Abstract
Five B-class MADS-box genes, including four APETALA3 (AP3)-like PeMADS2∼5 and one PISTILLATA (PI)-like PeMADS6, specify the spectacular flower morphology in orchids. The PI-like PeMADS6 ubiquitously expresses in all floral organs. The four AP3-like genes, resulted from two duplication events, express ubiquitously at floral primordia and early floral organ stages, but show distinct expression profiles at late floral organ primordia and floral bud stages. Here, we isolated the upstream sequences of PeMADS2∼6 and studied the regulatory mechanism for their distinct gene expression. Phylogenetic footprinting analysis of the 1.3-kb upstream sequences of AP3-like PeMADS2∼5 showed that their promoter regions have sufficiently diverged and contributed to their subfunctionalization. The amplified promoter sequences of PeMADS2∼6 could drive beta-glucuronidase (GUS) gene expression in all floral organs, similar to their expression at the floral primordia stage. The promoter sequence of PeMADS4, exclusively expressed in lip and column, showed a 1.6∼3-fold higher expression in lip/column than in sepal/petal. Furthermore, we noted a 4.9-fold increase in histone acetylation (H3K9K14ac) in the translation start region of PeMADS4 in lip as compared in petal. All these results suggest that the regulation via the upstream sequences and increased H3K9K14ac level may act synergistically to display distinct expression profiles of the AP3-like genes at late floral organ primordia stage for Phalaenopsis floral morphogenesis.
Introduction
In Arabidopsis thaliana and Antirrhinum majus, the development of different floral organs is controlled by several classes of floral-organ identity genes [1]. All these genes, except AP2 from A. thaliana, belong to the MADS-box family, with a highly conserved 180-bp sequence of the MADS domain that can bind to the conserved CArG-box [CC(A/T)6GG] sequence [2], [3]. These MADS-box genes were cloned from a wide range of plant species to explain the floral organ development [1], [4]–[6]. The diversification of MADS-box genes during evolution has been proposed to be a major driving force for floral diversity in land plant architecture [1], [7].
Various regulatory strategies have been reported for the expression of MADS-box genes in Arabidopsis, including transcriptional regulation on the upstream sequences or intron regions by transcription factors, with feedback and feed-forward loops, and epigenetic regulation by small RNAs [8]. Discrete cis-acting elements on the B-class APETALA3 (AP3) and PISTILLATA (PI) promoters are responsible for their expression in petal and stamen [9]–[11]. In addition, the first and second introns of FLOWERING LOCUS C (FLC) [12] and AGAMOUS (AG) [13], respectively, have a role in regulating spatial or temporal gene expression patterns. For epigenetic control of gene expression, both dimethyl histone H3 lysine-9 (H3K9me2) and dimethyl histone H3 lysine-27 (H3K27me2) are the gene repression markers. In contrast, both trimethyl histone H3 lysine-4 (H3K4me3) and acetyl histone H3 (H3Ac) are the active histone markers.
Orchidaceae is one of the largest families of flowering plants. The high species diversity in orchids is largely due to their adaptation to specialized insect pollination [14]. The orchid flower is spectacular with a gynostemium or column (a fusion of the male and female reproductive organs) and a highly modified petal, the labellum or lip, which offers a landing platform for pollinators [14]–[16].
In Phalaenopsis orchids, four AP3-like and one PI-like B-class MADS-box genes, PeMADS2∼6, have been isolated and characterized for their roles in flower morphogenesis [17]–[19]. Two duplication events resulted in the four AP3-like PeMADS2∼5. The first, occurring early in the evolutionary history of Orchidaceae, resulted in AP3A and AP3B clades, and the second resulted in four subclades, AP3A1 (PeMADS3), AP3A2 (PeMADS4), AP3B1 (PeMADS2), and AP3B2 (PeMADS5) [20]–[22]. Fluorescence in situ hybridization revealed that the four AP3-like PeMADS2∼5 genes are located on different chromosomes of P. equestris, so the four orchid AP3 paralogs may have been resulted from genome duplication [20]. The effects of gene duplication and their differences on gene regulation are important in the diversity and evolution of flowering plants [23]–[25]. At the floral primordia and early floral organ primordia stages, the transcripts of PeMADS2∼5 were detected ubiquitously, and then they are constrained to distinctively expressed organs at the late floral organ primordia stage and floral bud stage: PeMADS2 mainly expresses in sepal and petal, PeMADS3 predominantly expresses in petal and lip, PeMADS4 exclusively expresses in lip and column, and PeMADS5 is mainly expressed in petal [17], [20]. In contrast, the PI-like PeMADS6 is ubiquitously expressed in sepal, petal, lip, and column [18]. The ‘Orchid code’ assumes that the differential expression of B-class genes determined the development of sepal, petal, lip, and column [26], [27]. Moreover, the ‘homeotic orchid tepal’ (HOT) model are proposed for the dualistic features of duplicated B-class MADS-box genes involved in orchid perianth development and growth [20].
Although the five B-class MADS-box genes play important roles in the perianth development in orchids, the regulatory strategies for their distinct expression profiles in various floral organs have not been characterized. In this study, we identified the upstream promoter sequences of PeMADS2∼6 in Phalaenopsis orchids and used phylogenetic footprinting to identify conserved motifs among these promoter sequences. We analyzed the promoter activity of the upstream sequences of PeMADS2∼6 for driving GUS and luciferase gene expression in various floral organs. In addition, we examined the regulatory effects of the intron region, DNA methylation, and histone modification for their association with the high expression level of PeMADS4 in lip.
Materials and Methods
Plant materials
All upstream sequences of PeMADS genes were isolated from P. equestris with red sepal, petal and orange lip [17]. P. aphrodite subsp. formosana with white sepal, petal and yellow lip was purchased from Taiwan Sugar Corp. (Tainan, Taiwan) and used in particle bombardment experiments because the white sepal and petal made it easier for GUS staining. All plant materials were grown in the greenhouse at National Cheng Kung University (Tainan, Taiwan) under natural light and controlled temperature from 23°C to 27°C.
Isolation of the upstream promoter sequences of PeMADS2∼6
Genomic DNA was extracted from young flower buds by the cetyltriammonium bromide (CTAB) method [28]. The upstream promoter sequences of PeMADS2∼6 were isolated by use of the Universal GenomeWalker Kit (Clontech, Palo Alto, CA, USA). The desired DNA fragment was obtained by two successive PCR-based rounds of screening the GenomeWalker libraries and checked by agarose gel electrophoresis. The major bands were recovered from gels with use of the Gel DNA Fragment Extraction Kit (Geneaid, New Taipei City, Taiwan), and cloned into the pGEM-T Easy Vector (Promega, Madison, WI, USA). We randomly selected 10 to 12 colonies for sequencing. The promoter sequences were compared to all known DNA sequences with use of the default settings of BLASTN from NCBI (www.ncbi.nlm.nih.gov).
When genome walking could not extend the upstream regulatory sequences for PeMADS3∼5 genes, we used BAC clones constructed from P. equestris [29] for promoter identification. Southern blot hybridization was used to identify BAC clones containing various PeMADS genes with the gene-specific probes used in previous study [17], [18]. The BAC DNAs of each clone were isolated following the standard method [30], then digested with HindIII and separated by electrophoresis in 0.7% agarose gel. The resolved bands corresponding to promoter sequences with hybridized signals were recovered with use of the Gel DNA Fragment Extraction Kit (Geneaid) and cloned into the pGEM-T Easy vector (Promega). We randomly selected 10 to 12 colonies for sequencing. The promoter sequences characterized are deposited at the NCBI site under the accession numbers: PeMADS2 promoter (KJ127932), PeMADS3 promoter (KJ127933), PeMADS4 promoter (KJ127931), PeMADS5 promoter (KJ127934), and PeMADS6 promoter (KJ127935).
Promoter sequence analysis
We used the PLACE software (A database of Plant cis-acting Regulatory DNA Elements, http://www.dna.affrc.go.jp/PLACE/index.html) to predict the putative CArG box having 10-nt matches with the C(A/T)8G sequence [31]. In addition, the CArG box sequence was predicted for a standard of 9 of 10 matches with the core consensus binding site CC(A/T)6GG [9] by using a homemade software (designed by Dr. Chih-Hsiung Fu, Department of Engineering Science, National Cheng Kung University).
Since only 1.3-kb upstream regulatory sequence of PeMADS3 was cloned, we then chose the 1.3-kb upstream sequences of PeMADS2∼5 for phylogenetic footprinting analysis by FOOTPRINTER [32]. This tool takes into account the evolutionary relationships and the phylogenetic tree analysis. The prediction of a conserved 10-bp or 11-bp motif with a 0-bp mutation allowance was performed and the motif losses were allowed to identify the conserved motif within two promoter sequences. For all other parameters, default values were used.
Construction of chimeric reporter gene fusions
All the promoter sequences of PeMADS2∼6 were amplified directly from the genomic DNA of P. equestris by using PCR with the length of 3,249, 1,293, 3,303, 2,062, and 1,514 bp, respectively, and constructed in-frame translational fusions with the GUS reporter gene in pBI221 vector. Serial deletion fragments of PeMADS2∼6 promoters were also cloned by PCR-amplification with a series of forward primers and a reverse primer (S1 Table). The resulting PCR products were cloned into pGEM-T Easy vector (Promega) and then digested with SphI and BamHI. pBI221 containing the GUS reporter gene was digested with the same enzymes to replace the cauliflower mosaic virus (CaMV) 35S promoter with the serial deletion fragments of promoter sequences. All constructs were confirmed by sequencing to eliminate possible PCR-introduced mutations. Both the pBI221 vector containing the CaMV 35S promoter-GUS fusion (pBI221) and pBI221 vector containing a promoterless GUS cassette (pBI-PL) were recruited and considered as positive and negative controls, respectively.
