Abstract
Background
Caviomorph rodents, some of the oldest Leishmania spp. hosts, are widely dispersed in Brazil. Despite both experimental and field studies having suggested that these rodents are potential reservoirs of Leishmania parasites, not more than 88 specimens were analyzed in the few studies of natural infection. Our hypothesis was that caviomorph rodents are inserted in the transmission cycles of Leishmania in different regions, more so than is currently recognized.
Methodology
We investigated the Leishmania infection in spleen fragments of 373 caviomorph rodents from 20 different species collected in five Brazilian biomes in a period of 13 years. PCR reactions targeting kDNA of Leishmania sp. were used to diagnose infection, while Leishmania species identification was performed by DNA sequencing of the amplified products obtained in the HSP70 (234) targeting. Serology by IFAT was performed on the available serum of these rodents.
Principal findings
In 13 caviomorph rodents, DNA sequencing analyses allowed the identification of 4 species of the subgenus L. (Viannia): L. shawi, L. guyanensis, L. naiffi, and L. braziliensis; and 1 species of the subgenus L. (Leishmania): L. infantum. These include the description of parasite species in areas not previously included in their known distribution: L. shawi in Thrichomys inermis from Northeastern Brazil and L. naiffi in T. fosteri from Western Brazil. From the four other positive rodents, two were positive for HSP70 (234) targeting but did not generate sequences that enabled the species identification, and another two were positive only in kDNA targeting.
Conclusions/Significance
The infection rate demonstrated by the serology (51.3%) points out that the natural Leishmania infection in caviomorph rodents is much higher than that observed in the molecular diagnosis (4.6%), highlighting that, in terms of the host species responsible for maintaining Leishmania species in the wild, our current knowledge represents only the “tip of the iceberg.”
Author Summary
Leishmaniasis is a major public health problem expanding in Brazil and one of the reasons is that we still have poor knowledge of some aspects of the biology and epidemiology of Leishmania species, including the role of wild mammals. Caviomorph rodents, some of the oldest Leishmania spp. hosts, are widely dispersed in Brazil and reported as potential reservoirs of Leishmania parasites. Spleen fragments of 373 brazilian caviomorph rodents from 20 species were investigated for Leishmania infection. The molecular algorithm proposed to diagnose the infection associate the sensitivity of a molecular target with multiple copies with the specificity of another marker with discriminatory taxonomic ability between species. These demonstrated their usefulness in identifying most of the parasite species infecting the rodents, including the description of species in previously unknown hosts and in areas not previously included in their known distribution, such as L. shawi in Thrichomys inermis from Northeastern Brazil and L. naiffi in T. fosteri from Western Brazil. Although the percent of infection by molecular diagnosis was 4.6%, the serology demonstrated that about 51% of them had been exposed to Leishmania parasites pointing that caviomorph rodents are inserted in enzootic cycles of Leishmania, to a higher extent than currently recognized.
Introduction
More than 30 species within the Leishmania genus (Trypanosomatidae, Trypanosomatida) are recognized, including both extremely specific as well as generalist species transmitted by a variety of Phlebotomine vectors worldwide [1]–[2] In fact, several Leishmania species are multi-host parasites that infect mammal species of different orders, including humans [3]–[4]. The diversity of Leishmania species, mammal hosts and environments in which the transmission occurs reveals a complex system. Leishmaniasis has an important impact on public health since it results in a spectrum of debilitating diseases, which can progress to very severe, even fatal cases. In Brazil, human cutaneous leishmaniasis is associated with at least six different species of the subgenus Leishmania (Viannia) besides Leishmania (Leishmania) amazonensis, while the visceral form is exclusively associated with Leishmania (L.) infantum [5]–[7].
Although usually interpreted as such, the description of Leishmania DNA in a given mammal species is not sufficient to consider this species a reservoir host. Reservoir is better defined not as a single species, but as an assemblage of species responsible for the long lasting maintenance of a parasite in a given environment [8]. In reservoir systems, each species of mammal host plays a unique role in the maintenance of the parasite, which means that these systems should always be considered in a restricted spatio-temporal scale, particular to each site and each moment [9]. Failures in interrupting human transmission and preventing new epidemics are probably related to the lack of knowledge of various aspects of the natural transmission cycles of these parasites. In this sense, the involvement of synanthropic hosts, such as caviomorph rodents and their potential to act as reservoirs cannot be ignored.
Although the diagnosis of Leishmania sp. infection in mammal tissues through molecular assays has been conducted by several groups, the identification of the Leishmania species is still a great challenge [10]–[12]. The parasite isolation in wild mammals is complex to due to the difficulties in performing aseptic culture during field expeditions. Besides, the usually observed low parasite load and irregular distribution of parasites among host tissues impair even more the isolation efficiency [13]–[17]. To overcome such limitations, molecular approaches have been developed and applied aiming to detect and identify Leishmania species directly in biological samples. The advantage of molecular approaches based on PCR is that they combine high sensitivity for direct detection of the infecting parasites in various human, animal and sand fly tissues, with species specificity [18]. The PCR followed by either Restriction Fragment Polymorphism (RFLP) or DNA sequencing of distinct targets have already been employed in biological samples. Not all of them are, however, useful for identification at the species level. Molecular targets directed to the conserved region of the kinetoplast minicircles of Leishmania are the most used for diagnosis due to their sensitivity (which is related to the high number of copies in a single parasite (∼10.000) but detects only the subgenerum level. The PCR-RFLP of the internal transcribed spacer 1 (ITS1) is the assay commonly used for direct detection and identification of Leishmania species in the Old World, but for Brazilian species presents a lower resolution. Among the targets that result in intraspecific variability, the Heat Shock Protein of 70 kDa is encoded by a polymorphic gene which has regions allowing diagnosis of the Leishmania species circulating in Brazil [19].
Rodents comprise more than two hundred species distributed in many different habitats. In fact, in nature, we found semi-aquatic, terrestrial and semi-fossorial species [20]. This trait is probably the main reason explaining their exposure to the different transmission cycles of several species of Leishmania in the wild [8]. The South American rodents are divided into two sub orders (Sciurognathi and Hystricognathi). The first Caviomorph rodents (Rodentia: Hystricognathi) arrived in the Americas about 35mya (million years ago), much earlier than other rodent groups [21]. Along with the first caviomorph migrants, during the Oligocene, it has already been proposed that new species of the Leishmania (Leishmania) subgenus may have arrived and subsequently diversified in the native mammal fauna [22].
