Altering the number of kinetochores revealed that chromosomal attachments set the length of the metaphase spindle and the number of microtubules within it. Reducing the number of kinetochores increases length, whereas adding extra kinetochores shortens it, suggesting that kinetochore-generated inward forces help set spindle length in budding yeast.
Abstract
The length of the mitotic spindle varies among different cell types. A simple model for spindle length regulation requires balancing two forces: pulling, due to microtubules that attach to the chromosomes at their kinetochores, and pushing, due to interactions between microtubules that emanate from opposite spindle poles. In the budding yeast Saccharomyces cerevisiae, we show that spindle length scales with kinetochore number, increasing when kinetochores are inactivated and shortening on addition of synthetic or natural kinetochores, showing that kinetochore–microtubule interactions generate an inward force to balance forces that elongate the spindle. Electron microscopy shows that manipulating kinetochore number alters the number of spindle microtubules: adding extra kinetochores increases the number of spindle microtubules, suggesting kinetochore-based regulation of microtubule number.
INTRODUCTION
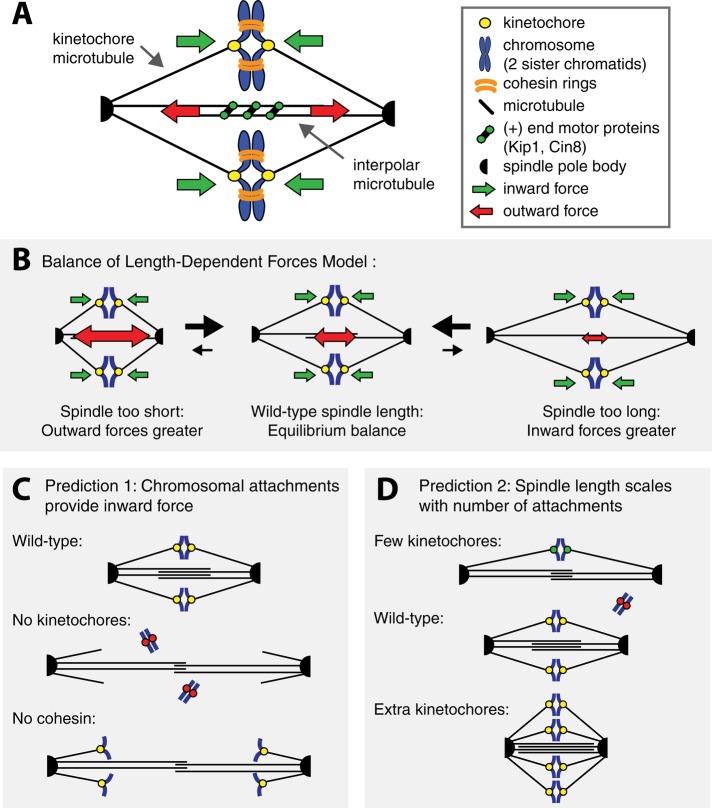
Cells must regulate the size of their internal structures. Some, such as viral capsids, are determined by specific molecular interactions. In others, such as the muscle sarcomere, molecular rulers set the length. The mitotic spindle is a dynamic structure whose size exceeds that of any of its individual components, and its length is set by rules that govern its self-assembly. The spindle is a bipolar array of microtubules that segregates chromosomes (Bloom and Joglekar, 2010). Microtubules attach to chromosomes through kinetochores—protein complexes assembled on centromeric DNA (Westermann et al., 2007). In metaphase, bioriented sister chromatids attach to microtubules from opposite poles and are held together by cohesin rings (Figure 1A). Anaphase chromosome segregation occurs when cohesin is cleaved and sister chromatids are pulled to opposite poles (Tanaka, 2008). Spindle lengths vary from 2 μm in budding yeast (Winey et al., 1995; Straight et al., 1997) to 60 μm in single-celled Xenopus embryos (Wühr et al., 2008), but the spindle of each cell type has a characteristic length (Goshima and Scholey, 2010).
FIGURE 1.
(A) Spindle components and forces. The kinetochores of sister chromatids attach to microtubules emanating from opposite spindle pole bodies (biorientation) generating inward force (green arrows). Interpolar microtubules interact with the plus-end motor proteins Cin8 and Kip1, generating outward force (red arrows). (B) Force balance model. If the spindle is too short, outward force exceeds inward force, and the spindle elongates, reducing the outward force. The opposite happens if the spindle is too long. (C, D) Force balance predictions. (C) If kinetochores are inhibited (red dots) or cohesion is removed, the spindle will elongate. (D) Spindle length scales with the number of chromosomal attachments. Each attachment contributes inward force; increasing attachment number will shorten spindles, and reducing it will elongate spindles.
What sets spindle length? Generally, proposed mechanisms fall into four categories: molecular rulers, gradients, limiting components, and force balance. Molecular rulers are molecules that set the length of other cellular structures (Marshall, 2004), such as proteins that dictate the length of bacteriophage tails (Katsura, 1987, 1990) or actin polymers in sarcomeres (Labeit et al., 1991; Labeit and Kolmerer, 1995). The spindle matrix—a polymeric hydrogel found surrounding spindles in some species—has been proposed to act as a molecular ruler of spindle length (Johansen et al., 2011). Protein gradients have been proposed to regulate spindle length in animals by establishing spatial regions with differing spindle assembly activity (Bastiaens et al., 2006; Kalab and Heald, 2008; Greenan et al., 2010). It has been shown that the microtubule nucleation profile varies spatially within the spindle, and this nonuniform nucleation, along with microtubule transport, could determine spindle length (Brugués et al., 2012). Spindle length has also been shown to scale with cell size during development (Wühr et al., 2008; Greenan et al., 2010; Courtois et al., 2012; Hara and Kimura, 2013), scaling particularly with cytoplasmic volume, leading to the proposal that limiting cytoplasmic components dictate spindle length (Good et al., 2013; Hazel et al., 2013).
In the budding yeast, evidence suggests that spindle length can be seen as a balance between two forces: an outward force generated by motor proteins that push overlapping microtubules apart, opposed by an inward force pulling the kinetochores toward the poles. Because cohesin links the sister chromatids, the kinetochore-generated force pulls the poles toward each other (Figure 1, A and B). Manipulations support this simple model; increasing the activity of Cin8, a kinesin-5 outward-pushing motor, lengthens the spindle (Saunders et al., 1997), and removing Cin8 shortens it (Straight et al., 1998), and removing cohesin elongates the spindle (Stephens et al., 2011).
This simple model for length regulation makes two predictions: the spindle will shorten as the number of kinetochores increases, increasing the inward force, and at least one force must vary with spindle length (Figure 1, C and D). For example, the outward force exerted by motors might fall as the distance between the poles, and thus the extent of microtubule overlap, falls. Without length-dependent force, spindles would have two problems. First, they could not maintain a constant length. Even if the mean level of the opposing forces was perfectly matched, stochastic fluctuations in the two forces would switch a spindle between growing and shrinking. As a result, a spindle's length would fluctuate as a one-dimensional random walk, and the variance in spindle length would increase linearly with the time. Second, the spindle could not cope with systematic variations in the two forces due to genetic or environmental variation. If the inward force exceeded its outward counterpart, the spindle would shrink indefinitely, and if the outward force was larger, the spindle would elongate until it hits the boundaries of the cell. If either force is length dependent, both problems vanish (Figure 1B). In their simplest forms, the ruler-, gradient-, and limiting component–based models predict that varying kinetochore number will not change spindle length. It is important to note that our force balance model based on outward pushing by motors and inward pulling of kinetochores is a simplified account of the forces in the spindle. Forces are generated by microtubule polymerization and depolymerization, friction, and elasticity, as well as from astral microtubules and other sources (reviewed in Dumont and Mitchison, 2009). Our model is a simplification designed to assess the role of kinetochores and chromosomal attachments, but all of these forces must be considered in a detailed model for spindle structure and function.
We verified the key predictions of our simple force balance model in the budding yeast Saccharomyces cerevisiae. Removing kinetochores elongated spindles, and adding kinetochores shortened them, demonstrating a kinetochore-dependent inward force and at least one length-dependent force. Adding extra kinetochores to wild-type cells increased spindle microtubule number and spindle pole body size, demonstrating that kinetochore number regulates microtubule number.
RESULTS
We tested two predictions of the force balance model for spindle length in budding yeast: 1) attaching chromosomes to the spindle pulls the poles toward each other (Figure 1C), and 2) at least one force is length dependent, predicting that kinetochore number regulates spindle length (Figure 1D). Eliminating kinetochores will make the spindle longer, and adding them will make it shorter.
To test these predictions, we changed the number of functional kinetochores. Kinetochores were eliminated using a temperature-sensitive allele of Ndc10 (ndc10-1), a component of the inner kinetochore complex CBF3, which binds to the centromeric DNA and connects the chromosome to the remainder of the kinetochore. The ndc10-1 allele was the only temperature-sensitive mutant of the CBF3 complex that fully eliminated kinetochore function as assayed by the inability of ndc10-1 cells to activate the spindle checkpoint, measured by accumulation of large-budded cells (Supplemental Figure S1, A and B; Gardner et al., 2001). Other temperature sensitive alleles in CBF3, ctf13-30, cep3-2, and ndc10-2, displayed residual kinetochore activity at 37°C and thus were not suitable for our experiments (Supplemental Figure S1B).
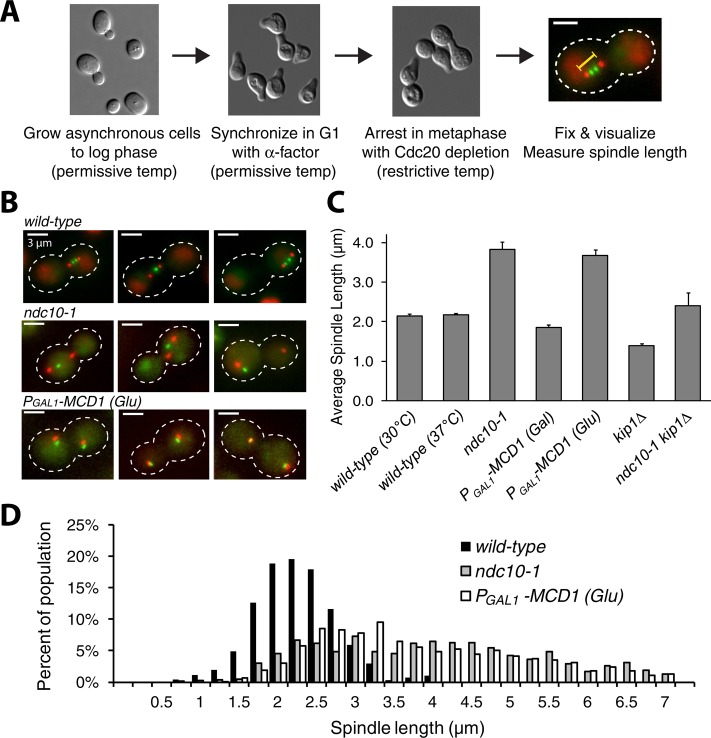
Does kinetochore inactivation lead to spindle elongation? We used the ndc10-1 mutant to produce cells with no chromosomal attachments and two manipulations to control the progress of the cell cycle: treating cells with a mating pheromone, α-factor, to arrest them in G1, and removing Cdc20, an activator of the anaphase-promoting complex (APC), to arrest cells in metaphase (Hartwell et al., 1973; Hartwell and Smith, 1985). Wild-type and ndc10-1 cells were grown to log phase at 23°C, synchronized in G1 with α-factor, and released into a Cdc20 depletion–induced metaphase arrest at 37°C (Figure 2A). Spindle pole bodies were labeled by fusing a red fluorescent protein (RFP) variant to a spindle pole body component (SPC42-mCherry), and chromosome XV was labeled with green fluorescent protein (GFP) by inserting a Lac operator (LacO) array ∼200 base pairs downstream of CEN15 (see Supplemental Table S1 for exact position; Shonn et al., 2000) and expressing a GFP-Lac repressor fusion (GFP-LACI; Straight et al., 1996). Spindle length was measured as the three-dimensional (3D) distance between the two spindle poles and reported as the average length of measured spindles; error bars are SDs of average spindle length over all trials. Figure 2B shows examples of fluorescent spindles.
