Abstract
Small bowel (SBNETs) and pancreatic neuroendocrine tumors (PNETs) often present with liver metastases. Although liver biopsy establishes a neuroendocrine diagnosis, the primary tumor site is frequently unknown without exploratory surgery. Gene expression differences in metastases may distinguish primary SBNETs and PNETs. This study sought to determine expression differences of four genes in neuroendocrine metastases and to create a gene expression algorithm to distinguish the primary site. Nodal and liver metastases from SBNETs and PNETs (n=136) were collected at surgery under an Institutional Review Board-approved protocol. Quantitative PCR measured expression of bombesin-like receptor-3, opioid receptor kappa-1, oxytocin receptor, and secretin receptor in metastases. Logistic regression models defined an algorithm predicting the primary tumor site. Models were developed on a training set of 21 nodal metastases and performance was validated on an independent set of nodal and liver metastases. Expression of all four genes was significantly different in SBNET compared to PNET metastases. The optimal model employed expression of bombesin-like receptor-3 and opioid receptor kappa-1. When these genes did not amplify, the algorithm used oxytocin receptor and secretin receptor expression, which allowed classification of all 136 metastases with 94.1% accuracy. In the independent liver metastasis validation set, 52/56 (92.9%) were correctly classified. Positive predictive values were 92.5% for SBNETs and 93.8% for PNETs. This validated algorithm accurately distinguishes SBNET and PNET metastases based on their expression of four genes. High accuracy in liver metastases demonstrates applicability to the clinical setting. Studies assessing this algorithm’s utility in prospective clinical decision-making are warranted.
Keywords: Neuroendocrine tumors, small bowel, pancreas, gene expression classifier, liver metastasis, diagnosis
Introduction
Small bowel (SBNETs) and pancreatic neuroendocrine tumors (PNETs) have an annual incidence of around 1.2 per 100,000 people in the United States, which has increased significantly since the 1970s.[1] These tumors present with metastases in 50–85% of cases, often to the liver.[2] While the neuroendocrine diagnosis can be established from tissue obtained from percutaneous liver biopsy, the primary site of origin generally cannot be determined from histology alone. Endoscopic and radiologic investigations, including CT, MRI, and peptide-receptor-based imaging strategies can assist in locating the primary tumor, but even after an optimal workup, the primary site of origin remains unknown prior to surgery in up to 20% of cases.[3–5] As primary gastric, duodenal, rectal, and colonic NETs can be identified on endoscopy, and bronchial NETs have radiographically detectable lung lesions, most NETs of unknown primary arise from either the small bowel or the pancreas.[3,6,7]
Knowing the primary site of origin of neuroendocrine liver metastases can alter clinical management. Despite the high incidence of metastases at diagnosis, NETs are quite treatable, and patients with stage IV disease may survive for many years.[8,9] Resection of the primary tumor is beneficial even in the setting of metastatic disease, and surgical planning is improved when the location of the primary tumor is known.[10–16] Even when surgery is not being considered, medical treatments differ depending on the primary site.[17] Targeted therapeutics, such as the mammalian target of rapamycin (mTOR) inhibitor everolimus and the multi-kinase inhibitor sunitinib are approved for PNET treatment based on improved progression-free survival in randomized trials, but are less effective in SBNETs.[18,19] Similarly, the chemotherapeutics streptozocin and temozolomide show greater effectiveness in PNETs than SBNETs, and are rarely prescribed for the latter.[17,20,21,4,22] Due to the importance of identifying the primary site, methods to distinguish the source of NETs from biopsy of metastases have been investigated. Immunohistochemistry (IHC) has been studied extensively for this purpose, but while sensitive candidate markers exist, the low specificity of individual markers may limit overall accuracy.[23,24]
Differences in gene expression between metastases arising from SBNETs and PNETs can be exploited to distinguish the site of unknown primaries. Gene expression signatures differentiating NETs have been reported, but suffer from small sample sizes, inclusion of specimens from primary tumors and metastases, or lack of sufficient validation, and have yet to find widespread clinical application.[25–27] Our own group has investigated gene expression profiles of SBNETs and PNETs to identify markers that could indicate the primary site of origin. Initial studies of G protein-coupled receptor and exon expression arrays in a limited number of clinical specimens suggested that a formula based on expression of the oxytocin receptor (OXTR) and secretin receptor (SCTR) in metastases might discriminate between metastases of small bowel versus pancreatic origin.[28] However, validation of this formula in 45 SBNET and PNET liver metastases revealed an accuracy of only 71% for determining the primary site.[29]
More recently we studied a panel of 13 genes, chosen from earlier expression array experiments, in a large number of primary and metastatic SBNET and PNET specimens to define novel therapeutic targets. In addition to identifying the gastric inhibitory polypeptide receptor (GIPR) as a promising target, this study found large differences between primary SBNET and PNET tumors in expression of the bombesin-like receptor-3 (BRS3) and the opioid receptor kappa-1 (OPRK1).[30] This suggested that expression of these genes might allow determination of the primary site of a liver metastasis of unknown origin. The aim of the present study was therefore to further characterize expression of these genes in SBNET and PNET metastases, and to develop and validate an algorithm to determine the primary tissue of origin based on gene expression in metastatic tissue.
Materials and Methods
Patients and Tumor Samples
Tissue specimens from liver and lymph node SBNET and PNET metastases in addition to primary tumors were collected at surgery under an Institutional Review Board-approved protocol. SBNETs were defined as tumors arising between the ligament of Treitz and the ileocecal valve. All patients provided informed consent. Tissue samples were preserved in RNAlater solution at −20°C (Life Technologies, Grand Island, NY). The primary site of origin was confirmed at the time of surgery. Clinical correlations employed the University of Iowa Neuroendocrine Tumor Database.[31]
Quantitative PCR
Expression levels of four genes (BRS3, OPRK1, OXTR, and SCTR) shown previously to have significant expression differences between SBNETs and PNETs were assessed as described.[29,30] Briefly, total RNA was extracted by the TRIzol method and reverse transcribed to cDNA (Life Technologies). Quantitative PCR (qPCR) was performed in triplicate with Taqman probes and primers,[29] using the 7900 HT-Fast Analyzer System (Life Technologies). Mean expression by threshold cycles (Ct) was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and polymerase (RNA) II polypeptide A (POLR2A), which are internal control genes that are uniformly and highly expressed, to give dCT. Lower dCT indicates higher expression, with fold changes calculated as 2(dCT1-dCT2).
