Abstract
Genome editing by Cas9, which cleaves double-stranded DNA at a sequence programmed by a short single-guide RNA (sgRNA), can result in off-target DNA modification that may be detrimental in some applications. To improve DNA cleavage specificity, we generated fusions of catalytically inactive Cas9 and FokI nuclease (fCas9). DNA cleavage by fCas9 requires association of two fCas9 monomers that simultaneously bind target sites ~15 or 25 base pairs apart. In human cells, fCas9 modified target DNA sites with >140-fold higher specificity than wild-type Cas9 and with an efficiency similar to that of paired Cas9 ‘nickases’, recently engineered variants that cleave only one DNA strand per monomer. The specificity of fCas9 was at least 4-fold higher_than that of paired nickases at loci with highly similar off-target sites. Target sites that conform to the substrate requirements of fCas9 occur on average every 34 bp in the human genome, suggesting the broad versatility of this approach for highly specific genome-wide editing.
The recent development of robust, predictable and user-friendly methods for the generation of sequence-specific DNA-binding proteins has accelerated the use of genome editing for biological research1 and development of therapeutics2. The CRISPR-Cas9 system is an especially convenient approach for genome editing as an agent for a new target site of interest can be created simply by generating the corresponding sgRNA. The 3′ end of the sgRNA forms a scaffold that binds Cas9 protein,3 whereas the ~17 to 20 bases4 at the 5′ end of the sgRNA pair with the target DNA to determine DNA cleavage specificity (Fig. 1). Provided that the target sequence is adjacent to a short 3′ motif—the protospacer adjacent motif (PAM) required for initial binding and Cas9 activation5—any DNA locus can in principle be targeted. In cells, double-strand breaks induced by targeted Cas9:sgRNA complexes enable either functional gene knockout through non-homologous end joining (NHEJ) or alteration of a target locus to virtually any sequence through homology-directed repair with an exogenous DNA template.6,3,7
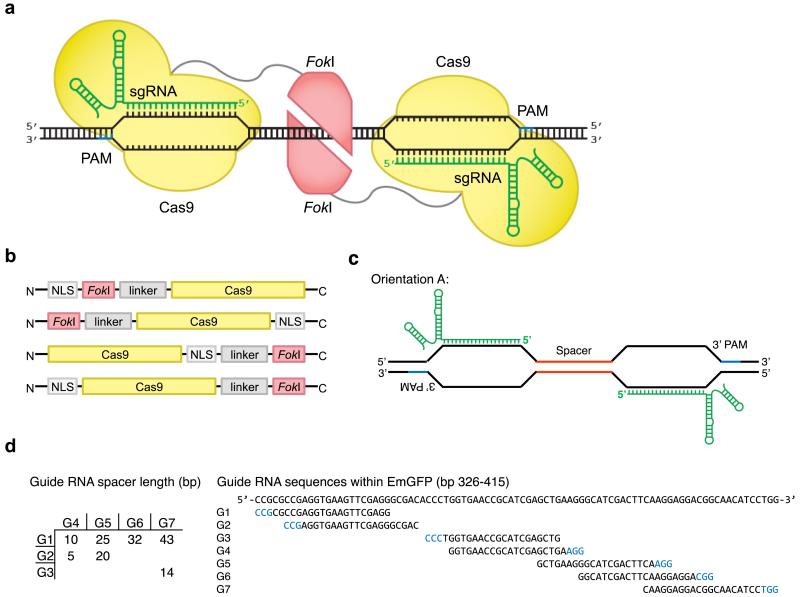
Figure 1. Architectures of Cas9 and FokI-dCas9 fusion variants.
(a) Two monomers of FokI nuclease (red) fused to dCas9 (yellow) bind in complex with guide RNAs (sgRNA, green) to separate sites within the target locus. Only adjacently bound FokI-dCas9 monomers can assemble a catalytically active FokI nuclease dimer, triggering dsDNA cleavage. (b) FokI-dCas9 fusion architectures tested. Four distinct configurations of NLS, FokI nuclease, and dCas9 were assembled. 17 protein linker variants were also tested (see main text). (c) sgRNA orientation and (d) target sites tested within the EmGFP gene. Seven sgRNA target sites were chosen to test FokI-dCas9 activity in an orientation in which the PAM is distal from the cleaved spacer sequence (orientation A). Together, these seven sgRNAs enabled testing of FokI-dCas9 fusion variants across seven spacer lengths ranging from 5 to 43 bp. See Supplementary Figure 1 for guide RNAs used to test orientation B, in which the PAM is adjacent to the spacer sequence.
The usefulness of Cas9 for research and therapeutic applications may be limited by its ability to cleave off-target genomic sites1-5. We hypothesized that engineering Cas9 variants to cleave DNA only when two simultaneous, adjacent Cas9:DNA binding events take place could substantially improve specificity because the likelihood of two adjacent off-target binding events is much smaller than the likelihood of a single off-target binding event (approximately 1/n2 vs. 1/n). This approach is analogous to that previously developed for dimeric zinc-finger nucleases (ZFNs) and TALENs. Based on those examples, we speculated that fusing the FokI restriction endonuclease cleavage domain to a catalytically dead Cas9 (dCas9) could create an obligate dimeric Cas9 that would cleave DNA only when two distinct FokI-dCas9:sgRNA complexes bind to adjacent sites (“half-sites”) with particular spacing constraints (Figure 1a).
