Abstract

A group of microbial retinal proteins most closely related to the proton pump xanthorhodopsin has a novel sequence motif and a novel function. Instead of, or in addition to, proton transport, they perform light-driven sodium ion transport, as reported for one representative of this group (KR2) from Krokinobacter. In this paper, we examine a similar protein, GLR from Gillisia limnaea, expressed in Escherichia coli, which shares some properties with KR2 but transports only Na+. The absorption spectrum of GLR is insensitive to Na+ at concentrations of ≤3 M. However, very low concentrations of Na+ cause profound differences in the decay and rise time of photocycle intermediates, consistent with a switch from a “Na+-independent” to a “Na+-dependent” photocycle (or photocycle branch) at ∼60 μM Na+. The rates of photocycle steps in the latter, but not the former, are linearly dependent on Na+ concentration. This suggests that a high-affinity Na+ binding site is created transiently after photoexcitation, and entry of Na+ from the bulk to this site redirects the course of events in the remainder of the cycle. A greater concentration of Na+ is needed for switching the reaction path at lower pH. The data suggest therefore competition between H+ and Na+ to determine the two alternative pathways. The idea that a Na+ binding site can be created at the Schiff base counterion is supported by the finding that upon perturbation of this region in the D251E mutant, Na+ binds without photoexcitation. Binding of Na+ to the mutant shifts the chromophore maximum to the red like that of H+, which occurs in the photocycle of the wild type.
The opsin-based light-driven ion pumps, like their prototype bacteriorhodopsin,1 are small (∼25 kDa) proteins2,3 constructed of seven transmembrane helices that surround a retinal chromophore at the center of the hydrophobic core.4 The retinal is attached through a protonated Schiff base (C=NH+) to a lysine residue. In the initial state, the retinal is typically in the all-trans configuration, but upon light absorption, it undergoes photoisomerization to the 13-cis,15-anti form.5,6 The isomerization initiates intraprotein transfer of an ion, H+ in bacteriorhodopsin7 and related proton pumps8,9 and Cl– in halorhodopsin.10 This initial internal transfer is followed by further ion transfer steps, which ultimately result in the translocation of the transported ion across the width of the membrane.11 The relative simplicity of such ion pumps, and the benefits that they provide for cell survival as light energy transducers, explains the abundance of these proteins in many bacteria and fungi.12−14
Recently, a new group of microbial retinal proteins capable of light-driven Na+ transport was identified.15−17 The genome sequences of Dokdonia donghaensis,16Krokinobacter sp.,18Gillisia limnaea,19 and other organisms16,20 revealed the presence of unusual retinal proteins in which the Schiff base counterion and proton acceptor, homologous to Asp85 in bacteriorhodopsin, is substituted with Asn, the residue homologous to Thr89 is an Asp, and the residue at the location of the proton donor, Asp96 in bacteriorhodopsin, is a Gln (Figure S1 of the Supporting Information). This characteristic “NDQ” motif is unlike the motifs of proton pumps, which are DTD in bacteriorhodopsin and DTE in the eubacterial rhodopsins.21 In D. donghaensis, the expression of the protein with the NDQ motif was reported to be highly sensitive to Na+ concentration.16 The protein (labeled DDR2) in the presence of Na+ produced light-induced pH changes that were not abolished but rather enhanced by CCCP, an ionophore that increases membrane permeability for protons. This indicated that proton uptake is a secondary event caused by the primary Na+ transport.15 As one might expect, the photocycle reactions of DDR2 were sensitive to Na+ concentration.22 A functional study of a similar protein KR2 from Krokinobacter eikastus(17) uncovered several similar and additional features. The most important are as follows. (i) Ilumination of Krokinobacter cells or Escherichia coli cells with KR2 protein expressed caused alkalinization of the medium in the presence of NaCl, which was eliminated by TPP, a membrane-penetrating cation, indicating that Na+ transport is electrogenic. (ii) In the absence of NaCl (which was replaced by KCl), light-induced acidification was observed, which was eliminated in turn by CCCP, indicating that in the absence of Na+ the protein can function as a proton pump.17 (iii) The absorption maximum was not affected by Na+ (as compared with K+), but the rate of photocycle turnover was greater in the presence of this cation. Evidence of the interaction of Na+ with the protein in the initial state was obtained from FTIR bands that appeared in the initial state upon addition of NaCl, with a dissociation constant Kd of 11.4 mM.17 (iv) Mutations of the carboxyl residues near the Schiff base (Asp116 and Asp251), as well as mutation of Arg109, eliminated both Na+ and H+ transport, suggesting that they share common elements of the transport mechanism. On the other hand, some mutations that eliminated the FTIR bands that arose upon Na+ binding did not eliminate Na+ transport.
In this work, we report on a similar retinal protein from G. limnaea, abbreviated GLR, which transports Na+ only. We present evidence of a high-affinity Na+ binding site that is formed transiently during the photocycle. We show that proton and Na+ transfer reactions are in competition: Na+ inhibits H+ uptake and release from the bulk, whereas lowering the pH, inhibits the sodium ion dependence of reactions of the photocycle. Evidence of the involvement of the Schiff base counterion region in Na+ binding was obtained: we found that unlike in the wild-type protein, in the D251E mutant (Asp251 being the homologue of Asp212 in bacteriorhodopsin) Na+ binds in the initial state. The binding of Na+ must be near the counterion to the Schiff base, as inferred from the comparable effects of Na+ and H+ on the chromophore spectrum. It should be pointed out that the mechanism of Na+ translocation and the relationship of the binding of Na+ and H+ have been widely discussed with respect to numerous Na+ pumps that have been investigated, including Na+/K+ ATP-ases,23−28 and constitute a fundamental problem not restricted to the retinal-based Na+ pumps like GLR. The advantage of retinal proteins as a model system is that the ion transfer reactions can be initiated with a pulse of light, and the entire reaction sequence can be followed spectroscopically, in real time. Further, there is a great deal of relevant mechanistic background in other versions of such proteins that function as pumps for protons6,20,29−34 and for chloride ions.35,36
Materials and Methods
The gene of GLR encoded Gilli_234019 was expressed in outer membrane protease-deficient E. coli strain UT5600. Overnight cultures of E. coli bearing the GLR gene were grown in LB (Luria-Bertani) medium (Fisher Scientific Inc.) in the presence of 100 μg/mL ampicillin at 30 °C. Protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) (Zymo Research) and 10 μM all-trans-retinal (Sigma). Cells were harvested 3 h after induction. Cell membranes were prepared by sonication as described previously37 and solubilized with 1% DDM. The six-His-tagged protein was purified on a Ni-NTA agarose column (Qiagen). GLR was eluted with 0.02% DDM and washed from imidazole and buffers by overnight dialysis in Spectra/Por moleculaporous membrane tubing (Spectrum Laboratories, Inc.) at 4 °C while being gently stirred in a solution containing 10 mM KCl and 1 mM buffer (pH 8). For further washing from salts, 0.5 mL Amicon Ultra centrifugal filter units (Ultracel 30K, Merck Millipore Ltd.) were used.
Light-induced pH changes in the suspension of E. coli cells were measured as described previously.9,38 For pH measurements, 30 mL of a cell suspension was washed four times by sedimenting at 3000g for 10 min and resuspending in an unbuffered salt solution (100 mM NaCl, 10 mM MgCl2, and 2.5 mM KCl), with intervals of several hours between washes to allow exchange of ions inside the cell with the bulk. In a similar way, replacement of 100 mM NaCl with 100 mM KCl was done by washing cells in 100 mM KCl and 10 mM MgCl2.
All mutants were generated from a plasmid containing the wild-type GLR gene using the QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies). Absorption spectra and their pH dependence were measured as previously described.38 The samples typically contained 0.02–0.1% DDM and one or several buffers (citric acid, MES, MOPS, BICINE, HEPES, CHES, and CAPS). Fitting of titration curves in which the absorption maximum is plotted versus the ion concentration assumes the existence of two species with different maxima, with the intermediate maxima being proportional to the extent of interconversion between the species. The difference spectra from the same data, a more rigorous parameter, yielded very similar titration curves and fits. Transient absorption changes during the photocycle were examined on a single-beam laser photometer as described previously.9,38 Kinetics of light-induced proton release and uptake was assayed with pH sensitive dye pyranine, as described previously.34 Excitation was produced with 532 nm, 7 ns laser flashes, with an appropriate duration between flashes (2–12 s). The kinetic traces were fit with FitExp.39
Samples for FTIR were prepared by drying the DDM-solubilized protein40 on a CaF2 window. Drying was estimated to increase the concentration by a factor of ∼30. Thus, the final concentration of DDM was ∼3%. For measurement at low Na+ concentrations, the samples were dried with a solution containing ∼30 nM NaCl (pH 8.6) (estimate from the number of repetitive washes). For high-Na+ conditions, the liquid sample was in 5 mM NaCl and reached 150 mM in the dried film. After being dried and rehumidified to approximately 80% relative humidity, the samples were sealed with a second CaF2 window. The FTIR spectra were measured for the phototrapped state formed at the end of the photocycle, using either continuous illumination by a lamp/filter/waveguide combination at 500 ± 20 nm41 or 10 Hz flashes from a 532 nm laser (MiniLite-2 by Continuum). Both illumination regimes allowed trapping of transients decaying slower than ∼1 s and gave essentially the same result. The spectra were recorded on a Bruker IFS 66/S FTIR spectrometer at 2 cm–1 resolution for a 20 min on/off illumination, averaging four to eight cycles. Correspondence with measurements in solubilized samples was confirmed for each sample by measuring the kinetics of the photocycle of the films in the visible range.
