.2,3.5,3.7 | 4.1,3.3,3.7 | 4.1,3.9,4.4 | 3.5, 3.5, 5.9 | 7.7, 7.7, 7.9 |
| Refinement | ||||||
| Resolution (Å) | 49.00–3.24 | 30.00–3.25 | 30.00–3.30 | 20.00–3.90 | 83.08–3.53 | |
| No. of reflections | 65917 | 66749 | 54779 | 45520 | 39983 | |
| Rwork/Rfree (%)c | 25.00/28.68 | 27.02/31.61 | 24.25/28.90 | 26.82/30.35 | 28.07/32.05 | |
| No. of atoms total | 22388 | 23011 | 23451 | 22259 | 22440 | |
| Ligand | 104 | 180 | 192 | 166 | 102 | |
|
Average B-factor (A2) |
184.73 | 147.51 | 102.22 | 175.60 | 225.74 | |
| Protein | 185.13 | 147.74 | 102.32 | 175.70 | 226.13 | |
| Ligand | 100.58 | 118.40 | 89.62 | 162.01 | 139.88 | |
| R.m.s. deviations | ||||||
| Bond lengths (Å) | 0.004 | 0.007 | 0.003 | 0.005 | 0.004 | |
| Bond angles (°) | 0.917 | 0.849 | 0.819 | 1.021 | 0.957 | |
| Ramachandran plot | ||||||
| Favored (%) | 97.34 | 97.58 | 97.35 | 97.55 | 98.31 | |
| Allowed (%) | 2.66 | 2.42 | 2.65 | 2.45 | 1.69 | |
| Disallowed (%) | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
The number in parenthesis is the shell at conventional cut off using I/σI =2 as criterion.
Anisotropy truncation was performed using the anisotropy server (http://services.mbi.ucla.edu/anisoscale/).
5% of reflections were used for calculation of Rfree.
Because we obtained crystal structures of full-length GluA2 receptor in apo, KA+(R,R)-2b bound and FW+(R,R)-2b-bound states in the same crystal form A with similar crystal packing environment, we assume that the changes between these three structures are due to conformational change in response to different ligands rather than crystal packing artifacts. Furthermore, because the extent of LBD domain closure that we observed in those full-length structures are apo<KA<FW, we propose the structural transitions from the apo to KA+(R,R)-2b and then to FW+(R,R)-2b structures are along GluA2 receptor gating trajectories, from a resting state to an agonist-bound, activated state.
Domain arrangement in full-length GluA2 receptor structures
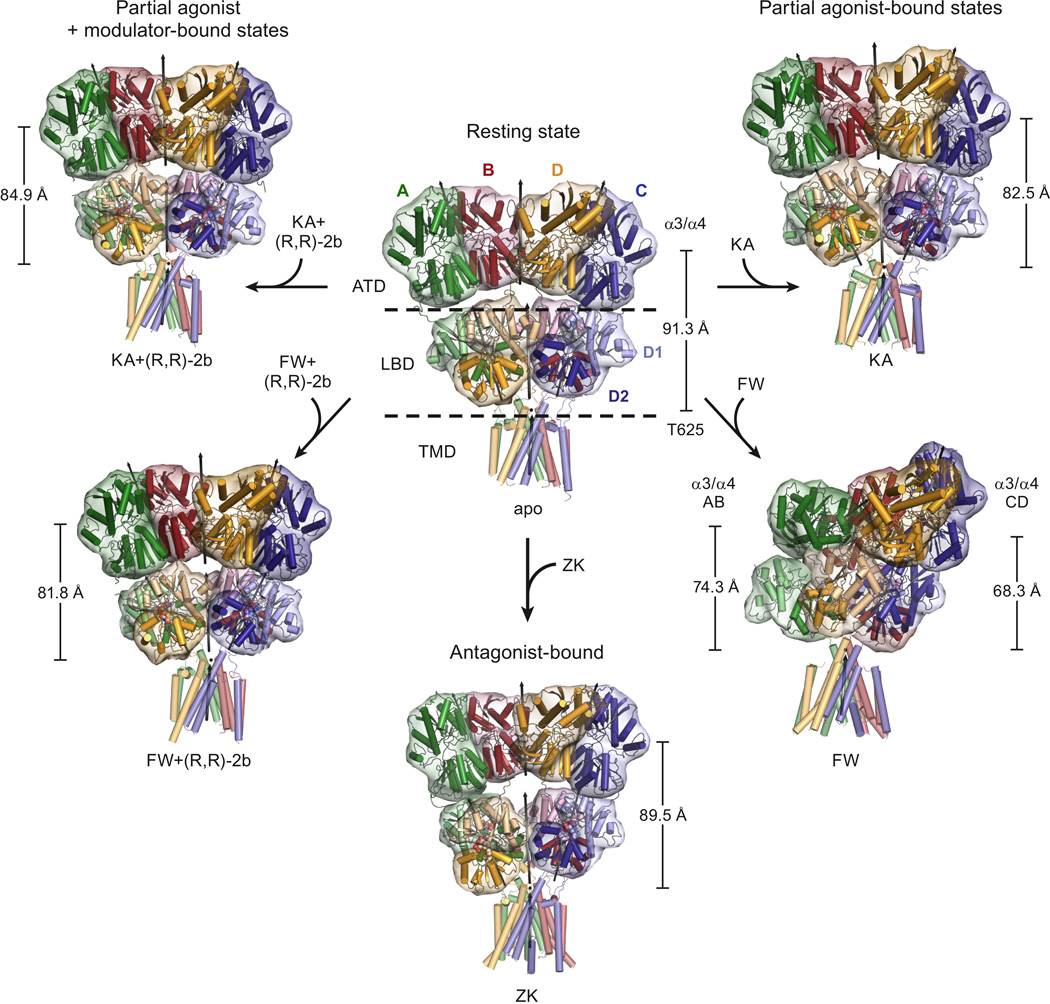
The Y-shaped appearance previously described for the receptor bound with the antagonist ZK200775 (‘ZK’) is maintained in the resting state and all partial agonist-bound structures, except for the FW-bound structure (Figure 1). The “vertical” dimension of the receptor, parallel to the long axis of the receptor, is comparable for the apo and antagonist-bound states, but substantially shortened for partial agonist-bound structures. To quantify the reduction in vertical dimension, we chose a vector between the center of masses (COMs) of helices α3 and α4 in the ATD layer and the COMs of the Cα atoms of residues Thr625 in the TMD-LBD linker region (see vertical scale bars in Figure 1). Because of the asymmetrical domain arrangement of the FW-bound structure, two of these vectors were calculated using COMs for each individual ATD dimer (AB or CD) and their respective projections onto the 4-fold axis (defined by the TMD region) were determined.
Figure 1. Structures of intact GluR2 AMPA receptor.
Views are approximately perpendicular to the overall 2-fold axes of symmetry, except for the FW-bound structure, which is oriented to match the TMD regions of the other structures. Axes of local 2-fold symmetry for the ATD and LBD dimers are indicated as black arrows. For each structure, the vertical scale bar represents the distance between the center of mass of residues Thr 625 and the center of mass of helices α3 and α4 in the ATD layer, using Cαs of all four chains. For the FW structure, the two scale bars indicate the length of the projection of this distance, calculated for the AB and CD pair individually, onto the 4-fold axis of symmetry defined by the TMD region. See also Figure S2.
The vertical compression observed for the partial agonist-bound states can be largely attributed to a shortening of the LBD and LBD-TMD linker region, which in turn is caused by increased domain closure induced by these ligands compared to the apo and antagonist-bound structures. Interestingly, the general organization of the ATD layer as pairs of dimers is maintained in all structures, whereas the local LBD dimers seem to be intact only for the Y-shaped structures (details below).
