Abstract
Background
As 2 important SNPs located in the promoter region of VEGF gene, the roles of rs833061 (−460C>T) and rs699947 (−2578C>A) in lung cancer susceptibility and survival remain inconclusive and controversial.
Material/Methods
For better understanding of these 2 SNPs in lung cancer risk and survival, a meta-analysis was performed to pool findings of previous studies and to generate large-scale evidence.
Results
Based on the 10 eligible studies included, this study observed that the −460C>T polymorphism generally had no significant effect on lung cancer risk. However, subgroup analysis found that −460TT homozygote variant might confer significantly increased cancer risk for Asians (TT vs. CC: OR=1.69, 95% CI 1.08–2.63, p=0.02), but not in Caucasians. Similar results were observed in −2578C>A in Asians (AA vs. CC: OR=3.00, 95% CI 1.51–5.95, p=0.002; AA vs. AC: OR=3.15, 95% CI 1.00–9.91, p=0.05; AA vs. (AC+CC): OR=2.92, 95% CI 1.51–5.65, p=0.001). In lung cancer survival, 4 trials included had conflicting results. One found −460C>T polymorphism had no effect on survival, 1 observed risk increasing, while the remaining 2 observed risk decreasing. This inconsistency was closely related to the different therapeutic practices applied in different studies, the effects of which were significantly affected by VEGF expression.
Conclusions
−460TT and −2578AA homozygote might lead to significantly increased cancer risk for Asians, but the effects on survival remain to be explored. These 2 SNPs might be potential indicators of lung cancer risk for Asians and should be considered when planning chemotherapy and radiotherapy for lung cancer patients.
MeSH Keywords: Lung Neoplasms; Meta-Analysis as Topic; Polymorphism, Single Nucleotide; Vascular Endothelial Growth Factor A
Background
Globally, lung cancer is one of the most frequently diagnosed cancers and is a leading cause of cancer-related death [1]. Although the mechanism of tumorigenesis of lung cancer is still not well defined, it is generally considered to be a complex process related to both epigenetic and genetic changes. Aberrant angiogenesis is one of the most studied genetic alterations in cancer development. In fact, angiogenesis is a critical process in the development, growth, and metastasis of malignant tumors [2]. Vascular endothelial growth factor (VEGF) and VEGF-receptor (VEGFR) are the key regulators of angiogenesis, playing an essential role in regulating neovascularization. Overexpression of VEGF was observed in lung cancer [3,4]. Inhibition of the VEGF signaling pathway could partly suppress tumor-induced angiogenesis and tumor growth [5,6]. During the past 2 decades, Bevacizumab, an agent recognizing and blocking the pathway of VEGF-A, has shown some efficacy in combination regimens to improving progression-free survival and overall survival of some solid malignancies, such as breast cancer, renal cell carcinoma, and non-small-cell lung cancer (NSCLC) [7].
The VEGF gene is located on chromosome 6p21.3 and has 8 exons [8]. Previous studies confirmed that this gene has at least 30 single-nucleotide polymorphisms (SNPs). Among them, −2578C>A (rs699947), −634G>C (rs2010963), −460C>T (rs833061), and +936C>T (rs3025039) were demonstrated to regulate VEGF expression [9,10]. Therefore, genetic variability of the VEGF gene could also modulate lung carcinogenesis through affecting angiogenesis. rs833061 and rs699947 are 2 important SNPs located in the promoter region of the VEGF gene, which might influence promoter activity [9]. Previous clinical studies observed that these 2 SNPs might contribute to altered risk of lung cancer [11,12]. In addition, some studies also observed altered lung cancer survival related to −460C>T variation [13,14]. High VEGF expression and associated enhanced tumor vasculature might facilitate the delivery of chemotherapy agents to target tumor issues and also inhibit tumor radioresistance caused by radiation-induced hypoxia, leading to a better synergistic effect between chemotherapy and radiotherapy. However, the results of previous studies remain inconclusive and controversial. For better understanding of the effects of these 2 SNPs on lung cancer, findings of previous studies were pooled to generate large-scale evidence in this meta-analysis.
Material and Methods
Search strategy
Relevant studies published from January 2000 to April 2014 were searched in PubMed, Web of Science, Embase, and Cochrane Library. The following terms and strategies were used for the search: (“rs833061” OR “rs699947” OR “single nucleotide polymorphism” OR “SNP” OR “genetic variation” OR “genetic polymorphism”) AND (“lung cancer” OR “lung neoplasms” OR “lung tumor”) AND (“Vascular endothelial growth factor” OR “VEGF” OR “VEGFA”). To avoid missing qualified trials, the backward snowballing method was used to manually screen reference lists of eligible trials. No language restriction was applied when searching.