FIGURE 2:
Chromosomes restrain spindle elongation. (A) Spindle length measurement. Cells were released from G1 into a metaphase arrest. Spindle length (yellow bar), labeled spindle pole bodies (Spc42-mCherry), and GFP CEN15 are indicated. Scale bar, 2 μm. Temperature-sensitive strains and their control were grown and G1 arrested at 23°C and then arrested in metaphase at 37°C; other strains were grown at 30°C. (B) Spindles elongate without kinetochores or cohesin. Fluorescence images of wild-type, ndc10-1, and PGAL1-MCD1 spindles. PGAL1-MCD1 cells were grown in glucose (Glu) to repress cohesin. Spindle pole bodies (red, Spc42-mCherry) and CEN15 (green, GFP-LacI bound to LacO array) are visible. Scale bar, 3 μm. (C) Effect of kinetochores, cohesin, or motors on spindle length. Spindle length in ndc10-1 strains and glucose-grown PGAL1-MCD1 strains is compared with wild-type cells grown at both temperatures and galactose-grown PGAL1-MCD1 strains. Elongation was statistically significant (p < 0.001, Student's t test). Deleting Kip1, a kinesin, shortens the spindle, whereas eliminating both Kip1 and Ndc10 allows the spindle to approach wild-type length. Spindle length was measured as the 3D distance between spindle pole bodies (n >120 cells). Error bars are SDs in average spindle length. (D) Distribution of spindle lengths. Wild type has a tight distribution compared with ndc10-1 and glucose-grown PGAL1-MCD1.
Inactivating kinetochores elongates the spindle
The mean spindle length of ndc10-1 cells was 1.75 times that of wild-type cells (3.82 ± 0.20 vs. 2.18 ± 0.02 μm; Figure 2C). Because Ndc10 plays a role in spindle midzone integrity along with the kinetochore, it is possible that the elongated pole–pole distance is due to broken spindles whose spindle pole bodies are no longer connected by microtubules (Pearson et al., 2003). To ensure that such broken spindles did not distort our measurements of spindle length, we expressed fluorescently tagged tubulin (GFP-TUB1) and measured spindle lengths along the long axis of the continuous bar of fluorescence in metaphase-arrested cells (Supplemental Figure S2A). Spindle lengths measured in this way were slightly shorter than when measured as the distance between mCherry-tagged spindle pole bodies (SPC42-mCherry) for both ndc10-1 cells (3.49 ± 0.29 vs. 3.82 ± 0.20 μm) and wild-type cells (1.94 ± 0.21 vs. 2.18 ± 0.02 μm; Supplemental Figure S2A). The spindle length estimated from tubulin fluorescence was ∼10% less than that estimated from the separation of fluorescently tagged spindle poles and was most likely due to spindle pole bodies sitting beyond the distal ends of GFP-tubulin (Supplemental Figure S2B). This comparison reveals that measuring the spindle pole body separation gives an estimate of spindle length that is not compromised by breakage of long spindles in ndc10-1 cells. Although broken spindles were found in our ndc10-1 populations (Supplemental Figure S2Bf), their inclusion in our spindle length measurements does not bias spindle length. Spindle length, measured by spindle pole body separation, is not statistically different between cells with continuous GFP spindles (Supplemental Figure S2B, d and e) and all ndc10-1 cells, including those with broken spindles (p = 0.3, Student's t test; Supplemental Figure S2C).
Ndc10 is also reported play a role in spindle pole body maturation (Romao et al., 2008), which could affect spindle length, so we removed cohesin as another way of eliminating inward forces on the spindle (Stephens et al., 2011). The inward force depends on the linkage between sister chromatids: without cohesin, forces on the kinetochores drag the chromosomes to the spindle poles but cannot pull the poles toward each other. Although the mechanisms are different, inactivating kinetochores and decoupling sister chromatids should equally elongate the spindle since both defects eliminate any inward force contributed by chromosomes (Figure 1C). Cohesin was inhibited by placing MCD1, the gene encoding one of its subunits, under the galactose-inducible GAL1 promoter (PGAL1). These cells produce cohesin when grown in galactose but not when grown in glucose. The mean spindle length of galactose-grown cells (cohesin expressed) was 1.85 ± 0.07 μm, but glucose-grown cells had spindles that were 3.67 ± 0.15 μm long, similar to those of ndc10-1 cells (Figure 2C). Because PGAL1-MCD1 strains were grown at 30°C and ndc10-1 strains were grown at 37°C, we measured wild-type cells grown at both temperatures. Temperature did not significantly affect wild-type spindle length (2.14 ± 0.02 μm at 30°C vs. 2.18 ± 0.02 μm at 37°C). Because spindle length in ndc10-1 and PGAL1-MCD1 cells is statistically indistinguishable, the elongated spindle length of ndc10-1 cells is most likely due to the reduction in inward force caused by inactivating the kinetochore rather than to defects in the spindle pole bodies.
Inhibiting kinetochores or cohesin increased the variation in spindle length. Spindle length in a population of wild-type cells has a mean around 2 μm with SD = ±0.5 μm (Figure 2D). In ndc10-1 cells, the distribution of spindle lengths is broad, with a mean length of 3.82 μm and SD = ±1.4 μm (Figure 2D). Removing cohesin produces a similarly broad distribution, with a mean length of 3.67 μm and SD = ±1.3 μm (Figure 2D). The high variation in spindle length could arise from residual kinetochore function (but Supplemental Figure S1B suggests that kinetochores are nonfunctional) or because the balance between outward and inward forces, which depends on kinetochores and cohesin, restrains the variation in spindle length. Without an inward force, spindle length is more variable, as expected if spindles now elongate to the point at which outward force is zero. The elongated spindle phenotype was not specific to ndc10-1; the other CBF3 mutants, ctf13-30 and cep3-2, also had longer spindles (Supplemental Figure S1C). Both cep3-2 and ctf13-30 spindles were longer than wild-type but shorter than ndc10-1 spindles, most likely due to incomplete abolition of kinetochore function (Supplemental Figure S1B), suggesting that the presence of a few functional kinetochores can partially restrain spindle elongation.
The elongation produced by detaching chromosomes or decoupling sister chromatids supports the prediction that chromosomal attachment opposes outward forces. These outward forces are generated by the kinesin-5 motors Cin8 and Kip1; kinesins of this class exert outward forces by cross-linking antiparallel microtubules and walking toward microtubule plus ends. Removing either Kip1 or Cin8 shortens the metaphase spindle and slows spindle elongation in anaphase (Saunders and Hoyt, 1992; Hoyt et al., 1992; Straight et al., 1998); increasing Cin8 expression elongates spindles (Saunders et al., 1997). Both Kip1 and Cin8 play a role in polymerization at microtubule plus ends in mitosis (Gardner et al., 2008). To show that chromosomal attachment and plus end–directed motors create opposing forces, we compared spindles in ndc10-1 and kip1∆ single mutants and the ndc10-1 kip1∆ double mutant. Deleting KIP1 shortens mean spindle length to 1.39 ± 0.05 μm (p < 0.001 relative to wild type, Student's t test; Figure 2C). Simultaneously deleting KIP1 and inactivating kinetochores with ndc10-1 yields a mean spindle length of 2.41 ± 0.3 μm, which slightly exceeds that of wild-type cells. showing that plus end–directed motors and kinetochores generate opposing forces.
To confirm that kinetochores were inactivated in ndc10-1 cells and cohesin function was lost in glucose-grown PGAL1-MCD1 cells, we scored the position of sister chromatids. When a chromosome is bioriented, its sister chromatids come under tension as they are pulled toward opposite poles but kept from separating by cohesin (Figure 1A). This conflict visibly separates the sister kinetochores and their associated centromeric DNA from each other, as seen by placing a LacO array near the centromere (200 base pairs downstream of CEN15 (Supplemental Table S1) and labeling it with GFP-LacI (Figure 3A; Goshima and Yanagida, 2000). The presence of two GFP dots separated by 0.2–1 μm indicates a bioriented chromosome, and a single GFP dot indicates a bioriented chromosome whose sister kinetochores have not stretched far enough apart to be resolved by light microscopy or a mono-oriented chromosome (Figure 3A). In wild-type cells with functional kinetochores and cohesin, 49 ± 2.8% of cells with GFP-labeled chromosome XV had two dots, indicating biorientation. In ndc10-1, only 4 ± 3.7% of cells had two GFP dots, and 96% had a single GFP dot, confirming kinetochore inactivation (Figure 3B). Similar to ndc10-1, only 14 ± 2.6% of the glucose-treated PGAL1-MCD1 cells had the two dots separated by the small distances (0.2–1.0 μm) that indicate a bioriented chromosome, but unlike ndc10-1, 77 ± 2.6% of PGAL1-MCD1 cells have two GFP dots localized near either spindle pole body. These dots are >1 μm apart and are located close to the spindle poles, showing that the sister chromatids were prematurely separated in metaphase, as expected if kinetochores were functional but cohesin was not (Figure 2C).
FIGURE 3:
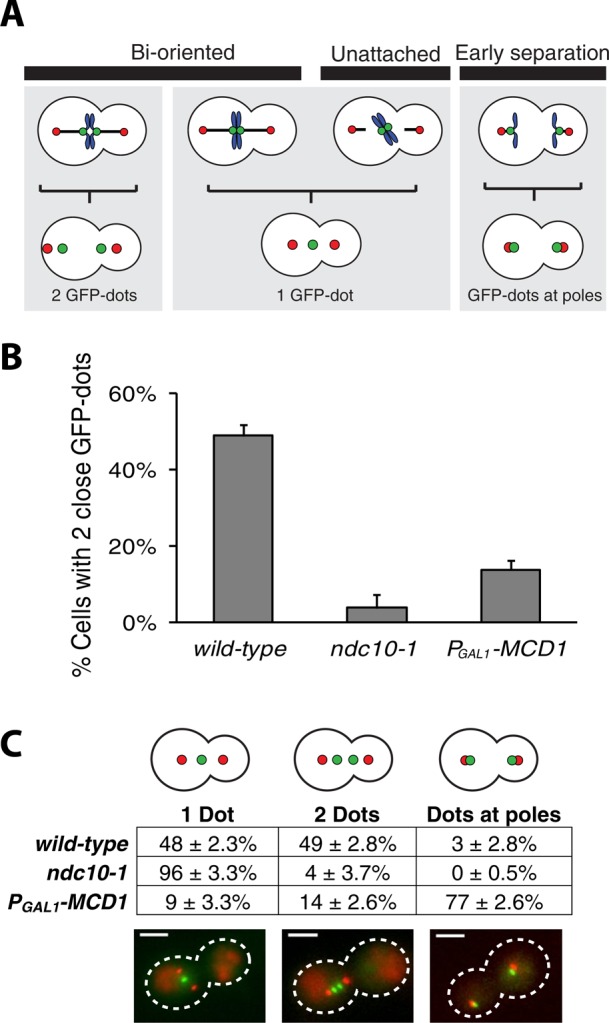
Confirming kinetochore and cohesin inactivation. (A) Chromosome attachment schematic. Correct attachment of a chromosome to the spindle results in biorientation, visualized as two GFP dots separated by ≤1 μm. A single GFP dot is a kinetochore pair separated by <0.3 μm or a chromosome attached to only one or neither spindle pole (red dots). Cohesin-less chromosomes separate prematurely during metaphase. GFP dots at poles are separated by >1 μm. (B) Chromosome biorientation. ndc10-1 and PGAL1-MCD1 strains have few bioriented chromosomes compared with wild type. Error bars are SDs. (C) Chromosome position. Positions of GFP dots were scored to distinguish between unattached (one dot, ndc10-1) and prematurely separated sister chromatids (two dots, PGAL1-MCD1). Error bars are SDs. Scale bar, 3 μm.
