Cds1 is an evolutionarily conserved CDP-DG synthase. Fission yeast are used to demonstrate that cells deficient in its function exhibit markedly increased triacylglycerol content and assemble unusual ER-associated lipid droplets that recruit the triacylglycerol synthesis machinery and grow by expansion.
Abstract
Excess fatty acids and sterols are stored as triacylglycerols and sterol esters in specialized cellular organelles, called lipid droplets. Understanding what determines the cellular amount of neutral lipids and their packaging into lipid droplets is of fundamental and applied interest. Using two species of fission yeast, we show that cycling cells deficient in the function of the ER-resident CDP-DG synthase Cds1 exhibit markedly increased triacylglycerol content and assemble large lipid droplets closely associated with the ER membranes. We demonstrate that these unusual structures recruit the triacylglycerol synthesis machinery and grow by expansion rather than by fusion. Our results suggest that interfering with the CDP-DG route of phosphatidic acid utilization rewires cellular metabolism to adopt a triacylglycerol-rich lifestyle reliant on the Kennedy pathway.
INTRODUCTION
Lipid droplets (LDs), the cellular neutral lipid storage organelles, are present in most eukaryotes. Triacylglycerols (TGs) and sterol esters (SEs) that constitute the LD core are mobilized to produce precursors for membrane lipid synthesis and to provide energy. The importance of LDs and LD-associated proteins is underscored by a host of human diseases related to abnormalities in neutral lipid storage. Furthermore, understanding the mechanisms underlying TG metabolism and storage in microorganisms and plants is crucial for biofuel and food security applications (reviewed in Fujimoto and Parton, 2011; also see Walther and Farese, 2009).
The organelle's central core is surrounded by a phospholipid monolayer (Tauchi-Sato et al., 2002; Bartz et al., 2007). A number of proteins localize to the outer lipid layer, with a subset of associated proteins executing the LD-specific functions, from neutral lipid biosynthesis to utilization (Athenstaedt et al., 1999; Buhman et al., 2001; Beller et al., 2006; Daum et al., 2007; Goodman, 2009). The LDs also appear to function as storage depots for cellular proteins that are not directly involved in lipid metabolism (Cermelli et al., 2006). Specific surface-associated proteins operate in LD trafficking (Savage et al., 1987; Welte et al., 2005) and promote interactions with other cellular organelles, including the endoplasmic reticulum (ER), peroxisomes, mitochondria, and vacuoles (Beller et al., 2006; Binns et al., 2006; Sturmey et al., 2006; Murphy et al., 2009; van Zutphen et al., 2014).
It is generally believed that LDs are generated at the ER, although the exact mechanism of their biogenesis is debated. A popular model proposes that neutral lipids accumulate as discrete “lenses” between the leaflets of the ER membrane bilayer, eventually budding outward into the cytoplasm and forming mature LDs coated with a phospholipid monolayer originating from the outer leaflet (Murphy and Vance, 1999). Alternative hypotheses envisage LD biogenesis through a “bicelle” intermediate, with both inner and outer membrane leaflets contributing to the phospholipid shell (Ploegh, 2007), or LD budding from the ER with the involvement of the vesicle trafficking machinery (Guo et al., 2008; Walther and Farese, 2009).
At least two mechanisms could account for LD enlargement: fusion of smaller LDs and de novo expansion of an individual LD. The former mechanism has been proposed to depend on the phospholipid content of the droplet shell, with higher levels of phosphatidic acid (PA) or lower amounts of phosphatidylcholine (PC) promoting fusion events (Guo et al., 2008; Fei et al., 2011; Krahmer et al., 2011; Penno et al., 2013). However, LD growth by expansion appears to be a more commonly realized scenario, with neutral lipids synthesized directly at the sites of the LD-ER contact or delivered into the lipid core of LDs after being produced in the ER (Fujimoto et al., 2007; Kuerschner et al., 2008; Jacquier et al., 2011; Xu et al., 2012). Although the molecular details of this process have not been worked out in sufficient detail, recent evidence suggests that enzymes responsible for neutral lipid production could relocate from the ER to LDs and synthesize neutral lipids in situ (Jacquier et al., 2011; Xu et al., 2012; Wilfling et al., 2013). Of interest, the emergence and dynamics of LDs appear to depend on the cell cycle stage (Long et al., 2012).