Algorithm development and statistical analysis
Progression-free and overall survival were estimated by Kaplan-Meier method from the date of surgery. Continuous clinical variables were compared by Wilcoxon Rank-Sum test and categorical variables by Fisher Exact test. Gene expression (dCT) in nodal and liver metastases was compared by Welch’s t-test. Model development employed multivariate logistic regression on a training set of nodal metastases without associated liver metastases (n=21). Models were developed with combinations of informative genes, and an overall prediction algorithm prioritized the best-performing models first. The algorithm was locked and its performance was assessed on all metastases (n=136) and on the independent validation set of liver metastases (n=56). Accuracy, sensitivity, specificity, and positive predictive values for primary site predictions were determined. All calculations were performed in R v.3.0.1 (Vienna, Austria). A web-based metastasis primary site prediction tool incorporating the prediction algorithm was developed using JavaScript.
Results
Clinical characteristics of patients with metastatic tumors
In total, 136 metastases from 86 patients were analyzed, representing 61 primary SBNETs and 25 primary PNETs. Thirty patients had metastatic tissue collected from lymph nodes only, while six had metastatic tissue collected from liver only. The remaining patients had tissue collected from both liver and lymph node metastases. Age at surgery ranged from 32 to 85 years old, with a median age of 60.6 in SBNET patients and 55.1 in PNET patients (p=0.07). Female patients comprised 45.3% of the cohort, but there were significantly fewer women in the SBNET than the PNET group (38 vs. 64%, p=0.03). With a median follow-up of 3.3 years, there was no difference in estimated median progression-free (PFS) or overall survival (OS) by primary tumor location (small bowel vs. pancreatic primary, PFS 2.5 vs. 2.0 years, p=0.7; OS not yet reached vs. 6.1 years, p=0.4). The primary tumor site was unknown prior to surgery in 24 patients (28%).
Gene expression in metastases
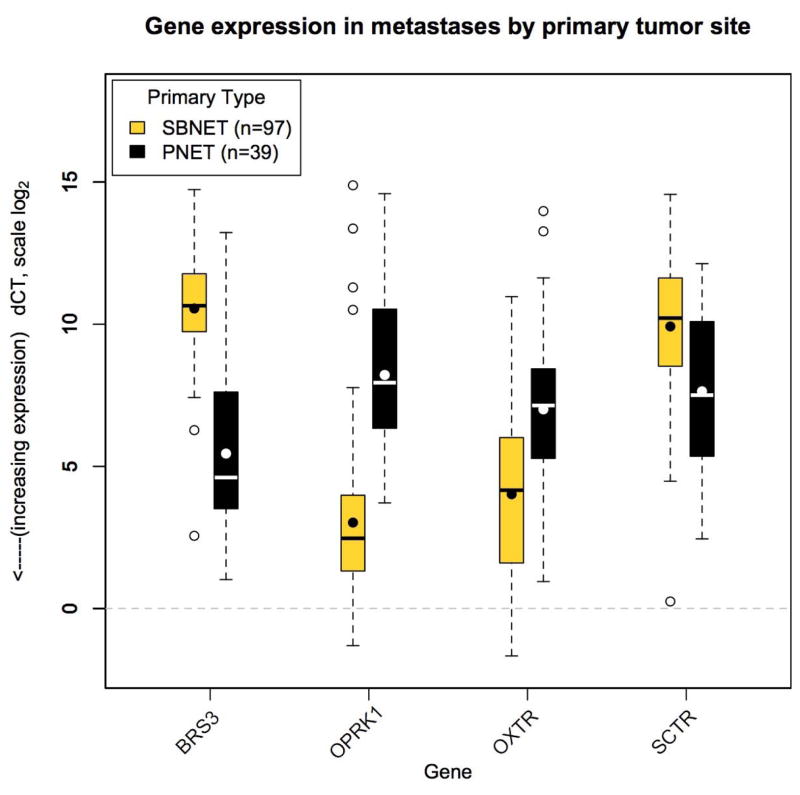
Four genes, BRS3, OPRK1, OXTR, and SCTR were previously shown to have significantly different expression in primary SBNETs compared to PNETs.[30] To determine whether expression differences by primary tumor type were also present in metastatic tissues, the dCTs of these four genes were measured in 97 SBNET and 39 PNET liver and lymph node metastases (Figure 1). All four genes showed significantly different expression in metastases based on primary type (Table 1). Fold-expression differences between the metastases of the two primary tumor types ranged from 4.9-fold for SCTR to 36.5-fold for OPRK1. Two genes, BRS3 and SCTR, showed lower dCTs in PNET metastases while the other two, OPRK1 and OXTR, had lower dCTs in SBNET metastases. The large differences in gene expression between metastases from different primary sites suggested that measuring their expression could help distinguish the primary site of origin.
Figure 1.
Gene expression by primary tumor site. Expression of BRS3, OPRK1, OXTR, and SCTR in small bowel (light boxes) and pancreatic (dark boxes) neuroendocrine tumor metastases is significantly different by primary tumor site. Gene expression shown by log-scale dCT. Lower dCT indicates higher expression. Boxes indicate 25th to 75th percentile of expression (interquartile range, IQR). The bars show median expression, the dots show the mean. Whiskers indicate 1.5*IQR and open circles show outlying observations. The dotted line at zero indicates expression level of GAPDH and POLR2A internal control genes.
Table 1.
Expression of four genes in small bowel and pancreatic neuroendocrine tumor metastases is significantly different by primary tumor type.
| SBNET mets (n=97) | PNET mets (n=39) | Mean Fold Difference | P value SBNET vs. PNET | |
|---|---|---|---|---|
| Gene | Mean dCT (IQR) | Mean dCT (IQR) | ||
| BRS3 | 10.6 (9.7–11.8) | 5.4 (3.5–7.3) | 34.4 | <0.0001 |
| OPRK1 | 3.0 (1.3–4.0) | 8.2 (6.3–10.5) | 36.5 | <0.0001 |
| OXTR | 4.0 (1.6–6.0) | 7.0 (5.3–8.4) | 7.9 | <0.0001 |
| SCTR | 9.9 (8.5–11.6) | 7.6 (5.3–10.1) | 4.9 | <0.0001 |
Abbreviations: SBNET: small bowel neuroendocrine tumor; PNET: pancreatic neuroendocrine tumor; mets: metastases; n: number of samples; IQR: interquartile range
Algorithm development
Predictive model development generally involves choosing a subset of the total dataset for model training, with the remaining cases reserved for validation. For neuroendocrine tumors, a model’s performance in discriminating the primary site of liver metastases is of greatest interest, because these are the tissues most accessible to percutaneous biopsy. To reserve the maximum number of liver metastases for validation, models were developed using a training set of nodal metastases. However, since some patients had both nodal and liver metastases, employing a training set comprised of all available nodal metastases would compromise the independence of the liver metastasis validation set. To solve this problem, the training set for model development used only “independent” nodal metastases – those without associated liver metastases (n=21 at time of model development). This approach provided a suitable training set, while preserving all liver metastases for performance assessment.