LBD domain closure in full-length receptor versus soluble LBD structures
To compare the LBD domain closure in the full-length GluA2 receptor structures and the respective soluble LBD (sLBD) structures, we measure two distances, ξ1 and ξ2, which span the D1 and D2 lobes of the LBD, on opposite sides of the ligand binding pocket (Lau and Roux, 2011) (Figure S3 A , B). The ξ1 and ξ2 distances in the full-length structures are: ZK>apo>KA>FW, which is consistent with previous findings from studies with sLBDs showing that the domain closure increases with increasing ligand efficacy (Jin et al., 2003). When comparing different agonist-bound structures with each other, we observe that the changes in ξ1 are much smaller than the changes in ξ2. This is likely because the agonists we used share similar α-carboxyl and γ-anionic moieties that bind to the binding pocket via the same binding mode and ξ1 measures changes at this binding region. In contrast, ξ2 measures the changes where the efficacy-determining variable groups of these different agonists interact with the receptor.
Upon comparison of sLBD structures to the corresponding full-length receptor structures, some differences in domain closure are observed, mainly in the ξ2 distance: ξ2 in sLBDs is always smaller than in the full-length structures, which indicates that the sLBD adopts a more closed conformation. This might be because in the full-length receptor the D2 lobe of the LBD is physically connected to the TMD by the D2-M3 linker, whereas in the sLBD domain, the D2 lobe is not restrained or because the LBD is coupled to a closed pore. The domain closures of each of the four LBDs in the full-length structures are slightly different even in the presence of the same agonists which might be due to crystal packing or inherent structural plasticity and asymmetry of a full-length receptor. This interpretation is in agreement with previous molecular dynamics (MD) simulation studies using sLBD structures showing that agonist-bound LBDs can sample different conformations (Lau and Roux, 2011).
Inter-domain movements in full-length receptor structures
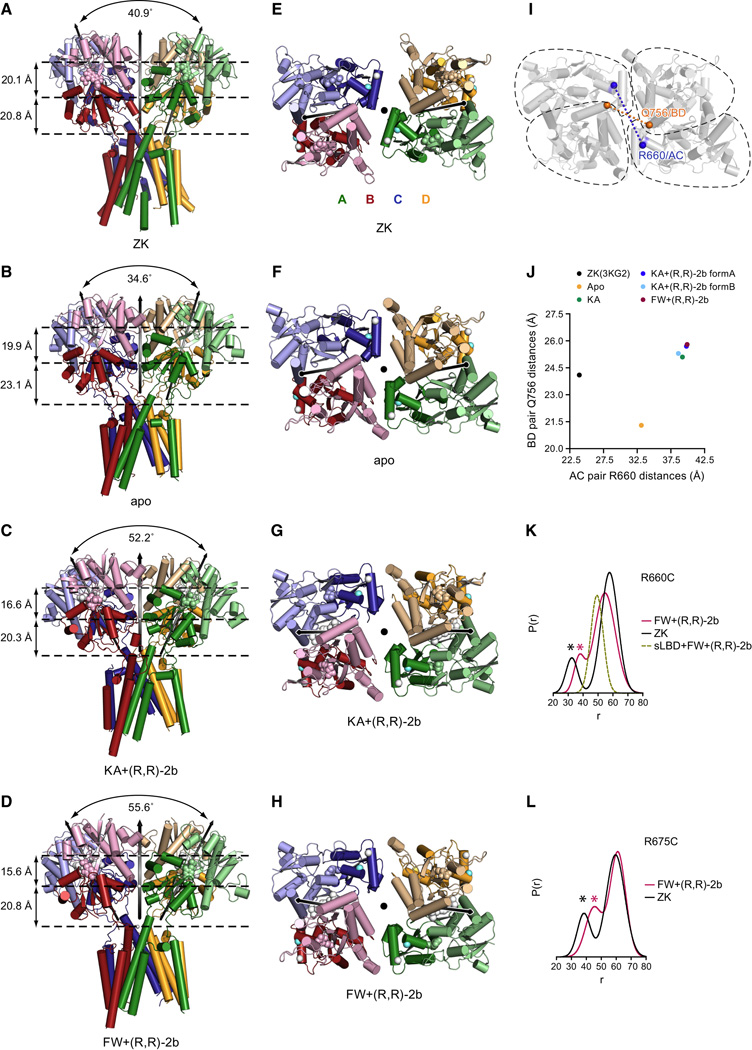
There are large inter-domain motions in the LBD layer which can only be investigated in the context of an intact receptor structure. To describe the relative orientation of the two local LBD dimers - AD and BC - within each structure we defined a Cartesian coordinate system for each dimer, using the COM of each dimer as an origin (Figure S3C). The z-axis (shown in red) of this coordinate system was chosen to be parallel to the local 2-fold axis of the dimer and the y-axis (in green) is parallel to the vector defined by the COMs of each subunit. After translating the COM of dimer BC onto the COM of dimer AD, three subsequent rotations align dimer BC onto AD (Figure S3C and Movie M1), yielding three Euler angles of rotation (also called “roll”, “pitch”, “yaw”), as listed in Table S2. Because in all Y-shaped structures the local 2-fold axes of both dimers and the global 2-fold axis nearly reside in the same plane, the “roll” roughly corresponds to the angle between these local 2-fold axes. This “roll” angle is slightly larger for the ZK-bound structure compared to the apo structure (40.9° versus 34.6°, see Figure 2A,B), and as shown in panels C and D, these angles are much larger for the partial agonist + modulator-bound structures (52.2° and 55.6° for KA+(R,R)-2b and FW+(R,R)-2b, respectively). Likewise, there is only a small difference in the vertical compression of the LBD layer between apo and ZK-bound structures (in total ≈2 Å, arrows between dotted lines in Figure 2A,B), whereas the TMD-LBD layers of the two partially activated structures in panels C and D of Figure 2 are shortened by more than 6 Å, compared to apo. The vertical “compression” concomitant with receptor activation is in agreement with a previous modeling study (Sobolevsky et al., 2009) and an MD simulation (Dong and Zhou, 2011) based on the ZK-bound full-length structure.
Figure 2. Inter-domain movements of LBD ‘gating ring’.
A–H. LBD layer arrangement of full-length GluR2 structures in complex with the competitive antagonist ZK (A,E; PDB code: 3KG2), in the apo state (B,F), in complex with partial agonist KA + (R,R)-2b (C, G), and in complex with partial agonist FW+(R,R)-2b (D, H). A-D. Views of the LBD layer perpendicular to the global 2-fold axes of symmetry. Angles between the local 2-fold rotation axes, shown as tilted black arrows, of LBD dimers BC and AD are indicated. Dashed lines indicate the layers defined by the positions of D1 centers of mass, D2 centers of mass and centers of mass of Thr 625 respectively. Vertical arrows on the left denote the respective distances between layers. E-H. Top down views of the LBD gating ring from the extracellular side, parallel to the overall 2-fold axes. The modulator (R,R)-2b is shown in white space-filling representation. Cα atoms of Arg 660 and Arg 675 are shown as aquamarine and white spheres, respectively. I. Same view showing the location of marker positions Arg 660 (AC pair) and Gln 756 (BD pair), whose Cα atoms were used to describe the enlargement of the central opening upon activation, with the respective distances plotted in panel J. K-L. DEER distance distributions of MTSSL-labeled GluA2 receptor constructs R660C (panel K) or R675C (panel L) measured with bound ZK (black) and FW+(R,R)-2b (magenta), respectively. DEER decays and fits are provided in Figure S3 D and, E respectively. In panel L, the distance distribution of the respective soluble MTSSL-labeled sLBD R660C construct in presence of FW+(R,R)-2b is superposed as a yellow dashed line. See also Figure S3, Table S2, Table S3, Movie M1, Movie M2, Movie M3.
When inspecting the LBD layer of the full-length structures from the extracellular side along the global 2-fold axis of symmetry, differences in the lateral placement of the two LBD dimers are revealed (Figure 2E–H). Comparison between apo and ZK-bound structures shows that the two dimers are more staggered in the antagonist-bound structure (Figure 2E) than in the apo state (Figure 2F). This can be seen from the relative orientation of the local 2-fold axes of the two dimers (represented as black arrows) which are almost in one plane in the apo structure (Figure 2F) but are moved “out-of-plane” in the ZK structure (Figure 2E). The tilting of the local dimers with respect to each other also affects the positioning of the global 2-fold axis of the LBD layer and is therefore reflected by a larger angle between this global axis and the 4-fold axis of the TMD layer for the ZK-bound structure compared to the apo structure (6.8° versus 3.8°, see Table S2). This change in the relative orientation of the LBD dimers is illustrated in Movie 2, showing a morph transitioning between apo and ZK-bound structures.
