Abstract
Background
Cystic hydatid disease (CHD) is caused by the larval stages of the cestode and affects humans and domestic animals worldwide. Protoscoleces (PSCs) are one component of the larval stages that can interact with both definitive and intermediate hosts. Previous genomic and transcriptomic data have provided an overall snapshot of the genomics of the growth and development of this parasite. However, our understanding of how PSCs subvert the immune response of hosts and maintains metabolic adaptation remains unclear. In this study, we used Roche 454 sequencing technology and in silico secretome analysis to explore the transcriptome profiles of the PSCs from E. granulosus and elucidate the potential functions of the excretory-secretory proteins (ESPs) released by the parasite.
Methodology/Principal Findings
A large number of nonredundant sequences as unigenes were generated (26,514), of which 22,910 (86.4%) were mapped to the newly published E. granulosus genome and 17,705 (66.8%) were distributed within the coding sequence (CDS) regions. Of the 2,280 ESPs predicted from the transcriptome, 138 ESPs were inferred to be involved in the metabolism of carbohydrates, while 124 ESPs were inferred to be involved in the metabolism of protein. Eleven ESPs were identified as intracellular enzymes that regulate glycolysis/gluconeogenesis (GL/GN) pathways, while a further 44 antigenic proteins, 25 molecular chaperones and four proteases were highly represented. Many proteins were also found to be significantly enriched in development-related signaling pathways, such as the TGF-β receptor pathways and insulin pathways.
Conclusions/Significance
This study provides valuable information on the metabolic adaptation of parasites to their hosts that can be used to aid the development of novel intervention targets for hydatid treatment and control.
Author Summary
The successful infection establishment of parasites depends on their ability to combat their host's immune system while maintaining metabolic adaptation to their hosts. The mechanisms of these processes are not well understood. We used the protoscoleces (PSCs) of E. granulosus as a model system to study this complex host-parasite interaction by investigating the role of excretory-secretory proteins (ESPs) in the physiological adaptation of the parasite. Using Roche 454 sequencing technology and in silico secretome analysis, we predicted 2280 ESPs and analyzed their biological functions. Our analysis of the bioinformatic data suggested that ESPs are integral to the metabolism of carbohydrates and proteins within the parasite and/or hosts. We also found that ESPs are involved in mediating the immune responses of hosts and function within key development-related signaling pathways. We found 11 intracellular enzymes, 25 molecular chaperones and four proteases that were highly represented in the ESPs, in addition to 44 antigenic proteins that showed promise as candidates for vaccine or serodiagnostic development purposes. These findings provide valuable information on the mechanisms of metabolic adaptation in parasites that will aid the development of novel hydatid treatment and control targets.
Introduction
Cystic hydatid disease (CHD) is a serious parasitic zoonosis that is caused by the larval stages of Echinococcus granulosus, a cestode that poses a threat to public health as well as significant economic losses [1], [2], [3]. At present, more than 3 million people are infected with this parasite [4], [5], and the prevalence reaches 10% in some areas [6], [7]. The disease is difficult to control because appropriate diagnostic procedures are lacking and the available drugs are inefficient [8].
E. granulosus has a complex developmental cycle, involving eggs, oncospheres, protoscoleces (PSCs), and adult stages. Adult parasites live in the small intestine of dogs. After sexual maturation, numerous eggs are produced by the adult parasites and are then excreted with the dog feces. Infections occur in an intermediate host, when eggs containing larvae are ingested. Hydatid cysts (the larval stage or metacestode) develop in the internal organs (primarily in liver and lungs) of intermediate hosts. The larval stages of E. granulosus are comprised of two layers of cyst wall: cyst fluid and PSCs [9].
As the only infectious form of the larval stages, PSCs can interact with both definitive and intermediate hosts. They mature into adult parasites when the hydatid cysts are ingested by the definitive host. They can also differentiate into new cysts when released into the body cavity of intermediate hosts upon cyst rupture [10]. Mouse models of CHD are often established via the intraperitoneal inoculation with PSCs, a method that has been widely applied to drug screening and vaccine development [11], [12]. Overall, the PSC is an important infectious reagent that contributes to the transmission of CHD and also an excellent model system in which many aspects of the host-parasite interaction can be studied.
Understanding the elaborate immune evasion strategies and mechanisms of physiological adaptation of the PSCs is critical to ascertain effective intervention targets to control the prevalence of the parasite. In this study, we focus on the role of excretory-secretory products (ESPs) that are released by parasites, as these compounds are exposed directly to the immune system of the hosts and are engaged at the host-parasite interface [13]. The mechanism by which PSCs can subvert the immune environment via ESPs is the key to successful infection. Recently, we found that ESPs from adult E. granulosus could downregulate host immune responses by preventing dendritic cells (DC) from maturing, by impairing DC function and by inducing the generation of CD4+ CD25+ FoxP3+ T cells (unpublished data). Previous studies have shown that cystic fluids produced in the intermediate hosts can modulate DC differentiation and cytokine secretion [14], while antigen B released by the germinal cells of E. granulosus can direct immature DCs towards the maturation of a Th2 cell response [15]. Moreover, the ESPs from E. multilocularis larvae have been found to induce apoptosis and tolerogenic properties in DC in vitro [16]. To date, studies have focused primarily on the immune regulation of ESPs by the host, with little work undertaken to investigate the influence of ESPs on the physiological adaptation of parasites to their hosts. Interestingly, several intracellular proteins that were not previously thought to be exposed to the immune system of hosts have recently been identified in the ESPs of PSCs [9], [17]. This finding suggests that parasite-derived ESPs are incorporated in the metabolites of the host [18], [19].
Further investigations into the mechanisms of physiological adaptation of ESPs released by PSC have been hampered due to the paucity of information regarding ESPs. Although studies have utilized proteomics to identify the constituents of ESPs [9], [20]–[22], very few have been identified. This is largely because of interference from host proteins [20]–[21] and because of technical limitations of the methodologies used. In recent years, however, the combination of transcriptomics and proteomics has enabled the identification of an increasing number of parasitic proteins [23], [24].
In this study, we used Roche 454 sequencing technology and in silico secretome analysis to explore the transcriptome profiles of E. granulosus PSCs and to elucidate the potential functions of the ESPs released by the parasite.
Materials and Methods
Ethics statement
This study was performed in strict accordance with the recommendations provided in the Guide for the Care and Use of Laboratory Animals of the National Institute of Parasitic Diseases, Chinese Center for Disease Control and Prevention. The protocol was approved by the Laboratory Animal Welfare & Ethics Committee (LAWEC), National Institute of Parasitic Diseases, Chinese Center for Diseases Control and Prevention (Permit Number: IPD 2011-006).
Sample collection
Hydatid cysts were collected from the livers of a naturally infected sheep in a slaughterhouse in Qinghai, China. Cyst fluids containing PSCs were sucked out of the cysts using a sterile syringe. After natural sedimentation for 10 min, PSCs were carefully collected from the sediment of cyst fluids and washed 10 times with saline solution. We then added 2 mL of Trizol reagent (Invitrogen, USA) to the well-washed PSCs. After continuous mixing with a pipette, the PSCs were stored at −80°C prior to use.
Genotyping the PSCs
Genomic DNA from the PSCs was extracted using the DNeasy tissue kit (Qiagen, Hilden, Germany) and used as a template for a polymerase chain reaction (PCR) [25]. The following two primer pairs were used to amplify the mitochondrial genes of Echinococcus species: cytochrome coxidase subunit 1 (cox1) gene (F: 5′-TTGAATTTGCCACGTTTGAATGC-3′; and R: 5′-GAACCTAACGACATAACATAATGA-3′) and cytochrome b (cytb) gene (F: 5′-GTCAGATGTCTTATTGGGCTGC-3′; R: 5′-TCTGGGTGACACCCACCTAAATA-3′). Each 25-µL reaction mixture contained 1 µL of template DNA, 12.5 µL Premix Taq® mix (TaKaRa Biomedicals, Tokyo, Japan), l µL of 10 µM of each primer, and 9.5 µL nuclease-free water. The procedure of PCR amplification consisted of 94°C for 1 min, 30 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min, followed by 72°C for 10 min, with a final holding step at 4°C. The PCR products were directly sequenced with a Dye Terminator Cycle Sequencing Kit (Amersham Biosciences, Tokyo, Japan) and ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, USA).
cDNA library preparation, Roche 454 sequencing and sequence assembly
The total RNA was extracted from the PSCs in TRIzol reagent, and RNA quality was performed by gel electrophoresis with a 2100 BioAnalyzer (Agilent Technology, Santa Clara, USA). The sequencing protocol followed that described in Liao et al. [26], and was carried out at the Shanghai OE Biotech Company. cDNA was synthesized using 2 µg of total RNA with the SMART cDNA synthesis kit (Clontech Laboratories, Mountain View, USA) according to the manufacturer's instructions. The cDNA library was constructed using a GS-FLX Titanium General Library Preparation Kit (Roche, Branford, USA) without normalization [27], and then sequenced using a half run on the Roche 454 GS-FLX Titanium platform. The modules built-in Newbler 2.5.3 (a de novo sequence assembly software, Roche, USA) was used to remove low quality sequences and assemble the remaining sequences. Briefly, the quality score trimming filter trims back from the 3′ end of reads and was based on estimated quality scores (not the final quality scores) derived from an internal calibrated signal histogram. The error rate in a sliding window (default size of 40 bp) was calculated from the estimated quality scores and multiplied by an empirical scaling factor (default of 1.1). The window was moved leftwards until the estimated error rate in the window was <1.0% (by default). If the resulting read was less than 40 bp (default), the read was discarded and not counted (numTrimmedTooShortQuality metric). After removing low quality sequences and sequencing adaptors, the remaining sequencing reads were assembled using the Newbler 2.5.3 with the ‘extend low depth overlaps’ parameter. All of the ESTs from the Roche 454 were used to run the final assembly. The resulting isotig consensus sequences and singletons were referred as ‘unigenes’ in the following study.
Bioinformatic analyses of transcriptomic sequence data
The software SOAP2 was used to map the raw sequence reads to the nonredundant sequence data [28]. Briefly, raw reads were aligned to the assembled, nonredundant transcriptomic data, to ensure that each read was mapped to a unique transcript. Reads mapped to more than one transcript were randomly assigned to one unique transcript, to ensure that they were recorded only once. Reads per kilobase per million reads (RPKM), the evaluation index of relative assessment of transcript abundance, was calculated using the standard formula [29].
