Summary
N-methyl-d-aspartate (NMDA) receptors are Hebbian-like coincidence detectors, requiring binding of glycine and glutamate in combination with the relief of voltage-dependent magnesium block to open an ion conductive pore across the membrane bilayer. Despite the importance of the NMDA receptor in the development and function of the brain, a molecular structure of an intact receptor has remained elusive. Here we present x-ray crystal structures of the GluN1/GluN2B NMDA receptor with the allosteric inhibitor, Ro25-6981, partial agonists and the ion channel blocker, MK-801. Receptor subunits are arranged in a 1-2-1-2 fashion, demonstrating extensive interactions between the amino terminal and ligand binding domains. The transmembrane domains harbor a closed-blocked ion channel, a pyramidal central vestibule lined by residues implicated in binding ion channel blockers and magnesium, and a ~2-fold symmetric arrangement of ion channel pore loops. These structures provide new insights into the architecture, allosteric coupling and ion channel function of NMDA receptors.
Glutamate is the primary excitatory neurotransmitter in the brain, acting at ionotropic and metabotropic glutamate receptors. Rapid excitation by glutamate, in turn, solely involves action at AMPA, kainate and NMDA ionotropic glutamate receptors1. The NMDA receptor is central to the development and function of the nervous system and to neurotoxicity 2. As a linchpin of synaptic plasticity, blockade of the NMDA receptor interferes with memory formation and recall3. Moreover, mutations within the coding regions of NMDA receptor subunit genes are associated with a spectrum of neurological diseases and neuropsychiatric disorders, from schizophrenia to epilepsy4. Autoimmune responses to the NMDA receptor, and presumed disruption in NMDA receptor organization on neural cell surfaces, likely underlie NMDA receptor encephalitis5. In keeping with the profound roles of the NMDA receptor in brain function, the receptor is a target of small molecules for the treatment of cognitive impairment, depression, schizophrenia and pain2.
Whilst AMPA and kainate receptors can be activated solely by glutamate6–8, NMDA receptors are Hebbian-like coincidence detectors, requiring the binding of glycine and glutamate to GluN1 and GluN2 subunits9, respectively, combined with membrane depolarization to relieve magnesium block 10, 11. Activation of the receptor opens a cation-selective, calcium permeable channel, thus causing further depolarization of the cell membrane and influx of calcium 12. NMDA receptors are obligatory heterotetrameric assemblies13, 14, typically composed of two glycine-binding GluN1 subunits and two glutamate-binding GluN2A-D subunits, with the GluN1/GluN2A/GluN2B complex the predominate receptor at hippocampal synapses15. Glycine-and d-serine-binding GluN3 subunits are additional subunits, expressed throughout the nervous system but with roles less well defined in comparison to the GluN1/GluN2 assemblies. A hallmark of NMDA receptors, by contrast with AMPA and kainate receptors, is a wide spectrum of allosteric modulation, from nanomolar concentrations of zinc, to the small molecule ifenprodil, polyamines and protons16 and to voltage-dependent ion channel block by MK-801, ketamine and memantine17.
The GluN1, GluN2 and GluN3 NMDA receptor subunits are related in amino acid sequence and, like AMPA and kainate receptor subunits, possess a modular domain architecture, with amino terminal domains (ATDs) and ligand binding domains (LBDs) residing on the extracellular side of the membrane, a transmembrane domain (TMD) spanning the membrane and defining the ion channel pore, and an intracellular carboxy terminal domain (CTD) within the cytoplasm1. Multiple high resolution crystal structures of the isolated LBDs from NMDA, AMPA and kainate receptors show that these domains adopt similar clamshell-like structures that are organized in an approximately dimeric, back-to-back fashion18–20. While crystal structures of isolated ATDs illustrate that they too possess a clamshell-like structure 21–23, in NMDA receptors not only is the organization of each clamshell lobe distinct from that in AMPA and kainate receptors, but the interactions between subunits are also different24. The functional properties of the NMDA ion channel pore, which harbors binding sites for magnesium and small molecule blockers, are also distinct from AMPA and kainate receptors1.
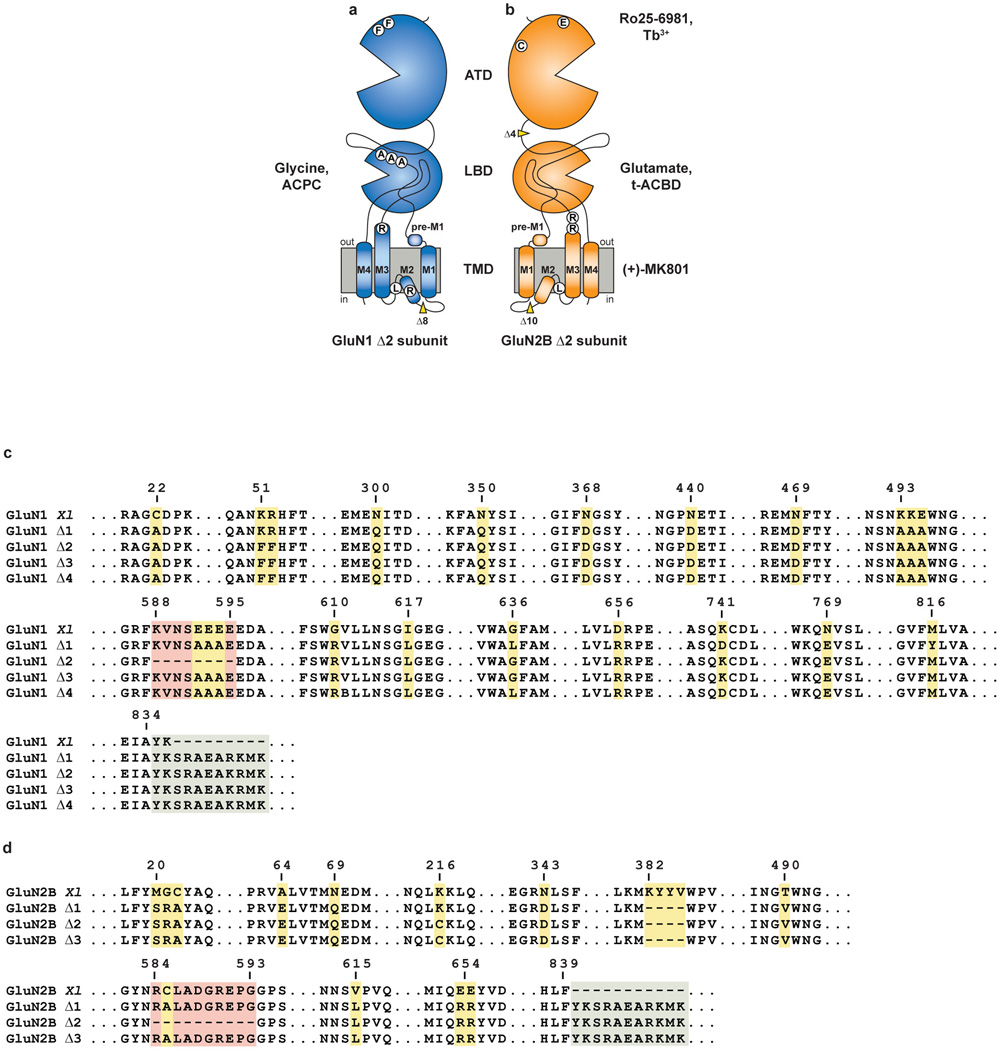
Here we report crystal structures of the GluN1/GluN2B NMDA receptor from Xenopus laevis in complex with the GluN2B-specific allosteric inhibitor, Ro25-698125, the GluN1 and GluN2B partial agonists 1-aminocyclopropane-1-carboxylic acid (ACPC) 26 and trans-1-aminocyclobutane-1,3-dicarboxylic acid (t-ACBD)27, respectively, and the ion channel blocker, MK-801. To enhance the stability of the receptor in detergent micelles and to reduce conformational surface entropy, we replaced the cytoplasmic C-terminus of the GluN1 and GluN2B subunits with 11 residues from the GluA2 C-terminus28 and we introduced a number of mutations into each subunit, ultimately finding a NMDA receptor complex that preserved binding of full and partial agonists and Ro25-6981, together with small but measurable conductance activated by glycine and glutamate, and with channel block by magnesium. To decrease conformational mobility of the extracellular domains, we substituted GluN2B Lys216 to Cys (K216C), resulting in spontaneous disulfide bond formation between GluN2B subunits, improving crystal quality yet reducing agonist-induced ion channel activity (Extended Data Table 1; Extended Data Figs. 1–4). We determined crystal structures of the GluN1/GluN2B K216C receptor at resolutions of 3.7 Å (Structure 1) and 3.9 Å (Structure 2) and refined the structures to reasonable crystallographic residuals and good stereochemistry. In addition, we mapped cation sites in the ATD by exploiting anomalous scattering from a Tb3+ derivative and probed the mobility of the ATD and LBD layers by comparing a non K216C crosslinked structure to the higher resolution K216C structures (Extended Data Table 2; Supplementary Discussion).
Architecture and symmetry
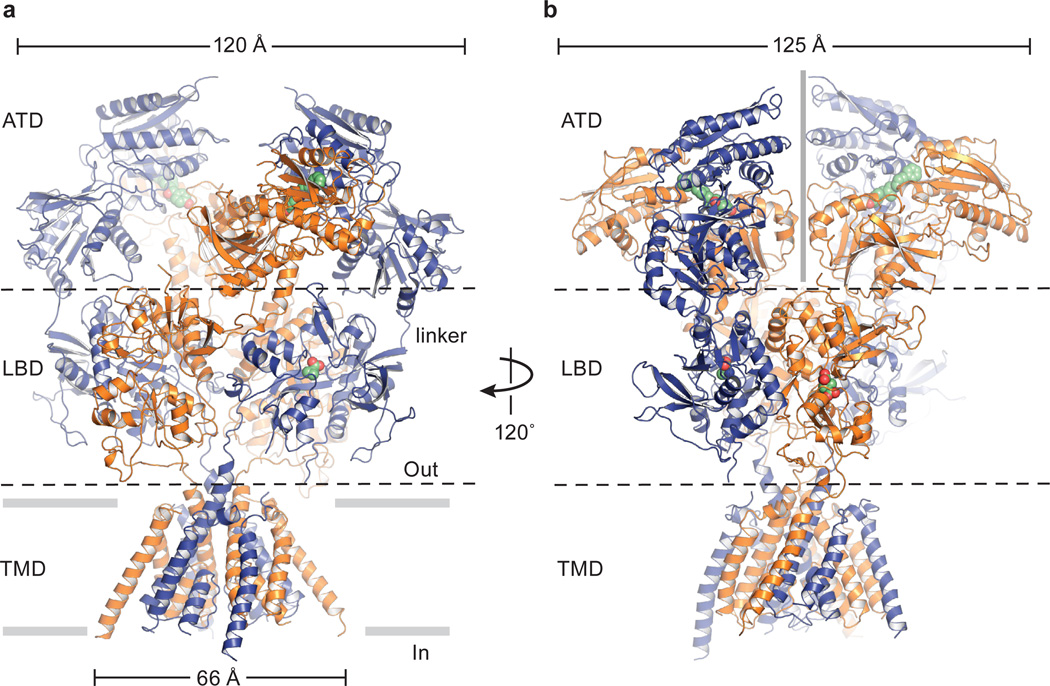
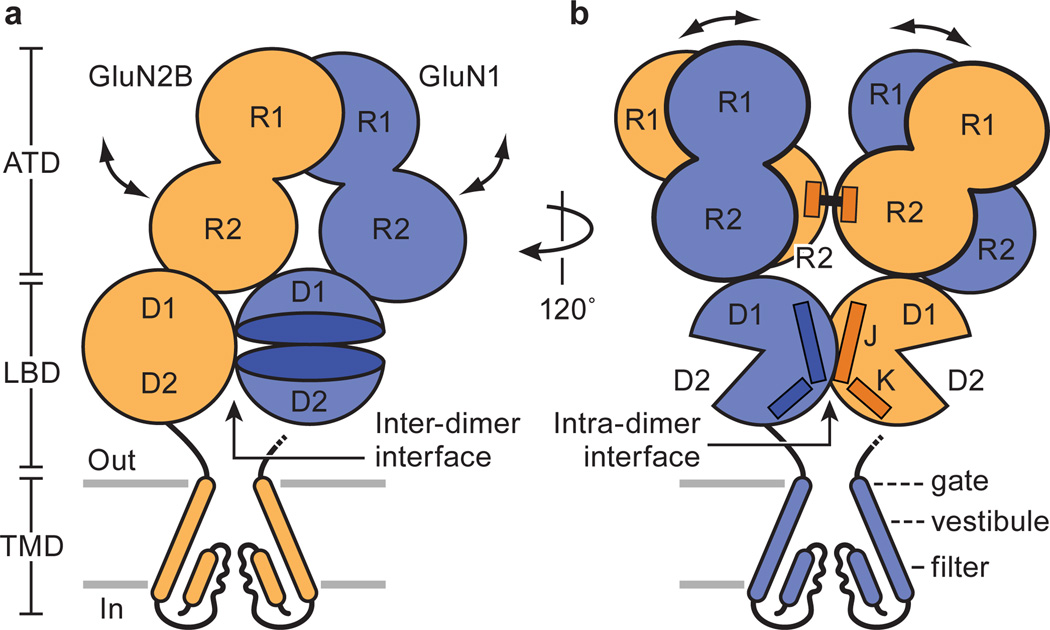
The structure of the GluN1/GluN2B NMDA receptor resembles that of a mushroom with a height of ~150 Å and widths of ~125 × 115 Å (Fig. 1a, b; ,Supplementary Video 1). Endowed with an overall 2-fold axis of symmetry, reminiscent of the intact GluA2 AMPA receptor architecture and symmetry28, the receptor domains are organized into three layers with the ATD layer at the ‘top’, the LBD layer in the ‘middle’ and the TMD layer at the ‘bottom’. By contrast with the AMPA receptor, the extracellular layers are more compact, with the ATD layer adopting an entirely different structure, interdigitated within the crevices of the LBD layer. The LBD layer caps the extracellular end of the transmembrane domain, with loops from the GluN2B LBDs drooping toward the extracellular leaflet of the membrane bilayer (Fig. 1a). The TMD hews to an AMPA-like topology and arrangement of helices28, yet with electron density for the M2 segments and pore loops in Structure 2, allowing us to define the structure of nearly the entire ion channel pore. Structure 1 and Structure 2 are similar, nevertheless, with an overall rmsd on main chain atoms of 0.6 Å. Here we primarily use Structure 1 to discuss the ATDs, LBDs and LBD to TMD linkages and Structure 2 to describe the TMD.