Changes in inter-domain arrangement upon receptor activation
The same top view of the two new structures obtained under non-desensitizing conditions (KA+(R,R)-2b and FW+(R,R)-2b, see Figure 2G and 2H respectively) reveals an enlargement of the central opening between LBD dimers, which is reminiscent of Ca2+ activated BK channel gating ring (Yuan et al., 2012). This enlargement results from a concerted structural rearrangement within dimers and between dimers upon receptor activation from a resting state conformation (apo state in Figure 2F), with the D2 lobes from both diagonal pairs (“short” AC and “long" BD) simultaneously undergoing outward movements (illustrated in Movie M3). To quantify this diagonal separation, we measured inter-subunit distances between Cα atoms of two marker residues (Arg 660, located on helix F of the D2 lobe for the AC pair and Gln 756 on helix J for the BD pair, respectively, see Figure 2I) and plotted the respective diagonal distances against each other for each structure (Figure 2J). Note that the AC inter-subunit distance between these marker atoms increases from 33.1 Å in the apo structure to 39.8 Å and 39.9 Å for the modulator-bound structures KA+(R,R)-2b and FW+(R,R)-2b, respectively. Similarly, we can also observe a separation of D2 lobes belonging to the BD pair, as reported by the second diagonal marker atom distance (Cα atoms of Q756), which increases from 21.3 Å to 25.7 Å and 25.8 Å, respectively (apo vs. KA+(R,R)-2b and FW+(R,R)-2b). The dual increase in both AC and BD marker atom distances upon activation from a resting state is reflected by distinct clusters of data points for apo, partial agonist-bound and antagonist-bound structures (Figure 2J).
Detection of LBD inter-domain movements by DEER
To further investigate the increased AC distances derived from the partial agonist-bound structures, we performed double electron-electron resonance (DEER) experiments (Jeschke, 2012; McHaourab et al., 2011) with spin-labeled full-length receptor in detergent micelles, using a modified receptor construct without free cysteines. We introduced cysteine substitutions for the aforementioned marker residue Arg 660 (Figure 2E–H) and, separately, for an additional residue Arg 675, located on the adjacent helix G, to create reporter sites for spin labeling. Probability distributions calculated from the DEER decays (Figure S3D,E) of spin-labeled R660C and R675C receptors in presence of competitive antagonist ZK and under non-desensitizing conditions (FW+(R,R)-2b) are shown in Figure 2K and 2L, respectively. Note that the short distance peaks around 30–35 Å reflect dipolar interactions between labels attached to the AC subunits, whereas the larger peaks at longer distances (around 50–60 Å) can be attributed to dipolar interactions of labeled R660C within a dimer (BC and AD distances in Table S3). This interpretation is supported by the DEER spectrum we obtained in presence of FW and (R,R)-2b for spin-labeled R660C in context of the soluble LBD (sLBD) fragment, showing a single peak around 50 Å (dotted yellow trace in Figure 2K).
For both reporter constructs R660C and R675C, these short-distance peaks are shifted toward the right for the non-desensitizing conditions (FW+(R,R)-2b, shown as magenta traces in Figure 2K and 2L) compared to the antagonist-bound condition (black traces in Figure 2K and 2L), thus confirming the separation of the short diagonal AC pair upon receptor activation. Notably, these results are also in line with previous functional studies demonstrating that crosslinking of introduced cysteine mutations at positions 664 and 665 traps the GluA2 receptor in a low-activity state (Lau et al., 2013; Sobolevsky et al., 2009).
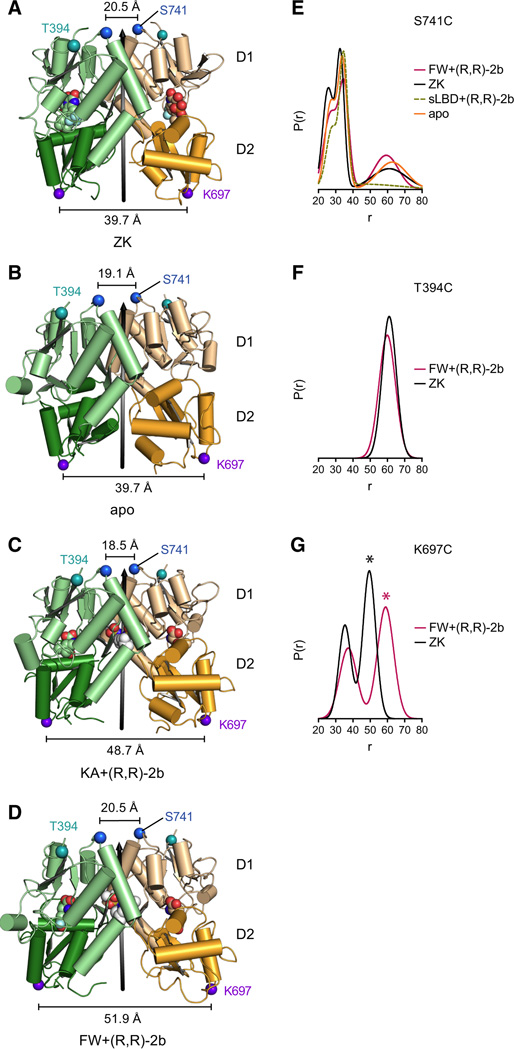
State of the LBD intra-dimer interface in full-length receptor structures
The intact LBD dimer interface is a key feature of non-desensitized agonist-bound states as well as the apo/resting state, as demonstrated by numerous crystallographic and functional studies (Sun et al., 2002) and by apo state structures of the sLBD (Armstrong and Gouaux, 2000). Nevertheless, luminescence resonance energy transfer (LRET) experiments with the full-length AMPA receptor suggest that the interface might be decoupled under apo conditions (Gonzalez et al., 2010). However, side views of the LBD dimer (perpendicular to the global 2-fold axis) show that the D1-D1 dimer interface is intact in the apo full-length structure (Figure 3B) and is similar to the previously published ZK-bound structure (Figure 3A). The interface is also intact for the two modulator-bound full-length receptor structures, and stabilized by the presence of (R,R)-2b (Figure 3C,D). The observation of an intact dimer interface for apo, antagonist and modulator-bound structures is further confirmed by DEER measurements of spin-labeled full-length receptor (Figures S4E,F), using labeling positions Thr 394 and Ser 741 (located on top of the D1 lobes, shown in Figure 3 A–D). Indeed, labeled Ser 741C receptor molecules under apo, ZK-bound and under non-desensitizing conditions share a similar distribution profile between 20–40 Å (Figure 3E). Because this position matches well with the distribution observed for the sLBD S741C mutant in the presence of FW and (R,R)-2b (yellow trace in Figure 3E), we conclude that this distance reflects intra-dimer (AD or BC) spin interactions which are characteristic for LBD dimers with an intact D1-D1 interface. Furthermore, the distance distribution of labeled mutant T394C in presence of antagonist ZK is similar to the spectrum obtained under non-desensitizing conditions (compare black and magenta traces in Figure 3F).
Figure 3. Intra-dimer LBD interfaces in apo and partial agonist-bound states.
A–D Side views of LBD dimers from chain A (green) and chain D (yellow) of full-length structures in complex with the competitive antagonist ZK (A; PDB code: 3KG2), in the apo state (B), in complex with partial agonist KA + modulator (R,R)-2b (C) and in complex with partial agonist FW + modulator (R,R)-2b (D). D1 domains are colored in lighter shades. LBD local 2-fold axes are shown as black arrows. Distances between Cα atoms (blue spheres) of residues Ser 741 are indicated by black scale bars. Distances between Cα atoms (purple spheres) of residues Lys 697 are indicated by black scale bars. Cα atoms of marker position T394C are shown as teal spheres. Modulator (R,R)-2b is shown in white space-filling representation. E-G. Probability distributions of DEER distances calculated from DEER decays, provided in Figure S4 E–G, of MTSSL-labeled receptor constructs S741C (E), T394C (F) and K697C (panel G) measured under apo conditions (orange), with ZK (black), or with FW+(R,R)-2b (magenta), respectively. Note that the long-distance peak of ~60 Å present under all conditions in the DEER distribution of mutant S741C (in F) is dependent on the background correction. The distance distribution of the respective MTSSL-labeled sLBD S741C construct in presence of FW+(R,R)-2b is superposed as yellow dashed line (E). The asterisk in panel G indicates putative inter-dimer distances.