Our results support the force balance model. Eliminating kinetochore-dependent inward forces elongates the metaphase spindle (Figure 1), by removing either the connection between chromosomes and poles (kinetochore inactivation) or the linkage between sister chromatids (cohesin removal; Figure 2C).
Spindle length scales with kinetochore number
If at least one of the forces in the spindle is length dependent, spindle length should scale with the number of chromosomes. Two types of length dependence would produce this effect: an increase in outward force or a decrease in the inward force generated at each kinetochore as the spindle poles come closer together. In either case, increasing kinetochore number would increase the initial inward force, leading to a new balance point at a shorter spindle length (Figure 1D). We altered the number of chromosomal attachments by varying the number of functional kinetochores in two ways: 1) by knocking out natural kinetochores using ndc10-1 and then introducing defined numbers of synthetic kinetochores to reattach a few chromosomes, and 2) by adding centromeric plasmids to put extra kinetochores in cells with functional kinetochores.
Synthetic kinetochores can replace natural kinetochores by biorienting and segregating chromosomes (Kiermaier et al., 2009; Lacefield et al., 2009). Recruiting a fusion of the Lac repressor and Ask1, a component of the microtubule-binding Dam1 complex, to a LacO array creates a synthetic kinetochore that bypasses the need for CBF3 and thus does not require Ndc10 or Ctf13 (Figure 4A; Lacefield et al., 2009). We introduced the synthetic kinetochore (sKT) to ndc10-1 cells by integrating LacO arrays on a one, two, or three chromosomes, thus reattaching them to the mitotic spindle (Figure 4B).
FIGURE 4:
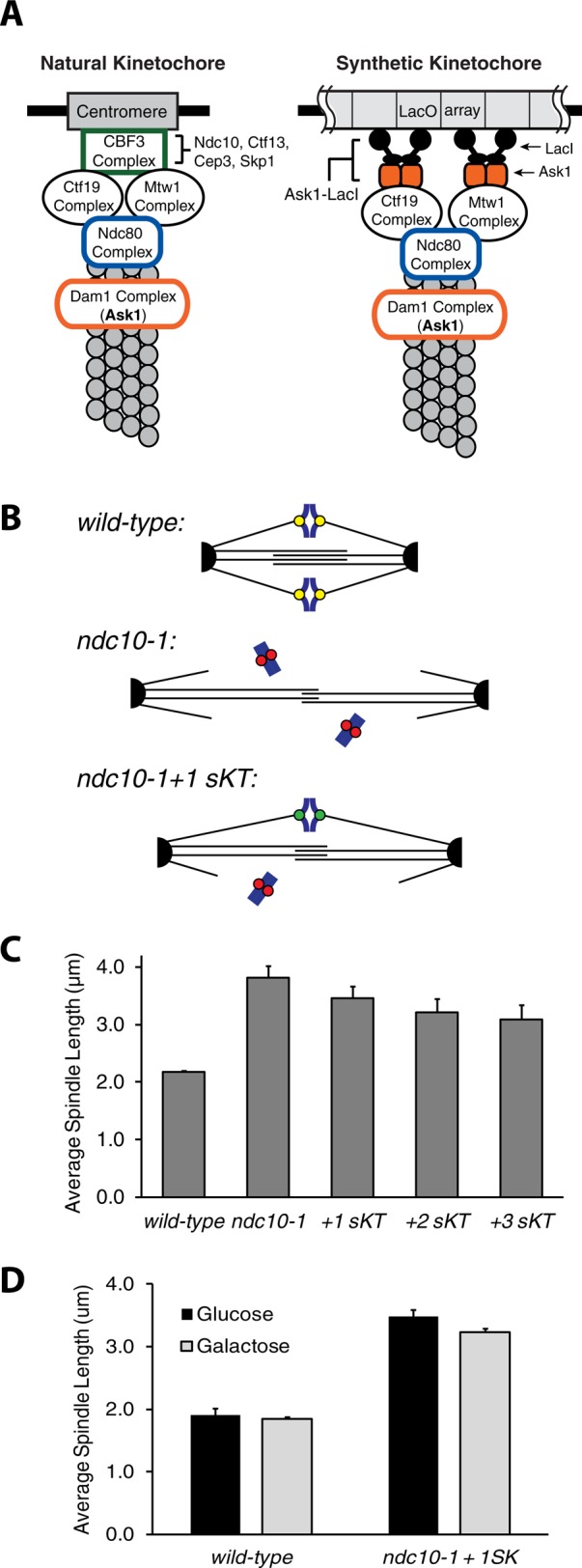
Synthetic kinetochores shorten ndc10-1 spindles. (A) Natural and synthetic kinetochores. The budding yeast kinetochore contains many proteins and binds the 125–base pair centromeric DNA. The CBF3 complex (Ndc10, Ctf13, Cep3, and Skp1) binds centromere DNA. The synthetic kinetochore depends on binding of an Ask1-LacI fusion to a LacO array, recruiting other kinetochore components independently of CBF3. (B) Prediction. Adding synthetic kinetochores (sKT) to ndc10-1 cells will shorten their spindles. (C) Synthetic kinetochores shorten ndc10-1 spindles. Each additional synthetic kinetochore shortened spindles significantly (p < 0.04, Student's t test). Error bars are SDs in average spindle length. (D) Shorter spindles require Ask1-LacI. Cells expressing PGAL1-ASK1-LacI were grown in raffinose, synchronized in G1, and released into metaphase arrest in glucose (repressing) or galactose (inducing). Wild-type cells had similar-length spindles in glucose and galactose (p = 0.36, Student's t test), but repressing Ask1-LacI lengthened spindles in ndc10-1 cells carrying a synthetic kinetochore (p = 0.02, Student's t test). Error bars are SDs in average spindle length.
The ndc10-1 allele was reported to impair spindle pole body maturation; in reconstructed ndc10-1 spindles, the new spindle pole often nucleated few or no microtubules (Romao et al., 2008). If ndc10-1 cells cannot nucleate microtubules from both spindle pole bodies, chromosomal attachments between the two poles cannot be added back. We found a similar asymmetry between the spindle poles and a reduction in microtubule number in ndc10-1 spindles (Supplemental Figure S4B and Supplemental FigS4video1) compared with wild type (Supplemental Figure S4A and Supplemental FigS6video1). However, when three synthetic kinetochores were introduced into ndc10-1 cells, spindles contained more microtubules and had a more symmetric distribution, suggesting that both spindle pole bodies in ndc10-1 cells are capable of nucleating microtubules that can attach chromosomes to the spindle (Supplemental Figure S4C and Supplemental FigS4video2).
The average spindle length in ndc10-1 cells decreased with the addition of each synthetic kinetochore (Figure 4C); the addition of one, two, and three synthetic kinetochores decreases the mean length of the ndc10-1 spindle from 3.8 ± 0.20 to 3.5 ± 0.22, 3.2 ± 0.24, and 3.1 ± 0.25 μm, respectively (p < 0.04 for each additional synthetic kinetochore, Student's t test). Spindle shortening depends on Ask1-LacI expression. When the Ask1-LacI fusion is placed under the inducible galactose promoter and cells are shifted to 37°C, the average spindle length of cells with a single synthetic kinetochore is 3.2 μm when grown on galactose (Ask1-LacI expressed) and 3.5 μm when grown on glucose (no Ask1-LacI; p = 0.02, Student's t test), but wild-type spindle length is not statistically different between the two sugars (Figure 4D).
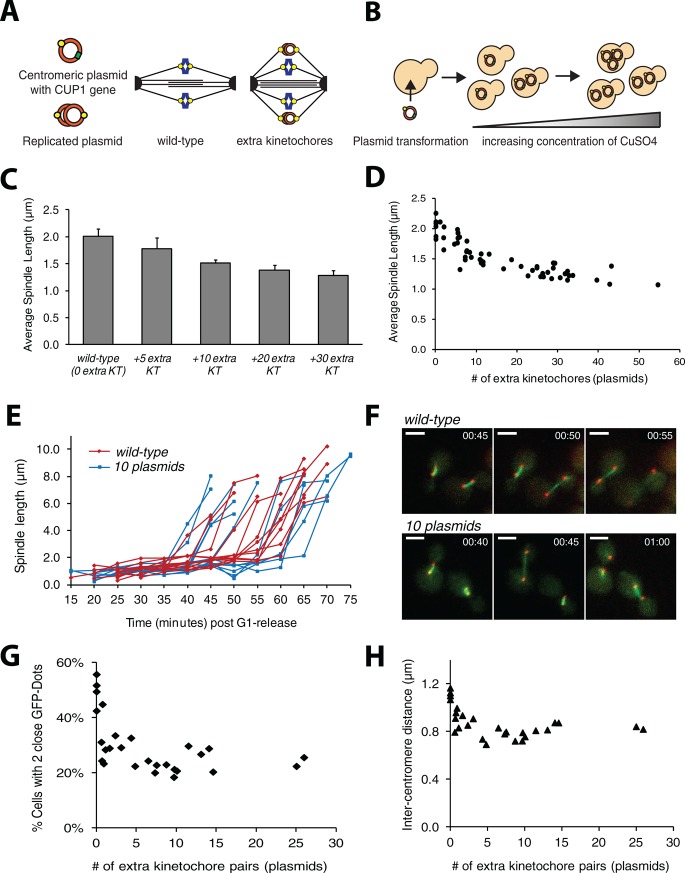
Adding extra kinetochores to wild-type cells should shorten their spindles by creating more attachments between the spindle poles and the chromosomes (Figure 1D). We introduced additional kinetochores in the form of minichromosomes or centromeric plasmids (Figure 5A), which assemble a fully functional kinetochore (Clarke and Carbon, 1980). Multiple centromeric plasmids slow the cell cycle (Futcher and Carbon, 1986), but this delay depends on the spindle checkpoint and is completely eliminated in mad2∆ strains (Wells and Murray, 1996), which lack the checkpoint. We therefore used checkpoint-deficient cells to allow us to add substantial numbers of extra kinetochores without delaying progress through mitosis.
FIGURE 5:
Extra kinetochores shorten spindles. (A) Plasmid schematic. The centromeric plasmid assembles a kinetochore (yellow circle) and carries the CUP1 copper resistance gene (green box). Replicated plasmids attach to the spindle via their kinetochores. (B) Driving up plasmid copy number. Plasmid-bearing cells were grown in increasing concentrations of CuSO4 to select for increased plasmid number. (C) Spindle length in strains containing an average of 5, 10, 20, or 30 centromeric plasmids. Spindles are shorter in cells with extra kinetochores and shorten with increasing kinetochore number (p < 0.001, Student's t test). Error bars are SDs in average length. (D) Spindle length scales with attachment number. Each data point is the average spindle length in an independent trial. (E) Extra kinetochores do not alter anaphase dynamics. Wild-type cells (red lines, n = 12) and cells with 10 plasmids (blue lines, n = 12) were synchronized in G1 and released to proceed through mitosis; spindle length was tracked in individual live cells. The time to enter anaphase, maximum length of the anaphase spindle, and rate of anaphase spindle elongation were statistically indistinguishable between wild-type and plasmid-containing cells (p > 0.5 for all parameters, Student's t test). (F) Frames from anaphase movies. Wild type and cells containing 10 plasmids were released from G1 at T = 0:00 (time represented as hours:minutes) and allowed to proceed through mitosis. Frames displayed from the 10-plasmid movie jumps from T = 0:45 to 1:00 to show the second cell entering anaphase. Spindle pole bodies are labeled with mCherry, and spindles are labeled with GFP-tubulin; scale bars, 3 μm. (G, H) Intercentromere separation in the presence of extra kinetochores. (G) Percentage of cells with two close GFP dots signifying a correctly attached chromosome in cells containing a range of plasmids. Error bars are SDs. (H) Mean distance between sister chromatids. Error bars are SDs in average separation.