Selection criteria
The following criteria are used to screen eligible trials evaluating SNP and lung cancer risk for this meta-analysis: (1) a case-control study, cross-sectional study, or cohort study that explored the association between VEGF rs833061 and rs699947 variants and susceptibility to lung cancer; (2) the genotype distribution of the controls were as expected by Hardy-Weinberg equilibrium (HWE); (3) sufficient data were available for calculation of allele/genotype frequency. Only studies that simultaneously meet these criteria were included for analysis. For studies evaluating SNP and lung cancer survival, genotype and correspondent overall survival time and hazard ratio (HR) needed to be reported.
Data extraction
Two authors (JT and SW) independently extracted data from original studies. Data extracted included first author, year of publication, country of origin, total number of cases and controls, source of participants, their ethnicity, histological types of lung cancer, smoking status, method of genotyping, SNP, genotype distributions, and HWE information in controls. Disagreement in data extraction was resolved through group discussion by referring to original studies with a third reviewer.
Quality assessment
The quality of eligible studies assessing SNP and lung cancer risk was assessed with the modified Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) quality score system. This system appraises a trial’s quality score based on 40 assessment items with score ranging from 0 to 40. Based on scoring of this system, the quality of a study could be classified into 3 levels: low quality (0–19), moderate quality (20–29), and high quality (30–40).
Statistical analysis
For trials evaluating the 2 SNPs and lung cancer risks, the odds ratios (OR) and corresponding 95% confidence intervals (CI) were calculated to assess the association between genetic variants and lung cancer risks with 5 genetic models, including allelic, homozygote, heterozygote, dominant model, and recessive model. The significance of pooled estimates was assessed with the Z test. Hardy-Weinberg equilibrium of genotype frequency in the control group was assessed by chi-square test. Between-studies heterogeneity was assessed by the chi-square-based Q test and I2. p<0.1 or I2>50% was considered as significant heterogeneity. A primary test was performed with a fixed-effects model. If no significant heterogeneity was observed, the fixed-effects model with Mantel-Haenszel method was used to make estimates. However, if significant heterogeneity was observed, the sources of heterogeneity were be further analyzed. If there were no significant clinical or methodological differences, the random-effects model based on DerSimonian-Laird method would be used. If significant clinical or methodological differences were observed, descriptive analysis was used. Subgroup analysis was performed based on ethnicity of participants. For studies evaluating the SNP and lung cancer survival, due to significant heterogeneity in clinical features, including patient baseline and therapeutic practices, descriptive analysis was applied.
Results
Characteristics of trials included
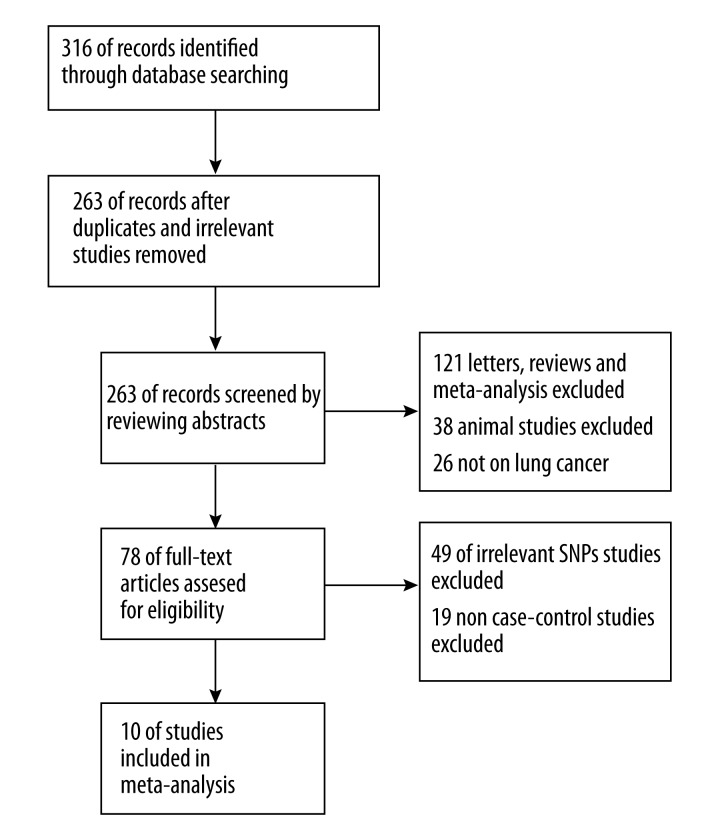
After searching and screening with preset criteria, a total of 10 studies were included for this meta-analysis. Four trials explored the association between rs833061 (−460C>T) and lung cancer risk [11,12,14,15]. Four trials explored the association between rs699947 (−2578C>A) and lung cancer risk [14,16–18]. Four studies studied the association between rs833061 (−460C>T) and lung cancer survival [13,14,19,20]. The key characteristics and quality scoring of the studies were given in Table 1. The search and screening process was summarized in Figure 1. A total of 6,614 patients and controls were involved in this study. Seven studies were conducted in Asians (5 in China, 1 in Korea, and 1 in Japan) and 3 were in Caucasians (2 in the US and 1 in Portugal). Six studies used polymerase chain reaction-restriction fragment length polymorphism (PCRRFLP) for genotyping, 3 studies used TaqMan method, and 1 used MassARRAY. The quality score of all studies was between 20 and 35, suggesting moderate-to-high quality. The genotype distribution of cases and controls is summarized in Table 2. All studies had p>0.05 in HWE test of control group, suggesting no population stratification.