D2-D2 separation upon GluA2 receptor activation
Because (R,R)-2b stabilizes the LBD D1-D1 dimer interface in the presence of agonists, the major structural changes within the dimer induced by agonist binding are separation of the D2-D2 lobes (Figures 3A–D). To monitor this separation in DEER experiments, we engineered position K697C, located on D2, close to the D2-M3 linker, for spin labeling (Figure3A–D). The DEER spectra of spin-labeled K697C under non-desensitizing conditions indicate a shift of the probability distributions to longer distances compared to spectra obtained in presence of the competitive antagonist ZK (Figure 3G and Figure S4G), thus confirming the D2-D2 distance increases shown in Figure 3A–D.
Mechanism of GluA2 receptor modulation by (R,R)-2b
In accord with a previous structural study characterizing the related modulator (R,R)-2a (Kaae et al., 2007), the full-length GluA2 structures in complex with KA (or FW) confirm the location of modulator (R,R)-2b within the D1-D1 interface, as shown in Figure S4A–B. To further characterize the molecular details of GluA2 receptor modulation by (R,R)-2b, we crystallized the sLBD (Chen et al., 1998) with (R,R)-2b under saturating partial agonist concentrations and obtained high resolution X-ray crystal structures (Table S1). Similar to the closely related (R,R)-2a compound, (R,R)-2b also binds at the LBD dimer interface in a symmetric way (Figure S4CD), with the sulfonamide moiety mimicking the norbornene moiety of cyclothiazide (CTZ) (Bertolino et al., 1993; Yamada and Tang, 1993). Interestingly, the substitution of the methyl group of (R,R)-2a with the isopropyl group of (R,R)-2b causes a rotation of the sulfonamide group, resulting in a disruption of a hydrogen bond between the sulfonamide oxygen of (R,R)-2a and hydroxyl group of Ser 754 (Figure S4C–D). However, this new rotamer conformation of (R,R)-2b allows for van der Waals interactions between the isopropyl group and Leu 751 and Ile 481 of the receptor, which might contribute to the slightly higher affinity of (R,R)-2b compared to (R,R)-2a (Kaae et al., 2007).
The two phenyl rings of (R,R)-2b make van der Waals interaction with Pro 494, Ser 497 and Ser 729 located at the inter-domain hinge region of LBD (Figure S4B). These residues are involved in the binding of another set of AMPA receptor positive modulators, such as aniracetam and CX614 (Jin et al., 2005). Modulators that bind at this site slow the deactivation time course by stabilizing the LBD in an agonist-bound conformation. Indeed, both 3H KA binding or 3H FW binding by scintillation proximity assay (SPA) using purified receptor in detergent micelles show an increase in agonist affinity in the presence of (R,R)-2b (Figure S2A–D). Moreover, co-application of (R,R)-2b with different agonists shifts the receptor activation curve left and decreases the EC50 of agonists (Figure S2E–G). These data suggest (R,R)-2b also slows the deactivation of GluA2 receptor. Therefore from both a structural and a functional perspective, (R,R)-2b behaves like a hybrid of CTZ and aniracetam: it blocks desensitization and slows deactivation, thus favoring trapping of the receptor in an activated state.
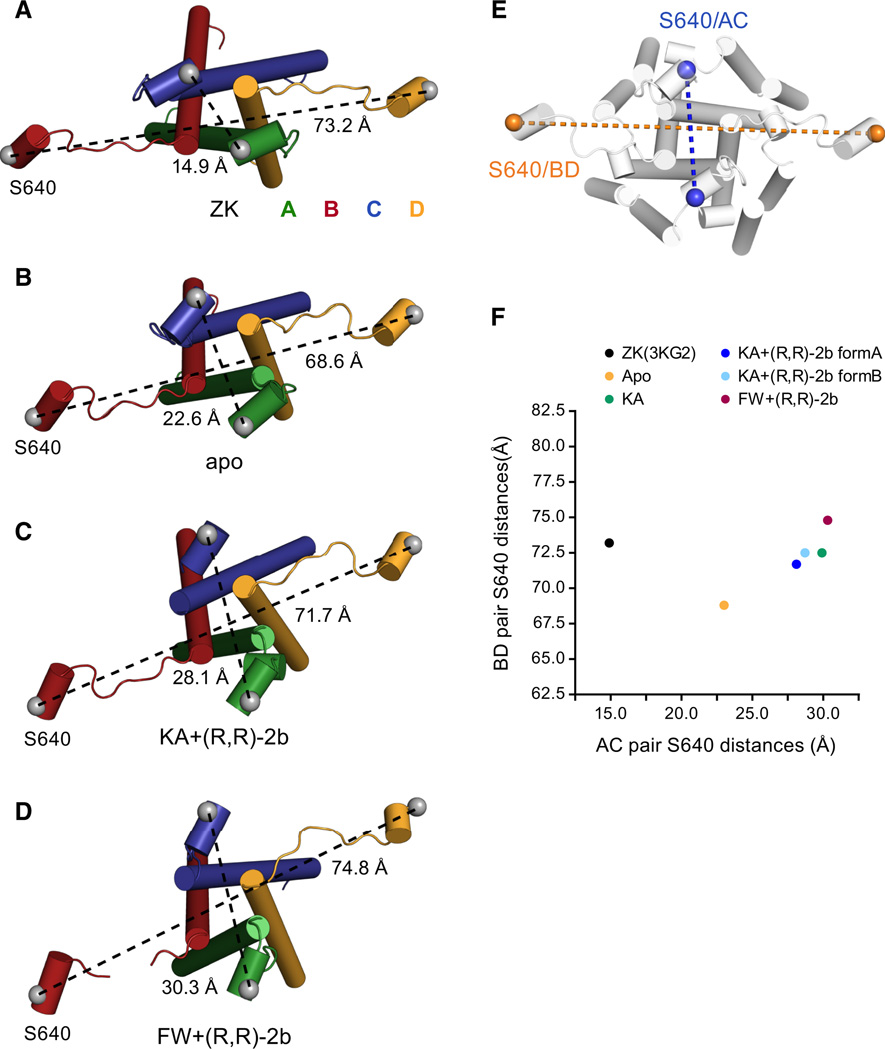
Structural changes of the LBD-TMD linker region
As a consequence of the separation of the D2 lobes in the partial agonist-bound non-desensitized structures, we also observe conformational changes in the D2-TM3 linker region upon activation from a resting state conformation (compare Figure 4C and D to Figure 4B). This can be described by the distance changes between marker atoms Cα of Ser 640 on helix E, which direct connect to TM3 gating helix (Figure 4E). When comparing these distances in the apo structure (Figure 4B) to the partially activated KA+(R,R)-2b structure (Figure 4C), there is an increase in the AC and BD distances by 5 Å and 3 Å, respectively. For the FW+(R,R)-2b structure in Figure 4D, these distances increase by additional 2 Å and 3 Å, respectively, which correlates with the reported differences in agonist efficacy and LBD domain closure of FW > KA (Armstrong and Gouaux, 2000; Jin et al., 2003). These distance changes in the LBD-TMD linker correlate well with the respective changes in D2-D2 distances (comparing Figure 2J and Figure 4F), further emphasizing that LBD is the driving force for TMD movements. It should be mentioned that there are also some differences in the AC and BD distances when comparing the apo structure to the ZK-bound structure (compare panels A and B in Figure 4). Whereas the AC distance is larger for the apo structure than for the antagonist-bound structure (22.6 Å versus 14.9 Å), the BD distance is shorter (68.6 Å versus 73.2 Å). These changes might originate from the difference in LBD clamshell closure (Figure S3B) or from different crystal packing environment of the LBD layer.