Transient transformation by particle bombardment
The promoter deletion-GUS fusion plasmids were isolated by use of the High-Speed Plasmid Mini kit (Geneaid) and coated on gold particles 1.6 µm in diameter by coprecipitation as described [33]. Before particle bombardment, each floral organ was separated from the floral buds and placed on a central core 2 cm in diameter on solid agar medium. Promoter constructs were bombarded into various floral organs by use of Model Biolistic PDS-1000/He (BioRad, Hercules, CA, USA) at 1,100 psi helium gas pressure, 28.5-inch Hg vacuum and 9-cm target distance. After bombardment, floral organs were incubated at 23°C to 27°C for 2 days in an incubator with a 10-h/14-h light-dark photoperiod until analyzed by GUS histochemical staining and quantitative dual luciferase assays.
GUS histochemical staining assay
Histochemical staining of GUS activity was performed as described [34]. Tissues for GUS staining were vacuum-infiltrated in GUS assay buffer (1 mg/ml 5-bromo-4-chloro-3-indoyl glucuronide [X-Gluc]; 50 mM sodium phosphate, pH 7.0; 10 mM EDTA, pH 8.0; 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide and 0.1% Triton X-100) and incubated at 37°C overnight. Stained tissues were cleared of chlorophyll in 70% ethanol and then photographed under a microscope (TMS-F, Nikon). For each promoter-GUS fusion, the GUS staining pattern was analyzed in four independent bombarded buds, and repeated three times independently.
Quantitative dual luciferase assay
Serial-deleted promoter fragments were obtained by digestion of PeMADS4 and PeMADS6 promoters in pBI221 with SphI and BamHI, and then ligated into pJD301, containing a firefly (Photinus pyralis) luciferase gene, to replace the CaMV 35S promoter for the serial pJD-Pe4p and pJD-Pe6p constructs. pJD301_R, with Renilla luciferase gene driven by the CaMV 35S promoter, was an internal control to normalize transfection efficiency.
On day 2 after bombardment, each sample was ground, and then 1 X Passive Luciferase Buffer (Promega, Madison, WI, USA) was added. Luciferase activity was measured by use of the dual-luciferase reporter assay system (Promega) with a Lumat LB 9507 Luminometer (Berthold Technologies, Bad Wildbad, Germany), a 10-sec pre-measurement delay and a 10-sec measurement period for each assay. The relative luciferase activity was calculated as the ratio of firefly to Renilla luciferase activity. For each analysis, two independent buds were bombarded and analyzed, and the bombardments were repeated three times independently. Statistical analysis was performed by T-test, and the differences were considered significant at p<0.01.
Southern blot hybridization
Genomic DNA isolated from sepals, petals, lips, and columns of P. equestris was digested with restriction enzymes HpaII and MspI or DraI and HpaII, resolved on 0.8% agarose gel, and transferred to nylon filters (Amersham Pharmacia Biotech, Piscataway, NJ, USA) by use of a vacuum transfer system (Amersham Pharmacia Biotech). The recognition sites of HpaII and MspI are both CCGG, but digestion of HpaII is blocked by cytosine methylation and MspI is blocked by only cytosine methylation within the first cytosine (CpCGG). Two probes of PeMADS4 for methylation Southern blot assay were used: probe 1 contained the promoter and 5′ UTR sequences, and probe 2 contained the 5th intron regions. The primers were listed in S1 Table. Southern blot hybridization was performed and followed the standard protocol [30] with the 32P-labeled probes prepared by a PCR strategy.
Bisulfite sequencing
Bisulfite sequencing analysis was carried out with the EpiTect Bisulfite kit (QIAGEN, Hilden, Germany). 2-µg genomic DNAs from the petal and lip of P. equestris were treated with conversion reagents and then cleaned up as described in the manufacturer's instruction. The DNA was served as a template for PCR amplification with the incubation at 94°C for 5 min, thermocycling for 35 cycles (94°C for 30 s, 55°C for 30 s, and 72°C for 1 min), and finally at 72°C for 7 min with the primers listed in S1 Table. The PCR products were cloned into pGEM-T Easy vector (Promega) and transformed into Escherichia coli. We randomly selected 10 colonies for sequencing and analysis.
Chromatin immunoprecipitation (ChIP) and real-time PCR analyses
ChIP assay was as described [35]. Chromatin extracts were prepared from the petal and lip of P. equestris treated with formaldehyde. Chromatin was sheared to an average length of 500–1500 bp by sonication and immunoprecipitated with the antibodies anti-H3K4me3 (catalogue no. 04-745, Millipore, Billerica, MA, USA), anti-H3K9me2 (catalogue no. 04-768), or anti-H3K9K14ac (catalogue no. 06-599). The immunocomplexes were harvested with Protein G agarose beads (Millipore) and heated at 65°C for 5 hours to release DNA cross-linked to the immunoprecipitated proteins. The DNA cross-linked to the immunoprecipitated proteins was analyzed by real-time PCR with the primers listed in S1 Table. The immunoprecipitations were performed twice.
The DNA template was mixed with 2X SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA) in an ABI Prism 7000 sequence detection system (Applied Biosystems), and each sample was analyzed in triplicate. Reactions involved incubation at 95°C for 10 min, and thermocycling for 40 cycles (95°C for 15 s and 60°C for 1 min). After amplification, melting curve analysis was used to verify amplicon specificity and primer dimer formation. The amount of DNA after ChIP was quantified and normalized to an internal control ACTIN2 for H3K4me3 and H3K9K14ac or Ta3 for H3K9me2 [36]. Data are mean ± SD calculated from three technological and two biological replicates.
Results
Cloning of the upstream sequences of PeMADS2∼6
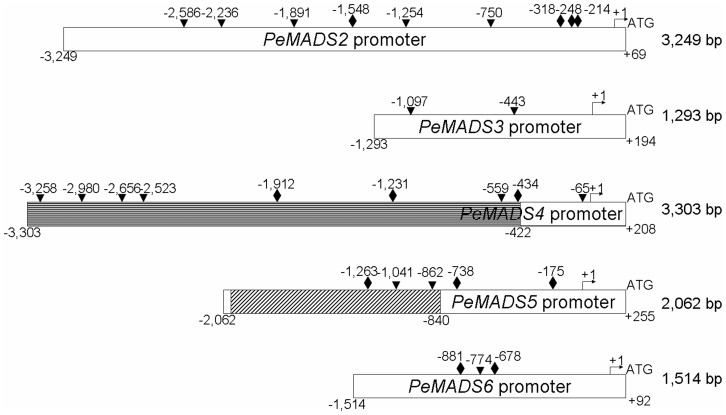
The five PeMADS promoter regions were cloned from genomic DNA by use of GenomeWalker. The PeMADS2∼6 promoter fragments obtained were 3,249, 1,293, 422, 2,121, and 1,514 bp, respectively (Fig. 1). However, the 2,121-bp promoter sequence of PeMADS5 could not be amplified from the genomic DNA of P. equestris by using PCR, and both PeMADS3 and PeMADS4 with the promoter fragments <1.5 kb could not be extended. We then used BAC clones of P. equestris for promoter identification of PeMADS3∼5 by Southern blot hybridization with probes from the coding sequences of PeMADS3∼5 (Table 1). BAC DNA was isolated and confirmed by restriction enzyme digestion. The resolved bands corresponding to the hybridized signals were recovered and sequenced. After assembly, the PeMADS3 promoter remained at 1,293 bp. For the PeMADS4 promoter, a 4.8-kb DNA fragment from BAC clones was recovered and sequenced, which extended its upstream sequence to 3,303 bp (Fig. 1, horizontal line box). For PeMADS5, the promoter fragment was replaced with a 2,062-bp fragment (Fig. 1).
Figure 1. Promoter sequences of PeMADS2∼6 and the putative CArG boxes.
Length of promoter sequences of PeMADS2, PeMADS3, and PeMADS6 were 3,249, 1,293 and 1,514 bp, respectively. The promoter sequence of the PeMADS4 was extended from −422 bp to −3,303 bp (horizontal line box), and the original fragment of −2,121 to −840 bp of PeMADS5 promoter was replaced by a 1,227-bp fragment (diagonal line box). The rhombus and the triangles indicate the putative CArG boxes predicted by consensus CC(A/T)6GG and C(A/T)8G motifs, respectively. “+1” means the transcription start site and ATG was the translation start site. The lengths of the promoters are show from the transcription start site to the upstream sequences.
Table 1. BAC clones containing various PeMADS genes.
| Genes | Clone no. | BAC clones |
| PeMADS3 | 3 | NCKU-PE-btBAC-2050 C15 |
| NCKU-PE-btBAC-2065 P3 | ||
| NCKU-PE-btBAC-3081 A24 | ||
| PeMADS4 | 1 | NCKU-PE-btBAC-1105 H24 |
| PeMADS5 | 4 | NCKU-PE-btBAC-2016 E10 |
| NCKU-PE-btBAC-2022 D13 | ||
| NCKU-PE-btBAC-2035 M21 | ||
| NCKU-PE-btBAC-2049 A22 |
Prediction of CArG box in the promoter sequences of PeMADS2∼6
We used an in-house developed software to predict the presence of the putative CArG box with a standard of a 9-in-10-nt match with the core consensus CC(A/T)6GG motif [9]. Four CArG boxes were detected at nucleotides −1,548, −318, −248, and −214 in the PeMADS2 promoter region; three at nucleotides −1,912, −1,231, and −434 in the PeMADS4 promoter region; three at nucleotides −1,263, −738, and −175 in the PeMADS5 promoter region, and two at nucleotides −881 and −678 in the PeMADS6 promoter region (Fig. 1, Table 2). In contrast, no CArG-box-like sequence was detected in the PeMADS3 promoter sequence.