Results
Na+-Dependent Light-Induced pH Changes Produced by GLR in a Cell Suspension: Evidence of Electrogenic Na+ Transport
Figure 1 shows light-induced pH changes produced by GLR expressed in E. coli. Illumination of the cells in the absence of Na+ (in 100 mM KCl) does not produce significant pH changes, which suggests that unlike KR2,17 GLR does not transport protons, or if there is such transport it is extremely weak. In 100 mM NaCl, the light-induced increase in pH is observed, and it is not abolished by CCCP as in the H+ pumps, BR,42 XR,9 or ESR,38 but is enhanced by this proton conductor. This indicates that the light-induced alkalinization is from passive H+ uptake in response to the active electrogenic transport of Na+, similar to what was observed for KR2 from Krokinobacter,17 and the main difference from KR2 is that the latter transports protons in the absence of Na+ whereas in GLR no such activity is seen. The light-induced pH changes were suppressed by 10 mM TPP (not shown), a membrane penetrant cation, confirming the electrogenic nature of the Na+ transport, as in KR2.17 These are the same kind of observations that had identified halorhodopsin as a light-driven pump not for protons but for chloride ions.10
Figure 1.
Light-induced pH changes in a suspension of E. coli cells with G. limnaea rhodopsin expressed and reconstituted with all-trans-retinal: trace 1, absence of pH change in sodium free medium [in 100 mM KCl (pH 7.5)]; trace 2, proton uptake (alkalinization) in 100 mM NaCl; trace 3, same as trace 2 but after addition of a protonophore (50 μM CCCP), which increases the rate and extent of passive proton influx.
Photocycle of GLR in the Absence of Na+
At low concentrations of Na+ (≤1 μM), the turnover of the GLR photocycle is very slow (Figure 2): the absorption changes produced by flash illumination relax fully after ∼8 s. The first detected intermediate is a red-shifted photoproduct, which in analogy with BR and other retinal proteins can be called K. Its relaxation back to the initial state involves at least six or seven distinctive transitions. In <6 μs, a fraction of K is replaced by a blue-shifted, M-like intermediate (see the increase in absorbance at 410 nm and the simultaneous decrease in absorbance at 550 and 590 nm). The absorption maximum of this state is at or below 400 nm, which suggests that the retinal Schiff base is deprotonated. It accumulates in only small amounts, as in other eubacterial rhodopsins.31,38,43 In the subsequent transition, with a time constant of ∼60 μs, an intermediate absorbing at ∼470 nm is formed (X470). This state and M may be in equilibrium. In KR2, an equilibrium between L and M had been suggested,17 but we find that the intermediate in question arises after M, rather than before as L would. In the following steps, these two blue-shifted intermediates decay to a red-shifted species that we tentatively identify as an N-like state [see the increase in absorbance at 550 and 590 nm with time constants at 2.7 and 274 ms, the latter coincident with the major component in M decay (see the 410 nm trace)]. The last two transitions involve a strongly red-shifted intermediate (O-like) that arises in 1 s and decays with a time constant of 2.6 s (Figure 2, 590 nm trace), which determines the rate of recovery of the initial state (Figure 2, 510 nm trace). As shown below, this intermediate contains a reisomerized but distorted all-trans-retinal, as O of BR. The formation of the M-like intermediate and its decay to the N intermediate slow in D2O by ∼2.5- and ∼8-fold, respectively, consistent with deprotonation and reprotonation of the Schiff base during these steps, and the movement of protons limiting the rates.
Figure 2.
Flash-induced absorption changes of GLR at pH 8.5 in 10 mM KCl at several characteristic wavelengths, 410, 510, 550, and 590 nm. The concentration of NaCl is <1 μM. The traces were globally fit with six kinetic components with time constants shown at the top.
Na+ Dependence of the GLR Photocycle
The rise or decay of the photocycle intermediates, if they are dependent on the bulk Na+ concentration, can serve as intrinsic reporters of transient Na+ binding. We gained insight into the dynamics of sodium ion transport and the possible location of the principal binding site by exploring the Na+ concentration dependence of the photocycle transitions monitored at characteristic wavelengths (Figure 3A). It appears that addition of 0.1, 1, and 3 mM NaCl progressively replaces the slowly forming and slowly decaying red-shifted (O-like) intermediate with a rapidly rising and decaying O-like intermediate (Figure 3A, traces 2–4, respectively) and shortens the lifetime of the M state by linearly increasing the rate constant of its decay (Figure 3B). The existence of two parallel pathways is evident from traces 2 and 3 of Figure 3A, which show that the rapid and slow O intermediates coexist. We refer to the two cycles as “Na+-dependent” and “Na+-independent”, respectively. The transition from the Na+-independent photocycle to the Na+-dependent photocycle is best characterized by the pH dependence of the yield of the long-lived O-like intermediate assayed as absorption changes at 590 nm (Figure 3C). The data can be fit by a simple equation derived from a scheme for the photocycle that branches at M, where some of the molecules in the blue-shifted intermediates M and X470 undergo transitions to the red-shifted intermediates. The time constants are k0 and kb[Na+] for the Na+-independent and Na+-dependent branches, respectively, where kb is the rate constant for Na+ binding in M. This leads to the following equation: ΔA590 = ΔA0/(1 + K[Na+]), where ΔA590 is the maximal amplitude of the absorption change at 590 nm for the long-lived O intermediate, K = kb/k0, and ΔA0 is the ΔA590 in the absence of Na+. The concentration of Na+ at which the level of accumulation of the long-lived O intermediate is decreased by half is ∼60 μM (Figure 3C). We note that the value of 60 μM is not the equilibrium binding constant for Na+, but the Na+ concentration at which the photocycle branching is at its midpoint. Its relatively low value indicates a high affinity of the transiently formed binding site for sodium. Further additions of NaCl not only completely eliminate the long-lived O intermediate but also continue to accelerate the decay of the blue-shifted intermediate M and the corresponding formation of the red-shifted state(s), N/O (Figure 3A, traces 5–8). The rate of M decay is accelerated by 3 orders of magnitude in 100 mM NaCl and continues to accelerate up to 1 M Na+ (latter not shown). The linear dependence of the rate of its conversion on Na+ concentration (Figure 3B) indicates that entry of Na+ into a transiently formed binding site, which arises during the photocycle, becomes rate-limiting. The subsequent reactions of the photocycle are similarly affected (Figure 3A).
Figure 3.

Effect of sodium chloride on the kinetics of the GLR photocycle at two selected wavelengths, 590 nm, representing formation and decay of the red-shifted intermediate(s) K and O, and 410 nm, tracking the blue-shifted intermediate(s) M. (A) Traces 1–8 are the absorption changes at 0.3 μM, 100 μM, 300 μM, 1 mM, 3 mM, 10 mM, 30 mM, and 100 mM NaCl. (B) Rate constant of the decay of the blue-shifted intermediate vs NaCl concentration (in a single-component fit; k0 is reciprocal of the half-decay time at 410 nm at 0.3 μM NaCl). (C) Decrease in the amplitude of absorption changes at 590 nm from the long-lived O-like intermediate (time constant of 2.6 s) upon addition of NaCl, which indicates a switch from a “Na+-independent” to a “Na+-dependent” photocycle. The data were fit with the equation ΔA590([Na+]) = ΔA0/(1 + K[Na+]) (see the text), from which it was determined that the Na+ concentration at which 50% of the molecules proceed through the sodium ion-dependent cycle is K–1 = 60 ± 7 μM.