Figure 4. Conformational ‘expansion’ of LBD-TMD linker region upon receptor activation.
A–D. Top down views of the D2 (helix E) -M3 linker region of full-length structures in complex with competitive antagonist ZK (A; PDB code 3KG2), in the apo state (B), in complex with partial agonist KA+(R,R)-2b (C) and in complex with partial agonist FW+(R,R)-2b (D). E. Location of marker residue Ser 640 on helix E where Cαs are shown as orange spheres for the distal BD pair and as blue spheres for the short diagonal A/C pair, respectively. F. Plot of AC versus BD distances, measured between Cαs of Ser 640, for the indicated full-length structures, showing the increase in "pulling force" caused by the D2 separation of GluA2 receptor with partial agonists + (R,R)-2b compared to antagonist-bound and apo structures. See also Figure S4.
In summary, there is 6–7 Å increase in marker atom separation in both AC and BD pairs, transitioning from apo to FW+(R,R)-2b structure (see Figure 4F), which reflects an enhanced mechanical pulling force exerted by bound agonists (+modulator) in the context of a full-length receptor. Despite these movements, the gating residues on TM3 are still in the same position as in the ZK-bound state, which indicates that the pore is essentially closed, reminiscent of the pre-open state of the fully liganded GIRK2 channel (Whorton and MacKinnon, 2013). This might be because the pulling force generated by partial agonists is insufficient to maintain the gate constantly open (Jin et al., 2003) or because the conditions of crystallization do not stabilize an open state.
Diverse ensemble of extracellular domain conformations in desensitized state
In the FW-bound, desensitized state, the ATDs and LBD undergo large conformational rearrangements as suggested by a molecular replacement (MR) solution derived from an ~8 Å dataset (Figure 5; see Table 1). Using soluble X-ray structures of ATD dimers, the TMD portion of the ZK structure and of individual LBDs in complex with FW (PDB 1MQI) as search models, we obtained a partial model with all receptor domains except for two missing LBDs (chains B and C). Further MR search with the S729C structure (PDB 2I3W) as probe yielded the full-length model shown in Figure 5B.
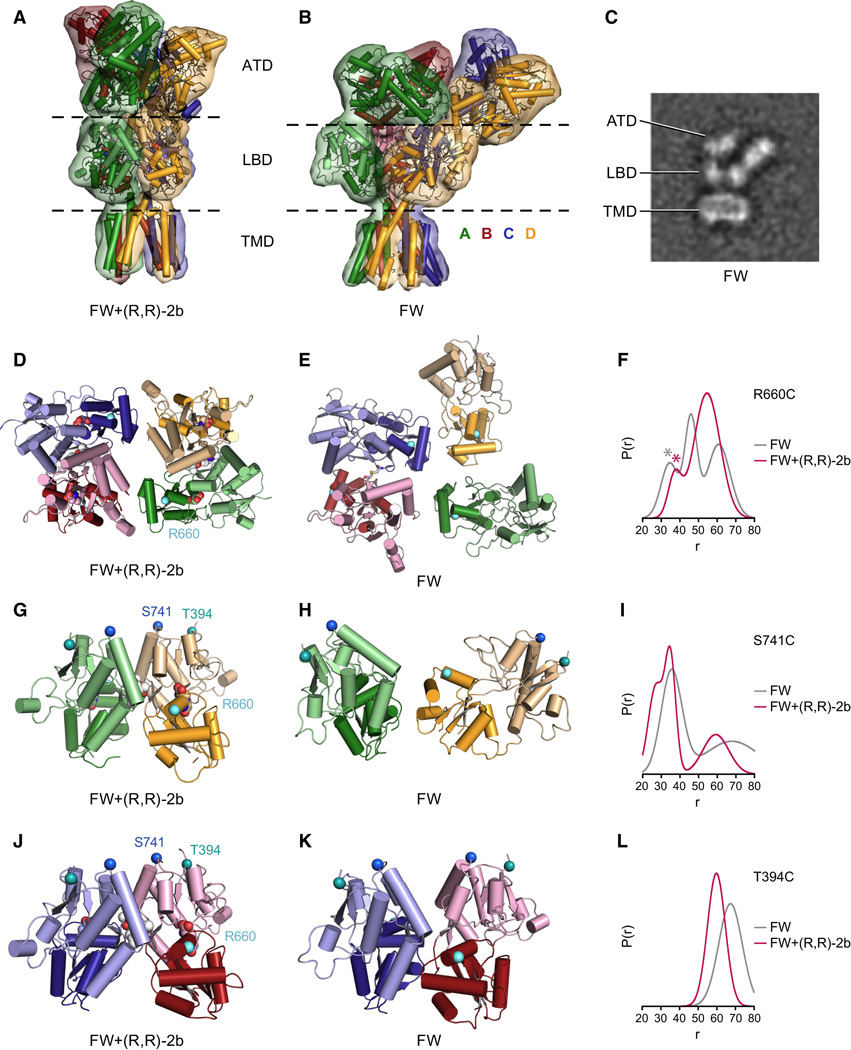
Figure 5. Conformational rearrangements upon receptor desensitization.
Comparison of the non-desensitized full-length structure in complex with FW and (R,R)-2b (A,D,G,J) and low resolution X-ray (B,E,H,K) or cryo-EM (C) structures complexed with FW alone. A. X-ray structure in complex with FW+(R,R)-2b. B. Molecular replacement solution obtained from an 8.0 Å dataset collected from a crystal grown in presence of FW. C. Selected cryo-EM class average of the same receptor construct/ligand combination as in B (see Figure S5 for all class averages). The side length of the panel is 29.1 nm. D,E. View of LBD layers from the top of the receptor, Cα atoms of Arg 660 are shown as white spheres. In panel E, the disulfide link between S729C side chains in chains B and C is shown in yellow stick representation. F. Distance distributions of MTSL-labeled R660C receptor measured with FW (grey) or FW+(R,R)-2b (magenta). G,H. Side view of LBD dimer AD. Cα atoms of Ser 741 are shown as blue spheres, Cα atoms of Thr 394 are shown as teal spheres. I. Distance distributions of MTSSL-labeled S741C receptor measured with FW (grey) or FW+(R,R)-2b (magenta). J,K. Side view of LBD dimer BC. Cα atoms of Ser 741 are shown as blue spheres, Cα atoms of Thr 394 are shown as teal spheres. L. Distance distributions of MTSSL-labeled T394C receptor measured with FW (grey) or FW+(R,R)-2b (magenta). DEER decays of the shown mutants R660C, S741C and T394C are provided in Figure S5 C–E). See also Figure S5. Movie M4 and Movie M5.
Despite the limited resolution, the domain arrangement proposed by this MR solution is plausible because the general three-layer ATD-LBD-TMD architecture is maintained and the distances between N- and C-termini of interdomain polypeptide segments are reasonable. Nevertheless, we suggest cautious interpretation of our structural model, especially in the LBD layer, because of the limited resolution.
In comparison to the other Y-shaped structures, the structure shows remarkable changes in the orientation of both ATDs and LBDs. Compared to the FW-bound structure with modulator (shown in Figure 5A), one of the ATD dimers (AB) is tilted downwards by about 89°, which causes the top of the dimer to approach the side of the other ATD dimer (CD) (see Figure 5B, Figure S6A and movie M5). To accommodate the tilted ATD AB dimer, the other ATD dimer CD slides laterally outwards by about 40 Å, away from the central 2-fold axis.
According to the MR solution, the D1-D1 interfaces are disrupted in both dimers (Figure 5E, H and K). It should be noted that the BC pair is derived from the dimeric S729C search model, whereas the positions of A and D chains result from MR searches using monomeric LBD complexed with FW. The latter two LBD monomers are even more separated from each other than suggested by the S729C structure, with the LBD of chain D being rotated outward by ~105° (see Figure 5E,H and Figure S6B). Interestingly, a similar separation of the LBD monomers upon desensitization was concluded from cryo-electron microscopy (cryo-EM) analysis of desensitized kainate receptors (Schauder et al., 2013).