Table 2. Putative CArG boxes at the PeMADS promoter regions predicted by homemade software and the PLACE database.
| PeMADS promoter (bp) | CArG-box sequences | Location (nt at the promoter regions) |
| PeMADS2 | CCCTAAATGG | −214a |
| (3,249) | CCATTCTAGG | −248a |
| CTTTAAATGG | −318a | |
| CTATATTAAG | −750b | |
| CATAATTTTG | −1,254b | |
| CCAAAATTTG | −1,548a | |
| CTAATTTTAG | −1,891b | |
| CAAAATTTAG | −2,236b | |
| CATATTAAAG | −2,586b | |
| PeMADS3 | CAAAAAAAAG | −443b |
| (1,293) | CTTTTATAAG | −1,097b |
| PeMADS4 | CTTATAAAAG | −65b |
| (3,303) | CTATTATAGG | −434a |
| CATATTATAG | −559b | |
| CATATTTTGG | −1,231a | |
| CCTATGTAGG | −1,9128a | |
| CATATATTAG | −2,523b | |
| CTTTTTTATG | −2,656b | |
| CAAAATTTTG | −2,980b | |
| CAAAATTTTG | −3258b | |
| PeMADS5 | GCTTAATTGG | −175a |
| (2,062) | TCAAAATTGG | −738a |
| CATAAATATG | −862b | |
| CTTTATATTG | −1,041b | |
| CGATTTAAGG | −1,263a | |
| PeMADS6 | CCAAATTTGA | −678a |
| (1,514) | CAAATTTAAG | −774b |
| GCAAAATAGG | −881a |
CArG boxes predicted with a homemade software.
CArG boxes predicted with the PLACE database.
We used the PLACE database with a standard of 10-nt match with the C(A/T)8G sequence to further examine the CArG box [31]. Five CArG boxes were detected at nucleotides −2,586, −2,236, −1,891, −1,254, and −750 in the PeMADS2 promoter region; two at nucleotides −1,097 and −443 in the PeMADS3 promoter region; six at nucleotides −3,258, −2,980, −2,656, −2,523, −559, and −65 in the PeMADS4 promoter region; and two at nucleotides −1,041 and −862 of the PeMADS5 promoter region; one at nucleotide −774 in the PeMADS6 promoter region (Fig. 1, Table 2).
The conserved regulatory motifs predicted within the promoter regions of PeMADS2∼5
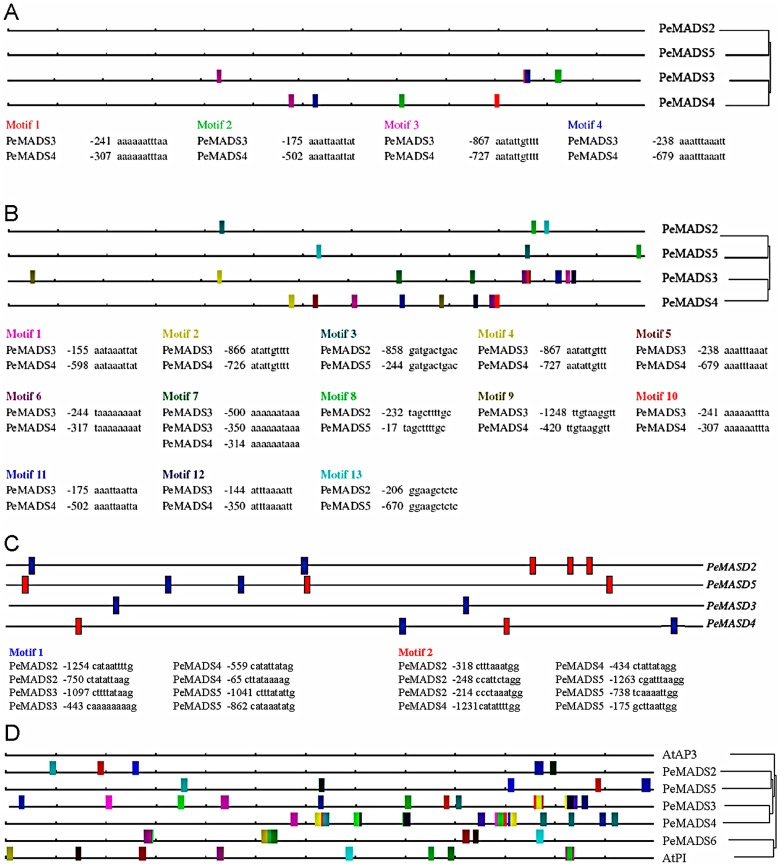
The conserved regulatory motifs are thought to be functional important for gene expression profiles [37]–[40]. However, multiple alignment of these promoter sequences using global alignment procedures was failed because the inversions often cause rearrangements of the regulatory elements [41]. Therefore, we examined the 1.3-kb upstream sequences of PeMADS2∼5 by phylogenetic footprinting, a method for discovering regulatory elements in a set of regulatory regions [32] and has been used to promoter analysis for MADS-box genes in Arabidopsis and Orchis italica and for bZIP genes in rice and sorghum [40], [42], [43]. With prediction of a conserved 11-bp motif with a 0-bp mutation allowance, conservation of four 11-bp motifs was identified between the promoter regions of PeMADS3 and PeMADS4 in different order (Fig. 2A), while no motifs were conserved between promoter regions of PeMADS2 and PeMADS5 (Fig. 2A). With prediction of conserved 10-bp motifs, increased conserved motifs were identified between the promoter regions of PeMADS3 and PeMADS4, and three motifs were lined up in those of PeMADS2 and PeMADS5 (Fig. 2B). Interestingly, we noticed that differential conserved 10-bp motif sets were detected between the promoter regions of PeMADS2/PeMADS5 and PeMADS3/PeMADS4 (Fig. 2B), which suggests that the two lineage of PeMADS2/5 and PeMADS3/4 have diverged for their subfuncationalization after gene duplication. Moreover, the CArG boxes were broadly distributed in the promoter regions of the four PeMADS genes, and no clear correlations of these CArG boxes were detected with their distinct expression profiles (Fig. 2C). Furthermore, with prediction of a conserved 12-bp motif with a 1-bp mutation allowance, motifs were identified between the promoter regions of PeMADS2 and PeMADS5, PeMADS3 and PeMADS4, and PeMADS6 and AtPI, but no motifs were present between the promoter sequences of AtAP3 and PeMADS2∼5 (Fig. 2D).
Figure 2. Visual representation of motifs in the 1.3-kb promoter sequences of PeMADS2∼5.
FOOTPRINTER parameters: (A) motif size: 11, allowed mutations: 0, (B) motif size: 10, allowed mutations: 0. (C) Putative CArG boxes in the PeMADS promoter regions predicted with the homemade software (red box) and the PLACE database (blue box).
Functional analysis of PeMADS2∼6 promoter sequences
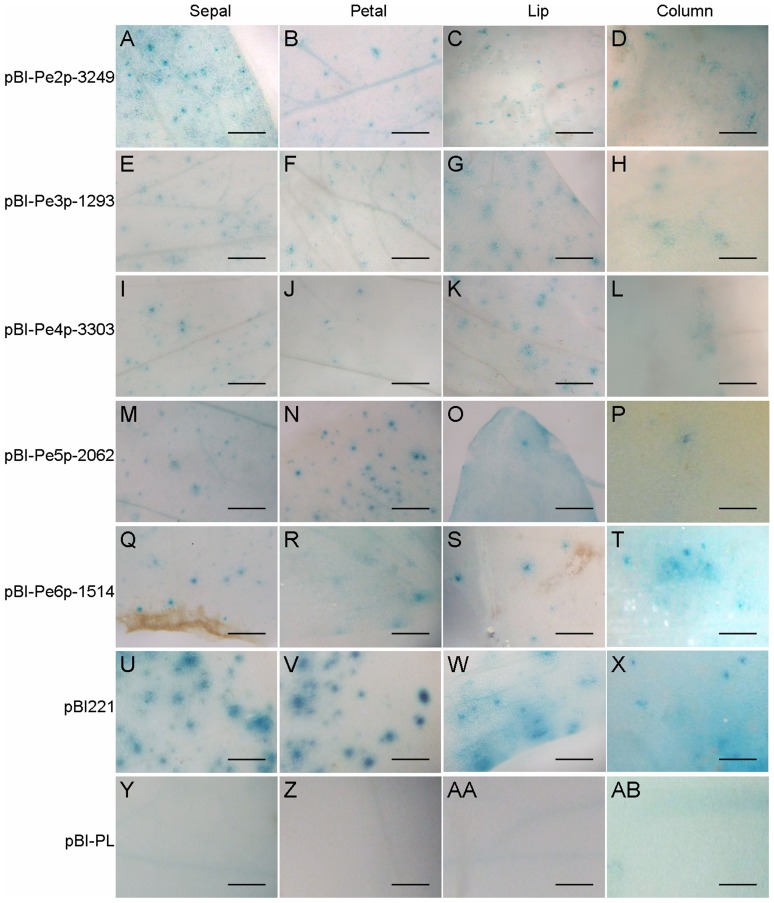
We examined the promoter activities of PeMADS2∼6 fragments with length of 3,249 bp, 1,293 bp, 3,303 bp, 2,062 bp, and 1,514 bp, respectively, for their ability to drive GUS expression by bombarding them into 1.5-cm floral buds of P. aphrodite subsp. formosana. Notably, all five PCR-amplified promoter fragments could drive GUS expression in the floral organs examined (Fig. 3A–T), similar to their expression patterns at the early floral primordia stage [20]. Moreover, serial deletion clones of the upstream sequences of PeMADS2∼6 were constructed for GUS expression assay.