Transitions in the Na+-Dependent Photocycle
The recovery of the initial state in 100 mM NaCl is ∼50-fold faster than in the Na+-independent cycle (∼50 ms vs 2.6 s). To better resolve the rapid early steps involving the formation of M and X470, we used D2O instead of H2O, which slows them by a factor of ∼2.5. Subsequent stages are less affected. In 100 mM NaCl, the kinetics of M decay in D2O slows by <1.4-fold, much lower than the value of 8-fold in the absence of Na+, suggesting that binding of a sodium ion facilitates reprotonation of the Schiff base in the M to N transition and determines its rate. Figure 4A shows the kinetics at selected wavelengths. In panels B–D, the spectra of the initial (first) absorption changes produced by light (labeled Initial) and the seven subsequent kinetic steps obtained from a global fit are shown. The spectra, given here as differences between consecutive intermediates, represent conversion of each intermediate state into the next. In the first transition (τ1 = 7.7 μs), a fraction (∼30%) of K with a maximum at 550 nm is replaced by the short wavelength M-like intermediate, with a maximum at ≤400 nm. In the subsequent step (τ2 = 48 μs), most of the remaining K is replaced by X470. In the third step (τ3 = 124 μs), the M-like state and the rest of K disappear and a red-shifted species (maximum in the difference spectrum is at 550 nm) is produced. The fourth and fifth steps involve transitions of the X470 intermediate with τ4 = 0.9 ms and τ5 = 4.3 ms into species designated as N and O, with maxima in the difference absorption spectra at 570 and 600 nm, respectively. The initial state is recovered in the sixth and seventh kinetic steps with τ6 = 12 ms and τ7 = 54 ms through decay of O. The Na+-dependent and Na+-independent cycles differ therefore mainly in the time constants of the interconversions. The mixtures of species associated with single kinetic steps arise most likely from back reactions that produce transient equilibria of several states, as in other retinal proteins.44−48
Figure 4.
Kinetics of the GLR photocycle in 100 mM NaCl in D2O (pD 7.6). (A) Absorption changes at four selected wavelengths. (B) Initial, difference spectrum “K minus initial GLR” (1 μs after the flash). The numerals 1 and 2 denote the first two components of the decay of K to an M-like and X470 intermediates with τ1 = 7.7 μs and τ2 = 48 μs. (C and D) Difference spectra of the subsequent third, fourth, fifth, sixth, and seventh transitions that occur with times of 124 μs, 0.9 ms, 4.3 ms, 12 ms, and 54 ms, respectively.
Light-Induced pH Changes in the Suspension of GLR
We measured the kinetics of transient H+ concentration changes in the bulk after flash photoexcitation, using pyranine as a pH sensitive probe.34,49 The increase in absorbance at 455 nm from the dye (after subtraction of the trace from a sample without the dye) corresponds to proton uptake, whereas a decrease indicates proton release. As shown in Figure 5A, in the absence of Na+, formation of the intermediate M, with two time constants of 35 μs (60%) and 150 μs (40%), is followed by proton release with a time constant of ∼2 ms. The decay of M and the formation of N/O-like states, with τ ≈ 400 ms, virtually coincide with the uptake of protons (that reverses the earlier release and causes net alkalinization of the bulk). At the end of the cycle, the decay of the long-lived O state with τ = 2.7 s is accompanied by a coincident release (with τ = 2.6 ± 0.1 s) of the net proton gained earlier during the decay of M and formation of N/O.
Figure 5.
Light-induced pH changes produced by GLR assayed with pyranine (pH 7.2–7.4). (A) In 100 mM KCl: 1, pyranine response; 2, ΔA at 410 nm; 3, ΔA at 590 nm. Proton release occurs with two time constants, 0.7 and 9.9 ms; the subsequent proton uptake with one (430 ms) and slow release one (2.6 ± 0.1 s). The decay of M (ΔA at 410 nm) and the rise of the red-shifted intermediate (ΔA at 590 nm) occurred with a time constant of 400 ms, similar to that of H+ uptake. The decay of ΔA at 590 nm occurs with a time constant of 2.7 s, similar to that of slow proton release. (B) In 100 mM NaCl: 1, pyranine response; 2, ΔA at 410 nm; 3, ΔA at 450 nm; 4, ΔA at 590 nm. Proton release occurs with a time constant of ∼1 ms and uptake with a time constant of 50 ms. (C) Comparison of the pyranine response in 100 mM KCl and 100 mM NaCl. A decrease in pyranine absorbance corresponds to proton release.
In the presence of 100 mM NaCl, the movement of protons into and out of the protein, and especially its relationship to the decay of M and the formation of the red-shifted N/O intermediate(s), is quite different (Figure 5B). The proton uptake that correlates with formation of the red-shifted species in the absence of NaCl is missing in 100 mM NaCl (Figure 5C). Instead, proton release and uptake occur with kinetics similar to those of the rise and decay of the O-like intermediate, respectively (Figure 5B). Together with the Na+ dependence of the rate of N/O formation (Figure 3A), this suggests that in the Na+-dependent cycle Na+ eliminates the need for H+ uptake in the formation of this species, probably by substitution of H+ at the counterion. However, decay of M must include reprotonation of the Schiff base, in spite of a proton being released to the bulk at the same time. Thus, the H+ for the reprotonation is likely to originate from inside the protein, and in that case, Na+ binding is associated with acceleration of this internal proton transfer.
FTIR Evidence of Protonation of a Carboxylate Residue in the Absence of Na+ but Not in Its Presence
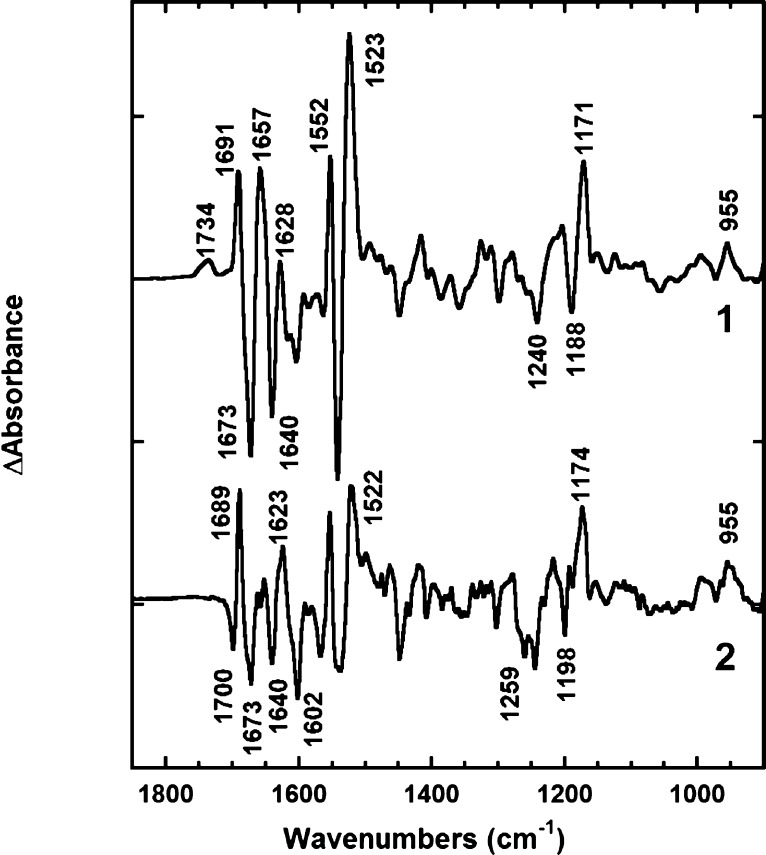
FTIR spectra measured under photostationary conditions at 270 K (Figures 6) revealed that the phototrapped state(s) are O-like. The ethylenic stretch frequency at 1523 cm–1 indicates that this state is red-shifted by ∼76 nm (from the ethylenic to λmax correlation), consistent with the strongly red-shifted O state observed in the visible range. The HOOP band at 955 cm–1 and the pattern in the fingerprint region (1150–1270 cm–1) indicate that the chromophore is in a distorted all-trans conformation like the O state of bacteriorhodopsin. Shifts in the amide I (1630–1680 cm–1) and amide II (at 1552 cm–1) bands, comparable to those in the N state of BR,50,51 indicate strong conformational perturbation of the protein.
Figure 6.
Light minus dark FTIR spectra of photostationary states at 270 K: spectrum 1, in the absence of NaCl (<1 μM); spectrum 2, in 150 mM NaCl (pH8.6). The ethylenic stretch, the fingerprint, and the HOOP regions of spectra in the infrared and the corresponding spectra in the visible range of the same samples (not shown) indicate that the intermediate trapped is like the O state of bacteriorhodopsin. The Na+ dependence of the C=O stretch region is discussed in the text.