Figure 1. Architecture, symmetry and domain organization of the GluN1/GluN2B NMDA receptor.
a, View of the receptor complex, parallel to the membrane, with the GluN1 subunits in blue and the GluN2B subunits in orange. The ligands Ro25-6981, ACPC and t-ACBD are in space filling representation. b, View of complex rotated ~120° around overall the 2-fold axis of the receptor. The approximate position of the overall 2-fold axis is shown by a vertical gray bar in the center of the ATD layer. Structure 2 is shown.
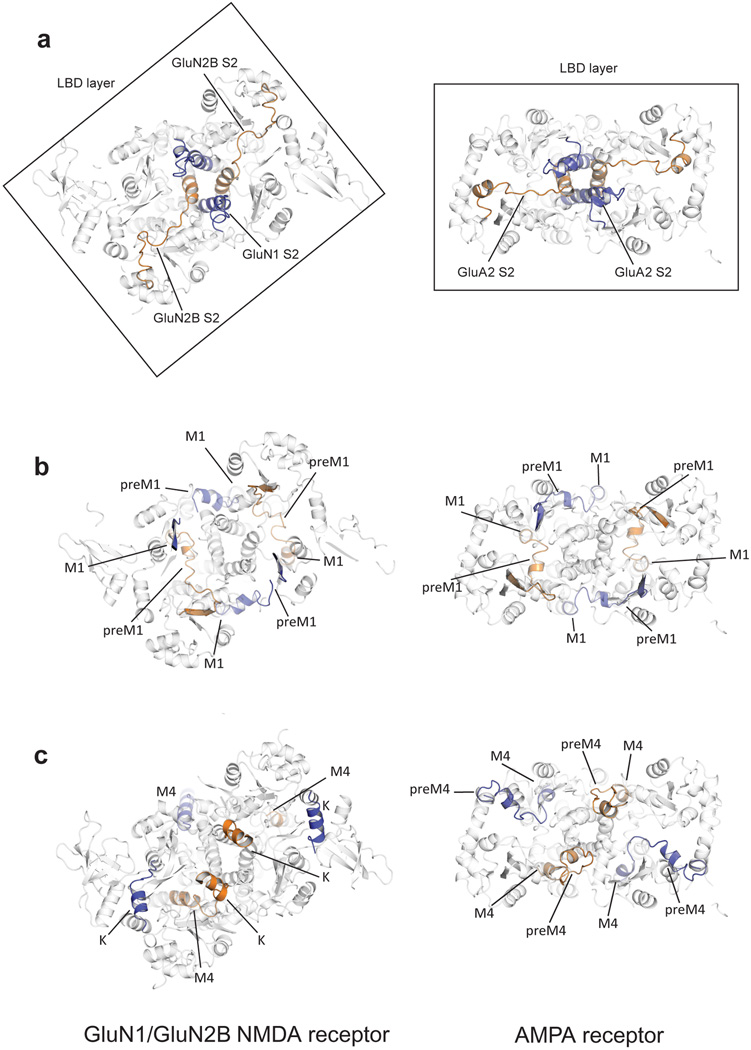
Subunit arrangement within the GluN1/GluN2B NMDA receptor adheres to the organization of the AMPA receptor28, with the glycine-binding GluN1 subunits occupying the A/C subunit positions and the glutamate-binding GluN2B subunits situated in the B/D subunit sites (Fig. 2a). In harmony with cross-linking studies on the GluN1/GluN2A receptor28–30 and isolated ATDs31 and in agreement with crystal structures of the GluN1/GluN2A LBDs32 and the GluN1/GluN2B ATDs24, the ATDs and the LBDs are organized as local GluN1/GluN2B heterodimers. Like the AMPA receptor28, there is subunit ‘cross over’ between ATD and LBD layers such that the subunits of a given ATD heterodimer are connected to subunits in a different LBD heterodimer, thus knitting together the extracellular domain superstructure. The TMDs are further stitched together by the M4 helices interacting nearly exclusively with TM segments from an adjacent subunit. The arrangement of subunits within this NMDA receptor complex illustrates how the subunit non equivalence first described for the homomeric AMPA receptor28 has been exploited in an obligatory heteromeric assembly.
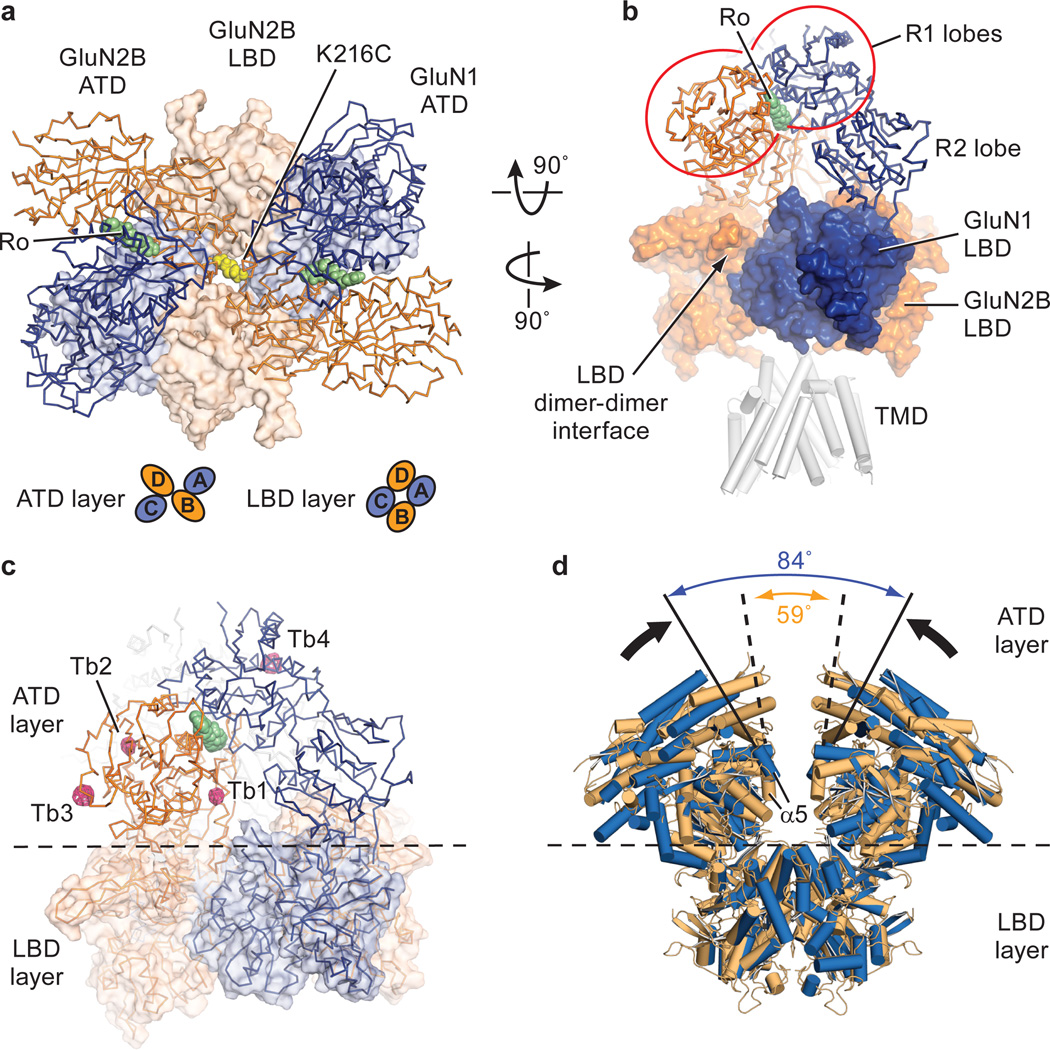
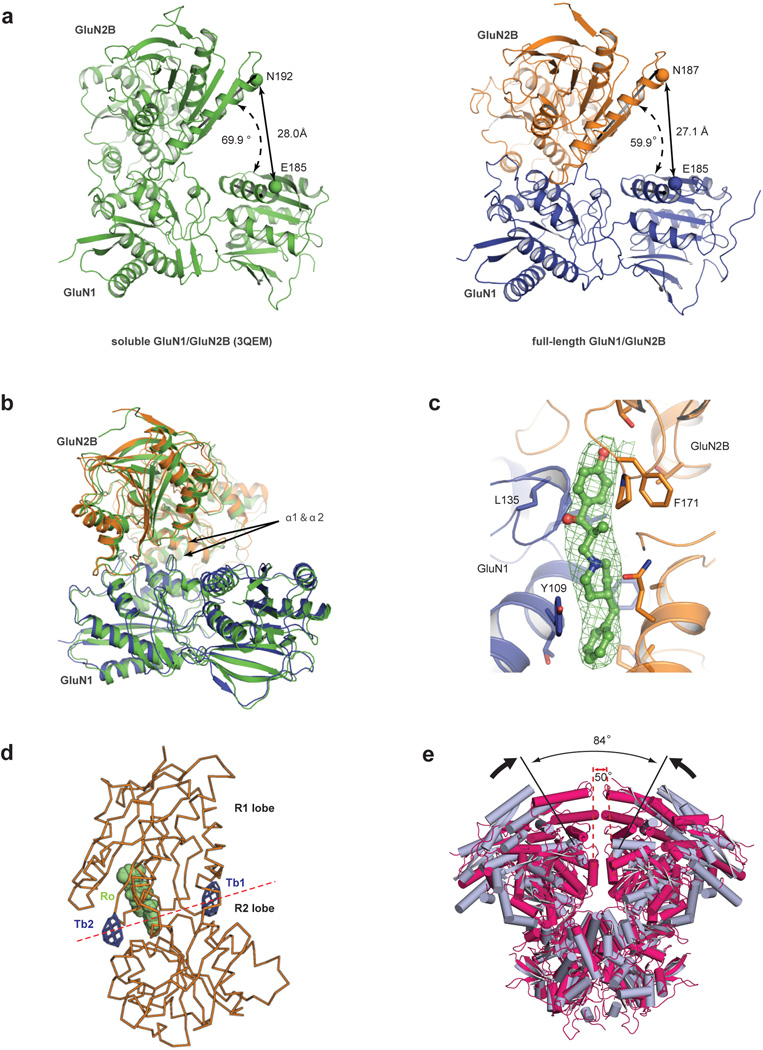
Figure 2. ATD arrangement, cation binding sites and conformational mobility.
a, View of the ATD layer along the overall 2-fold axis, from the extracellular side of the membrane, centered on the overall 2-fold axis, and showing the relative location of the underlying LBD layer. Ro25-6981 is green and the K216C disulfide is yellow. The arrangements of subunits for ATD and LBD layers are shown as insets. b, The inverted ATD heterodimeric ‘V’ straddles GluN1 and GluN2B LBD subunits on different local LBD heterodimers. Whereas the ATD R2 lobes interact with the LBDs, the R1 lobes cradle bound Ro25-6981 at an ATD subunit interface. Structure 1 is shown in panels (a) and (b) c, Tb3+ binding sites. Shown is an anomalous difference electron density map, contoured at 3.5 σ (pink mesh). Sites Tb1 and Tb2 are located at the ‘hinge’ between the R1 and R2 lobes whereas sites Tb3 and Tb4 are at receptor-receptor contacts in the crystal lattice. d, Shown are the ATD and LBD extracellular domains derived from the two low resolution GluN1/GluN2B receptor structures (Extended Data Table 2; Data set 4/Structure 4) where the GluN2B subunits do not harbor the K216C disulfide bridge, illustrating the conformational mobility of the ATD layer. The angles between the α5 helices of the GluN2B subunits for each of the two independent receptor complexes in the asymmetric unit illustrate the conformational mobility of the ATD layers.
Arrangement of amino terminal domains
The GluN1/GluN2B ATDs are perched above the LBD layer, with the R2 lobes of the GluN2B subunits proximal to each other and near the overall 2-fold axis of symmetry while the GluN1 ATDs reside at the periphery of the receptor assembly (Fig. 2a, b). The ATD heterodimer is shaped like an inverted ‘V’, where the open end of the ‘V’ straddles the GluN1 LBD beneath it, with loops and residues of the GluN1 R2 lobe interacting with its GluN1 LBD and the GluN2B R2 lobe wedged into the interdimer LBD interface. The conformation of an individual ATD heterodimer from the intact receptor structure is slightly ‘contracted’ compared to the structure of the isolated heterodimer, perhaps because of interactions with the LBD layer or due to lattice contacts (Extended Data Fig. 5a, b; Extended Data Table 3). There is prominent electron density at the interface between the GluN1 and GluN2B subunits for the allosteric antagonist, Ro25-6981, where it stabilizes the intersubunit interface (Extended Data Fig. 5c) 24 . A small interface, formed between the ATD heterodimers, is centered at the engineered disulfide cross link at residue 216 of the α5 helix on the GluN2B R2 lobe (Fig. 2a).