Table 1.
Key characteristics of trials included.
| Study | Country | Ethnicity | SNP | Genotyping method | Source of Controls | Age (Case/control or case) | Patient | Sample size | Quality Score | |
|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | |||||||||
| *de Mello et al. 2013 | Portugal | Caucasian | rs833061 | MassARRAY | HB | 61.5/48 | NSCLC | 144 | 144 | 34 |
| Lee et a. 2005 | Korea | Asian | rs833061 | PCR–RFLP | PB | 61.6/60.9 | Lung cancer | 432 | 432 | 28 |
| Sun et al. 2013 | China | Asian | rs833061 | PCR–RFLP | PB | 56.7/55.2 | Lung cancer | 126 | 160 | 32 |
| Zhai et al. 2008 | USA | Caucasian | rs833061 | TaqMan | HB | 65/58 | NSCLC | 1,900 | 1,458 | 32 |
| *Heist et al. 2008 | USA | Caucasian | rs833061 | TaqMan | – | 69 | Early NSCLC | 462 | – | – |
| *Masago et al. 2009 | Japan | Asian | rs833061 | TaqMan | – | 67 | Advanced NSCLC | 126 | – | – |
| *Guan et al. 2010 | China | Asian | rs833061 | PCR–RFLP | 35 to 88 | Advanced NSCLC | 124 | – | – | |
| de Mello et al. 2013 | Portugal | Caucasian | rs699947 | MassARRAY | HB | 61.5/48 | NSCLC | 144 | 144 | 34 |
| Deng et al. 2014 | China | Asian | rs699947 | PCR–RFLP | PB | 55.8/53.8 | Lung cancer | 65 | 110 | 33 |
| Li et al. 2012 | China | Asian | rs699947 | PCR–RFLP | PB | N.A. | Lung cancer | 150 | 150 | 25 |
| Liang et al. 2009 | China | Asian | rs699947 | PCR–RFLP | PB | 57.5/56.9 | Lung cancer | 171 | 172 | 31 |
HB – hospital based control; PB – population based control; PCR-RFLP – polymerase chain reaction-restriction fragment length polymorphism; NSCLC – non-small-cell lung cancer;
studies assessing SNP and lung cancer survival; vacancy means not applicable.
Figure 1.
Flow diagram of the search process.
Table 2.
Genotype distribution of cases and controls in studied included.
| Study | SNP | Case | Control | p of HWE | ||||
|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | CC | CT | TT | |||
| de Mello et al. 2013 | rs833061 | 28 | 79 | 37 | 31 | 72 | 41 | 0.95 |
| Lee et al. 2005 | rs833061 | 18 | 184 | 228 | 27 | 168 | 237 | 0.70 |
| Sun et al. 2013 | rs833061 | 22 | 43 | 61 | 38 | 69 | 53 | 0.10 |
| Zhai et al. 2008 | rs833061 | 439 | 922 | 539 | 342 | 694 | 422 | 0.09 |
| Heist et al. 2008 | rs833061 | 112 | 205 | 145 | – | – | – | – |
| Masago et al. 2009 | rs833061 | 11 | 54 | 61 | – | – | – | – |
| Guan et al. 2010 | rs833061 | 24 | 67 | 33 | – | – | – | – |
| CC | CA | AA | CC | CA | AA | p of HWE | ||
| de Mello et al. 2013 | rs699947 | 43 | 75 | 26 | 44 | 73 | 27 | 0.25 |
| Deng et al. 2014 | rs699947 | 26 | 33 | 6 | 62 | 41 | 7 | 0.95 |
| Li et al. 2012 | rs699947 | 93 | 45 | 12 | 98 | 49 | 3 | 0.27 |
| Liang et al. 2009 | rs699947 | 129 | 28 | 14 | 112 | 56 | 4 | 0.33 |
HWE – Hardy-Weinberg equilibrium; vacancy means not applicable.
