Abstract
AMPA-sensitive glutamate receptors are crucial to the structural and dynamical properties of the brain, to the development and function of the central nervous system and to the treatment of neurological conditions from depression to cognitive impairment. However, the molecular principles underlying AMPA receptor activation have remained elusive. Here we determine multiple x-ray crystal structures of the GluA2 AMPA receptor in complex with a Conus striatus cone snail toxin, a positive allosteric modulator and orthosteric agonists at 3.8 – 4.1 Å resolution. We show how the toxin acts like a ‘straight jacket’ on the ligand-binding domain (LBD) “gating ring”, restraining the domains via both intra and interdimer cross links such that agonist-induced closure of the LBD “clamshells” is transduced into an iris-like expansion of the gating ring. By structural analysis of activation-enhancing mutants, we show how the expansion of the LBD gating ring results in ‘pulling’ forces on the M3 helices that, in turn, are coupled to ion channel gating.
Introduction
Fast excitatory signal transduction at the chemical synapses of the brain is largely mediated by the activation of ionotropic glutamate receptors (iGluRs) by glutamate released from presynaptic neurons(1). Within the iGluR family of receptors are AMPA, kainate and NMDA subtypes, receptors that are all activated by glutamate, related in amino acid sequence, subunit stoichiometry and domain structure yet distinct in overall architecture, pharmacology and biophysical characteristics. AMPA receptors undergo fast, submillisecond activation by glutamate, leading to depolarization of the postsynaptic membrane and relief of magnesium block of NMDA receptors, followed by rapid desensitization and deactivation on a millisecond time scale (1). AMPA receptor activity is fundamental to synaptic transmission, synaptic plasticity and homeostatic scaling (2), and allosteric potentiators that slow desensitization and/or deactivation enhance synaptic currents and task-dependent neuron firing and show promise in the treatment of mild cognitive impairment and depression (3). By contrast, allosteric antagonists that diminish glutamate-induced AMPA receptor activity reduce glutamate-induced synaptic currents and are promising agents for the treatment of seizure disorders (4). Thus, understanding the molecular principles of AMPA receptor activation and action by allosteric modulators is important for continued development of new therapeutic strategies.
AMPA receptors are tetrameric complexes composed of subunits with a modular domain arrangement, beginning with the amino terminal domain (ATD), the ligand or agonist-binding domain (LBD) and the pore-forming transmembrane domain (TMD) (5). Whereas the ATDs and LBDs are organized as dimers-of-dimers, the TMD harbors approximate 4-fold symmetry, thus yielding a symmetry mismatch between the LBD – TMD layer and giving rise to two non equivalent subunit pairs, A/C and B/D (6). Hence, the mechanism and efficacy of transduction of agonist binding to ion channel gating may differ between the A/C and B/D subunit pairs. Because AMPA receptors undergo rapid and nearly complete desensitization in the continued presence of agonist (7), it has proven difficult to elucidate high resolution structures of agonistbound, activated states and to define mechanisms by which the chemical potential of agonist binding is transduced into the mechanical force of ion channel gating.
Here we report x-ray crystal structures of an intact, rat GluA2 AMPA receptor in complex with a homodimeric Conus striatus cone snail toxin, con-ikot-ikot (8), with (R,R)-2b, a high affinity, 2-fold symmetric positive allosteric modulator (9), and with the partial agonist kainate (10) or the potent partial agonist fluorowillardiine (11). To do this, we expressed, purified and solved the structure of con-ikot-ikot, a disulfide-bond-rich polypeptide, previously shown to induce paralysis in fish and potently and selectively block desensitization of AMPA receptors (8) (Fig. S1). We next screened single amino acid mutants of the GluA2 receptor for enhanced thermostability in detergent micelles by fluorescence-detection size exclusion chromatography (12) because the native, agonist-bound receptor is unstable in detergent micelles. Substitution of 10 amino acids in the TMD (construct GluA2cryst1) gives rise to a receptor with enhanced stability and with retention of agonist-induced ion channel gating activity (Fig. S2). By combining con-ikot-ikot toxin with the GluA2cryst1 receptor, together with the nanomolar affinity modulator (R,R)-2b and either kainate or FW, we were able to grow single crystals of the receptor complex that diffract x-rays to between 3.5 and 4 Å resolution. To further address how agonist binding is coupled to ion channel gating, we resolved the structures of two Lurcher-like mutants (13) within the M3 ion channel gate region (14).