Figure 3. GUS histochemical staining for the promoter activities of PeMADS2∼6.
Histochemical assay of GUS expression in floral organs shown in the order of pBI-Pe2p-3249 (A–D), pBI-Pe3p-1293 (E–H), pBI-Pe4p-3303 (I–L), pBI-Pe5p-2062 (M–P), pBI-Pe6p-1514 (Q–T), pBI221 (U–X) and pBI-PL (Y-AB). Constructs were bombarded into four independent floral buds, and results are representative of three independent bombardment experiments. Scale bar = 0.5 mm.
The 208- and 375-bp promoter sequences of PeMADS6 and PeMADS4, respectively, were sufficient to drive GUS expression
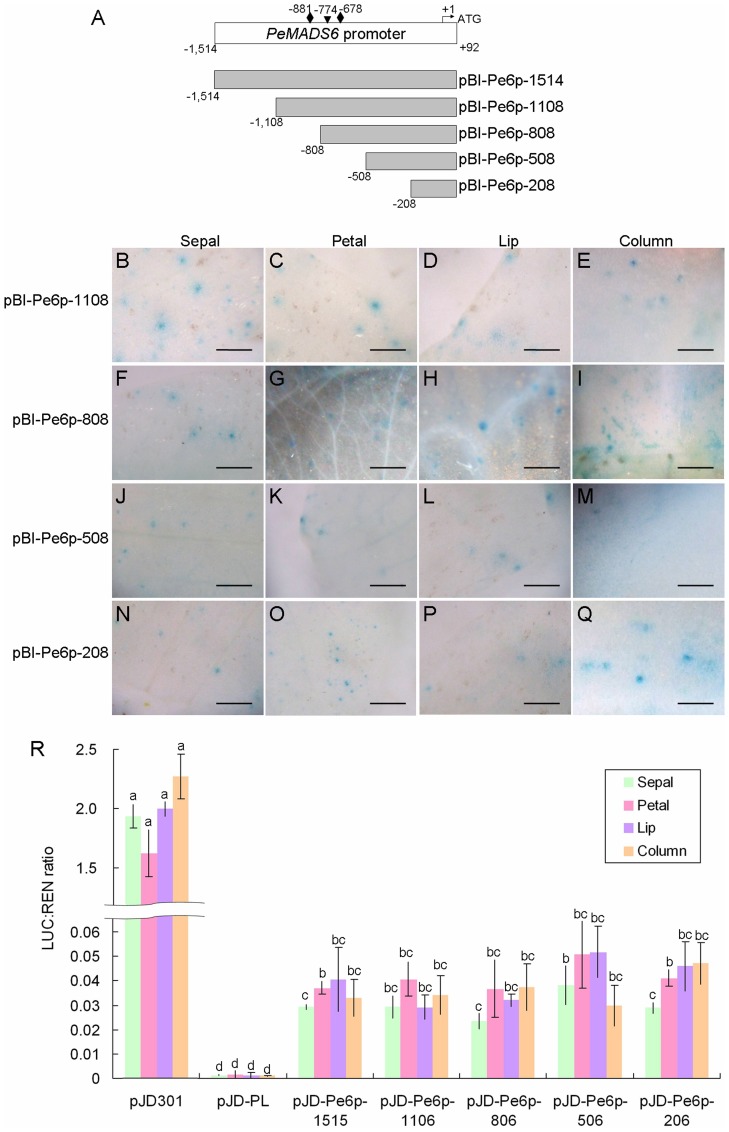
The PI-like PeMADS6 was expressed ubiquitously in all floral organs. To assess the minimal promoter region of PeMADS6, four deletion clones of the PeMADS6 promoter sequence were resulted, including 1,108-bp, 808-bp, 508-bp, and 208-bp fragments containing 3, 2, 0, and 0 CArG boxes, respectively. Similar to the full length PeMADS6 promoter construct, pBI-Pe6p-1514 (Fig. 3Q-T), all four deletion promoter sequences of PeMADS6 could drive GUS expression in all the floral organs examined (Fig. 4B–Q), although the expression was slightly decreased in pBI-Pe6p-508 and pBI-Pe6p-208 constructs (Fig. 4J–Q). Moreover, quantitative dual luciferase assay was performed to further examine the differential promoter activities of these serial deletion constructs. The pJD-Pe6p-208 construct was sufficient to drive luciferase expression in all floral organs (Fig. 4R). Extension of the upstream sequence from the pJD-Pe6p-508 to pJD-Pe6p-1514 constructs conferred similar GUS expression in all floral organs (Fig. 4R). Therefore, the 208-bp promoter sequence was a minimal promoter for PeMADS6 expression in all four floral organs.
Figure 4. Functional analysis of serial deletions of PeMADS6 promoter.
(A) Serial deletion constructs of PeMADS6 promoter. (B-Q) Histochemical assay of flower organs bombarded with serial deletions of PeMADS6 promoter shown in the order of pBI-Pe6p-1108 (B–E), pBI-Pe6p-808 (F–I), pBI-Pe6p-508 (J–M) and pBI-Pe6p-208 (N–Q). Constructs were bombarded into four independent floral buds, and results are representative of three independent bombardment experiments. Scale bar = 0.5 mm. (R) Dual luciferase assay of serial deletions of PeMADS6 promoter. The same letters above the bars are not statistically different by T-test analysis (p<0.01). Data are mean ± SD (n = 6). All constructs were analyzed for promoter activities of driving luciferase expression by bombardment into two floral buds, and results are representative of three independent bombardment experiments.
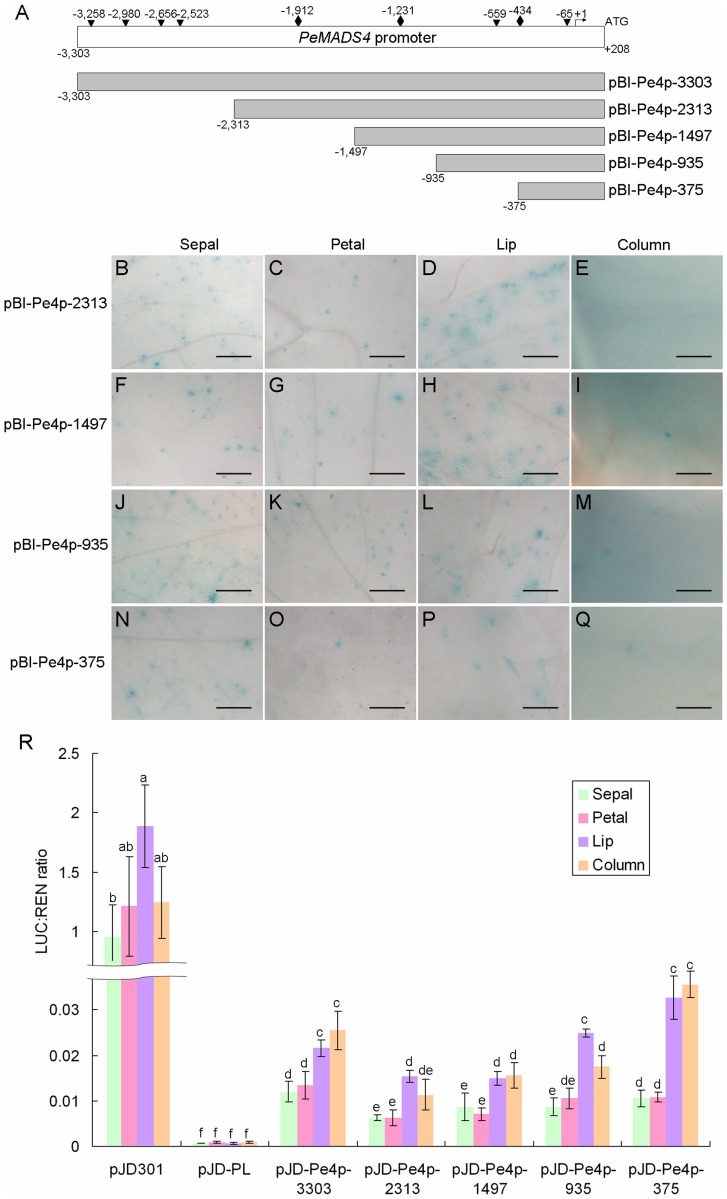
The four AP3-like PeMADS2∼5 genes express differentially in the floral organs with distinct patterns [17]. Two to five serial deletion clones for the upstream regulatory sequences of PeMADS2∼5 were constructed for GUS expression assay. Similarly, most serial deletion constructs could drive GUS expression in all four floral organs (Fig. 5B–Q, S1–S3 Figures), resembled to the gene expression patterns at the early floral organ primordia stage. Among them, the minimal promoters of PeMADS2∼5 were found to be 291 bp, 407 bp, 375 bp, and 122 bp of their upstream regulatory sequences, respectively (Fig. 5N–Q, S1–S3 Figures).
Figure 5. Functional analysis of serial deletions of PeMADS4 promoter.
(A) Serial deletion constructs of PeMADS4 promoter. (B–Q) Histochemical assay of flower organs bombarded with serial deletions of PeMADS4 promoter shown in the order of pBI-Pe4p-2313 (B–E), pBI-Pe4p-1497 (F–I), pBI-Pe4p-935 (J–M) and pBI-Pe4p-375 (N–Q). Constructs were bombarded into four independent floral buds, and results are representative of three independent bombardment experiments. Scale bar = 0.5 mm. (R) Dual luciferase assay of the serial deletions of PeMADS4 promoter. The same letters above the bars are not statistically different by T-test analysis (p<0.01). Data are mean ± SD (n = 6). All constructs were analyzed for promoter activities of driving luciferase expression by bombardment into two floral buds, and results are representative of three independent bombardment experiments.