This approach is distinct from the use of ‘nickases’, mutant Cas9 proteins that cleave only a single strand of dsDNA. Paired nickases can be used to nick opposite strands of two nearby target sites, generating what is effectively a double-strand break, and can effect substantial on-target DNA modification with reduced off-target modification.4,8-10 Because each of the component Cas9 nickases remains catalytically active5,6,11 and single-stranded DNA cleavage events are weakly mutagenic,12,13 nickases can induce genomic modification even when acting as monomers5-8,14. Moreover, because paired Cas9 nickases can efficiently induce dsDNA cleavage-derived modification events when bound up to ~100 bp apart,8,9 the statistical number of potential off-target sites for paired nickases is larger than that of a more spatially constrained dimeric Cas9 cleavage system. In contrast, DNA cleavage by FokI-dCas9 requires simultaneous binding of two distinct FokI-dCas9 monomers because monomeric FokI nuclease domains are not catalytically competent.14 In principle, this approach should increase the specificity of DNA cleavage relative to wild-type Cas9 by doubling the number of specified target bases. The use of fCas9 should also result in improved specificity compared to nickases due to inactivity of monomeric FokI-dCas9:sgRNA complexes and due to the more stringent spatial requirements for assembly of a FokI-dCas9 dimer.
RESULTS
Development of an active FokI-dCas9 fusion architecture
We began by constructing and characterizing a wide variety of FokI-dCas9 fusion proteins with distinct configurations of a FokI nuclease domain, a dCas9 containing inactivating mutations D10A and H840A, and a nuclear localization sequence (NLS). We fused wild-type, homodimeric, FokI to either the N- or C-terminus of dCas9, and varied the location of the NLS to be at either terminus or between the two domains (Figure 1b and Supplementary Notes). We further varied the length of the linker sequence as either one or three repeats of Gly-Gly-Ser (GGS) between the FokI and dCas9 domains. As previously developed dimeric nuclease systems are sensitive to the length of the spacer sequence between half-sites, 15,16 we also tested a wide range of spacer sequence lengths between two sgRNA binding sites within a test target gene, Emerald GFP (Life Technologies) (referred to hereafter as GFP) (Figures 1c-1d and Supplementary Figure 1).
We chose two sets of sgRNA binding-site pairs with different orientations within GFP. One set placed the pair of NGG PAM sequences distal from the spacer sequence, with the 5′ end of the sgRNA adjacent to the spacer (orientation A) (Figure 1c), whereas the other placed the PAM sequences immediately adjacent to the spacer (orientation B) (Supplementary Figure 1). In total, seven pairs of sgRNAs were suitable for orientation A, and six were suitable for orientation B. By pairwise combination of the sgRNA targets, we tested spacer lengths in both dimer orientations, ranging from 5 to 43 bp in orientation A, and 4 to 42 bp in orientation B. In total, 216 pairs of FokI-dCas9:sgRNA complexes were generated and tested, exploring four fusion architectures, 17 protein linker variants (described below), both sgRNA orientations and 13 spacer lengths between half-sites.
To assay the activities of these candidate FokI-dCas9:sgRNA pairs, we used a previously described flow cytometry-based fluorescence assay4,17 in which DNA cleavage and NHEJ of a stably integrated constitutively expressed GFP gene in HEK293 cells leads to loss of cellular fluorescence (Supplementary Figure 2). For comparison, we assayed the initial set of FokI-dCas9 variants side-by-side with the corresponding Cas9 nickases and wild-type Cas9 in the same expression plasmid across both sgRNA spacer orientation sets A and B. Cas9 protein variants and sgRNA were generated in cells by transient co-transfection of the corresponding Cas9 protein expression plasmids together with the appropriate pair of sgRNA expression plasmids. The FokI-dCas9 variants, nickases and wild-type Cas9 all targeted identical DNA sites using identical sgRNAs. Although this assay showed a consistent ~5% background signal in the absence of sgRNA, it enabled the rapid assessment of many fusion constructs, sgRNA orientations and DNA spacer lengths to identify active constructs for further optimization.
Most of the initial FokI-dCas9 fusion variants were inactive or very weakly active (Supplementary Figure 3). Only the NLS-FokI-dCas9 architecture resulted in a frequency of GFP-negative cells that was substantially higher than what was observed in the corresponding no-sgRNA control when used in orientation A (with PAMs distal from the spacer). (Supplementary Figure 3a). By contrast, NLS-FokI-dCas9 activity above background was not detected when used with sgRNA pairs in orientation B, with PAMs adjacent to the spacer (Supplementary Figure 3b). Examination of the recently reported Cas9 structures18,19 reveals that the Cas9 N-terminus protrudes from the RuvC domain, which contacts the 5′ end of the sgRNA:DNA duplex. We speculate that this arrangement places an N-terminally fused FokI distal from the PAM, resulting in a preference for sgRNA pairs with PAMs distal from the cleaved spacer (Figure 1a). Further, examination of the structure of the C-terminus of dCas9 suggests that access to the spacer by FokI C-terminal fusions may require longer linkers in order to span the PI and RuvC Cas9 domains with paired sgRNAs in orientation A, or to span the REC1 domain of Cas9 and sgRNA stem-loops with sgRNAs in orientation B. Although other FokI-dCas9 fusion pairings and the other sgRNA orientation in some cases showed modest activity (Supplementary Figure 3), we chose NLS-FokI-dCas9 with sgRNAs in orientation A for further development.
Optimizing and validating the NLS-FokI-dCas9 architecture
Next we optimized the protein linkers between the NLS and FokI domain and between the FokI domain and dCas9 in the NLS-FokI-dCas9 architecture. We tested 17 linkers with a wide range of amino acid compositions, predicted flexibilities, and lengths varying from 9 to 21 residues (Supplementary Figure 4a). For linker between the FokI domain and dCas9 the highest levels of genomic GFP modification was observed for the proteins with a flexible 18-residue linker (GGS)6 and a 16-residue “XTEN” linker, which was based on a previously reported engineered protein with an open, extended conformation20 (FokI-L8 in Supplementary Figure 4a; Supplementary Figure 4b; Supplementary Results). Many of the FokI-dCas9 linkers tested including the optimal XTEN linker resulted in nucleases with a marked preference for spacer lengths of ~15 and ~25 bp between half-sites, with all other spacer lengths, including 20 bp, showing substantially lower activity (Supplementary Figure 4b). This pattern of linker preference is consistent with a model in which the FokI-dCas9 fusions must bind to opposite faces of the DNA double helix to cleave DNA, with optimal binding taking place ~1.5 or 2.5 helical turns apart. The variation of the NLS-FokI linkers did not strongly affect nuclease performance, especially when combined with the XTEN FokI-dCas9 linker (Supplementary Figure 4b and Supplementary Results).