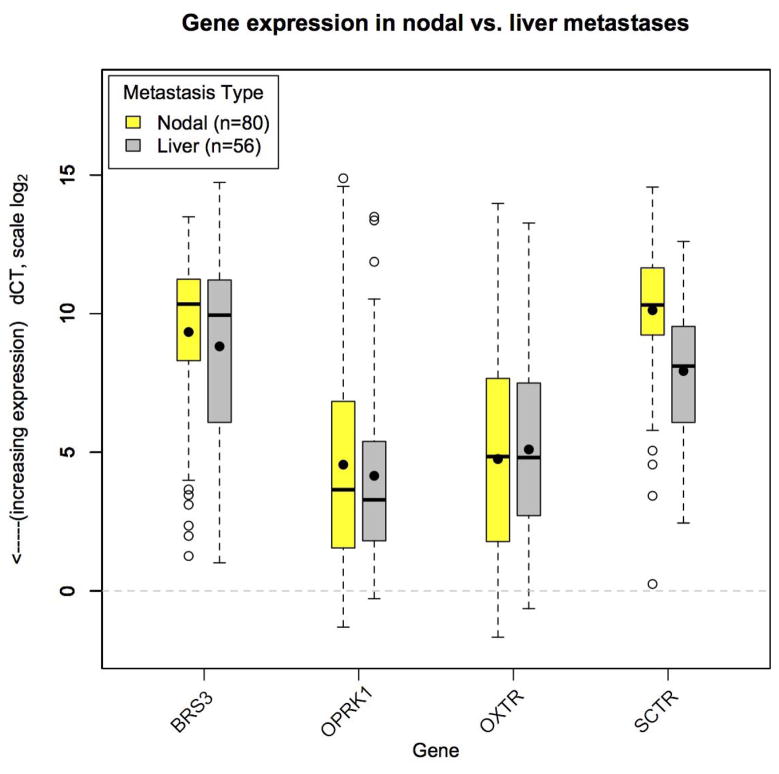
To ensure validity of a strategy treating nodal metastases as equivalent to liver metastases, expression of BRS3, OPRK1, OXTR, and SCTR was examined in 80 nodal metastases as compared to 56 liver metastases (Figure 2). Expression levels of three genes (BRS3, OPRK1, and OXTR) were not significantly different in nodal compared to liver metastases (p>0.05 for all)(Table 2). Unexpectedly, SCTR had significantly higher expression (lower dCT) in liver compared to nodal metastases from both primary tumor types (mean +3.5-fold in SBNET and +7.0-fold in PNET metastases, p<0.001 for each). From these results we conclude that BRS3, OPRK1, and OXTR have similar expression in SBNET and PNET nodal and liver metastases, and that these genes represent the strongest candidates for inclusion in nodal metastasis-derived models to predict the primary site of liver metastases.
Figure 2.
Gene expression in nodal versus liver metastases. Expression of BRS3, OPRK1, and OXTR are similar in nodal (light boxes) and liver (dark boxes) metastases. Gene expression shown by log-scale dCT. Lower dCT indicates higher expression. Boxes indicate 25th to 75th percentile of expression (interquartile range, IQR). The bars show median expression, the dots show the mean. Whiskers indicate 1.5*IQR and open circles show outlying observations. Dotted line at zero indicates expression level of GAPDH and POLR2A internal control genes.
Table 2.
Expression of BRS3, OPRK1, and OXTR is similar in nodal and liver metastases of both small bowel and pancreatic neuroendocrine tumors.
| SBNET | PNET | |||||
|---|---|---|---|---|---|---|
| Nodal Mets (n=59) | Liver Mets (n=38) | P value Nodal vs. Liver | Nodal Mets (n=21) | Liver Mets (n=18) | P value Nodal vs. Liver | |
| Gene | Mean dCT (IQR) | Mean dCT (IQR) | Mean dCT (IQR) | Mean dCT (IQR) | ||
| BRS3 | 10.6 (9.7–11.3) | 10.9 (9.9–11.8) | 0.92 | 5.9 (3.6–7.8) | 5.0 (3.6–5.9) | 0.42 |
| OPRK1 | 3.2 (1.3–4.4) | 2.8 (1.4–3.5) | 0.44 | 8.9 (6.9–11.1) | 7.4 (4.6–9.5) | 0.15 |
| OXTR | 4.0 (1.5–6.1) | 4.0 (1.8–5.5) | 0.91 | 6.6 (5.0–8.4) | 7.5 (5.5–9.0) | 0.38 |
| SCTR | 10.6 (9.6–11.9) | 8.8 (7.5–10.6) | <0.001 | 8.9 (7.5–10.4) | 6.1 (5.2–7.2) | <0.001 |
Abbreviations: SBNET: small bowel neuroendocrine tumor; PNET: pancreatic neuroendocrine tumor; mets: metastases; n: number of samples; IQR: interquartile range
Models were developed using combinations of two, or all three of the strongest candidate genes. When tested for performance in the training set, two models correctly distinguished 100% of metastases (21/21). One of these incorporated expression of BRS3, OPRK1, and OXTR, while the other used BRS3 and OPRK1 only. The two-gene model was selected as the optimal model due to its lower Akaike information criterion score (6 vs. 19), and the greater separation in the distribution of OPRK1 dCT values between SBNET and PNET metastases compared to OXTR (Figure 1). The optimal model is defined by the equation:
The result ranges from 0 to 1, with values of Prediction ≤ 0.5 indicating an SBNET primary and Prediction > 0.5 indicating a PNET primary.
For the model to return a prediction, dCT values for both BRS3 and OPRK1 must be available. Occasionally, a gene will not amplify and its Ct value cannot be determined. This occurs most often because of expression below the limit of detection in that specimen. Due to the expected failure of the optimal genes to amplify in all samples, the less-favored two-gene models, and a model employing SCTR expression, were incorporated as backups into an overall algorithm (Figure 3). The algorithm predicts the primary site based first on the optimal BRS3 and OPRK1 model (Step 1). When either of these genes do not amplify, models using OXTR and either BRS3 or OPRK1 determine the primary site (Step 2). Finally, for metastases without amplification of BRS3 or OPRK1, the less-preferred OXTR and SCTR model assigns the primary site (Step 3). If none of these models applies, the sample is deemed a technical failure.