Unigene sequences were compared (using BLASTn with a cutoff E-value of 1e-5) to public sequences available in NCBI non-reductant (Nr) and STRING databases, and to five entire genome sequences (E. multilocularis [30], E. granulosus [31], Schistosoma hematobium [32], S. japonicum [33], S. mansoni [34]).
After conceptual translation from the predicted coding domains of individual transcriptomic sequences, the functions of the potential proteins were predicted using InterProScan [35], employing the default parameters. According to their homology with conserved domains and with protein families, proteins inferred for E. granulosus PSC (EgPSC) were assigned to three gene ontology (GO) categories, including molecular function, cellular component and biological process [36]. The pathway analysis of inferred proteins was carried out using the KEGG (Kyoto Encyclopedia of Genes and Genomes) database [37].
In silico secretome analysis
Excretory-secretory proteins (ESPs) were predicted according to the methods described by Garg and Ranganathan [38], [39]. Briefly, the secretory proteins were predicted utilizing the following five tools: ESTScan 3.0.3 [40] to translate the unigenes into putative proteins; SecretomeP 1.0 [41] for non-classical secreted proteins; SignalP 4.1 [42] for classical secreted proteins; TargetP 1.1 [43] for trimming mitochondrial proteins; and TMHMM 2.0 [44] for trimming transmembrane proteins. The predicted proteins with no transmembrane helices were thought to be ESPs.
In addition to traditional computational approaches for ESPs prediction, we also predicted E. granulosus ESPs (EgESPs) using BLASTP [45]. Based on their homology, a list of ESP sequences that included 478 nucleotides and 1,126 proteins was obtained to extract ESPs from the proteins that were predicted to be non-secretory by SecretomeP. Those ESPs had been identified in experiments in other species (S. mansoni, S. japonicum, Brugia malayi, Ancylostoma caninum, Teladorsagia circumcinta, Fasciola hepatica and Clonorchis sinesis) [46]–[59]. In this approach, a correct match for protein (Query) to protein (Subject) was designated when the query ratio was>80% of their length and identity ≥60, while a correct match for protein (Query) to nucleic acids (Subject) was designated when the query ratio was>80% of their length and identity ≥90.
All potential ESPs were blasted with known ESP sequences from E. granulosus (including nucleotide and protein sequences [9], [7], [20]–[22] and our unpublished data) to validate the in silico secretome analysis. They were then annotated against GO, KEGG, Reactome (http://www.reactome.org/ReactomeGWT/entrypoint.htm1) and Panther (http://www.patherdb.org/) databases to identify functional groups and pathway annotations. Enrichment of KEGG pathways for genes with significant expression was calculated utilizing a classical hypergeometric distribution statistical comparison of the query gene list against all predicted E. granulosus genes. Caenorhaditis elegans pathways were used as a reference. Calculated P-values were subjected to FDR correction, with p<0.05 taken as the threshold for significance.
Accession number
The transcriptome data is stored in Sequence Read Archive (SRA, No. SRP040541, http://www.ncbi.nlm.nih.gov/sra/?term=SRP040541).
Results/Discussion
Genotyping of E. granulosus PSCs
The genotype of E. granulosus PSCs used in this study was sheep G1, as the PCR fragment amplified from cytb gene showed the highest identity (99%) to the E. granulosus G1 genotype referenced in GenBank (accession AF297617, S1 Figure). This was consistent with the fact that sheep G1 strain is the most common strain worldwide [60].
Roche 454 transcriptome sequencing and reads assembly
A total of 330,188 raw reads (mean length = 411.8 bp) were generated. The data is stored in Sequence Read Archive (SRA, No. SRP040541). After trimming to remove adaptors, low quality reads and polyN tail sequences, 329,927 clean reads remained (mean length = 400.3 bp; Table 1). Clean reads were assembled and produced about 26,514 unigenes ranging in size form 150–3,357 bp (mean = 501.5 bp). These included 4,175 isotigs ranging in size from 154 to 3,357 bp and 22,339 singletons of 150 to 1,710 bp. Approximately 84% of the isotigs were>500 bp, while most singletons (85.97%) were between 300 and 800 bp in size (Table 1, S2 Figure). The numbers of EgPSCs unigenes matching known sequences are listed in Table 1. In summary, 26,514 unigenes were inferred from our transcriptome. The large majority of these (17,861, 67.4%) exhibited the highest level of homology to proteins in E. multilocularis, followed by proteins from E. granulosus (17,732; 66.9%), Caenorhabditis elegans (8,946; 33.7%) and S. mansoni (2,159; 17.5%). Moreover, 22,910 (86.4%) contigs were mapped to the E. granulosus genome and 17,705 (66.8%) of these were distributed within the coding sequence (CDS) region, which suggested that our results were reliable.
Table 1. Summary of the nucleotide sequence data for EgPSCs prior to and following assembly, with detailed bioinformatic annotation and analyses.
| Raw reads | 330188 |
| Unigenes (average length; min-max length) | 26514 (510.5; 150–3357) |
| Containing an open reading frame (%) | 19576 (73.8) |
| With homologues in E. granulosus (%) | 17732 (66.9) |
| E. multilocularis | 17861 (67.4) |
| Caenorhabditis elegans | 8946(33.7) |
| Clonorchis sinensis | 2540 (20.6) |
| Schistosoma mansoni | 2159 (17.5) |
| Schistosoma japonicum | 1485 (12.1) |
| Escherichia coli | 159 (1.3) |
| Returning STRING results (%) | 3188 (12.0) |
| Returning NCBI NR results (%) | 12408 (46.8) |
| Gene Ontology (%) | 5846 (22.0) |
| Number of biological process terms (level 2) | 24 |
| Cellular component | 20 |
| Molecular function | 14 |
| Returning a KOBAS result (%) | 5657 (21.3) |
| Number of predicted biological pathways | 306 |
Annotation of the transcriptome
Proteins predicted from EgPSCs transcriptome were categorized using Blast2Go [61]. A total of 5,846 were assigned at least one GO term involved in 56 GO assignments. The predominant terms for ‘biological process’ were ‘cellular process’ and ‘metabolic process’ (19.69% and 17.42%, respectively), for ‘cellular component’ were ‘cell part’ and ‘cell’ (21.65% and 21.65%, respectively), and for ‘molecular function’ were ‘catalytic activity’ and ‘binding’ (43.41% and 40.89%, respectively) (S3 Figure).
Of the proteins predicted for EgPSCs, 5,657 proteins were assigned to 306 biological pathway terms in the KEGG database (Table S1), including ‘endocytosis’ (n = 144 molecules), ‘oocyte meiosis pathway’ (n = 120), and ‘focal adhesion pathway’ (n = 118). We obtained 25 KOG clusters (S4 Figure), with 1,590 of the identified unigenes involved in at least one cluster. The largest functional group represented ‘translation, ribosomal structure and biogenesis’ (n = 214, 13.45%), followed by proteins associated with ‘post-translational modification, protein turnover, chaperones’ (n = 206, 12.95%). We also identified a further 220 (13.84%) peptidases and proteins that were linked to metabolism in eight functional categories.
Potential secretome database
PSCs are an important, infectious component of the larval stages of E. granulosus that can interact with both definitive and intermediate hosts [10]. The adaptive mechanisms that facilitate this interaction between host and parasite is of great interest to our understanding of the transmission of this widespread disease. Preliminary investigations suggest that parasites secrete certain molecules to assist in host tissue colonization [13]. We therefore focused on the components of ESPs released by PSCs and their potential roles in the physiological adaptation to their hosts and/or themselves.
Of the 26,514 unigenes identified, 19,576 were translated into proteins by ESTScan, 437 proteins were predicted to be classical secreted proteins using SignalP, while 592 were predicted to be non-classical secreted proteins according to SecretomeP. The classical and non-classical proteins were then analyzed using TargetP software for mitochondrial proteins, which resulted in the removal of 25 proteins. A further 123 transmembrane proteins were removed from the secretory protein dataset by TMHMM. In total, we obtained 881 ESPs using the four tools. A further 1,399 proteins that showed a high degree of similarity to experimentally identified secreted proteins were added by the Blast program. Thus, a total of 2,280 proteins were finally predicted as secretory proteins (Table 2).
Table 2. Prediction of secretory-excretory proteins (ESP) from the transcriptome of EgPSCs.
| Classfication | No. of predicted proteins | Prediction tools |
| Unigene | 26514 | Newbler |
| Protein | 19576 | ESTScan-3.0.3 |
| Classic secreted proteins | 437 | SignalP 4.1, Web |
| Non-classical secretory proteins | 592 | SecretomeP 1.0 |
| Mitochondrial proteins | 25 | TargetP 1.1, Web |
| Transmembrane proteins | 123 | TMHMM 2.0, Web |
| Homologues of experimentally verified proteins | 1399 | Blast-2.2.27 |
| Total secreted proteins predicted | 2280 |
To validate the in silico secretome analysis, we compiled a list of all experimentally identified ESP sequences of E. granulosus from the NCBI database and from previous studies (47 nucleotides and 77 proteins) [9], [17], [20]–[22], and then blasted the putative ESP sequences with the known ESP sequences (see Table S2). Ninety-one proteins were successfully mapped to the known ES proteins, of which 18 shared 100% identity and 33 shared 95%–99% identity. In addition, most known ESPs from other parasites [62] were matched successfully to those identified in our study. More importantly, domains in ESPs of Teladorsagia circumcincta (including metridin-like ShK toxin, lectin, proteinase inhibitor I29, and allergen V5/Tpx-1) were also found in the ESPs of EgPSC, which strengthens the concept that parasites employ universal ESPs to mediate parasite-host interplay [55]. Overall, these data suggest that the ESPs of EgPSCs identified in this study were reliable.
To date, there have been five proteomic studies regarding E. granulosus that have identified just 157 ESPs among them [9], [17], [20]–[22]. In this study, approximately 500 ESP domains were found, including known proteins (Table S3), a result that significantly expands the known ES components of EgPSCs. For example, WD40 repeats [63], [64], G-protein-coupled receptor (GPCR) [65] and Cadherin [66] all presented novel ESPs that were involved in parasite development-related processes. Recent studies using genome-wide and transcriptome data provide comprehensive information about the growth and development of E. granulosus [31], [67]. The results of this study extend this information and pave the way to a greater understanding of how PSCs utilize ESPs to survive in hosts.
ES proteins annotation
The putative ESPs were allocated to functional categories based on InterPro domains and GO categories. Of the 2,280 proteins predicted from EgPSC, the largest functional group represented ‘binding’ (n = 201, GO: 0005488), followed by ‘catalytic activity’ (n = 196, GO: 0003824) for ‘molecular function’, ‘metabolic process’ (n = 190, GO: 0008152) and ‘cellular process’ (n = 181, GO: 0009987) for ‘biological process’, and ‘cell part’ (n = 200, GO: 0044464) and ‘cell’ (n = 200, GO: 0005623) for ‘cellular component’ (S5 Figure).