Architecture of AMPA receptor-toxin complex
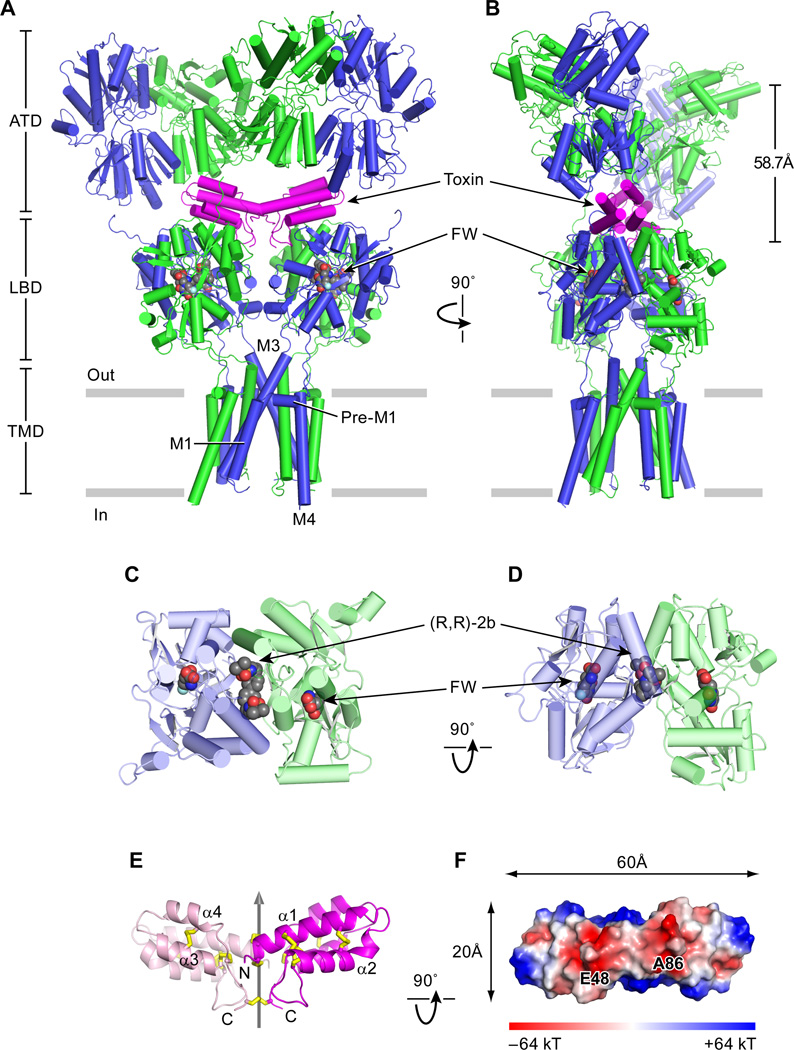
The overall shape of the GluA2 receptor complex with con-ikot-ikot, (R,R)-2B and either kainate or FW resembles the letter ‘Y’, reminiscent of the preceding GluA2 antagonist complex (Fig. 1A, B,C,D) (6). The toxin is entirely enshrouded within a large solvent-filled chamber between the ATD and LBD layers, thus providing molecular explanations for its slow on rate and potent activity (8), and requiring movement of the ATDs and/or ATD – LBD portals for binding or unbinding. By wedging between the ATD and LBD layers, the toxin buttresses the receptor complex, extending the height of the ATD to LBD layer by nearly 10 Å, as measured from the centers of masses (COMs) between the ATD and the D1 lobe of the LBD. Strikingly, the boomerang-shaped toxin spans LBD dimers, with interactions both within and between LBD dimers, burying 2128 Å2 of surface area in the interface and aligning its molecular 2-fold axis with the overall 2-fold axis of the receptor. Despite proximity to the ‘overhanging’ ATD layer, the toxin participates in few contacts with the ATD, thus explaining why ΔATD AMPA receptor complexes retain toxin sensitivity (8).
Fig. 1.
Architecture of GluA2-toxin complex. (A)View of the GluA2-toxin-(R,R)-2b-FW complex, parallel to the membrane, with the A/C subunits in blue, the B/D subunits in green and the toxin in magenta. The (R,R)-2b allosteric modulator and the agonist FW are in space-filling representation. (B) View of the complex rotated 90° around the overall two-fold axis of receptor. The distance between the ATD layer and the LBD layer D1 lobes was defined by measuring the distance between the center of mass (COM) of the ATD layer and the COM of the LBD layer D1 lobes. (C) Close-up top view of LBD AD dimer. (D) Close-up side view of LBD AD dimer. (E) Side view of con-ikot-ikot toxin. Cartoon representation of the dimeric toxin, with one subunit pink and one purple, showing how the subunits are related by a non crystallographic 2-fold axis of symmetry, shown as a gray vertical arrow. Disulfide bonds are shown as yellow sticks. (F) “Bottom view” of the toxin, looking ‘up’ from the LBD layer, showing the electrostatic surface of the toxin. Negatively charged patches on the LBD-facing surface are implicated in receptor-toxin interactions.
The 1.6 Å resolution crystal structure of the isolated toxin unambiguously shows that it is a dimeric complex, in contrast to previous suggestions that it was a dimer-of-dimers (Fig. 1E,F) (8). The size exclusion chromatography profile of the toxin confirms that it behaves as dimer in solution (Fig S1A, B). Each subunit of the homodimeric toxin harbors a four helix bundle that is extensively cross-linked by 5 disulfide bonds and is, in turn, linked to the second subunit by way of 3 intersubunit disulfide bonds (Fig. S1C), yielding a robustly rigidified complex in which the only region of flexibility resides in the subunit-subunit interface. The boomerang shaped homodimer has a conspicuous pattern of charged residues and non polar residues on its surface, with a prominent acidic patch defined by residues E48 and carboxyl terminal of A86 (Fig. 1F), and hydrophobic regions due to exposed non polar residues of F34 and I61. The con-ikot-ikot structure provides a framework to understand the structure and function of other family members.
Complex web of receptor – toxin interactions
Con-ikot-ikot toxin spans the tetrameric LBD ‘gating ring’, participating in extensive interactions with all 4 subunits such that it simultaneously fortifies intradimer interfaces and spans the interdimer cleft (Fig. 2). The electrostatic surface of the toxin “bottom” (Fig. 1F) shows extensive negative patches which are complementary to multiple positively charged residues on the receptor surface, suggesting that ionic and polar contacts are crucial for receptor-toxin interaction (Fig. 2A). Furthermore, the convex face of the boomerang-shaped toxin is complementary to the concave receptor binding site, demonstrating that shape is also central to nano-molar toxin binding affinity (8).