Nowadays, caviomorph rodents are widely distributed in Brazil and contribute significantly to the animal biomass of these habitats. They are also found in peridomestic environments that report the circulation of Leishmania parasites among other species of mammals, humans and/or dogs [23]–[26]. To date in Brazil, there are few studies of natural Leishmania infection in caviomorph rodents and not more than 88 specimens were analyzed [27]–[29]. Furthermore, experimental studies pointed to the putative role of two caviomorph species as Leishmania reservoirs: Thrichomys laurentius infected by L. infantum and L. braziliensis [17] and Proechimys semispinosus infected with L. infantum [30]. In this sense, the analysis of different caviomorph rodent species from different regions and collected at different periods of time, may define the role of these taxa in the reservoir host system of Leishmania sp. In the present study we investigate the infection by Leishmania spp. in 373 caviomorph rodents from 20 different species and discuss the role of this group in the maintenance of the transmission cycles of Leishmania spp. in different regions of Brazil.
Materials and Methods
Animal samples
We evaluated spleen fragments from caviomorph rodents (n = 373) collected in five out six Brazilian biomes between 1999 and 2012 (S1 Table). These animals were captured using Tomahawk and Shermann “live-traps” during studies conducted by our laboratory [31]–[33]. In these studies, whenever the euthanasia of animals were realized, usually for taxonomic identification and/or diagnosis of parasite's infection, spleen fragments were collected in plastic tubes containing ethanol and stored at −20°C.
Molecular diagnosis
All tissue fragments were re-hydrated with Nuclease-free water and the DNA extraction was performed using the Wizard Genomic DNA Purification Kit (Promega, Madison, USA) according to the manufacturer's recommendations. Positive and negative controls were derived from fragments of spleen and liver from infected (Leishmania braziliensis–IOC-L2483) and non-infected hamsters provided by the animal facilities of the Oswaldo Cruz Foundation. PCR were conducted using the pureTaq Ready-To-Go PCR beads (Amersham Biosciences, Buckinghamshire, UK) and primers directed to the conserved region of the Leishmania -kDNA minicircle, as follows: forward: 5′ – GGGAGGGGCGTTCTGCGAA-3′ and reverse: 5′ – GGCCCACTAT ATTACACCAACCCC – 3′ [17]. The PCR products were visualized after electrophoresis on 8% polyacrylamide gel and silver staining using a specific kit (DNA Silver Staining, GE Healthcare). Positive samples were submitted to a new PCR directed to a fragment of 234 bp of the hsp70 gene, using the following primers (5′ - GGA CGA GAT CGA GCG CAT GGT - 3′) and (5′- TCC TTC GAC GCC TCC TGG TTG - 3′) [19]. The PCR amplifications were performed in a final volume of 50 µL containing 5 µL of DNA, 0.2 pmol of each primer, 0.2 µM dNTPs, 1.5 mM MgCl2 and 1 U GoTaq®DNA polymerase (Promega). The PCR assays used the following amplification cycle: 94°C for 5 min followed by 30 cycles of 94°C for 30 sec, 63°C for 1 min and 72°C for 1 min and a final extension at 72°C for 10 min. To increase the number of DNA copies, products obtained in the first reaction were submitted to a second PCR with the same primers and conditions described.
The PCR products obtained for HSP70 (234) targeting and the products positive only in kDNA targeting were purified using the Wizard SV Gel kit and PCR Clean-up System kit (Promega). The both products were sequenced with the same primers used for the PCR assay using the ABI PRISM BigDyeTerminator v3.1 Cycle Sequencing Kit. Sequencing was performed on an automated DNA sequencer (ABI PRISM®BigDye™ Terminator Cycle Sequencing) at the Genomic Platform – DNA Sequencing (PDTIS – FIOCRUZ). Consensus sequences were aligned and edited with the BioEdit Version v7.1.11 [34] and Phred/Phrap/Consed package Version: 0.020425.c [35] from two forward and two reverse strands. Sequences with Phred values below ten over their extent were discarded and only sequence segments with values above twenty were used for contig construction. Contigs from all samples were manually assembled and aligned in MEGA4 [36]. For that samples that not generated good-quality consensus sequence, the PCR reaction and the DNA sequencing was repeated two or more times until we obtain sequence segments equal to the previously established criteria. Species identification was performed by similarity analysis obtained by Alignment Search Tool (BLAST) algorithm hosted by NCBI, National Institute of Health, USA (http://www.ncbi.nlm.nih.gov), against sequences available on GeneBank after calculation of the statistical significance of matches. Besides, we also compared our sequences with a panel of sequences for HSP70 (234) gene obtained from representative strains of different Leishmania species circulating in Brazil available in the Leishmania collection of the Oswaldo Cruz Institute/CLIOC (Table 1). This table was allowed the identification of the positions of the polymorphic site that differentiates the parasite species.
Table 1. Main polymorphisms along the HSP70 (234) sequences that allow the distinction of Leishmania species from reference strains.