The pathway enrichment analysis for identified ESPs was performed using KOBAS v2.0 software and more than 400 pathways were identified, of which 33 were statistically significant (Table 3). The term for ‘Huntington disease’ represented the most significant group (39, corrected p<0.0001), followed by Phagosome (37, p<0.0001), Protein folding (22, p<0.0001) and Chaperonin-mediated protein folding (16, p<0.0001).
Table 3. Pathway enrichment analysis of 1406 ESPs in the EgPSCs transcriptomes.
| Category | Terma | Pathway databaseb | Pathway Idc | Sample numberd | Background numbere | P-Valuef | Corrected P-valuef |
| Carbohydrate metabolism | |||||||
| Pentose phosphate pathway | KEGG | cel00030 | 13 | 18 | 9.41E-10 | 4.21E-08 | |
| Glycolysis/Gluconeogenesis | KEGG | cel00010 | 19 | 40 | 2.62E-08 | 8.80E-07 | |
| Gluconeogenesis | Reactome | — | 12 | 19 | 4.87E-08 | 1.51E-06 | |
| Glycolysis | Reactome | — | 7 | 8 | 9.66E-08 | 2.60E-06 | |
| Starch and sucrose metabolism | KEGG | cel00500 | 10 | 26 | 0.000253 | 0.004432 | |
| Fructose and mannose metabolism | KEGG | cel00051 | 8 | 23 | 0.001801 | 0.0226766 | |
| Amino sugar and nucleotide sugar metabolism | KEGG | cel00520 | 10 | 32 | 0.001974 | 0.0241093 | |
| Glucose metabolism | Reactome | — | 18 | 30 | 2.63E-10 | 1.32E-08 | |
| Carbon metabolism | KEGG | cel01200 | 24 | 78 | 1.50E-05 | 0.0003552 | |
| Biosynthesis of amino acids | KEGG | cel01230 | 17 | 65 | 0.001585 | 0.0212919 | |
| Signal transduction | |||||||
| Heterotrimeric G-protein signaling pathway | PANTHER | P00026 | 7 | 8 | 9.66E-08 | 2.60E-06 | |
| Calcium signaling pathway | KEGG | cel04020 | 13 | 37 | 0.000157 | 0.0030182 | |
| IFN-alpha/beta pathways | Reactome | — | 3 | 5 | 0.001394 | 0.0193684 | |
| TGF-beta receptor signaling | Reactome | — | 3 | 6 | 0.003739 | 0.0367554 | |
| Apoptosis signaling pathway | PANTHER | P00006 | 7 | 18 | 0.00122 | 0.0182041 | |
| Proteins metabolism | |||||||
| Protein folding | Reactome | — | 22 | 23 | 6.32E-21 | 8.49E-19 | |
| Metabolism of proteins | Reactome | — | 64 | 293 | 1.75E-05 | 0.0003918 | |
| Mitochondrial protein import | Reactome | — | 9 | 22 | 0.000238 | 0.0043566 | |
| Chaperonin-mediated protein folding | Reactome | — | 16 | 19 | 1.57E-13 | 1.59E-11 | |
| Post-chaperonin tubulin folding pathway | Reactome | — | 8 | 9 | 1.28E-08 | 5.16E-07 | |
| Activation of chaperones by ATF6-alpha | Reactome | — | 5 | 7 | 3.41E-05 | 0.000723 | |
| Calnexin/calreticulin cycle | Reactome | — | 5 | 12 | 0.002474 | 0.0293186 | |
| Gene expression | |||||||
| MicroRNA (miRNA) biogenesis | Reactome | — | 6 | 16 | 0.002776 | 0.0310737 | |
| Genetic information processing | |||||||
| Spliceosome | KEGG | cel03040 | 25 | 103 | 0.000786 | 0.0121906 | |
| Transport and catabolism | |||||||
| Phagosome | KEGG | cel04145 | 37 | 55 | 2.53E-21 | 5.10E-19 | |
| Disease pathway | |||||||
| Huntington disease | PANTHER | P00029 | 39 | 41 | 2.31E-34 | 9.29E-32 | |
| Parkinson disease | PANTHER | P00049 | 15 | 31 | 3.76E-07 | 9.46E-06 | |
| Others | |||||||
| Cytoskeletal regulation by Rho GTPase | PANTHER | P00016 | 15 | 18 | 1.07E-12 | 7.16E-11 | |
| CCT/TriC | Reactome | — | 15 | 18 | 1.07E-12 | 7.16E-11 | |
| mRNA splicing - minor pathway | Reactome | — | 13 | 39 | 0.0003 | 0.0050358 | |
| N-glycan trimming in ER and CNX/CRT | Reactome | — | 6 | 13 | 0.000599 | 0.0096585 | |
| Adenine and hypoxanthine salvage pathway | PANTHER | P02723 | 3 | 6 | 0.003739 | 0.0367554 | |
KEGG enrichment analysis was performed by KOBAS 2.0 (http://kobas.cbi.pku.edu.cn/home.do).
Caenorhaditis elegans pathways were used as a reference. The ESP corresponding to each pathway can be found in Table S9.
Pathway databases mapped by KOBAS including KEGG pathway: http://www.genome.jp/kegg/pathway.htm1; reactome: http://www.reactome.org/ReactomeGWT/entrypoint.htm1; PANTHER: http://www.patherdb.org/.
Pathway identified in specific database.
“-” means not given.
The number of input proteins mapped to the particular pathway.
The number of identified proteins mapped to the particular pathway.
Only significant results (p<0.05) were shown.
The statistical method was a hypergeometric test, whereas the FDR correction method was from Benjamini and Hochberg (1995).
Of the 2,280 putative ESPs, only 1,406 were mapped to known functions (Table S3). These proteins included not only many common and abundant ‘house-keeping proteins’ (e.g., ribosome proteins, cytochrome subunit proteins, and enzymes involved in carbohydrate and protein metabolism), but also some rare but interesting proteins (e.g., putative receptor and antigenic proteins). This highlights the important roles of ESPs in parasite survival and development within hostile host environments. Below, we characterize these potential ESPs in greater detail.
Metabolism of carbohydrates for parasite energy and nutrition
The interaction of pathogens with mammalian hosts leads to a variety of physiology responses that drive the adaptation of the interacting partners to their new environments and conditions [19]. The ESPs released by parasites might be important actors in this process of adaptation, because they are involved in the metabolism of carbohydrates [68]. We identified a total of 122 domains (summarized in Table S4), of which, 32 proteins were identified to have a higher level of expression in the parasite (Table 4).
Table 4. The potential functional proteins with a high abundance in the ESPs from EgPSCs transcriptome.
| GI Number | Description | Speciesa | Functionb |
| Proteases | |||
| 116242320 | Lysosomal pro-X carboxypeptidase | C. sinensis | |
| 111036376 | Cathepsin L-like proteinase | E. multilocularis | A |
| 498980202 | Lysosome membrane protein 2-like isoform X1 | M. zebra | |
| 226478810 | Cytochrome c-type heme lyase | S. japonicum | |
| Protease inhibitor | |||
| 223037336 | Kunitz protein 8 | E. granulosus | |
| Structural | |||
| 124783098 | Ribosomal protein S18 | T. asiatica | |
| 56753617 | Ribosomal protein L21 | S. japonicum | |
| 226483022 | Putative small subunit ribosomal protein S27Ae | S. japonicum | |
| 256074063 | 60S ribosomal protein L9 | S. mansoni | |
| 392495090 | Ribosomal protein S13 | S.erinaceieuropaei | |
| 421975923 | 60S ribosomal protein L7 | S.erinaceieuropaei | |
| 29841212 | Putative ribosomal protein L27A protein | S. japonicum | |
| 60692924 | Ribosomal protein | S. japonicum | |
| 421975956 | Putative ribosomal protein S25 | S.erinaceieuropaei | |
| 358340304 | U1 small nuclear ribonucleoprotein A | C. sinensis | |
| 358332789 | Ribosomal RNA-processing protein 9 | C. sinensis | |
| 256078860 | U3 small nucleolar ribonucleoprotein protein imp4 | S. mansoni | |
| 55976640 | Actin-1/4 actin | T. solium | |
| 207298859 | Beta-actin | A.transmontanus | |
| 543766 | Actin-1 | E. granulosus | |
| 133721998 | Actin | G. viridula | |
| 29337144 | Tubulin beta-2 chain | E. multilocularis | |
| 29337143 | Tubulin beta-3 chain | E. multilocularis | |
| 29337145 | Tubulin beta-1 chain | E. multilocularis | |
| 410897689 | Tubulin alpha-1C chain-like | T. rubripes | |
| 311992220 | Tropomyosin 2 high molecular weight isoform | M. corti | A |
| 29337029 | Tropomyosin | E. multilocularis | A |
| 168071448 | Tropomyosin B | E. granulosus | A |
| 256086965 | Myosin heavy chain | S. mansoni | A |
| 547974 | Paramyosin | E. granulosus | A |
| 432897369 | Dynein light chain 2, cytoplasmic-like | O. latipes | A |
| 171473974 | Dynein light chain LC6 | S. japonicum | A |
| 405970739 | Dynein light chain 2, cytoplasmic | C. gigas | A |
| 68071557 | Dynein light chain 1 | P. berghei | A |
| 29467010 | Dynein light chain | E. multilocularis | A |
| 226487996 | Nucleolar protein 5 | S. japonicum | |
| 226487430 | Myophilin | S. japonicum | A |
| 29336625 | Myophilin | E. granulosus | A |
| 256086246 | Histone H3 | S. mansoni | |
| 344240017 | Histone H2A type 1 | C. griseus | |
| 358338242 | Histone H2A.V | C. sinensis | |
| 405975240 | Histone H2A | C. gigas | |
| 358331974 | PHD finger protein 7 | C. sinensis | |
| Molecular chaperone | |||
| 343887008 | Heat shock protein 90 alpha | K. marmoratus | A, D |
| 1661112 | Heat shock 70 kDa protein, partial | M. corti | A |
| 29336623 | Heat shock cognate 70 kDa protein | E. granulosus | A |
| 124783198 | Heat shock protein gp96 | T. asiatica | |
| 124783152 | 40, partial | T. asiatica | A |
| 124783287 | Chaperonin | T. asiatica | |
| 256082744 | T-complex protein 1 epsilon subunit | S. mansoni | |