Zinc acts as an antagonist at nanomolar concentrations on GluN2A-containing receptors and at micromolar concentrations on receptors harboring the GluN2B subunit33. Because lanthanum also antagonizes the NMDA receptor in a voltage-independent manner34 and lanthanides can bind to zinc sites 35, we exploited the anomalous scattering signal of terbium and measured x-ray diffraction data near its f” maximum. Anomalous difference electron density maps show two peaks near the interdomain ‘hinge’ of the R1-R2 lobes of the GluN2B subunit (Fig. 2c; Extended Data Fig. 5d). The Tb2 site overlaps with the previously determined Zn1 site23, while the other site (Tb1), near residues Glu 146, Asp176 and Asp349, is unique. These data support the notion that ions and small molecules can bind to the ATD ‘clamshells’36 in a position to modulate ATD conformation, although future experiments are required to establish the roles of these sites in allosteric regulation of GluN2B-containing NMDA receptors.
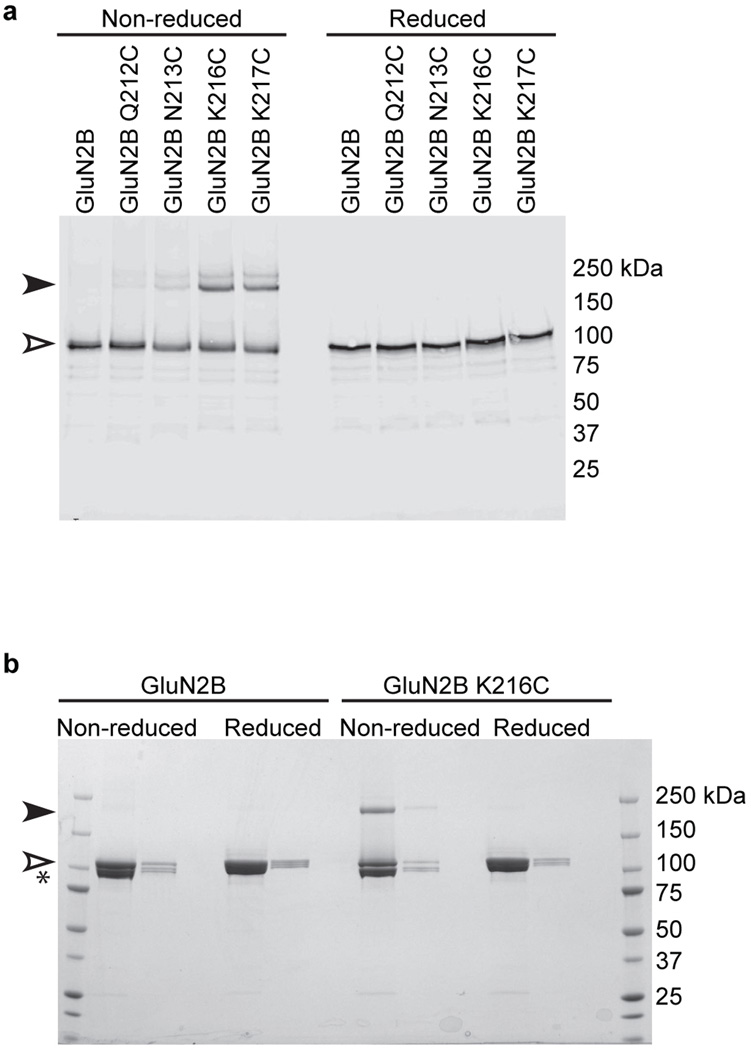
The first structure of the NMDA receptor was derived from the low resolution Data set 4 (Extended Data Table 2) and involved a construct lacking the GluN2B K216C mutant. In this crystal form, there are two halves of a receptor in the asymmetric unit and application of crystal symmetry creates two intact receptors, each with a different conformation of the ATDs in which the angles of the ATD domains range from 59° to 84° across the overall 2-fold axis (Fig. 2d). We further observed that helix α5 of the GluN2B R2 lobes face each other, proximal to the overall 2-fold axis of symmetry. Because we hypothesized that these structures were indicative of substantial mobility in the ATD layer, we made single cysteine substitutions on the exposed face of helix α5 and screened for redox dependent cross linking of GluN2B subunits. Indeed, the K216C mutant, as well as other residues on the face of α5, spontaneously form subunit-subunit cross links (Extended Data Figs. 1, 2, 4), bringing the GluN2B ATDs in close apposition (Extended Data Fig. 5e), diminishing ion channel activity and increasing the resolution to which the crystals diffract. In two electrode voltage clamp experiments, reduction of oocytes using dithiothreitol enhances current responses from the K216C mutant, suggesting that movements of the ATDs allosterically modulate the activity of the ion channel (Extended Data Fig. 2).
Ligand binding domain layer
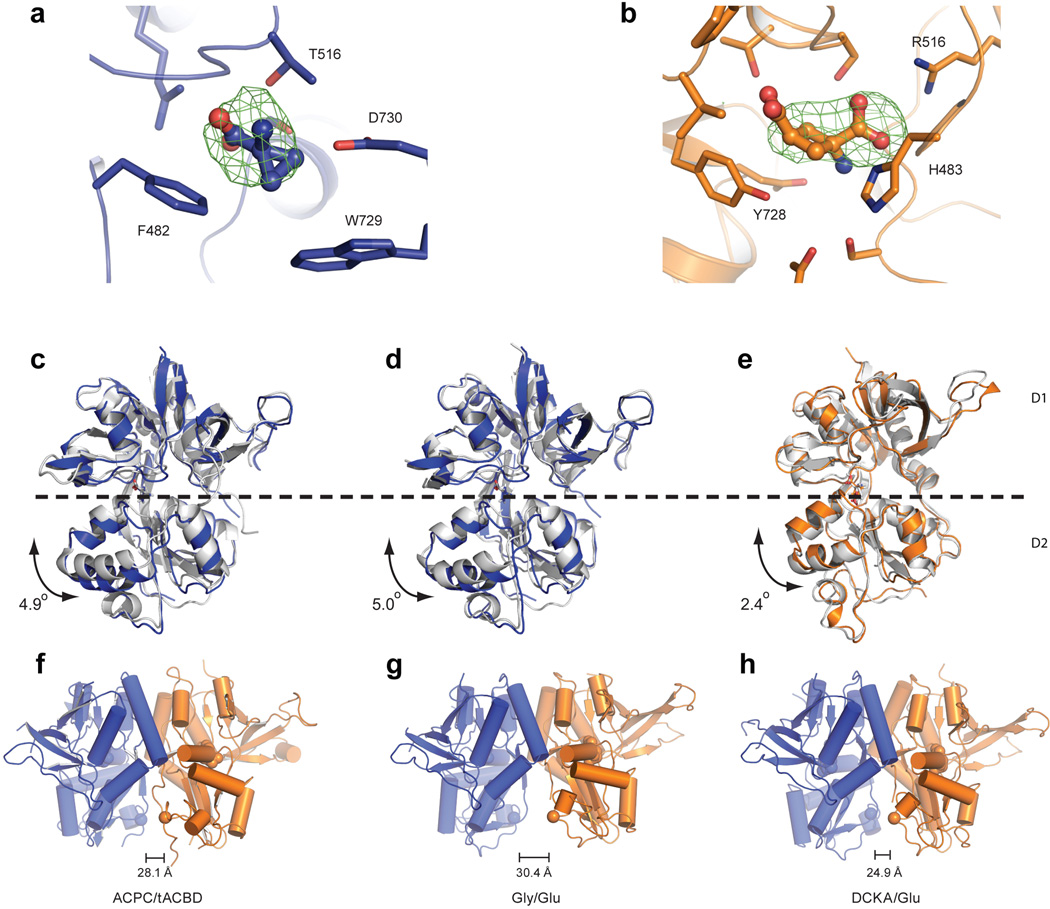
The agonist-binding LBDs of the NMDA receptor are organized as a nearly equivalent pair of GluN1/GluN2B heterodimers where each GluN1/GluN2B heterodimer (Figs. 3a, 3c) closely resembles the water-soluble heterodimers of the isolated GluN1/GluN2A LBDs32 and the homodimeric assemblies of AMPA18 and kainate receptor37 LBDs in non desensitized conformations. Moreover, the arrangement is similar to that previously observed in the structure of the full length AMPA receptor (Figs. 3b, 3d)28, although here the electron density for the GluN1 and GluN2B LBDs in chains B and C is weak, perhaps due to an absence of lattice contacts. In comparing this NMDA receptor structure to the antagonist-bound state of the AMPA receptor, the extent to which the local 2-fold axes of each LBD dimer are tipped off of the overall molecular 2-fold axis of symmetry differ (Figs. 3a, b). In addition, inspection of the GluN1/GluN2B and AMPA receptor LBD layers, viewed from the ‘top’ (Figs. 3c, d), shows that there is a relative translation, or shift, of the LBD dimers along the interdimer interface (Fig. 3e). Using helix J to align the B/C LBDs, the A/D LBD dimer in the AMPA receptor has undergone a translational ‘shift’ of ~15 Å relative to the A/D NMDA receptor LBD heterodimer. While we do not know if these differences in LBD dimer ‘roll’ angle (Figs. 3a, 3b) and translational ‘shift’ (Fig. 3e) are due to inherent differences between NMDA and AMPA receptors or to the closed-blocked state of the NMDA receptor versus the competitive antagonist-bound form of the AMPA receptor, or to both factors, this analysis illustrates conformational mobility of the LBD dimers perhaps related to how the LBD couples agonist-binding to the TMD.
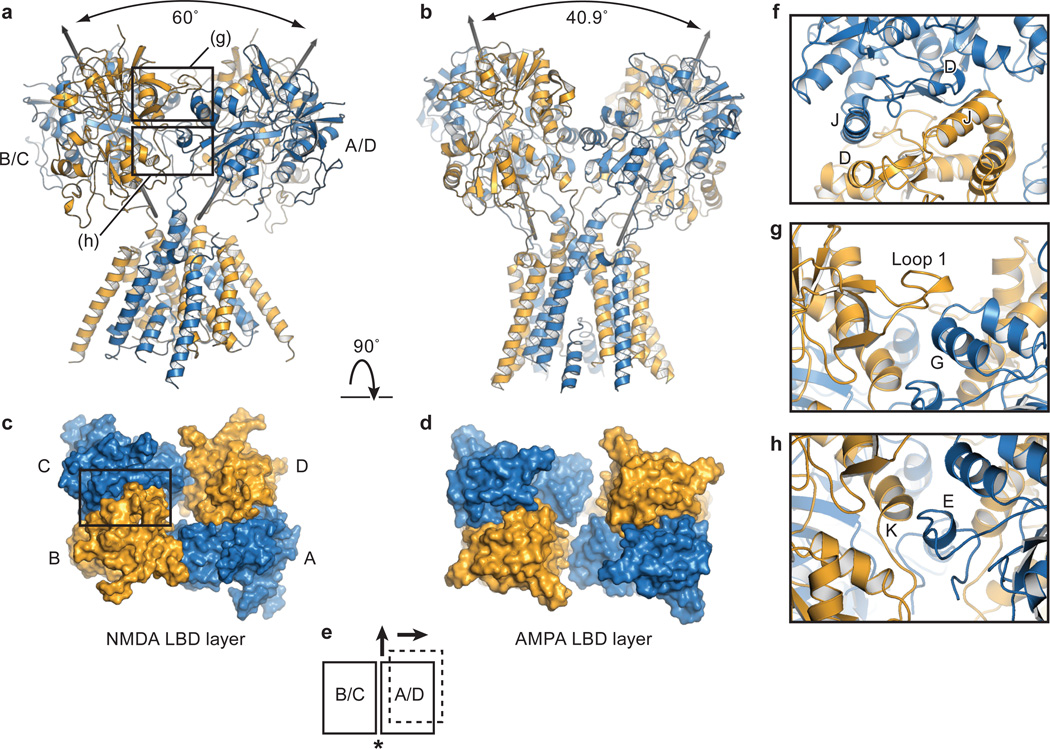
Figure 3. LBD layer forms a ring-like structure.
a, The GluN1/GluN2B LBD and TMD, showing that the pseudo 2-fold axes of the B/C and A/D LBD heterodimers diverge with an angle of 60°. The boxed areas define regions of LBD dimer-dimer contacts shown in panels (g) and (h). b, View of the antagonist-bound state of the GluA2 AMPA receptor, which shows that the 2-fold axes of the LBD dimers diverge by an angle of 40.9°c, View from the extracellular side of the membrane, along the overall 2-fold axis of the receptor, showing the LBDs of the GluN1 and GluN2B subunits, with the LBD heterodimer interface of the B/C subunits emphasized by a box. d, GluA2 LBD layer, illustrating how the interface between the B/C and A/D subunits has increased in comparison to the NMDA receptor LBD layer. e, Schematic of the LBD layer, showing the NMDA receptor B/C and A/D heterodimers as rectangles (solid lines) and illustrating the translational shift of the A/D subunits in the AMPA receptor (dotted lines). The asterisk indicates the dimer-dimer interface. f,g,h, Closeup view of the canonical D1-D1 intradimer interface32, together with views of the interactions at the interdimer interfaces in panels (g) and (h). The domains from Structure 2 are shown, with GluN1 subunits in blue and GluN2B subunits in orange.
Within the LBD layer there are two major interfaces, one within a heterodimer, at the D1-D1 interface of GluN1 and GluN2B subunits, and the second between heterodimers. The intradimer D1-D1 interface is a region of allosteric modulation in NMDA receptors 38, 39 and, within one heterodimer, buries ~1100 Å2 of solvent accessible surface area on each subunit (boxed region Fig. 3c; Fig. 3f). There are two nearly equivalent interdimer interfaces between the heterodimeric LBDs, each burying ~600 Å2 of solvent-accessible surface area, and comprising two loci (boxed regions in Fig. 3a). One involves helix G on GluN1 interacting with residues on loop 1 of GluN2B (Fig. 3g) and the second includes residues on helix K (GluN2B) making contacts with residues on helix E (GluN1; Fig. 3h). Both sets of interactions involve contacts between residues on the D1 and D2 lobes of the GluN1 and GluN2B subunits, providing a direct route by which modulation of LBD ‘clamshell’ closure could be translated into rearrangement of the LBD layer. As previously suggested, both the NMDA receptor LBD intradimer interface and the dimer-dimer interface may adopt different conformations depending on the functional state of the receptor.