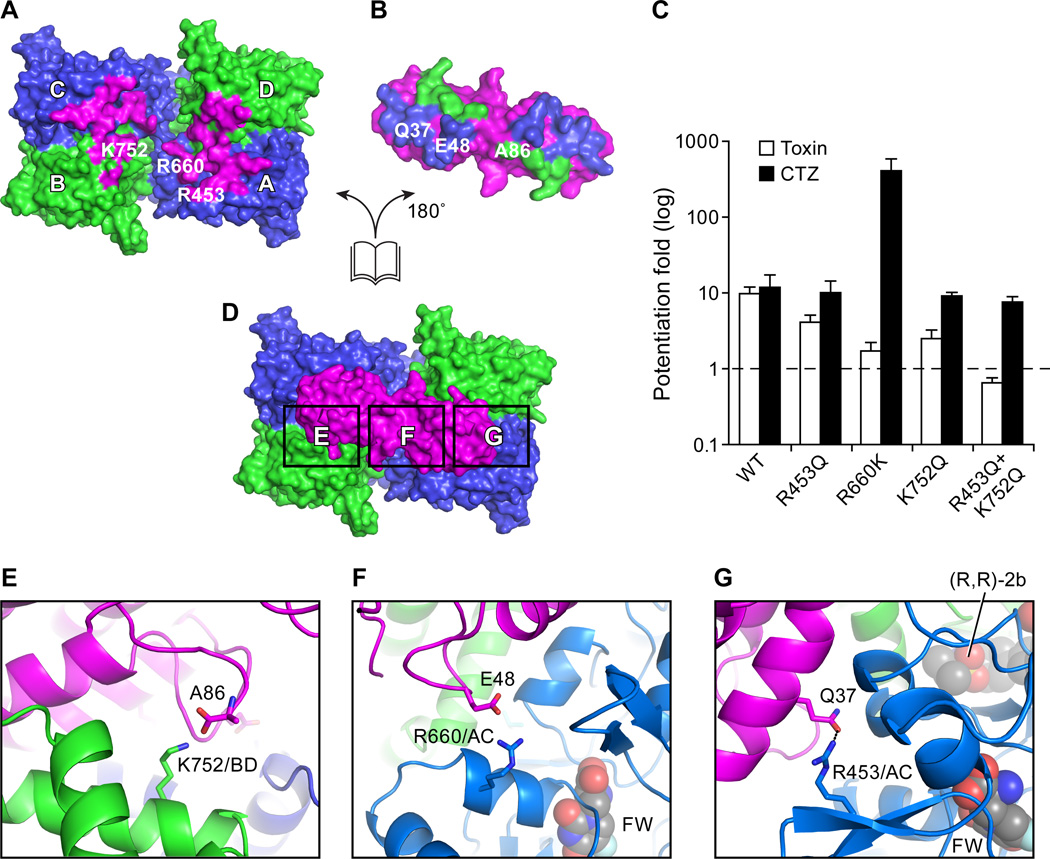
Fig. 2.
Complex web of receptor – toxin interactions. (A,B,D) An “open-book” view of the GluA2 LBD - toxin interactions shown in surface representation. (A) View of the GluA2 LBD layer from the ATD, showing the residues that interact with toxin in magenta. (B) View of the toxin residues that interact with the GluA2 A/C subunits in blue and residues that interact with the B/D subunits in green. (C) Extents of potentiation by toxin (hollow bar) and CTZ (black bar) on the “steady state” currents of wild-type GluA2 and GluA2 mutants, elicited by application of 10 mM glutamate. Data are ±S.E.M (n=3). (D) Top view onto the LBD layer from the extracellular side, highlighting key regions of toxin – receptor interactions (shown as black rectangles). (E) Close-up view of the salt bridge between K752 (GluA2 B/D subunit) and the carboxy terminus of the toxin (A86) (D). (F) Close-up view of the R660 (GluA2 A/C subunit) - E48 (toxin) interaction, boxed in (D). (G) Close-up view of the R453 (GluA2 A/C subunit) - Q37 (toxin) interaction, boxed in (D).
To more precisely understand the mechanism by which the toxin blocks desensitization, we peeled away the toxin from its LBD binding site to give an ‘open book’ view of the receptor - toxin interface (Fig. 2A, 2B). The receptor-toxin interactions can be classified into two types of contacts based on the positions of subunits in the receptor complex (6), namely LBD “proximal” A/C subunit -toxin interactions and “distal” B/D subunit - toxin interactions. Crucial contacts are between the extreme C-terminal carboxyl group of the toxin (A86) and the ammonium group of K752 from LBD B/D pair D1 lobe (Fig. 2E), and between the side chain carboxyl group of toxin reside E48 and the side chain guanidine group of R660 from the LBD A/C pair D2 lobe (Fig. 2F). Augmenting these interactions is a polar contact between the side chain carbonyl group of toxin Q37 with the side chain guanidine group of R453 from LBD A/C pair D1 lobe. These residues on the GluA2 receptor are conserved in AMPA-subtype iGluRs but not in kainate receptors or NMDA receptors (Fig.S3A), which is consistent with previous findings that conikot- ikot only potentiates the activity of AMPA receptors (8). To validate the interactions we observed in the receptor - toxin crystal structure, we mutated the residues on GluA2 to the corresponding residues on kainate receptors and measured the extent to which toxin potentiates the activity of the mutants in comparison with cyclothiazide (CTZ) (15). While all of the mutants respond to CTZ, potentation by toxin is reduced and, for the double mutant of R453Q/K752Q, toxin potentiation is ablated (Fig. 2C). Interestingly, the R660K mutant desensitizes profoundly, based on the magnitude of CTZ potentiation. This might be because R660 indirectly decreases LBD dimer stability, and thus promotes desensitization.
On the basis of extensive structural and functional studies of soluble LBDs (sLBD), the LBD undergoes clamshell-like domain closure upon agonist binding and the degree of domain closure is correlated with agonist efficacy (11, 16), although partial agonists may nevertheless occupy the fully closed state, albeit with reduced probability (17). Because the toxin interacts with both the D1 and D2 lobes of the LBD A/C pair of subunits, we compared the KA-bound structure with the FW-bound structure by superposing the toxin domains (Fig. S3B). While the D2 lobes of the two structures are almost super imposable, the D1 lobe shows an “outward” displacement in the KA-bound structure compared to the FW-bound structure, as evidenced by a 3 Å shift of the marker Cα atom of K458 of the A/C pair, which is on β4 in D1 lobe and in close proximity to R453. This conformational difference is consistent with the degree of clamshell closure both in full length structures (FigS3E,F) and in sLBDs (18) (19) (11) which show that the KA clamshells are more open in comparison to the FW subunits. Nevertheless, a change in rotamer conformation of toxin residue Q37 allows it to maintain its interaction with receptor residue R453 in both complexes (FigS3C), thus showing how the toxin can interact with the receptor in the presence of KA or FW, despite differences in the degree of LBD clamshell closure. However, if we model a hypothetical complex of the toxin with an apo receptor subunit by superposing the D2 lobe of the apo sLBD (19) (1FTO chain A) structure onto the KA and toxin bound structure, there is a further 7 Å outward shift of the K458 Cα marker atom so that Q37 of the toxin can no long interact with R453 of the LBD (Fig. S3C). Indeed, in a toxin binding assay we show that receptor - toxin complex formation requires the presence of agonist (Fig. S3D). Thus, receptor-toxin binding requires ligands that induce clamshell closure. Because the toxin binds to both D1 and D2 lobes of the LBD A/C pair, the LBD is stabilized in a ligand bound conformation. Consistent with the mechanism, toxin increases apparent agonist affinity in a 3H FW binding assay (Fig. S2F) and previous studies have shown that toxin can modestly retard the rate of receptor deactivation (8).