The spectra for samples with and without NaCl are nearly the same between 900 and 1665 cm–1. The main difference is in the region of 1665–1750 cm–1, where the C=O stretch bands of protonated carboxyls of Asp and Glu and the side chain C=O stretch bands of Asn and Gln are found.52 The frequency below 1700 cm–1 is usually considered to be too low for the carbonyl vibrations53,54 of Asp or Glu. The spectrum with NaCl, but not the spectrum without NaCl, contains a triple band at 1700/1689/1673 cm–1 indicating, most probably, a perturbation of the side chain C=O stretch of Asn or Gln. For example, a shift from 1700 to 1689 cm–1 would be consistent with a Na+ coordinated by a side chain C=O group, e.g., from Asn112 near the Schiff base. In the absence of NaCl, the negative band at 1700 cm–1 disappears and the magnitude of the positive band at 1689 cm–1 is decreased, but a positive carboxylic C=O stretch band of Asp/Glu at 1734 cm–1 appears instead. The appearance of a new band in this region indicates that in the absence of Na+ a carboxylate becomes protonated.53,55 This band, however, seems to be too wide to originate from a single protonated COOH stretching vibration, which on average has a line width of 10–15 cm–1.55 The complex band might be due either to protonation of two (or more) carboxylates or, more probably, to the protonated C=O stretch of a single carboxyl that is present in two or more red-shifted (sub)states at the end of the photocycle. The latter is in accord with the visible kinetics and time-resolved IR measurements of the films: both indicate the presence of more than one red-shifted transient state at the end of the photocycle (not shown). The position of the band at 1734 cm–1 is similar to that of the protonated Asp115 band in BR,56 which is hydrogen-bonded to a threonine.57,58 Assignment of the C=O stretch band(s), using the D116E and D251E mutants, is underway, but their photocycles are greatly changed. Importantly, the protonation of an anionic carboxylate group in the O-like state in the absence of Na+, but not in its presence, correlates to the observation of net proton uptake during the formation of this intermediate under the same conditions (Figure 5) and may account for one of the protons taken up.
pH Dependence of the GLR Photocycle in NaCl: Correlation with the Protonation State of the Counterion and Its Competition with the Na+-Independent Pathway
Between pH 7.0 and 8.5, no significant pH dependence in the photocycle reactions is seen in the presence of 100 mM NaCl. However, at lower pH, the amplitudes of transient changes at all three characteristic wavelengths that reflect M rise and decay (410 nm), N/O rise and decay (590 nm), and depletion of the initial state (510 nm) decrease (Figure 7). This decrease correlates with a decrease in the fraction of initial GLR with a deprotonated counterion, as follows from the titration curve in Figure 8B. At pH 6.1 and especially pH 5.1, M decay is slowed in the same way as when the concentration of Na+ is lowered and the long-lived O intermediate from the “Na+-independent” cycle appears, providing evidence of the competition of H+ and Na+ in diverting the photocycle into the “Na+-independent” and “Na+-dependent” pathways, respectively. At pH 3.5, where the counterion is expected to be nearly fully protonated (see below), both M and O are absent, as in bacteriorhodopsin at acidic pH.
Figure 7.
pH dependence of transient absorption changes of GLR in the presence of 100 mM NaCl at selected wavelengths: (A) 410 nm and (B) 590 and 510 nm. Numerals 1–4 correspond to pH 8.3, 6.1, 5.1, and 3.5, respectively.
Figure 8.
pH dependence of the absorption spectra of GLR and its D116N and D251N mutants. (A) Chromophore absorption bands of (1) the wild type (WT) at pH 8.0, (2) WT at pH 3.3, (3) D116N at pH 7.l, and (4) D251N at pH 8.0. (B) Titration of WT from pH 9.0 to 3.0 in 100 mM KCl (⊞) and 100 mM NaCl (●). (C) Difference spectrum from a decrease in pH from 7.0 to 3.5 in (1) WT, (2) D251N, and (3) D116N and (4 and 5) absorption changes that accompany recovery of the initial state in the photocycle in the absence of Na+ (4, ●) and in the presence of 100 mM NaCl (5, ○) taken with an inverse sign. (D) pH dependence of the absorption maximum in (1) WT (fit with pKa values of 4.8 and 6.5), (2) D116N (pKa value of 4.8), and (3) D251N (pKa value of 5.1). Spectra were measured in 10 mM KCl. The pH was adjusted with HCl.
Evidence That Asp116 and Asp251 Are Components of the Counterion to the Schiff Base: pH-Dependent Transitions in Wild-Type GLR and Its D116N and D251N Mutants
The absorption band of GLR exhibits a maximum at ∼520 nm at pH 8.0 (Figure 8A, spectrum 1). The maximum shows a strong pH dependence as in many other bacterial rhodopsins. Lowering the pH to 3.0 causes a shift to ∼550 nm (Figure 8A and Figure S2 of the Supporting Information). There is very little difference between the titration curves in 100 mM KCl and 100 mM NaCl (Figure 8B), as reported for KR2.17 The absorption changes with a maximum of ∼580 nm in the difference spectrum (shown in Figure 8C) can be described with a minimum of two transitions, with apparent pKa values of 6.7 and 4.5 (Figure 8B), suggesting the presence of at least two ionizable residues that affect the chromophore absorption band. A large red shift at low pH in the microbial retinal proteins is a common indicator of the protonation of the counterion to the Schiff base. Interestingly, the absorption changes that are observed in pH titration of GLR (Figure 8C, spectrum 1) coincide with the absorption changes accompanying the decay of the red-shifted O intermediate into the initial state in the Na+-independent and Na+-dependent photocycles (Figure 8C, spectra 4 and 5, respectively). In GLR, the homologue of Asp85 in bacteriorhodopsin is an asparagine, and one would expect that this protein would exhibit a strongly red-shifted spectrum as the D85N mutant of BR59 or ESR.38 As shown in Figure 8A, this is not so. The most likely reason is that the aspartic acid residue in GLR (and KR2) located a helical turn (four residues) downstream, in place of Thr89 in BR, is apparently able to function as part of the Schiff base counterion. Indeed, substitution of this residue, Asp116, with asparagine causes a large (52 nm) red shift of the spectrum, to 572 nm, at neutral pH (Figure 8A, spectrum 3). Replacement of the second aspartate near the Schiff base, Asp251, the homologue of Asp212 in BR, conserved in all pumps, with Asn causes a smaller (14 nm) red shift to 534 nm at pH 8.0 (Figure 8A, spectrum 4). With a decrease in pH, the D251N mutant exhibits an additional 14 nm red shift (Figure 8D, curve 3), apparently from protonation of Asp116. As expected, at pH ≤4.0 the maximum of D251N matches that of the wild type (Figure 8D).
At high pH (>8.5), where the counterion is fully deprotonated, the absorption maximum of wild-type GLR is not affected by an increase in NaCl concentration from essentially zero (5 μM) to 3 M (data not shown), suggesting that GLR does not bind Na+ in the unphotolyzed state, consistent with the observation (Figure 8B) that the titration is nearly unaffected by replacing NaCl with KCl. At lower pH, however, addition of salt (NaCl or KCl) causes a blue shift in the absorption maximum, by affecting either surface pH or the pKa of protonation of the counterion (Figure S3 of the Supporting Information). A divalent cation is more efficient than monovalent cations (see a comparison of the effects of Mg2+ and NaCl at pH 6.6 in Figure S3 of the Supporting Information), suggesting that the salt effects originate from the increased level of shielding of surface charges at the higher ionic strengths. Under these conditions, the surface concentration of cations would approach that in the bulk, shifting the observed pKa toward its actual value, as in BR.60,61An alternative model would contain a nonspecific low-affinity cation binding site, probably near the protein surface, which would affect the pKa of the counterion.
The behavior of the D116N mutant is somewhat surprising. As mentioned above, in BR the absorption maximum in the mutant of the Schiff base counterion D85N62 (615 nm) is strongly red-shifted at neutral pH compared to that of the wild type (560 nm).59 If Asp116 is the main part of the counterion, it might have been expected that D116N be red-shifted also and show a smaller additional red shift as the pH is lowered, from protonation of the other part of the counterion, Asp251. The absorption maximum of D116N is indeed red-shifted relative to that of the wild type at high pH, but the red shift is much greater than what would be expected from the total red shift in the wild type when the counterion charge is eliminated at low pH. With a decrease in pH, D116N shows a 25 nm blue shift, with a pKa of ∼4.8, and at pH 4.0, its maximum roughly matches the maximum of the wild type. Three factors could contribute to this unexpected blue shift at low pH: (i) binding of chloride ions upon protonation of Asp251, as in BR at low pH63,64 and in its D85N mutant where a blue shift was also observed at pH <5,59 (ii) protonation of an unidentified acidic residue, possibly Glu160, which in GLR is near the ionone ring of the retinal, and (iii) pH-dependent thermal isomerization of the retinal in D116N that favors isomers other than the all-trans form. We examined the effect of chloride at low pH, and while the blue shift is smaller (by ∼30%) in the absence of chloride, it is not eliminated (not shown). Protonation of Glu160, a residue in GLR near the retinal ionone ring, would be expected to cause a blue shift, but the titration of the E160Q mutant did not exhibit a significant difference from that of the wild type (not shown), ruling out this possibility. The retinal isomeric composition does appear to change at low pH: the long wavelength component of its band decreases with a decrease in pH, whereas the short wavelength β-bands increase (data not shown), which is characteristic for a transition of trans-retinal to other isomers.65 Wild-type GLR is stable in DDM at pH ≥3.0; the D116N mutant exhibits signs of instability below pH 3.5, but this cannot account for the observed blue shift between pH 6.0 and 4.0, which appears to be from a combination of Cl– binding and chromophore isomerization.