| Leishmania Species | Position of Polymorphyc Site | ||||||||||||||||||||||||||||||||||
| 19 | 22 | 23 | 24 | 35 | 36 | 39 | 45 | 47 | 48 | 58 | 70 | 78 | 79 | 113 | 116 | 117 | 122 | 145 | 148 | 150 | 156 | 158 | 160 | 161 | 163 | 168 | 170 | 171 | 174 | 181 | 182 | 183 | 185 | 201 | |
| L. (V) braziliensis | T | G | T | C | C | A | C | A | A | T | G | G | G | T | G | G | C | A | G | G | G | G | A | C | G | G | A | T | C | C | C | A | C | G | A |
| L. (V) shawi | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | G | . | . | . | . | . | . | . | . | G | . | . | . |
| L. (V) guyanenis | . | . | . | . | . | . | . | . | . | . | . | . | . | . | A | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| L. (V) naiffi | . | . | . | . | . | . | . | . | . | . | . | . | . | C | . | . | . | . | . | . | . | . | . | . | A | . | C | . | . | . | . | G | . | . | . |
| L. (V) lainsoni | . | . | . | . | . | . | . | . | . | . | . | . | C | . | A | T | . | . | . | . | . | A | G | . | . | . | . | . | . | . | . | . | . | . | . |
| L. (L) infantum | C | . | A | T | G | C | A | G | G | C | C | A | . | C | C | . | G | T | C | T | . | A | G | . | A | . | . | A | A | A | . | G | T | A | G |
| L. (L) amazonensis | C | A | A | T | G | C | A | . | G | C | C | A | . | . | C | . | G | T | C | C | C | A | T | G | A | A | . | A | A | A | G | G | . | . | G |
Serological diagnosis
The immunofluorescence assay (IFAT) was performed on 130 available serum samples from the evaluated rodents, as described by Camargo (1966) [37]. A mixture of promastigotes from cultures of L. (V.) braziliensis (IOC/L566) and L. (L.) infantum (IOC/L579) obtained from CLIOC was used as antigen. In order to detect possible cross-reaction with Trypanosoma cruzi, sera were also tested using reference strains of T. cruzi (TcI M000/BR/1974/F and TcII MHOM/BR/1950/Y). The reactions were conducted using an in-house intermediary antibody anti-Thrichomys serum produced in rabbits [38]. The reaction was visualized using a commercial anti-rabbit IgG-FITC (Sigma-Aldrich, St. Louis, USA). Rodents that displayed serological titers 1/10 and 1/20 for Leishmania spp. infection were considered positive only if these titers were equal or higher than the titers observed in the same rodent for T. cruzi infection. Rodents that displayed Leishmania serological titers equal or higher than 1∶40 were considered positive independent of T. cruzi results.
Ethics statement
All the procedures carried out with these animals were authorized by the Brazilian Institute of Environment and Renewable Natural Resources (IBAMA) and followed protocols approved by the Ethics Committee of Animal Use Fiocruz (P0007-99, P0179-03; P0292/06; L0015-07).
Results
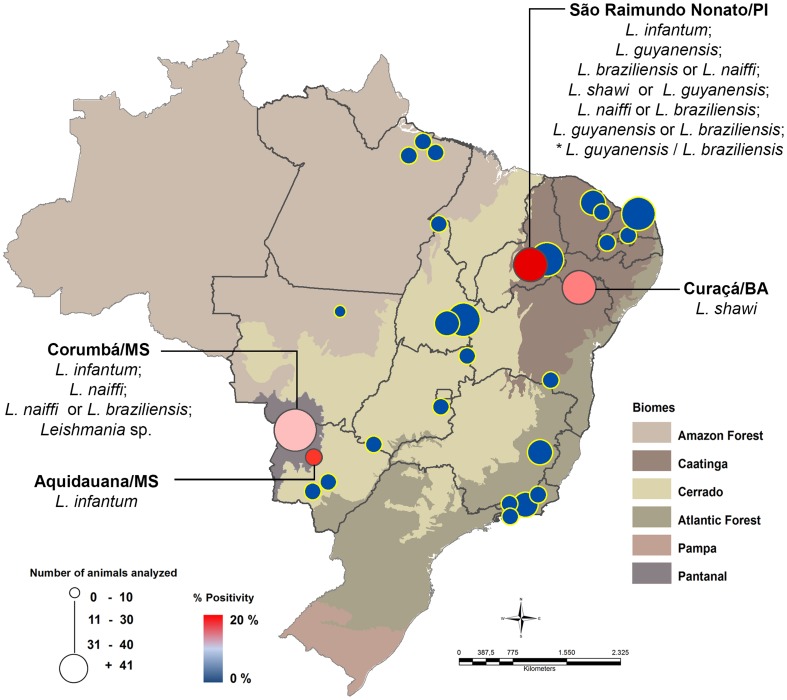
We found 17 caviomorph rodents (4.6%) positive in the PCR directed to Leishmania sp. kDNA. From these, 15 samples were also positive when tested for HSP70 (234) primers, two directly after the PCR and 13 only after the re-amplification of the product obtained in the first PCR reaction (Fig. 1). DNA sequencing analyses allowed the identification of −4 species of the subgenus Leishmania (Viannia) and −1 species of the subgenus Leishmania (Leishmania) in samples of 13 caviomorph rodents (Fig. 2, Table 2). In five situations, the DNA sequence analysis revealed identity values above 95% for more than one Leishmania species and we opted not to define one of them as the etiological agent. A single sample (one Thrichomys laurentius from Piauí) displayed different DNA sequences that presented high values of similarity with distinct species of Leishmania in two consecutive reactions. This was considered as result of a hybrid population or mixed infection. Leishmania infection was observed in five caviomorph rodent species captured in two municipalities belonging to Pantanal and in two belonging to Caatinga biomes (Fig. 2). In the Pantanal, 7 animals were positive for Leishmania spp and the identification of the Leishmania species was possible in 4 of them: one T. fosteri captured in Corumbá, was found infected by L. (V). naiffi while L. (L). infantum was found infecting one Dasyprocta azarae and two Clyomys laticeps in Aquidauna and Corumbá, respectively. Also in Corumbá, we were unable to identify the Leishmania species infecting one T. fosteri because the analysis of the parasite DNA sequence revealed similarity with two Leishmania species (L. (V). naiffi and L. (V). braziliensis).
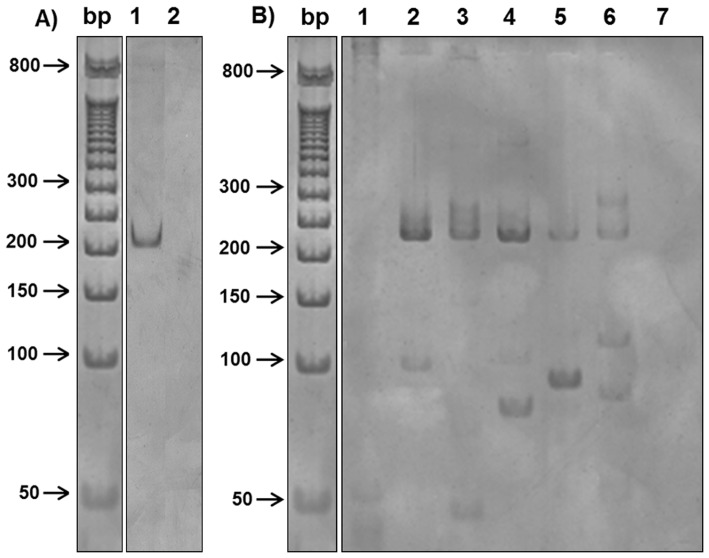
Figure 1. Illustrative representation of HSP 70 (234) amplification for Leishmania sp. before and after re-amplification of the PCR products.