Association between rs833061 and rs699947 in VEGF and susceptibility to lung cancer
Pooled results and stratified analysis of the association between the 2 SNPs and the risk of lung cancer are summarized in Table 3. The results of meta-analysis showed that rs833061 −460C>T polymorphism generally had no influence on lung cancer susceptibility. (T vs. C: OR=1.07, 95% CI 0.91–1.26, p<0.41; TT vs. CC: OR=1.08, 95% CI 0.91–1.27, p=0.40; TT vs. TC: OR=1.01, 95% CI 0.79–1.29, p=0.95; (TT+TC) vs. CC: OR=1.08, 95% CI 0.93–1.24, p=0.99; TT vs. (TC+CC): OR=1.05, 95% CI 0.82–1.35, p=0.67) (Table 3). However, subgroup analysis showed that although −460C>T variants were not related to lung cancer risk among Caucasians, the homozygote TT variant might contribute to significantly increased lung cancer risk in Asians (TT vs. CC: OR=1.69, 95% CI 1.08–2.63, p=0.02) (Table 3).
Table 3.
Association between rs833061, rs699947 in VEGF and susceptibility to lung cancer.
| VEGF SNP | No. studies | T vs. C | TT vs. CC | TT vs. TC | (TT+TC) vs. CC | TT vs. (TC+CC) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs833061 (Overall) | 4 | OR (95% CI) | P | P-H | I2 | OR (95% CI) | P | P-H | I2 | OR (95% CI) | P | P-H | I2 | OR (95% CI) | P | P-H | I2 | OR (95% CI) | P | P-H | I2 |
| 1.07 [0.91, 1.26] | 0.41 | 0.09 | 54% | 1.08 [0.91, 1.27] | 0.40 | 0.17 | 41% | 1.01 [0.79, 1.29] | 0.95 | 0.09 | 54% | 1.08 [0.93, 1.24] | 0.99 | 0.42 | 0% | 1.05 [0.82, 1.35] | 0.67 | 0.06 | 60% | ||
| Population | |||||||||||||||||||||
| Asians | 2 | 1.14 [0.95, 1.37] | 0.15 | 0.03 | 79% | 1.69 [1.08, 2.63] | 0.02 | 0.48 | 0% | 1.23 [0.60, 2.54] | 0.58 | 0.01 | 83% | 1.50 [0.98, 2.29] | 0.06 | 0.93 | 0% | 1.29 [0.64, 2.59] | 0.48 | 0.01 | 85% |
| Caucasian | 2 | 0.99 [0.91, 1.09] | 0.91 | 0.96 | 0% | 1.00 [0.83, 1.20] | 0.96 | 0.99 | 0% | 0.95 [0.81, 1.11] | 0.51 | 0.59 | 0% | 1.03 [0.88, 1.20] | 0.73 | 0.72 | 0% | 0.96 [0.83, 1.11] | 0.62 | 0.68 | 0% |
| A vs. C | AA vs. CC | AA vs. AC | (AA+AC) vs. CC | AA vs. (AC+CC) | |||||||||||||||||
| rs699947 (Overall) | 4 | OR (95% CI) | P | P-H | I2 | OR (95% CI) | P | P-H | I2 | OR (95% CI) | P | P-H | I2 | OR (95% CI) | P | P-H | I2 | OR (95% CI) | P | P-H | I2 |
| 1.11 [0.92, 1.35] | 0.27 | 0.17 | 41% | 1.76 [1.10, 2.81] | 0.02 | 0.14 | 45% | 2.17 [0.78, 6.01] | 0.14 | 0.01 | 73% | 1.06 [0.68, 1.65] | 0.8 | 0.03 | 67% | 1.94 [0.89, 4.25] | 0.10 | 0.06 | 59% | ||
| Population | |||||||||||||||||||||
| Asians | 3 | 1.20 [0.84, 1.72] | 0.32 | 0.11 | 55% | 3.00 [1.51, 5.95] | 0.002 | 0.72 | 0% | 3.15 [1.00, 9.91] | 0.05 | 0.08 | 61% | 1.08 [0.57, 2.04] | 0.81 | 0.01 | 78% | 2.92 [1.51, 5.65] | 0.001 | 0.40 | 0% |
| Caucasian | 1 | 1.00 [0.72, 1.39] | 1.00 | – | – | 0.99 [0.50, 1.95] | 0.97 | – | – | 0.94 [0.50, 1.76] | 0.84 | – | – | 1.03 [0.62, 1.71] | 0.9 | – | – | 0.95 [0.53, 1.73] | 0.88 | – | – |
P – p value; P-H – P value of Q for heterogeneity test; I2 >50% – high heterogeneity; Random effects model was used when P value of Q for heterogeneity test P-H >0.1 or I2>50%; otherwise, fixed effect model was used; Bold was used for highlight statistical significance.