The initial trigger for the eventual opening of the ion channel gate resides in agonist binding to the LBD clamshells. NMDA receptors require binding by agonists at both the GluN1 and GluN2 sites9, and here we have crystallized the receptor in complex with the partial agonists ACPC40 and t-ACBD41. Agonist binding results in closure of the LBD clamshell42 and separation of the region proximal to the M3 transmembrane helix18. Analysis of the GluN1 and GluN2B LBDs demonstrates that each of the two GluN1 and GluN2B clamshells adopt similar conformations (Extended Data Table 3). Moreover, the degree of closure is similar to that observed for the isolated LBDs (Extended Data Fig. 6), except that they are both slightly more open in comparison to the isolated domains, perhaps due to direct linkage to the ion channel. Separation of the region proximal to the M3 helices is similar between the equivalent residues in the LBD dimers of the full length receptor and in the glycine/glutamate complex of the isolated GluN1/GluN2A LBDs yet longer than in an LBD antagonist (DCKA)/glutamate complex (Extended Data Fig. 6f-h). Thus by this metric the LBD dimers adopt an agonist-bound, activated conformation.
ATD - LBD interactions and allosteric coupling
The molecular puzzle of how allosteric inhibitors such as Ro25-6981 and ifenprodil promote closure of the ion channel gate despite the binding of agonists to their cognate LBD dimers must be resolved, at least in part, through communication between the ATD and LBD layers and perturbation of the LBD layer from an ‘active’ conformation to an ‘inactive’ state. In the GluN1/GluN2B structure, the ATD heterodimers ‘straddle’ LBD subunits (Fig. 4a, b), with the R2 lobe of each GluN1 subunit making extensive contacts with the D1 lobe of the cognate GluN1 LBD and the R2 lobe of the GluN2B subunit insinuated into the LBD dimer-dimer interface, positioned to make contacts with its cognate GluN2B LBD and with the neighboring GluN1 LBD. Thus we see that the ATDs are judiciously positioned to mediate conformational changes at both LBD intra dimer18, 38, 39 and interdimer interfaces. Nevertheless, because the intradimer LBD interface is ‘intact’, the action of Ro25-6981 and related compounds may not necessarily involve rupture of this interface and other conformational changes with the LBD layer may be involved in rendering the LBD layer in the apparently ‘inactive’ conformation observed in the present structures. However, additional studies with robustly active receptor constructs will help to resolve these questions.
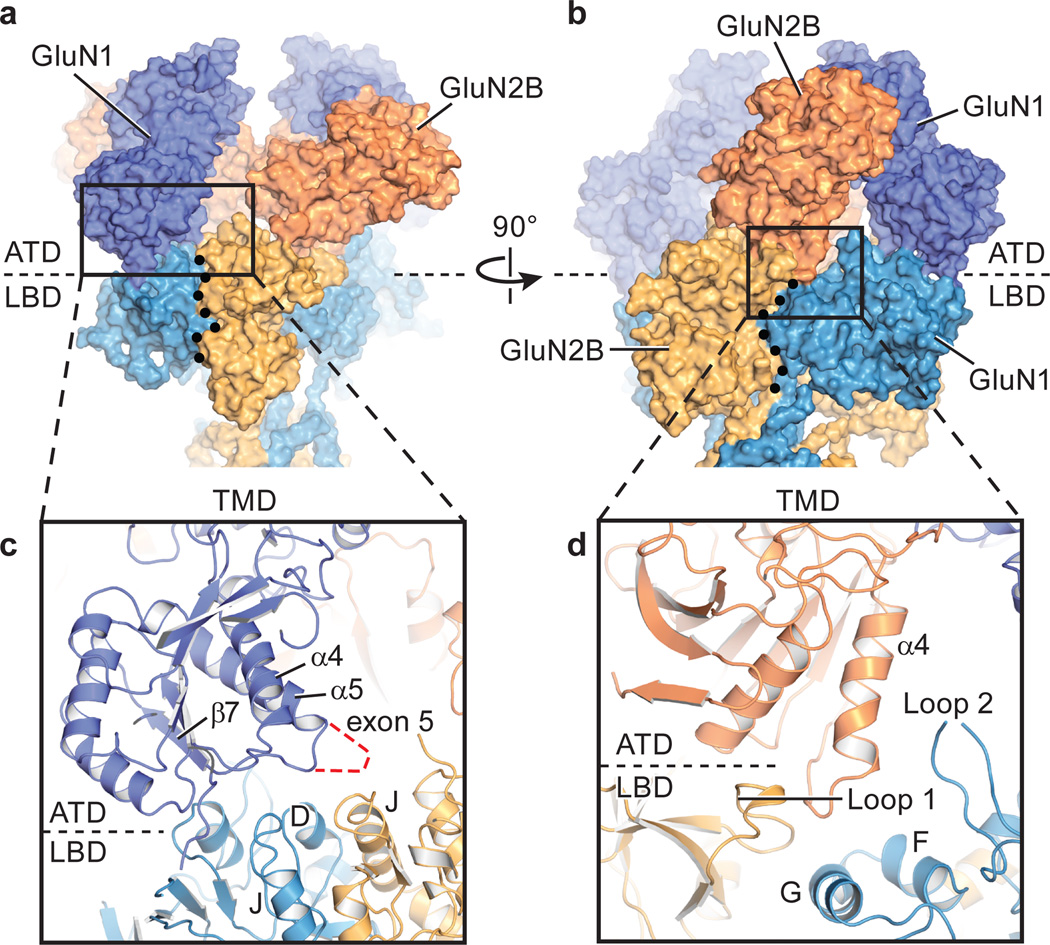
Figure 4. The ATDs participate in extensive contacts with the LBD layer.
a. Surface representation of the ATD and LBD domains, illustrating how the R2 lobe of the GluN1 subunit is poised above its cognate GluN1 LBD and also near the D1-D1 LBD dimer interface and in (b) how the R2 lobe of the GluN2B subunit participates in contacts with its cognate GluN2B LBD, near an inter LBD dimer interface. c. Close up views of potential interactions between the GluN1 R2 lobe and the GluN1 LBD and (d) between the GluN2B R2 lobe with regions on its GluN2B LBD. Note that the GluN2B R2 lobe is also near helices G and F and loop 2 of the GluN1 LBD. In (a) and (b) the black dots define the approximate intra- and interdimer LBD interfaces, respectively. Structure 1 is shown in all panels.
In the GluN1 subunit, the ATD α5 helix C-terminus, which harbors exon 5 in an alternatively spliced form of the gene43, in combination with the α4-β7 loop, resides close to the LBD dimer interface, near the C-terminus of helix J, and in a plausible position to perturb the conformation of the LBD layer (Fig. 4c). The GluN2B α4 helix C-terminus, along with the loop connecting α4 to β7, a region implicated in regulation of the NMDA receptor by polyamines44, rest on top of the F and G helices of the GluN1 LBD and close to residues in loop 1 of the GluN2B LBD (Fig. 4d). Thus, while the linking peptides connecting the ATDs to the LBDs play an important role in the transduction of conformational changes between the two layers45, direct contacts that harness the predicted large-scale motions of the ATDs36 also play a central role in transmitting changes to the transmembrane, ion channel domain.
Transmembrane domain
NMDA receptors are calcium permeable and toxic to cells upon overexpression and therefore we introduced mutations known in AMPA receptors to increase receptor desensitization46, finding that in the context of the GluN1/GluN2B NMDA receptor, they lead to decreased current amplitudes and enhanced stability of the receptor in detergent micelles. Because native GluN1/GluN2B receptors have a low open probability (Po)47 and the modifications we have introduced further reduce Po, the functional state of the receptor should be an Ro25-6981, partial agonist bound, closed-blocked channel state.
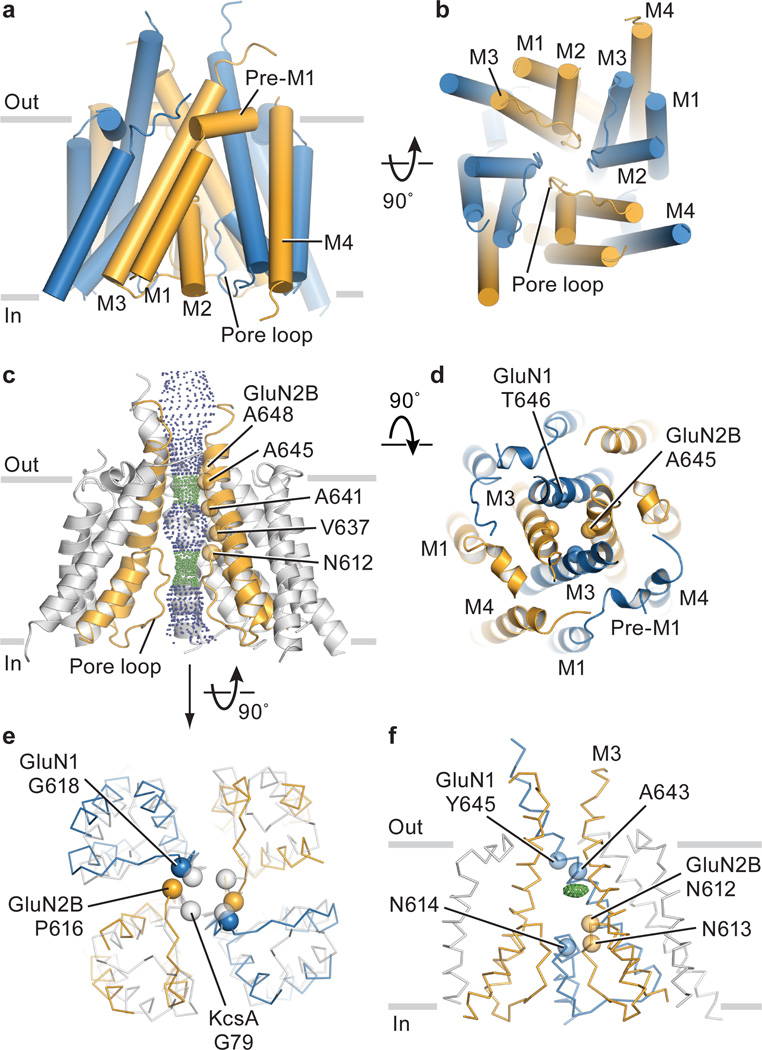
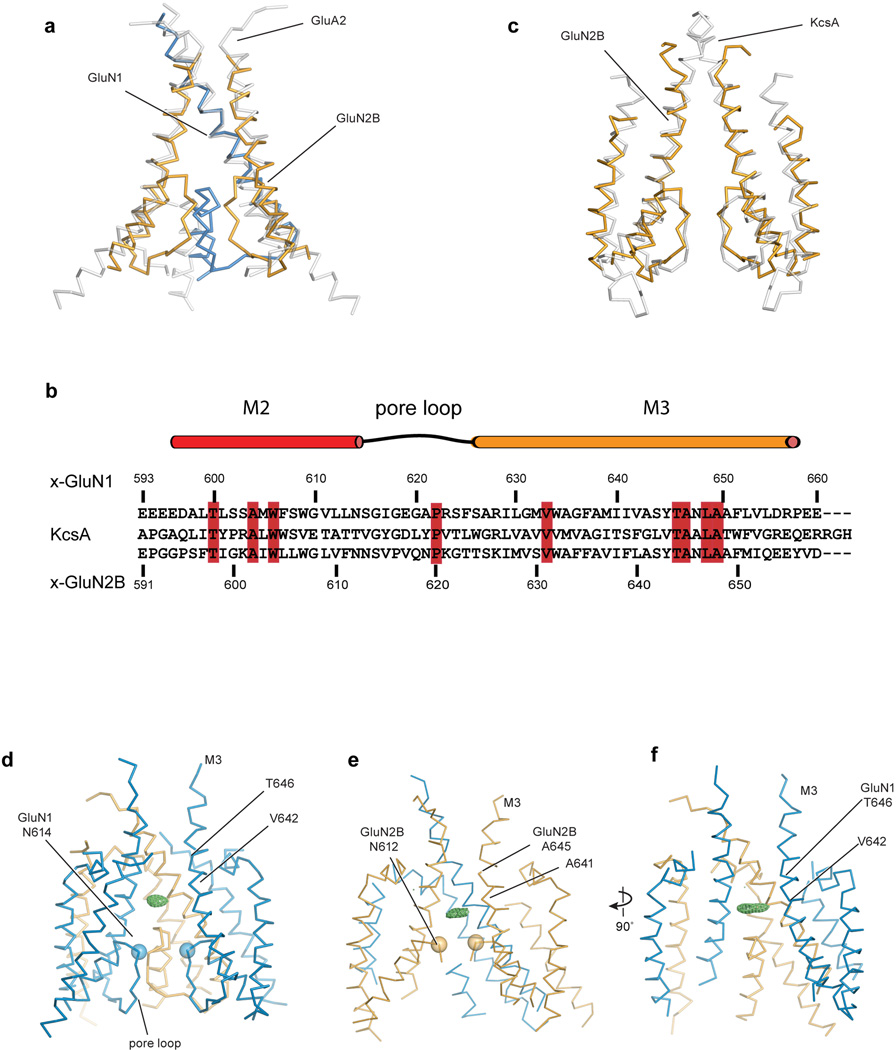
The electron density associated with Data set 2/Structure 2 allowed us to position the polypeptide main chain for the M1-M4 helices of all subunits (Supplementary Video 1). To trace the polypeptide associated with the pore loop, we exploited the continuous electron density for this region in the GluN2B subunit D and, by applying non crystallographic symmetry defined by the transmembrane segments of the other subunits, we traced the three remaining pore loops (Fig. 5a, 5b). The arrangement of transmembrane helices is like that of the GluA2 AMPA receptor28 (Extended Data Fig. 7a), although in the NMDA receptor we have a more complete representation of the ion channel pore and putative selectivity filter. The pre-M1 region of the NMDA receptor forms a ‘collar’ around the extracellular regions of the M3 helices, residing near the boundary of the extracellular side of the membrane. The M1 helix descends across the membrane and makes interactions with the pore-lining M3 helix of the same subunit and the M4 helix of a neighbor. Electron density for the cytoplasmic loop connecting M1 to M2 is weak or missing, and thus this region is absent from the structure. We can visualize the M2 pore helix and most of the extended region of the pore loop forming the selectivity filter and its connection to the N-terminus of M3.