| 421975972 | T-complex protein 1 subunit alpha | S.erinaceieuropaei | |
| 349934375 | T-complex protein 1 subunit zeta | C. sinensis | |
| 358342604 | Molecular chaperone GrpE | C. sinensis | |
| 318064648 | DnaJ-like protein subfamily b member 11 | I. punctatus | |
| 312065499 | Protein disulfide isomerase | L. loa | |
| 256081230 | Ubiquitin-conjugating enzyme E2r | S. mansoni | |
| 29841024 | 26S proteasome regulatory complex subunit p42A | S. japonicum | |
| 226470558 | Proteasome subunit beta type 4 | S. japonicum | |
| 56754539 | 20S proteasome subunit alpha 8 | S. japonicum | |
| 29336773 | Putative growth regulator 14-3-3 | E. granulosus | A, ST |
| 62178030 | Putative 14-3-3 protein | E. granulosus | A, ST |
| 148613837 | Calreticulin | E. granulosus | A |
| 444792465 | Calcineurin B | E. granulosus | A |
| 353530026 | Calcineurin B | E. granulosus | A |
| Carbohydrate metabolism | |||
| 167541050 | Phosphoglycerate mutase | C. sinensis | |
| 358333945 | Phosphoglycerate kinase | C. sinensis | |
| 262192839 | Enolase | E. granulosus | A |
| 62178020 | Putative glucose phosphate isomerase | E. granulosus | A |
| 29336626 | 78 kDa glucose-regulated protein,GRP-78 | E. multilocularis | |
| 328789193 | UTP–glucose-1-phosphate uridylyltransferase isoform 1 | A. mellifera | |
| 6016079 | Glyceraldehyde-3-phosphate dehydrogenase | E. multilocularis | A |
| 338827784 | Glucose-6-phosphatase | E. granulosus | |
| 470364276 | UDP-glucose dehydrogenase | C. owczarzaki | |
| 470610058 | Cyclophilin B | T. truncatus | A, S, M |
| 31077167 | Cyclophilin | T. truncatus | A, S, M |
| 358252886 | Dehydrodolichyl diphosphate synthase | C. sinensis | |
| 338827788 | Phosphoenolpyruvate carboxykinase | E. granulosus | A |
| 358334589 | Dolichyl-phosphate beta-glucosyltransferase | C. sinensis | |
| 256090534 | Phosphoglucomutase | S. mansoni | |
| 46406288 | Malate dehydrogenase | E. granulosus | A |
| 29841093 | Citrate synthase | S. japonicum | |
| 29336561 | Fructose-bisphosphate aldolase | E. multilocularis | A |
| 56682906 | Hypoxanthine-guanine phosphoribosyltranferase | S. japonicum | |
| 256082514 | Uridine cytidine kinase I | S. mansoni | |
| 358336324 | Sterol O-acyltransferase | C. sinensis | |
| 256085769 | Methyltransferase | S. mansoni | |
| 170579277 | Lysyl-tRNA synthetase | B. malayi | |
| 256071828 | Polyadenylate binding protein | S. mansoni | |
| Oxidation/reduction | |||
| 29337026 | Thioredoxin peroxidase | E. granulosus | A |
| 1004227 | Glutathione transferase | E. multilocularis | A |
| 341616326 | Peroxiredoxin 3 | C. sinensis | A |
| 347948498 | Cu2+/Zn2+ superoxide dismutase (SOD1) | T. solium | A, T |
| 29337032 | Thioredoxin | E. granulosus | A |
| 358340540 | Thioredoxin domain-containing protein 9 | C. sinensis | A |
| 94556988 | Neuronal nitric oxide synthase protein inhibitor | T. solium | PI |
| 256070830 | Peroxidasin | S. mansoni | A |
| Transporters | |||
| 256080958 | Multidrug resistance protein | S. mansoni | |
| 85701472 | Trans-Golgi network vesicle protein 23A | M. musculus | |
| 226478102 | Secretory carrier-associated membrane protein 2 | S. japonicum | |
| 124782903 | Phosphatidylinositol transfer protein alpha | T. asiatica | |
| 358336646 | F-type H+-transporting ATPase subunit c | C. sinensis | |
| 226468748 | Voltage-dependent anion-selective channel protein 2 | S. japonicum | |
| 392495096 | Sorting nexin SNX11 | S. japonicum | |
| Translation | |||
| 148717323 | Elongation factor 1 alpha | E. granulosus | A |
| 148717331 | Elongation factor 1 alpha | E. vogeli | A |
| 148717335 | Elongation factor 1 alpha | E. shiquicus | A |
| 159138037 | RNA polymerase II elongation factor | C. sinensis | A |
| 358334689 | Elongation factor 2 | C. sinensis | A |
| Transcription | |||
| 221509352 | Zinc finger (C3HC4 type) protein | T. gondii | |
| 358332148 | Eukaryotictranslation initiation factor, TFIIA | C. sinensis | |
| Engery conversion | |||
| 256077755 | ATP synthase beta subunit | S. mansoni | |
| 226478810 | Putative cytochrome c-type heme lyase (CCHL) | S. japonicum | |
| RNA Processing | |||
| 358334450 | ATP-dependent RNA helicase FAL1, partial | C. sinensis | |
| Cell cycle | |||
| 353230502 | Mitotic phosphoprotein 44 | S. mansoni | |
| Others | |||
| 5051948 | Antigen B8/1 | E. granulosus | A |
| 7339849 | Immunogenic protein Ts11 | T. solium | A |
The full names of species can be seen in Table S8.
Abbreviations: A, antigenic protein; D, drug gene; ST, signal transduction; S, structural; M, molecular chaperone; T, transporters; PI, protease inhibitor.
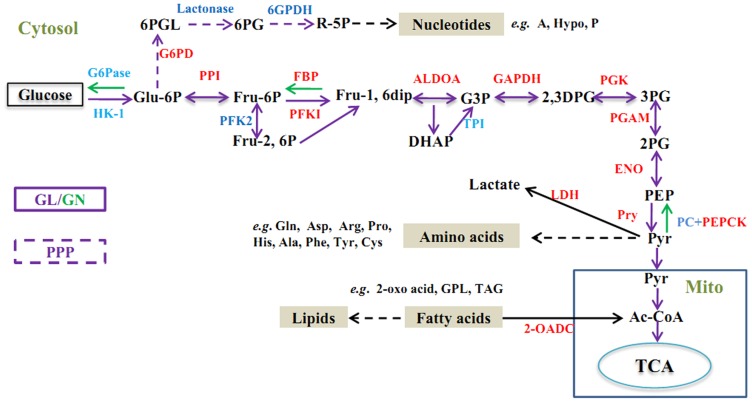
E. granulosus has evolved an optimal strategy to gain energy and nutrition from its host using ESPs (Fig. 1). Firstly, the parasite can regulate glycolysis (GL). We identified nine enzymes associated with GL, including the rate-limiting enzymes PFK1 and pyruvate kinase. Through GL, non-essential amino acids (e.g., glutamine, aspartic acid, arginine, proline, histidine, alanine, tyrosine and cysteine), fatty acids, adenine and hypoxanthine nucleotides, as well as pyrimidine, could be synthesized to support parasite development and growth. Alternatively, glucose and other carbohydrates could be synthesized via gluconeogenesis (GN) when alternative carbon sources (e.g., glucogenic amino acids, lactate, and glycerol) were available. In addition to the reversible enzymatic GL steps, several reactions are essential in the GN pathway from pyruvate via oxaloacetate to glucose: the reactions catalyzed by pyruvate carboxylase, phosphoenolpyuvate carboxykinase (PEPCK), fructose-1, 6-bisphosphatase, and glucose-6-phosphatase leading to oxaloacetate, phosphoenolpyruvate (PEP), fructose-6-phosphate, and glucose. Finally, tricarboxylic acid (TCA) enzymes, such as aconitate hydratase, succinate dehydrogenase complex, malate dehydrogenase, were identified in the TCA cycle. Other enzymes involved in carbohydrate metabolism are shown in Table 4.
Figure 1. Schematic diagram showing the carbohydrate metabolic pathways involved in the ESPs of EgPSCs transcriptome (Reference from Eisenreich W et al. [19] with some modifications).
Glycolysis (GL, purple arrows) and gluconeogenesis (GN, grass green arrows); pentose-phosphate pathway (PPP, broken purple arrows); tricarboxylic acid cycle (TCA, blue circle) other catabolic reactions that occur in the mitochondrion and in the cytosol (black arrows). Anabolic reactions leading to amino acids, nucleotides, and lipids are indicated by broken thick black arrows. Metabolites are marked in black. Enzymes identified in our study are marked in red, while other enzymes are marked in blue. Abbreviations: HK, hexokinase; PFK, phosphofructokinase; FBP, fructose bisphosphatase; PK, pyruvate kinase; PDH, pyruvate dehydrogenase complex; PCK, PEP-carboxylase; PPI, phosphohexose isomerase; TPI, triose phosphate isomerase; PGK, phosphoglycerate kinase; GAPDH, glyceraldehydes 3-phosphate dehydrogenase; PC, pyruvate carboxylase; LDH, lactate dehydrogenase; PEPCK, phosphoenol pyruvate carboxykinase; G6PD, glucose-6-phosphate dehydrogenase; ALDOA, fructose-biphosphate aldolase; PGK, phosphoglycerate kinase; PGAM, phosphoglycerate mutase; ENO, enolase; G6Pase, glucose 6-phosphatase; G6PD, glucose-6-phosphate dehydrogenase; 6GPDH, 6-phosphogluconatedehydrogenase. Gln, Glutanine; Asp, aspartic acid; Arg, arginine; Pro, proline; His, histidine; Ala, alanine; Tyr, tyrosine; Cys, cysteine; Ade, adenine; Hyp, hypoxanthine.
Certain enzymes have been recognized to play key roles in the development of parasites. Phosphoglucose isomerase (PGI), one of glycolytic enzymes, has been found to stimulate parasite growth and the formation of novel blood vessels nearby the developing metacestode [69]. Vaccinating mice with recombinant PGI increases their resistance towards a secondary infection challenge [69]. Similarly, PEPCK is a novel egg antigen of S. mansoni [70] and an abundant protein in adult parasites that is related to numerous metabolic pathways (e.g., endocrine function, excretion and carbohydrate metabolism [22].
To date, only five ESPs have been identified to participate in this metabolic process [17]. The results of this study support the role of these proteins in metabolic adaptation to their hosts and, more importantly, demonstrate that many more ESPs may be used by E. granulosus to regulate carbohydrate metabolism. Further work is required to identify these additional ESPs and establish their functions.