For rs699947, meta-analysis showed that the AA homozygote carriers had significantly higher risk of lung cancer, but this difference was not observed in heterozygote carriers (A vs. C: OR=1.11, 95% CI 0.92–1.35, p=0.27; AA vs. CC: OR=1.76, 95% CI 1.10–2.81, p=0.02; AA vs. AC: OR=2.17, 95% CI 0.78–6.01, p=0.14; (AA+AC) vs. CC: OR=1.06, 95% CI 0.68–1.65, p=0.80; AA vs. (AC+CC): OR=1.94, 95% CI 0.89–4.25, p=0.10) (Table 3). Subgroup analysis showed that this trend was not observed in Caucasians. In Asians, typically in Chinese, −2578C>A variant conferred increased lung cancer susceptibility, but significant association was only observed in AA homozygote carriers (A vs. C: OR=1.20, 95% CI 0.84–1.72, p=0.32; AA vs. CC: OR=3.00, 95% CI 1.51–5.95, p=0.002; AA vs. AC: OR=3.15, 95% CI 1.00–9.91, p=0.05; AA vs. (AC+CC): OR=2.92, 95% CI 1.51–5.65, p=0.001) (Table 3).
Association between rs833061 −460C>T variants and lung cancer survival
Through retrieval in databases, only 4 studies reporting the association between rs833061 −460C>T polymorphism and lung cancer survival were available. Due to significant heterogeneity in patient baseline, therapeutic methods, and data form, it was inappropriate to pool the survival results directly, so we used descriptive analysis. Survival results of the studies included are presented in Table 4. A total of 4 studies explored −460C>T polymorphism and lung cancer survival. Heist et al. [19] study based on 904 early-stage NSCLC patients found no significant association between −460C>T polymorphism and lung cancer survival. Masago et al. [13] study observed homozygote −460CC was associated with poorer survival among Japanese patients with NSCLC. The median survival time of CC and CT+TT groups were 257 (95%CI: 125–351) and 643 (95%CI: 342–801) days, respectively. The HR ratio of CC compared with CT+TT was 1.719 (95% CI 1.166–2.390, p=0.0084) (Table 4).
Table 4.
Association between rs833061 −460C>T variants and lung cancer survival.
| Trials | N | SNP variant | Reference genotype (No.) | Survival | Variant genotype (No.) | Survival | HR (95% CI) | P | Variant genotype (No.) | Survival | HR (95% CI) | P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Heist et a. 2008 | 462 | −460T/C | TT (145) | *57 (47–65) | CC (112) | *52 (41–61) | 1.25 (0.89–1.77) | 0.20 | TC (205) | *61 (53–67) | 0.99 (0.73 to 1.36) | 0.98 |
| Guan et al. 2010 | 124 | −460T/C | TT (33) | †16.0 (11.0–25.0) | CC (24) | †27.0 (10.0–36.0) | 0.67 (0.36–1.26) | 0.212 | CC+CT (91) | †21.0 (17.0–31.0) | 0.58 (0.37–0.92) | 0.022 |
| Masago et al. 2009 | 126 | −460T/C | CT+TT (115) | ‡643 (342–801) | CC (11) | ‡257 (125, 351) | 1.719 (1.166–2.390) | 0.0084 | – | – | – | – |
| de Mello et al. 2013 | 144 | −460T/C | CC (28) | †9 (5.38–12.61) | CT (79) | †11 (8.47–13.52) | 1.028 (0.331–3.196) | 0.036 | CT+TT (116) | †10 (8.14–11.86) | 1.011 (0.336–3.039) | 0.011 |
5-Year OS (%)(95% CI);
Median Survival Time (month)(95% CI);
Median Survival Time (day) (95% CI);
HR – hazard ratio; CI – confidence interval; Bold was used for highlight statistical significance.