To vary the number of additional kinetochores, we used copper resistance as a graded selection for plasmid copy number (Figure 5B). CUP1 encodes metallothionein and confers copy number–dependent copper resistance (Karin et al., 1984); laboratory strains of budding yeast with one or two copies of CUP1 can grow in 0.15 mM CuSO4, but industrial strains have been isolated that can grow in 2 mM CuSO4 and have up to 10 copies of CUP1 (Welch et al., 1983). After we deleted the chromosomal copies of CUP1 (CUP1-1 and CUP1-2), we transformed Spc42-mCherry mad2∆ cells with a centromeric plasmid bearing CUP1. The plasmid carries the only copy of CUP1, and thus copper resistance increases with increasing plasmid number. The transformants were transferred to successively higher concentrations of CuSO4 to drive up the plasmid copy number (Figure 5B). Strains resistant to 1.0–3.0 mM CuSO4 contained on average 20–30 plasmids. The mean plasmid number was determined in each experiment by quantitative PCR (qPCR) of the plasmid's centromere sequence relative to ALG9, a house-keeping gene present in one copy per cell (Teste et al., 2009). The number of plasmids is reported as the prereplication number of plasmids as qPCR was performed on samples synchronized in G1 at the beginning of every experiment.
We measured the spindle length in cells with 0–30 plasmids (0–30 extra kinetochores) that had been grown at 30°C (Figure 2A). The average spindle length scaled with the number of kinetochores, shortening with additional kinetochores (Figure 5C). The spindles of cells containing an average of 5, 10, 20, and 30 plasmids were 1.8 ± 0.20, 1.5 ± 0.7, 1.4 ± 0.10, and 1.3 ± 0.09 μm long, respectively, compared with 2.0 ± 0.14 μm for plasmid-free cells. Spindles with extra kinetochores were significantly shorter than wild type (p < 0.001, Student's t test) and shorter with each increase in the number of extra kinetochores (p < 0.001, Student's t test, for each comparison). We probed the minimum spindle length by growing cells in a large range of CuSO4 concentrations to drive up the number of plasmids; the average spindle length appeared to plateau around a length of 1.2 μm. The shortest spindles recovered had an average length of 1.1 μm and were measured in cells contained an average of 54 plasmids (Figure 5D).
The presence of extra kinetochores shortened the spindle but did not affect its stability or dynamics. Wild-type cells and cells with an average of 10 centromeric plasmids were synchronously released from G1 into a metaphase arrest, and spindle length was measured by video microscopy beginning 2 h after release from G1. During the 60 min in which spindles were measured, spindle length in both individual wild-type cells (red lines) and cells containing 10 plasmids (blue lines) fluctuated by ∼18% of their length. Supplemental Figure S3A shows the fluctuations in the lengths of representative spindles from cells with and without 10 extra kinetochores. The fluctuations in spindle length were calculated by measuring the mean spindle length of 18 individual cells of each population and calculating its SD over the period of observation (T = 120–180 min); the SD of spindle length was divided by mean length to give fluctuation as a percentage of spindle length. The fluctuations of wild-type spindles and spindles with extra kinetochores were statistically indistinguishable (p = 0.3, Student's t test). A frame (T = 2:50 postrelease from G1) from a representative movie is shown in Supplemental Figure S3B (Supplemental FigS3video1); the cell walls of wild-type cells were labeled with Texas Red to distinguish them from cells containing 10 plasmids.
The presence of extra kinetochores does not affect the kinetics of anaphase. In synchronized cells that were allowed to proceed through mitosis and imaged by live microscopy, wild type (red lines) and cells with extra kinetochores (blue lines) both enter anaphase on average 50 min after release from G1 (50 ± 8 min for wild-type and 49 ± 10 min for cells with extra kinetochores; Figure 5E). There was also no statistical difference in the maximum anaphase spindle length (wild type 7.62 ± 1.48 μm vs. extra kinetochores 7.50 ± 1.28 μm, p = 0.84, Student's t test) and the rate of anaphase spindle elongation (wild type 0.52 ± 0.13 μm/min vs. extra kinetochores 0.56 ± 0.23 μm/min, p = 0.54, Student's t test). Figure 5F shows several frames from representative movies of wild-type cells (Supplemental Fig5video1) and cells with 10 extra kinetochores (Supplemental Fig5video2) going through mitosis.
To investigate the effect of the extra kinetochores on the behavior of a cell's chromosomes, we tagged the centromere of chromosome III with a LacO array and scored it for the ability to see two discrete dots—one associated with each sister chromatid (Figure 5G) as previously described (Figure 3A). Seeing two dots demonstrates that a chromosome is bioriented, but a single dot can reflect two states: a bioriented chromosome whose sister kinetochores are not far enough to allow the two dots to be resolved by light microscopy or a mono-oriented chromosome. The viability of our strains is inconsistent with a high frequency of mono-orientation and suggests that we are measuring differences in the amount of interkinetochore stretch. The percentage of cells with two GFP dots and the distance these chromatids are separated (intercentromere distance) quickly decreased with the addition of plasmids. Wild-type, plasmid-free cells have ∼50% of chromosomes pulled far enough apart to produce two resolvable GFP dots, whereas the presence of just one to five plasmids or extra kinetochores causes a drop to ∼25% paired dots (Figure 5G), but the percentage of cells with two dots does not significantly change with increased numbers of extra kinetochores; for example, 22% of cells have a visibly bioriented chromosome with either 5 or 25 extra kinetochores. Similarly, the distance chromatids separate plateaus around 0.8 μm with the presence of a few extra kinetochores. Wild-type, plasmid-free cells have an average intercentromere distance of 1.1 μm, but the presence of 1 extra kinetochore yielded an average intercentromere distance of 0.8 μm, 4 extra kinetochores gave 0.7 μm, and 25 extra kinetochores gave 0.8 μm (Figure 5H). These plateaus suggest that whereas the presence of plasmids with their extra kinetochores alters the distance between chromatids, increasing the number of plasmids does not prevent attachment and biorientation of natural chromosomes. The cells containing many centromeric plasmids are viable and able to biorient a labeled chromosome, suggesting that every chromosome attaches and biorients on the spindle.
We asked whether the extra plasmids were attaching to microtubules. If extra centromeres disrupted plasmid or chromosome attachment to the spindle, both plasmids and chromosomes should be lost more frequently in mitosis, as kinetochores on plasmids and chromosomes are functionally indistinguishable (Clarke and Carbon, 1980; Bloom and Carbon, 1982). A marked tester plasmid containing a centromere was retained and segregated at rates that were statistically indistinguishable regardless of the number of plasmids present (∼90% plasmid retention after 9 h in nonselective media, p > 0.20, Student's t test; Supplemental Figure S5A), whereas tester plasmids that did not contain a centromere were lost at much higher rates (∼10% plasmid retention, p < 10−5, Student's t test), suggesting that the extra kinetochores are attached to microtubules, and their presence does not disrupt segregation of other centromeres. Both retention levels are similar to those reported by others (Clarke and Carbon, 1980). Whereas Figure 5, G and H, showed that extra centromeres do not inhibit the attachment and biorientation of natural chromosomes on the spindle, we also measured cellular growth rate to determine whether their presence disrupted the timing of the cell cycle, perhaps by creating mitotic delays. Growth rate is unaffected by the number of plasmids; there is no statistical difference in doubling time between wild-type cells with no plasmids and cells with plasmids (124 ± 4 min for wild-type cells vs. 125 ± 8 min for plasmid-containing cells, p = 0.79, Student's t test; Supplemental Figure S5B). These results demonstrate that the presence of extra centromeres does not impede the cell cycle, and it also shows that the vast majority of natural chromosomes are segregated correctly. Although this assay is not a sensitive quantification of chromosome loss rate, it does reveal that individual chromosomes are not lost at >0.007/chromosome per cell per division, because this level of loss would slow the growth rate of a culture by >10%, since yeast has 16 chromosomes. At this threshold level, only e−(16*0.007) = 0.89 of the cell divisions would produce two viable daughter cells. Earlier work by Schulman and Bloom (1993) showed that putting part of a centromere on a very high copy plasmid could increase the frequency of chromosome loss, but their study differed from ours in two ways: they used only part of the centromere sequence (CDEIII), and they used cells whose spindle checkpoint is intact. Increasing the number of centromeric plasmids in cells with an intact spindle checkpoint can cause profound mitotic delays (Wells and Murray, 1996), which may impair chromosome segregation. Our data would not allow us to detect more subtle elevations in rate at which the natural chromosomes missegregate, which is 10−5/cell per division in unperturbed cells, and we cannot exclude the possibility that shortening the spindle impairs the accuracy of chromosome segregation to some extent.
In summary, spindle length scales with kinetochore number. Introducing synthetic kinetochores to cells whose kinetochores have been inactivated shortens spindles, as does adding more kinetochores to wild-type cells, supporting the argument for length-dependent spindle forces (Figure 1D).
Extra kinetochores increase microtubule number
Unlike animal and plant spindles, in which most microtubules do not interact with kinetochores, >70% of the budding yeast spindle microtubules are believed to terminate at kinetochores (Winey et al., 1995). This observation, combined with the size and microtubule packing of spindle pole bodies, has led to speculation that they have a limited ability to nucleate microtubules (Winey and Bloom, 2012). We created strains with more than three times the endogenous number of kinetochores, allowing us to test this idea. Haploid, metaphase yeast spindles contain 44 ± 8 microtubules, with 36 ± 7 identified as kinetochore microtubules available to attach the 16 chromosomes (Winey et al., 1995). Postreplication chromosomes consist of two chromatids each with a centromere that recruits a kinetochore (Figure 1A), so 2 microtubules are required to attach a single replicated chromosome to the spindle, and 32 are required to attach all 16 replicated chromosomes. Our strains would require many more microtubules; for example, cells with 20 replicated plasmids would require at least 80 microtubules: 32 to attach the 16 replicated chromosomes, 40 to attach the 20 replicated plasmids, and 8 interpolar microtubules.
How do cells survive with all these extra kinetochores? Their ability to grow normally (Supplemental Figure S5B), attach and biorient chromosomes (Figure 5, G and H), and segregate plasmids (Supplemental Figure S5A) admits two explanations: their spindle pole bodies support many more microtubules than are present in unperturbed cells, or single microtubules can attach to and segregate multiple kinetochores. To distinguish these possibilities, we performed electron tomography on cells containing many copies of the centromeric plasmids. Cells were grown in CuSO4 to select for high plasmid copy number, arrested in G1, released into a metaphase arrest, fixed, and imaged.