As shown by the large red shifts and the blue shifts in D116N at various pH values, the titrations of the wild type and the D116N and D251N mutants do suggest that Asp116 and Asp251 are both ionized at pH 8.0 and together constitute the counterion to the Schiff base. Moreover, the titration curve can be fit assuming interaction (coupling) of the protonation states of these two residues in such a way that protonation of one decreases the pKa of the other by ∼2 pH units.
Binding of H+ and Na+ in the D251E Mutant in the Unphotolyzed State
This mutant at neutral pH exhibits a red-shifted maximum compared to that of the wild type, 562 nm (Figure 9A, spectrum 1), which implies that the counterion is protonated. Indeed, at higher pH, the absorption band undergoes a 28 nm blue shift to 534 nm (with the titration largely complete at pH 10.6), indicating deprotonation of the counterion (Figure 9A, spectrum 3). In 100 mM KCl, the pKa of this transition is 8.8 (Figure 9B, curve 1). In contrast, in 100 mM NaCl, the blue shift at high pH is much smaller, only 7 nm (Figure 9B, curve 2). Titration in 10 mM NaCl is accompanied by a larger shift (Figure 9B, curve 3), but smaller than in 100 mM KCl. In the wild type, the pH dependence was similar in KCl and NaCl except for a small (∼2 nm) blue shift in 100 mM KCl (Figures 8B and 9B). The different effects of KCl and NaCl are further illustrated by the absorption changes in panels C and D of Figure 9. Addition of 10 mM NaCl to a sample with a deprotonated counterion at pH 10.3 and a low ionic strength (3 mM KCl) causes absorption changes virtually identical to those that are caused by an increase in proton concentration (Figure 9C). Via the addition of more NaCl, the blue shift caused by the increase in pH from 7 to 10.3 can be largely reversed by Na+ as shown in Figure 9D. The apparent binding constant for Na+ at this pH is ∼2 mM. It appears that Na+ ions can substitute for protons and bind to the counterion in the initial state of the D251E mutant, causing the red shift of the absorption spectrum.
Figure 9.
Properties of the D251E mutant in the initial state. (A) Shift of the absorption spectrum from an increase in pH at low salt concentrations (1–3 mM KCl): (1) pH 5.6, (2) pH 9.0, and (3) 10.6. (B) Different pH dependence of the absorption maximum of the D251E mutant (curves 1–3) and the WT (curves 4 and 5): (1) 100 mM KCl, (2) 100 mM NaCl, (3) 10 mM NaCl, (4) 100 mM NaCl, and (5) 100 mM KCl. (C) Absorption changes produced by (1) binding of H+ with a decrease in pH from 10.4 to 9.7 in 3 mM KCl, (2) addition of 10 mM NaCl to 3 mM KCl at pH 9.7, and (3) subsequent addition of 20 mM KCl (note that the latter change is the opposite of the others). (D) Red shift of the absorption maximum of the D251E mutant produced by the addition of NaCl at pH 10.3 (in the presence of 3 mM KCl). Such a shift does not occur in the WT under the same conditions.
As the wild type, the mutant shows also nonspecific salt effects on the absorption maximum (Figure S4 of the Supporting Information) and apparent pKa. Compared to low-salt conditions (1 mM KCl and buffer only), 100 mM KCl causes a distinct shift of the pKa to a lower value (Figure S5 of the Supporting Information). As in the wild type (Figure S3 of the Supporting Information), the origin of the KCl effect must be that it screens charged groups on the surface of the protein. This effect is not specific to a particular cation, and as in the wild type, a divalent cation (Mg2+) is more efficient, as in BR. If K+ and Mg2+ act by binding to a distinct binding site at the surface or in a more buried location, this site would be additional to that which binds Na+ selectively. We interpret the specific effect of Na+ in the D251E mutant as follows. In D251E, Na+ and H+ bind competitively with one another. Na+ binds to, or near, the anionic counterion, and then its charge replaces the charge of H+: Asp-H ↔ Asp– ↔ Asp-Na. The designation “Asp” simply indicates that the binding site contains an ionizable residue, i.e., at Asp116 and/or Asp251. This model yields a simple formula for the titration of the counterion with H+ and Na+. The fraction of the ionized (unbound) counterion, Fu, depends on both H+ and Na+ concentrations:
| 1 |
where KNa is the ratio of the rate constants of Na+ dissociation and association at the binding site (KNa = kd/ka). The fraction of the red-shifted species with a neutralized counterion (the sum of protonated and sodium ion-bound species) (Fb = FbH+ + FbNa+) is equal to 1 – Fu. This formula describes the titration curves in the presence of Na+ shown in Figure 9B and the red shift upon addition of NaCl to the protein with a deprotonated counterion (Figure 9D). Fits of the data in panels B and D of Figure 9 with this model yield the pKa and KNa. Both are apparent and depend on salt concentration, changing in concert, indicating that the capability to bind H+ and Na+ is similarly affected by salt concentration from its shielding of surface charges. Thus, in 100 mM KCl, the values of pKa and KNa are 8.8 and 20 mM, changed from 9.8 and 2 mM in 3 mM KCl, respectively.
Discussion
Titration experiments (Figure 8B) indicate that the resting state of wild-type GLR does not appear to bind a cation other than H+ at the Schiff base. This observation hints at the possibility that Na+ transport is different from H+ transport in bacteriorhodopsin, where the transported ion is bound to the Schiff base before photoexcitation.
A High-Affinity Binding Site for Na+ Is Transiently Formed in the GLR Photocycle
In GLR, the transported ion, Na+, binds during the photocycle. The observed linear Na+ dependence of the rate constants of some of the photocycle transitions (Figure 3) leads to three conclusions. (i) The absorption changes identify two parallel reaction sequences that diverge after a branching point in the cycle. The switch from a slow “Na+-independent” to a rapid “Na+-dependent” photocycle occurs at a Na+ concentration of 60 μM, which demonstrates that a high-affinity Na+ binding site is formed in the M intermediate state and possibly the X470 intermediate state. The discovery of a high-affinity binding site formed transiently during the photocycle is important for the functioning of GLR as an electrogenic generator. The magnitude of the light-induced electrochemical gradient created by a pump is limited by the difference in the pK for binding of an ion during uptake and the pK for release. The higher the former and the lower the latter, the larger the difference, and the pump will function against greater gradients. The virtually complete lack of binding in the initial (and therefore the final) state of the cycle state ensures full Na+ release, and the transient binding with high affinity during the transport cycle ensures efficient operation at relatively low (submillimolar) concentrations for the light-driven sodium pumps. Further, the lack of binding in the initial state prevents the leakage of Na+ through the pump when it is not photoactivated. (ii) Na+ is captured from the bulk during the lifetime of the M to N transition, and this remains the limiting step in the photocycle at Na+ concentrations of at least 1 M. (iii) The results suggest that the likely site for Na+ binding that arises transiently in the photocycle is the Schiff base–counterion region because (a) formation of the red-shifted intermediate is expected to reflect neutralization of a negative charge in the vicinity of the protonated Schiff base either by H+ (in the “Na+-independent” cycle) or by Na+ (in the “Na+-dependent” cycle) and (b) protonation of the counterion at low pH eliminates the characteristic features of the Na+-dependent cycle. This conclusion does not necessarily exclude an additional, transient cytoplasmic binding site if it functions as a rapid donor of Na+ to the central binding site. The conclusion that the counterion is the site at which Na+ binds is supported by the properties of the D251E mutant. Perturbation of the Schiff base region in this mutant (Asp251 being the homologue of Asp212 in BR) creates a Na+ binding site in the unphotolyzed state. As shown in Figure 9, shifts of the retinal absorption maximum in titration experiments indicate that Na+ binds at the counterion when it is anionic. The location of Na+ must be near the location of where H+ binds because (a) the difference spectra for the binding of the two ions agree (Figure 9C) and thus at saturating Na+ concentrations the red shift of the maximum is as great as with protonation of the counterion and (b) Na+ binds only when the counterion is anionic.
The implication of the results with D251E is that the Schiff base region has the potential to bind Na+ in its vicinity. Perturbation of this region in the D251E mutant and from the changed shape of the photoisomerized retinal66 and/or any ensuing electrostatic changes in the wild type could be similar and create a binding site for Na+. We propose that the key factor in creating such a site during the wild-type photocycle is a large increase in the pKa of the counterion, as in BR,67,68 the consequence of retinal isomerization and cleavage of the salt bridge between the Schiff base and the counterion in M.69 The affinity for Na+ will change in parallel to the affinity for protons (as follows from titration of the D251E mutant). With the higher affinities, binding of Na+ to the counterion will occur at concentrations of tens of micromolar. Reisomerization and re-formation of the salt bridge at the end of the photocycle would restore the initial low pKa of the counterion and “push” the bound Na+ to the extracellular side, in the same way as the proton is ejected from the counterion in bacteriorhodopsin during O decay.70−73 In Na+ pumps, such a creation of the binding site must be combined with optimization of the geometry of the site for sodium ion binding, which in contrast to H+, is usually coordinated not by a single group but by five to six atoms,27,74 which can be provided by Asp116, Asp251, Asn112, and any bound water.