Work on the model budding yeast Saccharomyces cerevisiae has contributed greatly to our understanding of LD biogenesis and metabolism (for review, see Kohlwein et al., 2013). Yeast LD core consists of virtually equal amounts of TGs and SEs (Czabany et al., 2008; Connerth et al., 2009), with dynamic distribution of fatty acid species that vary depending on carbon source, growth conditions, and the presence of externally supplemented fatty acids (Connerth et al., 2009; Klose et al., 2012). Electron microscopy (EM) studies showed that budding yeast LDs exhibit a narrow size distribution centering around 400 nm in diameter, although they often deviate from this size in mutants defective in various aspects of lipid metabolism (Czabany et al., 2008; Fei et al., 2008, 2011; Wolinski et al., 2011). In budding yeast, neutral lipids accumulate toward the end of the logarithmic phase of growth, and the neutral lipid content is particularly high in stationary phase. TGs are synthesized by the two acyltransferases, the acyl-CoA-dependent Dga1 (Oelkers et al., 2002) and Lro1, which uses phospholipids as an acyl donor (Oelkers et al., 2000). The individual contributions of the two enzymes may depend on growth conditions. Conversion of sterols to SEs is carried out by two paralogous acyltransferases, Are1 and Are2 (Yang et al., 1996). The storage lipids are rapidly mobilized when cells are returned to growth in fresh glucose-containing medium (Kurat et al., 2006). Although lipolysis is not required during the exponential phase of growth, yeast cells lacking TG lipases exhibit defects in sporulation, transition to growth from the stationary phase, and synthesis of phosphatidylinositol (PI) and sphingolipids (Kurat et al., 2009; Rajakumari and Daum, 2010; Rajakumari et al., 2010; Gaspar et al., 2011).
Triacylglycerols, one of the two main components of the LDs, are synthesized from the precursor lipid diacylglycerol (DG), which is also utilized in the so-called Kennedy pathway for production of PC and phosphatidylethanolamine (PE). DG, in turn, is synthesized from PA, a precursor for all major phospholipid species. Thus PA and the enzymes controlling its utilization may regulate the balance between production of storage lipids and phospholipids: PA could be either dephosphorylated by the PA phosphatases to yield DG or used to produce CDP-diacylglycerol (CDP-DG) by condensation with cytidine triphosphate (reviewed in Carman and Han, 2009). The enzyme that catalyzes the latter reaction, the CDP-DG synthase (also known as phosphatidate cytidylyltransferase), was first discovered in Escherichia coli and later identified in all major domains of life (Carter, 1968; Carman and Kelley, 1992; Saito et al., 1997; Zhou et al., 2013).
Using the fission yeast species Schizosaccharomyces japonicus and Schizosaccharomyces pombe as a model, we show that deficiency in CDP-DG synthase function triggers a pronounced increase in TG abundance and formation of unusually large LDs exhibiting abnormal morphology. We investigate dynamics and possible mechanisms underlying biogenesis of these structures and discuss our data in light of current hypotheses of LD formation and expansion.
RESULTS
Isolation of the S. japonicus mutant exhibiting high triacylglycerol content and abnormal lipid droplets
The fission yeast S. japonicus, an emerging model system, differs from a related species, S. pombe, in several important aspects of cellular physiology, including mitotic nuclear envelope (NE) remodeling (Aoki et al., 2011; Yam et al., 2011, 2013; Gu et al., 2012; Niki, 2014). We set out to obtain a collection of mutagenized S. japonicus strains conditionally exhibiting a wide range of NE/ER structure- and mitotic progression–related phenotypes. The heterothallic S. japonicus fission yeast strain that was “wild type” except for a uracil biosynthesis auxotrophic marker was modified to express the ER marker Tts1 mCherry and the actomyosin division ring marker Rlc1-GFP under the control of their native regulatory elements.
The resulting strain was mutagenized using ultraviolet light and screened for a wide range of temperature-sensitive phenotypes (see Materials and Methods for details).
One of the mutants, #185, exhibited unusually large spherical ER structures after incubation at the restrictive temperature of 36°C, prompting us to name it bubble1 (bbl1; Figure 1A). By backcrossing the #185 bbl1 strain to the wild-type strain and scoring segregation of phenotypes of progeny, we verified that the bbl phenotype was due to a mutation at a single genomic locus. On shift to the restrictive temperature, bbl1 colonies exhibited an increase in phloxine B staining suggestive of some cell death, but most cells growing in liquid cultures were able to divide normally in the rich, yeast extract–based medium at both 24°C (2.1 vs. 2 h for wild-type and bbl1 cells, respectively) and 36°C (2.2 vs. 2.1 h for wild-type and bbl1 cells, respectively).