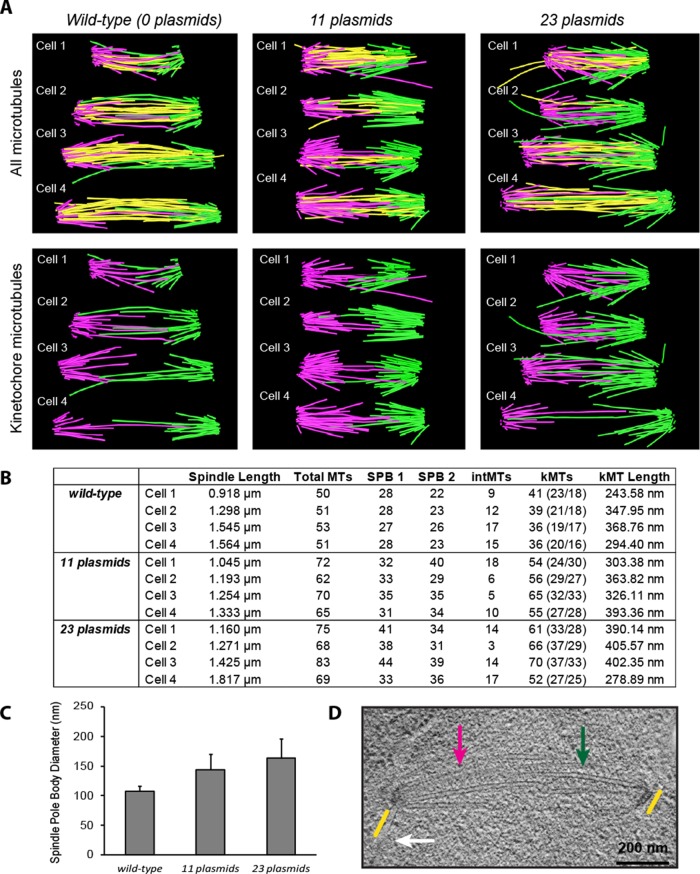
Wild-type, plasmid-free cells contained an average of 51 ± 1 microtubules, within the SD of microtubules previously observed (Winey et al., 1995). Cells containing an average of 23 plasmids as determined by qPCR contained significantly more microtubules, with an average of 74 ± 7 microtubules (p = 0.006, Student's t test; Figure 6, A and B). Similarly, cells containing an average of 11 plasmids contained an average of 67 ± 5 microtubules, a statistically greater number than that of wild type (p = 0.004, Student's t test) and intermediate between that of wild type and the cells with 23 plasmids (Figure 6, A and B; see Supplemental Fig6video1–Fig6video3 for representative 3D movies of spindles). Because budding yeast chromosomes do not condense to structures that are visible in EM, we used programs described in Winey et al. (1995) to identify the central, “core bundle” (interpolar microtubules) and presumptive kinetochore microtubules. The interpolar microtubules of the central spindle are defined as microtubules that are separated by up to 45 nm for lengths of ≥300 nm; they are shown in yellow in Figure 6A and form an obvious central spindle. Microtubules that do not meet this criterion are displayed in magenta and green and represent the putative kinetochore microtubules from each pole; the difference in microtubule number in both 11- and 23-plasmid-containing cells comes from additional kinetochore microtubules (p = 0.001 and 0.005 respectively, Student's t test). Figure 6A, bottom, shows only the putative kinetochore microtubules, to highlight the difference in number and spindle organization. Spindles with 11 plasmids have 58 ± 5 kinetochore microtubules, and those with 23 plasmids have 62 ± 8, compared with 38 ± 3 for wild type. The number of interpolar microtubules (yellow microtubules) is not significantly different between wild-type and plasmid-containing spindles (Figure 6B). Adding kinetochores increases the size of the spindle pole body's central plaque. Wild-type spindle pole bodies had a mean diameter of 107 ± 10 nm (n = 7). Spindle pole bodies from cells containing 11 plasmids (average diameter of 144 ± 27 nm, n = 7, p = 0.009, Student's t test) and 23 plasmids (average diameter of 163.4 ± 32.8, n = 8, p = 0.001, Student's t test) were both significantly larger than those from wild-type cells (Figure 6C). Figure 6D shows an example, indicating the width of spindle pole bodies, microtubules from opposite poles, and the nuclear envelope.
FIGURE 6:
Extra kinetochores increase microtubule number. (A) Spindle reconstruction by electron tomography. Microtubules are categorized as kinetochore microtubules or interpolar microtubules based on their length and pitch angle from the spindle (see Methods and Materials). Kinetochore microtubules from the spindle pole bodies are green (SPB1) or magenta (SPB2); interpolar microtubules are yellow. Top, all microtubules; bottom, only kinetochore microtubules. (B) Microtubule numbers in spindles. The total number of microtubules (Total MTs) with classification as interpolar or kinetochore microtubules number from each spindle pole body (SPB1 and SPB2), with kinetochore microtubule number in parentheses. (C) Spindle pole bodies enlarge with extra microtubules. The diameter of the central plaque of the spindle pole body in cells with 11 or 23 plasmids exceeds that in wild-type cells. (D) Electron micrograph. Microtubules (green and magenta arrows), nuclear envelope (white arrow), and spindle pole bodies (yellow lines) are visible. Scale bar, 200 nm.
Adding extra synthetic kinetochores also increases microtubule number. ndc10-1 cells grown at 37˚C had only 32 ± 6 microtubules (Supplemental Figure S4), compared with 51 ± 1 in wild type (p = 0.03, Student's t test). Introducing three synthetic kinetochores increased the number of microtubules to an average of 50 ± 12 microtubules (p = 0.05, Student's t test). The synthetic kinetochore is created by Ask-LacI binding an array of 256 repeats of the Lac operator, and it may be capable of binding and stabilizing more than one microtubule per kinetochore, which would explain how three synthetic kinetochores could stabilize as many microtubules as wild-type cells.
We measured spindle length in the cells imaged by electron microscopy; the average spindle length of the cells with and without centromeric plasmids is not significantly different (p = 0.48 for 11 plasmids and 0.69 for 23 plasmids, Student's t test). At first glance, this may seem surprising, since the spindle lengths measured by light microscopy for the control cells and those that have extra kinetochores are significantly different (p << 10−6, Student's t test). The difference in significance arises because the number of spindles is so different in the two samples—four for each strain for electron microscopy, and hundreds for light microscopy. If we repeatedly take a sample of four spindle lengths for the two conditions (plasmids and no plasmids) from the light microscopy data, the difference is statistically significant (p < 0.05, Student's t test) in only 29% of the comparisons. Similarly, we failed to see any shortening of the kinetochore microtubules as extra kinetochores were added to wild-type cells, although the shortening due to the decrease in spindle length would be partially offset by the reduction in interkinetochore distance (Figure 5, G and H). The difficulty in collecting serial sections and reconstructing spindles means that an enormous effort would be required to collect enough cells to be reasonably sure of seeing a significant difference in the length of spindles or kinetochore microtubules between control cells and those with extra kinetochores.
DISCUSSION
We manipulated kinetochore number to test a simple model for spindle length regulation. Removing and adding kinetochores elongated and shortened spindles, respectively, showing that kinetochores generate inward force and at least one spindle force is length dependent. Adding extra kinetochores to wild-type spindles showed that kinetochore number regulates spindle microtubule number.
Force balance regulates spindle length
We tested the relationship between kinetochore number and spindle length. Destroying all kinetochores (ndc10-1) or removing cohesin led to similar spindle elongation (Figure 2C; Stephens et al., 2011). The elongation of the kinetochore-deficient spindle was suppressed when Kip1 (a kinesin-5 motor) was deleted (Figure 2C). Adding kinetochores shortened spindles; synthetic kinetochores shortened ndc10-1 spindles (Figure 4C), and extra kinetochores shortened wild-type spindles (Figure 5, C and D), suggesting that paired sister kinetochores provide the inward force that restrains spindle elongation. Our work extends studies in fission yeast (Goshima et al., 1999) and human cells (DeLuca et al., 2002) that show that inhibiting kinetochores or cohesin elongates spindles.
Other studies on budding yeast reached similar conclusions. Bouck and Bloom (2007) proposed a balance between outward force generated by motors and inward force from spring-like pericentric chromatin. Both cohesin and condensin were reported to control the stiffness (Stephens et al., 2011) of a nonlinear spring (Stephens et al., 2013). Showing that spindle length varies with kinetochore number provides further evidence for the existence of length-dependent forces and their role in setting spindle length.
Early cytological studies on meiotic cells support the role of kinetochores and chromosomal attachments in regulating spindle assembly and length. In insect spermatocytes, chromosomes trigger spindle assembly (Zhang and Nicklas, 1995), control microtubule number (Zhang and Nicklas, 1995), and affect spindle length (LaFountain, 1972), even though assembled spindles can survive without chromosomes (Zhang and Nicklas, 1996). Studies on chromosome positions suggested the existence of length-dependent forces (Hays et al., 1982) whose size depended on microtubule number (Hays and Salmon, 1990).
Earlier force balance models proposed antagonism between plus and minus end–directed microtubule motors as the source of opposing forces (Saunders et al., 1997; Troxell et al., 2001; Civelekoglu-Scholey et al., 2010). Our results show that reducing and increasing kinetochore number lengthens and shortens the spindle, revealing that models that rely only on opposing classes of motors acting on interpolar microtubules are too simple and that forces exerted by kinetochore microtubules must also be considered.
Our results do not reveal which forces are length dependent or the molecular mechanism that produces length dependence. The pushing force generated by the action of plus end–directed motors, such as Cin8 and Kip1, should depend on the length of the overlap zone between antiparallel microtubules in the central spindle, but roles have also been proposed for other molecules, including Kip3, a plus end–directed motor (Varga et al., 2006, 2009; Syrovatkina et al., 2013), and Ase1, a microtubule-bundling protein (Braun et al., 2011; Syrovatkina et al., 2013).
The congression of chromosomes to a position equally far from either pole suggests that length- or position-dependent forces must also act on chromosomes; if they did not, a pair of sister chromatids would show a random walk along the axis between the spindle poles. Length-dependent microtubule depolymerization by Kip3 is one model for a length-dependent poleward force on kinetochores, but other possibilities also exist (Gardner et al., 2005, 2008).
Different force models are likely to apply to different spindles. In budding yeast, kinetochore microtubules are five times more common than interpolar microtubules, but in many animal cells, most microtubules do not contact kinetochores, implying that other interactions regulate spindle length. A balance between dynein, a minus end–directed motor, and Eg5, a plus end–directed motor, helps set the length of Xenopus (Uteng et al., 2008) and human (Tanenbaum et al., 2008) spindles, whereas embryonic Drosophila spindles are regulated by a balance between two antagonistic kinesins that act on antiparallel microtubules (Sharp et al., 2000).
Other models of spindle length regulation
We verified the predictions of a model based on length-dependent forces (Figure 1). Although we did not attempt to explicitly rule out models based on molecular rulers, gradients, or limiting components, our results argue against simple forms of these models. A homodimeric coiled-coil that reached from the spindle pole to the spindle midpoint 1 μm away would need to be ∼7000 amino acids long, whereas the largest yeast protein, Rea1, is only 4910 amino acids long (Garbarino and Gibbons, 2002); thus no one yeast protein is long enough to serve as a ruler. The spindle matrix, which has been described as a ruler (Johansen et al., 2011), is a poorly understood polymeric structure. There is no proposal for what gives it a defined size, and no matrix has been found in yeast. The spindle's ability to elongate upon kinetochore destruction (Figure 2C and Supplemental Figure S1C) and continued nuclear envelope synthesis in mitosis (Witkin et al., 2012) argue against physical restraint by a matrix or the nuclear envelope.