The promoter sequence of PeMADS4 was chosen for further examination by using quantitative dual luciferase assay to delineate its exclusive expression in lip and column at the late floral primordia stage [20]. All five serial deletion promoter constructs of PeMADS4 with various lengths conferred a 1.6∼3-fold increase of luciferase activities in lip and column than in sepal and petal (Fig. 5R), in accordance with the high expression of PeMADS4 in lip and column. The shortest fragment, pJD-Pe4p-375 conferred a 3-fold higher promoter activity in lip and column than in sepal and petal (Fig. 5R), which suggests that the upstream sequences of PeMADS4 was necessary, but not sufficient for its exclusive high expression in lip and column. It is possible that other factors are also required for regulating the lip- and column-specific expression of PeMADS4 at the late floral organ primordia stage of Phalaenopsis orchids.
The 5th intron of PeMADS4 had no effect on its organ-specific expression pattern
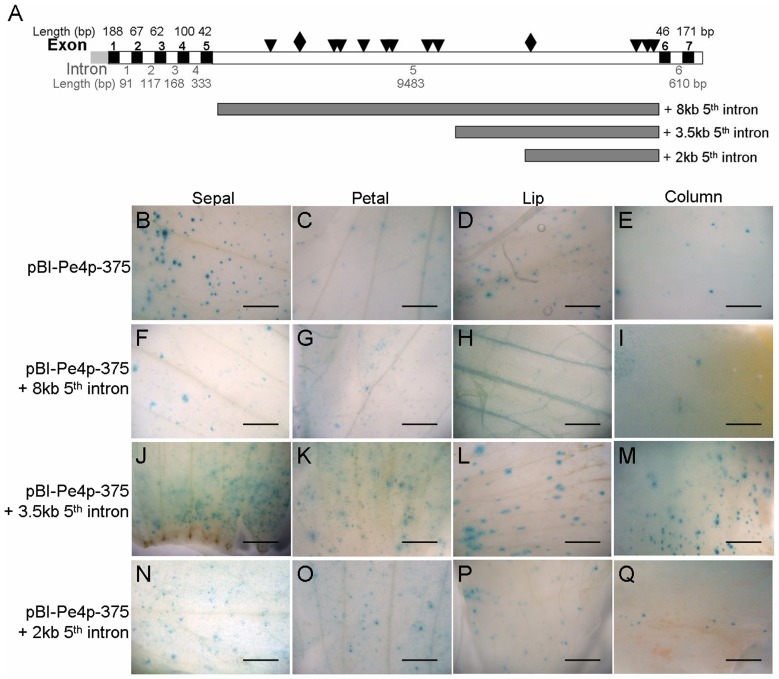
The longest introns of AG and FLC in Arabidopsis play a regulatory role for their gene expression [12], [13]. It was intriguing to know whether the introns of PeMADS4 may regulate its distinct expression at the late floral organ primordia stage of Phalaenopsis orchids. To test this, we first sequenced the genomic sequence of BAC clones containing PeMADS4, NCKU-PE-btBAC-1105 H24. Then, the genomic sequence was compared to its cDNA sequence, and seven exons and six introns were identified for PeMADS4 with a long 5th intron of 9,483 bp (Fig. 6A). The 5th intron contains two conserved CC(A/T)6GG motifs (Fig. 6A, rhombus) and 11 C(A/T)8G motifs (Fig. 6A, triangles) (Fig. 6A).
Figure 6. Histochemical assay of the 5th intron of PeMADS4.
(A) Genomic structure of PeMADS4. Gray, black, and white boxes indicate the promoter, exon, and intron regions of PeMADS4 gene, respectively. Numbers above the black boxes are the number and length (bp) of exons, respectively. Numbers beneath the white boxes are the number and length (bp) of introns, respectively. Two CC(A/T)6GG sequences (rhombus) and 11 C(A/T)8G sequences (triangles) are located in the 5th intron. Three serial deletions of the 5th intron were designed for 2-, 3.5- and 8-kb sequences, respectively, and inserted into the upstream region of the Pe4pF1 promoter sequence in the pBI-Pe4p-375 construct. (B-Q) Histochemical assay of the serial deletions of the 5th intron of PeMADS4 were in the order of pBI-Pe4p-375 (B–E), pBI-Pe4p-375-8- (F–I), pBI-Pe4p-375-3.5- (J–M) and pBI-Pe4p-375-2-kb 5th intron constructs (N–Q). Constructs were bombarded into four independent floral buds, and results are representative of three independent bombardment experiments. Scale bar = 0.5 mm.
To assess the effect of the 5th intron on PeMADS4 expression, we generated three subfragments of 8-kb, 3.5-kb, and 2-kb fragments by PCR amplification, cloned into the upstream region of the promoter sequence in the pBI-Pe4p-375 construct, and named as pBI-Pe4p-375+8 kb, pBI-Pe4p-375+3.5 kb, and pBI-Pe4p-375+2 kb 5th intron constructs, respectively (Fig. 6A). The addition of the 8-kb fragment intron resulted a sharp decrease of GUS expression in all four floral organs (Fig. 6F–I) as compared with the pBI-Pe4p-375 native construct (Fig. 6B–E), which suggests that the 8-kb 5th intron may have a negative effect on PeMADS4 expression. Alternatively, the addition of the 8-kb fragment was too long to affect the transformation efficiency. Otherwise, the GUS expression in all floral organs was not significantly different with the addition of either the 2- or 3.5-kb fragments of the 5th intron (Fig. 6J–Q), so the 3.5- or 2-kb 5th-intron sequence showed little or no effects for the exclusive PeMADS4 expression in lip and column.
DNA methylation was not responsible for regulation of PeMADS4 expression
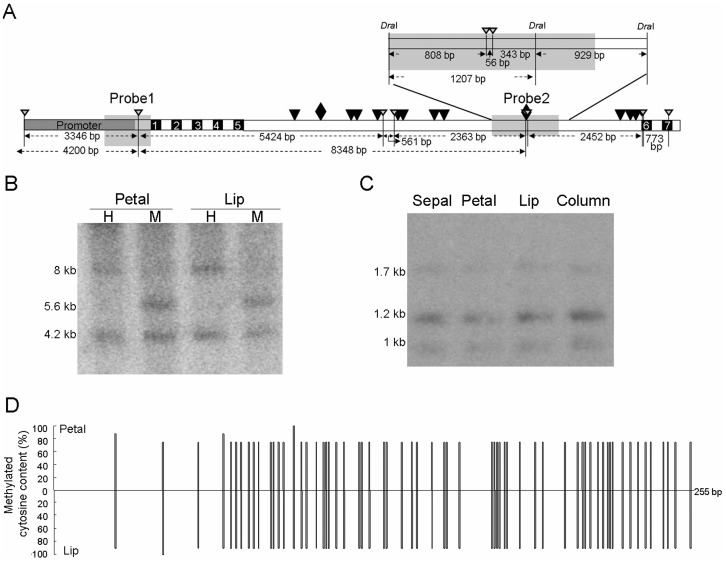
To examine whether the specific expression of PeMADS4 in lip and column was caused by DNA methylation in the regulatory sequences in planta, Southern blot hybridization was performed to analyze the DNA methylation status in the promoter, translation start site, and intron regions of PeMADS4. DNA samples isolated from petal and lip of P. equestris were digested with the methylation-sensitive endonucleases HpaII or MspI (H or M) (Fig. 7B) and DraI/HpaII (Fig. 7C). Probe 1 containing a 582-bp fragment including a 375-bp promoter sequence and a 207-bp 5′-UTR region of PeMADS4 (Fig. 7A, B) was used for DNA samples from petal or lip and similar methylation status were obtained with digestion of HpaII (Fig. 7B, 4.2- and 8-kb fragments) and MspI (Fig. 7B, 4.2- and 5.6-kb fragments). Moreover, hybridization with probe 2, containing a 2,136-bp fragment of the 5th intron of PeMADS4, gave the same results for all DraI/HpaII-digested DNA samples from sepal, petal, lip, and column (Fig. 7A, C, 1- and 1.2-kb fragments), so the HpaII site within probe 2 region was methylated in all four floral organs. Thus, the DNA methylation status was the same in the promoter region, translation start site, and the 5th intron regions of the PeMADS4 gene for both petal and lip of Phalaenopsis flowers.
Figure 7. Methylation status in the promoter and 5th intron regions of PeMADS4.
(A) Locations of the probes used for Southern blot analysis and methylation status in the promoter (B) and 5th intron (C) regions of PeMADS4. Gray, black, and white boxes indicate the promoter, exon, and intron regions of PeMADS4 gene, respectively. The rhombus and black triangles are the predicted CArG boxes. The white triangles point to the HpaII/MspI sites. Probes used in this study are shown in gray located in the 5′ UTR (Probe 1) or in the 5th intron (Probe 2). (B) Southern blot analysis was performed with genomic DNA extracted from petal and lip of P. equestris, digested with methylation-sensitive enzymes, HpaII (H) and MspI (M), and hybridized with probe 1. (C) Southern blot analysis was performed with the genomic DNA extracted from sepals, petals, lips and columns of P. equestris, double-digested with DraI and HpaII, and hybridized with probe 2. Probe 2 was a 2,136-bp fragment between three DraI restriction enzyme cleavage sites and contained two HpaII site. (D) Bisultife sequencing for the methylation status within the promoter region of PeMADS4.
To have a single-base resolution of methylation status, bisulfite sequencing technology was performed. Highly methylated cytosine residues were detected within the promoter region and translation start site of PeMADS4 in DNA samples from both petal and lip (Fig. 7D). Therefore, DNA methylation may not play a role if any in tissue specificity of PeMADS4 expression in lip and column.