FIGURE 1:
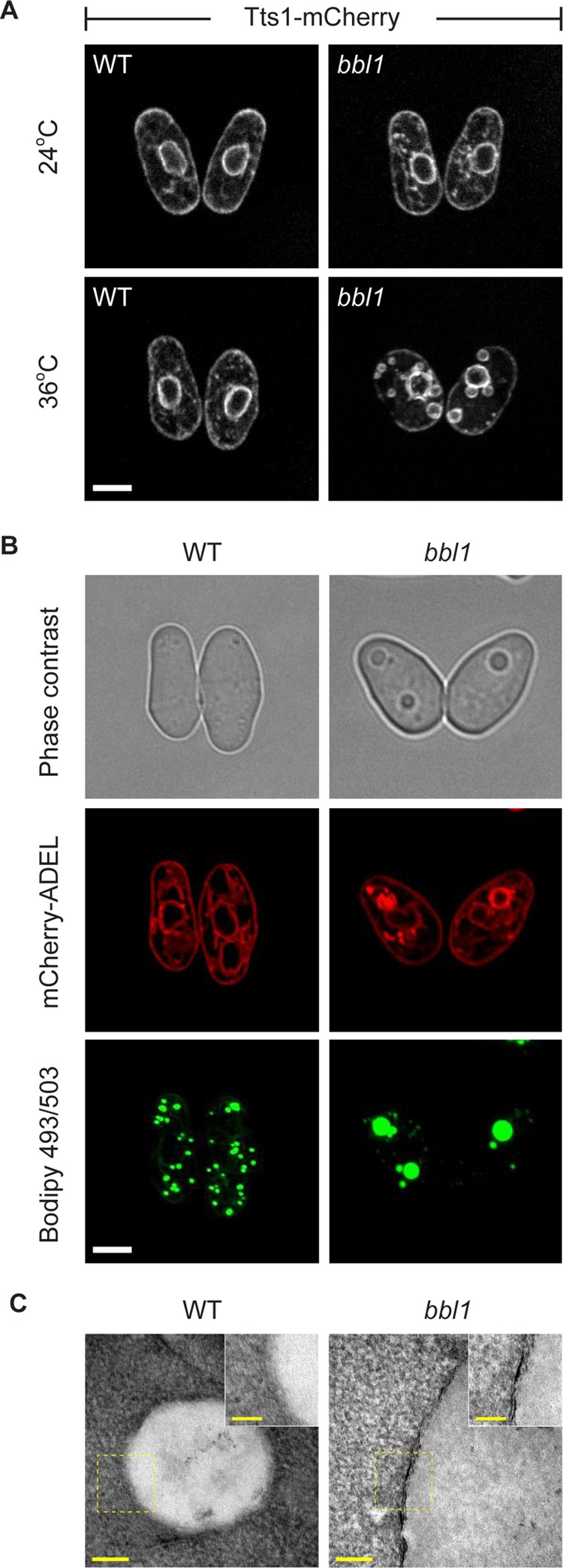
S. japonicus bbl1 mutant exhibits large LDs associated with the ER. (A) The S. japonicus bbl1 mutant cells expressing Tts1-mCherry exhibit large circular ER structures at the restrictive temperature (36°C). Scale bar, 5 μm. (B) BODIPY 493/503–stained neutral lipid deposits (green) are surrounded by the ER (mCherry-ADEL, red). Also shown is a corresponding phase contrast image. Scale bar, 5 μm. (C) ER is associated with large LDs in bbl1 mutant cells, as shown in this transmission electron microscopy image (n = 10 cells). (A–C) Cells grown in rich yeast extract–based medium. Scale bar, 100 nm (overview), 50 nm (magnification).
We noticed that unusual bubble-like structures were visible by phase contrast microscopy, appearing as highly refractile bodies (Supplemental Figure S1A and Figure 1B). The lipid droplet—a neutral lipid storage organelle—is known to be the only organelle in yeast capable of considerable light refraction. Indeed, these structures stained intensely for the neutral lipid probe BODIPY 493/503, suggesting that bbl1 cells accumulated large, neutral-lipid–containing bodies at the restrictive temperature (Figure 1B). Whereas the LD size dramatically increased in bbl1 cells, their number per cell dropped, as compared with the wild-type control (Supplemental Figure S1B). Abnormal LDs in bbl1 cells were found in proximity to ER membranes (Figure 1B; the ER is visualized by a luminal marker mCherry-ADEL; and Supplemental Figure S1, C and D). Furthermore, we observed the ER close to LDs using transmission electron microscopy (Figure 1C; n = 10 cells).