The NLS-GGS-FokI-XTEN-dCas9 construct consistently exhibited the highest activity among the tested candidates, inducing loss of GFP in ~10% of cells over background, compared to ~15% and ~25% for Cas9 nickases and wild-type Cas9 nuclease, respectively (Figure 2a). All subsequent experiments were performed using this construct, hereafter referred to as fCas9. To confirm the ability of fCas9 to efficiently modify genomic target sites, we used the T7 endonuclease I Surveyor assay to measure the amount of mutation at each of seven target sites within the integrated GFP gene in HEK293 cells treated with fCas9, Cas9 nickase or wild-type Cas9 and either two distinct sgRNAs in orientation A or no sgRNAs as a negative control. Consistent with our flow cytometry-based studies, fCas9 was able to modify the GFP target sites with optimal spacer lengths of ~15 or ~25 bp at a rate of ~20%, comparable to the efficiency of nickase-induced modification and approximately two-thirds that of wild-type Cas9 (Figure 2a-c).
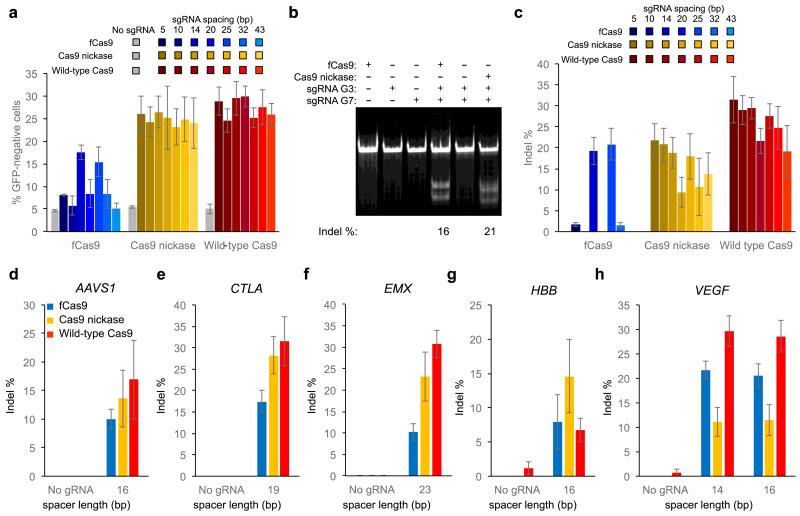
Figure 2. Genomic DNA modification by fCas9, Cas9 nickase, and wild-type Cas9.
Detection of genomic modification by loss of GFP signal or Surveyor assay at either an integrated GFP gene, or at endogenous genomic targets within the AAVS1, CLTA, EMX, HBB, or VEGF genes (Supplementary Figure 5) (a) GFP disruption activity of fCas9, Cas9 nickase, or wild-type Cas9 with either no sgRNA, or sgRNA pairs of variable spacer length targeting the GFP gene in orientation A. (b) Indel modification efficiency from PAGE analysis of a Surveyor cleavage assay of renatured target-site DNA amplified from cells treated with fCas9, Cas9 nickase, or wild-type Cas9 and two sgRNAs spaced 14 bp apart targeting the GFP site (sgRNAs G3 and G7; Figure 1d), each sgRNA individually, or no sgRNAs. The indel modification percentage is shown below each lane for samples with modification above the detection limit (~2%). (c-h) Indel modification efficiency for (c) two pairs of sgRNAs spaced 14 or 25 bp apart targeting the GFP site, (d) on pair of sgRNAs spaced 16 bp apart targeting the AAVS1 site, (e) one pair of sgRNAs spaced 19 bp apart targeting the CLTA site, (f) one pair of sgRNAs spaced 23 bp apart targeting the EMX site (g) one pair of sgRNAs spaced 16 bp apart targeting the HBB site, and (h) two pairs of sgRNAs spaced 14 or 16 bp apart targeting the VEGF site. Error bars reflect standard error of the mean from three biological replicates performed on different days.
Modification of endogenous genomic targets by optimized fCas9
Next, we evaluated the ability of the optimized fCas9 to modify 14 distinct endogenous genomic loci in five genes by Surveyor assay. AAVS1 (one site), CLTA (two sites), EMX (two sites), HBB (six sites), and VEGF (three sites) were targeted with two sgRNAs per site in orientation A spaced at various lengths (Supplementary Figure 5). Consistent with the results of the experiments targeting GFP, at appropriately spaced target half-sites fCas9 induced efficient modification of all five genes, with efficiencies ranging from 8% to 22% (Figure 2d-h and Supplementary Figure 6). With the sgRNA spacer lengths resulting in the highest modification at each of the six genes targeted (including GFP), fCas9 induced on average 14.9% (± 6.0% s.d.) modification, whereas Cas9 nickase and wild-type Cas9 induced on average 20.6% (± 5.6% s.d.) and 28.2% (± 6.2% s.d.) modification, respectively. Because decreasing the amount of Cas9 expression plasmid and sgRNA expression plasmid during transfection generally did not proportionally decrease genomic modification activity for Cas9 nickase and fCas9 (Supplementary Figure 7a-c), expression was likely not limiting under the conditions tested.
Stringent spatial requirements of fCas9-mediated DNA cleavage
As the sgRNA requirements of fCas9 potentially reduce the number of potential off-target substrates of fCas9, we compared the effect of guide RNA orientation on the ability of fCas9, Cas9 nickase, and wild-type Cas9 to cleave target GFP sequences. Consistent with previous reports8-10, Cas9 nickase efficiently cleaved targets when guide RNAs were bound either in orientation A or orientation B, similar to wild-type Cas9 (Supplementary Figure 8a, b). In contrast, fCas9 only cleaved the GFP target when guide RNAs were aligned in orientation A (Figure 2a-c and Supplementary Figure 8a, b). This orientation requirement further limits opportunities for undesired off-target DNA cleavage.