Control of parasite homeostasis
Following infection with E. granulosus, the intermediate host produces a significant immune response that affects the growth and development of parasites [71], [72], while the parasites initiate effective evasion mechanisms to counteract adverse host environments.
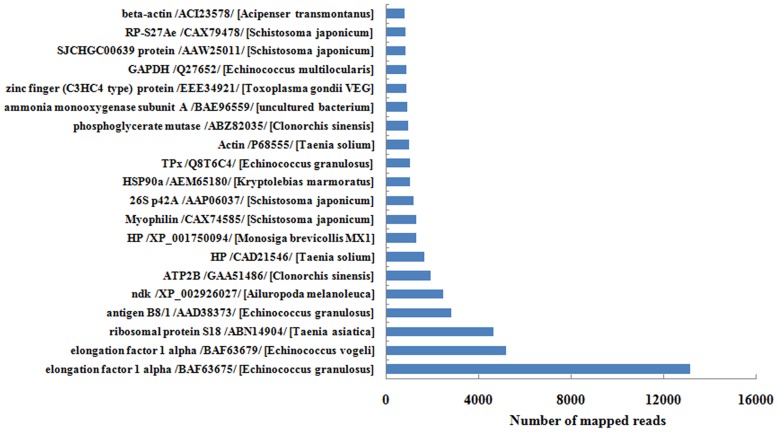
In this study, we found that 36 ESP domains were molecular chaperones (Table S5), and identified a further 25 proteins that were present with high levels of abundance (Table 4), including several novel molecules (heat shock proteins, HSP90 and HSP40, universal stress protein [Usp], calreticulin, calcineurin B, GrpE in the HSP60 family and Gp96). HSP90 was the most strongly expressed of all the molecular chaperons (Fig. 2), suggesting it is one of the key molecules in mediating parasite development. This is supported by the fact that nitration of HSP90 is known to induce cell death [73], and HSP90 has been used as a drug target in protozoa intervention [74]. Previous studies have also shown that UspA and Usp8 are associated with stress resistance and growth in bacterial species [75]. ESPs might disrupt the expression of intracellular 70 protein in the host immune cells, while the parasite itself might release HSP70 to prevent damage from those same cells [76]. These molecular chaperone-like proteins may be released to regulate the stress responses that arise in the extremely harsh intestinal environments of definitive hosts (e.g., numerous highly active proteases, variable pH levels).
Figure 2. The transcription profiling of putative ESPs in EgPSCs transcriptome.
The 20 most abundant ESPs encoded in the transcriptome are shown. Abbreviations: RP-S27Ae, putative small subunit ribosomal protein S27Ae; GAPDH, glyceraldehydes-3-phosphate dehydrogenase; ndk, nucleoside diphosphate kinase B-like; ATP2B, Ca2 + transporting ATPase plasma membrane; HSP90α, heat shock protein alpha; 26S p42A, 26S proteasome regulatory complex subunit p42A.
E. granulosus may secrete proteases or inhibitors to digest host proteins, or to protect itself from digestion by endogenous or host-derived proteinases. In this study, 39 proteases, including serine, aspartic, metallo- and cysteine proteinases, and five inhibitors, were inferred among the set of ESPs (see Table S6). Several of these (serine, cysteine, and the proteinase inhibitors) are likely to be important targets for parasite intervention and control [77]–[79]. However, only three proteases and two protease inhibitors were strongly expressed in the set of ESPs (Table 4). More sensitive technologies will therefore be required to identify other proteases that were expressed at lower levels of abundance.
In contrast, the action of antioxidant enzymes is a key component of parasite survival during infection. In this study, seven ESPs were identified as antioxidant enzymes, including glutathione transferase, peroxiredoxin, thioredoxin, Cu2 +/Zn2 + superoxide dismutase, and neuronal nitric oxide synthase protein inhibitor. These molecules might be utilized by the parasite to detoxify the reactive oxygen species produced by the host environments [80].
Direct regulation of host immunological responses
In previous experiments we demonstrated that following infection with EgPSCs the microenvironment of the murine peripheral immune system undergoes several changes. These included T cell activation and the accumulation of immunosuppressive cells, such as myeloid-derived suppressor cells (MDSC) and CD4+CD25+FoxP3+ T cells (Treg) [71]. Such alterations might occur via the action of ESPs as many ESPs have been found to redirect host immune responses [13], [17]. In this study, we found several ESPs that contribute to immune regulation following infection (Table 4). Tegument protein (Teg) is known to induce a biased Th2 cell immune response related to chronic infection [81], while 14-3-3 proteins are associated with resistance to the immune responses mediated by local cells [82]. In addition, the antigen B (AgB) family are important in immune evasion because the antigen is secreted at variable amounts [83], and have also been demonstrated to direct immature DC maturation towards a preferential Th2 immune response [15].
Notably, cysteine proteinases have been reported to inhibit Th1 immune response via the induction of IL-4, which is the main cytokine responsible for Th2 differentiation [84]. HSP70 has been shown to stimulate both of types of response in CHD patients [85]. Also, the intraperitoneal injection of calreticulin (CRT) significantly influences Th1/Th2 balance [86]. Hence, these proteins might be novel immunoregulatory molecules that contribute to immune evasion.
Signaling pathways
We found that EgPSC possesses many signaling pathways such as P13K-Akt, mitogen-activated protein kinase (MAPK), Wnt, calcium, HIF-1, insulin, estrogen and chemokine signaling (Table S1). However, in the putative set of ESPs, only G-protein, calcium, IFN-α/β, TGF-β receptor and apoptosis signaling pathways were dominant (Table S7), which indicated their importance in parasite-host interactions and physiological processes.
Notably, we found that G-protein-coupled receptors (GPCRs), TGF-β and insulin signaling pathways might closely associate with the development of EgPSCs. For example, GPCRs can activate the G-proteins located within the cell. They work cooperatively to deliver varied signals, which in turn regulate various physiological processes [87]. However, the exact function of G-protein signaling in parasites remain unclear.
Studies have shown that TGF-β and insulin signaling pathways in C. elegans can trigger an ‘alternative’ developmental pathway, and can regulate and transit the environmental stresses on the first larval stage of the parasite [88], [89]. In particular, the disruption of both signaling pathways leads to arrested development in this species [90], [91]. Indeed, the TGF-β pathway is speculated to regulate developmental events in parasitic nematodes [92], as molecules involved in the TGF-β pathway have been found in several parasitic nematodes including Brugia pahangi, Brugia malayi and Parastrongyloides trichosuri [93]–[95]. The role of TGF-signaling in E. granulosus development and growth warrants further investigation. A recent study revealed that host insulin acts as a stimulant for parasite development within the host liver and that E. multilocularis senses the hormones of hosts through an evolutionary-conserved insulin signaling pathway, which demonstrates the importance of insulin signaling for parasite survival [96].
Potential targets for diagnosis and vaccine development
CHD has a global distribution and causes high rates of morbidity and has a high socio-economic burden in several countries [97]. The Eg95 vaccine induces a high antibody titer in sheep and goats, which protects them against CHD [98]. However, due to antigenic variation caused by genotypic diversity [99], the common Eg95 vaccine does not bind the antibodies of all E. granulosus species, which limits its utility. We suggest that the ESPs of EgPSCs are an excellent alternative candidate for a vaccine, as they are easy to prepare and safer for human health. More importantly, the ESPs obtained by in vitro culture have shown a 92.07% protection rate against a high dose of egg infection in sheep (1,000 eggs per sheep) [100].
Using in silico secretome analysis, we identified 44 antigenic proteins present at high abundance in our set of ESPs (Table 4). Of these, elongation factor 1 alpha, antigen B8/1, myophilin, thioredoxin peroxidase, phosphoglycerate mutase, heat shock protein 90a and actin, were the most abundant. In addition, HSP70, enolase, 14-3-3, phosphate glucose isomerase, malate dehydrogenase, glutathione S-transferase were also present at high abundance in the set of ESPs (Table S8). These abundant proteins hold enormous potential as diagnostic markers or intervention targets. Indeed, malate dehydragenase (MDH) has been tested for the immunodiagnosis of E. granulosus, while thiredoxin peroxidase (TPx) has been used for the immunodiagnosis of human CHD [101]. Likewise, the 14-3-3 molecule has been demonstrated to be a candidate vaccine against E. granulosus in mice [12], while recombinant GST protein has been used in the diagnosis of echinococcosis [102].
Proteins that are present at lower levels of abundance might also be relevant as diagnostic markers or target molecules for vaccine development. In this study, these include antigen 5 (Ag5), calreticulin, calcineurin B, thioredoxin, phosphoglucomutase, fructose-bisphosphate aldolase and gp96 (Table S8). Many of these have already shown promise for serodiagnostic purposes. For example, Ag5 is a dominant immunogenic and diagnostic antigen of the E. granulosus metacestode in both adults and PSCs [22]. Similarly, calcineurin B has been previously identified as a candidate for a vaccine or drug target [103]. Surprisingly, the E. granulosus-specific protein domain antigen B (EgAgB) family, which are well known as diagnostic targets, were undetectable in this study. This result was consistent with previous observations that little or no AgB is secreted by in vitro cultured PSCs [17], [104]. Previous studies have demonstrated that the germinal layer, but not the PSC, contributes to the primary secretion of AgB [17]. Thus, serological examination based on the AgB antibody would not be useful in early-stage PSC infection as only minute amounts of AgB antibody are produced at that time.
There are currently just two methods for the treatment of hydatid disease: surgery and the use of benzimidazole, both of which give unsatisfactory results. Hence, novel treatment compounds are urgently needed. In this study, we have identified several secretory drug targets for echinococcosis (Table 4, Table S3), including GPCRs, threonine and tyrosine protein kinase and nuclear hormones, which have been the targets of successful new drug discoveries [65]. Insulin signaling [96], thyrotropin-releasing hormone receptor, pancreatic hormone-like or transforming growth factor-β (TFG-β) families have been linked to the larval developmental of E. multilocularis. Thus, interventions that utilize these molecules could also arrest parasite growth. In addition, GL enzymes could be drug targets for parasites that rely on the GL pathway for growth and development [22]. Finally, HSP90 has been used as a drug target in protozoa intervention programs [74].