We did not observe large neutral lipid deposits surrounded by the ER in bbl1 cells at 24°C, although the BODIPY 493/503–stained LDs exhibited some degree of clustering (Figure 2A). Thin-layer chromatography (TLC) analysis indicated that the amount of TGs was higher in bbl1 cells than in the wild-type control even at 24°C but increased further after shifting to 36°C (Figure 2B). Direct measurements of the TG content by mass spectrometry showed an approximately twofold increase in the amount of the entire spectrum of TGs but not sterol esters in bbl1 cells as compared with the control (Figure 2C and Supplemental Figure S2). On the basis of these observations, we concluded that bbl1 mutant cells exhibited a considerable increase in TG abundance and abnormal LD morphology.
FIGURE 2:
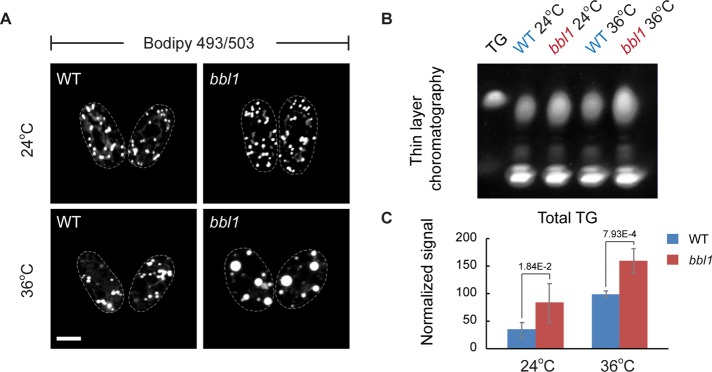
S. japonicus bbl1 mutant cells accumulate triacylglycerols. (A) LDs in bbl1 mutant cells are considerably larger than that in the wild type at the restrictive temperature of 36°C. Scale bar, 5 μm. (B) TLC shows that bbl1 mutant cells contain higher amounts of triacylglycerols than the wild type (WT) at both permissive (24°C) and restrictive temperatures (36°C). (C) Mass spectrometry analysis of the triacylglycerol content in WT (blue) and bbl1 (red) mutant cells is consistent with TLC data. Error bars, SDs (n = 5). (A–C) Cells grown in rich yeast extract–based medium.
Loss-of-function mutation in CDP-DG synthase triggers abnormal lipid droplet biogenesis and triacylglycerol accumulation in S. japonicus bbl1 mutant cells
We sought to identify the mutation underlying the bbl1 phenotype using a whole-genome resequencing approach (see Materials and Methods for experimental details). Two adjacent CC nucleotides in the SJAG_00426.4 open reading frame (ORF) were found mutated to TT, which resulted in substitution of the evolutionarily conserved proline 363 residue to serine (P363S; Figure 3A and Supplemental Figure S3A). SJAG_00426.4 encodes a lipid biosynthesis enzyme from the CDP-DG synthase family highly conserved across all domains of life (Figure 3A and Supplemental Figure S3B). The budding yeast S. cerevisiae orthologue of SJAG_00426.4 is known as Cds1 (Shen et al., 1996).
FIGURE 3:
The proline 363-to-serine (P363S) mutation in the ER-resident CDP-DG synthase Cds1 is responsible for bbl1 phenotype. (A) Cds1 is an evolutionarily conserved protein. The P363S mutation identified in S. japonicus bbl1 cells is highlighted in red. (B) The bbl1 phenotype is rescued by reintroduction of the WT cds1 copy to the native chromosomal locus, whereas replacement of the WT version with the reconstructed PP363S mutated version induces mutant phenotype de novo. (C) Second-copy introduction of the WT cds1 gene in bbl1 mutant cells rescues the bbl1 phenotype, whereas a second copy of cds1bbl1 mutant in WT cells does not induce it. (D) Both WT and mutant Cds1 proteins tagged with GFP localize to the ER. (B–D) Cells grown in rich yeast extract–based medium. (E) A time course of a typical enzyme activity assay shows that catalytic activity of mutant Cds1bbl1 (red) is lower than with the WT protein (blue). Error bars, SDs (n = 3). (B–D) Scale bar, 5 μm.