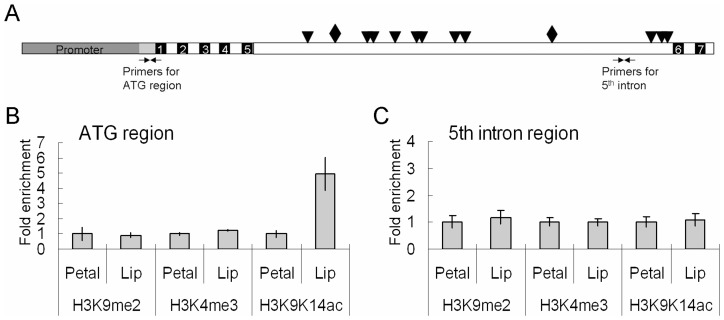
Concomitant differential histone acetylation for PeMADS4 expression
It is possible that the tissue-specific expression profiles of PeMADS genes not only reside in the DNA-level promoter sequences, but also in the protein-level histone modification. To address this, various histone modifications were analyzed by ChIP assay with antibodies against the gene repression marker H3K9me2 and gene activation markers H3K4me3 and H3K9K14ac. The precipitated DNA samples from both petal and lip were analyzed by real-time PCR with the primer sequences located at the translation start site (ATG) and the 5th intron regions of PeMADS4 (Fig. 8A). Notably, we detected a 4.9-fold higher H3K9K14ac at the translation start site in lip than in petal (Fig. 8B). In contrast, no differential levels of H3K9me2 and H3K4me3 were detected in the translation start site in both petal and lip (Fig. 8B). Furthermore, no substantial differential levels of H3K9me2, H3K4me3, and H3K9K14ac within the 5th intron region of PeMADS4 were detected in petal and lip (Fig. 8C). Thus, the increased level of H3K9K14ac on the translation start site of PeMADS4 gene may allow for more access of the transcription factor and the further increased gene expression in lip, thus leading to its optimized expression in Phalaenopsis.
Figure 8. Histone modification on the ATG and 5th intron regions of PeMADS4.
(A) Locations of the primers used in ChIP assay. Gray, black, and white boxes indicate the promoter, exon, and intron regions of PeMADS4 gene, respectively. The rhombus and black triangles are the predicted CArG boxes. ChIP assay of histone modification of dimethyl-H3K9 (H3K9me2), trimethyl-H3K4 (H3K4me3), acetyl-H3K9 and H3K14 (H3Ac) was analyzed on the ATG (B) and 5th intron regions (C) of PeMADS4 in the petal and lip of P. equestris. The amount of DNA after ChIP was quantified and normalized to an internal control ACTIN2 for H3K4me3 and H3K9K14ac or Ta3 for H3K9me2. Data are mean ± SD calculated from three technological and two biological replicates. X = fold.
Discussion
In model species Arabidopsis thaliana and Antirrhinum majus, the molecular genetic studies on flower morphogenesis indicate that homeotic B-class MADS-box genes determine petals and stamen identities. A shift model has been proposed through several comparative expression analyses suggesting that a shift expression of B-class genes to the outer perianth is associated with the petal-like organs on the first flower whorl in several monocot species, such as Tulipa gesneriana and Lilium longiflorum [44], [45]. To understand the development and evolution of orchid flowers, extensive studies of the molecular phylogeny and expression patterns of candidate B-class MADS-box genes have been performed in several genus, species, and hybrids of orchids. Four ancient orchid-specific clades of AP3-like and one major lineage of PI-like B-class genes were identified and characterized [17], [26], [46]. Differential expression of AP3-like genes has been associated with the identity of distinct perianth organs, that are the basis of several models used to explain the morphogenesis and evolution of the orchid flowers [20], [27].
Histone acetylation and promoter sequences act synergistically to regulate PeMADS4 expression in lip
The minimum promoter sequence necessary for a wild-type AP3 expression pattern in Arabidopsis is localized within the 727-bp fragment upstream of the transcriptional start site [9]. In this study, we showed that the minimal promoter sequences for PeMADS2∼6 were 291 bp, 407 bp, 375 bp, 122 bp, and 208 bp of their upstream regulatory sequences, respectively. In addition, the regulation of the promoter sequence and the increased H3K9K14ac level may act synergistically to result in the exclusive high expression of PeMADS4 in lip and column at the late floral organ primordia stage and floral bud stage of Phalaenopsis orchids. A similar regulatory mechanism may be adopted for regulation of the distinct expression profiles of the other three AP3-like paralogs, PeMADS2, PeMADS3, and PeMADS5, in Phalaenopsis flowers to complete the diversified subfunctionalization for orchid floral morphogenesis.
Transient expression assay for promoter analysis by use of particle bombardment
Stable transformation of Phalaenopsis orchids is time-consuming and requires considerable human resources, because their long life cycles of two to three years for the transition from the seed germinative to reproductive stages. Transient expression assays, which were carried out by using particle bombardment [47], [48] and protoplast transfection [49], have been used to reduce the time for analyzing gene functions in Arabidopsis [50], [51], rice [49], [52], maize [53], potato [54], soybean [55], tomato [56], wheat [55], and white spruce [57]. Protoplasts retain many signal transduction pathways from the cells which they are derived [58], but the process of protoplast cultivation may change the mRNA expression profiles [59]. In contrast, particle bombardment permits the transient expression within intact tissues of entire plants. However, an important argument for transient expression is that it is easy to overexpress gene constructs because of high copy numbers of plasmid DNA, strong promoters, long expression times, and the independence of the transgene expression on the genomic site of integration [60]. While in stable transgenic plants, a high transgene copy is frequently accompanied with gene silencing and the position effect affects the expression of promoter constructs by flanking sequences in the genome. In Orchidaceae, particle bombardment with histochemical GUS staining have been used to analyze the promoter activities of disease resistance response protein (OnDRRP), Expansin (OnExpansin), and three trypsin inhibitor (OnTI1∼3) in leaves and flowers of Oncidium Gower Ramsey, and cytokinin oxidase (DSCKX1) in protocorm-like-bodies of Dendrobium Sonia, respectively [61], [62]. Here, we analyzed the promoter activities of PeMADS2∼6 for driving GUS and luciferase reporter genes by particle bombardment, and the ubiquitous expression in all floral organs may be caused by the high copies of bombarded plasmid DNA and/or the naked DNA lack of chromatin modification. However, the serial deletion sequences of PeMADS4 promoter showed a higher luciferase activity in lip and column than in sepal and petal in contrast to the more-or-less similar lucifease activity detected in all four floral organs driven by PeMADS6 promoter. Moreover, we further examined the DNA methylation and histone modification within the translation start site of PeMADS4 to verify the regulatory strategy for its differential expression pattern. All these results suggested that the transient expression assay by particle bombardment accompanying with DNA methylation and histone modification analyses provides a basic information about the regulatory strategies of these PeMADS2∼6 genes exhibiting distinct expression profiles.
375-bp promoter sequence of PeMADS4 was required for its lip and column expression
The 375-bp promoter sequence of PeMADS4 conferred higher luciferase activity in lip and column, which meant that the PeMADS4 promoter was regulated by a lip- and column-specific transcription factor. Several transcription factors have been shown to dominantly express in lip, such as MADS, ARF, C3H, HB-other, YABBY, ZF-HD, bZIP, CO, TALE, HD-ZIP, MYB, and AP2-like families [63]. The cis-acting regulatory elements on the upstream region of PeMADS4 were analyzed by PLACE software, and the CARGCW8GAT for MADS and MYBCOREATCYCB1 or MYBST1 for MYB-binding motifs were predicted within this sequence (S2 Table). It is possible that unidentified motifs were resided in the 375-bp fragment of PeMADS4 promoter for the interaction with above mentioned transcription factors. The exact transcription factors responding for the activation on the 375-bp fragment of PeMADS4 promoter were required for further studies.
The promoter sequences of OrcPI and PeMADS6 showed little similarity
Comparing the differential expression patterns of AP3-like genes, PI-like genes showed uniformly and highly conserved expression patterns in all four floral organs in several species of various subfamily of Orchidoideae, such as Cypripedioideae, Epidendroideae, Orchidoideae, and Vanilloideae [20]. OrcPI is a PI-like MADS gene from Orchis italica, a species of Orchidoideae, and expresses in young inflorescences and all floral organs [42]. Various serial deletion of promoter sequence of OrcPI with 1324-bp, 854-bp, 577-bp, and 356-bp upstream sequences can drive the GUS expression in petal tissue in the white Rosa hybrid [42]. However, the upstream regulatory sequences of PeMADS6 and OrcPI could not be aligned together, even for their minimal promoter with 208- and 356-bp fragments, respectively. However, it is intriguing that two 11-bp motifs were detected between nucleotide −249 and −173 bp of PeMADS6 promoter and between −937 and −641 bp of OrcPI promoter by using the BLAST2 algorithm (S4 Figure). Whether this element plays any roles in the ubiquitous expression of the PI-like genes in orchid flowers awaits further studies.