Conclusions
The larval stages of E. granulosus are pathogenic to human, which therefore have become the research focus of CHD. Parkinson et al. [2012] first reported genes with features that reflect physiological adaptations of different parasite stages, including PSCs, and revealed abundant long non-protein coding transcripts, upregulated fermentative pathways, candidate apomucins and a set of platyhelminth-specific gene products, which greatly increased the quality and the quantity of the molecular information regarding E. granulosus [67]. The most newly published genome of the parasite also uncovered several key events of the parasites, including the species-specific genes AgB family, bile salt pathways and Cavβ1 gene variation associated with praziquantel sensitivity [31]. Those studies have provided a molecular understanding of the growth and development of E. granulosus. In this study, we focused on the transcriptome of PSCs, which is the only infective component of the larval stages. We present novel and urgently needed information regarding the components of ESPs released by PSCs and their potential roles in the metabolic adaptation of parasites to their hosts. We suggest that intracellular ESPs are essential to the metabolism of carbohydrates within their hosts and that various molecular chaperones with a high level of expression may play a role in resisting harsh host environments. We also reveal a set of antigenic ESPs that show promise as candidates for vaccine development or in the development of serodiagnostic markers. Such findings will encourage more novel strategies for the treatment and control of CHD.
Although the coverage of the transcriptome data in this study was not deep as the genome-wide study [31], [67], these findings are novel and hold importance for understanding the mechanisms of parasite metabolic adaptations within their hosts. Overall, this study adds supplementary knowledge regarding the genomics of E. granulosus, and deepens our understanding of host-parasite interactions.
Supporting Information
Genotype identification of E. granulosus . (A) PCR amplification. M, DNA maker; Cytb, 601 bp; Cox1, 885 bp. (B) Sequence alignment of the cytochrome b (cytb) gene. Bases that differed are marked with red boxes.
(TIF)
Length distribution of singletons and isotigs of the Eg PSCs transcriptome.
(TIF)
Gene ontology (GO) analysis of the Eg PSCs transcriptome. BLASTP against SwissProt and GO mapping of identified proteins (performed with BLAST2GO) [61].
(TIF)
Distribution of the KOG functional categories of the proteins identified from the Eg PSCs transcriptome. Percentages and numbers of proteins in each functional category are indicated in the sectors of the circle. KOG functional categories: (A) RNA processing and modification; (B) Chromatin structure and dynamics; (C) Energy production and conversion; (D) Cell cycle control, cell division, chromosome partitioning; (E) Amino acid transport and metabolism; (F) Nucleotide transport and metabolism; (G) Carbohydrate transport and metabolism; (H) Coenzyme transport and metabolism; (I) Lipid transport and metabolism; (J) Translation, ribosomal structure and biogenesis; (K) Transcription; (L) Replication, recombination and repair; (M) Cell wall/membrane/envelope biogenesis; (N) Cell motility; (O) Posttranslational modification, protein turnover, chaperones; (P) Inorganic ion transport and metabolism; (Q) Secondary metabolites biosynthesis, transport and catabolism; (R) General function prediction only; (S) Function unknown; (T) Signal transduction mechanisms; (U) Intracellular trafficking, secretion, and vesicular transport; (V) Defense mechanisms; (W) Extracellular structures; (Y) Nuclear structure; (Z) Cytoskeleton. The number of proteins in the graphic might exceed the total of predicted ESP because some were grouped in more than one functional category.
(TIF)
Gene ontology (GO) analysis of the identified ESPs from the Eg PSCs transcriptome. The figure shows the number of mapped proteins identified in this study as a function of all the available GO terms of level 2 for (A) biological process, (B) cellular component, and (C) molecular function.
(TIF)
KEGG pathway analysis of the Eg PSCs transcriptome sequences.
(XLSX)
Validation evaluation of the predicted ESPs from the Eg PSCs transcriptome.
(XLS)
Overview of the predicted ESPs from the Eg PSCs transcriptome. ESPs were conceptually translated and inferred from the coding domains of transcriptomic sequences. Domain analysis of ESPs was then carried out using InterProScan.
(XLS)
Domains associated with carbohydrate metabolism in the ESP.
(XLSX)
Domains related to post-translational modification, protein turnover, and chaperones in the ESPs.
(XLSX)
Domains of the proteases and protease inhibitors in the ESPs.
(XLSX)
Overview of the KEGG pathways involved in the predicted ESPs.
(XLSX)
The most abundant transcripts in the ESPs of the Eg PSCs based on RPKM (reads per kilobase per million reads).
(XLSX)
The proteins that were significantly enriched in the KEGG pathways of the predicted ESPs.
(XLSX)
Acknowledgments
We would like to thank Ms Ling Wang (OE company, Shanghai) for helping in data analysis, and Professor Werner Goebel for allowing the quote of the figure that described the metabolic pathways in this study.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. The raw data mentioned in our manuscript are available at http://www.ncbi.nlm.nih.gov/sra/?term=SRP040541.
Funding Statement
This study is supported by grants from the National Natural Science Foundation of China (Nos. 81371841 to JC, 81371842 to YS) and the National S & T Major Program (Nos. 2012ZX10004-201, 2013ZX10004805 to JC). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Carmena D, Sánchez-Serrano LP, Barbero-Martínez I (2008) Echinococcus granulosus infection in Spain. Zoonoses Public Health 55: 156–165. [DOI] [PubMed] [Google Scholar]
- 2. Battelli G (2009) Echinococcosis: costs, losses and social consequences of a neglected zoonosis. Vet Res Commun 33: 47–52. [DOI] [PubMed] [Google Scholar]
- 3. Hotez PJ, Savioli L, Fenwick A (2012) Neglected tropical diseases of the Middle East and North Africa: review of their prevalence distribution, and opportunities for control. PLoS Negl Trop Dis 6: e1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McManus DP, Zhang W, Li J, Bartley PB (2003) Echinococcosis. Lancet 362: 1295–1304. [DOI] [PubMed] [Google Scholar]
- 5. Craig PS, McManus DP, Lightowlers MW, Chabalgoity JA, Garcia HH, et al. (2007) Prevention and control of cystic echinococcosis. Lancet Infect Dis 7: 385–394. [DOI] [PubMed] [Google Scholar]
- 6. Li T, Ito A, Pengcuo R, Sako Y, Chen X, et al. (2011) Post-treatment follow-up study of abdominal cystic echinococcosis in tibetan communities of northwest Sichuan Province, China. PLoS Negl Trop Dis 5: e1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moro PL, Gilman RH, Verastegui M, Bern C, Silva B, et al. (1999) Human hydatidosis in the central Andes of Peru: evolution of the disease over 3 years. Clin Infect Dis 29: 807–812. [DOI] [PubMed] [Google Scholar]
- 8. McManus DP, Gray DJ, Zhang W, Yang Y (2012) Diagnosis, treatment, and management of echinococcosis. BMJ 344: e3866. [DOI] [PubMed] [Google Scholar]
- 9. Monteiro KM, de Carvalho MO, Zaha A, Ferreira HB (2010) Proteomic analysis of the Echinococcus granulosus metacestode during infection of its intermediate host. Proteomics 10: 1985–1999. [DOI] [PubMed] [Google Scholar]
- 10.Thompson RCA, Lymbery AJ. (1995) Echinococcus and Hydatid Disease. CAB International, Wallingford, 1–50. [Google Scholar]
- 11. Pensel PE, Castro S, Allemandi D, Bruni SS, Palma SD, et al. (2014) Enhanced chemoprophylactic and clinical efficacy of albendazole formulated as solid dispersions in experimental cystic echinococcosis. Vet Parasitol 203: 80–86. [DOI] [PubMed] [Google Scholar]
- 12. Li ZJ, Wang YN, Wang Q, Zhao W (2012) Echinococcus granulosus 14-3-3 protein: a potential vaccine candidate against challenge with Echinococcus granulosus in mice. Biomed Environ Sci 25: 352–358. [DOI] [PubMed] [Google Scholar]
- 13. Dzik JM (2006) Molecules released by helminth parasites involved in host colonization. Acta Biochim Pol 53: 33–64. [PubMed] [Google Scholar]
- 14. Kanna JH, Chain BM (2006) Modulation of dendritic cell differentiation and cytokine secretion by the hydatid cyst fluid of Echinococcus granulosus . Immunology 118: 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Riganò R, Buttari B, Profumo E, Ortona E, Delunardo F, et al. (2007) Echinococcus granulosus antigen B impairs human dendritic cell differentiation and polarizes immature dendritic cell maturation towards a Th2 cell response. Infect Immun 75: 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nono JK, Pletinckx K, Lutz MB, Brehm K (2012) Excretory/Secretory-products of Echinococcus multilocularis larvae induce apoptosis and tolerogenic properties in dendritic cells in vitro . PLoS Negl Trop Dis 6: 1516–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Virginio VG, Monteiro KM, Drumond F, de Carvalho MO, Vargas DM, et al. (2012) Excretory/secretory products from in vitro-cultured Echinococcus granulosus protoscoleces. Mol Biochem Parasitol 183: 15–22. [DOI] [PubMed] [Google Scholar]
- 18. Rosenzvit MC, Camicia F, Kamenetzky L, Muzulin PM, Gutierrez AM (2006) Identification and intra-specific variability analysis of secreted and membrane-bound proteins from Echinococcus granulosus . Parasitol Int 55: S63–67. [DOI] [PubMed] [Google Scholar]
- 19. Eisenreich W, Heesemann J, Rudel T, Goebel W (2013) Metabolic host responses to infection by intracellular bacterial pathogens. Front Cell Infect Microbiol 3: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aziz A, Zhang W, Li J, Loukas A, McManus DP, et al. (2011) Proteomic characterization of Echinococcus granulosus hydatid cyst fluid from sheep, cattle and humans. J Proteomics 74: 1560–1572. [DOI] [PubMed] [Google Scholar]
- 21. Chemale G, van Rossum AJ, Jefferies JR, Barrett J, Brophy PM, et al. (2003) Proteomic analysis of the larval stage of the parasite Echinococcus granulosus: the causative agent of cystic hydatid disease. Proteomics 3: 1633–1636. [DOI] [PubMed] [Google Scholar]
- 22. Cui SJ, Xu LL, Zhang T, Xu M, Yao J, et al. (2013) Proteomic characterization of larval and adult developmental stages in Echinococcus granulosus reveals novel insight into host-parasite interactions. J Proteomi 84: 158–175. [DOI] [PubMed] [Google Scholar]