To confirm that the SNP located in SJAG_00426.2/cds1 was responsible for the bbl1 phenotype, we attempted complementation of bbl1 by replacing the mutant cds1 with its wild-type allele. The resulting bbl1::cds1+ strain was phenotypically wild type with respect to LD formation at all temperatures tested (Figure 3B, top). Conversely, we used targeted mutagenesis to introduce the bbl1-specific C1226T and C1227T mutations into the wild-type Cds1 ORF, thus causing the P363S amino acid substitution. The resulting wt::cds1bbl1 strain indeed exhibited abnormal LDs at 36°C, similar to the original bbl1 mutation (Figure 3B, bottom). We concluded that the P363S mutation in cds1 was responsible for accumulation of TGs and abnormal LD biogenesis in bbl1 cells.
The cds1bbl1 mutation was recessive, since introduction of the wild-type gene as an extra copy fully rescued the phenotype of bbl1 cells (Figure 3C, top). In line with this, an additional copy of the mutated gene in an otherwise wild-type genetic background failed to induce abnormal LDs (Figure 3C, bottom). The cds1bbl1 mutation did not lead to aberrant protein localization or changes in its overall abundance (Figure 3D and Supplemental Figure S3C). Both wild-type and mutant Cds1 proteins tagged with GFP and expressed from the native chromosomal locus localized to the ER membranes at 24 and 36°C (Figure 3D). In Cds1bbl1-GFP cells grown at 36°C, the mutant protein was also found at the ER domains surrounding the abnormal LDs, similar to other ER proteins (Figure 3D; compare with Figure 1 and Supplemental Figure S1).
CDP-DG synthase is responsible for a rate-limiting step in the production of phospholipids. It catalyzes condensation of PA and cytidine triphosphate, releasing the CDP-diacylglycerol and pyrophosphate products (Figure 3E, top). Measurements of phosphatidate cytidylyltransferase activity at 36°C in wild-type and bbl1 cell lysates (determined as transfer of radioactively labeled CDP to PA) showed that the bbl1 mutant enzyme was catalytically deficient (Figure 3E and Supplemental Figure S3D).
It was previously shown that the loss of the Cds1 function in budding yeast triggered an approximately twofold increase in the cellular amount of its substrate PA (Fei et al., 2011). We did not observe an increase in cellular PA levels in S. japonicus cells deficient in Cds1 function (Figure 4A). However, we detected a noticeable increase in the levels of diacylglycerols in these cells, likely due to rapid dephosphorylation of available PA (Figure 4B and Supplemental Figure S4).
FIGURE 4:
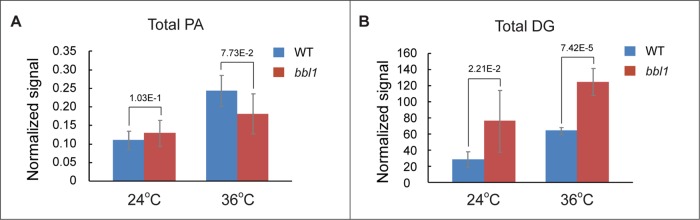
S. japonicus cds1bbl1 mutant cells exhibit significant increase in levels of diacylglycerols (DGs). (A) Results of mass spectrometry of total PA content in WT (blue) and bbl1 (red) mutant cells. (B) Results of mass spectrometry of total DG content in WT (blue) and bbl1 (red) mutant cells. (A, B) Cells grown in rich yeast extract–based medium. Error bars, SDs (n = 5).
Together with the genetic data, these results suggest that the P363S mutation in the Cds1 enzyme leads to a reduction in its CDP-DG synthase activity and potential shunting of a bulk of cellular PA into TG biosynthesis through the DG pathway.
Genetic modification of the Cds1 orthologue in S. pombe yields a large number of mutants exhibiting abnormal accumulations of neutral lipids
S. japonicus is highly photosensitive, which complicates long-term live-cell imaging approaches required to probe formation of abnormal LDs (Okamoto et al., 2013). With this in mind, we decided to transfer our research to the sister organism, S. pombe, a well-studied model system with an extensive experimental toolbox. We randomly mutagenized the C-terminus of the S. pombe Cds1 orthologue and screened for mutants with abnormal LD biogenesis and accumulation of neutral lipids (see Materials and Methods for experimental details). We readily identified a series of S. pombe bbl1-like mutants, both conditional (temperature sensitive) and constitutive, containing large refractile circular structures (Supplemental Figure S5).