A Tentative Scheme of Transitions To Account for the Observed Uptake and Release of Proton and Sodium Ion in the Photocycle
Taking advantage of the considerable body of mechanistic information from bacteriorhodopsin and similar proteins, we propose a minimal scheme that is consistent with all the data (Figure 10). We need to assume three sites: the retinal Schiff base, a counterion that is also part of a binding site for H+ and Na+, and an additional site for H+ that could be analogous to the extracellular proton release site of BR.49,68,75,76 A model with fewer sites cannot be reconciled with the observations. The first steps in the photocycle do not depend on Na+. For the K state of KR2, this was explicitly shown.77 M formation is very rapid, on a time scale of a few microseconds, suggesting the existence of a ready acceptor for the Schiff base proton. Asp116 and Asp251 are the likely candidates, but a state in which this could be examined using FTIR spectroscopy could not be trapped. The initial H+ release occurs at approximately the same time in the presence and absence of Na+ (Figure 5C). It is delayed compared to M rise (Figure 5A), suggesting that the proton is not directly from the Schiff base, providing further support for the involvement of a proton acceptor. This means also that there are two M states: one before proton release and another after, as in BR.49,68,75
Figure 10.

Tentative scheme for internal ion transfer and binding that accounts for the observed uptake of H+ from the bulk and its release. The top surface of the schematic representation of the protein is the cytoplasmic side. For the sake of clarity, not all detected intermediates are shown. The top sequence after the branch is the “Na+-independent” cycle and the bottom the “Na+-dependent” cycle. SBH+ and SB refer to the protonated and unprotonated retinal Schiff base, respectively. “Asp” refers to the proton acceptor group and the ionizable part of the Na+ binding site, without commitment as to whether it is Asp116, Asp251, or both. The dashed circle is a postulated proton binding site analogous to the proton release site of bacteriorhodopsin, made necessary by the observation of proton release in the O state of the Na+-dependent cycle.
In the absence of Na+, the further reactions of the photocycle are limited by the rate of reprotonation of the Schiff base. Reprotonation of the Schiff base to produce the red-shifted intermediate would be from the counterion, reversing the first proton transfer, and the counterion itself would then be reprotonated, as shown by the FTIR spectrum in the absence of Na+ (Figure 6), apparently from the bulk. This accounts for one of the two protons taken up at this time. The second proton taken up would reprotonate the vacant release site. Deprotonation of the counterion and release of this proton at the end of the cycle complete the recovery of the initial state. Because net translocation of H+ in this system has not been detected (Figure 1), all proton release and uptake are assumed to be from the same side (extracellular side).
In the presence of Na+, reprotonation of the unprotonated Schiff base (decay of M) occurs order(s) of magnitude faster and starts before proton uptake (Figure 5B). Electrostatics dictates that uptake of Na+ and its binding at the counterion, which occurs at this time, would accelerate the transfer of the proton from the counterion to the Schiff base. In this case, the red shift of the N and O-like intermediates is from the Na+ bound at the anionic counterion. This conclusion is drawn from (a) the ability of sodium ion binding to cause a red shift, as demonstrated in the experiments with D251E, and (b) the large red shift of the absorption maximum of GLR upon titration to low pH, characteristic of a counterion protonation, and the good agreement of the difference spectrum of this red shift with the absorption change that occurs during the last step of the “Na+-dependent” photocycle (Figures 4D and 8C). In the presence of NaCl, proton release is coincident with rapid conversion of the M intermediate to the O intermediate, and an M state with a released H+ does not accumulate. The released H+ cannot originate from either the Schiff base or the counterion and must be from the site that is invoked also for the Na+-dependent cycle (Figure 10). The protonated state of the Schiff base and the neutralized counterion will increase the extent of delocalization of π-electrons in the chromophore to facilitate its reisomerization.78 Thus, uptake of the sodium ion to the binding site will correlate with both reprotonation of the Schiff base and reisomerization of the retinal to the all-trans form (FTIR spectrum in the presence of Na+ shown in Figure 6). The release of the Na+ in the last reaction of the cycle results in restoration of the salt bridge of the Schiff base with the anionic counterion. For net Na+ translocation, uptake and release have to be postulated to take place at the cytoplasmic and extracellular surfaces, respectively.
Glossary
Abbreviations
- GLR
retinal protein from G. limnaea
- BR
bacteriorhodopsin
- WT
wild type
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
- TPP
tetraphenylphosphonium chloride
- DDM
n-dodecyl β-d-maltopyranoside
- MES
2-(N-morpholino)ethanesulfonic acid
- MOPS
3-(N-morpholino)propanesulfonic acid
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- CHES
N-cyclohexyl-2-aminoethanesulfonic acid
- CAPS
N-cyclohexyl-3-aminopropanesulfonic acid
- FTIR
Fourier transform infrared.
Supporting Information Available
Alignment of amino acid sequences of three sodium pumps, GLR, HRR, and KR2, with proton pumps BR and XR (Figure S1), absorption changes observed upon titration of GLR (Figure S2), the dependence of the absorption maximum of GLR on the concentrations of NaCl and MgCl2 at pH 6.6 (Figure S3), the dependence of the absorption maximum of the D251E mutant of GLR on the concentrations of KCl and MgCl2 (Figure S4), and titration of the D251E mutant of GLR in 1 mM KCl, 100 mM KCl, and 10 mM MgCl2 (Figure S5). This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported in part by grants from the National Institutes of Health (GM29498) and the U.S. Department of Energy (DEFG03-86ER13525) to J.K.L. and S.P.B., by a Korea Research Foundation Grant (KRF2004-042-C00113) to K.-H.J., and by the 21C Frontier Microbial Genomics and Application Center Program, Ministry of Education, Science and Technology, Korea (K.H.J.).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Oesterhelt D.; Stoeckenius W. (1971) Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature 233, 149–152. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov Y. A.; Abdulaev N. G.; Feigina M. Y.; Kiselev A. V.; Lobanov N. A. (1979) The structural basis of the functioning of bacteriorhodopsin: An overview. FEBS Lett. 100, 219–224. [DOI] [PubMed] [Google Scholar]
- Khorana H. G.; Gerber G. E.; Herlihy W. C.; Gray C. P.; Anderegg R. J.; Nihei K.; Biemann K. (1979) Amino acid sequence of bacteriorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 76, 5046–5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigorieff N.; Ceska T. A.; Downing K. H.; Baldwin J. M.; Henderson R. (1996) Electron-crystallographic refinement of the structure of bacteriorhodopsin. J. Mol. Biol. 259, 393–421. [DOI] [PubMed] [Google Scholar]
- Tsuda M.; Glaccum M.; Nelson B.; Ebrey T. G. (1980) Light isomerizes the chromophore of bacteriorhodopsin. Nature 287, 351–353. [DOI] [PubMed] [Google Scholar]
- Mathies R. A.; Lin S. W.; Ames J. B.; Pollard W. T. (1991) From femtoseconds to biology: Mechanism of bacteriorhodopsin’s light-driven proton pump. Annu. Rev. Biophys. Biophys. Chem. 20, 491–518. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D.; Stoeckenius W. (1973) Functions of a new photoreceptor membrane. Proc. Natl. Acad. Sci. U.S.A. 70, 2853–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béjà O.; Aravind L.; Koonin E. V.; Suzuki M. T.; Hadd A.; Nguyen L. P.; Jovanovich S. B.; Gates C. M.; Feldman R. A.; Spudich J. L.; Spudich E. N.; DeLong E. F. (2000) Bacterial rhodopsin: Evidence for a new type of phototrophy in the sea. Science 289, 1902–1906. [DOI] [PubMed] [Google Scholar]