Validation of desensitized state structure by cryo-EM and DEER measurements
Because of the limited resolution of the FW X-ray structure, we investigated the desensitized state using cryo-EM as an additional structural approach. Figure S5 shows single-particle averages of vitrified GluA2 receptors under non-desensitizing (GluA2+FW+R,R)-2b, Figure S5A) and desensitizing conditions (GluA2+FW, Figure S5B). Whereas the particles under non-desensitizing conditions represent a relatively uniform population of receptor molecules, resembling the Y-shaped X-ray structure obtained under the same conditions, the particles under desensitizing conditions are conformationally heterogeneous, suggesting that the receptor assumes a variety of different conformations. Notably, some particle projections exhibit a striking overall similarity to the x-ray MR solution (compare panel B and C in Figure 5). Furthermore, for a considerable number of the particles under desensitizing conditions, the two ATD dimers are splayed apart to varying extents, indicating a large degree of conformational flexibility of the extracellular domains (see Figure S5B and movie M4). These results suggest that the low-resolution crystal structure of the FW-bound state captured one conformation from an ensemble of desensitized receptor states with multiple conformers.
To further validate the disruption of the dimer interface under desensitizing conditions, we also performed DEER experiments with spin-labeled reporter constructs R660C, S741C and T394C (the latter two being located on top of the D1 lobe). The distance distributions obtained from spin-labeled R660C receptor molecules showed a shift towards shorter distances under desensitizing conditions compared to the spectrum in the presence of modulator (Figure 5F), which matches the predicted distance change between spin-labeled AC subunits (compare Figure 5D and E). On the other hand, distances calculated from DEER decays (Figure S5) of spin-labeled T394C and S741C receptors in the presence of FW alone are both shifted towards longer distances compared to the same condition plus modulator (R,R)-2b (Figure 5I and L), thus confirming the increase in the intra-dimer distance suggested by the MR solution (Figure 5H and K) in comparison to the FW+ (R,R)-2b X-ray structure with an intact interface (Figure 5G and J). To be noted, we used construct 5M (see extended experimental procedures) to obtain the desensitized state crystal structure, as well as for the cryo-EM studies. Although this altered receptor construct shows normal opening and desensitization properties in TEVC recordings, the ensemble of conformational states may depend on the details of such modifications.
X-ray structure in complex with KA without modulator (R,R)-2b
We also solved an X-ray structure of full-length GluA2 receptor in the presence of KA alone (see Figure 1 and Table 1), which looks almost identical to the KA+(R,R)-2b structure (form A) with an overall RMSD of 1.3 Å. Although based on the ratio of steady-state currents with and without modulator (R,R)-2b, our crystallization construct desensitizes in response to KA, the extent of current potentiation by the modulator in response to KA is lower than in response to the full agonist glutamate (Figure S2H,I). A reduced extent of desensitization for KA-induced currents has been described in the literature (Levchenko-Lambert et al., 2011) and it was also reported that the extent of desensitization drops substantially with decreasing agonist efficacy for other partial agonists like substituted willardiines (Jin et al., 2003). Thus, it is conceivable that in the KA alone condition, more receptors are populated in a “pre-open" state than in presence of the more potent agonist FW and thus explaining why, in the crystal, we have isolated a kainate-bound, non-desensitized conformation of the receptor.
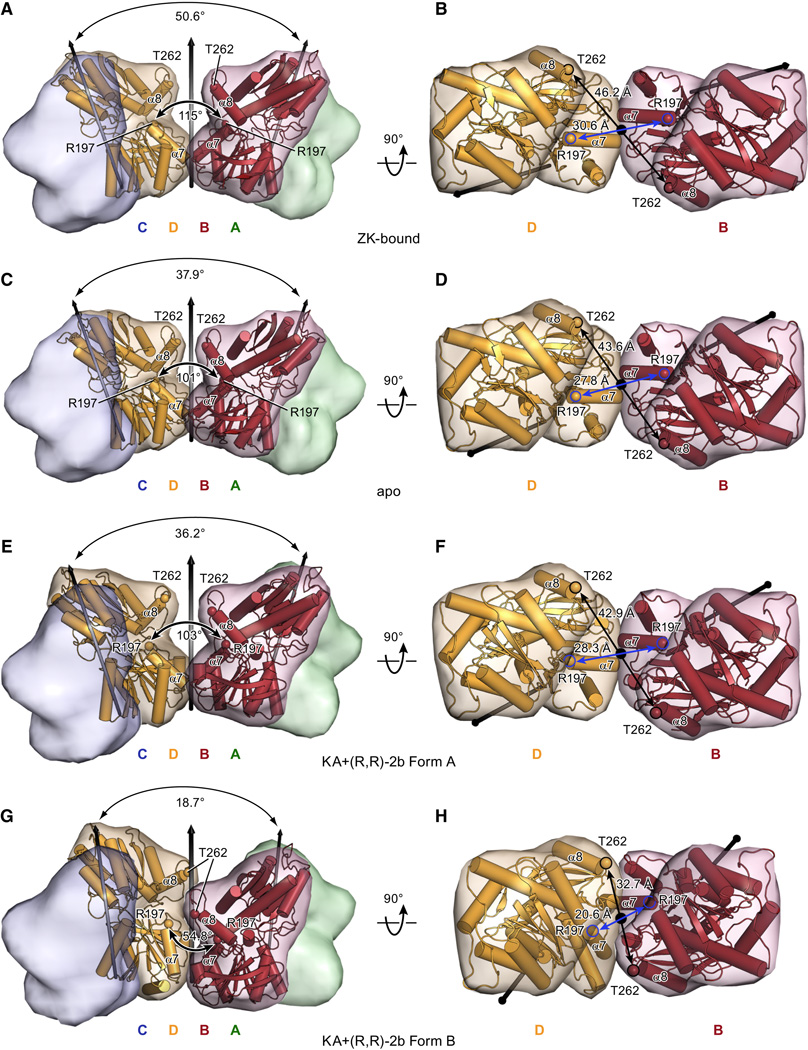
ATD movements in X-ray structures under non-desensitizing conditions
Apart from these large ATD rearrangements observed under desensitizing conditions, there are also smaller movements in our Y-shaped X-ray structures, as shown in Figure 6. Compared to the previously published ZK-bound structure, the two ATDs of the apo structure are tilted towards each other. The angle between the local 2-fold axes of each ATD dimer decreases from 50.6° to 37.9° (see Figure 6A and C), thereby decreasing the distance of Cα atoms of Thr 262 (in helix α8) and Arg 197 (helix α7) located on the proximal BD pair (46.2 Å versus 43.6 Å and 30.6 Å versus 27.8 Å respectively, see Figure 6B and D).
Figure 6. ATD movements in X-ray structures under non-desensitizing conditions.
Relative orientation of ATD dimers AB and CD for the ZK-bound structure (A,B), the apo structure (C,D) and the KA+(R,R)-2b-bound structures form A (E,F) or form B (G,H). Front views perpendicular to the overall 2-fold axes of symmetry (left panels A,C,E,G) and top view from the extracellular side, parallel to overall 2-fold axes, showing only subunits B and D (right panels B,D,F,H). Local and global 2-fold axes of symmetry are indicated as black arrows. Angles between the local 2-fold axes of dimer AB and CD are indicated below the larger arc, angles between α7-helices of subunits B and D are indicated below the smaller arc. Cα atoms of Thr 262 and R197 are shown as spheres. See also Figure S6.
Moreover, we solved X-ray structures of two different crystal forms in presence of KA+(R,R)-2b (3.5 Å and 3.8 Å resolution for form A and B respectively; Table 1) which exhibit even more pronounced differences in the relative orientation of the ATD dimers, as illustrated in Figure 6E–H. Whereas the angle between the local 2-fold axes in Form A is comparable to the apo structure (36.2° versus 37.9°), it decreases dramatically in form B (18.7°, Figure 6G) and the distances of the aforementioned marker atoms Thr 262 and Arg 197 in the proximal subunits B and D are further shortened (42.9 Å and 28.3 Å in form A versus 32.7 Å and 20.6 Å in form B).