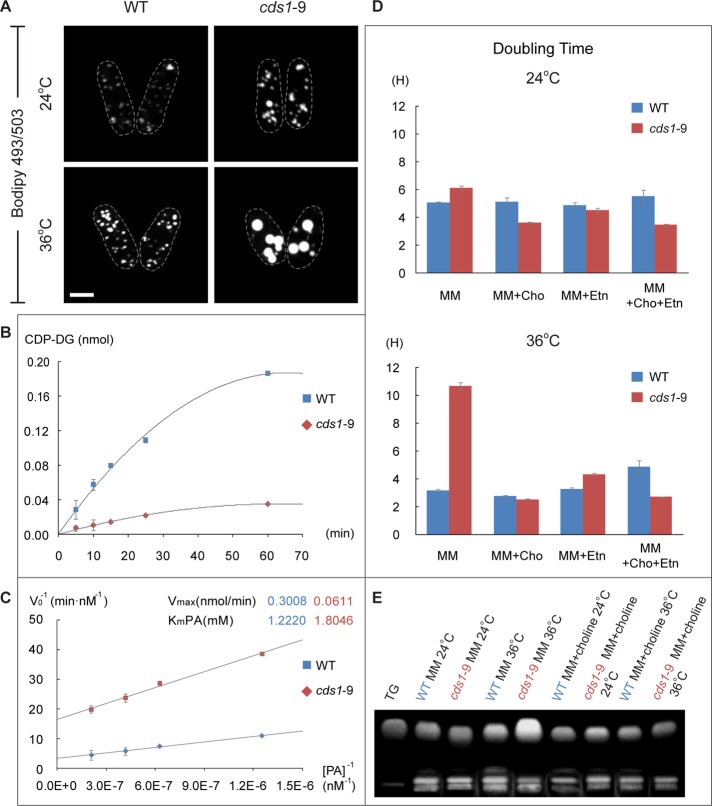
The cds1-9 allele was chosen for further characterization because it had a single amino acid substitution, changing cysteine at position 287 to arginine (C287R), and because it exhibited a tight conditional phenotype with respect to abnormal LD formation (Supplemental Figure S5 and Figure 5A). The CDP-DG synthase activity was markedly decreased in cds1-9 cells at 36°C (Figure 5B). The mutant protein appeared to bind PA nearly as well as the wild-type enzyme but exhibited a strongly decreased rate of catalysis (Figure 5C).
FIGURE 5:
A loss-of-function mutation in S. pombe cds1 leads to bbl1-like phenotype. (A) S. pombe cds1-9 mutant cells grown in rich yeast extract–based medium exhibit large BODIPY 493/503–stained lipid droplets (green) at the restrictive temperature of 36°C. Scale bar, 5 μm. (B) Typical time courses of CDP-DG synthesis reactions in crude extracts obtained from wild WT (blue) and cds1-9 mutant cells. (C) Measurements of KmPA and Vmax show that catalytic activity of the mutant enzyme (red) is significantly than that of WT (blue), although the proteins exhibit comparable PA binding. Error bars, SDs (n = 3). (D) Patterns of doubling time of WT (blue) and cds1-9 (red) mutant cells in chemically defined minimal medium with indicated supplements. Cho, choline; Etn, ethanolamine. Error bars, SDs (n = 3). (E) TLC shows that supplementation with choline largely rescues TG accumulation in cds1-9 mutant cells grown in the minimal medium.
Cells carrying the cds1-9 allele grew normally in the rich yeast extract–based medium at both 24°C (3.5 vs. 3.1 h for wild-type and cds1-9 cells, respectively) and 36°C (2.5 vs. 2.7 h for wild-type and cds1-9 cells, respectively), yet they showed a vastly decreased growth rate in the chemically defined minimal medium in a temperature-sensitive manner (Figure 5D). Supplementing the minimal medium with choline or ethanolamine, the precursors required for phospholipid biosynthesis through the de novo Kennedy pathway (reviewed in Carman and Han, 2009), restored the doubling time of cds1-9 cultures at 36°C to almost wild-type levels (Figure 5D). Of interest, supplementation with choline also alleviated abnormal accumulation of TG in the absence of Cds1 activity (Figure 5E), suggesting that it permitted rerouting of the bulk lipid synthesis away from the storage pathway. Analysis of total cellular fatty acid (FA) content showed that cds1-9 cells grown in the minimal medium accumulated lipid species containing stearic acid (C18:0) at the restrictive temperature of 36°C. Similar rise in FA content mirroring an increase in total TG was previously observed for S. pombe cells lacking several TG lipases (Yazawa et al., 2012). Choline supplementation brought absolute quantities of stearic acid in cds1-9 cells close to the wild-type values and also decreased the abundance of other major FAs (Table 1).