Figure 5. Transmembrane domain architecture, symmetry and coupling to LBD.
a, View of the TMD parallel to the membrane. GluN1 subunits are blue and the GluN2B subunits are orange. b, View of the TMD, along the pore axis, from the cytoplasmic side of the membrane. c, View of a solvent accessible surface carved along the pore axis using the computer program HOLE, parallel to the membrane, showing that the M3 bundle crossing near the extracellular side of the membrane and the entry into the selectivity filter region, from the central aqueous vestibule, form constrictions in the pore. The color coding for the dots that indicate the pore radius is 1.15 Å < green < 2.3 Å < blue. Because a number of side chains are not included in the structure, due to the moderate resolution of the diffraction data, the size of the pore is approximate. d, View of the extracellular ends of the M3 helices of the NMDA receptor. We have highlighted as spheres the α-carbon atoms for residues Thr 646 and Ala 645 in the GluN1/GluN2B structure, respectively. The distances between neighboring atoms are 6.2, 8.0, 5.4 and 7.1 Å, starting from the α-carbon of GluN2B on the left and going clockwise. e, View of the intracellular ends of the TMD of the NMDA receptor in comparison with KcsA. Here, the M2 helices of the NMDA receptor were superimposed on the corresponding helices in KcsA, showing the deviation from 4-fold symmetry. f, Side view of the TMD showing a positive electron density feature (green mesh) in the central vestibule, calculated using Fo-Fc coefficients and phases from the refined structure. The map is contoured at 2.8 σ. Data set 2 and Structure 2 were employed in all panels (Extended Data Table 2).
The conformation of the polypeptide chain throughout the M2 helix and the pore loop are reminiscent of a potassium channel48, although there are differences in the local conformation of the NMDA pore loops, perhaps due to non glycine residues, ‘…SVP…’ within the canonical ‘…GYG…’ motif of potassium channels in the GluN2B subunit (Extended Data Fig. 7b, 7c) or to the lower resolution of the present structures. Asn residues implicated in voltage-dependent magnesium block49 are situated at the turn between the end of M2 and the beginning of the extended filter sequence, in a position to project their side chains into an aqueous vestibule (Figs. 5c, 5f). Forming the core of the ion channel is the M3 segment, in a similar conformation as the M3 segment in the GluA2 receptor28 (Extended Fig. 7a). The extracellular ends of the M3 segments adopt a pyramid-like shape, forming a physical constriction to the ion channel permeation pathway (Fig. 5c, d). The M4 segment resides on the periphery of the transmembrane domain, interacting primarily with the M1 and M3 helices of a neighboring subunit and extending for several more turns into the cytoplasmic-space than that seen in the GluA2 receptor.
Ion channel gate and central vestibule
The solvent accessible pathway through the ion channel pore from the extracellular side of the membrane to the cytoplasm shows an occlusion near the predicted extracellular boundary of the membrane bilayer, a central vestibule, a second occlusion at the beginning of the selectivity filter, followed by a solvent accessible pathway to the cytoplasm (Fig. 5c). We hypothesize that the physical gate of this closed-blocked state of the NMDA receptor is at the bundle crossing of the M3 helices, in a position similar to that of the shut gate of the antagonist-bound GluA2 receptor28. The narrowest constriction is defined by Thr 646 of GluN1 and Ala 645 of GluN2B, within the highly conserved ‘..SYTANLAAF…’ motif crucial to ion channel gating, near the extracellular boundary of the M3 helices50. Flanking the constriction is a narrow region too small for ion permeation, spanning Val 642-Leu 653 of GluN1 and Ala 641-Ile 652 of GluN2. While the arrangement of the M3 helices at the ion channel gate diverges from the approximate 4-fold symmetry of the AMPA receptor (Fig. 5d) 28, we do not yet know if this distinction is inherent to NMDA receptors or is a consequence of their different ligand-bound and functional states.
The pore expands immediately ‘below’ the M3 bundle crossing to the central vestibule, a cavity flanked by the M3 helices on the ‘sides’ and the ends of the M2 helices together with the tips of the pore loops on the ‘bottom’. A second constriction of the pore is immediately below the central vestibule and is defined by the residues at the beginning of the pore loop (Fig. 5c). Because of the limited resolution of the diffraction data, we are unable to position side chain groups and to precisely define the location of main chain carbonyl oxygen atoms, and thus are not able to determine if this region of the pore is large enough to conduct ions. Following this constriction, the pore expands to the cytoplasmic space. By contrast with 4-fold symmetric potassium channels, the pore loops of this specific ligand-bound state of the NMDA receptor are arranged with approximate 2-fold symmetry (Fig. 5e).
In electron density maps derived separately from Data set 1 and Data set 2 and their respective structures (Extended Data Table 2), we found positive difference electron density within the central vestibule (Fig. 5f; Extended Data Fig. 7d-7f). Because we crystallized the receptor in the presence of 1 mM MK-801, we suggest this electron density feature could represent a trapped MK-801 molecule that occupies multiple positions or orientations within the central vestibule. Unfortunately, we have been unable to validate MK-801 binding by direct binding assays or by electrophysiology experiments, perhaps because of the very low Po of the receptor construct. Nevertheless, several residues including Ala 643 and Tyr 645 on the GluN1 M3 helix, and Asn 614 (GluN1), Asn 612 (GluN2B) and Asn 613 (GluN2B) on the pore loop ‘tips’, are near the electron density feature and have been implicated in MK-801 binding17. We speculate that ion channel blockers, such as MK-801, occupy the central vestibule and block ion conduction by preventing ions from entering the pore loop-lined selectivity filter.
Coupling of ligand binding and transmembrane domains
The coupling of the LBD to the TMD is similar, in principle, to the AMPA receptor28, with the crucial M3 connections to the LBDs proximal or distal to the overall 2-fold axis for the GluN1 A/C subunits or the GluN2B B/D subunits, respectively (Extended Data Fig. 8). However, the relative orientation of the NMDA receptor TMD with respect to the LBD is distinct from the GluA2 receptor. Specifically, the LBD layer of the NMDA receptor is rotated by ~35° around an axis that is approximately coincident with the overall 2-fold axis of the receptor. At this juncture we do not know if this difference is due to inherent differences between NMDA and AMPA receptors or because this specific NMDA receptor complex is trapped in an ATD antagonist-bound, LBD partial-agonist bound, closed-blocked state.
Conclusion
The GluN1/GluN2B structure harbors an overall 2-fold symmetry, a layered dimer-of-dimers arrangement of subunits and a positioning of NR1 and NR2B subunits in the A/C and B/D positions defined by the full length GluA2 receptor28 (Supplementary Video 1). The rich interdigitations and covalent linkage of the R2 lobes of the ATDs to the LBDs provides molecular routes for transmission of allosteric signals to the glycine and glutamate-binding LBD layer, which is organized as a ring of heterodimeric units above the transmembrane ion channel (Fig. 6). Despite closure of the LBD ‘clamshells’ around partial agonists and an intact ‘non desensitized’ D1-D1 interface of the LBD heterodimers, the ion channel gate is in a closed-blocked state, providing the first insights into the structural basis for allosteric inactivation of a NMDA receptor and suggesting that plasticity of the LBD layer may provide a mechanism for modulation of receptor gating. Departing from the 4-fold symmetry of the GluA2 transmembrane domain, the pore loops of the NMDA receptor are approximately 2-fold symmetric. The allosteric antagonist-bound GluN1/GluN2B structure provides a molecular blueprint for the development of new therapeutic agents and a structural framework for biophysical mechanisms of allosteric modulation, gating and ion channel function, as well as a spring board for future studies directed toward determining structures of resting, open and desensitized states and defining locations of ion binding sites.
Figure 6. Schematic of the NMDA receptor.
a, Shown is a single ATD heterodimer, two LBD ‘clamshells’ residing in different LBD heterodimers, and the TMD of GluN2B subunits, emphasizing only the M2, pore loop and M3 elements. The line connecting the M3 helix on the right is ‘broken’ to illustrate that it is connected to the GluN2B LBD ‘behind’ the shown GluN1 LBD. Double-headed arrows suggest possible movements of ATDs within an ATD heterodimer. b, Rotation of the receptor schematic shown in panel (a) by ~120° showing two ATD heterodimers, a single LBD heterodimer and the TMD of GluN1 subunits. Double-headed arrows show conformational movements between ATD heterodimers observed in the structures described here. The α5 helices, harboring the K216C crosslink, are shown as rectangles at the R2-R2 interface. In both schematics, we emphasize how the R2 lobes of the ATDs are positioned such they could modulate inter- and intradimer LBD interfaces and, in turn, the ion channel gate.
Methods
Receptor constructs
The constructs are detailed in Extended Data Table 1. Residues 1-834 of Xenopus laevis GluN1 (FJ571597.1) and 1-839 of Xenopus laevis GluN2B (NM_001110721) were cloned into pEG BacMam51, 52 for analytical-scale transient transfection in adherent cells or for large-scale virus-mediated expression in suspension cells. At the C-terminus of both constructs the 3C cleavage site (Leu-Glu-Val-Leu-Phe-Gln-Gly-Pro), enhanced green fluorescent protein (eGFP) and either an octa-histidine tag (at the C-terminus of GluN1) or StrepII tag (at the C-terminus of GluN2B) were placed for purification and fluorescence- detection size exclusion chromatography (FSEC) 53 and FSEC-thermostability (FSEC-TM) 54 analysis.
Expression and purification
HEK293S GnTI (−) cells55 were grown in suspension and transduced using P2 BacMam virus at a multiplicity of infection (MOI) of 1:1 (GluN1:GluN2) and incubated at 37 °C. After 14hr post transduction, 10mM sodium butyrate and 2.5 µM MK-801 were added to the cultures. Cells were harvested 60 hours post-transduction, collected by centrifugation and disrupted by sonication in 150mM NaCl, 20mM Tris-HCl pH8.0. The homogenized material was clarified by centrifugation, membranes were resuspended and homogenized with 50 ml per gram of membrane in 150mM NaCl, 20mM Tris-HCl pH8.0 and solubilized in a buffer containing 1 % MNG-3, protease inhibitors, 1 mM glutamate, 1 mM glycine, and 2 mM cholesteryl hemisuccinate (CHS) for 1.5 hr, 4 °C. The soluble fraction was bound to streptactin resin and eluted with buffer containing 5 mM desthiobiotin. The receptor was concentrated and digested with 3C protease and endoglycosidase H treatment. Prior to size-exclusion chromatography (SEC), the K216C containing receptor was treated with 500 uM copper phenanthroline (CuP) to enhance cysteine cross-linking. The concentrated GluN1/GluN2B receptor was further purified by size exclusion chromatography in a buffer composed of 400 mM NaCl, 20 mM MES pH 6.5, 1 mM C12M, 0.2 mM CHS. Peak fractions were pooled and concentrated to 2.2mg/ml.
Crystallization and cryoprotection
Initial crystals of the GluN1/GluN2B NMDA receptor constructs diffracted to ~7 Å resolution. Prior to crystallization, 28 mM n-dodecyl β-D-maltoside (DDM), 300 µg cholesterol, 5mM 1-aminocyclopropane-1-carboxylic acid (ACPC), 1 mM trans-1-aminocyclobutane-1,3-dicarboxylic acid (t-ACBD), 1mM Ro25-6981 and 1 mM MK-801 were added to the GluN1/GluN2B protein, incubated for 14–16 hrs56. Crystals appeared in 16–18% PEG 3350, 200 mM potassium nitrate and 100 mM MES pH7.1. Crystals were cryoprotected by mother liquor supplemented with 20% glycerol. Crystals used to measure Data sets 1 and 2 were obtained with the GluN1 Δ2 and GluN2B Δ2 constructs and by adding 10 mM 6-cyclohexyl-hexyl-β-d-maltoside (Cymal6), together with ACPC, t-ACBD, Ro25-6981 and MK-801 to the protein. Crystals appeared in 19–21% pentaerythritol ethoxylate, 100mM NaCl, 100mM MgCl2, and 100mM HEPES pH 7.5 and were cryoprotected by 12% ethylene glycol. To obtain terbium-complexed crystals, 10 mM Cymal6, 1 mM glutamate, 1mM glycine, 1 mM Ro25-6981 and 1 mM MK-801 were added to the protein. Crystals appeared in 21–22% PEG400, 50 µM terbium nitrate and 100 mM MES pH 6.5. Crystals were cryoprotected using a reservoir solution that include PEG400 at 25 % and supplemented with 5% ethylene glycol.