Various gradients are proposed to regulate spindle length. In C. elegans, a gradient of the TPXL-1 protein emanates from centrosomes (Greenan et al., 2010); reducing centrosome size alters the gradient and shortens the spindle. In our study, however, cells with larger spindle poles bodies have shorter spindles (Figure 6D). Our results also suggest that gradients formed by chromatin, such as the Ran-GTP gradient (Carazo-Salas et al., 2001; Wilde et al., 2001; Bastiaens et al., 2006), are unlikely to play an important role in budding yeast, since spindle length varies widely in cells containing similar amounts of chromatin. Inactivating kinetochores elongates the spindle, even though the amount of chromatin and chromosome number were unchanged (Figures 2C and 4C), and adding 20 centromeric plasmids increased the amount of nuclear DNA by only 2.5% but shortened the spindle by 31% (Figure 5C). We cannot rule out gradients emanating from kinetochores, since we manipulated kinetochore number. Eliminating this possibility would require manipulating every molecule that might create such a gradient. One known kinetochore-based signaling pathway, the spindle checkpoint, was broken in the experiments with extra kinetochores, arguing that the final output of the checkpoint does not control spindle length or microtubule number.
Spindle length has been shown to scale with cell size in developing organisms (Wühr et al., 2008; Hara and Kimura, 2009), and this is believed to be due to limiting amounts of spindle components. In vitro–assembled spindles scale with the volume of cytoplasm, with shorter spindles built in smaller volumes (Good et al., 2013; Hazel et al., 2013). Whereas limiting amounts of spindle materials, such as tubulin, may dictate how large a spindle can be assembled in Xenopus, in which >50% of cytoplasmic tubulin is polymerized into spindles (Good et al., 2013), this does not appear to be the case for budding yeast. Reducing total tubulin levels by half does not affect S. cerevisiae’s ability to grow, establish a spindle, or segregate chromosomes, and these cells do not up-regulate tubulin expression in response to tubulin depletion (Katz et al., 1990). Reducing cellular tubulin by 80% is required to produce a reduction in anaphase spindle length (Lacefield et al., 2006). In addition, studies with Kip3 also demonstrate that tubulin is not limiting; when Kip3 is deleted, the late-anaphase spindle elongates dramatically compared with wild-type cells (Rizk et al., 2014). Limited amounts of tubulin are not being redistributed from astral microtubules to the spindle, because astral microtubules also increase fourfold in length in kip3∆ cells (Cottingham and Hoyt, 1997; Miller et al., 1998; Rizk et al., 2014). In our experiments, the total mass of tubulin polymerized into microtubules increased in cells containing many minichromosomes: cells with 23 plasmids contained 40% more tubulin polymer than wild type (p = 0.05, Student's t test), demonstrating that extra tubulin is available to incorporate into spindle microtubules. In addition, our cells with extra kinetochores showed normal cell cycle progress with no detectable pause to accumulate additional protein (Supplemental Figure S5B). It is unknown what percentage of available tubulin is polymerized into the budding yeast spindle, but given our results and previous studies on tubulin depletion (Katz et al., 1990) and Kip3 (Rizk et al., 2014), it is unlikely that limiting levels of tubulin set spindle length in this organism. In addition, because our spindles did not show a conservation of spindle mass in the presence of extra kinetochores, it is unlikely that spindle length models based on mass balance and depletion of other limiting components (Reber et al., 2013) regulate the length of the budding yeast spindle.
We measured the effect of kinetochore number on spindle length without altering ploidy. Previous work showed that haploid, diploid, triploid, and tetraploid cells all had the same spindle length despite different numbers of chromosomal attachments (Lin et al., 2001; Storchová et al., 2006). Perhaps the number of kinetochore and interpolar microtubules both increase with ploidy, maintaining the balance between inward and outward forces on the spindle without changing its length. By manipulating the number of attachments in haploid cells, we avoided any possible confounding variables produced by changing ploidy.
Kinetochores increase microtubule number
We generated yeast cells with up to 30 extra centromeric plasmids per cell. Although early studies led to the idea that extra centromeres titrate away limited amounts of kinetochore proteins or microtubules (Futcher and Carbon, 1986; Runge et al., 1991), Wells and Murray (1996) showed that deleting the spindle checkpoint abolished the cell cycle delay and lethality caused by extra centromeres. We showed that cells with >30 extra centromeric plasmids can still biorient their chromosomes (Figure 5, G and H), go through anaphase (Figure 5E), segregate plasmids (Supplemental Figure S5A), grow at normal rates, and produce viable daughters (Supplemental Figure S5B), suggesting that kinetochore and spindle pole body components do not limit the number of functional kinetochores that a yeast cell can segregate. High-resolution electron tomography showed that haploid yeast spindle pole bodies could increase in size and produce more microtubules to meet this higher demand (Figure 6C). This phenomenon is different from the increase in spindle pole body size and microtubule number seen on prolonged arrest of cdc20 mutants at 37°C (O’Toole et al., 1997), since the increases we see are related to the number of kinetochores that cells contain rather than being constitutive.
It has been suggested that the spindle checkpoint regulates the number of microtubules in the budding yeast spindle; since the checkpoint senses unattached kinetochores, it could induce extra microtubule nucleation to increase the chance of kinetochore capture (Winey and Bloom, 2012). Our cells lacked Mad2, a checkpoint protein, but increased kinetochore microtubule number to roughly match kinetochore number. We can imagine three ways of explaining the correlation between kinetochore and microtubule number: spindle pole bodies nucleate more microtubules than are seen in wild-type spindles, but only those that attach to kinetochores are stable and seen by electron microscopy; kinetochores communicate with spindle pole bodies via a checkpoint-independent mechanism that regulates microtubule nucleation; or there is selection on epigenetically heritable variation in the nucleation capacity. We cannot exclude the last possibility, since we gradually elevated the copy number of the centromeric plasmids and could have been selecting the small fraction of cells that could expand their spindle pole bodies and transmit this larger size during cell division.
Our work supports a simple model for spindle length regulation by a balance of length-dependent forces and shows that kinetochores regulate microtubule number in the budding yeast spindle. Budding yeast spindles are small and have close to the minimal number of microtubules required to segregate the cell's chromosomes, whereas animals and plants have larger spindles with many more microtubules. Balancing length-dependent forces that pull kinetochores toward the spindle poles and those that push the poles away from each other may be the ancestral solution to regulating spindle length, and higher eukaryotes may have evolved additional regulation to organize their larger and more complex spindles.
MATERIALS AND METHODS
Yeast strains and cell culturing
Strains used in this study are listed in Supplemental Table S1. All yeast strains were constructed in W303 background (ade2-1 his3-11,15 leu2-3112 trp1-1 ura3-1 can1-100) and were constructed using standard genetic techniques. Arrays containing 256 repeats of the lac operator used to label chromosomes or create synthetic kinetochores were placed near centromeres at the following genetic locations: chromosome III, 116002–117002; chromosome VIII, 123761–124110; and chromosome XV, 325382–326387 (Supplemental Table S1). High-copy centromeric plasmid strains were constructed by transformation of the plasmid, followed by growth in CuSO4. All experiments with extra kinetochores were conducted in checkpoint-deficient (mad2∆) cells to allow the addition of up to 30 extra kinetochores without delaying progress through mitosis. Growth in successively higher CuSO4 concentration drives up the copy number of the plasmid; cultures began in 0.01 mM CuSO4 and were passaged until resistant to 2.0–4.0 mM CuSO4. All media was prepared by standard recipes (Sherman et al., 1986) and contained 2% (wt/vol) of specified sugar. Cells were grown in either yeast extract/peptone/dextrose (YPD; 2% glucose) or synthetic complete medium (2% glucose) without methionine (SC-Met) at 23°C for temperature-sensitive strains or 30°C for all others. PGAL1-MCD1 cells were grown in SC-Met plus 2% galactose. YPD containing 1-(butylcarbamoyl)-2-benzimidazolecarbamate (benomyl) and nocodazole was prepared by heating YPD to 65°C and adding dimethyl sulfoxide (DMSO) 10 mg/ml stocks of benomyl dropwise to a final concentration of 30 μg/ml; medium was cooled to 37°C for dropwise addition of DMSO 10 mg/ml stock of nocodazole to a final concentration of 30 μg/ml. All drugs and chemicals were purchased from Sigma-Aldrich.
Fluorescence microscopy imaging
Images were acquired at room temperature (25°C) using a Nikon Eclipse Ti-E inverted microscope with a 60× Plan Apo VC, 1.4 numerical aperture oil objective lens with a Photometrics CoolSNAP HQ camera (Roper Scientific). MetaMorph 7.7 (Molecular Devices) was used to acquire z-series image stacks with z-step size of 200 nm and 21 total z-planes. Spindle pole bodies were labeled with mCherry (Spc42-mCherry), chromosomes were labeled with GFP by a 256 LacO repeat array inserted 200 base pairs downstream from CEN15 (Supplemental Table S1), and expression of monomeric yeast optimized GFP fused to LacI. Some strains (Supplemental Table S1) carried monomeric yeast optimized GFP fused to the TUB1 gene to label spindles. Fixed samples were imaged in 1.2 M sorbitol plus 0.1 M KH2PO4, pH 8.5, buffer on concanavalin A–coated coverslips (VWR) adhered to glass slides (Corning). Exposure times were 10 ms for differential interference contrast images and 300 ms for fluorescence images.
Live-cell imaging
Video microscopy was performed as described, except that cells were not fixed and were imaged in low-fluorescence SC medium. G1-arrested cells were washed three times with SC medium to remove α-factor, adhered to concanavalin A–coated coverslips, and imaged for up to 3 h. For anaphase experiments (Supplemental Figure S3), images were collected every 5 min. For metaphase arrest experiments (Figure 5, E and F), images were collected every 10 min. for experiments in which wild-type and plasmid-containing cells were imaged together (Supplemental Figure S3), wild-type cells walls were labeled with Texas red dye (Life Technologies). Cells were washed twice with 1× phosphate-buffered saline, resuspended in 0.1 M NaHCO3, pH 8.3, incubated for 5 min with 10 mg/ml DMSO stocks of Texas red at a final concentration of 10 μg/ml, quenched by washing three times with 0.1 M NaHCO3, pH 8.3, plus 200 μM lysine, and washed twice with SC medium. Plasmid-containing cells were mock labeled by incubating with DMSO alone and washed as described.
Electron microscopy and 3D spindle reconstruction
Strains were prepared for electron microscopy as previously described (Giddings et al., 2001). Aliquots from liquid cultures were collected onto a 0.45-μm Millipore filter by vacuum filtration, loaded into freezer hats, and frozen using a Wohlwend Compact 02 high-pressure freezer. The frozen samples were freeze substituted in 0.25% glutaraldehyde and 0.1% uranyl acetate in acetone for 3 d at −90°C. The samples were then warmed to −20°C, rinsed in acetone, and embedded in Lowicryl HM20 resin. Serial sections (250 nm) were collected onto Formvar-coated slot grids and poststained using 2% aqueous uranyl acetate, followed by lead citrate. Colloidal gold particles (15 nm) were affixed to the sections to serve as alignment markers.
Tomography was performed as previously described (Giddings et al., 2001; O’Toole et al., 2002). Dual-axis tilt series data were collected using a Tecnai F20 or F30 intermediate electron microscope operating at 200 or 300 kV, respectively. The SerialEM program (Mastronarde, 2005) was used to automatically acquire images every 1° over a ±60° range. Data were acquired at a pixel size of 1–1.5 nm using a Gatan charge-coupled device camera. The tilt series images were aligned and tomograms computed using the IMOD software package (Mastronarde, 1997). The mitotic spindles examined spanned two to five serial sections. Tomograms were computed from each section and then joined to produce the final volume of the spindle. In total, spindles from four wild-type, four 11-plasmid, four 23-plasmid, three ndc10-1, and three ndc10-1 + 3 synthetic kinetochores cells were reconstructed.
Tomograms were displayed and modeled using the 3dmod program in the IMOD software package (Kremer et al., 1996). Spindle microtubules originating from each pole were modeled in the tomographic volume. The core bundle microtubules were identified using the mtpairing program in the IMOD software package described in Winey et al. (1995). Supplemental movies show example raw EM images and 3D spindle reconstruction from these images.