Histone modification regulated the plant development and stress response
The epigenetic regulation is dynamic and varies between cell types and in response to development stages or environmental stimuli [64]. H3K9me2 is mainly detected in heterochromatin regions and associated with transposable elements (TEs) [65]. In contrast, H3K4me3 is enriched in euchromatic regions and associated with transcribed regions of non-TE genes. Expectedly, typical activating histone modification, such as H3K4me3 and H3K9ac is detected in the same genomic regions [66]. For example, during the vernalization response (exposure to a prolonged period of low temperature), the gene repression marks, H3K9me2 and H3K27me2, are enriched at FLC locus and thereby controlling flowering time, in contrast to the activated state of FLC chromatin with active histone marks, H3K4me3 and H3ac, before prolonged cold exposure [67]. Moreover, several epigenetic regulators are involved in the regulation of floral homeotic genes. Mutation in a H3K4 methyltransferase ATX1 results down-regulation of AP1, AP2, PI, and AG, but not of AP3 and SEP3 [68]. The PRC2-like complexes containing CLF, FIE, EMF2 and MSI1 act on repression of AG by regulating H3K27me3 [69], [70], and the emf2 mutant results ectopic overexpression of AG, AP3, AP1, PI, SEP2, and SEP3 [71]. Furthermore, the gene repression marker, H3K27me3, have been shown that its release results in tissue-specific gene activation [72]. Here, we showed that an increased level of H3K9K14ac on the translation start site of PeMADS4 gene may enhance the exclusive gene expression in lip to decipher the lip morphogenesis in Phalaenopsis orchids, although its mechanism is still needed to be investigated.
Transgenic plants by use of the PeMADS2∼5 promoter
In Arabidopsis, the promoter sequences of AP3 and PI have been used for the floral organ-specific expression in the transgenic approach. The 288-bp promoter sequence of AP3 with a petal-specific domain [9] and the 300-bp fragment of PI with expression in petal and stamen [11] were used to drive an RNAi vector targeting the GUS reporter gene and introduced into a line constitutively expressing GUS, which resulted in reduced GUS expression in petal [73]. However, the promoter sequences of PeMADS2∼5 drove the expression of GUS and luciferase reporter genes in the whole flower but not exclusively in distinct floral organs. Thus, the amplified upstream regulatory sequences of PeMADS2∼5 could be used as flower-specific, but not practically for floral organ-specific promoters in the application for transgenic plants.
Supporting Information
Functional analysis of serial deletions of PeMADS2 promoter. (A) Serial deletion constructs of PeMADS2 promoter. (B–Q) Histochemical assay of flower organs bombarded with serial deletions of PeMADS2 promoter shown in the order of pBI-Pe2p-2224 (B-E), pBI-Pe2p-1823 (F-I), pBI-Pe2p-1312 (J-M), pBI-Pe2p-750 (N–Q), and pBI-Pe2p-291 (R–U). Constructs were bombarded into four independent floral buds, and results are representative of three independent bombardment experiments. Scale bar = 0.5 mm.
(TIF)
Functional analysis of serial deletions of PeMADS3 promoter. (A) Serial deletion constructs of PeMADS3 promoter. (B–Q) Histochemical assay of flower organs bombarded with serial deletions of PeMADS3 promoter shown in the order of pBI-Pe3p-1007 (B–E) and pBI-Pe3p-407 (F–I). Constructs were bombarded into four independent floral buds, and results are representative of three independent bombardment experiments. Scale bar = 0.5 mm.
(TIF)
Functional analysis of serial deletions of PeMADS5 promoter. (A) Serial deletion constructs of PeMADS5 promoter. (B–Q) Histochemical assay of flower organs bombarded with serial deletions of PeMADS5 promoter shown in the order of pBI-Pe5p-1507 (B-E), pBI-Pe5p-1053 (F–I), pBI-Pe5p-441 (J–M), and pBI-Pe5p-122 (N–Q). Constructs were bombarded into four independent floral buds, and results are representative of three independent bombardment experiments. Scale bar = 0.5 mm.
(TIF)
Alignment of the promoter sequences of PeMADS6 and OncPI .
(TIF)
Primers used in this study.
(DOC)
Cis -acting regulatory elements on the upstream region of PeMADS4 .
(DOC)
Acknowledgments
We thank Dr. Chih-Hsiung Fu (Department of Engineering Science, National Cheng Kung University) for the design of a homemade software for analysis of CArG box.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Science Council, Taiwan (Grant no.: NSC 102-2811-B-006-022). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, et al. (2000) A short history of MADS-box genes in plants. Plant Mol Biol 42:115–149. [PubMed] [Google Scholar]
- 2. Dolan JW, Fields S (1991) Cell-type-specific transcription in yeast. Biochim Biophys Acta 1088:155–169. [DOI] [PubMed] [Google Scholar]
- 3. Treisman R (1992) The serum response element. Trends Biochem Sci 17:423–426. [DOI] [PubMed] [Google Scholar]
- 4. Kramer EM, Dorit RL, Irish VF (1998) Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149:765–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Whipple CJ, Ciceri P, Padilla CM, Ambrose BA, Bandong SL, et al. (2004) Conservation of B-class floral homeotic gene function between maize and Arabidopsis . Development 131:6083–6091. [DOI] [PubMed] [Google Scholar]
- 6. Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, et al. (2006) Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa . Plant Cell 18:15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Irish VF, Litt A (2005) Flower development and evolution: gene duplication, diversification and redeployment. Curr Opin Genet Dev 15:454–460. [DOI] [PubMed] [Google Scholar]
- 8. Gramzow L, Theissen G (2010) A hitchhiker's guide to the MADS world of plants. Genome Biol 11:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hill TA, Day CD, Zondlo SC, Thackeray AG, Irish VF (1998) Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3 . Development 125:1711–1721. [DOI] [PubMed] [Google Scholar]
- 10. Tilly JJ, Allen DW, Jack T (1998) The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125:1647–1657. [DOI] [PubMed] [Google Scholar]
- 11. Honma T, Goto K (2000) The Arabidopsis floral homeotic gene PISTILLATA is regulated by discrete cis-elements responsive to induction and maintenance signals. Development 127:2021–2030. [DOI] [PubMed] [Google Scholar]
- 12. Sheldon CC, Conn AB, Dennis ES, Peacock WJ (2002) Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14:2527–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deyholos MK, Sieburth LE (2000) Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12:1799–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dressler RL (1993) Phylogeny and classification of the orchid family. University of Cambridge: 320 p.
- 15. Rudall PJ, Bateman RM (2002) Roles of synorganisation, zygomorphy and heterotopy in floral evolution: the gynostemium and labellum of orchids and other lilioid monocots. Biol Rev Camb Philos Soc 77:403–441. [DOI] [PubMed] [Google Scholar]
- 16. Cozzolino S, Widmer A (2005) Orchid diversity: an evolutionary consequence of deception. Trends Ecol Evol 20:487–494. [DOI] [PubMed] [Google Scholar]
- 17. Tsai WC, Kuoh CS, Chuang MH, Chen WH, Chen HH (2004) Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol 45:831–844. [DOI] [PubMed] [Google Scholar]
- 18. Tsai WC, Lee PF, Chen HI, Hsiao YY, Wei WJ, et al. (2005) PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant Cell Physiol 46:1125–1139. [DOI] [PubMed] [Google Scholar]
- 19. Su CL, Chen WC, Lee AY, Chen CY, Chang YC, et al. (2013) A modified ABCDE model of flowering in orchids based on gene expression profiling studies of the moth orchid Phalaenopsis aphrodite . PLoS One 8:e80462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pan ZJ, Cheng CC, Tsai WC, Chung MC, Chen WH, et al. (2011) The duplicated B-class MADS-box genes display dualistic characters in orchid floral organ identity and growth. Plant Cell Physiol 52:1515–1531. [DOI] [PubMed] [Google Scholar]
- 21. Mondragon-Palomino M, Hiese L, Harter A, Koch MA, Theissen G (2009) Positive selection and ancient duplications in the evolution of class B floral homeotic genes of orchids and grasses. BMC Evol Biol 9:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mondragon-Palomino M, Theissen G (2009) Why are orchid flowers so diverse? Reduction of evolutionary constraints by paralogues of class B floral homeotic genes. Ann Bot 104:583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tautz D (2000) Evolution of transcriptional regulation. Curr Opin Genet Dev 10:575–579. [DOI] [PubMed] [Google Scholar]
- 24. Kellogg EA (2004) Evolution of developmental traits. Curr Opin Plant Biol 7:92–98. [DOI] [PubMed] [Google Scholar]
- 25. Nam J, dePamphilis CW, Ma H, Nei M (2003) Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol Biol Evol 20:1435–1447. [DOI] [PubMed] [Google Scholar]
- 26. Mondragon-Palomino M, Theissen G (2008) MADS about the evolution of orchid flowers. Trends Plant Sci 13:51–59. [DOI] [PubMed] [Google Scholar]
- 27. Mondragon-Palomino M, Theissen G (2011) Conserved differential expression of paralogous DEFICIENS- and GLOBOSA-like MADS-box genes in the flowers of Orchidaceae: refining the ‘orchid code'. Plant J 66:1008–1019. [DOI] [PubMed] [Google Scholar]
- 28. Carlson JE, Tulsieram LK, Glaubitz JC, Luk VW, Kauffeldt C, et al. (1991) Segregation of random amplified DNA markers in F1 progeny of conifers. Theor Appl Genet 83:194–200. [DOI] [PubMed] [Google Scholar]
- 29.Hsu CC, Chung YL, Chen TC, Lee YL, Kuo YT, et al. (2011) An overview of the Phalaenopsis orchid genome through BAC end sequence analysis. BMC Plant Biol 11. [DOI] [PMC free article] [PubMed]
- 30.Sambrook J, Fritsch EF, Maniatis T (2001) Molecular cloning: a laboratory manual, 3rd ed.Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31. Tang W, Perry SE (2003) Binding site selection for the plant MADS domain protein AGL15: an in vitro and in vivo study. J Biol Chem 278:28154–28159. [DOI] [PubMed] [Google Scholar]