However, Guan et al. found opposite finding in Caucasian patients. They found −460 C allele was associated with better survival among patients with locally advanced NSCLC [20]. The median survival time of TT homozygote carriers was 16.0 months (95% CI: 11.0–25.0), while that of CC homozygote carriers and both CC+CT carriers were 27.0 months (95% CI: 10.0–36.0) and 21.0 months (9% CI: 17.0–31.0), respectively. Compared with CC, HR of TT and CC+CT were 0.67 (95% CI: 0.36–1.26, p=0.212) and 0.58 (95% CI: 0.37–0.92, p=0.022), respectively (Table 4). de Mello et al. [14] recent study observed that −460T conferred increased lung cancer survival. Compared with CC (9 months, 95% CI: 05.38–12.61), the median survival time and HR of the CT group and TT+CT group was 11 months (95% CI: 8.47–13.52) and 1.028 (95% CI: 0.331–3.196, p=0.036) versus 10 months (95% CI: 8.14–11.86) and 1.011 (95% CI: 0.336–3.039, p=0.011), respectively (Table 4).
Discussion
Angiogenesis is a critical process of tumor development, growth, and metastasis [21]. As one of the essential regulators of angiogenesis, the molecular basis of the VEGF pathway has been explored by a series of studies. Previous studies have already identified several functional polymorphisms of the VEGF gene that might affect serum VEGF level, including −634G>C, −1154G>A, 936C>T, −1498C>T, −2578C>A, and −460C>T [10,22]. Previous studies also confirmed that functional genetic polymorphisms could alter mRNA or protein expression, thus generating significant influence on disease development, including cancer [23–25]. As 2 important SNPs in the promoter region of the VEGF gene, previous meta-analyses observed that −2578C>A and −460C>T polymorphisms were related to risk of gastric cancer, colorectal cancer, and breast cancer [26,27]. However, in lung cancer, previous studies reported conflicting results about the function of these 2 SNPs.
This meta-analysis involved 2602 cases and 2194 controls in 4 trials to evaluate the risk of −460C>T polymorphism in lung cancer development and 1297 patients in another 4 trials to evaluate the effect of this SNP on lung cancer survival. Results from this meta-analysis showed that −460C>T polymorphism generally had no significant effect on lung cancer risk. However, subgroup analysis found that −460TT homozygote variant might be associated with significantly increased cancer risk in Asians but not in Caucasians. In lung cancer survival, 4 trials had conflicting results. One found −460C>T had no effect on survival, 1 observed a risk-increasing effect, and the remaining 2 observed a risk-decreasing effect. As to −2578C>A polymorphism, this meta-analysis based on 530 cases and 576 controls found −2578AA homozygote carriers might have significantly higher risk of lung cancer. However, significant between-studies heterogeneity was observed. Subgroup analysis found Asian −2578A/A homozygote carriers might have significantly higher risk of lung cancer, but this trend was not observed in Caucasians. Therefore, these 2 SNPs might be potential indicators of lung cancer risk for Asians.
In lung cancer, 4 trials reported inconsistent findings in the effect of −460C>T on patient survival. This inconsistency might be caused by several factors. Firstly, the ethnic discrepancy might be caused by a series of factors, including different gene-gene interaction due to different genetic background and different gene-environmental interaction due to different lifestyles. Thus, the inconsistency could be partly explained by divergences in the genomic expression and gene-environmental interaction of different populations [14]. Secondly, there were clinical differences in trials. This study found that even within the same ethnicity, different findings were also observed in different studies. de Mello et al. [14] study observed that −460C allele conferred decreased lung cancer survival in Caucasians but Heist et al. [19] study in Caucasians failed to demonstrate any significant association between −460C allele and survival among NSCLC patients. One point worth noticing is the different therapeutic practices applied in these studies. For example, in Heist et al. [19] study, most of the 462 early-stage NSCLC patients had received resection, only 32 patients (7%) had radiation, and 3 patients (0.6%) received chemotherapy. However, Guan et al. study involved 124 patients, all treated with radiotherapy. In de Mello’s study, all patients received gefitinib therapy and all EGFR-negative patients had a platinum-based regimen [14]. Previous study observed that C variants of −460C>T (rs833061) were associated with increased VEGF promoter activity [9]. Thus, it is possible of VEGF −460 C allele might enhance tumor angiogenesis. For patients who received both radiotherapy and chemotherapy, it is possible that enhanced tumor vasculature might facilitate the delivery of chemotherapy agents to target tumor issues and also inhibit tumor radioresistance caused by radiation-induced hypoxia, leading to a better synergistic effect between chemotherapy and radiotherapy. This might be why Guan et al. [20] and de Mello et al. [14] observed better survival of lung cancer patients with −460C allele who received a combination of chemotherapy and radiotherapy or even only chemotherapy. Therefore, when planning chemotherapy and radiotherapy for patients, it is necessary to consider their VEGF genetic variances and VEGF expression.