No modification was observed by GFP disruption or Surveyor assay when any of four single sgRNAs were expressed individually with fCas9, as expected because two simultaneous binding events are required for FokI activity (Figure 2b and Supplementary Figure 9). By contrast, GFP gene disruption resulted from expression of any single sgRNA with wild-type Cas9 (as expected) and, in the case of two single sgRNAs, with Cas9 nickase (Figure 3a). High-throughput sequencing to detect indels at the GFP target site in cells treated with paired sgRNAs and fCas9, Cas9 nickase, or wild-type Cas9 revealed the expected substantial level of modification ranging from 2.3% to 14.3% of sequence reads. Modification by fCas9 in the presence of any of the four single sgRNAs was not detected above background signal (ranging from < 0.01% to 0.073% modification), consistent with the requirement of fCas9 to engage two sgRNAs in order to cleave DNA. By contrast, Cas9 nickases in the presence of single sgRNAs resulted in modification levels ranging from 0.05% to 0.16% at the target site (Figure 3a). The detection of bona fide indels at target sites following Cas9 nickase treatment with single sgRNAs confirms the mutagenic potential of genomic DNA nicking, consistent with previous reports.3,8,10,12,13 These results collectively demonstrate that Cas9 nickase can induce genomic DNA modification in the presence of a single sgRNA, in contrast with the absence of single-sgRNA modification by fCas9 .
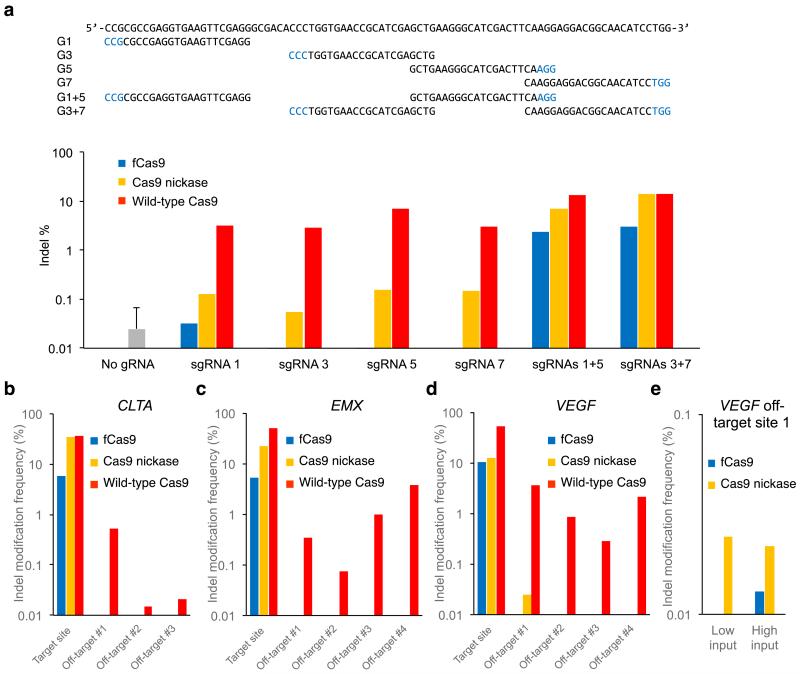
Figure 3. DNA modification specificity of fCas9, Cas9 nickase, and wild-type Cas9.
(a) Results from high-throughput sequencing of GFP on-target sites amplified from 150 ng genomic DNA isolated from human cells treated with a plasmid expressing wild-type Cas9, Cas9 nickase, or fCas9; and either a plasmid expressing a single sgRNA (G1, G3, G5, or G7), or two plasmids each expressing a different sgRNA (G1+G5 or G3+G7). As a negative control, transfection and sequencing were performed in triplicate as above without any sgRNA expression plasmids. Sequences with more than one insertion or deletion at the GFP target site (the start of the G1 binding site to the end of the G7 binding site) were considered indels. Indel percentages are the number of indels observed divided by the total number of sequences. While wild-type Cas9 produced indels across all sgRNA treatments, fCas9 and Cas9 nickase produced indels efficiently (> 1%) only when paired sgRNAs were present. Indels induced by fCas9 and single sgRNAs were not detected at a frequency above that of the no-gRNA control, whereas Cas9 nickase and single sgRNAs modified the target GFP sequence at an average rate of 0.12%. (b-e) The indel mutation frequency from high-throughput DNA sequencing of amplified genomic on-target sites and off-target sites from human cells treated with fCas9, Cas9 nickase, or wild-type Cas9 and (b) two sgRNAs spaced 19 bp apart targeting the CLTA site (sgRNAs C1 and C2), (c) two sgRNAs spaced 23 bp apart targeting the EMX site (sgRNAs E1 and E2), or (d, e) two sgRNAs spaced 14 bp apart targeting the VEGF site (sgRNAs V1 and V2). (e) Two in-depth trials to measure genome modification at VEGF off-target site 1. Trial 1 used 150 ng of genomic input DNA and > 8 × 105 sequence reads for each sample; trial 2 used 600 ng of genomic input DNA and > 23 × 105 sequence reads for each sample. In (b-e), all significant (P value < 0.005 Fisher’s Exact Test) indel frequencies are shown. P values are listed in Supplementary Table 3. For (b-e) each on- and off-target sample was sequenced once with > 10,000 sequences analyzed per on-target sample and an average of 76,260 sequences analyzed per off-target sample (Supplementary Table 3).