Structure determination
X-ray diffraction data sets were collected at the Advanced Light Source on beamlines 8.2.1 and 5.0.2. Diffraction sets were indexed, integrated, and scaled by XDS57 or HKL200058 together with the microdiffraction assembly method59. The best diffraction data for Data set 1 were derived from merging data from three crystals60. A single crystal was used for Data set 2. Structure 1 was determined by molecular replacement with Phaser61 using the isolated Xenopus/rat GluN1/GluN2B ATD domains in complex with Ro25-6981 (PDB code 3QEM)24 and the isolated rat/rat GluN1/GluN2A LBD (PDB code 2A5T)32 structures as search probes. The molecular replacement solutions were robust with the highest best log likelihood gain and translation function Z-score of 3071.7 and 31.9, respectively. Initial maps were improved by density modification62. A partial model of the transmembrane domain was manually built into ‘omit’ style electron density maps. Cycles of manual model building and crystallographic refinement were carried out using the computer graphic program Coot63 and the crystallographic refinement software package Phenix64. During the course of model building and refinement, the amino acid sequence and corresponding structure within the ATDs and LBDs were adjusted to the Xenopus amino acid sequences. The model was refined to a nominal resolution of 3.7 Å with reasonable R-factors. Structure 2 derived from Data set 2 was solved by molecular replacement using Structure 1 as a search probe. Upon inspection of electron density maps, density for the pore loops was visible, along with additional residues in the other TM segments. The final Structure 2 was obtained by cycles of manual model building and crystallographic refinement, as described above. Stereochemistry of the model was evaluated by MolProbity 65, pore dimensions were estimated using HOLE66 and figures were created using Pymol67. Important information on the qualities of the structures is provided in Supplementary Information.
Two-electrode voltage clamp electrophysiology and Western blotting
Oocytes were injected with RNA (20 ng; 1:1 ratio, GluN1:GluN2B) and stored at 16 °C in the presence of 30uM DCKA. Recordings were made using a bath solution containing 5 mM HEPES pH 8.0, 100 mM NaCl, 2.8 mM KCl, 10 mM Tricine and 0.3 mM BaCl2. NMDA receptor constructs were activated with a perfusion solution containing 100 uM glycine and 100uM glutamate with or without1 mM MgCl2. The holding potential of these recordings is −60 mV. For studies under reducing conditions, oocytes were treated with 5mM DTT for at least 15 min before recording. For Western blots, oocytes were solubilized in 1% MNG-3 buffer (20 mM Tris pH 8.0, 150 mM NaCl, 1% MNG-3 plus protease inhibitors and 1 mM PMSF), and lysates were resolved by SDS-PAGE under non-reducing conditions followed by Western analysis using anti-GluN2B antibody.
Ligand binding assays
Binding constants were determined by the scintillation proximity assay (SPA)68. SPA experiments were set up in triplicate wells of a 96-well plate at a final volume of 100 µl in SPA buffer (20 mM Tris pH 8, 150 mM NaCl, 0.02 mM CHS and 0.01% MNG-3). Affinity-purified GluN1 Δ2/ GluN2B Δ2 NMDA receptor (2–5 nM) was incubated with 0.5mg/ml of Ysi-Cu (for 3H-Ro25-6981) or PVT-Cu (for 3H-glutamate and 3H-glycine binding) SPA beads. Nonspecific binding was determined by the addition of 1 mM ifenprodil (for 3H-Ro25-6981), 1 mM DCKA (for 3H-glycine), or 1 mM NMDA (for 3H-L-glutamate). 3H-Ro25-6981 binding was performed in the presence of unlabelled 100 µM L-glutamate and 100 µM glycine.
Inhibition constants were determined by the SPA assay using 5 nM GluN1 Δ2/ GluN2B Δ2 NMDA receptor, 0.5 mg/ml PVT-Cu SPA beads, 200 nM 3H-glycine or 70 nM 3H-glutamate, and varying concentrations of ACPC (for competition with 3H-glycine at the GluN1 LBD) or t-ACBD (for competition with 3H-glutamate at the GluN2B LBD). Samples were incubated at room temperature for 2h after which the counts were measured. Data was analyzed using GraphPad Prism using a one-site binding model.
Extended Data
Extended Data Figure 1. Summary of Xenopus laevis NMDA crystallization constructs.
a, b, Cartoon representation of amino terminal domain (ATD), ligand binding domain (LBD) and transmembrane domain (TMD) for (a) GluN1 Δ2 and (b) GluN2B Δ2 subunit constructs. Location of point mutations are highlighted in white circles. Location of deletions are highlighted with a yellow wedge. Mutated glycosylation sites are not shown and are listed in Extended Data Table 1. c, d, Select amino acid sequences of constructs used in these studies compared to wildtype sequence to highlight mutations in (c) GluN1 and (d) GluN2B. Mutations are numbered and the purpose of each is detailed in Extended Data Table 1.
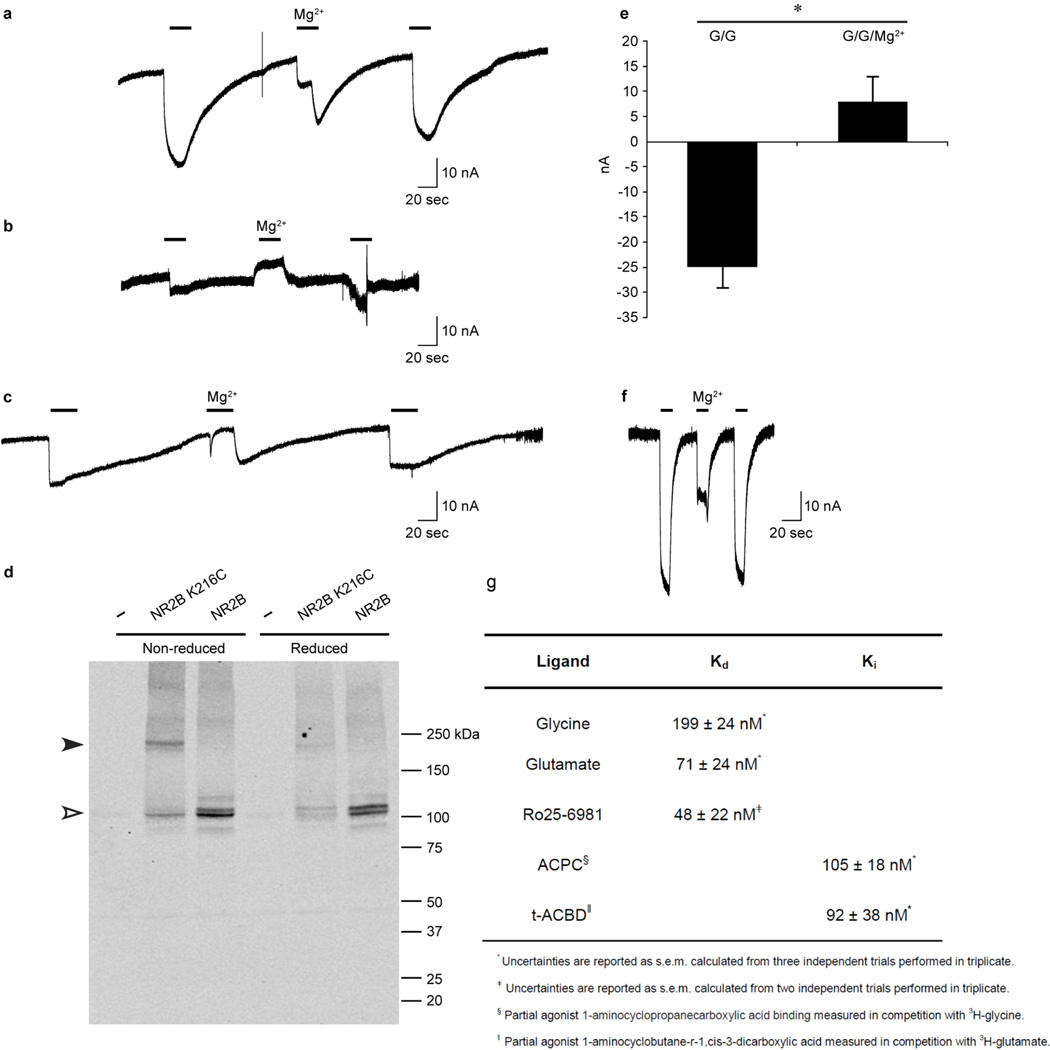
Extended Data Figure 2. Electrophysiology and Western analysis of GluN1 Δ/GluN2B Δ receptor combinations.
a, b, c, Representative TEVC currents recorded for oocytes expressing GluN1 Δ4 and (a) GluN2B Δ1 or (b, c) GluN2B Δ3 receptors in response to agonist (100µM glycine and 100 µM glutamate, bars, 20 sec) or agonist plus 1 mM MgCl2 (indicated) after soaking oocytes in the (a, b) absence or (c) presence of 5mM DTT. d, Western blot analysis of oocytes demonstrating spontaneously crosslinking cysteines (Lys216Cys) introduced at the GluN2B Δ3 intersubunit interface. Oocytes were soaked in the absence (left lanes) or presence of 5mM DTT (right lanes) before processing for Western analysis using an anti-GluN2B antibody. Filled and open triangles indicate positions of crosslinked and monomeric GluN2B, respectively. e, Graph of mean agonist-induced inward currents from four reduced oocytes expressing GluN1 Δ4 and GluN2B Δ3 in the absence (G/G, −25 ± −4 nA) or presence of 1mM MgCl2 (G/G/Mg2+, 8 ± 5 nA). Error bars represent s.e.m. The p value is <0.001 for the paired T-test (asterisk). f, Representative TEVC currents recorded in response to agonist (100µM glycine and 100 µM glutamate bars, 10 sec) or agonist plus 1 mM MgCl2 for oocytes expressing constructs similar to the GluN1 Δ2/GluN2B Δ2 receptor combination with the following exceptions: GluN1 subunit, Asp656 (wt), Gly636Arg and Lys741Asp; and GluN2B subunit, Glu654 (wt), Glu655 (wt), and Lys216 (wt). g, Binding constants for the GluN1 Δ2/GluN2B Δ2 construct.
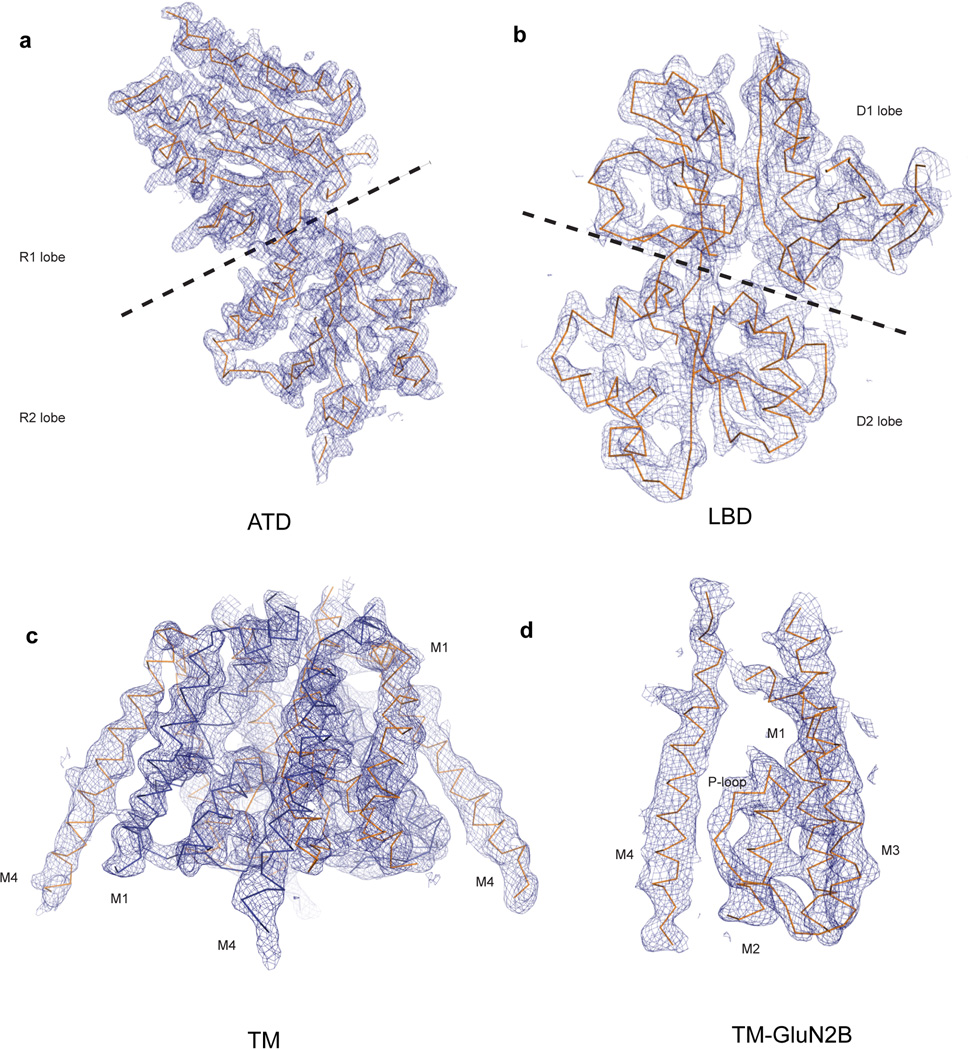
Extended Data Figure 3. 2Fo-Fc electron density maps of the GluN1/GluN2B NMDA structure.
a, The electron densities associated with the GluN1 ATD (chain A) contoured at 1.7 σ, (b) the GluN1 LBD (chain A) contoured at 1.6 σ, (c) the TMD of the entire tetrameric receptor contoured at 1.0 σ and (d) the TMD of a single GluN2B subunit (chain D), showing the pore loop, also contoured at 1.0 σ. Electron density maps and structures were derived from Data set 1/Structure 1 for panels (a) and (b) and from Data set 2/Structure 2 for panels (c) and (d) (see Extended Data Table 2).
Extended Data Figure 4. Analysis of spontaneous crosslinking of single cysteine point mutants introduced in the GluN2B ATD of the GluN1/GluN2B receptor complex.
a, Western blot analysis of single cysteine mutants in the α5 helix of the GluN2B subunit. Solubilized extracts of HEK293S GnTI cells expressing a C-terminal GFP-StrepII tag GluN2B construct (GluN2B Δ1) containing mutants as indicated with untagged GluN1 (GluN1 Δ1) were analyzed by Western blot using an anti-GFP polyclonal antibody. The open and filled arrows correspond to monomeric and dimeric GluN2B bands, respectively. b, Coomassie stained SDS-PAGE analysis of spontaneous crosslinking of GluN2B K216C containing receptor. Left and right lanes illustrate samples with different concentrations of protein for GluN1/GluN2B and GluN1/GluN2B K216C receptors. The asterisk indicates GluN1 monomer while the open and filled arrows correspond to monomeric and dimeric GluN2B bands, respectively.