(A) PCR products of first amplification of HSP 70 (234) targeting analyzed by electrophoresis polyacrylamide gel stained with silver. Lanes: bp. molecular-weight marker (50 bp DNA ladder); 1. Infected Thrichomys laurentius from São Raimundo Nonato/PI; 2. Negative control of PCR reaction. (B) PCR products of re-amplification of the product obtained in the first PCR reaction. Lanes: bp. molecular-weight marker (50 bp DNA ladder); 1. Negative Thrichomys fosteri from Corumbá/MS (Positive only in kDNA); 2–5. Infected T. laurentius from São Raimundo Nonato/PI; 6. Infected T. fosteri from Corumbá/MS; 7. Negative control of PCR after re-amplification.
Figure 2. Map of the distribution of Leishmania species infecting caviomorph rodents in Brazil.
Blue markers indicate municipalities where caviomorph rodents were collected, but were all negative in the molecular assay. Red markers indicate municipalities with animals positive for Leishmania infection. * Sample with high value between distinct species of Leishmania was considered as a result of hybrid or mixed infection.
Table 2. Infected caviomorph rodents and their respective Leishmania species identified by the analysis of similarity between the DNA sequences from the PCR products targeting HSP70 (234) and available sequences from GeneBank.
| Locality | Positive mammal | Leishmania identification | Max Score | Identity (%) |
| Bahia | Thrichomys inermis | Leishmania (V). shawi | 422 | 99 |
| Mato Grosso do Sul | Thrichomys fosteri | Leishmaia (V). naiffi | 390 | 99 |
| Dasyprocta azarae | Leishmania (L). infantum | 185 | 88 | |
| Clyomys laticeps | Leishmania (L). infantum | 427 | 99 | |
| Clyomys laticeps | Leishmania (L). infantum | 361 | 93 | |
| Thrichomys fosteri | Leishmaia (V). naiffi OR | 435 | 100 | |
| Leishmania (V). braziliensis | 424 | 99 | ||
| Piauí | Thrichomys laurentius | Leishmania (V). guyanensis | 433 | 100 |
| Thrichomys laurentius | Leishmania (L). infantum | 303 | 97 | |
| Thrichomys laurentius | Leishmania (V). braziliensis OR | 422 | 99 | |
| Leishmaia (V). naiffi | 422 | 99 | ||
| Thrichomys laurentius | Leishmania (V). shawi OR | 390 | 95 | |
| Leishmania (V). guyanensis | 385 | 95 | ||
| Thrichomys laurentius | Leishmaia (V). naiffi OR | 435 | 100 | |
| Leishmania (V). braziliensis | 424 | 99 | ||
| Thrichomys laurentius | Leishmaia (V). naiffi OR | 422 | 99 | |
| Leishmania (V). braziliensis | 411 | 98 | ||
| Thrichomys laurentius * | Leishmania (V). guyanensis | 416 | 100 | |
| Leishmania (V). braziliensis | 416 | 99 |
* Sample with high value between distinct species of Leishmania was considered as a result of hybrid or mixed infection.
From Caatinga biome, in the municipality of Curacá/Bahia, one Thrichomys inermis was found infected by L. (V). shawi. In São Raimundo Nonato/Piauí, two T. laurentius were found infected, respectively, by L. (V). guyanensis and L. (L.) infantum. In four T. laurentius, the identification of the Leishmania species was not possible because the parasite DNA sequencing analyses showed high similarity for two Leishmania species (Table 2). The specific diagnosis in the seventh T. laurentius was only possible after two attempts and showed the presence of two Leishmania species, L. (V). braziliensis and L. (V). guyanensis (S2 Table). This was interpreted as the result of a mixed infection and/or a possible hybrid of them.
The prevalence of antibodies against Leishmania sp. in IFAT was 51.3% (59/115). In 15 samples the results were inconclusive and were therefore not included in the final analysis. Among the samples analyzed (S1 Table), 56 animals were positive in IFAT but negative by PCR. From these, the majority of them (n = 34) displayed titers equal or higher than 1∶20 in the IFAT. Of the six animals positive for Leishmania kDNA, three were also positive by IFAT. Other 53 rodents were negative for both tests.
Discussion
A clear expansion of leishmaniasis around the country has been observed in the last decades [23], [39]–[41]. From the different reasons already proposed to explain this expansion, a common point is the recognition that we still have poor knowledge of some aspects of the biology and epidemiology of Leishmania species, which, in the end, result in inefficient control strategies. The mammalian hosts of most Leishmania species are still poorly understood, which reinforces the need for studies that verify the distribution of these parasites in other mammalian taxa beyond those classically reported as reservoirs. Caviomorph rodents comprise an enormous group of species that exploit different habitats, and also include some species already domesticated by humans, such as the chinchilla and the guinea pig (Cavia spp.) [42].
Among the rodent species found infected in this study, only species from genus Dazyprocta and Thrichomys were already found infected by Leishmania spp. in Brazil. Dasyprocta sp., was found infected by L. guyanensis in Pará state [27] while T. apereoides, was founded infected by L. braziliensis, L. guyanensis, and L. amazonensis in Minas Gerais state [28]–[29]. Morevoer, other Thrichomys species were reported as potential reservoirs of trypanosome species in different regions of Brazil [33], [43]. We presented here the first description of Leishmania infection in Clyomys laticeps, a rodent species already found naturally infected by T. evansi and T. cruzi in Pantanal region” [33], [44].
Thrichomys inermis was the most abundant small mammal captured during the field expedition in Curaçá/Bahia, which points to its importance as a potential reservoir host. We described here for the first time, L. shawi in rodents, as well as this Leishmania species outside the Amazon region. Up to know, L. shawi is known to be transmitted only by Lutzomyia whitmani associated to arboreal and/or scansorial mammals (primates, sloths and coatis) suggesting a transmission cycle restricted to this forest strata [7], [45]. In a first view, our findings led to the suggestion that this transmission could also occur near the ground, since T. inermis is a terrestrial rodent that only rarely explores the understory strata [20]. However, we recently revealed through camera traps another species of this genus (T. fosteri) invading a coati nest about 12 meters high in the Pantanal region (Guilherme Mourão, personal communication). This finding reflects the lack of knowledge about the biology of some of the most widely distributed mammal species and the risk of incorrectly interpreting the parasite transmission cycle based on misconceptions of the biology of their mammal hosts and/or vectors.