The observed rate of nickase-induced DNA modification did not account for the much higher GFP disruption signal in the flow cytometry assay (Supplementary Figure 9b). Because the sgRNAs that induced GFP signal loss with Cas9 nickase (sgRNAs G1 and G3) both target the non-template strand of the GFP gene, and because targeting the non-template strand with dCas9 in the coding region of a gene is known to mediate efficient transcriptional repression,21 we speculate that Cas9 nickase combined with the G1 or G3 single guide RNAs induced substantial transcriptional repression, in addition to a low level of genome modification. The same effect was not seen for fCas9, suggesting that fCas9 may be more easily displaced from DNA by transcriptional machinery. Taken together, these results indicate that fCas9 can modify genomic DNA efficiently and in a manner that requires simultaneous engagement of two guide RNAs targeting adjacent sites, unlike the ability of wild-type Cas9 and Cas9 nickase to cleave DNA when bound to a single guide RNA.
The above results collectively reveal much more stringent spacer, sgRNA orientation, and guide RNA pairing requirements for fCas9 compared with Cas9 nickase. In contrast with fCas9 (Supplementary Figure 10), Cas9 nickase cleaved sites across all spacers assayed (5- to 47- bp in orientation A and 4 to 42 bp in orientation B in this work) (Figure 2a, c and Supplementary Figure 8a, b). These observations are consistent with previous reports of Cas9 nickases modifying sites targeted by sgRNAs with spacer lengths up to 100 bp apart.8,9 The more stringent spacer and sgRNA orientation requirements of fCas9 compared with Cas9 nickase reduces the number of potential genomic off-target sites of the former by approximately 10-fold (Supplementary Table 1). Although the more stringent spacer requirements of fCas9 also reduce the number of potential targetable sites, sequences that conform to the fCas9 spacer and dual PAM requirements exist in the human genome on average once every 34 bp (9.2 × 107 sites in 3.1 × 109 bp) (Supplementary Table 1). We also anticipate that the growing number of Cas9 homologs with different PAM specificities22 will further increase the number of targetable sites using the fCas9 approach.
Improved specificity of fCas9 at endogenous off-target sites
To evaluate the DNA cleavage specificity of fCas9, we measured the modification of known Cas9 off-target sites of CLTA, EMX, and VEGF genomic target sites.4,9,17,23 The target site and its corresponding known off-target sites (Supplementary Table 2) were amplified from genomic DNA isolated from HEK293 cells treated with fCas9, Cas9 nickase, or wild-type Cas9 and two sgRNAs spaced 19 bp apart targeting the CLTA site, two sgRNAs spaced 23 bp apart targeting the EMX site, two sgRNAs spaced 14 bp apart targeting the VEGF site, or two sgRNAs targeting an unrelated site (GFP) as a negative control. In total 11 off-target sites were analyzed by high-throughput sequencing (Supplementary Notes). Sequences containing insertions or deletions of two or more base pairs in potential genomic off-target sites and present in significantly greater numbers (P value < 0.005, Fisher’s exact test) in the target sgRNA-treated samples versus the control sgRNA-treated samples were considered Cas9 nuclease-induced genome modifications. For all 11 off-target sites initially assayed, fCas9 did not result in any detectable genomic off-target modification within the sensitivity limit of our assay (< 0.002%, Supplementary Results; see further discussion of VEGF off-target site 1 below), while demonstrating substantial on-target modification efficiencies of 5% to 10% (Figure 3b-d and Supplementary Table 3a-c). The detailed inspection of fCas9-modified VEGF on-target sequences (Supplementary Figure 11a) revealed a prevalence of deletions ranging from two to dozens of base pairs consistent with cleavage occurring in the DNA spacer between the two target binding sites. For each target site at CLTA, EMX, and VEGF, fCas9 predominantly induces deletions with insertions representing < 5% of all modifications. This prevalence of deletions is also observed with TALENs, which have similar spacer length preferences.24
By contrast, genomic off-target DNA cleavage was observed for wild-type Cas9 at all 11 sites assayed. Using the detection limit of the assay as an upper bound for off-target fCas9 activity, we calculated that fCas9 has a much lower off-target modification rate than wild-type Cas9 nuclease. At the 11 off-target sites modified by wild-type Cas9 nuclease, fCas9 resulted in on-target:off-target modification ratios at least 140-fold higher than that of wild-type Cas9 (Figure 3b-d).
Consistent with previous reports,4,9,10 paired Cas9 nickases also induced substantially fewer off-target modification events (1/11 off-target sites modified at a detectable rate) than wild-type Cas9. An initial high-throughput sequencing assay revealed significant (P value < 10−3, Fisher’s Exact Test) modification induced by Cas9 nickases in 0.024% of sequences at VEGF off-target site 1. This genomic off-target site was not modified by fCas9 despite similar VEGF on-target modification efficiencies of 12.3% for Cas9 nickase and 10.4% for fCas9 (Figure 3d and Supplementary Table 3c). Because Cas9 nickase-induced modification levels were within an order of magnitude of the limit of detection and fCas9 modification levels were undetected, we repeated the experiment with a larger input DNA samples and a greater number of sequence reads (low input of 150 ng versus high input 600 ng genomic DNA and > 8 × 105 versus > 23 × 105 reads for the initial and second trial, respectively) to detect off-target cleavage at this site by Cas9 nickase or fCas9 (Supplementary Notes). From this deeper interrogation, we observed Cas9 nickase and fCas9 to both significantly modify (P value < 10−5, Fisher’s Exact Test) VEGF off-target site 1 (Figure 3e, Supplementary Table 3d, Supplementary Figure 11b). For both experiments interrogating the modification rates at VEGF off-target site 1, fCas9 exhibited a greater on-target:off-target DNA modification ratio than that of Cas9 nickase (> 5,150 and 1,650 for fCas9, versus 510 and 1,230 for Cas9 nickase, Figure 3e).