Extended Data Figure 5. Structural analyses and electron density maps of GluN1/GluN2B ATD heterodimer in the full-length NMDA structure.
a, Intersubunit distance between the indicated marker atoms and angle of domain closure in the soluble ATD structure (PDB 3QEM, left panel) or full-length ATD structure (right panel). b, Superposition of the full-length GluN1 (blue)/GluN2B (orange) ATD heterodimer onto the soluble heterodimer structure (PDB 3QEM, light grey) by aligning the indicated helices (green) in the R1 lobe of GluN2B. c, Fo-Fc omit electron density map for Ro25-6981 bound at the GluN1/GluN2B ATD heterodimer interface (chains A and B), contoured at 3σ (Data set 1/Structure 1). d, Anomalous difference electron density of Tb3+ (blue mesh) near the R1-R2 hinge of a single GluN2B ATD (chain B, Data set 3), contoured at 3.5 σ. e, Superposition of the LBD layer of the low resolution GluN1/GluN2B receptor (light blue, Data set 4/Structure 4) onto the LBD layer of the high resolution K216C receptor (magenta, Data set 1/Structure 1) illustrates the relative difference in ATD conformations between the two receptor structures (see Extended Data Table 2). Shown is the most 'open' conformation of the ATDs derived from one of the two independent receptors in the asymmetric unit of Data set 4/Structure 4.
Extended Data Figure 6. LBD ligand electron densities and conformations.
Fo-Fc omit electron density maps for (a) ACPC bound to GluN1 LBD (chain A) and (b) t-ACBD bound to GluN2B LBD (chain D), contoured at 3 σ and 2.5 σ, respectively (Data set 1/Structure 1). c, d, e, Comparison of LBD in the full-length GluN1/GluN2B structure to isolated structures by aligning the D1 lobe. The angle of rotation relative to beta strand 10 is indicated for each. c, The ACPC-bound GluN1 LBD of the full-length structure (chain A, blue) is more open than the ACPC-bound isolated GluN1 LBD structure (PDB 1Y20, grey). d, The ACPC-bound GluN1 LBD of the full-length structure (chain A, blue) is more open than the glycine-bound isolated GluN1 LBD structure (PDB 2A5T, chain A, grey). e, The t-ACBD-bound GluN2B LBD of the full-length structure (chain D, orange) has a similar domain closure to the glutamate-bound isolated GluN2B LBD (PDB 2A5T, chain B, grey). f, GluN1/GluN2B LBD heterodimer (chains A and D) from the full-length receptor structure showing the separation of the D2 lobes, measured using the α-carbon atoms of residues Gly 664 and Gly 662, respectively. g, A similar measurement as in (f) using the equivalent residues in the context of the rat glycine/glutamate bound isolated GluN1/GluN2A LBDs (PDB 2A5T). h, The same measurement as in (g), except in the GluN1 antagonist/Glu2A glutamate-bound conformation (PDB 4NF4). Structures shown in panels c-f were derived from Data set 1/Structure 1 and are similar in conformation to the related domains derived from Data set 2/Structure 2 (see Extended Data Table 2).
Extended Data Figure 7. Structural analyses of the transmembrane domain of NMDA receptor.
a, Alpha-carbon superposition of the M3 helices of the GluN1/GluN2B NMDA receptor (Data set 2/Structure 2) onto the corresponding M3 regions of GluA2 receptor (PDB 3KG2; grey). Rmsd is 1.89 Å for 144 aligned α-carbon atoms. The GluN1 subunits are blue and the GluN2B subunits are yellow. b, Amino acid sequence alignment of the NMDA receptor and the KcsA channel in the M2 and M3 regions using Promals3D (http://prodata.swmed.edu/promals3d/promals3d.php). c, Superposition of the four M2 helices of the NMDA receptor onto the corresponding four M2 regions of the KcsA channel (PDB 1K4C; residues 61–75). Rmsd is 1.86 Å. Only chains B and D of the NMDA GluN2B subunits are shown. d, Residual electron density in the central vestibule. Fo-Fc electron density in the central vestibule is shown for the GluN1/GluN2B receptor from Data set 2/Structure 2. For clarity, chain C is removed. e, Fo-Fc electron density map in the central vestibule derived from Data set 1/Structure 1. For clarity, chain B is removed. f, The same electron density map as shown in panel (e) except that the structure has been rotated by ~90° around the pore axis and chain C of the GluN1 subunit has been removed for clarity. All maps are contoured at 2.8 σ.
Extended Data Figure 8. Comparison of LBD layers and LBD-TMD linkers between the NMDA receptor and the GluA2 receptor structures.
a, View from the extracellular side of the membrane of the connections between the TMD and LBD domains of the GluN1/GluN2B structure and of the GluA2 structure (PDB 3KG2), showing the relative rotation of GluA2 layer by ~35°. The S2 segment resides within the LBD. The LBD-M3 linkers are highlighted. b, The LBD-M1 linkers are highlighted. c, The LBD-M4 linkers are highlighted. Shown in all panels are structures derived from Data set 1/Structure 1.
Extended Data Table 1.
Constructs and mutations
| Basic GluN1 Δ construct |
Basic GluN2B Δ construct |
|||
|---|---|---|---|---|
| Residue substitutions | Purpose of mutation | Residue substitutions | Purpose of mutation | |
| C22A | Remove a potentially reactive cysteine |
M20S, G21R.C22A | Remove a potentially reactive cysteine and in so doing, change these three residues to their identities in the human GluN2B subunit |
|
| K493A, K494A, E495A║ | Reduce surface entropy | Δ64E | Remove potential protease site |
|
| G610R, 1617L | Improve thermostability | ΔK382-V385 | Remove flexible regions | |
| N300Q, N350Q, N368D N440D, N469D, N769E§ |
Remove predicted glycosylation sites |
V615L | Improve thermostability | |
| D656R* | Promote desensitized conformation, improve thermostability and expression level |
N69Q, N343D, T490V§ | Remove predicted glycosylation sites |
|
| residues 837–847 of AMPA (YKSRAEAKRMK; NP_058957) inserted at C- terminus‡ |
Improve thermostability and expression level |
E654R, E655R* | Promote desensitized conformation, improve thermostability and expression level |
|
| residues 837–847 of AMPA (YKSRAEAKRMK) inserted at C-terminus‡ |
Improve thermostability and expression level |
|||
| C585A | Remove a potentially reactive cysteine |
|||
| K216 (7.5 Å) |
GluN1 Δ1: Basic GluN1 Δ construct plus: |
GluN2B Δ1: Basi GluN2B Δ construct | ||
| Residue substitutions | Purpose of mutation | |||
| E592A, E593A, E594A║ | Reduce surface entropy | |||
| G636L, M816Y | Improve thermostability and expression level |
|||
| K741D | Improve expression level | |||
| K216C (3.7 Å) |
GluN1 Δ2: Basic GluN1 Δ construct plus: |
GluN2B Δ2: Basic GluN2B Δ construct plus: |
||
| Residue substitutions | Purpose of mutation | Residue substitutions | Purpose of mutation | |
| K51F, K52F | Improve crystal packing | K216C | Decrease flexibility | |
| ΔK588-E595 | Remove flexible regions | ΔR584-G593 | Remove flexible regions | |
|
K216C (Tb3+ complex 6.5 Å) |
GluN1 Δ3: Basic GluN1 Δ construct plus: |
GluN2B Δ3: Basic GluN1 Δ construct plus: |
||
| Residue substitutions | Purpose of mutation | Residue substitutions | Purpose of mutation | |
| K51F, K52F | Improve crystal packing | K216C | Decrease flexibility | |
| K493A, K494A, E495A║ | Reduce surface entropy | |||
| TEVC |
GluN1 Δ4: Basic GluN1 Δ construct plus: |
|||
| Residue substitutions | Purpose of mutation | |||
| K51 F, K52F | Improve crystal packing | |||
| E592A, E593A, E594A║ | Reduce surface entropy | |||
| G636L | Improve thermostability and expression level |
|||
| K741D | Improve expression level | |||
Yelshansky M.V. et al. Block of AMPA receptor desensitization by a point mutation outside the ligand-binding domain. J Neurosci. 24, 4728-4736 (2004).
Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 462745-56 (2009).
Potential glycosylation sites predicted using the NetNGlyc 1.0 Server (http://www.cbs.dtu.dk/services/NetNGlyc/).
Sites identified for Surface Entropy Reduction approach using the SERp Server (http://services.mbi.ucla.edu/SER/).
Extended Data Table 2.
Crystallographic and structure refinement statistics‡
| Data set 1 GluN1 Δ2/GIUN2B Δ2 Structure 1 ACPC/t-ACBD |
Data set 2 GluN1 Δ2/GIUN2B Δ2 Structure 2 ACPC/t-ACBD |
Data set 3 GluN1 Δ3/GIUN2B Δ3 Structure 3 Gly/Glu |
Data set 4 GIUN1 Δ1/GIUN2B Δ1 Structure 4 ACPC/t-ACBD |
|
|---|---|---|---|---|
| Data collection | ALS 8.2.1 | ALS 8.2.1 | ALS 5.0.2 | ALS 5.0.2 |
| Space group | C2 | C2 | C2 | C222 |
| Cell dimensions a, b, c (Å) | 201.5, 117.3, 218.8 | 203.5, 118.4, 226.6 | 203.4, 118.2, 232.9 | 128.0, 616.2, 152.3 |
| Cell angles α, β, γ (Å) | 90.0, 106.7, 90.0 | 90.0, 103.8, 90.0 | 90.0, 104.3, 90.0 | 90.0, 90, 90.0 |
| Wavelength (Å) | 1.00 | 1.00 | 1.2 | 1 |
| Resolution (Å)* | 48.1-3.70 (3.80-3.70) | 48.7-3.90 (4.00-3.90) | 49.3 (6.67-6.50) | 50 (7.63-7.50) |
| Completeness* | 97.0 (94.5) | 97.4 (95.0) | 90.5 (80.1) | 99 (96.5) |
| Multiplicity* | 5.62 (3.36) | 3.44 (2.74) | 1.87 (1.53) | 6.2 (5.8) |
| I/σI* | 6.69 (2.13) | 7.46 (2.22) | 5.1 (1.27) | 31 (1.79) |
| Rmeas (%)* | 14.4 (48.6) | 11.0 (49.9) | 13.6 (55.8) | 14.0 (>100)† |
| CC1/2 (%)* | 99.5 (48.9) | 99.7 (62.6) | 99.6 (16.7) | |
| Anisotropy (Å: a*/b*/c*)# | 3.5/3.5/3.9 | 3.5/3.5/4.8 | ||
| Refinement | ||||
| Resolution (Å)║ | 48.1-3.59 (3.66-3.59) | 30.0-3.77 (3.85-3.77) | ||
| Completeness (%) | 93.1 (56.4) | 83.2 (43.2) | ||
| No. of reflections | 53380 | 44017 | ||
| Rwork/Rfree (%) | 28.2 (35.7) / 32.0 (39.6) | 26.8 (1.9)/31.0 (48.6) | ||
| No. of atoms total | 19991 | 20704 | ||
| Ligand | 124 | 124 | ||
| Average B-factor (A2) | ||||
| Protein | 183 | 199 | ||
| Ligand | 133 | 113 | ||
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.002 | 0.006 | ||
| Bond angles (°) | 0.636 | 1.384 | ||
| Ramachandran plot | ||||
| Favored (%) | 95.1 | 94.2 | ||
| Allowed (%) | 4.8 | 5.6 | ||
| Disallowed (%) | 0.1 | 0.2 |
Data set 1 corresponds to GluN1 Δ2/GluN2B Δ2 structure at ~3.7 Å resolution; Data set 2 corresponds to the GluN1 Δ2/GluN2B Δ2 structure at ~3.9 Å resolution; Data set 3 is derived from the Tb3+-soaked crystals of the GluN1 Δ3/GluN2B Δ3 construct; and Data set 4 is corresponds to the low resolution GluN1 Δ1/GluN2B Δ1 molecular replacement solution at 7.5 Å resolution. The LBD ligands are shown under the column headings. All crystallizations included Ro25-6981 and MK-801.
Highest resolution shell in parentheses.
Estimates of anisotropy calculated using the anisotropy server (http://services.mbi.ucla.edu/anisoscale/).
Rsym is reported.
Due to crystal anisotropy and incompleteness at higher resolution, the resolution of the diffraction data used in refinement was decreased from that employed in diffraction data scaling for the Data set I and II structures.
5% of reflections were used for calculation of Rfree
Extended Data Table 3.