TABLE 1:
Absolute quantities of fatty acids in WT and cds1-9 cells.
| Minimal medium | Minimal medium + choline | |||||||
|---|---|---|---|---|---|---|---|---|
| 24°C | 36°C | 24°C | 36°C | |||||
| Fatty acida | WT (ng/1 OD) (n = 2) | cds1-9 (ng/1 OD) (n = 3) | WT (ng/1 OD) (n = 3) | cds1-9 (ng/1 OD) (n = 2) | WT (ng/1 OD) (n = 3) | cds1-9 (ng/ 1OD) (n = 3) | WT (ng/1 OD) (n = 2) | cds1-9 (ng/1 OD) (n = 3) |
| Palmitoleic acid (C16:1) | 132.0 ± 1.7 | 130.4 ± 12.4 | 159.3 ± 10.6 | 102.7 ± 3.89** | 138.1 ± 4.4 | 110.0 ± 9.1** | 104.5 ± 4.8 | 49.0 ± 7.0* |
| Oleic acid (C18:1) | 2663.7 ± 270.5 | 2093.5 ± 166.2 | 2753.1 ± 167.1 | 2063.2 ± 137.27 | 2676.8 ± 32.3 | 2284.4 ± 279.0 | 2380.4 ± 141.6 | 1239.4 ± 173.1* |
| Myristic acid (C14:0) | 238.3 ± 15.8 | 336.9 ± 39.4 | 172.5 ± 18.7 | 125.7 ± 1.55 | 183.5 ± 8.0 | 196.5 ± 24.8 | 147.7 ± 5.9 | 90.8 ± 13.9* |
| Palmitic acid (C16:0) | 4177.6 ± 426.1 | 3837.5 ± 252.5 | 5040.9 ± 320.3 | 4103.5 ± 132.50 | 4081.1 ± 10.2 | 4236.8 ± 421.2 | 4813.1 ± 258.0 | 2874.9 ± 398.6* |
| Stearic acid (C18:0) | 3300.5 ± 290.3 | 3655.5 ± 339.5 | 4054.8 ± 331.3 | 9653.3 ± 805.78*** | 3263.9 ± 5.7 | 3537.3 ± 404.1 | 3031.0 ± 96.5 | 3084.5 ± 412.8 |
| Total FAs | 10,512.1 ± 1004.4 | 10,053.8 ± 810.0 | 12,180.5 ± 848.0 | 16,048.4 ± 1081.0 | 10,343.4 ± 60.5 | 10,365.0 ± 1138.1 | 10,476.6 ± 506.7 | 7338.7 ± 1005.4 |
Fatty acid quantities are shown as nanograms per optical density unit of cells ± SD and represent the average of two or three replicates.
aLength of carbon chain and number of double bonds of each FA are indicated in parentheses (e.g., C16:1 denotes 16 carbons and 1 double bond). Values in red denote FA quantities that are significantly different between WT and cds1-9 within each temperature and medium condition.
*p < 0.05; **p < 0.01;***p < 0.002, t test with Holm–Sidak correction for multiple comparisons.
These results suggested that the CDP-DG–dependent phospholipid production route was inhibited in the cds1-9 genetic background and that these mutant cells relied on the Kennedy pathway to generate phospholipids required for their growth.