Spindle checkpoint activation by benomyl and nocodazole
Strains were grown in YPD at the permissive temperature of 23°C and maintained in log phase for 24 h before the experiment. Log-phase cells (∼5 × 106 cells/ml) were arrested in G1 with 10 μg/ml α-factor (Bio-Synthesis) for 2.5 h at 23°C and then moved to 37°C for 30 min. After confirmation of arrest by light microscopy, cells were washed three times with YPD at 37°C to remove α-factor and resuspended in YPD plus 30 μg/ml benomyl plus 30 μg/ml nocodazole. Cells were grown for 4 h at 37°C, sonicated to separate them, and then scored for cell morphology by light microscopy. Large-budded cells are indicative of spindle checkpoint–induced mitotic arrest.
Measuring metaphase spindle length, chromatin stretch, and biorientation
Strains were grown to log phase in SC-Met medium at either 23°C for experiments with temperature-sensitive alleles or 30°C for experiments with centromeric plasmids. For PGAL1-MCD1 experiments, cells were grown to log phase at 30°C in SC-Met plus 2% galactose. Log- phase cells (∼5 × 106 cells/ml) were arrested in G1 by α-factor treatment (10 μg/ml). In ndc10-1 experiments, cells were arrested at 23°C for 3 h, methionine was added (500 μg/1 ml) to induce Cdc20 deletion, and then cells were incubated for an additional 1 h at 37°C. Cells were washed three times at 37°C with YPD plus methionine and grown 3 h in YPD plus methionine at 37°C. For experiments with centromeric plasmids and PGAL1-MCD1, strains were arrested with α-factor at 30°C for 3 h, with methionine addition after 2.5 h. Cells were washed three times and released into YPD plus methionine and grown at 30°C for 3 h. In all experiments, after 3 h postrelease, cells were fixed with 10% Formalin (final concentration of 1%) for 10 min, washed with 0.1 M KH2PO4, pH 8.5, washed with 1.2 M sorbitol plus 0.1 M KH2PO4, pH 8.5, resuspended in 1.2 M sorbitol plus 0.1 M KH2PO4, pH 8.5, and stored at 4°C.
Samples were imaged as described. Spindle length was calculated as the 3D distance between mCherry-labeled spindle pole bodies, and chromatin stretch distance was calculated as the 3D distance between GFP-chromatin dots. In brief, we used the MOSAIC 3D Single-Particle Tracking ImageJ software (Sbalzarini and Koumoutsakos, 2005) to achieve subpixel and sub-z-plane resolution of spindle pole body or chromatid location in x, y, z dimensions. The subpixel x, y-location of the particle (spindle pole body or chromatid) was determined by fitting the pixel of peak fluorescence intensity and its surrounding pixels with a uniform Gaussian; x- and y- coordinates were the peak values of the Gaussian. To determine the subplane z-position of the particle, a uniform Gaussian was fitted to the peak plane and its surrounding planes (Sbalzarini and Koumoutsakos, 2005). Once the x-, y-, and z-coordinates of the particles were determined, a MATLAB (MathWorks) script was used to pair the nearest-neighbor particles and calculate distance between the two paired coordinates. Given the average signal-to-noise ratio in our fluorescence images, particle position can theoretically be determined with accuracy to within 1 nm (Sbalzarini and Koumoutsakos 2005); however this is under the assumption that the particle is smaller than the resolution of light. Spindle lengths >7 μm were not included in calculated averages; a small fraction of spindles escaped metaphase arrest, and these anaphase spindles were characterized by lengths ≥7 μm and were thus excluded from calculations. The percentage of biorientated chromosomes was calculated as the number of chromosomes with two resolvable GFP dots out of all chromosomes scored.
Quantifying plasmid copy number
Centromeric plasmid copy was determined by quantitative PCR. Genomic DNA was purified from cells and quantified using Qubit 2.0 Fluorometer (Invitrogen). Quantitative PCR was performed with 0.5 ng of template DNA, 0.4 mM of forward and reverse primers, and PerfeCta SYBR Green FastMix/Rox master mix in a 7900 Real-Time PCR machine (Applied Biosystems), with an annealing temperature of 55°C. Control primers (forward, 5′-GTTTAATCCGGGCTGGTTCCAT-3′; reverse, 5′-TAGACCCAGTGGACAGATAGCG-3′) amplified a 113–base pair fragment of gene ALG9, and experimental primers (forward:, 5′-GGAAAAAAAGCACTACCTAGGAGCGGCC-3′; reverse, 5′-CTGTGACGATAAAACCGGAAGGAAG-3′) amplified 127 base pairs near CEN4, the centromere on the plasmid.
Plasmid loss assay
Strains were created that contained a tester plasmid and multiple CUP1 centromeric plasmids (plasmids used to introduce extra kinetochores). High copy number of the CUP1 plasmid was selected by successive growth in CuSO4, and the presence of the tester plasmid (which contains the URA3 gene) was maintained by growth in medium lacking uracil. Cells were grown in SC-URA-MET plus CuSO4 and maintained in log phase for 24 h before the experiment. Cells were backdiluted to 1 × 106 cells/ml and grown in nonselective SC-MET for 9 h; ∼500 cells were plated on SC-MET and SC-MET-URA. The number of colonies on each plate type and the percentage of cells containing the plasmid were calculated by dividing the number of colonies on SC-MET-URA by the number of colonies on SC-MET (Tsai et al., 2006). The loss rate of a tester plasmid without a centromere was also calculated as a reference for high loss rates; this plasmid contained the LEU2 gene, so cells were grown selectively in medium lacking leucine and plated on SC-MET and SC-MET-LEU to calculate loss rate.
Growth curves and calculation of doubling time
Cells were grown in SC-MET and maintained in log phase for 24 h before the experiment. Log-phase cultures were backdiluted to 1 × 106 cells/ml and allowed to grow for 10 h. Time points were taken every 2 h, and culture density was measured with a Coulter counter. Doubling time was calculated using the linear regression of the log of cell density over time.
Supplementary Material
Acknowledgments
We thank T. H. Giddings, Jr., and C. Clarissa of the Molecular, Cellular, and Developmental Biology EM Suite for freezing and preparing samples for electron microscopy and the Boulder Laboratory for 3D Electron Microscopy of Cells for use of their equipment, as well as A. Amon, S. Biggins, V. Denic, T. Mitchison, E. Salmon, and members of the Murray lab for comments on the manuscript. This study was supported by a National Science Foundation Graduate Research Fellowship (DGE0946799 and DGE0644491) to N.J.N. and National Institute of General Medical Sciences Grants GM043987 to A.W.M. and 8P41GM103431 to the Boulder Laboratory for 3D Electron Microscopy of Cells to E.O.T.
Abbreviations used:
- APC
anaphase-promoting complex
- GFP
green fluorescence protein
- KT
kinetochore
- LacI
lactose repressor
- LacO
lactose operator
- sKT
synthetic kinetochore.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-01-0016) on October 15, 2014.
REFERENCES
- Bastiaens P, Caudron M, Niethammer P, Karsenti E. Gradients in the self-organization of the mitotic spindle. Trends Cell Biol. 2006;16:125–134. doi: 10.1016/j.tcb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Bloom KS, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–3017. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Bloom K, Joglekar A. Towards building a chromosome segregation machine. Nature. 2010;463:446–456. doi: 10.1038/nature08912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck DC, Bloom KS. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc Natl Acad Sci. 2007;102:5408–5413. doi: 10.1073/pnas.0405925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun M, Lansky Z, Fink G, Ruhnow F, Diez S, Janson ME. Adaptive braking by Ase1 prevents overlapping microtubules from sliding completely apart. Nat Cell Biol. 2011;13:1259–1264. doi: 10.1038/ncb2323. [DOI] [PubMed] [Google Scholar]
- Brugués J, Nuzzo V, Mazur E, Needleman DJ. Nucleation and transport organize microtubules in metaphase spindles. Cell. 2012;149:554–564. doi: 10.1016/j.cell.2012.03.027. [DOI] [PubMed] [Google Scholar]
- Carazo-Salas RE, Gruss OJ, Mattaj IW, Karsenti E. Ran–GTP coordinates regulation of microtubule nucleation and dynamics during mitotic-spindle assembly. Nat Cell Biol. 2001;3:228–234. doi: 10.1038/35060009. [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Tao L, Brust-Mascher I, Wollman R, Scholey JM. Prometaphase spindle maintenance by an antagonistic motor-dependent force balance made robust by a disassembling lamin-B envelope. J Cell Biol. 2010;188:49–68. doi: 10.1083/jcb.200908150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Cottingham FR, Hoyt MA. Mitotic spindle positioning in Saccharomyces cerevisiae is accomplished by antagonistically acting microtubule motor proteins. J Cell Biol. 1997;138:1041–1053. doi: 10.1083/jcb.138.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtois A, Schuh M, Ellenberg J, Hiiragi T. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J Cell Biol. 2012;198:357–370. doi: 10.1083/jcb.201202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon E. hNuf2 inhibition blocks stable kinetochore–microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–555. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Mitchison TJ. Force and length in the mitotic spindle. Curr Biol. 2009;19:R749–R761. doi: 10.1016/j.cub.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futcher B, Carbon J. Toxic effects of excess cloned centromeres. Mol Cell Biol. 1986;6:2213–2222. doi: 10.1128/mcb.6.6.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino JE, Gibbons I. Expression and genomic analysis of midasin, a novel and highly conserved AAA protein distantly related to dynein. BMC Genomics. 2002;3:18. doi: 10.1186/1471-2164-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Bouck DC, Paliulis LV, Meehl JB, O’Toole ET, Haase J, Soubry A, Joglekar AP, Winey M, Salmon ED. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 2008;135:894–906. doi: 10.1016/j.cell.2008.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MK, Pearson CG, Sprague BL, Zarzar TR, Bloom K, Salmon E, Odde DJ. Tension-dependent regulation of microtubule dynamics at kinetochores can explain metaphase congression in yeast. Mol Biol Cell. 2005;16:3764–3775. doi: 10.1091/mbc.E05-04-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner RD, Poddar A, Yellman C, Tavormina PA, Monteagudo MC, Burke DJ. The spindle checkpoint of the yeast Saccharomyces cerevisiae requires kinetochore function and maps to the CBF3 domain. Genetics. 2001;157:1493–1502. doi: 10.1093/genetics/157.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings T, O Toole ET, Morphew M, Mastronarde DN, McIntosh JR, Winey M. Using rapid freeze and freeze-substitution for the preparation of yeast cells for electron microscopy and three-dimensional analysis. Methods Cell Biol. 2001;67:27–41. doi: 10.1016/s0091-679x(01)67003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good MC, Vahey MD, Skandarajah A, Fletcher DA, Heald R. Cytoplasmic volume modulates spindle size during embryogenesis. Science. 2013;342:856–860. doi: 10.1126/science.1243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Saitoh S, Yanagida M. Proper metaphase spindle length is determined by centromere proteins Mis12 and Mis6 required for faithful chromosome segregation. Genes Dev. 1999;13:1664–1677. doi: 10.1101/gad.13.13.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Scholey JM. Control of mitotic spindle length. Annu Rev Cell Dev Biol. 2010;26:21–57. doi: 10.1146/annurev-cellbio-100109-104006. [DOI] [PubMed] [Google Scholar]
- Goshima G, Yanagida M. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 2000;100:619–633. doi: 10.1016/s0092-8674(00)80699-6. [DOI] [PubMed] [Google Scholar]
- Greenan G, Brangwynne CP, Hyman AA. Centrosome size sets mitotic spindle length. Curr Biol. 2010;20:353–358. doi: 10.1016/j.cub.2009.12.050. [DOI] [PubMed] [Google Scholar]