Significance of domain swapping for AMPA receptor desensitization
The differences in the ATD arrangement under identical ligand conditions (KA+(R,R)-2b form A versus form B) suggest that the native GluA2 receptor has a weak interface between ATD dimers and thus freedom of movement and that the major constraint preventing the two ATD dimers from separating widely, as seen under desensitizing conditions, is provided by the adjacent intact LBD dimers. As a direct consequence of the subunit crossover or domain swapping, it is logical that a disruption of the dimer interface in both LBD dimers will destabilize the Y-shaped structure characteristic of resting and non-desensitizing conditions because there is no significant additional stabilization by other inter-subunit interfaces, such as between proximal BD subunits in the ATD layer or between proximal AC pairs in the LBD layer. Conversely, the crossover between ATD and LBD layers is most likely responsible for the slower onset of desensitization and a faster recovery from desensitization for GluA1-4 receptors with deleted ATDs (Moykkynen et al., 2014). Because each subunit within a LBD dimer is connected to a different ATD dimer, additional strain maybe exerted on the D1-D1 interface during activation, hence promoting D1-D1 separation and the disruption of the dimer interface. By serving as “molecular amplifiers" for the destabilization of the dimer interface in response to agonist-induced LBD domain closure, the large ATDs facilitate transition into a multitude of desensitized states, rather than into an open state.
Conclusions
Using a multidisciplinary approach combining X-ray crystallography, cryo-EM and EPR spectroscopy, we studied GluA2 AMPA receptor structure and dynamics in resting, pre-open and desensitized states, expanding our knowledge into the molecular mechanisms of AMPA receptor gating in the context of the full-length receptor. Here, we observe the structural changes that happen not only in the LBD clamshell and within the LBD dimer but also between LBD dimers and the ATD layers. Furthermore, to analyze the complex movements between LBD dimers, we developed a ‘roll-pitch-yaw’ analysis of the GluA2 receptor structures in multiple states, and we suggest this approach might also be applicable for defining the complex motions in other multi-domain system.
AMPA receptor activation upon agonist binding to the apo/resting state occurs with an intact D1-D1 LBD dimer interface, enhancing the probability that domain closure in response to agonist binding is conveyed to separation of the D2 lobes within a LBD dimer. Accompanying the conformational changes within LBD dimers is a rotation, or ‘roll’, of LBD dimers together with translation of LBD dimers, thus resulting in a three-dimensional separation of the LBD D2 lobes. We speculate that this separation exerts a “pulling” force onto the D2-M3 linker region, opening the ion channel gate at or near the M3 bundle crossing. The transient opening of the ion channel gate is terminated by relaxation of D2 lobes to a contracted, resting state-like conformation, which in turn results from rupture of the D1-D1 LBD dimer interface and marked structural rearrangement and increased dynamics of the ATD layer. Our newly observed inter-domain movements during GluA2 gating transitions suggest mechanisms to modulate AMPA receptor gating by manipulating inter-domain interactions, either by genetic or pharmacological methods. These studies provide a nearly comprehensive mechanism of full-length AMPA receptor gating and shed new light on the gating mechanism of kainate receptors and NMDA receptors.
Experimental Procedures
Expression, purification and crystallization of AMPA receptor
The modified intact GluA2 AMPA receptor crystallization constructs were expressed in HEK293S GnTI− cells by baculovirus-mediated gene transduction of mammalian cells (BacMam) system (Baconguis et al., 2014). sLBD protein was expressed in Escherichia coli as previously described (Armstrong et al., 2006). Purified receptor protein was crystallized with different ligand combinations by hanging drop vapor diffusion. Diffraction data were collected using synchrotron radiation at ALS 5.0.2 or APS 24ID-E/C beam lines. The structures were solved by molecular replacement using individual domain structures as molecular replacement search models. Structures were further subjected to iterative crystallographic refinement. Data processing, model building and refinement were performed using XDS (Kabsch, 2010), COOT (Emsley and Cowtan, 2004) and Phenix (Adams et al., 2011) computer programs.
3H KA and 3H FW binding assays
The streptavidin-binding peptide (SBP) tag was added to the crystallization construct after the GFP-His8 tag to generate the construct used for scintillation proximity assay (SPA) ligand-binding experiments. All of the 3H KA and 3H FW binding assays were carried out at room temperature. The total counts were determined in triplicate and estimates of background readings were determined in duplicate with 2 minutes reading time per well. The results shown were obtained from readings taken at 12 h or 24 h.
Two electrode voltage clamp recordings
The crystallization constructs 5M and 10Mdel were subcloned as GFP-free open reading frames into the pCDNA3.1x plasmid for in vitro cRNA synthesis. Stage V-VI oocytes were selected and injected with 0.05–2 ng of cRNA. At 2–4 days after injection, currents in response to glutamate, KA or FW were recorded using the two electrode voltage clamp (TEVC) technique at a holding potential of −60 mV as previously described (Armstrong et al., 2006).
Electron microscopy and image processing
Detergent-solubilized AMPA receptors under desensitizing (FW alone) and non-desensitizing conditions (FW+(R,R)-2b) were plunge-frozen on holey carbon films using a Vitrobot (FEI). Grids were imaged with a Tecnai F20 electron microscope (FEI) operated at 200 kV and equipped with a K2 Summit camera (Gatan). Dose-fractionated image stacks were drift-corrected using the UCSF Image4 software (Li et al., 2013), and particles were picked using EMAN (Ludtke et al., 1999). Class averages were calculated using the iterative stable alignment and clustering (ISAC) procedure (Yang et al., 2012) implemented in SPARX (Hohn et al., 2007).
Spin-labeling and DEER experiments
For spin-labeling and subsequent DEER experiments, native cysteine not involve in disulfide bridges were removed by introducing additional cysteine knock-out mutations into the crystallization construct 5M, which exhibits glutamate-induced gating similar to the wild-type receptor and shows negligible labeling by MTSSL. Single cysteine mutations were introduced as labeling sites for the MTSSL spin label using site-directed mutagenesis. Electron paramagnetic resonance (EPR) spectra were obtained using continuous wave EPR as previously described (Mishra et al., 2014). DEER experiments were carried out using a standard four pulse protocol (Jeschke, 2002). DEER distributions were obtained from global analysis of DEER decays as described in (Mishra et al., 2014)
More detailed experimental procedures are described in Extended Experimental Procedures.
Supplementary Material
Highlights.
Ligand-binding domains (LBDs) define AMPA receptor gating ‘ring’.
Resting/apo state of AMPA receptor harbors intact LBD dimer interface.
Receptor activation involves movements within and between LBD dimers.
Broad spectrum of extracellular domain conformations in desensitized state.
Acknowledgments
We thank L. Vaskalis for figures, ALS BCSB and APS NE-CAT beamline staff for support and M. Suga for early work on thermostability mutation screening. L.C. is supported by an American Heart Association postdoctoral fellowship (13POST13960004). K.L.D. was supported by a Long-Term Fellowship of the European Molecular Biology Organization, an institutional National Research Service Award (NRSA) and is currently recipient of an individual NRSA (F32MH100331). H.S.M. and R.A.S. were supported by grant U54-GM087519 and S10 RR027091. The Orchestra High Performance Computer Cluster at Harvard Medical School is a shared facility partially supported by NIH grant NCRR 1S10RR028832-01. We thank P. Penczek for guidance in the use of SPARX and ISAC. E.G. and T.W. are investigators with the Howard Hughes Medical Institute.