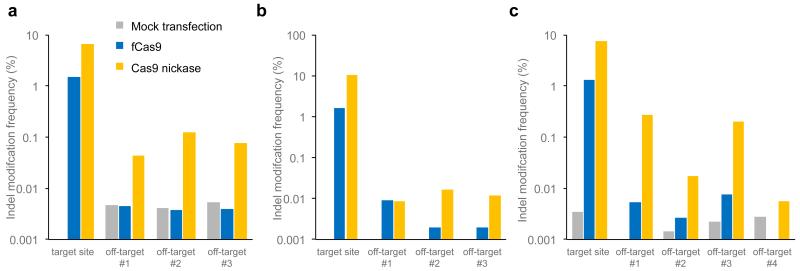
To further investigate differences in specificity between fCas9 and Cas9 nickases, three new target sites (sites A, B, and C) were chosen in the human genome that each had potential genomic off-target sites containing one identical or nearly identical half-site (Supplementary Table 4). The three on-target sites and their highly similar off-target sites were amplified and sequenced from cells treated with a mock transfection of a GFP expression plasmid, fCas9, or Cas9 nickases and either two sgRNAs spaced 15 bp apart targeting genomic site A, two sgRNA spaced 24 bp apart targeting genomic site B, or two sgRNA spaced 23 bp apart targeting genomic site C. At these three sites fCas9 demonstrated ≥ 4-fold average increase in on-target:off-target genome modification activity compared to Cas9 nickases (Figure 4 and Supplementary Table 5). These results suggest that for highly similar sets of on- and off-target sites, as may be found with repetitive genomic loci, pseudogenes, or homologous gene families, fCas9 can result in improved genome modification specificity over that of Cas9 nickases.
Figure 4. Genomic DNA modification specificity of fCas9 and Cas9 nickase at genomic sites with highly similar off-target sites.
(a-c) The indel mutation frequency from high-throughput DNA sequencing of amplified genomic on-target sites and off-target sites (Supplementary Table 4) from human cells treated with fCas9 or Cas9 nickase and (a) two sgRNAs (sgRNAs SA1 and SA2) spaced 15 bp apart targeting site A, human genomic locus chr1:21,655,401-21,655,461; (b) two sgRNAs (sgRNAs SB1 and SB2) spaced 24 bp apart targeting site B, human genomic locus chr2:31,485,447-31,485,516; or (c) two sgRNAs (sgRNAs SC1 and SC2) spaced 23 bp apart targeting the site C, human genomic locus chr3:48,747,484-48,747,552. P values are listed in Supplementary Table 5. Each on- and off-target sample was sequenced once with > 10,000 sequences analyzed per on-target sample and an average of 83,000 sequences analyzed per off-target sample (Supplementary Table 5). The mock transfection control represents the limit of detection for each site, determined from cells transfected with a GFP expression plasmid and no sgRNA or nuclease expression constructs.
DISCUSSION
In this work we developed fCas9, an obligate dimeric FokI-dCas9 nuclease architecture. The fCas9 nuclease modified all 11 genomic loci tested with sgRNA pairs spaced ~15 bp or ~25 bp apart, demonstrating the generality of using fCas9 to induce genomic modification in human cells. The use of fCas9 is straightforward, requiring only that PAM sequences be present with an appropriate spacing and orientation, and using the same sgRNA architecture as wild-type Cas9 or Cas9 nickases.
Although modification with fCas9 was generally less efficient than with wild-type Cas9, fCas9 was consistently more specific, producing substantially fewer off-target modification events. The observed low off-target:on-target modification ratios of fCas9 were at least 140-fold lower than that of wild-type Cas9 and from 1.3- to 8.8-fold lower than that of paired Cas9 nickases. These improvements likely arise from the distinct mode of action of dimeric FokI, which cleaves DNA only if two sites are occupied simultaneously by two FokI domains at a specified distance (here, ~15 bp or ~25 bp apart) and in a specific half-site orientation. Indeed, when presented with only a single sgRNA (Figure 3a), or a single matching half-site at off-target genomic loci (Figure 4), fCas9 was unable to modify DNA. Given our use of plasmid transfection in this study, more efficient gene delivery methods such as nucleofection may offer higher fCas9 modification efficiency.
The only observed off-target DNA modification by fCas9 in this study was the modification of VEGF off-target site 1. On either side of VEGF off-target site 1 there exist no other sites with six or fewer mutations from either of the two half-sites of the VEGF on-target sequence. We speculate that the first 11 bases of one sgRNA (V2) might hybridize to the single-stranded DNA freed by canonical Cas9:sgRNA binding within VEGF off-target site 1 (Supplementary Figure 11c). Through this sgRNA:DNA hybridization it is possible that a second Cas9 nickase or fCas9 could be recruited to modify this off-target site at an extremely rare, but detectable frequency.
A recent report25 demonstrates the promiscuity of dCas9 binding to sequences containing only an NGG PAM and a “seed” sequence as short at five bases immediately 5′ of the PAM. A search of the sequence surrounding VEGF off-target site 1 identified a single site of this type, with a single mismatch within the six bases 5′ of an NGG PAM (Supplementary Figure 11d). This potential site corresponds to an untested sgRNA pair orientation, and is likely a very inefficient substrate for fCas9 cleavage. Judicious sgRNA pair design could eliminate either of these potential modes of off-target DNA cleavage at sites similar to VEGF off-target site 1. The promiscuity of dCas9 binding at short seed-PAM sequences may be mitigated by the requirement of fCas9 to assemble on two dCas9 binding sites with very specific spacer constraints and PAM orientations.
The low off-target activity of fCas9 may enable applications of Cas9:sgRNA-based technologies that require a very high degree of target specificity, such as ex vivo or in vivo therapeutic modification of human cells. This work also provides a foundation for future studies to characterize in greater detail and further improve the DNA cleavage activity and specificity of fCas9 in vitro and in vivo. For example, the use of recently described orthogonal Cas9 homologs22 coupled with obligate heterodimeric FokI variants26 may offer additional specificity gains.
ONLINE METHODS
Oligonucleotides and PCR
All oligonucleotides were purchased from Integrated DNA Technologies (IDT). Oligonucleotide sequences are listed in Supplementary Notes. PCR was performed with 0.4 μL of 2 U/μL Phusion Hot Start Flex DNA polymerase (NEB) in 50 μL with 1× HF Buffer, 0.2 mM dNTP mix (0.2 mM dATP, 0.2 mM dCTP, 0.2 mM dGTP, 0.2 mM dTTP) (NEB), 0.5 μM of each primer and a program of: 98 °C, 1 min; 35 cycles of [98 °C, 15 s; 65 °C, 15 s; 72 °C, 30 s] unless otherwise noted.