- Hara Y, Kimura A. Cell-size-dependent spindle elongation in the Caenorhabditis elegans early embryo. Curr Biol. 2009;19:1549–1554. doi: 10.1016/j.cub.2009.07.050. [DOI] [PubMed] [Google Scholar]
- Hara Y, Kimura A. An allometric relationship between mitotic spindle width, spindle length, and ploidy in Caenorhabditis elegans embryos. Mol Biol Cell. 2013;24:1411–1419. doi: 10.1091/mbc.E12-07-0528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Mortimer RK, Culotti J, Culotti M. Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985;110:381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays T, Salmon E. Poleward force at the kinetochore in metaphase depends on the number of kinetochore microtubules. J Cell Biol. 1990;110:391–404. doi: 10.1083/jcb.110.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays TS, Wise D, Salmon E. Traction force on a kinetochore at metaphase acts as a linear function of kinetochore fiber length. J Cell Biol. 1982;93:374–382. doi: 10.1083/jcb.93.2.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazel J, Krutkramelis K, Mooney P, Tomschik M, Gerow K, Oakey J, Gatlin JC. Changes in cytoplasmic volume are sufficient to drive spindle scaling. Science. 2013;342:853–856. doi: 10.1126/science.1243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, He L, Loo KK, Saunders WS. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. J Cell Biol. 1992;118:109–120. doi: 10.1083/jcb.118.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen KM, Forer A, Yao C, Girton J, Johansen J. Do nuclear envelope and intranuclear proteins reorganize during mitosis to form an elastic, hydrogel-like spindle matrix. Chromosome Res. 2011;19:345–365. doi: 10.1007/s10577-011-9187-6. [DOI] [PubMed] [Google Scholar]
- Kalab P, Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Najarian R, Haslinger A, Valenzuela P, Welch J, Fogel S. Primary structure and transcription of an amplified genetic locus: the CUP1 locus of yeast. Proc Natl Acad Sci USA. 1984;81:337–341. doi: 10.1073/pnas.81.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura I. Determination of bacteriophage lambda tail length by a protein ruler. Nature. 1987;327:73–75. doi: 10.1038/327073a0. [DOI] [PubMed] [Google Scholar]
- Katsura I. Mechanism of length determination in bacteriophage lambda tails. Adv Biophys. 1990;26:1–18. doi: 10.1016/0065-227x(90)90004-d. [DOI] [PubMed] [Google Scholar]
- Katz W, Weinstein B, Solomon F. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: consequences of altered tubulin gene copy number. Mol Cell Biol. 1990;10:5286–5294. doi: 10.1128/mcb.10.10.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiermaier E, Woehrer S, Peng Y, Mechtler K, Westermann S. A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast. Nat Cell Biol. 2009;11:1109–1115. doi: 10.1038/ncb1924. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Labeit S, Gibson T, Lakey A, Leonard K, Zeviani M, Knight P, Wardale J, Trinick J. Evidence that nebulin is a protein-ruler in muscle thin filaments. FEBS Lett. 1991;282:313–316. doi: 10.1016/0014-5793(91)80503-u. [DOI] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B. The complete primary structure of human nebulinand its correlation to muscle structure. J Mol Biol. 1995;248:308–315. doi: 10.1016/s0022-2836(95)80052-2. [DOI] [PubMed] [Google Scholar]
- Lacefield S, Lau DT, Murray AW. Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast. Nat Cell Biol. 2009;11:1116–1120. doi: 10.1038/ncb1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacefield S, Magendantz M, Solomon F. Consequences of defective tubulin folding on heterodimer levels, mitosis and spindle morphology in Saccharomyces cerevisiae. Genetics. 2006;173:635–646. doi: 10.1534/genetics.105.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFountain J. Spindle shape changes as an indicator of force production in crane-fly spermatocytes. J Cell Sci. 1972;10:79–93. doi: 10.1242/jcs.10.1.79. [DOI] [PubMed] [Google Scholar]
- Lin H, de Carvalho P, Kho D, Tai C, Pierre P, Fink GR, Pellman D. Polyploids require Bik1 for kinetochore–microtubule attachment. J Cell Biol. 2001;155:1173–1184. doi: 10.1083/jcb.200108119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF. Cellular length control systems. Annu Rev Cell Dev Biol. 2004;20:677–693. doi: 10.1146/annurev.cellbio.20.012103.094437. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Dual-axis tomography: an approach with alignment methods that preserve resolution. J Struct Biol. 1997;120:343. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Miller RK, Heller KK, Frisen L, Wallack DL, Loayza D, Gammie AE, Rose MD. The kinesin-related proteins, Kip2p and Kip3p, function differently in nuclear migration in yeast. Mol Biol Cell. 1998;9:2051–2068. doi: 10.1091/mbc.9.8.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole E, Mastronarde D, Giddings T, Jr, Winey M, Burke D, McIntosh J. Three-dimensional analysis and ultrastructural design of mitotic spindles from the cdc20 mutant of Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:1. doi: 10.1091/mbc.8.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole ET, Winey M, McIntosh JR, Mastronarde DN. Electron tomography of yeast cells. Methods Enzymol. 2002;351:81–96. doi: 10.1016/s0076-6879(02)51842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Maddox PS, Zarzar TR, Salmon ED, Bloom K. Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol Biol Cell. 2003;14:4181–4195. doi: 10.1091/mbc.E03-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber SB, Baumgart J, Widlund PO, Pozniakovsky A, Howard J, Hyman AA, Jülicher F. XMAP215 activity sets spindle length by controlling the total mass of spindle microtubules. Nat Cell Biol. 2013;15:1116–1122. doi: 10.1038/ncb2834. [DOI] [PubMed] [Google Scholar]
- Rizk RS, Discipio KA, Proudfoot KG, Gupta ML The kinesin-8 Kip3 scales anaphase spindle length by suppression of midzone microtubule polymerization. J Cell Biol. 2014;204:965–975. doi: 10.1083/jcb.201312039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romao M, Tanaka K, Sibarita J, Ly-Hartig NTB, Tanaka TU, Antony C. Three-dimensional electron microscopy analysis of ndc10-1 mutant reveals an aberrant organization of the mitotic spindle and spindle pole body defects in Saccharomyces cerevisiae. J Struct Biol. 2008;163:18–28. doi: 10.1016/j.jsb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Runge KW, Wellinger RJ, Zakian VA. Effects of excess centromeres and excess telomeres on chromosome loss rates. Mol Cell Biol. 1991;11:2919–2928. doi: 10.1128/mcb.11.6.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders WS, Hoyt MA. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 1992;70:451–458. doi: 10.1016/0092-8674(92)90169-d. [DOI] [PubMed] [Google Scholar]
- Saunders W, Lengyel V, Hoyt M. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol Biol Cell. 1997;8:1025. doi: 10.1091/mbc.8.6.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. J Struct Biol. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Schulman IG, Bloom K. Genetic dissection of centromere function. Mol Cell Biol. 1993;13:3156–3166. doi: 10.1128/mcb.13.6.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DJ, Brown HM, Kwon M, Rogers GC, Holland G, Scholey JM. Functional coordination of three mitotic motors in Drosophila embryos. Mol Biol Cell. 2000;11:241–253. doi: 10.1091/mbc.11.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Shonn MA, McCarroll R, Murray AW. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science. 2000;289:300–303. doi: 10.1126/science.289.5477.300. [DOI] [PubMed] [Google Scholar]
- Stephens AD, Haase J, Vicci L, Taylor RM, Bloom K. Cohesin, condensin, and the intramolecular centromere loop together generate the mitotic chromatin spring. J Cell Biol. 2011;193:1167–1180. doi: 10.1083/jcb.201103138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens AD, Haggerty RA, Vasquez PA, Vicci L, Snider CE, Shi F, Quammen C, Mullins C, Haase J, Taylor RM. Pericentric chromatin loops function as a nonlinear spring in mitotic force balance. J Cell Biol. 2013;200:757–772. doi: 10.1083/jcb.201208163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchová Z., Breneman A, Cande J, Dunn J, Burbank K, O’Toole E, Pellman D. Genome-wide genetic analysis of polyploidy in yeast. Nature. 2006;443:541–547. doi: 10.1038/nature05178. [DOI] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein–protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Straight AF, Sedat JW, Murray AW. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AG, Marshall WF, Sedat JW, Murray AW. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Syrovatkina V, Fu C, Tran PT. Antagonistic spindle motors and MAPs regulate metaphase spindle length and chromosome segregation. Curr Biol. 2013;23:2423–2429. doi: 10.1016/j.cub.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU. Bi-orienting chromosomes: acrobatics on the mitotic spindle. Chromosoma. 2008;117:521–533. doi: 10.1007/s00412-008-0173-5. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Galjart N, Medema RH. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 2008;27:3235–3245. doi: 10.1038/emboj.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teste M, Duquenne M, François J, Parrou J. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol Biol. 2009;10:99. doi: 10.1186/1471-2199-10-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell CL, Sweezy MA, West RR, Reed KD, Carson BD, Pidoux AL, Cande WZ, McIntosh JR. pkl1 andklp2: Two kinesins of the Kar3 subfamily in fission yeast perform different functions in both mitosis and meiosis. Mol Biol Cell. 2001;12:3476–3488. doi: 10.1091/mbc.12.11.3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M, Wang S, Heidinger JM, Shumaker DK, Adam SA, Goldman RD, Zheng Y. A mitotic lamin B matrix induced by RanGTP required for spindle assembly. Science. 2006;311:1887–1893. doi: 10.1126/science.1122771. [DOI] [PubMed] [Google Scholar]
- Uteng M, Hentrich C, Miura K, Bieling P, Surrey T. Poleward transport of Eg5 by dynein-dynactin in Xenopus laevis egg extract spindles. J Cell Biol. 2008;182:715–726. doi: 10.1083/jcb.200801125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Welch J, Fogel S, Cathala G, Karin M. Industrial yeasts display tandem gene iteration at the CUP1 region. Mol Cell Biol. 1983;3:1353–1361. doi: 10.1128/mcb.3.8.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells W, Murray AW. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Wilde A, Lizarraga SB, Zhang L, Wiese C, Gliksman NR, Walczak CE, Zheng Y. Ran stimulates spindle assembly by altering microtubule dynamics and the balance of motor activities. Nat Cell Biol. 2001;3:221–227. doi: 10.1038/35060000. [DOI] [PubMed] [Google Scholar]
- Winey M, Bloom K. Mitotic spindle form and function. Genetics. 2012;190:1197–1224. doi: 10.1534/genetics.111.128710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, McDonald KL, McIntosh JR. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin KL, Chong Y, Shao S, Webster MT, Lahiri S, Walters AD, Lee B, Koh JL, Prinz WA, Andrews BJ. The budding yeast nuclear envelope adjacent to the nucleolus serves as a membrane sink during mitotic delay. Curr Biol. 2012;22:1128–1133. doi: 10.1016/j.cub.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M, Chen Y, Dumont S, Groen AC, Needleman DJ, Salic A, Mitchison TJ. Evidence for an upper limit to mitotic spindle length. Curr Biol. 2008;18:1256–1261. doi: 10.1016/j.cub.2008.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Nicklas RB. Chromosomes initiate spindle assembly upon experimental dissolution of the nuclear envelope in grasshopper spermatocytes. J Cell Biol. 1995;131:1125–1131. doi: 10.1083/jcb.131.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Nicklas RB. “Anaphase”and cytokinesis in the absence of chromosomes. Nature. 1996;382:466–468. doi: 10.1038/382466a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.