Conformation of the LBD layer in an activated state
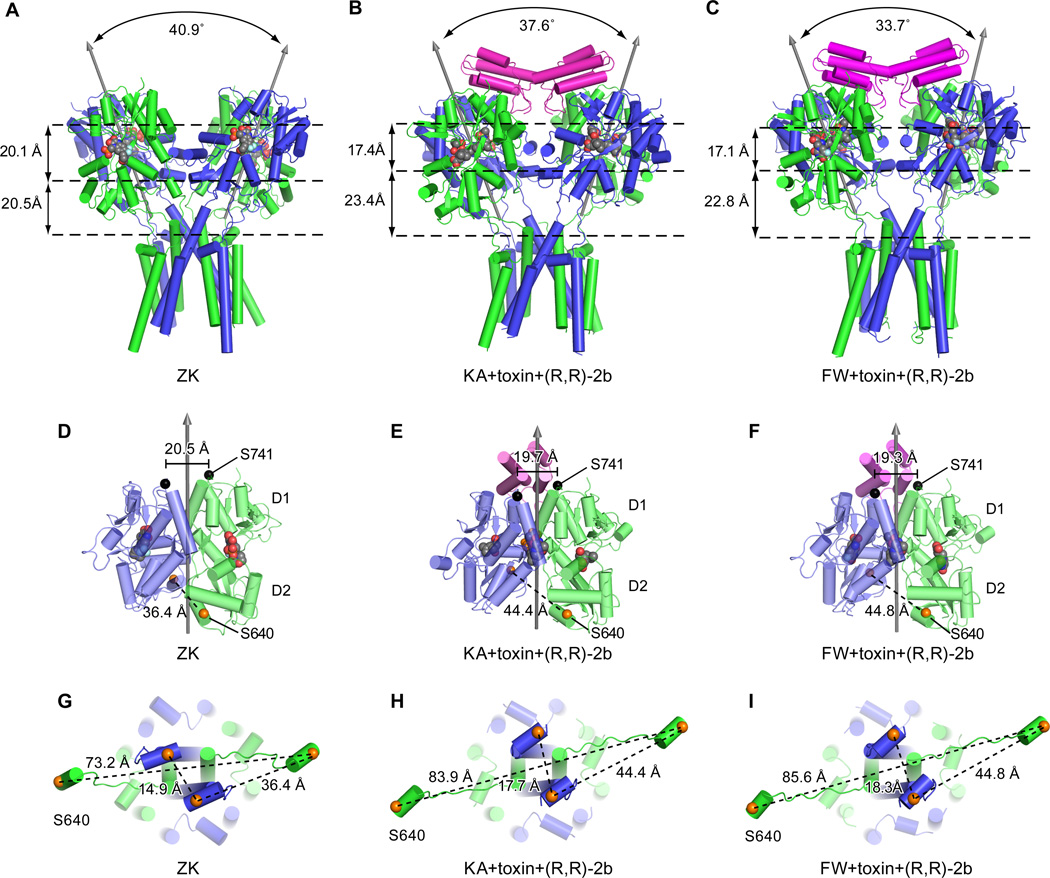
The inter axis or “roll” angle (20) between the 2-fold axes of the LBD dimers is smaller in the toxin complex than in the ZK antagonist-bound structure (Fig. 3 A, B, C), largely because the toxin ‘locks’ the LBD gating ring in a fixed conformation. Furthermore, the toxin structure with FW bound has a smaller inter axis angle than the KA complex (Fig. 3B, C) because the D2 lobes, which are coupled to the ion channel, share a common position but the FW complex has a greater LBD domain closure. Thus there is a corresponding difference in the position of the D1 lobes (Fig. S3E, F). By binding both subunits within a LBD dimer, the toxin stabilizes the LBD dimer D1 – D1 interface in a non desensitized conformation, yielding a distance between S741 Cα marker atoms on the D1 – D1 interface of 19.3–19.7 Å (21, 22). By using the distance between Cα atoms of S741 residues in the D1 – D1 interface as a metric, we show that the D1 – D1 interfaces adopt non desensitized conformations in all of the structures (Fig. 3D, E, F). The toxin complexes reported here also have the modulator (R,R)-2b bound because inclusion of modulator improves diffraction quality, presumably by stabilizing the receptor in a conformationally homogenous population.
Fig. 3.
Conformation of the LBD layer in an activated state. Structures of the GluA2 receptor in the antagonist-bound state “ZK” (A, D, G). Structures of the GluA2 receptor in the KA+toxin+(R,R)-2b bound state (B, E, H) and in complex with FW+toxin+(R,R)-2b (C, F, I). In panels A, B, and C are side views of the LBD-TMD layers showing the angles between the two local two-fold rotation axes of the LBD dimers. The distances between the LBD D1 lobes and the D2 lobes and the distances between the D2 lobes and T625 are measured between the COMs of each layer and are shown on the left. In panels D, E and F are side views of the A/D LBD dimers. Black arrows depict the local two-fold axes of LBD dimers. The Cα atoms of S741 are shown as black spheres, along with the inter-subunit distances. The Cαs atoms of S640 are shown as orange spheres. In panels G, H, and I are views of helix E in the LBD layer and the LBD D2-M3 linker, seen from the LBD layer. The Cα atoms of S640 are also shown as orange spheres, together with inter subunit distances.