Construction of FokI-dCas9, Cas9 Nickase and sgRNA Expression Plasmids
The human codon-optimized streptococcus pyogenes Cas9 nuclease with NLS and 3×FLAG tag (Addgene plasmid 43861)17 was used as the wild-type Cas9 expression plasmid. PCR (72 °C, 3 min) products of wild-type Cas9 expression plasmid as template with Cas9_Exp primers listed in Supplementary Notes were assembled with Gibson Assembly Cloning Kit (New England Biolabs) to construct Cas9 and FokI-dCas9 variants. Expression plasmids encoding a single sgRNA construct (sgRNA G1 through G13) were cloned as previously described.17 Briefly, sgRNA oligonucleotides listed in Supplementary Notes containing the 20-bp protospacer target sequence were annealed and the resulting 4-bp overhangs were ligated into BsmBI-digested sgRNA expression plasmid. sgRNA expression plasmids encoding expression of two separate sgRNA constructs from separate promoters on a single plasmid were cloned in a two-step process depicted in Supplementary Notes. First, one sgRNA (sgRNA E1, V1, C1, C3, H1, G1, G2 or G3) was cloned as above and used as template for PCR (72 °C, 3 min) with PCR_Pla-fwd and PCR_Pla-rev primers, 1 μl DpnI (NEB) was added, and the reaction was incubated at 37 °C for 30 min and then subjected to QIAquick PCR Purification Kit (Qiagen) for the “1st sgRNA + vector DNA”. PCR (72 °C, 3 min) of 100 pg of BsmBI-digested sgRNA expression plasmid as template with PCR_sgRNA-fwd1, PCR_sgRNA-rev1, PCR_sgRNA-rev2 and appropriate PCR_sgRNA primer listed in Supplementary Notes was DpnI treated and purified as above for the “2nd sgRNA instert DNA”. ~200 ng of “1st sgRNA + vector DNA” and ~200 ng of “2nd sgRNA instert DNA” were blunt-end ligated in 1 × T4 DNA Ligase Buffer, 1 μl of T4 DNA Ligase (400 U/μl, NEB) in a total volume of 20 μl at room temperature (~21 °C) for 15 min. For all cloning, 1 μl of ligation or assembly reaction was transformed into Mach1 chemically competent cells (Life Technologies). Protein and DNA sequences are listed in Supplementary Notes. The optimized FokI-dCas9 (fCas9) expression plasmid is available from Addgene (52970).
Modification of Genomic GFP
HEK293-GFP stable cells (GenTarget) were used as a cell line constitutively expressing an Emerald GFP gene (GFP) integrated on the genome. Cells were maintained in Dulbecco’s modified Eagle medium (DMEM, Life Technologies) supplemented with 10% (vol/vol) fetal bovine serum (FBS, Life Technologies) and penicillin/streptomycin (1x, Amresco). 5 × 104 HEK293-GFP cells were plated on 48-well collagen coated Biocoat plates (Becton Dickinson). One day following plating, cells at ~75% confluence were transfected with Lipofecatmine 2000 (Life Technologies) according to the manufacturer’s protocol. Briefly, 1.5 μL of Lipofecatmine 2000 was used to transfect 950 ng of total plasmid (Cas9 expression plasmid plus sgRNA expression plasmids). 700 ng of Cas9 expression plasmid, 125 ng of one sgRNA expression plasmid and 125 ng of the paired sgRNA expression plasmid with the pairs of targeted sgRNAs listed in Figure 1d and Supplementary Figure 1a. Separate wells were transfected with 1 μg of a near-infrared iRFP670 (Addgene plasmid 45457)27 as a transfection control. Transfection efficiencies ranged from 70% to 80% per construct.. 3.5 days following transfection, cells were trypsinized and resuspended in DMEM supplemented with 10% FBS and analyzed on a C6 flow cytometer (Accuri) with a 488 nm laser excitation and 520 nm filter with a 20 nm band pass. For each sample, transfections and flow cytometry measurements were performed once.
T7 Endonuclease I Surveyor Assays of Genomic Modifications
HEK293-GFP stable cells were transfected with Cas9 expression and sgRNA expression plasmids as described above. A single plasmid encoding two separate sgRNAs was transfected. For experiments titrating the total amount of expression plasmids (Cas9 expression + sgRNA expression plasmid), 700/250, 350/125, 175/62.5, 88/31 ng of Cas9 expression plasmid/ng of sgRNA expression plasmid were combined with inert carrier plasmid, pUC19 (NEB), as necessary to reach a total of 950 ng transfected plasmid DNA.
Genomic DNA was isolated from cells 2 days after transfection using a genomic DNA isolation kit, DNAdvance Kit (Agencourt). Briefly, cells in a 48-well plate were incubated with 40 μL of tryspin for 5 min at 37 °C. 160 uL of DNAdvance lysis solution was added and the solution incubated for 2 hr at 55 °C and the subsequent steps in the Agencourt DNAdvance kit protocol were followed. 40 ng of isolated genomic DNA was used as template to PCR amplify the targeted genomic loci with flanking Survey primer pairs specified in the Supplementary Notes. PCR products were purified with a QIAquick PCR Purification Kit (Qiagen) and quantified with Quant-iT™ PicoGreen ® dsDNA Kit (Life Technologies). 250ng of purified PCR DNA was combined with 2 μL of NEBuffer 2 (NEB) in a total volume of 19 μL and denatured then re-annealed with thermocycling at 95 °C for 5 min, 95 to 85 °C at 2 °C/s; 85 to 20 °C at 0.2 °C/s. The re-annealed DNA was incubated with 1 μl of T7 Endonuclease I (10 U/μl, NEB) at 37 °C for 15 min. 10 μL of 50% glycerol was added to the T7 Endonuclease reaction and 12 μL was analyzed on a 5% TBE 18-well Criterion PAGE gel (Bio-Rad) electrophoresed for 30 min at 150 V, then stained with 1× SYBR Gold (Life Technologies) for 30 min. Cas9-induced cleavage bands and the uncleaved band were visualized on an AlphaImager HP (Alpha Innotech) and quantified using ImageJ software.28 The peak intensities of the cleaved bands were divided by the total intensity of all bands (uncleaved + cleaved bands) to determine the fraction cleaved which was used to estimate gene modification levels as previously described.29 For each sample, transfections and subsequent modification measurements were performed in triplicate on different days.