Figure 3.
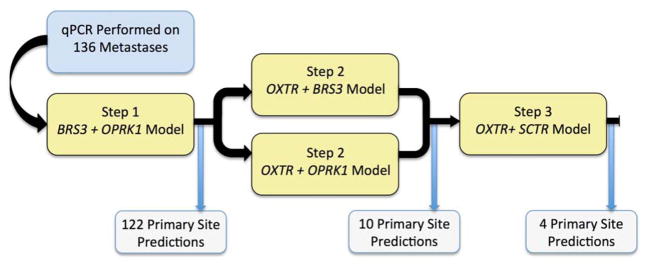
Flowchart demonstrating prediction algorithm and technical success. Predictions were possible for all 136 metastases.
Model performance
After development in the training set of nodal metastases, the algorithm’s formulae were locked and its performance was assessed in all available metastases (Table 3). The algorithm made primary site predictions in all 136 samples tested, with no technical failures. The optimal model (Step 1) made the prediction in 90% (122/136), with ten predictions made by Step 2 and the remaining four by Step 3 (Figure 3). In the combined training set and the independent set of nodal and liver metastases, the algorithm correctly classified the primary site in 128 of 136 metastases (94.1% overall accuracy). The model performed better in SBNET metastases (94/97, 96.9% sensitivity) than PNET metastases (34/39, 87.2% sensitivity, p=0.04). Overall positive predictive values were 94.9% for SBNETs and 91.9% for PNETs. Accuracy was not significantly different depending on which algorithm Step made the primary site prediction (p=0.22), however, low numbers of predictions by Steps 2 and 3 preclude full evaluation of these models’ individual performance. The optimal model (Step 1), correctly predicted 116/122 metastases (95.1%), while Step 2 correctly predicted 8/10 and Step 3 predicted 4/4.
Table 3.
The algorithm accurately classifies the primary site of small bowel and pancreatic neuroendocrine tumor metastases based on gene expression.
| All Metastases (n=136) | ||||
| Classification | ||||
| Primary Type | Correct | Incorrect | Sensitivity (95% CI) | PPV |
| SBNET | 94 | 3 | 96.9% (93.5–100.0) | 94.9% |
| PNET | 34 | 5 | 87.2% (76.7–97.7) | 91.9% |
| Liver Metastases (n=56) | ||||
| Classification | ||||
| Primary Type | Correct | Incorrect | Sensitivity (95% CI) | PPV |
| SBNET | 37 | 1 | 97.4% (92.3–100.0) | 92.5% |
| PNET | 15 | 3 | 83.3% (66.1–100.0) | 93.8% |
Abbreviations: SBNET: small bowel neuroendocrine tumor; PNET: pancreatic neuroendocrine tumor; n: number of samples; CI: confidence interval; PPV: positive predictive value
Model validation
A limitation of analyzing all metastases together is that it combines the training set and validation set, and also nodal and liver metastases arising in the same patient. To obtain the best understanding of the likely clinical performance of the algorithm, we next limited our analysis to the independent validation set of 56 liver metastases from 56 patients (Table 3). Among these metastases, the algorithm correctly assigned the primary site of origin in 52 of 56 (92.9% accuracy). Performance was again better in SBNET metastases (37/38, 97.4% sensitivity). Sensitivity in PNET liver metastases was lower at 83.3% (15/18, p=0.09), however, positive predictive values were greater than 92% for both tumor types (92.5% for SBNETs, 93.8% for PNETs). In the 24 patients with unknown primaries prior to surgery, the algorithm correctly classified the primary site in 23 (95.8%), including 11/12 liver metastases. From these results in an independent validation set of liver metastases, we conclude that the algorithm accurately discriminates SBNET and PNET metastases. The algorithm performs better for SBNET metastases, but high positive predictive values for both tumor types indicate that this validated algorithm’s results are clinically relevant.
Misclassified metastases
Closer examination of the four misclassified liver metastases revealed that all four had expression patterns of BRS3 and OPRK1 more consistent with the other primary tumor type, rather than aberrant expression of a single gene. The misclassified SBNET liver metastasis had dCTs for BRS3 and OPRK1 of 2.6 and 4.9, which with a low BRS3 dCT and high OPRK1 dCT, more closely matches the normal PNET expression pattern. The three misclassified PNET liver metastases had higher BRS3 dCTs and lower OPRK1 dCTs, which is the pattern seen in most SBNET metastases (BRS3 and OPRK1 dCTs: 8.8 and 4.6; 8.2 and 4.5; 10.7 and 5.2). All BRS3 and OPRK1 dCTs in misclassified liver metastases lay outside of the expected interquartile ranges for their true primary types, but only one of these (BRS3 in the misclassified SBNET) was a true outlier, falling outside of 1.5 times the interquartile range. From this we conclude that the Step 1 model is well calibrated to distinguish the primary site, but that variability in gene expression exists and precludes perfect primary site discrimination.
Performance in metastases from low-grade tumors was slightly better than in intermediate and high-grade metastases (low: 95/99, 95.9% correctly classified; intermediate: 25/28, 89.3%; high: 8/9, 88.9%), but these differences were not statistically significant (p=0.2). Likewise, metastases in which BRS3 or OPRK1 did not amplify (and therefore required Steps 2 or 3 of the algorithm to assign a primary site) were no less likely to be low-grade (10/14 were low-grade) than those in which both of these genes amplified (89/122 were low-grade, p=0.9). Thus, although non-low-grade metastases might be expected to show more variable gene expression than low-grade metastases, the algorithm performed well in metastases from all grades of primary tumors. A caveat to these results is that grade information abstracted from older pathology reports did not employ current WHO grading criteria.
Web-based metastasis calculator
To permit other researchers to use this algorithm to determine the most likely primary site of a neuroendocrine metastasis suspected to arise from an SBNET or PNET, a web-based metastasis calculator was developed (http://myweb.uiowa.edu/sksherman/NETCalc.html). For samples prepared following these methods, the user inputs mean Ct values for the informative genes and internal controls, and the calculator returns the most likely primary site of origin. The calculator features open-source code and freely shares all model formulae.
Discussion
The primary site of metastatic SBNETs and PNETs cannot be determined from biopsy specimens in a significant number of patients. The present study describes an algorithm based on expression of four informative genes in metastatic tissues that correctly determined the primary site in over 94% of metastases. Its excellent discriminatory ability in the independent validation set, where it correctly classified 52/56 liver metastases, constitutes its expected accuracy (92.9%) in clinically-relevant samples. Positive predictive values of greater than 92% for both SBNET and PNET primary site assignments, and the finding that the algorithm’s accuracy in classifying specimens from patients whose primary site was truly unknown prior to surgery (23/24, 96%) matches its overall performance (94%) further supports its potential clinical utility.