- 32. Blanchette M, Tompa M (2003) FootPrinter: a program designed for phylogenetic footprinting. Nucleic Acids Res 31:3840–3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sanford JC, Smith FD, Russell JA (1993) Optimizing the biolistic process for different biological applications. Methods Enzymol 217:483–509. [DOI] [PubMed] [Google Scholar]
- 34. Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 58:387–405. [Google Scholar]
- 35. Gendrel AV, Lippman Z, Martienssen R, Colot V (2005) Profiling histone modification patterns in plants using genomic tiling microarrays. Nat Methods 2:213–218. [DOI] [PubMed] [Google Scholar]
- 36. Luo M, Wang YY, Liu X, Yang S, Lu Q, et al. (2012) HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis . J Exp Bot 63:3297–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wasserman WW, Palumbo M, Thompson W, Fickett JW, Lawrence CE (2000) Human-mouse genome comparisons to locate regulatory sites. Nat Genet 26:225–228. [DOI] [PubMed] [Google Scholar]
- 38. Bulyk ML (2003) Computational prediction of transcription-factor binding site locations. Genome Biol 5:201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weitzman JB (2003) Tracking evolution's footprints in the genome. J Biol 2:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. De Bodt S, Theissen G, Van de Peer Y (2006) Promoter analysis of MADS-box genes in eudicots through phylogenetic footprinting. Mol Biol Evol 23:1293–1303. [DOI] [PubMed] [Google Scholar]
- 41. Chuzhanova NA, Krawczak M, Nemytikova LA, Gusev VD, Cooper DN (2000) Promoter shuffling has occurred during the evolution of the vertebrate growth hormone gene. Gene 254:9–18. [DOI] [PubMed] [Google Scholar]
- 42. Aceto S, Cantone C, Chiaiese P, Ruotolo G, Sica M, et al. (2010) Isolation and phylogenetic footprinting analysis of the 5'-regulatory region of the floral homeotic gene OrcPI from Orchis italica (Orchidaceae). J Hered 101:124–131. [DOI] [PubMed] [Google Scholar]
- 43. Xu F, Park MR, Kitazumi A, Herath V, Mohanty B, et al. (2012) Cis-regulatory signatures of orthologous stress-associated bZIP transcription factors from rice, sorghum and Arabidopsis based on phylogenetic footprints. BMC Genomics 13:497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kanno A, Saeki H, Kameya T, Saedler H, Theissen G (2003) Heterotopic expression of class B floral homeotic genes supports a modified ABC model for tulip (Tulipa gesneriana). Plant Mol Biol 52:831–841. [DOI] [PubMed] [Google Scholar]
- 45. Kramer EM, Jaramillo MA (2005) Genetic basis for innovations in floral organ identity. J Exp Zool B Mol Dev Evol 304:526–535. [DOI] [PubMed] [Google Scholar]
- 46. Chang YY, Kao NH, Li JY, Hsu WH, Liang YL, et al. (2010) Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol 152:837–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ueki S, Lacroix B, Krichevsky A, Lazarowitz SG, Citovsky V (2009) Functional transient genetic transformation of Arabidopsis leaves by biolistic bombardment. Nat Protoc 4:71–77. [DOI] [PubMed] [Google Scholar]
- 48. Zhang G, Lu S, Chen TA, Funk CR, Meyer WA (2003) Transformation of triploid bermudagrass (Cynodon dactylon x C. transvaalensis cv. TifEagle) by means of biolistic bombardment. Plant Cell Rep 21:860–864. [DOI] [PubMed] [Google Scholar]
- 49. Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, et al. (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7:417–427. [DOI] [PubMed] [Google Scholar]
- 50. Abel S, Theologis A (1994) Transient transformation of Arabidopsis leaf protoplasts: a versatile experimental system to study gene expression. Plant J 5:421–427. [DOI] [PubMed] [Google Scholar]
- 51. Blachutzik JO, Demir F, Kreuzer I, Hedrich R, Harms GS (2012) Methods of staining and visualization of sphingolipid enriched and non-enriched plasma membrane regions of Arabidopsis thaliana with fluorescent dyes and lipid analogues. Plant Methods 8:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang Y, Su J, Duan S, Ao Y, Dai J, et al. (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods 7:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hamilton DA, Roy M, Rueda J, Sindhu RK, Sanford J, et al. (1992) Dissection of a pollen-specific promoter from maize by transient transformation assays. Plant Mol Biol 18:211–218. [DOI] [PubMed] [Google Scholar]
- 54. Sidorov VA, Kasten D, Pang SZ, Hajdukiewicz PT, Staub JM, et al. (1999) Technical Advance: Stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J 19:209–216. [DOI] [PubMed] [Google Scholar]
- 55. Wang YC, Klein TM, Fromm M, Cao J, Sanford JC, et al. (1988) Transient expression of foreign genes in rice, wheat and soybean cells following particle bombardment. Plant Mol Biol 11:433–439. [DOI] [PubMed] [Google Scholar]
- 56. Baum K, Groning B, Meier I (1997) Improved ballistic transient transformation conditions for tomato fruit allow identification of organ-specific contributions of I-box and G-box to the RBCS2 promoter activity. Plant J 12:463–469. [DOI] [PubMed] [Google Scholar]
- 57. Li YH, Tremblay FM, Seguin A (1994) Transient transformation of pollen and embryogenic tissues of white spruce (Picea glauca (Moench.) Voss) resulting from microprojectile bombardment. Plant Cell Rep 13:661–665. [DOI] [PubMed] [Google Scholar]
- 58. Sheen J (2001) Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol 127:1466–1475. [PMC free article] [PubMed] [Google Scholar]
- 59. Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, et al. (2005) Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods 2:615–619. [DOI] [PubMed] [Google Scholar]
- 60. Denecke J, Aniento F, Frigerio L, Hawes C, Hwang I, et al. (2012) Secretory pathway research: the more experimental systems the better. Plant Cell 24:1316–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hsu CT, Liao DC, Wu FH, Liu NT, Shen SC, et al. (2011) Integration of molecular biology tools for identifying promoters and genes abundantly expressed in flowers of Oncidium Gower Ramsey. BMC Plant Biol 11:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang SH, Yu H, Goh CJ (2002) Isolation and characterization of the orchid cytokinin oxidase DSCKX1 promoter. J Exp Bot 53:1899–1907. [DOI] [PubMed] [Google Scholar]
- 63. Hsiao YY, Huang TH, Fu CH, Huang SC, Chen YJ, et al. (2013) Transcriptomic analysis of floral organs from Phalaenopsis orchid by using oligonucleotide microarray. Gene 518:91–100. [DOI] [PubMed] [Google Scholar]
- 64. Roudier F, Teixeira FK, Colot V (2009) Chromatin indexing in Arabidopsis: an epigenomic tale of tails and more. Trends Genet 25:511–517. [DOI] [PubMed] [Google Scholar]
- 65. Bernatavichute YV, Zhang X, Cokus S, Pellegrini M, Jacobsen SE (2008) Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana . PLoS One 3:e3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Charron JB, He H, Elling AA, Deng XW (2009) Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis . Plant Cell 21:3732–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dennis ES, Peacock WJ (2007) Epigenetic regulation of flowering. Curr Opin Plant Biol 10:520–527. [DOI] [PubMed] [Google Scholar]
- 68. Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, et al. (2003) ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol 13:627–637. [DOI] [PubMed] [Google Scholar]
- 69. Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, et al. (2006) Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25:4638–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hennig L, Taranto P, Walser M, Schonrock N, Gruissem W (2003) Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130:2555–2565. [DOI] [PubMed] [Google Scholar]
- 71. Moon YH, Chen L, Pan RL, Chang HS, Zhu T, et al. (2003) EMF genes maintain vegetative development by repressing the flower program in Arabidopsis. Plant Cell 15:681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lafos M, Kroll P, Hohenstatt ML, Thorpe FL, Clarenz O, et al. (2011) Dynamic regulation of H3K27 trimethylation during Arabidopsis differentiation. PLoS Genet 7:e1002040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Burgos-Rivera B, Dawe RK (2012) An Arabidopsis tissue-specific RNAi method for studying genes essential to mitosis. PLoS One 7:e51388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Functional analysis of serial deletions of PeMADS2 promoter. (A) Serial deletion constructs of PeMADS2 promoter. (B–Q) Histochemical assay of flower organs bombarded with serial deletions of PeMADS2 promoter shown in the order of pBI-Pe2p-2224 (B-E), pBI-Pe2p-1823 (F-I), pBI-Pe2p-1312 (J-M), pBI-Pe2p-750 (N–Q), and pBI-Pe2p-291 (R–U). Constructs were bombarded into four independent floral buds, and results are representative of three independent bombardment experiments. Scale bar = 0.5 mm.
(TIF)
Functional analysis of serial deletions of PeMADS3 promoter. (A) Serial deletion constructs of PeMADS3 promoter. (B–Q) Histochemical assay of flower organs bombarded with serial deletions of PeMADS3 promoter shown in the order of pBI-Pe3p-1007 (B–E) and pBI-Pe3p-407 (F–I). Constructs were bombarded into four independent floral buds, and results are representative of three independent bombardment experiments. Scale bar = 0.5 mm.
(TIF)
Functional analysis of serial deletions of PeMADS5 promoter. (A) Serial deletion constructs of PeMADS5 promoter. (B–Q) Histochemical assay of flower organs bombarded with serial deletions of PeMADS5 promoter shown in the order of pBI-Pe5p-1507 (B-E), pBI-Pe5p-1053 (F–I), pBI-Pe5p-441 (J–M), and pBI-Pe5p-122 (N–Q). Constructs were bombarded into four independent floral buds, and results are representative of three independent bombardment experiments. Scale bar = 0.5 mm.
(TIF)
Alignment of the promoter sequences of PeMADS6 and OncPI .
(TIF)
Primers used in this study.
(DOC)
Cis -acting regulatory elements on the upstream region of PeMADS4 .
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.