Footnotes
Author contributions
K.L.D. performed purification and crystallographic studies of structures without modulator and L.C. performed purification and crystallographic studies of structures with modulator (R,R)-2b. K.L.D and L.C. purified protein samples for DEER experiments carried out by R.A.S. and H. M. K.L.D. purified protein samples for cryo-EM studies performed by R.D.Z., I.M.F. and T.W. L.C. carried out radio ligand-binding experiments. K.L.D. performed electrophysiological studies. All authors contributed to experimental design and manuscript preparation.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Echols N, Headd JJ, Hung LW, Jain S, Kapral GJ, Grosse Kunstleve RW, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- Baconguis I, Bohlen CJ, Goehring A, Julius D, Gouaux E. X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na(+)-selective channel. Cell. 2014;156:717–729. doi: 10.1016/j.cell.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino M, Baraldi M, Parenti C, Braghiroli D, DiBella M, Vicini S, Costa E. Modulation of AMPA/kainate receptors by analogues of diazoxide and cyclothiazide in thin slices of rat hippocampus. Receptors & channels. 1993;1:267–278. [PubMed] [Google Scholar]
- Boulter J, Hollmann M, O'Shea-Greenfield A, Hartley M, Deneris E, Maron C, Heinemann S. Molecular cloning and functional expression of glutamate receptor subunit genes. Science. 1990;249:1033–1037. doi: 10.1126/science.2168579. [DOI] [PubMed] [Google Scholar]
- Chen G-Q, Sun Y, Jin R, Gouaux E. Probing the ligand binding domain of the GluR2 receptor by proteolysis and deletion mutagenesis defines domain boundaries and yields a crystallizable construct. Protein Sci. 1998;7:2623–2630. doi: 10.1002/pro.5560071216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie LA, Russell TA, Xu J, Wood L, Shepherd GM, Contractor A. AMPA receptor desensitization mutation results in severe developmental phenotypes and early postnatal lethality. Proc Natl Acad Sci USA. 2010;107:2010. doi: 10.1073/pnas.0908206107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhou HX. Atomistic mechanism for the activation and desensitization of an AMPA-subtype glutamate receptor. Nature communications. 2011;2:354. doi: 10.1038/ncomms1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gonzalez J, Du M, Parameshwaran K, Suppiramaniam V, Jayaraman V. Role of dimer interface in activation and desensitization in AMPA receptors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9891–9896. doi: 10.1073/pnas.0911854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Hibbs RE, Gouaux E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure. 2012;20:1293–1299. doi: 10.1016/j.str.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn M, Tang G, Goodyear G, Baldwin PR, Huang Z, Penczek PA, Yang C, Glaeser RM, Adams PD, Ludtke SJ. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Hollmann M, O'Shea-Greenfield A, Rogers SW, Heinemann S. Cloning by functional expression of a member of the glutamate receptor family. Nature. 1989;342:643–648. doi: 10.1038/342643a0. [DOI] [PubMed] [Google Scholar]
- Huganir RL, Nicoll RA. AMPARs and synaptic plasticity: The last 25 years. Neuron. 2013;30:704–717. doi: 10.1016/j.neuron.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson JS, Nicoll RA. Aniracetam reduces glutamate receptor desensitization and slows the decay of fast excitatory synaptic currents in the hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:10936–10940. doi: 10.1073/pnas.88.23.10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeschke G. Distance measurements in the nanometer range by pulse EPR. Chemphyschem : a European journal of chemical physics and physical chemistry. 2002;3:927–932. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Jeschke G. DEER distance measurements on proteins. Annual review of physical chemistry. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- Jin R, Banke T, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behavior of the GluR2 amino-terminal domain. EMBO J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaae BH, Harpsoe K, Kastrup JS, Sanz AC, Pickering DS, Metzler B, Clausen RP, Gajhede M, Sauerberg P, Liljefors T, et al. Structural proof of a dimeric positive modulator bridging two identical AMPA receptor-binding sites. Chemistry & biology. 2007;14:1294–1303. doi: 10.1016/j.chembiol.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinänen K, Wisden W, Sommer B, Werner P, Herb A, Verdoorn TA, Sakmann B, Seeburg PH. A family of AMPA-selective glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- Kumar J, Mayer ML. Functional insights from glutamate receptor ion channel structures. Annu Rev Physiol. 2013;75:313–337. doi: 10.1146/annurev-physiol-030212-183711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuusinen A, Abele R, Madden DR, Keinänen K. Oligomerization and ligand-binding properties of the ectodomain of the a-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid receptor subunit GluRD. J Biol Chem. 1999;41:28937–28943. doi: 10.1074/jbc.274.41.28937. [DOI] [PubMed] [Google Scholar]
- Lau AY, Roux B. The hidden energetics of ligand binding and activation in a glutamate receptor. Nature structural & molecular biology. 2011;18:283–287. doi: 10.1038/nsmb.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AY, Salazar H, Blachowicz L, Ghisi V, Plested AJ, Roux B. A conformational intermediate in glutamate receptor activation. Neuron. 2013;79:492–503. doi: 10.1016/j.neuron.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko-Lambert Y, Turetsky DM, Patneau DK. Not all desensitizations are created equal: physiological evidence that AMPA receptor desensitization differs for kainate and glutamate. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:9359–9367. doi: 10.1523/JNEUROSCI.6761-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Mooney P, Zheng S, Booth CR, Braunfeld MB, Gubbens S, Agard DA, Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Shi Y, Jackson AC, Bjorgan K, During MJ, Sprengel R, Seeburg PH, Nicoll RA. Subunit composition of synaptic AMPA receptors revealed by a single-cell genetic approach. Neuron. 2009;62:254–268. doi: 10.1016/j.neuron.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- McHaourab HS, Steed PR, Kazmier K. Toward the fourth dimension of membrane protein structure: insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19:1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Verhalen B, Stein RA, Wen PC, Tajkhorshid E, McHaourab HS. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. eLife. 2014;3:e02740. doi: 10.7554/eLife.02740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg P. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- Moriyoshi K, Masu M, Ishii T, Shigemoto R, Mizuno N, Nakanishi S. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354:31–37. doi: 10.1038/354031a0. [DOI] [PubMed] [Google Scholar]
- Moykkynen T, Coleman SK, Semenov A, Keinanen K. The N-terminal domain modulates alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor desensitization. The Journal of biological chemistry. 2014;289:13197–13205. doi: 10.1074/jbc.M113.526301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MJ, Bleakman D, Zimmerman DM, Nisenbaum ES. AMPA receptor potentiators for the treatment of CNS disorders. Curr Drug Targets: CNS Neurol Disord. 2004;3:181–194. doi: 10.2174/1568007043337508. [DOI] [PubMed] [Google Scholar]
- Patneau DK, Vyklicky L, Mayer ML. Hippocampal neurons exhibit cyclothiazide-sensitive rapidly desensitizing responses to kainate. J Neurosci. 1993;13:3496–3509. doi: 10.1523/JNEUROSCI.13-08-03496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Stern-Bach Y, Stevens RC. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- Saab AS, Neumeyer A, Jahn HM, Cupido A, Šimek AA, Boele HJ, Scheller A, Le Meur K, Götz M, Monyer H, et al. Bergmann glial AMPA receptors are required for fine motor coordination. Science. 2012;337:749–753. doi: 10.1126/science.1221140. [DOI] [PubMed] [Google Scholar]
- Schauder DM, Kuybeda O, Zhang J, Klymko K, Bartesaghi A, Borgnia MJ, Mayer ML, Subramaniam S. Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5921–5926. doi: 10.1073/pnas.1217549110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TC, Howe JR. Concentration-dependent substate behavior of native AMPA receptors. Nature Neurosci. 2000;3:992–997. doi: 10.1038/79931. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Olson RA, Horning M, Armstrong N, Mayer ML, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whorton MR, MacKinnon R. X-ray structure of the mammalian GIRK2-betagamma G-protein complex. Nature. 2013;498:190–197. doi: 10.1038/nature12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Tang C-M. Benzothiadiazines inhibit rapid glutamate receptor desensitization and enhance glutamatergic synaptic currents. J Neurosci. 1993;13:3904–3915. doi: 10.1523/JNEUROSCI.13-09-03904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada KA, Rothman SM. Diazoxide blocks glutamate desensitization and prolongs excitatory postsynaptic currents in rat hippocampal neurons. J Physiol. 1992;458:409–423. doi: 10.1113/jphysiol.1992.sp019424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan P, Leonetti MD, Hsiung Y, MacKinnon R. Open structure of the Ca2+ gating ring in the high-conductance Ca2+-activated K+ channel. Nature. 2012;481:94–97. doi: 10.1038/nature10670. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.