In comparison to the GluA2 ZK structure (PDB code 3KG2) (6), the toxin complex structures with either KA or FW bound have much larger degrees of clamshell closure and thus a larger separation of the D2 lobe within dimers. To quantify the separation of the D2 lobes, we compare the distances between S640 Cα marker atoms, which are on helix E of the D2 lobe and are directly connected to D2-M3 linker. There is 8.4 Å increase in separation between the D2 lobes upon transition from the ZK antagonist state to the FW agonist-bound structure, thus providing structural evidence for the long standing hypothesis that LBD domain closure is mechanically coupled to ion channel gating (19, 21). Moreover, there is an additional 3.4 Å separation in the proximal A/C pair and a 12.4 Å separation in distal B/D pair distance (Fig. 3G, H, I), thus suggesting that there is a non equivalent "pulling force" of the LBDs onto the LBD-TMD linker (23), consistent with the symmetry non equivalence of the A/C and B/D subunit pairs (6).
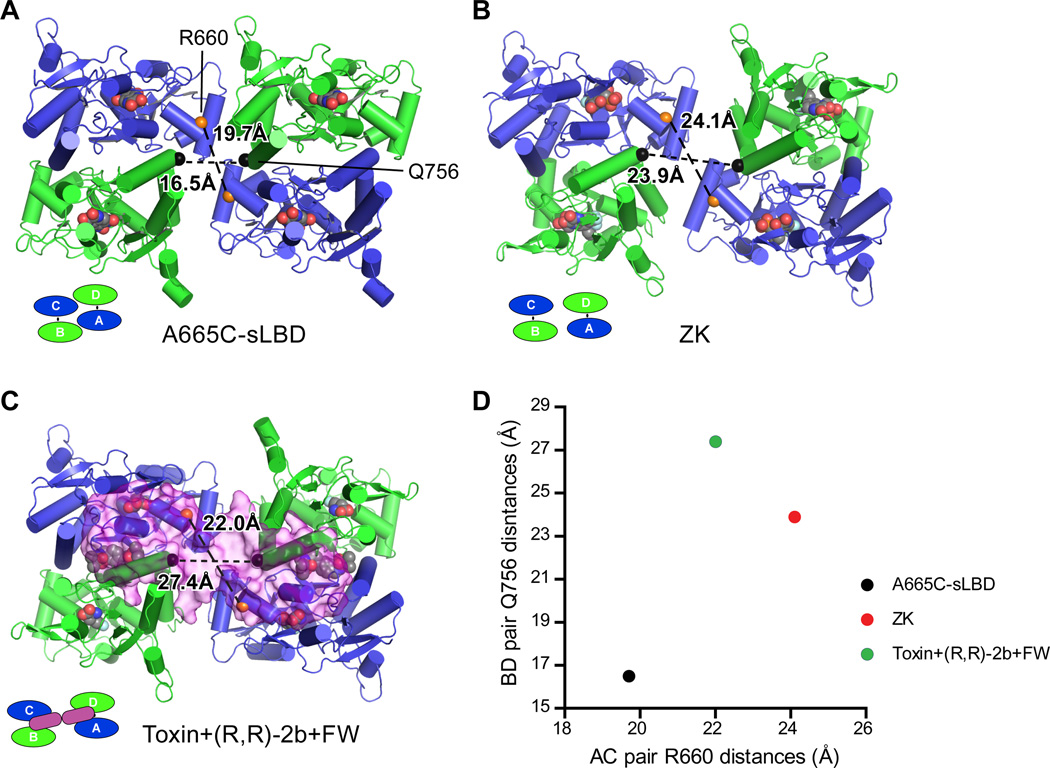
Toxin stabilization of LBD ‘gating ring’
A view of the LBD layer from the ATD shows how the toxin stabilizes the LBD layer in an expanded conformation (Fig. 4). By measuring the distances between Cα marker atoms of R660 on helix F, and Q756 on helix J, both of which are close to the central opening of the ‘gating ring’, we can compare the inter-dimer distances between selected structures. Beginning with the A665C cross-linked sLBD tetramer (Fig. 4A,D) (24), which is trapped in an inactive state yet has been suggested to represent an activation intermediate, and then to the ZK state (Fig.4B,D) and finally to the toxin and FW-bound state (Fig. 4C,D), we observe an increase of the B/D distance from 16.5 Å to 23.9 Å to 27.4 Å, respectively, while the A/C distance changes from 19.7 Å to 24.1 Å to 22.0 Å, respectively. The overall change in the B/D distances is larger in comparison to the change in the A/C pair, an observation which is correlated with the changes in the S640 Cα distances mentioned previously, and consistent with the hypothesis that the B/D pair may play a more important role in ion channel opening. We suggest that without toxin, the LBD dimers are more conformationally dynamic and can adopt different conformations because of the relatively small inter-dimer interface (25).
Fig. 4.
Toxin stabilizes the LBD ‘gating ring’ in an expanded conformation. Top view of the LBD layer in the A665C-crosslinked GluA2 sLBD structure (PDB 4L17) (A), the GluA2 ZK bound state (B) and the GluA2 FW+toxin+(R,R)-2b bound state (C). The distances between Cα atoms of GluA2 R660 (orange spheres) from the A/C subunits and the distances between the Cαs atoms of GluA2 Q756 (black spheres) from the B/D subunit are shown. Distances are plotted in (D).
Coupling of agonist-bind to channel gating via M3-D2 linker
The channel pore of the toxin-GluA2 complex structure with either KA or FW adopts the same closed conformation as the ZK bound structure (6), although there is a small conformational change at the upper region of the crucial M3 helix between residues 623–626 (Fig. 5A,5D). In the toxin complex structures, the LBD dimer interfaces are intact and stabilized by both toxin and modulator. In addition, partial agonists are also bound and induce LBD clamshell closure, thus we propose that these structures represent toxin-bound pre-open states. However, the ion channel gate is closed at the M3 bundle crossing, and we speculate that this is because either the crystallization conditions, the absence of lipid bilayer or the details of the receptor construct favor a closed-gate conformation. It is also possible that the use of partial agonists does not sufficiently populate the open gate conformation (11). To address the issue of the receptor construct, we solved the structure of GluA2cryst2 in complex with KA+toxin+(R,R)-2b, where the GluA2cryst2 construct only harbors the C815A mutation, located at the C-terminal end of the M4 helix. The structure of this GluA2cryst2 complex is similar to the corresponding GluA2cryst1 structure (Fig. S5C D; overall rmsd of 0.28 Å on main chain atoms), thus demonstrating that other factors are responsible for precluding the crystallization of an open gate conformation.