American visceral leishmaniasis (AVL) is widely distributed in the state of Mato Grosso do Sul. Currently, the Aquidauana municipality displays an increasing number of human and canine cases in the urban area [46] and here we report for the first time the presence of L. infantum in sylvatic areas of this municipality. It is worth mentioning that although Dazypropcta azarae is a wild rodent species, these animals are frequently found in urban environments such as parks and woodlands. In these localities, these animals have no natural predators and are usually provided with food by local visitors, resulting in propitious conditions for reproduction. The presence of synanthropic L. infantum hosts in areas of intense transmission is a factor usually not considered in the epidemiology of VL. Nevertheless, we can not rule out the participation of other wild hosts in this transmission cycle, which is a complicating factor for the success of control strategies.
Also in the Pantanal region, the Corumbá municipality is endemic for AVL, recording dog's and human's cases [47]–[48]. In this municipality, there were conducted four expeditions between 2003 and 2009, and Leishmania infected rodents were found in all of them. The captures occurred in two farms that present different land uses. The Alegria Farm has livestock activities whereas the Nhumirim Farm is the center for scientific studies and presents large conservation areas. Two Clyomys laticeps, each one captured in one farm were found infected by L. infantum. This is especially important in the Alegria Farm where the interaction between wild and domestic mammals is common. All other infected rodents were captured in the Nhumirim Farm, pointing out that richness of biodiversity, as encountered in preserved areas, also reflect parasite diversity. At least one Thrichomys fosteri were found infected by Leishmania naiffi and this represents the first description of Leishmania naiffi in rodents and also of this Leishmania species outside the Amazon region [49]–[50]. Although we found no reports of the known L. naiffi vectors in the Mato Grosso do Sul state, their presence cannot be discarded or, alternatively, this transmission is being maintained by other phlebotomine species [51]. Another T. fosteri was founded infected by L. naiffi although the sequence of the amplified products revealed that L. braziliensis could not be discarded as etiologic agent of this infection. Indeed, these species differ in solely four nitrogenous bases at positions 79, 161, 168 e 182 when sequenced to target HSP70 (234) (Table 1).
Our data from São Raimundo Nonato/Piauí revealed the higher diversity of Leishmania species in a single host species, Thrichomys laurentius. This diversity could be observed in two consecutive years of expeditions. T. laurentius are a wild mammal species that has synanthropic habits and displayed high relative abundance in both expeditions. These rodents displayed high prevalence of infection that can potentially be considered a risk of infection not only for other wildlife populations, but also for domestic animals and men that expose themselves in areas of transmission. One T. laurentius was found infected by L. infantum. Indeed, the Piauí state is endemic for AVL with records of infection in marsupials, dogs and wild carnivores [52]–[55]. Another T. laurentius was found infected by L. guyanensis. This parasite species is commonly found in the Amazon region and known to be transmitted mainly by Lutzomyia umbratilis, a anthropophilic species that is generally found in understory strata [56].
Despite the difficulties, specific diagnosis is crucial to better understand the complex network of transmission of Leishmania species, which was achieved in the above mentioned situations. Ideally, studies that focus on the description of Leishmania reservoirs in a given area should be performed in long-termed studies. When not possible, punctual studies may also be quite informative, but it is imperative that this must to be analysed within a broad methodological approach that includes parasitological, serological and molecular diagnoses in different tissues. These factors will define the role of a given mammal host as a reservoir of Leishmania parasites. Nevertheless, this study design not always is possible due to inherent difficulties in working with wild mammals in the field. The molecular algorithm proposed here sought to combine the sensitivity of a molecular target and the specificity of DNA sequencing analysis of a locus. In fact, PCR targeted to the conserved region of kDNA has proved to be the most sensitive, but only allows the identification up to the subgenus level [15], [57]. Although less sensitive to diagnose the infection, the variable region of HSP 70 enabled the identification of the parasite species [19]. It is worth mentioning that in most cases positive reactions were only visualized after the re-amplification of the sequences obtained in the first reaction. This is the first time this approach is applied directly in tissue samples from wild mammals.
Also in São Raimundo Nonato, the analysis of the molecular identification of four animals resulted in similar sequences for more than one Leishmania species. One T. laurentius showed similar results for L. shawi and L. guyanensis, an expected result since both Leishmania species belong to the same complex and differ only in two bases, in HSP70 (234) sequence (Table 1). Other three T. laurentius showed similar results for L. braziliensis e L. naiffi (Table 2). We also obtained a divergent result after repeating the PCR reaction of one T. laurentius sample, which was interpreted as a result of a mixed infection or a hybrid between L. braziliensis and L. guyanensis. These five situations of inconclusive species identification reflect an inherent limitation of the employed methodology. On the other hand, it highlights how complex the identification of Leishmania species is (especially when sylvatic samples are used) and reflects how challenging is the establishment of a universal methodology, unique for diagnosis of the infection. In nature, the evolutionary success of Leishmania populations depends on their ability to multiply and be transmitted in different microenvironments (vertebrate and invertebrate hosts), which are both influenced by the habitat where they live. This process of evolution species is a dynamic, mutable and still poorly known phenomenon. Our attempt to describe the parasitic populations resulting from such complex interactions into discrete (not continuous) taxonomic units, although epidemiologically important, is always subject to findings like these. These should not be interpreted as an indicative to search for new discriminatory molecular markers, but instead as a natural report of the complexity that exists in Leishmania taxa.