- 23. Moreno Y, Gros PP, Tam M, Segura M, Valanparambil R, et al. (2011) Proteomic analysis of excretory-secretory products of Heligmosomoides polygyrus assessed with next-generation sequencing transcriptomic information. PLoS Negl Trop Dis 5: e1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cantacessi C, Mulvenna J, Young ND, Kasny M, Horak P, Aziz A, Hofmann A, Loukas A, Gasser RB (2012) A deep exploration of the transcriptome and ‘excretory/secretory’ proteome of adult Fascioloides magna . Mol Cell Proteomics 11 (11) 1340–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xiao N, Qiu J, Nakao M, Nakaya K, Yamasaki H, et al. (2003) Short report: Identification of Echinococcus species from a yak in the Qinghai-Tibet plateau region of China. Am J Trop Med Hyg 69: 445–446. [PubMed] [Google Scholar]
- 26. Liao X, Cheng L, Xu P, Lu G, Wachholtz M, et al. (2013) Transcriptome analysis of crucian carp (Carassius auratus), an important aquaculture and hypoxia-tolerant species. PLoS One 8: e62308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stefanni S, Bettencourt R, Pinheiro M, Moro GD, Bonqirni L, et al. (2014) Transcriptome of the Deep-Sea Black Scabbardfis, Aphanopus carbo (Perciformes: Trichiuridae): Tissue-specific expression patterns and candidate genes associated to depth adaptation. Int J Genomics 2014: 267482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. (2009) SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- 29. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- 30. Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, et al. (2013) The genomes of four tapeworm species reveal adaptations to parasitism. Nature 496: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng H, Zhang W, Zhang L, Zhang Z, Li J, et al. (2013) The genome of the hydatid tapeworm Echinococcus granulosus . Nat Genet 45: 1168–1175. [DOI] [PubMed] [Google Scholar]
- 32. Young ND, Jex A R, Li B, Liu S, Yang L, et al. (2012) Whole-genome sequence of Schistosoma haematobium. . Nat Genet 44: 221–225. [DOI] [PubMed] [Google Scholar]
- 33. Zhou Yan, Zheng Huajun, Chen Xiangyi, Zhang Lei, Wang Kai, et al. (2009) The Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium (2009) The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature 460: 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berriman M, Haas BJ, LoVerde PT, Wilson RA, Dillon GP, et al. (2009) The genome of the blood fluke Schistosoma mansoni . Nature 60: 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, et al. (2009) InterPro: The integrative protein signature database. Nucleic Acids Res 37: D211–D215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie C, Mao X, Huang J, Ding Y, Wu J, et al. (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39: W316–W322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Garg G, Ranganathan S (2012) Helminth secretome database (HSD): a collection of helminth excretory/secretory proteins predicted from expressed sequence tags (ESTs). BMC Genomics 13: S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garg G, Ranganathan S (2011) In silico secretome analysis approach for next generation sequencing transcriptomic data. BMC Genomics 12: S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iseli C, Jongeneel CV, Bucher P (1999) ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol 138–148. [PubMed] [Google Scholar]
- 41. Bendtsen JD, Jensen LJ, Blom N, Von Heijne G, Brunak S (2004) Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel 17: 349–356. [DOI] [PubMed] [Google Scholar]
- 42. Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795. [DOI] [PubMed] [Google Scholar]
- 43. Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016. [DOI] [PubMed] [Google Scholar]
- 44. Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–971. [DOI] [PubMed] [Google Scholar]
- 45. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- 46. Bennuru S, Semnani R, Meng Z, Ribeiro JM, Veenstra TD, et al. (2009) Brugia malayi excreted/secreted proteins at the host/parasite interface: stage- and gender-specific proteomic profiling. PLoS Negl Trop Dis 3: e410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Craig H, Wastling JM, Knox DP (2006) A preliminary proteomic survey of the in vitro excretory/secretory products of fourth-stage larval and adult Teladorsagia circumcincta . Parasitology 132: 535–543. [DOI] [PubMed] [Google Scholar]
- 48. Gourbal BE, Guillou F, Mitta G, Sibille P, Theron A, et al. (2008) Excretory-secretory products of larval Fasciola hepatica investigated using a two-dimensional proteomic approach. Mol Biochem Parasitol 161: 63–66. [DOI] [PubMed] [Google Scholar]
- 49. Ju JW, Joo HN, Lee MR, Cho SH, Cheun HI, et al. (2009) Identification of a serodiagnostic antigen, legumain, by immunoproteomic analysis of excretory-secretory products of Clonorchis sinensis adult worms. Proteomics 9: 3066–3078. [DOI] [PubMed] [Google Scholar]
- 50. Moreno Y, Geary TG (2008) Stage- and gender-specific proteomic analysis of Brugia malayi excretory-secretory products. PLoS Negl Trop Dis 2: e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Smith SK, Nisbet AJ, Meikle LI, Inglis NF, Sales J, et al. (2009) Proteomic analysis of excretory/secretory products released by Teladorsagia circumcincta larvae early post-infection. Parasite Immunol 31: 10–19. [DOI] [PubMed] [Google Scholar]
- 52. Mulvenna J, Hamilton B, Nagaraj SH, Smyth D, Loukas A, et al. (2009) Proteomics analysis of the excretory/secretory component of the blood feeding stage of the hookworm, Ancylostoma caninum . Mol Cell Proteomics 8: 109–121. [DOI] [PubMed] [Google Scholar]
- 53. Knudsen GM, Medzihradszky KF, Lim KC, Hansell E, McKerrow JH (2005) Proteomic analysis of Schistosoma mansoni cercarial secretions. Mol Cell Proteomics 4: 1862–1875. [DOI] [PubMed] [Google Scholar]
- 54. Curwen RS, Ashton PD, Sundaralingam S, Wilson RA (2006) Identification of novel proteases and immunomodulators in the secretions of schistosome cercaria that facilitate host entry. Mol Cell Proteomics 5: 835–844. [DOI] [PubMed] [Google Scholar]
- 55. Liu F, Cui SJ, Hu W, Feng Z, Wang ZQ, et al. (2009) Excretory/secretory proteome of the adult development stage of human blood fluke, Schistosoma japonicum . Mol Cell Proteomics 8: 1236–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Robinson MW, Menon R, Donnelly SM, Dalton JP, Ranganathan S (2009) An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica. Mol Cell Proteomics 8: 1891–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hewitson JP, Harcus YM, Curwen RS, Dowle AA, Atmadja AK, et al. (2008) The secretome of the filarial parasite, Brugia malayi: Proteomic profile of adult excretory-secretory products. Mol Biochem Parasitol 160: 8–21. [DOI] [PubMed] [Google Scholar]
- 58. Robinson MW, Gare DC, Connolly B (2005) Profiling excretory/secretory proteins of Trichinella spiralis muscle larvae by two-dimensional gel electrophoresis and mass spectrometry. Vet Parasitol 132: 37–41. [DOI] [PubMed] [Google Scholar]
- 59. Vercauteren I, Geldhof P, Peelaers I, Claerebout E, Berx G, et al. (2003) Identification of excretory-secretory products of larval and adult Ostertagia ostertagi by immunoscreening of cDNA libraries. Mol Biochem Parasitol 126: 201–208. [DOI] [PubMed] [Google Scholar]
- 60. Ma SM, Maillard S, Zhao HL, Huang X, Wang H, et al. (2008) Assessment of Echinococcus granulosus polymorphism in Qinghai province, People's Republic of China. Parasitol Res 102: 1201–1206. [DOI] [PubMed] [Google Scholar]
- 61. Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 62. Menon R, Gasser RB, Mitreva M, Ranganathan S (2012) An analysis of the transcritome of Teladorasgia circumcincta: its biological and biotechnological implications. BMC Genomics 13: S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wolf DA, Jackson PK (1998) Cell cycle: oiling the gears of anaphase. Curr Biol 8: R636–639. [DOI] [PubMed] [Google Scholar]
- 64. Leipe DD, Koonin EV, Aravind L (2004) STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol 343: 1–28. [DOI] [PubMed] [Google Scholar]
- 65. Hopkins AL, Groom CR (2002) The druggable genome. Nat Rev Drug Discov 1: 727–730. [DOI] [PubMed] [Google Scholar]
- 66. Lauwaet T, Oliveira MJ, Mareel M, Leroy A (2000) Molecular mechanisms of invasion by cancer cells, leukocytes and microorganisms. Microbes Infect 2: 923–931. [DOI] [PubMed] [Google Scholar]
- 67. Parkinson J, Wasmuth JD, Salinas G, Bizarro CV, Sanford C, et al. (2012) A transcriptomic analysis of Echinococcus granulosus larval stages: implications for parasite biology and host adaptation. PLoS Negl Trop Dis 6: e1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Carmena D, Mart nez J, Benito A, Guisantes JA (2004) Characterization of excretory-secretory products from protoscoleces of Echinococcus granulosus and evaluation of their potential for immunodiagnosis of human cystic echinococcosis. Parasitology 129: 371–378. [DOI] [PubMed] [Google Scholar]
- 69. Stadelmann B, Spiliotis M, Müller J, Scholl S, Müller N, et al. (2010) Echinococcus multilocularis phosphoglucose isomerase: a glycolytic enzyme involved in metacestode growth and parasite-host cell interactions. Int J Parasitol 40: 1563–1574. [DOI] [PubMed] [Google Scholar]
- 70. Asahi H, Osman A, Cook RM, LoVerde PT, Stadecker MJ (2000) Schistosoma mansoni phosphoenolpyruvate carboxykinase, a novel egg antigen: immunological properties of the recombinant protein and identification of a T-cell epitode. Infect Immun 68: 3385–3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pan W, Zhou HJ, Shen YJ, Wang Y, Xu YX, et al. (2013) Surveillance on the status of immune cells after Echinococcus granulosus protoscoleces infection in Balb/c mice. PLoS One 8: e59746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang W, McManus DP (2006) Recent advances in the immunology and diagnosis of echinococcosis. FEMS Immunol Med Microbiol 47: 24–41. [DOI] [PubMed] [Google Scholar]