- Balashov S. P.; Imasheva E. S.; Boichenko V. A.; Antón J.; Wang J. M.; Lanyi J. K. (2005) Xanthorhodopsin: A proton pump with a light-harvesting carotenoid antenna. Science 309, 2061–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobert B.; Lanyi J. K. (1982) Halorhodopsin is a light-driven chloride pump. J. Biol. Chem. 257, 10306–10313. [PubMed] [Google Scholar]
- Lanyi J. K. (2006) Proton transfers in the bacteriorhodopsin photocycle. Biochim. Biophys. Acta 1757, 1012–1018. [DOI] [PubMed] [Google Scholar]
- Venter J. C.; Remington K.; Heidelberg J. F.; Halpern A. L.; Rusch D.; Eisen J. A.; Wu D.; Paulsen I.; Nelson K. E.; Nelson W.; Fouts D. E.; Levy S.; Knap A. H.; Lomas M. W.; Nealson K.; White O.; Peterson J.; Hoffman J.; Parsons R.; Baden-Tillson H.; Pfannkoch C.; Rogers Y.-H.; Smith H. O. (2004) Environmental genome shotgun sequencing of the Sargasso Sea. Science 304, 66–74. [DOI] [PubMed] [Google Scholar]
- Spudich J. L., and Jung K.-H. (2005) Microbial Rhodopsins: Phylogenetic and Functional Diversity. In Handbook of photosensory receptors (Briggs W. R., and Spudich J. L., Eds.) pp 1–23, Wiley-VCH, Darmstadt, Germany. [Google Scholar]
- Brown L. S.; Jung K.-H. (2006) Bacteriorhodopsin-like proteins of eubacteria and fungi: The extent of conservation of the haloarchaeal proton-pumping mechanism. Photochem. Photobiol. Sci. 5, 538–546. [DOI] [PubMed] [Google Scholar]
- Jung K. H. (2012) New type of cation pumping microbial rhodopsins in marine bacteria. 244th American Chemical Society National Meeting, Philadelphia, August 19–23, 2012, Abstract 271. [Google Scholar]
- Kwon S. K.; Kim B. K.; Song J. Y.; Kwak M. J.; Lee C. H.; Yoon J. H.; Oh T. K.; Kim J. F. (2013) Genomic makeup of the marine flavobacterium Nonlabens (Donghaeana) dokdonensis and identification of a novel class of rhodopsins. Genome Biol. Evol. 5, 187–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K.; Ono H.; Abe-Yoshizumi R.; Yoshizawa S.; Ito H.; Kogure K.; Kandori H. (2013) A light-driven sodium ion pump in marine bacteria. Nat. Commun. 4, 1678. [DOI] [PubMed] [Google Scholar]
- Klippel B.; Lochner A.; Bruce D. C.; Davenport K. W.; Detter C.; Goodwin L. A.; Han J.; Han S. S.; Hauser L.; Land M. L.; Nolan M.; Ovchinnikova G.; Pennacchio L.; Pitluck S.; Tapia R.; Woyke T.; Wiebusch S.; Basner A.; Abe F.; Horikoshi K.; Keller M.; Antranikian G. (2011) Complete genome sequences of Krokinobacter sp strain 4H-3-7-5 and Lacinutrix sp strain 5H-3-7-4, polysaccharide-degrading members of the family Flavobacteriaceae. J. Bacteriol. 193, 4545–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel T.; Held B.; Nolan M.; Lucas S.; Lapidus A.; Tice H.; Del Rio T. G.; Cheng J. F.; Han C.; Tapia R.; Goodwin L. A.; Pitluck S.; Liolios K.; Mavromatis K.; Pagani I.; Ivanova N.; Mikhailova N.; Pati A.; Chen A.; Palaniappan K.; Land M.; Rohde M.; Tindall B. J.; Detter J. C.; Goker M.; Bristow J.; Eisen J. A.; Markowitz V.; Hugenholtz P.; Kyrpides N. C.; Klenk H. P.; Woyke T. (2012) Genome sequence of the Antarctic rhodopsins-containing flavobacterium Gillisia limnaea type strain (R-8282(T)). Stand. Genomic Sci. 7, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L. S. (2014) Eubacterial rhodopsins: Unique photosensors and diverse ion pumps. Biochim. Biophys. Acta 1837, 553–561. [DOI] [PubMed] [Google Scholar]
- Béjà O.; Lanyi J. K. (2014) Nature’s toolkit for microbial rhodopsin ion pumps. Proc. Natl. Acad. Sci. U.S.A. 111, 6538–6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fen Y. (2011) Spectroscopic studies of novel microbial rhodopsins from fungi and bacteria. Ph.D. Thesis, pp 214, The University of Guelph, Guelph, ON. [Google Scholar]
- Page M. J.; Di Cera E. (2006) Role of Na+ and K+ in enzyme function. Physiol. Rev. 86, 1049–1092. [DOI] [PubMed] [Google Scholar]
- Nakanishi-Matsui M.; Sekiya M.; Nakamoto R. K.; Futai M. (2010) The mechanism of rotating proton pumping ATPases. Biochim. Biophys. Acta 1797, 1343–1352. [DOI] [PubMed] [Google Scholar]
- Gadsby D. C. (2009) Ion channels versus ion pumps: The principal difference, in principle. Nat. Rev. Mol. Cell Biol. 10, 344–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier T.; Polzer P.; Diederichs K.; Welte W.; Dimroth P. (2005) Structure of the rotor ring of F-type Na+-ATPase from Ilyobacter tartaricus. Science 308, 659–662. [DOI] [PubMed] [Google Scholar]
- Nyblom M.; Poulsen H.; Gourdon P.; Reinhard L.; Andersson M.; Lindahl E.; Fedosova N.; Nissen P. (2013) Crystal structure of Na+,K+-ATPase in the Na+-bound state. Science 342, 123–127. [DOI] [PubMed] [Google Scholar]
- Tsai J.-Y.; Kellosalo J.; Sun Y.-J.; Goldman A. (2014) Proton/sodium pumping pyrophosphatases: The last of the primary ion pumps. Curr. Opin. Struct. Biol. 27, 38–47. [DOI] [PubMed] [Google Scholar]
- Khorana H. G. (1993) Two light-transducing membrane proteins: Bacteriorhodopsin and the mammalian rhodopsin. Proc. Natl. Acad. Sci. U.S.A. 90, 1166–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupts U.; Tittor J.; Oesterhelt D. (1999) Closing in on bacteriorhodopsin: Progress in understanding the molecule. Annu. Rev. Biophys. Biomol. Struct. 28, 367–399. [DOI] [PubMed] [Google Scholar]
- Dioumaev A. K.; Brown L. S.; Shih J.; Spudich E. N.; Spudich J. L.; Lanyi J. K. (2002) Proton transfers in the photochemical reaction cycle of proteorhodopsin. Biochemistry 41, 5348–5358. [DOI] [PubMed] [Google Scholar]
- Lanyi J. K. (2005) A Structural View of Proton Transport in Bacteriorhodopsin. In Biophysical and Structural Aspects of Bioenergetics (Wikström M., Ed.) pp 227–248, The Royal Society of Chemistry, Cambridge, U.K. [Google Scholar]
- Freier E.; Wolf S.; Gerwert K. (2011) Proton transfer via a transient linear water-molecule chain in a membrane protein. Proc. Natl. Acad. Sci. U.S.A. 108, 11435–11439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashov S. P.; Petrovskaya L. E.; Imasheva E. S.; Lukashev E. P.; Dioumaev A. K.; Wang J. M.; Sychev S. V.; Dolgikh D. A.; Rubin A. B.; Kirpichnikov M. P.; Lanyi J. K. (2013) Breaking the carboxyl rule: Lysine-96 facilitates reprotonation of the Schiff base in the photocycle of a retinal protein from Exiguobacterium sibiricum. J. Biol. Chem. 288, 21254–21265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi J. K. (1990) Halorhodopsin: A light-driven electrogenic chloride transport system. Physiol. Rev. 70, 319–330. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D. (1995) Structure and function of halorhodopsin. Isr. J. Chem. 35, 475–494. [Google Scholar]
- Petrovskaya L. E.; Lukashev E. P.; Chupin V. V.; Sychev S. V.; Lyukmanova E. N.; Kryukova E. A.; Ziganshin R. H.; Spirina E. V.; Rivkina E. M.; Khatypov R. A.; Erokhina L. G.; Gilichinsky D. A.; Shuvalov V. A.; Kirpichnikov M. P. (2010) Predicted bacteriorhodopsin from Exiguobacterium sibiricum is a functional proton pump. FEBS Lett. 584, 4193–4196. [DOI] [PubMed] [Google Scholar]
- Balashov S. P.; Petrovskaya L. E.; Lukashev E. P.; Imasheva E. S.; Dioumaev A. K.; Wang J. M.; Sychev S. V.; Dolgikh D. A.; Rubin A. B.; Kirpichnikov M. P.; Lanyi J. K. (2012) Aspartate-histidine interaction in the retinal Schiff base counterion of the light-driven proton pump of Exiguobacterium sibiricum. Biochemistry 51, 5748–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dioumaev A. K. (1997) Evaluation of intrinsic chemical kinetics and transient product spectra from time-resolved spectroscopic data. Biophys. Chem. 67, 1–25. [DOI] [PubMed] [Google Scholar]
- Lorenz-Fonfria V. A.; Heberle J. (2014) Channelrhodopsin unchained: Structure and mechanism of a light-gated cation channel. Biochim. Biophys. Acta 1837, 626–642. [DOI] [PubMed] [Google Scholar]
- Dioumaev A. K.; Lanyi J. K. (2007) Bacteriorhodopsin photocycle at cryogenic temperatures reveals distributed barriers of conformational substates. Proc. Natl. Acad. Sci. U.S.A. 104, 9621–9626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel H.; Oesterhelt D. (1976) Light-induced changes of the pH gradient and the membrane potential in Halobacterium halobium. FEBS Lett. 65, 175–178. [DOI] [PubMed] [Google Scholar]