The serological survey showed that 51.3% of the caviomorph rodents evaluated had become exposed to Leishmania parasites, and, therefore, are expected to be infected. However, we found only 4.6% of positive rodents through the molecular analysis of spleen fragments. Leishmania sp. has non-uniform distribution in tissues of vertebrate hosts, and may be present in other fragments of the spleen as well as in different tissues as skin and/or liver. The three animals that were positive for the molecular diagnosis, but negative for IFAT may have been caught in an initial phase of infection when there was not yet production of detectable IgG in serological tests. Another factor, although less known in wild hosts, may be an inability (permanent or temporary) of some individuals to produce detectable antibodies in serological assays. The rate of infection demonstrated by the serology points out that the natural Leishmania infection in caviomorph rodents is much higher than that observed in the molecular diagnosis. Our results based on PCR of spleen fragments and serology reflects only the “tip of the iceberg” highlighting that the knowledge about the epidemiology of different Leishmania species that infect caviomorph rodents is just at its beginning. The diversity of Leishmania species found infecting different caviomorph rodent species reflects the dynamism and complexity of the transmission cycles of these parasites in nature.
Supporting Information
Origin of the analyzed caviomorph rodents, which were collected in Brazil between 1999 and 2012.
(DOCX)
Similarity levels observed by BLAST analysis among the DNA sequencing obtained for the target HSP70 (234) in the infected caviomorph rodents and available sequences from GeneBank with their respective accession numbers.
(DOCX)
Acknowledgments
The authors are thankful to Vitor Rademaker Martins and the staff of the Laboratory of Tripanosomatids Biology and the Laboratory of Biology and Parasitology of Wild Mammal Reservoirs for capture and supply of the spleen fragments from some caviomorph rodents. We also offer thanks to Dr. Cibele Bonvicino for taxonomic identification of some rodents; Dr. Vera Bongertz for insightful comments; and Rodrigo Mexas and Dr. Samanta Xavier for being helpful in the development of the figures. The authors also thank Daniela Ribeiro Vallim da Silva and Regina Coeli Rathunde Matos for assistance with the molecular diagnosis and the Genomic Platform – DNA Sequencing (PDTIS – FIOCRUZ).
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
FAPERJ and CNPq supported this study through research grants provided to EC, AMJ and ALRR, while RCP had scholarship supported by CAPES. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fraga J, Montalvo AM, De Doncker S, Dujardin JC, Van der Auwera G (2010) Phylogeny of Leishmania species based on the heat-shock protein 70 gene. Infect Genet Evol 10: 238–245. [DOI] [PubMed] [Google Scholar]
- 2. Bañuls AL, Hide M, Prugnolle F (2007) Leishmania and the leishmaniases: a parasite genetic update and advances in taxonomy, epidemiology and pathogenicity in humans. Adv Parasitol 64: 1–109. [DOI] [PubMed] [Google Scholar]
- 3. Ashford RW (1996) Leishmaniasis reservoir and their significance in control. Clin Dermat 14: 523–32. [DOI] [PubMed] [Google Scholar]
- 4. Rotureau B (2006) Ecology of the Leishmania species in the Guianan ecoregion complex. Am J Trop Med Hyg 74: 81–96. [PubMed] [Google Scholar]
- 5. Grimaldi Jr G, Tesh RB, McMahon-Pratt D (1989) A review of the geographic distribution and epidemiology of leishmaniasis in The New World. Am J Trop Med Hyg 41: 687–725. [DOI] [PubMed] [Google Scholar]
- 6.Ministério da Saúde (2006) Manual de vigilância e controle da Leishmaniose Visceral. 1st Edition. Brasília, DF; Brazil. 120 p. [Google Scholar]
- 7.Ministério Da Saúde (2010) Manual de vigilância e controle da Leishmaniose Tegumentar Americana. 2nd Edition. Brasília, DF; Brazil. 182 p. [Google Scholar]
- 8.Roque ALR, Jansen AM. Hospedeiros e reservatórios de Leishmania sp. e sua importância na manutenção dos Ciclos de Transmissão nos ambientes silvestre e Sinantrópico. In: Conceição-Silva F, De-Simone SG, Alves CR, Porrozzi R. Questões atuais em leishmanioses do continente americano. Editora Fiocruz. In press. [Google Scholar]
- 9.Roque ALR, Jansen AM (2010) Reservatórios do Trypanosoma cruzi e sua relação com os vetores. In: Vetores da doença de Chagas no Brasil. Editora Fiocruz, Rio de Janeiro. [Google Scholar]
- 10. Reithinger R, Dujardin JC (2007) Molecular diagnosis of leishmaniasis: Current status and future applications. J Clin Microbiol 45: 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goto H, Lindoso JA (2010) Current diagnosis and treatment of cutaneous and mucocutaneous leishmaniasis. Expert Rev Anti Infect Ther 8: 419–433. [DOI] [PubMed] [Google Scholar]
- 12. Schönian G, Kuhls K, Mauricio IL (2011) Molecular approaches for a better understanding of the epidemiology and population genetics of Leishmania . Parasitology 138: 405–425. [DOI] [PubMed] [Google Scholar]
- 13. Lopez M, Inga R, Cangalaya M, Echevarria J, Llanos-Cuentas A, et al. (1993) Diagnosis of Leishmania using the polymerase chain reaction: a simplified procedure for field work. Am J Trop Med Hyg 49: 348–356. [DOI] [PubMed] [Google Scholar]
- 14. Degrave W, Fernandes O, Campbell D, Bozza M, Lopes U (1994) Use of molecular probes and PCR for detection and typing of Leishmania - a mini-review. Mem Inst Oswaldo Cruz 89: 463–469. [DOI] [PubMed] [Google Scholar]
- 15. Volpini AC, Passos VMA, Oliveira GC, Romanha AJ (2004) PCR-RFLP to identify Leishmania (Viannia) braziliensis and L. (Leishmania) amazonensis causing American cutaneous leishmaniasis. Acta Trop 90: 31–37. [DOI] [PubMed] [Google Scholar]