- 73. Franco MC, Ye Y, Refakis CA, Feldman JL, Stokes AL, et al. (2013) Nitration of Hsp90 induces cell death. Proc Natl Acad USA 110: E1102–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Angel SO, Matrajt M, Echeverria PC (2013) A review of recent patents on the protozoan parasite HSP90 as a drug target. Recent Pat Biotechnol 7: 2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Seifart Gomes C, Izar B, Pazan F, Mohamed W, Mraheil MA, et al. (2011) Universal stress proteins are important for oxidative and acid stress resistance and growth of Listeria monocytogenes EGD-e in vitro and in vivo . PLoS One 6: e24965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zahoor Z, Davies AJ, Kirk RS, Rollinson D, Walker AJ (2010) Larval excretory-secretory products from the parasite Schisosoma mansoni modulate HSP70 protein expression in defence cells of its snail host, Biomphalaria glabrata . Cell Stress and Chaperones 15: 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Karanu FN, Rurangirwa FR, McGuire TC, Jasmer DP (1993) Haemonchus contortus: Identification of proteases with diverse characteristics in adult worm excretory-secretory products. Exp Parasitol 77: 362–371. [DOI] [PubMed] [Google Scholar]
- 78. Kovaleva ES, Masler EP, Skantar AM, Chitwood DJ (2004) Novel matrix metalloproteinase from the cyst nematodes Heterodera glycines and Globodera rostochiensis . Mol Biochem Parasitol 136: 109–112. [DOI] [PubMed] [Google Scholar]
- 79. Yatsuda AP, Bakker N, Krijgsveld J, Knox DP, Heck AJ, et al. (2006) Identification of secreted cysteine proteases from the parasitic nematode Haemonchus contortus detected by biotinylated inhibitors. Infect Immun 74: 1989–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li J, Zhang WB, Loukas A, Lin RY, Ito A, et al. (2004) Functional expression and characterization of Echinococcus granulosus thioredoxin peroxidase suggests a role in protection against oxidative damage. Gene 326: 157–165. [DOI] [PubMed] [Google Scholar]
- 81. Ortona E, Margutti P, Delunardo F, Nobili V, Profumo E, et al. (2005) Screening of an Echinococcus granulosus cDNA library with IgG4 from patients with cystic echinococcosis identifies a new tegumental protein involved in the immune escape. Clin Exp Immunol 142: 528–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Andrade MA, Siles-Lucas M, Espinoza E, Pérez Arellano JL, Gottstein B, et al. (2004) Echinococcus multilocularis laminated-layer components and the E14t 14-3-3 recombinant protein decrease NO production by activated rat macrophages in vitro. Nitric Oxide 10: 150–155. [DOI] [PubMed] [Google Scholar]
- 83. Kamenetzky L, Muzulin PM, Gutierrez AM, Angel SO, Zaha A, et al. (2005) High polymorphism in genes encoding antigen B from human infecting strains of Echinococcus granulosus . Parasitology 131: 805–815. [DOI] [PubMed] [Google Scholar]
- 84. Machadao DC, Horton D, Harrop R, Peachell PT, Helm BA (1996) Potential allergens stimulate the release of mediators of the allergic response from cells of mast cell lineage in the absence of sensitization with antigen specific IgE. Eur J Immuol 26: 2972–2980. [DOI] [PubMed] [Google Scholar]
- 85. Ortona E, Margutti P, Delunardo F, Vaccari S, Rigano R, et al. (2003) Molecular and immunological characterization of the C-terminal region of a new Echinococcus granulosus Heat Shock Protein 70. Parasite Immunol 25: 119–126. [DOI] [PubMed] [Google Scholar]
- 86. Hong C, Qiu X, Li Y, Huang Q, Zhong Z, et al. (2010) Functional analysis of recombinant calreticulin fragment 39–272: Implications for immunobiological activities of calreculin in health and disease. J Immunol 185: 4561–4569. [DOI] [PubMed] [Google Scholar]
- 87. Neves SR, Ram PT, Iyengar R (2002) G protein pathways. Science 296: 1636–1639. [DOI] [PubMed] [Google Scholar]
- 88. Patterson GI, Padgett RW (2000) TGF beta-related pathways. Roles in Caenorhabditis elegans development. Trends Genet 16: 27–33. [DOI] [PubMed] [Google Scholar]
- 89. Beall MJ, Pearce EJ (2002) Transforming growth factor-beta and insulin-like signaling pathways in parasitic helminthes. Int J Parasitol 32: 399–404. [DOI] [PubMed] [Google Scholar]
- 90. Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, et al. (1996) Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science 274: 1389–1351. [DOI] [PubMed] [Google Scholar]
- 91. Sze JY, Victor M, Loer C, Shi Y, Ruvkun G (2000) Food and metallic signaling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403: 560–564. [DOI] [PubMed] [Google Scholar]
- 92. Nisbet AJ, Redmond DL, Mattews JB, Watkins C, Yaga R, et al. (2008) Stage-specific gene expression in Teladorsagia circumcincta (Nematode: Strongylida) infective larvae and early parasitic stages. Int J Parasitol 38: 829–838. [DOI] [PubMed] [Google Scholar]
- 93. Gomez-Escobar N, van den Biggelaar A, Maizels R (1997) A member of the TGF-beta receptor gene family in the parasitic nematode Brugia pahangi . Gene 199: 101–109. [DOI] [PubMed] [Google Scholar]
- 94. Gomez-Escobar N, Gregory WF, Maizels RM (2000) Identification of tgh-2, a filarial nematode homolog of Caenorhabditis elegans daf-7 and human transforming growth factor beta, expressed in microfilarial and adult stages of Brugia malayi . Infect Immun 68: 6402–6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Crook M, Thompson FJ, Grant WN, Viney ME (2005) Daf-7 and the development of Strongyloides ratti and Parastrongyloides trichosuri . Mol Biochem Parasitol 139: 213–223. [DOI] [PubMed] [Google Scholar]
- 96. Hemer S, Konrad C, Spiliotis M, Koziol U, Schaack D, et al. (2014) Host insulin stimulates Echinococcus multilocularis insulin signaling pathways and larval development. BMC Biol 12: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Carmena D, Cardona GA (2013) Canine echinococcosis: global epidemiology and genotypic diversity. Acta Trop 128: 441–460. [DOI] [PubMed] [Google Scholar]
- 98. Heath DD, Robinson C, Shakes T, Huang Y, Gulnur T, et al. (2012) Vaccination of bovines against Echinococcus granulosus (cystic echinococcosis). Vaccine 30: 3076–3081. [DOI] [PubMed] [Google Scholar]
- 99. Alvarez Rojas CA, Gauci CG, Lightowlers MW (2013) Antigenic differences between the EG95-related proteins from Echinococcus granulosus G1 and G6 genotypes: implications for vaccination. Parasite Immunol 35: 99–102. [DOI] [PubMed] [Google Scholar]
- 100. Zhu XQ, Dou LQ, Sun XQ, Shi XH, Wang XH, et al. (1991) 21: 8–10. Studies on the secretory/excretory proteins of Echinococcus granulosus—the immunogenicity of protosolex secretory/excretory proteins. Chin J Vet Sci Tech 21: 8–10. [Google Scholar]
- 101. Margutti P, Ortona E, Delunardo F, Tagliani A, Profumo E, et al. (2008) Thiredoxin peroxidase from Echinococcus granulosus: a candidate to extend the antigenic panel for the immunodiagnosis of human cystic echinococcosis. Diagn Microbiol Infect Dis 60: 279–285. [DOI] [PubMed] [Google Scholar]
- 102. Harispe L, Garcia G, Arbildi P, Pascovich L, Chalar C, et al. (2010) Biochemical analysis of a recombinant glutathione transferase from the cestode Echinococcus granulosus . Acta Trop 114: 31–36. [DOI] [PubMed] [Google Scholar]
- 103. Hu M, Su Z, Yin Y, Li J, Wei Q (2012) Calcineurin B subunit triggers innate immunity and acts as a novel Engerix-B HBV vaccine adjuvant. Vaccine 30: 4719–4727. [DOI] [PubMed] [Google Scholar]
- 104. Zhang W, Li J, Jones MK, Zhang Z, Zhao L, et al. (2010) The Echinococcus granulosus antigen B gene family comprises at least 10 unique genes in five subclasses which are differentially expressed. PLoS Negl Trop Dis 4: e784. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genotype identification of E. granulosus . (A) PCR amplification. M, DNA maker; Cytb, 601 bp; Cox1, 885 bp. (B) Sequence alignment of the cytochrome b (cytb) gene. Bases that differed are marked with red boxes.
(TIF)
Length distribution of singletons and isotigs of the Eg PSCs transcriptome.
(TIF)
Gene ontology (GO) analysis of the Eg PSCs transcriptome. BLASTP against SwissProt and GO mapping of identified proteins (performed with BLAST2GO) [61].
(TIF)
Distribution of the KOG functional categories of the proteins identified from the Eg PSCs transcriptome. Percentages and numbers of proteins in each functional category are indicated in the sectors of the circle. KOG functional categories: (A) RNA processing and modification; (B) Chromatin structure and dynamics; (C) Energy production and conversion; (D) Cell cycle control, cell division, chromosome partitioning; (E) Amino acid transport and metabolism; (F) Nucleotide transport and metabolism; (G) Carbohydrate transport and metabolism; (H) Coenzyme transport and metabolism; (I) Lipid transport and metabolism; (J) Translation, ribosomal structure and biogenesis; (K) Transcription; (L) Replication, recombination and repair; (M) Cell wall/membrane/envelope biogenesis; (N) Cell motility; (O) Posttranslational modification, protein turnover, chaperones; (P) Inorganic ion transport and metabolism; (Q) Secondary metabolites biosynthesis, transport and catabolism; (R) General function prediction only; (S) Function unknown; (T) Signal transduction mechanisms; (U) Intracellular trafficking, secretion, and vesicular transport; (V) Defense mechanisms; (W) Extracellular structures; (Y) Nuclear structure; (Z) Cytoskeleton. The number of proteins in the graphic might exceed the total of predicted ESP because some were grouped in more than one functional category.
(TIF)
Gene ontology (GO) analysis of the identified ESPs from the Eg PSCs transcriptome. The figure shows the number of mapped proteins identified in this study as a function of all the available GO terms of level 2 for (A) biological process, (B) cellular component, and (C) molecular function.
(TIF)
KEGG pathway analysis of the Eg PSCs transcriptome sequences.
(XLSX)
Validation evaluation of the predicted ESPs from the Eg PSCs transcriptome.
(XLS)
Overview of the predicted ESPs from the Eg PSCs transcriptome. ESPs were conceptually translated and inferred from the coding domains of transcriptomic sequences. Domain analysis of ESPs was then carried out using InterProScan.
(XLS)
Domains associated with carbohydrate metabolism in the ESP.
(XLSX)
Domains related to post-translational modification, protein turnover, and chaperones in the ESPs.
(XLSX)
Domains of the proteases and protease inhibitors in the ESPs.
(XLSX)
Overview of the KEGG pathways involved in the predicted ESPs.
(XLSX)
The most abundant transcripts in the ESPs of the Eg PSCs based on RPKM (reads per kilobase per million reads).
(XLSX)
The proteins that were significantly enriched in the KEGG pathways of the predicted ESPs.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. The raw data mentioned in our manuscript are available at http://www.ncbi.nlm.nih.gov/sra/?term=SRP040541.