This study included only metastases arising from SBNET and PNET tumors, which is justified based on the clinical profile of NET liver metastases of unknown primary. A multi-institutional analysis of NETs with liver metastases reported that in 295 patients with metastases of known primary sites, 217 (74%) were from SBNETs or PNETs, while 47 (16%) were from endoscopically accessible sites (gastric, colorectal), and 20 (7%) were bronchial-primary NETs.[11] Bronchial NETs represent the most common NET in the United States,[1] but when metastatic to the liver, they produce identifiable lesions on chest x-ray or CT imaging, and their primary site is therefore usually known.[11,32,6] Among GI sources for NET liver metastases of unknown primary, SBNETs and PNETs are the most common. In a review of 92 patients with NET liver metastases whose tumors were ultimately determined to be of gastrointestinal origin, Wang et al. found that 43 had tumors arising from the pancreas, 33 from the small bowel, 15 from the colorectum, and 1 from the stomach. The colorectal and stomach NETs were nearly always identified by endoscopy.[3] Bartlett et al. reported that of 61 patients with metastatic NETs, all arose from the foregut or midgut, and non-pancreatic foregut NETs were usually identified by endoscopy.[7] Thus, in clinical practice, after an appropriate workup including chest X-ray, CT, and upper and lower endoscopy, NET liver metastases of unknown primary usually originate from the small bowel or pancreas. An algorithm tuned to differentiate these primary sites therefore offers valuable information.
Knowing the primary site of NET liver metastases impacts patient care in several ways. Unlike most solid tumors, NET patients benefit from surgical resection in the setting of metastatic disease. Surgical resection or ablation of liver metastases can reduce symptoms and may prolong survival.[10,11,33–35] During these procedures, resection of the primary tumor is performed when possible.[16,36,35] Even when liver metastases cannot be completely resected, retrospective studies suggest that resection of the primary tumor prolongs survival of patients with SBNETs and PNETs.[13,16,15,36,14] Knowing with high positive predictive value that an unknown metastasis arose from an SBNET primary therefore presents a strong indication for surgical exploration. A criticism of using such an algorithm for surgical planning is that because most PNETs are visualized on CT imaging, unknown-primary NETs are already likely to be of small bowel origin.[3,32,7] However, in patients predicted to have a pancreatic primary when none can be radiographically visualized, the algorithm could still impact surgical choices. In series of operative exploration for unknown-primary NETs, most tumors are localized to the small bowel, but some are identified in the pancreas, and 9–14% of primaries cannot be found.[32,3,7] Due to the morbidity of pancreatic resection, few surgeons will perform this without radiologic confirmation of a tumor. PNET size correlates with metastatic potential, but small PNETs (< 2cm), which may fail to appear on preoperative imaging, have nodal metastases in 27% of cases based on population-level data.[37] In a large institutional series of small PNETs, even highly-selected patients thought to be at very low risk had nodal metastases identified at surgery in 9%.[38] It is therefore possible that some unlocalizable tumors actually arise from PNETs too small to detect on imaging. In the setting of a metastasis whose primary site is unknown after a full workup, and which this algorithm predicts to arise from the pancreas, if surgical exploration fails to identify a small bowel tumor, exposure of the pancreas and intraoperative ultrasound should be performed to search for a small occult pancreatic primary tumor.
Treatment of patients presenting with widely metastatic disease too advanced for surgery could also be affected by application of the algorithm. In patients who will not undergo surgical exploration, optimal medical therapy for low and intermediate grade NETs depends on the type of primary tumor.[39] The algorithm could help inform decisions on whether to initiate everolimus, sunitinib, or other PNET-directed chemotherapeutics, while avoiding toxicity in SBNET patients, where these agents have lower response rates and are not recommended.[39] As targeted therapeutics with greater activity in either PNETs or SBNETs continue to be developed, accurate assignment of primary site will likely become even more important.
Strengths of this study include surgical determination of the primary site for all specimens and its large sample size. Other gene expression classifiers have included NETs where the primary sites were not verified by surgery.[26,27] The 92-gene classifier developed by Kerr et al. relies on primary site determinations made by a central pathology adjudication committee.[26] Although they applied rigorous methods to define the primary site, inclusion of biopsy specimens without surgical confirmation of the presumed primary site introduces uncertainty into classifier development and validation. This is particularly true of NET subtypes, which can be especially difficult to distinguish by histopathology alone. In the present study, surgical resection of the primary tumor for all specimens permits unambiguous assignment of the true primary site, thus avoiding this potential confounding factor.
The large number of samples tested by our algorithm ensures a thorough assessment of its performance. Posorski et al. reported a three-gene expression signature for distinguishing gastrointestinal NETs, but this was based on expression measured in primary and metastatic NETs from only 17 patients, including samples from gastric, colonic, and unknown primaries in addition to PNETs and SBNETs.[27] Due to the low numbers of individual tumor types and lack of additional validation specimens, the value of this gene expression signature remains unproven. The much larger study by Kerr et al. determined a 92-gene expression signature to classify cancers of unknown primary based on a database of 2094 tumors of all types and validated on 790 others, including 50 neuroendocrine tumors.[26] This classifier performed well in distinguishing neuroendocrine tumors from all other cancer types (49/50), but its performance in distinguishing sites of origin of neuroendocrine tumors was more difficult to assess due to low numbers. Out of 1 primary and 11 metastatic “gastrointestinal” NETs (site of origin not further specified), and 4 primary and 6 metastatic PNETs, it correctly classified 12 and 8, respectively.[25] The performance of the 92-gene classifier (100% for gastrointestinal and 80% for PNETs) is comparable to our results, but whether such results will persist in a larger sample of metastases is unknown. Moreover, using a gene classifier designed to address all possible cancers appears inefficient for NETs, as conventional pathology can nearly always define the NET diagnosis.
A limitation of the present study is that its methods are not currently standard clinical practice. Although none of the samples in this study were collected by percutaneous liver biopsy, isolation of adequate mRNA for qPCR requires less than 500 nanograms of tissue. This suggests that by dividing percutaneous liver biopsy specimens, saving half in RNAlater for gene expression and half in formalin for conventional pathology studies, primary site predictions could be obtained from such samples. Future research will establish the algorithm’s performance in mRNA recovered from frozen formalin-fixed paraffin-embedded tissues, but at present, it is validated only for tissues preserved in RNAlater.