Fig. 5.
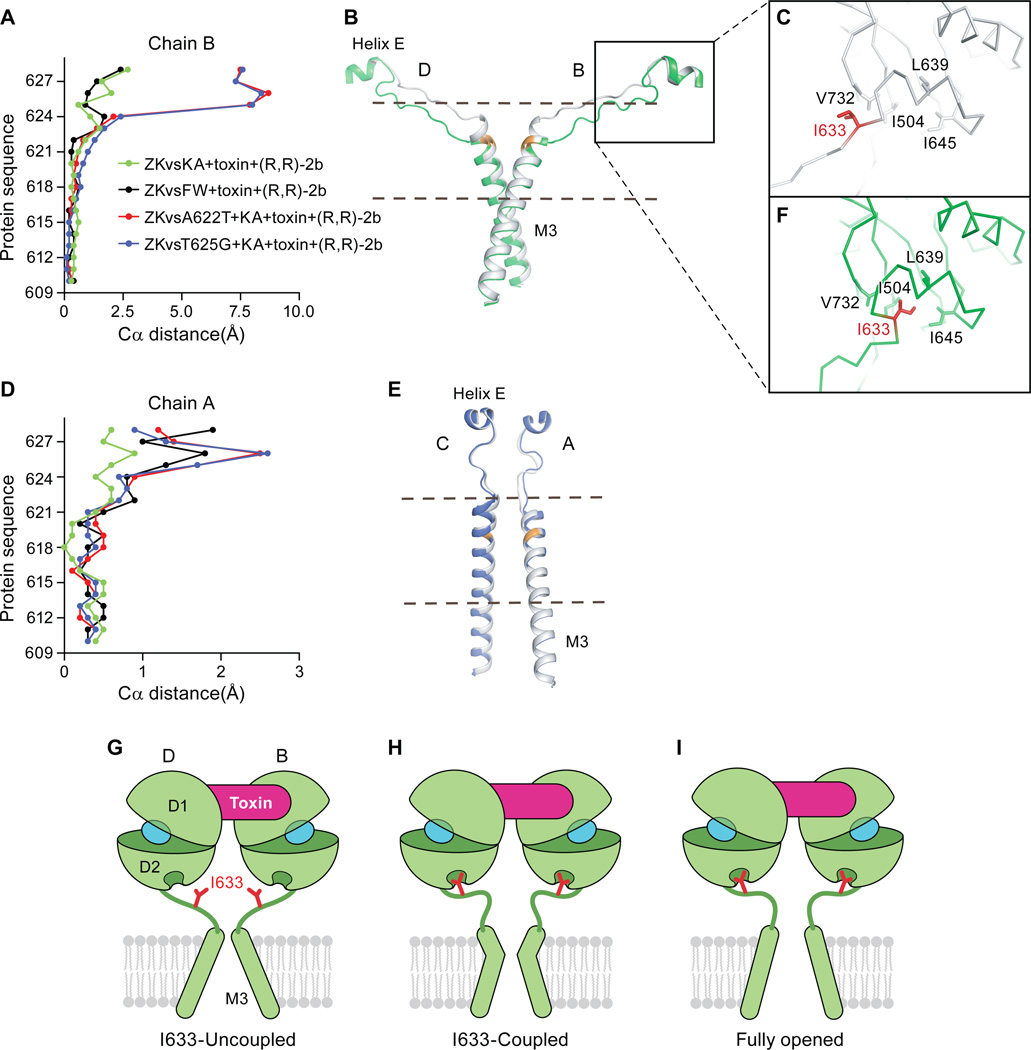
Coupling of agonist-binding to ion channel gating via the M3-D2 linker. (A) A plot of the differences in position of Cα atoms from residues in the M3 TMD helixes derived from chain B in the structures reported here following superposition of the M1, M3 and M4 helices. (B) Cα trace of the LBD D2 to M3 connections in the B/D subunits where the structure of the GluA2 KA+toxin+(R,R)-2b elements are shown in gray and those derived from the A622T GluA2 mutant KA+toxin+(R,R)-2b complex are shown in green. The main chain atoms of the site of mutation, A622, are illustrated in orange. The region used to generate panel (A) is shown between dash. (C) Close-up view of the “uncoupled” I633 site in the KA+toxin+(R,R)-2b structure subunit B. I633 is shown in red. The side chains of hydrophobic residues that line the I633-binding pocket are shown in sticks. (D) A plot analogous to that shown in panel (A), using the same structures, of the respective Cα distances from residues in the M3 helix from chain A. (E) The LBD D2 to M3 connections in A/C subunits. The structure of the GluA2 KA+toxin+(R,R)-2b complex is shown in gray and the structure of the A622T GluA2 KA+toxin+(R,R)-2b complex is shown in blue. At the mutation site, A622, the back bone is colored in orange. The region used to generate panel (D) is shown between dash. (F) Close-up view of the “coupled” I633 site in the KA+toxin+(R,R)-2b A622T mutant structure subunit B. I633 is in red. The side chains of hydrophobic residues that define the I633-binding pocket are shown in stick representation. Panels G, H and I depict schematic illustrations of a mechanism by which the toxin stabilizes an activated conformation of the LBD layer. Only LBD B/D subunits and M3 helices are shown for clarity. I633 residues are in shown red and agonists are in shown in cyan. (G) The toxin bound and I633-uncoupled state, as we observed in the partial agonist, toxin (R,R)-2b structures. (H) The toxin-bound and I633-coupled state. Here the M3 B/D helices expand via a kink in the helices, yet the ion channel gate remains closed, as we observed in the A622T or T625G structures. (I) A hypothetical model of the toxin bound, I633 coupled and open channel state. We speculate that even with toxin bound the channel probably flickers between G and I, perhaps via H as an intermediate state.