This meta-analysis had several limitations. Firstly, the sample size for each SNP was relatively small. Thus, the statistical power of genetic effects identified might be hampered, especially findings obtained from subgroup analysis. In addition, since only secondary data was used for analysis in this study, there might be selection bias. However, the combination of primary searching and backward snowballing method helped to minimize the possible bias. Therefore, this study cautiously made conclusions on the observed associations. Secondly, significant heterogeneity in trial features was observed when analyzing the effect of the SNP on cancer survival. Thus, the results were not pooled to evaluate the overall effect. Thirdly, tumorigenesis is a complex process modulated by a series of genetic factors beyond VEGF. However, this analysis only tried to explore the effect of 2 SNPs on VEGF promoter gene, which failed to link other gene variants that may be involved in pathophysiological pathways. Therefore, larger clinical trials are required to validate the hypothesis and findings obtained of this study. Once validated, these results can be very helpful in developing tailored therapeutics for individual patients.
Conclusions
This meta-analysis showed −460TT and −2578AA homozygote variants might confer significantly increased cancer risk in Asians but not in Caucasians. The effect of −460C>T on lung cancer survival was inconsistent in the studies included, which might be related to genetic and clinical differences. These 2 SNPs might be potential indicators of lung cancer risk for Asians and could be considered when planning chemotherapy and radiotherapy for lung cancer patients.
Footnotes
Source of support: Self financing
References
- 1.Jemal A, Bray F, Center MM, et al. Global cancer statistics. Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Kim KJ, Li B, Winer J, et al. Inhibition of vascular endothelial growth factor-induced angiogenesis suppresses tumour growth in vivo. Nature. 1993;362:841–44. doi: 10.1038/362841a0. [DOI] [PubMed] [Google Scholar]
- 3.Kaya A, Ciledag A, Gulbay BE, et al. The prognostic significance of vascular endothelial growth factor levels in sera of non-small cell lung cancer patients. Respir Med. 2004;98:632–36. doi: 10.1016/j.rmed.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 4.Nieder C, Andratschke N, Jeremic B, et al. Comparison of serum growth factors and tumor markers as prognostic factors for survival in non-small cell lung cancer. Anticancer Res. 2003;23:5117–23. [PubMed] [Google Scholar]
- 5.Ferrara N. VEGF and the quest for tumour angiogenesis factors. Nat Rev Cancer. 2002;2:795–803. doi: 10.1038/nrc909. [DOI] [PubMed] [Google Scholar]
- 6.Andersen S, Donnem T, Al-Shibli K, et al. Prognostic impacts of angiopoietins in NSCLC tumor cells and stroma: VEGF-A impact is strongly associated with Ang-2. PloS One. 2011;6:e19773. doi: 10.1371/journal.pone.0019773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welch S, Spithoff K, Rumble RB, et al. Bevacizumab combined with chemotherapy for patients with advanced colorectal cancer: a systematic review. Ann Oncol. 2010;21:1152–62. doi: 10.1093/annonc/mdp533. [DOI] [PubMed] [Google Scholar]
- 8.Vincenti V, Cassano C, Rocchi M, et al. Assignment of the vascular endothelial growth factor gene to human chromosome 6p21.3. Circulation. 1996;93:1493–95. doi: 10.1161/01.cir.93.8.1493. [DOI] [PubMed] [Google Scholar]
- 9.Stevens A, Soden J, Brenchley PE, et al. Haplotype analysis of the polymorphic human vascular endothelial growth factor gene promoter. Cancer Res. 2003;63:812–16. [PubMed] [Google Scholar]
- 10.Koukourakis MI, Papazoglou D, Giatromanolaki A, et al. VEGF gene sequence variation defines VEGF gene expression status and angiogenic activity in non-small cell lung cancer. Lung Cancer. 2004;46:293–98. doi: 10.1016/j.lungcan.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 11.Zhai R, Liu G, Zhou W, et al. Vascular endothelial growth factor genotypes, haplotypes, gender, and the risk of non-small cell lung cancer. Clin Cancer Res. 2008;14:612–17. doi: 10.1158/1078-0432.CCR-07-1655. [DOI] [PubMed] [Google Scholar]