Immunohistochemistry represents a promising approach to NET primary site assignment, yet despite high reported sensitivity of staining for caudal type homeobox 2 (CDX2) for SBNETs and insulin gene enhancer binding protein 1 (ISL1) for PNETs, most studies are small and few evaluate overall accuracy.[23] A larger study of 10 IHC markers in 70 PNETs and 107 SBNETs found that while 97% of SBNET primary tumors stained for CDX2, sensitivity fell to 83% in SBNET metastases, while 14% of PNET primary tumors were also CDX2-positive.[40] Although only 2% of SBNET metastases were positive for ISL1, its sensitivity in PNET metastases was only 85%. Similarly, although progesterone receptor (PR) and paired box gene 6 (PAX6) showed high specificity for PNETs versus SBNETs, each had sensitivity of only 69% in PNET metastases.[40] Incorporation of a panel of these and other IHC markers may improve overall accuracy, and our group continues to investigate the optimal combination of IHC and gene expression methodologies to efficiently classify metastases of unknown primary.
To increase access to this gene expression algorithm and facilitate future studies, we developed an online NET metastasis calculator. It requires amplification of two internal control genes and at least two informative genes to make a prediction. Since the genes for the optimal model, BRS3 and OPRK1, amplified in 90% of samples, a base assay would measure expression of as few as four total genes, although measuring expression of all six will allow a prediction to be made in nearly all specimens. In contrast to commercial gene classifiers that employ proprietary methods, its open-source code allows full evaluation of the algorithm’s predictive models by other investigators, and application to other groups’ data.
In summary, biopsy of liver metastases allows diagnosis of neuroendocrine tumors, but optimal treatment of metastatic SBNETs and PNETs requires knowledge of the primary site. An algorithm developed using nodal metastases and employing expression of four informative genes allowed for a primary site prediction in all 136 metastases tested. It correctly classified the primary site in 128/136 (94.1%) of all metastases, and in 52/56 (92.9%) of liver metastases in the independent validation set, with positive predictive values of 92.5% for SBNETs and 93.8% for PNETs. A web-based calculator (http://myweb.uiowa.edu/sksherman/NETCalc.html) makes the algorithm freely available. Based on its high accuracy in a group of metastases generalizable to clinical practice, and its potential to change management, we conclude that prospective evaluation of its impact on patient care in SBNETs and PNETs is warranted.
Acknowledgments
We gratefully acknowledge our patients for participating in this research.
Supported by NIH 5T32#CA148062-03 (SKS, JEM, JCC)
Abbreviations
- SBNETs
Small Bowel Neuroendocrine Tumors
- PNETs
Pancreatic Neuroendocrine Tumors
- NETs
Neuroendocrine Tumors
- mTOR
Mammalian Target of Rapamycin
- IHC
Immunohistochemistry
- OXTR
Oxytocin Receptor
- SCTR
Secretin Receptor
- GIPR
Gastric Inhibitory Polypeptide Receptor
- BRS3
Bombesin-like Receptor-3
- OPRK1
Opioid Receptor Kappa-1
- Ct
Threshold Cycles
- GAPDH
Glyceraldehyde-3-phosphate Dehydrogenase
- POLR2A
Polymerase (RNA) II Polypeptide A
- dCT
Delta Threshold Cycles
- PFS
Progression-free Survival
- OS
Overall Survival
- CDX2
Caudal Type Homeobox 2
- ISL1
Insulin Gene Enhancer Binding Protein 1
- PR
Progesterone Receptor
- PAX6
Paired Box Gene 6
- IQR
Interquartile Range
- CI
Confidence Interval
- PPV
Positive Predictive Value
Footnotes
An earlier version of this research was presented as an abstract at the Surgical Forum Session of the American College of Surgeons Clinical Congress in Washington, DC, October 7, 2013.
None of the authors has any potential conflicts of interest to disclose.
Disclosure/Conflict of Interest: None.
The authors declare that they have no conflict of interest.
References
- 1.Yao JC, Hassan M, Phan A, Dagohoy C, Leary C, Mares JE, Abdalla EK, Fleming JB, Vauthey JN, Rashid A, Evans DB. One hundred years after “carcinoid”: epidemiology of and prognostic factors for neuroendocrine tumors in 35,825 cases in the United States. J Clin Oncol. 2008;26(18):3063–3072. doi: 10.1200/JCO.2007.15.4377. [DOI] [PubMed] [Google Scholar]
- 2.Modlin IM, Oberg K, Chung DC, Jensen RT, de Herder WW, Thakker RV, Caplin M, Delle Fave G, Kaltsas GA, Krenning EP, Moss SF, Nilsson O, Rindi G, Salazar R, Ruszniewski P, Sundin A. Gastroenteropancreatic neuroendocrine tumours. Lancet Oncol. 2008;9(1):61–72. doi: 10.1016/S1470-2045(07)70410-2. [DOI] [PubMed] [Google Scholar]
- 3.Wang SC, Parekh JR, Zuraek MB, Venook AP, Bergsland EK, Warren RS, Nakakura EK. Identification of unknown primary tumors in patients with neuroendocrine liver metastases. Arch Surg. 2010;145(3):276–280. doi: 10.1001/archsurg.2010.10. [DOI] [PubMed] [Google Scholar]
- 4.Oberg K, Knigge U, Kwekkeboom D, Perren A. Neuroendocrine gastro-entero-pancreatic tumors: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2012;23(Suppl 7):vii124–130. doi: 10.1093/annonc/mds295. [DOI] [PubMed] [Google Scholar]
- 5.Dahdaleh FS, Lorenzen A, Rajput M, Carr JC, Liao J, Menda Y, O’Dorisio TM, Howe JR. The value of preoperative imaging in small bowel neuroendocrine tumors. Ann Surg Oncol. 2013;20(6):1912–1917. doi: 10.1245/s10434-012-2836-y. [DOI] [PubMed] [Google Scholar]