To favor the formation of an open state we generated the Lurcher-like mutants A622T (27) and the T625G (28) in the context of the GluA2cryst1 construct, mutations that are known to greatly facilitate ion channel gating. Structures of these mutants as complexes with KA+toxin+(R,R)-2b showed large conformational changes only within the M3-LBD linkers compared to the structure without the activation mutations (Fig. 5 and Fig. S6). Complexes with full agonists or with more efficacious partial agonists have so far not yielded crystals. Because the structures of the T625G and A622T variants are close to identical, we focus the discussion on the A622T structure unless indicated. In the mutant structures, the side chain of I633 from the B/D subunits binds within a hydrophobic pocket formed by V732, I504, L639 and I645 on the D2 lobe of the LBDs, adopting the same binding mode as I633 from the A/C pair or all subunits from the ZK structure (Fig. 5F), we term this the “coupled” conformation. By contrast, I633 from B/D pair in the toxin-bound structures without the M3 mutations is "pulled out” of the hydrophobic D2 pocket and adopts an “uncoupled” conformation (Fig. 5C).
To better define the register of the region flanking I633, we generated the S635C mutant in a GluA2cryst1 background and labeled the resulting protein with mercury. In anomalous difference electron density maps at the LBD-TMD linker region of the S635C mutant we see density at the A/C subunit positions but no prominent density for the B/D subunits (Fig. S5G). Assuming that the sites have similar labeling efficiencies, we can conclude that the S635C sites in the B/D pair are mobile, consistent with a dynamic linker region. However, the electron density is sufficiently strong to unambiguously trace the main chain which shows that the I633 side chain cannot adopt a ‘coupled’ conformation in the non M3 mutant structures. To further probe the role of I633 we generated I633A and I633E mutants in the background of the wild-type receptor, hypothesizing that these mutants would favor an uncoupled conformation and thus diminish agonist-induced gating. Indeed, the I633A mutant shows only very small currents compared to wild-type GluA2 (Fig. S4A, D, J) while the I633E mutant shows even smaller currents (Fig. S4G, J). Interestingly, both receptor assembly, as assessed by FSEC (Fig S4B, E, H), and receptor surface expression are similar to the wild-type receptor (Fig. S4C, F, I, K, L). These results are consistent with the conclusion that the I633 mutant has impaired gating, perhaps similar to the recently reported glycine insertion mutants of the NMDA receptor LBD-M3 linker (23). Indeed, the I633 position is conserved in iGluRs, with the exception that in the NMDA receptor GluN2 subunit there is a valine at this site (Fig. S4N). Because the uncoupled phenotype of the I633A mutant can be robustly rescued by coexpression with TARPγ2 (29) (Fig. S4M), an AMPA receptor auxiliary subunit that enhances receptor activation, we show that I633A mutant is properly folded and assembled yet likely has a low open probability in the absence of TARPγ2.
The activation mutant structures show that conformational changes of key gating residues near the M3 helix crossing are more prominent in the B/D subunits than in the A/C subunits (Fig. 5A, D). As identified in the ZK-bound structure, T617, A621, T625 and M629 form the physical shut gate of the channel (6). In the activation mutant structures, the outward movements of the Cα atoms in B/D subunits start at A620, increase in the direction of the extracellular ends of the M3 segments (Fig. 5A), and involve the unwinding of the last turn of M3 helix. By contrast with the large changes in the upper region of the M3 helix of the B/D subunits, there are only small changes in the lower region of the B/D subunits, N-terminal to T617, and only small changes in the entire M3 segment from the A/C subunits. Together, these results show that the pore is still shut and that the upper M3 helix transforms from 4-fold symmetry to 2-fold symmetry in transitioning from the cytoplasmic to the extracellular side of the membrane (30).
The coupled I633 position and the outward movements of the upper regions of the M3 segments in the B/D subunits are consistent with the mutant structures reflecting an activation intermediate state (Fig. 5H) between the toxin bound structure (Fig. 5G) and a hypothetical fully opened structure (Fig. 5I). Lurcher-like mutants may facilitate current flow by weakening the M3 helix bundle, allowing the ion channel gate to open more ‘easily’. In our structures, however, the ion channel has not opened, and instead the mutants have simply facilitated the distortion of the extracellular end of the M3 helix.
We suggest that I633 must be in a coupled position to transmit agonist binding and clamshell closure to gate opening and, secondly, that the unwinding of the M3 helix tip can absorb the "pulling force" generated by LBD. We note that if we model I633 in a coupled state, the linker is not of sufficient length to permit the combination of an activated LBD (closed clamshell) and a closed pore (Fig. S6C). Thus, we speculate that in the fully opened structure, I633 is coupled, the M3 tip preserves its helical structure and the M3 helix is flexed or kinked ‘outward’ to allow for an open channel gate.
Conclusion
Here we show that con-ikot-ikot toxin binds between ATD and LBD layers, forms extensive contacts with all four LBD subunits and stabilizes the LBD gating ring in an activation competent conformation. The cross linking of the LBDs precludes the rupture of the D1-D1 interface, locks the LBD dimers together and allows for agonist binding to be coupled to ion channel gating. By comparing agonist-bound toxin-receptor structures to the previously determined antagonist-receptor complex (Supplementary movie M1, M2), we show how agonist-binding is coupled to ion channel gating, thus validating decades of research on the isolated binding domains of iGluRs. We propose that agonist binding generates an asymmetric pulling force on the LBD-TMD linker and that a previously unidentified coupling switch defined by I633 is crucial to convey clamshell closure to channel opening. Taken together, the toxin bound GluA2 structures provide insight into the gating mechanism of AMPA receptors and shed light on mechanisms of closely related kainate and NMDA receptors. Insight into how AMPA receptor activity is modulated may also open new avenues for the development of therapeutic agents.