High-throughput Sequencing of Genomic Modifications
HEK293-GFP stable cells were transfected with Cas9 expression and sgRNA expression plasmids, 700 ng of Cas9 expression plasmid plus 250 ng of a single plasmid expression a pair of sgRNAs were transfected (high levels) and for just Cas9 nuclease, 88 ng of Cas9 expression plasmid plus 31 ng of a single plasmid expression a pair of sgRNAs were transfected (low levels). Genomic DNA was isolated as above and pooled from three biological replicates. 150 ng or 600 ng of pooled genomic DNA was used as template to amplify by PCR the on-target and off-target genomic sites with flanking HTS primer pairs specified in the Supplementary Notes. Relative amounts of crude PCR products were quantified by gel electrophoresis and samples treated with different sgRNA pairs or Cas9 nuclease types were separately pooled in equimolar concentrations before purification with the QIAquick PCR Purification Kit (Qiagen). ~500 ng of pooled DNA was run a 5% TBE 18-well Criterion PAGE gel (BioRad) for 30 min at 200 V and DNAs of length ~125 bp to ~300 bp were isolated and purified by QIAquick PCR Purification Kit (Qiagen). Purified DNA was PCR amplified with primers containing sequencing adaptors, purified and sequenced on a MiSeq high-throughput DNA sequencer (Illumina) as described previously.23
Data Analysis
Illumina sequencing reads were filtered and parsed with scripts written in Unix Bash as outlined in Supplementary Notes. DNA sequences will be deposited in NCBI’s Sequencing Reads Archive (SRA) and source code can be found in Supplementary Software. Sample sizes for sequencing experiments were maximized (within practical experimental considerations) to ensure greatest power to detect effects. Statistical analyses for Cas9-modified genomic sites in Supplementary Tables 3 were performed as previously described30 with multiple comparison correction using the Bonferroni method.
Supplementary Material
ACKNOWLEDGEMENTS
J.P.G., D.B.T., and D.R.L. were supported by Defense Advanced Research Projects Agency HR0011-11-2-0003 and N66001-12-C-4207, U.S. National Institutes of Health NIGMS R01 GM095501 (D.R.L.), and the Howard Hughes Medical Institute (HHMI). D.R.L. was supported as a HHMI Investigator. We thank Richard McDonald for technical assistance and Vikram Pattanayak for helpful comments.
Footnotes
ACCESSION CODES
SRP041161 [link should be http://www.ncbi.nlm.nih.gov/sra?term=SRP041161]
AUTHOR CONTRIBUTIONS
J.P.G., D.B.T. performed the experiments, designed the research, analyzed the data, and wrote the manuscript. D.R.L. designed the research, analyzed the data, and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: the co-authors have filed a provisional patent application related to this work. D.R.L. is a consultant for Editas Medicine, a company that applies genome editing technologies for human therapeutic applications.
REFERENCES
- 1.Shalem O, et al. Genome-Scale CRISPR-Cas9 Knockout Screening in Human Cells. Science. 2013;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mali P, et al. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014 doi: 10.1038/nbt.2808. doi:10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M, et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L, et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jinek M, et al. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471–e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ran FA, et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2013;24:132–141. doi: 10.1101/gr.162339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramirez CL, et al. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012;40:5560–5568. doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, et al. Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Res. 2012;22:1316–1326. doi: 10.1101/gr.122879.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanamee ÉS, Santagata S, Aggarwal AK. FokI requires two specific DNA sites for cleavage. J. Mol. Biol. 2001;309:69–78. doi: 10.1006/jmbi.2001.4635. [DOI] [PubMed] [Google Scholar]
- 15.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilinger JP, et al. Broad specificity profiling of TALENs results in engineered nucleases with improved DNA-cleavage specificity. Nat. Methods. 2014 doi: 10.1038/nmeth.2845. doi:10.1038/nmeth.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimasu H, et al. Crystal Structure of Cas9 in Complex with Guide RNA and Target DNA. Cell. 2014 doi: 10.1016/j.cell.2014.02.001. doi:10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinek M, et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science. 2014 doi: 10.1126/science.1247997. doi:10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schellenberger V, et al. A recombinant polypeptide extends the in vivo half-life of peptides and proteins in a tunable manner. Nat. Biotechnol. 2009;27:1186–1190. doi: 10.1038/nbt.1588. [DOI] [PubMed] [Google Scholar]
- 21.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y, Kweon J, Kim J-S. TALENs and ZFNs are associated with different mutation signatures. Nat. Methods. 2013;10:185–185. doi: 10.1038/nmeth.2364. [DOI] [PubMed] [Google Scholar]
- 25.Wu X, et al. Genome-wide binding of the crisPr endonuclease cas9 in mammalian cells. Nat. Biotechnol. 2014 doi: 10.1038/nbt.2889. doi:doi:10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyon Y, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8:74–9. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- 27.Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat. Methods. 2013;10:751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider CA, asband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guschin DY, et al. Mackay JP, Segal DJ. Eng. Zinc Finger Proteins. Vol. 649. Humana Press; 2010. pp. 247–256. [Google Scholar]
- 30.Sander JD, et al. In silico abstraction of zinc finger nuclease cleavage profiles reveals an expanded landscape of off-target sites. Nucleic Acids Res. 2013;41:e181–e181. doi: 10.1093/nar/gkt716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.