- Imasheva E. S.; Balashov S. P.; Wang J. M.; Lanyi J. K. (2006) pH-dependent transitions in xanthorhodopsin. Photochem. Photobiol. 82, 1406–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H.; Marti T.; Holtz M.; Mogi T.; Lindau M.; Khorana H. G.; Heyn M. P. (1989) Aspartic acid-96 is the internal proton donor in the reprotonaion of the Schiff base of bacteriorhodopsin. Proc. Natl. Acad. Sci. U.S.A. 86, 9228–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames J. B.; Mathies R. A. (1990) The role of back-reactions and proton uptake during the N → O transition in bacteriorhodopsin’s photocycle: A kinetic resonance Raman study. Biochemistry 29, 7181–7190. [DOI] [PubMed] [Google Scholar]
- Lozier R.; Xie A.; Hofrichter J.; Clore G. M. (1992) Reversible steps in the bacteriorhodopsin photocycle. Proc. Natl. Acad. Sci. U.S.A. 89, 3610–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Váró G.; Brown L. S.; Lakatos M.; Lanyi J. K. (2003) Characterization of the photochemical reaction cycle of proteorhodopsin. Biophys. J. 84, 1202–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chizhov I.; Schmies G.; Seidel R.; Sydor J. R.; Lüttenberg B.; Engelhard M. (1998) The photophobic receptor from Natronobacterium pharaonis: Temperature and pH dependencies of the photocycle of sensory rhodopsin II. Biophys. J. 75, 999–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimányi L.; Váró G.; Chang M.; Ni B.; Needleman R.; Lanyi J. K. (1992) Pathways of proton release in the bacteriorhodopsin photocycle. Biochemistry 31, 8535–8543. [DOI] [PubMed] [Google Scholar]
- Dioumaev A. K.; Lanyi J. K. (2008) Switch from conventional to distributed kinetics in the bacteriorhodopsin photocycle. Biochemistry 47, 11125–11133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda A. (1995) Application of FTIR spectroscopy to the structural study on the function of bacteriorhodopsin. Isr. J. Chem. 35, 387–400. [Google Scholar]
- Barth A. (2000) The infrared absorption of amino acid side chains. Prog. Biophys. Mol. Biol. 74, 141–173. [DOI] [PubMed] [Google Scholar]
- Nie B.; Stutzman J.; Xie A. (2005) A vibrational spectral maker for probing the hydrogen-bonding status of protonated Asp and Glu residues. Biophys. J. 88, 2833–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K.; Takahashi R.; Noguchi T. (2008) Correlation between the hydrogen-bond structures and the C=O stretching frequencies of carboxylic acids as studied by density functional theory calculations: Theoretical basis for interpretation of infrared bands of carboxylic groups in proteins. J. Phys. Chem. B 112, 6725–6731. [DOI] [PubMed] [Google Scholar]
- Dioumaev A. K. (2001) Infrared methods for monitoring the protonation state of carboxylic amino acids in the photocycle of bacteriorhodopsin. Biochemistry (Moscow) 66, 1269–1276. [DOI] [PubMed] [Google Scholar]
- Sasaki J.; Lanyi J. K.; Needleman R.; Yoshizawa T.; Maeda A. (1994) Complete identification of C=O stretching vibrational bands of protonated aspartic acid residues in the difference infrared spectra of M and N intermediates versus bacteriorhodopsin. Biochemistry 33, 3178–3184. [DOI] [PubMed] [Google Scholar]
- Luecke H.; Schobert B.; Richter H.-T.; Cartailler J.-P.; Lanyi J. K. (1999) Structure of bacteriorhodopsin at 1.55 Å resolution. J. Mol. Biol. 291, 899–911. [DOI] [PubMed] [Google Scholar]
- Lanyi J. K.; Schobert B. (2002) Crystallographic structure of the retinal and the protein after deprotonation of the Schiff base: The switch in the bacteriorhodopsin photocycle. J. Mol. Biol. 321, 727–737. [DOI] [PubMed] [Google Scholar]
- Turner G. J.; Miercke L. J. W.; Thorgeirsson T. E.; Kliger D. S.; Betlach M. C.; Stroud R. (1993) Bacteriorhodopsin D85N: Three spectroscopic species in equilibrium. Biochemistry 32, 1332–1337. [DOI] [PubMed] [Google Scholar]
- Szundi I.; Stoeckenius W. (1989) Surface pH controls purple-to-blue transition of bacteriorhodopsin. A theoretical model of purple membrane surface. Biophys. J. 56, 369–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas R.; Koutalos Y.; Ebrey T. G. (1990) Purple membrane: Surface charge density and the multiple effect of pH and cations. Photochem. Photobiol. 52, 1163–1177. [DOI] [PubMed] [Google Scholar]
- Subramaniam S.; Marti T.; Khorana H. G. (1990) Protonation state of Asp (Glu)-85 regulates the purple-to-blue transition in bacteriorhodopsin mutants Arg-82 → Ala and Asp-85 → Glu: The blue form is inactive in proton translocation. Proc. Natl. Acad. Sci. U.S.A. 87, 1013–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U.; Oesterhelt D. (1979) Chromophore equilibria in bacteriorhodopsin. Biophys. J. 28, 211–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen L.; Galajda P.; Száraz S.; Ormos P. (1999) Chloride ion binding to bacteriorhodopsin at low pH: An infrared spectroscopic study. Biophys. J. 76, 1951–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashov S. P.; Imasheva E. S.; Govindjee R.; Sheves M.; Ebrey T. G. (1996) Evidence that aspartate-85 has a higher pKa in all-trans than in 13-cis bacteriorhodopsin. Biophys. J. 71, 1973–1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schobert B.; Cupp-Vickery J.; Hornak V.; Smith S. O.; Lanyi J. K. (2002) Crystallographic structure of the K intermediate of bacteriorhodopsin: Conservation of free energy after photoisomerization of the retinal. J. Mol. Biol. 321, 715–726. [DOI] [PubMed] [Google Scholar]
- Braiman M. S.; Dioumaev A. K.; Lewis J. R. (1996) A large photolysis-induced pKa increase of the chromophore counterion in bacteriorhodopsin: Implications for ion transport mechanisms of retinal proteins. Biophys. J. 70, 939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balashov S. P. (2000) Protonation reactions and their coupling in bacteriorhodopsin. Biochim. Biophys. Acta 1460, 75–94. [DOI] [PubMed] [Google Scholar]
- Luecke H.; Schobert B.; Richter H.-T.; Cartailler J.-P.; Lanyi J. K. (1999) Structural changes in bacteriorhodopsin during ion transport at 2 Å resolution. Science 286, 255–260. [DOI] [PubMed] [Google Scholar]
- Hessling B.; Souvignier G.; Gerwert K. (1993) A model-independent approach to assigning bacteriorhodopsin’s intramolecular reactions to photocycle intermediates. Biophys. J. 65, 1929–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zscherp C.; Heberle J. (1997) Infrared difference spectra of the intermediates L, M, N and O of the bacteriorhodopsin photoreaction obtained by time-resolved attenuated total reflection spectroscopy. J. Phys. Chem. B 101, 10542–10547. [Google Scholar]
- Dioumaev A. K.; Brown L. S.; Needleman R.; Lanyi J. K. (1999) Fourier transform infrared spectra of a late intermediate of the bacteriorhodopsin photocycle suggest transient protonation of Asp-212. Biochemistry 38, 10070–10078. [DOI] [PubMed] [Google Scholar]
- Balashov S. P.; Lu M.; Imasheva E. S.; Govindjee R.; Ebrey T. G.; Othersen B. III; Crouch R. K.; Menick D. R. (1999) The proton release group in bacteriorhodopsin controls the rate of the final step of its photocycle at low pH. Biophys. J. 76(2), A147. [DOI] [PubMed] [Google Scholar]
- Harding M. M. (2002) Metal-ligand geometry relevant to proteins and in proteins: Sodium and potassium. Acta Crystallogr. 58, 872–874. [DOI] [PubMed] [Google Scholar]
- Garczarek F.; Brown L. S.; Lanyi J. K.; Gerwert K. (2005) Proton binding within a membrane protein by a protonated water cluster. Proc. Natl. Acad. Sci. U.S.A. 102, 3633–3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwert K.; Freier E.; Wolf S. (2014) The role of protein-bound water molecules in microbial rhodopsins. Biochim. Biophys. Acta 1837, 606–613. [DOI] [PubMed] [Google Scholar]
- Ono H.; Inoue K.; Abe-Yoshizumi R.; Kandori H. (2014) FTIR spectroscopy of a light-driven compatible sodium ion-proton pumping rhodopsin at 77 K. J. Phys. Chem. B 118, 4784–4792. [DOI] [PubMed] [Google Scholar]
- Balashov S. P.; Imasheva E. S.; Govindjee R.; Ebrey T. G. (1996) Titration of aspartate-85 in bacteriorhodopsin: What it says about chromophore isomerization and proton release. Biophys. J. 70, 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.