Supplementary Material
Acknowledgments
All members of the Gouaux laboratory and I. Baconguis are gratefully acknowledged for their support and assistance. We thank L.Vaskalis for assistance with figures. We thank the beam line staff at A.L.S BCSB and SSRL 12-2 for their support during data collection. We thank B. Olivera for providing crude Conus striatus venom. We thank David King and Larry David for mass spectrum analysis and M. Suga for early work on thermostability mutation screening. L. Chen is supported by an American Heart Association postdoctoral fellowship (13POST13960004). K.L. Duerr was supported by a Long-Term Fellowship of the European Molecular Biology Organization, an institutional National Research Service Award and is currently a recipient of an individual NRSA (F32MH100331). This work is supported by the NIH (E.G.) and E. Gouaux is an investigator with the Howard Hughes Medical Institute. The coordinates have been deposited in the PDB with codes 4U5B, 4U5C, 4U5D, 4U5E, 4U5F for the GluA2 receptor toxin complex and codes 4U5G, 4U5H for the isolated toxin.
Footnotes
Author contributions
L.C. contributed to all aspects of the research; K.L.D. contributed to thermostabilization mutation screening and initial characterization of native snail venom by TEVC. E.G. directed the research. All authors contributed to the preparation of the manuscript.
References
- 1.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 2010;62:405. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huganir RL, Nicoll RA. AMPARs and synaptic plasticity: The last 25 years. Neuron. 2013;30:704. doi: 10.1016/j.neuron.2013.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Neill MJ, Bleakman D, Zimmerman DM, Nisenbaum ES. AMPA receptor potentiators for the treatment of CNS disorders. Curr. Drug Targets: CNS Neurol. Disord. 2004;3:181. doi: 10.2174/1568007043337508. [DOI] [PubMed] [Google Scholar]
- 4.Rogawski MA. AMPA receptors as a molecular target in epilepsy therapy. Acta Neurol. Scand. Suppl. 2013;197:9. doi: 10.1111/ane.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar J, Mayer ML. Functional insights from glutamate receptor ion channel structures. Annu. Rev. Physiol. 2013;75:313. doi: 10.1146/annurev-physiol-030212-183711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raman IM, Trussell LO. The kinetics of the response to glutamate and kainate in neurons of the avian cochlear nucleus. Neuron. 1992;9:173. doi: 10.1016/0896-6273(92)90232-3. [DOI] [PubMed] [Google Scholar]
- 8.Walker CS, et al. A novel Conus snail polypeptide causes excitotoxicity by blocking desensitization of AMPA receptors. Curr. Biol. 2009;19:900. doi: 10.1016/j.cub.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaae BH, et al. Structural proof of a dimeric positive modulator bridging two identical AMPA receptor-binding sites. Chem. Biol. 2007;14:1294. doi: 10.1016/j.chembiol.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Patneau DK, Vyklicky L, Mayer ML. Hippocampal neurons exhibit cyclothiazide-sensitive rapidly desensitizing responses to kainate. J. Neurosci. 1993;13:3496. doi: 10.1523/JNEUROSCI.13-08-03496.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nat Neurosci. 2003;6:803. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- 12.Hattori M, Hibbs RE, Gouaux E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure. 2012;20:1293. doi: 10.1016/j.str.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuo J, et al. Neurodegeneration in Lurcher mice caused by mutation in d2 glutamate receptor gene. Nature. 1997;388:769. doi: 10.1038/42009. [DOI] [PubMed] [Google Scholar]
- 14.Kuner T, Seeburg PH, Guy HR. A common architecture for K+ channels and ionotropic glutamate receptors. Trends Neurosci. 2003;26:27. doi: 10.1016/s0166-2236(02)00010-3. [DOI] [PubMed] [Google Scholar]
- 15.Partin KM, Patneau DK, Winters CA, Mayer ML, Buonanno A. Selective modulation of desensitization at AMPA versus kainate receptors by cyclothiazide and concanavalin A. Neuron. 1993;11:1069. doi: 10.1016/0896-6273(93)90220-l. [DOI] [PubMed] [Google Scholar]
- 16.Armstrong N, Mayer ML, Gouaux E. Tuning activation of the AMPA-sensitive GluR2 ion channel by genetic adjustment of agonist-induced conformational changes. Proc Natl Acad Sci U S A. 2003;100:5736. doi: 10.1073/pnas.1037393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmed AH, Wang S, Chuang HH, Oswald RE. Mechanism of AMPA receptor activation by partial agonists: disulfide trapping of closed lobe conformations. J. Biol. Chem. 2011;286:35257. doi: 10.1074/jbc.M111.269001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Armstrong N, Sun Y, Chen G-Q, Gouaux E. Structure of a glutamate receptor ligand binding core in complex with kainate. Nature. 1998;395:913. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- 19.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 20.Lee CH, et al. NMDA receptor structures reveal subunit arrangement and pore architecture. Nature. 2014 doi: 10.1038/nature13548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y, et al. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 22.Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 23.Kazi R, Dai J, Sweeney C, Zhou HX, Wollmuth LP. Mechanical coupling maintains the fidelity of NMDA receptor-mediated currents. Nat. Neurosci. 2014;17:914. doi: 10.1038/nn.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau AY, et al. A conformational intermediate in glutamate receptor activation. Neuron. 2013;79:492. doi: 10.1016/j.neuron.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horning M, Mayer M. Regulation of AMPA receptor gating by ligand binding core dimers. Neuron. 2004;41:379. doi: 10.1016/s0896-6273(04)00018-2. [DOI] [PubMed] [Google Scholar]
- 26.Karakas E, Furukawa H. Crystal structure of a heteromeric NMDA receptor ion channel. Science. 2014;344:992. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wollmuth LP, et al. The Lurcher mutation identifies delta2 as an AMPA/Kainate receptor-like channel that is potentiated by Ca2+ J. Neurosci. 2000;20:5973. doi: 10.1523/JNEUROSCI.20-16-05973.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore BS, Mirshahi UL, Ebersole TL, Mirshahi T. A conserved mechanism for gating in an ionotropic glutamate receptor. J. Biol. Chem. 2013;288:18842. doi: 10.1074/jbc.M113.465187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson AC, Nicoll RA. The expanding social network of ionotropic glutamate receptors: TARPs and other transmembrane auxiliary subunits. Neuron. 2011;70:178. doi: 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sobolevsky AI, Yelshansky MV, Wollmuth LP. The outer pore of the glutamate receptor channel has 2-fold rotational symmetry. Neuron. 2004;41:367. doi: 10.1016/s0896-6273(04)00008-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.