- 16. Ampuero J, Rios AP, Carranza-Tamayo CO, Romero GA (2009) Genus-specific kinetoplast-DNA PCR and parasite culture for the diagnosis of localized cutaneous leishmaniasis: applications for clinical trials under field conditions in Brazil. Mem Inst Oswaldo Cruz 104: 992–997. [DOI] [PubMed] [Google Scholar]
- 17. Roque ALR, Cupolillo E, Marchevsky RS, Jansen AM (2010) Thrichomys laurentius (Rodentia; Echimydae) as a Putative Reservoir of Leishmania infantum and L. braziliensis: Patterns of Experimental Infection. PLoS Negl Trop Dis 4: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schönian G, Mauricio I, Cupolillo E (2010) Is it time to revise the nomenclature of Leishmania? Trends Parasitol 26: 466–469. [DOI] [PubMed] [Google Scholar]
- 19. da Graça GC, Volpini AC, Romero GA, de Oliveira Neto MP, Hueb M, et al. (2012) Development and validation of PCR-based assays for diagnosis of American cutaneous leishmaniasis and identification of the parasite species. Mem Inst Oswaldo Cruz 107: 664–674. [DOI] [PubMed] [Google Scholar]
- 20.Bonvicino CR, Oliveira JA, D'Andrea PS (2008) Guia dos Roedores do Brasil, com chaves para gêneros baseadas em caracteres externos. Rio de Janeiro: Centro Pan-Americano de Febre Aftosa – OPAS/OMS. 120 p. [Google Scholar]
- 21. Flynn JJ, Wyss AR (1998) Recent advances in South American mammalian paleontology. Trends Ecol. Evol 13: 449–454. [DOI] [PubMed] [Google Scholar]
- 22. Thomaz-Soccol V, Lanotte G, Rioux JÁ, Pratlong F, Martini-Dumas A, et al. (1993) Monophyletic origin of the genus Leishmania Ross, 1903. Ann Parasitol Hum Comp 68: 107–108. [PubMed] [Google Scholar]
- 23. Dantas-Torres F (2007) The role of dogs as reservoirs of Leishmania parasites, with emphasis on Leishmania (Leishmania) infantum and Leishmania (Viannia) braziliensis . Vet Parasitol 10: 139–146. [DOI] [PubMed] [Google Scholar]
- 24. Pita-Pereira D, Souza GD, Zwetsch A, Alves CA, Britto C, et al. (2009) Short Report: First Report of Lutzomyia (Nyssomyia) neivai (Diptera: Psychodidae: Phlebotominae) Naturally Infected by Leishmania (Viannia) braziliensis in a Periurban Area of South Brazil Using a Multiplex Polymerase chain Reaction Assay. Am J Trop Med Hyg 80: 593–595. [PubMed] [Google Scholar]
- 25. Shimabukuro PH, da Silva TR, Ribeiro FO, Baton LA, Galati EA (2010) Geographical distribution of American cutaneous leishmaniasis and its phlebotomine vectors (Diptera: Psychodidae) in the state of São Paulo, Brazil. Parasit Vectors 3: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Humberg RM, Oshiro ET, Cruz Mdo S, Ribolla PE, Alonso DP, et al. (2012) Leishmania chagasi in opossums (Didelphis albiventris) in an urban area endemic for visceral leishmaniasis, Campo Grande, Mato Grosso do Sul, Brazil. Am J Trop Med Hyg 87: 470–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lainson R, Shaw JJ, Povoa M (1981) The importance of edentates (sloths and anteaters) as primary reservoirs of Leishmania braziliensis guyanensis, causative agent of “pianbois” in north Brazil. Trans R Soc Trop Med Hyg 75: 611–612. [DOI] [PubMed] [Google Scholar]
- 28. Oliveira FS, Pirmez C, Pires MQ, Brazil RP, Pacheco RS (2005) PCR-based diagnosis for detection of Leishmania in skin and blood of rodents from an endemic area of cutaneous and visceral leishmaniasis in Brazil. Vet Parasitol 129(3–4): 219–227. [DOI] [PubMed] [Google Scholar]
- 29. Quaresma PF, Rêgo FD, Botelho HA, da Silva SR, Moura AJ Jr, et al. (2011) Wild, synanthropic and domestic hosts of Leishmania in an endemic area of cutaneous leishmaniasis in Minas Gerais State, Brazil. Trans R Soc Trop Med Hyg 105: 579–585. [DOI] [PubMed] [Google Scholar]
- 30. Travi BL, Arteaga LT, Leon AP, Adler GH (2002) Susceptibility of spiny rats (Proechimys semispinosus) to Leishmania (Viannia) panamensis and Leishmania (Leishmania) chagasi . Mem Inst Oswaldo Cruz 97: 887–892. [DOI] [PubMed] [Google Scholar]
- 31. Herrera HM, Norek A, Freitas TPT, Rademaker V, Fernandes O, et al. (2005) Domestic and wild mammals infection by Trypanosoma evansi in a pristine area of the Brazilian Pantanal region. Parasitol Res 96: 121–126. [DOI] [PubMed] [Google Scholar]
- 32. Roque ALR, D Andrea PS, Andrade GB, Jansen AM (2005) Trypanosoma cruzi: Distinct patterns of infection in the sibling caviomorph rodent species Thrichomys apereoides laurentius and Thrichomys pachyurus (Rodentia, Echimyidae). Exp Parasitol 111: 37–46. [DOI] [PubMed] [Google Scholar]
- 33. Rademaker V, Herrera HM, Raffel TR, D Andrea PS, Freitas TPT, et al. (2009) What is the role of small rodents in the transmission cycle of Trypanosoma cruzi and Trypanosoma evansi (Kinetoplastida Trypanosomatidae)? A study case in the Brazilian Pantanal. ActaTrop 111: 102–107. [DOI] [PubMed] [Google Scholar]
- 34. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Department of Microbiology, North Carolina State University. Nucleic Acids Symp Ser 41: 95–98. [Google Scholar]
- 35.Gordon D (2002) Viewing and Editing Assembled Sequences Using Consed. Current Protocols in Bioinformatics: John Wiley & Sons, Inc. [DOI] [PubMed] [Google Scholar]
- 36. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Molecular Biology and Evolution 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 37. Camargo ME (1966) Fluorescent antibody test for the serodiagnosis of American trypanosomiasis. Technical modification employing preserved culture forms of Trypanosoma cruzi in a slide test. Rev Inst Med Trop Sao Paulo 8: 227–235. [PubMed] [Google Scholar]