- 6.Prasad V, Ambrosini V, Hommann M, Hoersch D, Fanti S, Baum RP. Detection of unknown primary neuroendocrine tumours (CUP-NET) using (68)Ga-DOTA-NOC receptor PET/CT. Eur J Nucl Med Mol Imaging. 2010;37(1):67–77. doi: 10.1007/s00259-009-1205-y. [DOI] [PubMed] [Google Scholar]
- 7.Bartlett EK, Roses RE, Gupta M, Shah PK, Shah KK, Zaheer S, Wachtel H, Kelz RR, Karakousis GC, Fraker DL. Surgery for metastatic neuroendocrine tumors with occult primaries. J Surg Res. 2013;184(1):221–227. doi: 10.1016/j.jss.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Rinke A, Muller HH, Schade-Brittinger C, Klose KJ, Barth P, Wied M, Mayer C, Aminossadati B, Pape UF, Blaker M, Harder J, Arnold C, Gress T, Arnold R. Placebo-controlled, double-blind, prospective, randomized study on the effect of octreotide LAR in the control of tumor growth in patients with metastatic neuroendocrine midgut tumors: a report from the PROMID Study Group. J Clin Oncol. 2009;27(28):4656–4663. doi: 10.1200/JCO.2009.22.8510. [DOI] [PubMed] [Google Scholar]
- 9.Korse CM, Taal BG, van Velthuysen ML, Visser O. Incidence and survival of neuroendocrine tumours in the Netherlands according to histological grade: experience of two decades of cancer registry. Eur J Cancer. 2013;49(8):1975–1983. doi: 10.1016/j.ejca.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 10.Sarmiento JM, Heywood G, Rubin J, Ilstrup DM, Nagorney DM, Que FG. Surgical treatment of neuroendocrine metastases to the liver: a plea for resection to increase survival. J Am Coll Surg. 2003;197(1):29–37. doi: 10.1016/S1072-7515(03)00230-8. [DOI] [PubMed] [Google Scholar]
- 11.Mayo SC, de Jong MC, Pulitano C, Clary BM, Reddy SK, Gamblin TC, Celinksi SA, Kooby DA, Staley CA, Stokes JB, Chu CK, Ferrero A, Schulick RD, Choti MA, Mentha G, Strub J, Bauer TW, Adams RB, Aldrighetti L, Capussotti L, Pawlik TM. Surgical management of hepatic neuroendocrine tumor metastasis: results from an international multi-institutional analysis. Ann Surg Oncol. 2010;17(12):3129–3136. doi: 10.1245/s10434-010-1154-5. [DOI] [PubMed] [Google Scholar]
- 12.Zaknun JJ, Bodei L, Mueller-Brand J, Pavel ME, Baum RP, Horsch D, O’Dorisio MS, O’Dorisiol TM, Howe JR, Cremonesi M, Kwekkeboom DJ. The joint IAEA, EANM, and SNMMI practical guidance on peptide receptor radionuclide therapy (PRRNT) in neuroendocrine tumours. Eur J Nucl Med Mol Imaging. 2013;40(5):800–816. doi: 10.1007/s00259-012-2330-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Givi B, Pommier SJ, Thompson AK, Diggs BS, Pommier RF. Operative resection of primary carcinoid neoplasms in patients with liver metastases yields significantly better survival. Surgery. 2006;140(6):891–897. doi: 10.1016/j.surg.2006.07.033. discussion 897–898. [DOI] [PubMed] [Google Scholar]
- 14.Hill JS, McPhee JT, McDade TP, Zhou Z, Sullivan ME, Whalen GF, Tseng JF. Pancreatic neuroendocrine tumors: the impact of surgical resection on survival. Cancer. 2009;115(4):741–751. doi: 10.1002/cncr.24065. [DOI] [PubMed] [Google Scholar]
- 15.Capurso G, Bettini R, Rinzivillo M, Boninsegna L, Delle Fave G, Falconi M. Role of resection of the primary pancreatic neuroendocrine tumour only in patients with unresectable metastatic liver disease: a systematic review. Neuroendocrinology. 2011;93(4):223–229. doi: 10.1159/000324770. [DOI] [PubMed] [Google Scholar]
- 16.Capurso G, Rinzivillo M, Bettini R, Boninsegna L, Delle Fave G, Falconi M. Systematic review of resection of primary midgut carcinoid tumour in patients with unresectable liver metastases. Br J Surg. 2012;99(11):1480–1486. doi: 10.1002/bjs.8842. [DOI] [PubMed] [Google Scholar]
- 17.Strosberg JR. Systemic Treatment of Gastroenteropancreatic Neuroendocrine Tumors (GEP-NETs): Current Approaches and Future Options. Endocr Pract. 2013:1–30. doi: 10.4158/EP13262.RA. [DOI] [PubMed] [Google Scholar]
- 18.Yao JC, Shah MH, Ito T, Bohas CL, Wolin EM, Van Cutsem E, Hobday TJ, Okusaka T, Capdevila J, de Vries EG, Tomassetti P, Pavel ME, Hoosen S, Haas T, Lincy J, Lebwohl D, Oberg K. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med. 2011;364(6):514–523. doi: 10.1056/NEJMoa1009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raymond E, Dahan L, Raoul JL, Bang YJ, Borbath I, Lombard-Bohas C, Valle J, Metrakos P, Smith D, Vinik A, Chen JS, Horsch D, Hammel P, Wiedenmann B, Van Cutsem E, Patyna S, Lu DR, Blanckmeister C, Chao R, Ruszniewski P. Sunitinib malate for the treatment of pancreatic neuroendocrine tumors. N Engl J Med. 2011;364(6):501–513. doi: 10.1056/NEJMoa1003825. [DOI] [PubMed] [Google Scholar]
- 20.Oberg K, Casanovas O, Castano JP, Chung D, Delle Fave G, Denefle P, Harris P, Khan MS, Kulke MH, Scarpa A, Tang LH, Wiedenmann B. Molecular pathogenesis of neuroendocrine tumors: implications for current and future therapeutic approaches. Clin Cancer Res. 2013;19(11):2842–2849. doi: 10.1158/1078-0432.CCR-12-3458. [DOI] [PubMed] [Google Scholar]
- 21.Ganetsky A, Bhatt V. Gastroenteropancreatic neuroendocrine tumors: update on therapeutics. Ann Pharmacother. 2012;46(6):851–862. doi: 10.1345/aph.1Q729. [DOI] [PubMed] [Google Scholar]
- 22.Sherman SK, Howe JR. Translational research in endocrine surgery. Surg Oncol Clin N Am. 2013;22(4):857–884. doi: 10.1016/j.soc.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellizzi AM. Assigning site of origin in metastatic neuroendocrine neoplasms: a clinically significant application of diagnostic immunohistochemistry. Adv Anat Pathol. 2013;20(5):285–314. doi: 10.1097/PAP.0b013e3182a2dc67. [DOI] [PubMed] [Google Scholar]