ATD and LBD RMSD*
| ATD‡ | sGluN1 (chain A; 3QEM) |
SGlUN2 (chain B; 3QEM) |
GlUN1 (chain A) |
GluN1 (chain C) |
GlUN2B (chain B) |
GlUN2B (chain D) |
|
|---|---|---|---|---|---|---|---|
|
GlUN1 (chain A) |
0.8 | 3.1 | - | 0.7 | 2.9 | 3.0 | |
|
GluN1 (chain C) |
0.9 | 3.2 | 0.7 | - | 3.0 | 3.1 | |
|
GluN2B (chain B) |
2.9 | 1.2 | 2.9 | 3.0 | - | 0.9 | |
|
GlUN2B (chain D) |
3.0 | 1.3 | 3.0 | 3.1 | 0.9 | - | |
|
sGluN1/sGluNB (dimer AB; 3QEM) |
GlUN1/N2B (dimer AB) |
GlUN1/N2B (dimer CD) |
|||||
|
GlUN1/N2B (dimer AB) |
1.1 | - | 0.9 | ||||
|
GlUN1/N2B (dimer CD) |
1.2 | 0.9 | - | ||||
| LBD§ |
GluN1+ACPC (chainC) |
sGluN1+gly (chain A; 2A5T) |
sGluN1+ACPC (1Y20) |
||||
|
GluN1+ACPC (chain A) |
0.98 | 1.2 | 1.3 | ||||
|
GluN1+ACPC (chainC) |
- | 1.3 | 1.2 | ||||
|
sGluN1+gly (chain A; 2A5T) |
1.3 | - | 0.7 | ||||
|
sGluN1+ACPC (1Y20) |
1.2 | N.D.║ | - | ||||
|
GluN2B+tACBD (chainD) |
sGluN2A+Glu (chain B; 2A5T) |
||||||
|
GluN2B+tACBD (chainB) |
1.0 | 1.3 | |||||
|
GluN2B+tACBD (chainD) |
- | 1.3 | |||||
|
sGluN2A+ Glu (chain B; 2A5T) |
1.3 | - | |||||
RMSD were calculated using coot SSM superpose function, unit is Å.
RMSD values were determined from superpositions of indicated ATD of the full-length Structure 1 (GluN1, GluN2B) and of soluble ATDs (sGluN1 or sGluN2B) onto full-length structure ATDs. PDB codes for the soluble domains as indicated.
RMSD values determined from superpositions of indicated LBDs of the full-length Structure 1 (GluN1, GluN2B) and of soluble LBDs (sGluN1 or sGluN2B) onto full-length structure LBDs. PDB codes for the soluble domains as indicated.
N.D. Not determined.
Supplementary Material
Acknowledgements
All members of the Gouaux laboratory are gratefully acknowledged for their support and assistance, especially L. Chen, K. Duerr and K. Wang. L. Vaskalis is thanked for assistance with the figures, G. Westbrook and C. Jahr for comments on the manuscript, H. Owen for proofreading and I. Baconguis for making the animation. E.G. acknowledges the generous support of R. LaCroute, B. LaCroute and J. LaCroute. This work was also supported by an Oregon Brain Institute Graduate Student Fellowship (C.H.L), the NIH (E.G.) and the Vollum Institute (E.G.). E.G. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Author contributions
C.H.L. contributed to all aspects of the research; W.L. carried out crystallographic analysis; J.C.M. carried out molecular biology, cell culture, electrophysiology and ligand binding experiments; A.G. performed molecular biology, cell culture, receptor purification and crystallization studies; J.D. and X.S. analyzed the structures; E.G. directed the research; all authors contributed to the preparation of the manuscript.
Author information
The coordinates and structure factors for the structure have been deposited in the Protein Data Bank under accession code 4TLL and 4TLM for Structure 1 and Structure 2, respectively.
Reprints and permissions information are available at www.nature.com/reprints. The authors declare no competing financial interests.
References
- 1.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paoletti P, Bellone C, Zhou Q. NMDA receptor subunit diversity: impact on receptor properties, synaptic plasticity and disease. Nat. Rev. Neurosci. 2013;14:383–400. doi: 10.1038/nrn3504. [DOI] [PubMed] [Google Scholar]
- 3.Bliss TVP, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 4.Soto D, Altafaj X, Sindreu C, Bayes A. Glutamate receptor mutations in psychiatric and neurodevelopmental disorders. Commun. Integrative Biology. 2014;7:e27887. doi: 10.4161/cib.27887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peery HE, et al. Anti-NMDA receptor encephalitis. The disorder, the diagnosis and the immunobiology. Autoimmun. Rev. 2012;11:863–872. doi: 10.1016/j.autrev.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Keinänen K et al. A family of AMPA-selective glutamate receptors. Science. 1990;249:556–560. doi: 10.1126/science.2166337. [DOI] [PubMed] [Google Scholar]
- 7.Bettler B, et al. Cloning of a novel glutamate receptor subunit, GluR5: Expression in the nervous system during development. Neuron. 1990;5:583–595. doi: 10.1016/0896-6273(90)90213-y. [DOI] [PubMed] [Google Scholar]
- 8.Werner P, Voigt M, Keinänen K, Wisden W, Seeburg PH. Cloning of a putative high-affinity kainate receptor expressed predominatly in hippocampal CA3 cells. Nature. 1991;351:742–744. doi: 10.1038/351742a0. [DOI] [PubMed] [Google Scholar]
- 9.Johnson JW, Ascher P. Glycine potentiates the NMDA response in cultured mouse brain neurons. Nature. 1987;325:529–531. doi: 10.1038/325529a0. [DOI] [PubMed] [Google Scholar]
- 10.Mayer ML, Westbrook GL, Guthrie PB. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature. 1984;309:261–263. doi: 10.1038/309261a0. [DOI] [PubMed] [Google Scholar]
- 11.Nowak L, Bregestovski P, Ascher P, Herbet A, Prochiantz A. Magnesium gates glutamate-activated channels in mouse central neurones. Nature. 1984;307:462–465. doi: 10.1038/307462a0. [DOI] [PubMed] [Google Scholar]
- 12.Mayer ML, Westbrook GL. Permeation block of N-methyl-D-aspartic acid receptor channels by divalent cations in mouse cultured central neurones. J. Physiol. (Lond.) 1987;394:501–527. doi: 10.1113/jphysiol.1987.sp016883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moriyoshi K, et al. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354:31–37. doi: 10.1038/354031a0. [DOI] [PubMed] [Google Scholar]
- 14.Monyer H, et al. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- 15.Tovar KR, McGinley MJ, Westbrook GL. Triheteromeric NMDA receptors at hippocampal synapses. J. Neurosci. 2013;33:9150–9160. doi: 10.1523/JNEUROSCI.0829-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen KB, Furukawa H, Traynelis SF. Control of assembly and function of glutamate receptors by the amino terminal domain. Mol. Pharmacol. 2010;78:535–549. doi: 10.1124/mol.110.067157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kashiwagi K, et al. Channel blockers acting at N-methyl-D-asparate receptors: Differential effects of mutations in the vestibule and ion channel pore. Mol. Pharmacol. 2002;61:533–545. doi: 10.1124/mol.61.3.533. [DOI] [PubMed] [Google Scholar]
- 18.Sun Y, et al. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- 19.Mayer ML. Emerging models of glutamate receptor ion channel structure and function. Structure. 2011;19:1370–1380. doi: 10.1016/j.str.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pøhlsgaard J, Frydenvang K, Madsen U, Kastrup JS. Lessons from more than 80 structures of the GluA2 ligand-binding domain in complex with agonists, antagonists and allosteric modulators. Neuropharmacology. 2011;60:135–150. doi: 10.1016/j.neuropharm.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Jin R, et al. Crystal structure and association behavior of the GluR2 amino-terminal domain. EMBO J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat. Struct. Mol. Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009;28:3910–3920. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2 NMDA receptors. Nature. 2011;475:249–253. doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer G, et al. Ro 25-6981, a highly potent and selective blocker of N-methyl-D-asparate receptors containing the NR2B subunit. Characterization in vitro. J. Pharmacol. Exp. Ther. 1997;283:1285–1292. [PubMed] [Google Scholar]
- 26.Watson GB, Lanthorn TH. Pharmacological characteristics of cyclic homologues of glycine at the N-methyl-D-aspartate receptor-associated glycine site. Neuropharmacology. 1990;29:727–730. doi: 10.1016/0028-3908(90)90125-b. [DOI] [PubMed] [Google Scholar]
- 27.Allan RD, et al. Synthesis and activity of a potent N-methyl-D-aspartic acid agonist, trans-1-aminocyclobutane-1,3-dicarboxylic acid, and related phosphonic and carboxylic acids. J. Med. Chem. 1990;33:2905–2915. doi: 10.1021/jm00172a036. [DOI] [PubMed] [Google Scholar]
- 28.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salussolia CL, Prodromou ML, Borker P, Wollmuth LP. Arrangement of subunits in functional NMDA receptors. J. Neurosci. 2011;31:11295–11304. doi: 10.1523/JNEUROSCI.5612-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riou M, Stroebel D, Edwardson JM, Paoletti P. An alternating GluN1-2-1-2 subunit arrangement in mature NMDA receptors. PLoS One. 2012;7:e35134. doi: 10.1371/journal.pone.0035134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee CH, Gouaux E. Amino terminal domains of the NMDA receptor are organized as local heterodimers. PLoS One. 2011;6:e19180. doi: 10.1371/journal.pone.0019180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furukawa H, Singh S, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 33.Mony L, Kew JN, Gunthrope MJ, Paoletti P. Allosteric modulators of NR2B-containing NMDA receptors: molecular mechanisms and therapeutic potential. Br. J. Pharmacol. 2009;157:1301–1317. doi: 10.1111/j.1476-5381.2009.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reichling DB, MacDermott AB. Lanthanum actions on excitatory amino acid-gated currents and voltage-gated calcium currents in rat dorsal horn neurons. J. Physiol. 1991;441:199–218. doi: 10.1113/jphysiol.1991.sp018746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherry AD, Newman AD, Gutz CG. The activation of concavalin A by lanthanide ions. Biochemistry. 1975;14:2191–2196. doi: 10.1021/bi00681a023. [DOI] [PubMed] [Google Scholar]
- 36.Zhu S, Stroebel D, Yao CA, Taly A, Paoletti P. Allosteric signaling and dynamics of the clamshell-like NMDA receptor GluN1 N-terminal domain. Nat. Struct. Mol. Biol. 2013;20:477–485. doi: 10.1038/nsmb.2522. [DOI] [PubMed] [Google Scholar]
- 37.Weston MC, Schuck P, Ghosal A, Rosenmund C, Mayer ML. Conformational restriction blocks glutamate receptor desensitization. Nat. Struct. Mol. Biol. 2006;13:1120–1127. doi: 10.1038/nsmb1178. [DOI] [PubMed] [Google Scholar]
- 38.Gielen M, et al. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen KB, Ogden KK, Traynelis SF. Subunit-selective allosteric inhbition of glycine binding to NMDA receptors. J. Neurosci. 2012;32:6197–6208. doi: 10.1523/JNEUROSCI.5757-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inanobe A, Furukawa H, Gouaux E. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron. 2005;47:71–84. doi: 10.1016/j.neuron.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Erreger K, et al. Subunit-specific agonist activity at NR2A-, NR2B-, NR2C-, and NR2D-containing N-methyl-D-aspartate glutamate receptors. Mol. Pharmacol. 2007;72:907–920. doi: 10.1124/mol.107.037333. [DOI] [PubMed] [Google Scholar]
- 42.Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- 43.Sugihara H, Moriyoshi K, Ishii T, Masu M, Nakanishi S. Structures and properties of seven isoforms of the NMDA receptor generated by alternative splicing. Biochem. Biophys. Res. Commun. 1992;185:826–832. doi: 10.1016/0006-291x(92)91701-q. [DOI] [PubMed] [Google Scholar]
- 44.Mony L, Zhu S, Carvalho S, Paoletti P. Molecular basis of positive allosteric modulation of GluN2B NMDA receptors by polyamines. EMBO J. 2011;30:3134–3146. doi: 10.1038/emboj.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yelshansky MV, Sobolevsky AI, Jatzke C, Wollmuth LP. Block of AMPA receptor desensitization by a point mutation outside the ligand-binding domain. J Neurosci. 2004;24:4728–36. doi: 10.1523/JNEUROSCI.0757-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erreger K, Dravid SM, Banke TG, Wyllie DJ, Traynelis SF. Subunit-specific gating controls rat NR1/NR2A and NR1/NR2B NMDA channel kinetics and synaptic signalling profiles. J. Physiol. 2005;563:345–358. doi: 10.1113/jphysiol.2004.080028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 49.Burnashev N, et al. Control by asparagine residues of calcium permeability and magnesium blockade in the NMDA receptor. Science. 1992;257:1415–1419. doi: 10.1126/science.1382314. [DOI] [PubMed] [Google Scholar]
- 50.Kuner T, Seeburg PH, Guy HR. A common architecture for K+ channels and ionotropic glutamate receptors. Trends Neurosci. 2003;26:27–32. doi: 10.1016/s0166-2236(02)00010-3. [DOI] [PubMed] [Google Scholar]
- 51.Dukkipati A, Park HH, Waghray D, Fischer S, Garcia KC. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr. Purif. 2008;62:160–170. doi: 10.1016/j.pep.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baconguis I, Gouaux E. Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature. 2012;489:400–405. doi: 10.1038/nature11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 54.Hattori M, Hibbs RE, Gouaux E. A fluorescence-detection size-exclusion chromatography-based thermostability assay for membrane protein precrystallization screening. Structure. 2012;20:1293–1299. doi: 10.1016/j.str.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reeves PJ, Callewaert N, Contreras R, Khorana HG. Structure function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline-inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. Proc. Natl. Acad. Sci. USA. 2002;99:13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gourdon P, et al. HiLiDe--Systematic approach to membrane protein crystallization in lipid and detergent. Cryst. Growth Des. 2011;11:2098–2106. [Google Scholar]
- 57.Kabsch WXDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 59.Hanson MA, et al. Crystal structure of a lipid-G protein-coupled receptor. Science. 2012;335:851–855. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Strong M, et al. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis . Proc. Natl. Acad Sci. USA. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D. Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D. Biol. Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 65.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smart OS, Neduvelil JG, Wang X, Wallace BA, Samsom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 67.DeLano WL. DeLano Scientific. San Carlos, CA, USA; 2002. [Google Scholar]
- 68.Hart HE, Greenwald EB. Scintillation proximity assay (SPA) - a new method of immunoassay Direct and inhibition mode detection with human albumin and rabbit antihuman albumin. Mol. Immunol. 1979;16:265–267. doi: 10.1016/0161-5890(79)90065-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.