- 12.Lee SJ, Lee SY, Jeon HS, et al. Vascular endothelial growth factor gene polymorphisms and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:571–75. doi: 10.1158/1055-9965.EPI-04-0472. [DOI] [PubMed] [Google Scholar]
- 13.Masago K, Fujita S, Kim YH, et al. Effect of vascular endothelial growth factor polymorphisms on survival in advanced-stage non-small-cell lung cancer. Cancer Sci. 2009;100:1917–22. doi: 10.1111/j.1349-7006.2009.01253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Mello RA, Ferreira M, Soares-Pires F, et al. The impact of polymorphic variations in the 5p15, 6p12, 6p21 and 15q25 Loci on the risk and prognosis of portuguese patients with non-small cell lung cancer. PloS One. 2013;8:e72373. doi: 10.1371/journal.pone.0072373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun SF, Huang DB, Cao C, et al. Polymorphism of VEGF-460C/T associated with the risk and clinical characteristics of lung cancer in Chinese population. Med Oncol. 2013;30:410. doi: 10.1007/s12032-012-0410-x. [DOI] [PubMed] [Google Scholar]
- 16.Deng ZC, Cao C, Yu YM, et al. Vascular endothelial growth factor −634G/C and vascular endothelial growth factor −2578C/A polymorphisms and lung cancer risk: a case-control study and meta-analysis. Tumour Biol. 2014;35:1805–11. doi: 10.1007/s13277-013-1241-x. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Liang J, Liu X, et al. Correlation of polymorphisms of the vascular endothelial growth factor gene and the risk of lung cancer in an ethnic Han group of North China. Exp Ther Med. 2012;3:673–76. doi: 10.3892/etm.2012.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang J, Yu X, Liu X, et al. Vascular endothelial growth factor polymorphisms and risk of lung cancer. Chinese-German Journal of Clinical Oncology. 2009;8:269–72. [Google Scholar]
- 19.Heist RS, Zhai R, Liu G, et al. VEGF polymorphisms and survival in early-stage non-small-cell lung cancer. J Clin Oncol. 2008;26:856–62. doi: 10.1200/JCO.2007.13.5947. [DOI] [PubMed] [Google Scholar]
- 20.Guan X, Yin M, Wei Q, et al. Genotypes and haplotypes of the VEGF gene and survival in locally advanced non-small cell lung cancer patients treated with chemoradiotherapy. BMC Cancer. 2010;10:431. doi: 10.1186/1471-2407-10-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicklin DJ, Ellis LM. Role of the vascular endothelial growth factor pathway in tumor growth and angiogenesis. J Clin Oncol. 2005;23:1011–27. doi: 10.1200/JCO.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Closas M, Malats N, Real FX, et al. Large-scale evaluation of candidate genes identifies associations between VEGF polymorphisms and bladder cancer risk. PLoS Genet. 2007;3:e29. doi: 10.1371/journal.pgen.0030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 24.Yang R, Burwinkel B. A bias in genotyping the miR-27a rs895819 and rs11671784 variants. Breast Cancer Res Treat. 2012;134:899–901. doi: 10.1007/s10549-012-2140-3. [DOI] [PubMed] [Google Scholar]
- 25.Flego V, Ristic S, Devic Pavlic S, et al. Tumor necrosis factor-alpha gene promoter –308 and −238 polymorphisms in patients with lung cancer as a second primary tumor. Med Sci Monit. 2013;19:846–51. doi: 10.12659/MSM.889554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao C, Ying T, Fang JJ, et al. Polymorphism of vascular endothelial growth factor −2578C/A with cancer risk: evidence from 11263 subjects. Med Oncol. 2011;28:1169–75. doi: 10.1007/s12032-010-9613-1. [DOI] [PubMed] [Google Scholar]
- 27.Wang K, Liu L, Zhu ZM, et al. Five polymorphisms of vascular endothelial growth factor (VEGF) and risk of breast cancer: a meta-analysis involving 16,703 individuals. Cytokine. 2011;56:167–73. doi: 10.1016/j.cyto.2011.06.018. [DOI] [PubMed] [Google Scholar]