- 38. Herrera L, Xavier SCC, Viegas C, Martinez C, Cotias PM, et al. (2004) Trypanosoma cruzi in a caviomorph rodent: parasitological and features of the experimental infection of Thrichomys apereoides (Rodentia: Echimidae). Exp Parasitol 107: 78–88. [DOI] [PubMed] [Google Scholar]
- 39. Barreto ML, Teixeira MG, Bastos FI, Ximenes RAA, Barata RB, et al. (2011) Sucessos e Fracassos no Controle de Doenças Infecciosas no Brasil: O Contexto Social e Ambiental, Políticas, Intervenções e Necessidades de Pesquisa. Lancet. Saúde no Brasil 3: 47–60 Available at: http://download.thelancet.com/flatcontentassets/pdfs/brazil/brazilpor3.pdf. Accessed 20 March, 2014. [Google Scholar]
- 40. Harhay MO, Olliaro PL, Costa DL, Costa CH (2011) Urban parasitology: visceral leishmaniasis in Brazil. Trends Parasitol 27: 403–409. [DOI] [PubMed] [Google Scholar]
- 41. Marlow MA, da Silva Mattos M, Makowiecky ME, Eger I, Rossetto AL, et al. (2013) Divergent Profile of Emerging Cutaneous Leishmaniasis in Subtropical Brazil: New Endemic Areas in the Southern Frontier. PLoS One 8: e56177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Voloch CM, Vilela JF, Loss-Oliveira L, Schrago CG (2013) Phylogeny and chronology of the major lineages of New World hystricognath rodents: insights on the biogeography of the Eocene/Oligocene arrival of mammals in South America. BMC Res Notes 6: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Herrera L, D'Andrea PS, Xavier SCC, Mangia RH, Fernandes O, et al. (2005) Trypanosoma cruzi infection in wild mammals of the National Park ‘Serra da Capivara’ and its surroundings (Piauí, Brazil), an area endemic for Chagas disease. Trans R Soc Trop Med Hyg 99: 379–388. [DOI] [PubMed] [Google Scholar]
- 44. Herrera HM, Rademaker V, Abreu UGP, D'Andrea PS, Jansen AM (2007) Variables that modulate the spatial distribution of Trypanosoma cruzi and Trypanosoma evansi in the Brazilian Pantanal. Acta Trop 102: 55–62. [DOI] [PubMed] [Google Scholar]
- 45. Lainson R, Braga RR, de Souza AA, Povoa MM, Ishikawa EA, et al. (1989) Leishmania (Viannia) shawi sp. n., a parasite of monkeys, sloths and procyonids in Amazonian Brazil. Ann Parasitol Hum Comp 64: 200–207. [DOI] [PubMed] [Google Scholar]
- 46. Mato Grosso do Sul (2012) Governo do Estado de Mato Grosso do Sul. Secretaria de Saúde de Saúde do Estado Estadual de Vigilância Epidemiológica Coordenadoria. Estadual de Zoonoses Gerência. Informe epidemiológico das leishmanioses n° 4/2012. Campo Grande Available: http://www.saude.ms.gov.br/controle/ShowFile.php?id=123472. Acessed 20 March 2014. [Google Scholar]
- 47. Antonialli SAC, Torres TG, Paranhos Filho AC, Tolezano JE (2007) Spatial analysis of American Visceral Leishmaniasis in Mato Grosso do Sul State, Central Brazil. J Infect 54: 509–514. [DOI] [PubMed] [Google Scholar]
- 48. Pita-Pereira D, Cardoso MA, Alves CR, Brazil RP, Britto C (2008) Detection of natural infection in Lutzomyia cruzi and Lutzomyia forattinii (Diptera: Psychodidae: Phlebotominae) by Leishmania infantum chagasi in an endemic area of visceral leishmaniasis in Brazil using a PCR multiplex assay. Acta Trop 107: 66–9. [DOI] [PubMed] [Google Scholar]
- 49. Lainson R, Shaw JJ, Silveira FT, Braga RR, Ishikawa EA (1990) Cutaneous leishmaniasis of man due to Leishmania (Viannia) naiffi. Lainson and Shaw, 1989. Ann Parasitol Hum Comp 65: 282–284. [PubMed] [Google Scholar]
- 50. Naiffi RD, Freitas RA, Naiffi MF, Arias JR, Barret TV, et al. (1991) Epidemiological and nosological aspects of Leishmania naiffi. Lainson & Shaw, 1989. Mem Inst Oswaldo Cruz 86: 317–321. [DOI] [PubMed] [Google Scholar]
- 51. Azpurua J, De La Cruz D, Valderama A, Windsor D (2010) Lutzomyia sand fly diversity and rates of infection by Wolbachia and an exotic Leishmania species on Barro Colorado Island, Panama. PLoS Negl Trop Dis 4: 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Werneck GL, Costa CH (2005) Utilização de dados censitários em substituição a informações socioeconômicas obtidas no nível individual: uma avaliação empírica. Epidemiol Serv Saúde 14: 143–150. [Google Scholar]
- 53. Nascimento EL, Martins DR, Monteiro GR, Barbosa JD, Ximenes MF, et al. (2008) Forum: geographic spread and urbanization of visceral leishmaniasis in Brazil. Postscript: new challenges in the epidemiology of Leishmania chagasi infection. Cad Saude Publica 24: 2964–2967. [DOI] [PubMed] [Google Scholar]
- 54. Werneck GL (2008) Forum: geographic spread and urbanization of visceral leishmaniasis in Brazil. Introduction. Cad Saude Publica 24: 2937–2940. [DOI] [PubMed] [Google Scholar]
- 55. de Almeida AS, Medronho Rde A, Werneck GL (2011) Identification of risk areas for visceral leishmaniasis in Teresina, Piaui State, Brazil. Am J Trop Med Hyg 84: 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ready PD, Arias JR, Freitas RA (1985) A pilot study to control Lutzomyia umbratilis (Diptera: Psychodidae), the major vector of Leishmania braziliensis guyanensis, in a peri-urban rainforest of Manaus, Amazonas state, Brazil. Mem Inst. Oswaldo Cruz 80: 27–36. [DOI] [PubMed] [Google Scholar]
- 57. Volpini AC, Marques MJ, dos Santos SL, Machado-Coelho GL, Mayrink W, Romanha AJ (2006) Leishmania identification by PCR of Giemsa-stained lesion imprint slides stored for up to 36 years. Clin Microbiol Infect 12: 815–818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Origin of the analyzed caviomorph rodents, which were collected in Brazil between 1999 and 2012.
(DOCX)
Similarity levels observed by BLAST analysis among the DNA sequencing obtained for the target HSP70 (234) in the infected caviomorph rodents and available sequences from GeneBank with their respective accession numbers.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.