- 24.Koo J, Mertens RB, Mirocha JM, Wang HL, Dhall D. Value of Islet 1 and PAX8 in identifying metastatic neuroendocrine tumors of pancreatic origin. Mod Pathol. 2012;25(6):893–901. doi: 10.1038/modpathol.2012.34. [DOI] [PubMed] [Google Scholar]
- 25.Kerr SE, Schnabel CA, Sullivan PS, Zhang Y, Huang VJ, Erlander MG, Brachtel EF, Dry SM. A 92-gene cancer classifier predicts the site of origin for neuroendocrine tumors. Mod Pathol. 2013 Jul 12; doi: 10.1038/modpathol.2013.105. EPub. [DOI] [PubMed] [Google Scholar]
- 26.Kerr SE, Schnabel CA, Sullivan PS, Zhang Y, Singh V, Carey B, Erlander MG, Highsmith WE, Dry SM, Brachtel EF. Multisite validation study to determine performance characteristics of a 92-gene molecular cancer classifier. Clin Cancer Res. 2012;18(14):3952–3960. doi: 10.1158/1078-0432.CCR-12-0920. [DOI] [PubMed] [Google Scholar]
- 27.Posorski N, Kaemmerer D, Ernst G, Grabowski P, Hoersch D, Hommann M, von Eggeling F. Localization of sporadic neuroendocrine tumors by gene expression analysis of their metastases. Clin Exp Metastasis. 2011;28(7):637–647. doi: 10.1007/s10585-011-9397-5. [DOI] [PubMed] [Google Scholar]
- 28.Carr JC, Boese EA, Spanheimer PM, Dahdaleh FS, Martin M, Calva D, Schafer B, Thole DM, Braun T, O’Dorisio TM, O’Dorisio MS, Howe JR. Differentiation of small bowel and pancreatic neuroendocrine tumors by gene-expression profiling. Surgery. 2012;152(6):998–1007. doi: 10.1016/j.surg.2012.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carr JC, Sherman SK, Wang D, Dahdaleh FS, Bellizzi AM, O’Dorisio MS, O’Dorisio TM, Howe JR. Overexpression of membrane proteins in primary and metastatic gastrointestinal neuroendocrine tumors. Ann Surg Oncol. 2013;20(Suppl 3):739–746. doi: 10.1245/s10434-013-3318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherman SK, Carr JC, Wang D, O’Dorisio MS, O’Dorisio TM, Howe JR. Gastric inhibitory polypeptide receptor (GIPR) is a promising target for imaging and therapy in neuroendocrine tumors. Surgery. 2013;154(6):1206–1214. doi: 10.1016/j.surg.2013.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahdaleh FS, Calva-Cerqueira D, Carr JC, Liao J, Mezhir JJ, O’Dorisio TM, Howe JR. Comparison of clinicopathologic factors in 122 patients with resected pancreatic and ileal neuroendocrine tumors from a single institution. Ann Surg Oncol. 2012;19(3):966–972. doi: 10.1245/s10434-011-1997-4. [DOI] [PubMed] [Google Scholar]
- 32.Begum N, Hubold C, Buchmann I, Thorns C, Bouchard R, Lubienski A, Schloricke E, Zimmermann M, Lehnert H, Bruch HP, Burk CG. Diagnostics and Therapy for Neuroendocrine Neoplasia of an Unknown Primary - A Plea for Open Exploration. Zentralbl Chir. 2013 Mar 18; doi: 10.1055/s-0032-1327962. Epub. [DOI] [PubMed] [Google Scholar]
- 33.Taner T, Atwell TD, Zhang L, Oberg TN, Harmsen WS, Slettedahl SW, Kendrick ML, Nagorney DM, Que FG. Adjunctive radiofrequency ablation of metastatic neuroendocrine cancer to the liver complements surgical resection. HPB (Oxford) 2013;15(3):190–195. doi: 10.1111/j.1477-2574.2012.00528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eriksson J, Stalberg P, Nilsson A, Krause J, Lundberg C, Skogseid B, Granberg D, Eriksson B, Akerstrom G, Hellman P. Surgery and radiofrequency ablation for treatment of liver metastases from midgut and foregut carcinoids and endocrine pancreatic tumors. World J Surg. 2008;32(5):930–938. doi: 10.1007/s00268-008-9510-3. [DOI] [PubMed] [Google Scholar]
- 35.Falconi M, Bartsch DK, Eriksson B, Kloppel G, Lopes JM, O’Connor JM, Salazar R, Taal BG, Vullierme MP, O’Toole D. ENETS Consensus Guidelines for the management of patients with digestive neuroendocrine neoplasms of the digestive system: well-differentiated pancreatic non-functioning tumors. Neuroendocrinology. 2012;95(2):120–134. doi: 10.1159/000335587. [DOI] [PubMed] [Google Scholar]
- 36.Pape UF, Perren A, Niederle B, Gross D, Gress T, Costa F, Arnold R, Denecke T, Plockinger U, Salazar R, Grossman A. ENETS Consensus Guidelines for the management of patients with neuroendocrine neoplasms from the jejuno-ileum and the appendix including goblet cell carcinomas. Neuroendocrinology. 2012;95(2):135–156. doi: 10.1159/000335629. [DOI] [PubMed] [Google Scholar]
- 37.Kuo EJ, Salem RR. Population-level analysis of pancreatic neuroendocrine tumors 2 cm or less in size. Ann Surg Oncol. 2013;20(9):2815–2821. doi: 10.1245/s10434-013-3005-7. [DOI] [PubMed] [Google Scholar]
- 38.Lee LC, Grant CS, Salomao DR, Fletcher JG, Takahashi N, Fidler JL, Levy MJ, Huebner M. Small, nonfunctioning, asymptomatic pancreatic neuroendocrine tumors (PNETs): role for nonoperative management. Surgery. 2012;152(6):965–974. doi: 10.1016/j.surg.2012.08.038. [DOI] [PubMed] [Google Scholar]
- 39.Pavel M, Baudin E, Couvelard A, Krenning E, Oberg K, Steinmuller T, Anlauf M, Wiedenmann B, Salazar R. ENETS Consensus Guidelines for the management of patients with liver and other distant metastases from neuroendocrine neoplasms of foregut, midgut, hindgut, and unknown primary. Neuroendocrinology. 2012;95(2):157–176. doi: 10.1159/000335597. [DOI] [PubMed] [Google Scholar]
- 40.Stashek KM, Czeczok TW, Bellizzi AM. Extensive Evaluation of Immunohistochemistry to Assign Site of Origin in Well-Differentiated Neuroendocrine Tumors: A Study of 10 Markers in 265 Tumors. Mod Pathol. 2014;27(Suppl 2):160A. [Google Scholar]