ER-associated lipid droplets in S. pombe cells deficient in CDP-DG synthase activity are functional
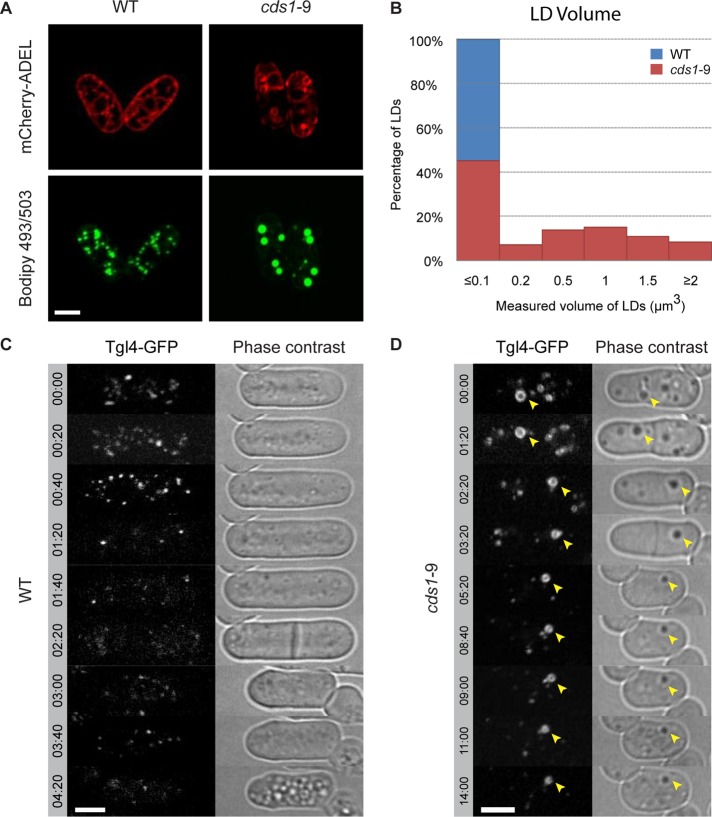
Similar to the cds1bbl1 mutant in S. japonicus, cds1-9 S. pombe cells formed large LDs associated with the ER membranes upon shift to the restrictive temperature of 36°C (Figure 6A). In the rich yeast extract–based medium, the average droplet volume dramatically increased, whereas the number of droplets decreased (Figure 6B; number of LDs per cell at 36°C was 23 ± 4 and 6 ± 2 for the wild type and cds1-9 mutant, respectively). Of note, we observed higher number of LDs when cells were grown in minimal medium (number of LDs per cell at 36°C was 34 ± 5 and 29 ± 7 for the wild type and cds1-9 mutant, respectively). Of interest, the overall abundance of TG in both wild-type and cds1-9 cells also strongly depended on growth conditions, with cells grown in the rich yeast extract medium showing considerably higher TG content (Supplemental Figure S6A).
FIGURE 6:
Large ER-associated neutral lipid deposits in cds1-9 S. pombe cells are lipid droplets. (A) The BODIPY 493/503–stained neutral lipid deposits (green) in S. pombe cds1-9 mutant cells are associated with ER membranes marked by the artificial luminal marker mCherry-ADEL (red). (B) Histogram demonstrating LD size distribution in WT (blue) and cds1-9 (red) mutant cells. LDs were binned according to volume (μm3) as follows: ≤0.1, between 0.1 and 0.2, between 0.2 and 0.5, between 0.5 and 1, between 1 and 1.5, between 1.5 and 2, and ≥2. n = 309 LDs for WT and 303 LDs for cds1-9. (C, D) Time-lapse sequences showing that the Tgl4-GFP-marked lipid droplets are metabolized in both WT and cds1-9 mutant cells grown in the presence of the fatty acid synthesis inhibitor cerulenin. (A–D) Cells grown in rich yeast extract–based medium. Time is in hours and minutes. Scale bar, 5 μm.
It has been proposed that abnormal LD morphology and ER-LD association could result in changes to metabolic state of LDs and deficient lipolysis (Guo et al., 2008; Wolinski et al., 2011). We wondered whether TGs contained in the mutant LDs could be mobilized to yield fatty acids and DG, supporting membrane biosynthesis and viability, when de novo fatty acid synthesis was inhibited. The major TG lipase Tgl4 (Kurat et al., 2009) was enriched at the periphery of these structures, similar to the wild type (Supplemental Figure S6B). Wild-type S. pombe cells treated with the fatty acid synthase inhibitor cerulenin exhibited progressive reduction in LD number and Tgl4-GFP fluorescence intensity and ceased growth after one or two rounds of division, presumably after the available reservoir of fatty acids had been exhausted (Figure 6C; n = 4 cells). On the other hand, cds1-9 cells continued dividing for considerably longer periods of time (Figure 6D; n = 8 cells), showing progressive shrinkage and eventual disappearance of Tgl4-GFP–marked abnormal LDs (Figure 6D). Thus the TG stores in cds1-9 cells can be used and promote survival of cells in the absence of fatty acid biosynthesis.
TG synthases function at the sites of persistent TG production in S. pombe cells deficient in CDP-DG function
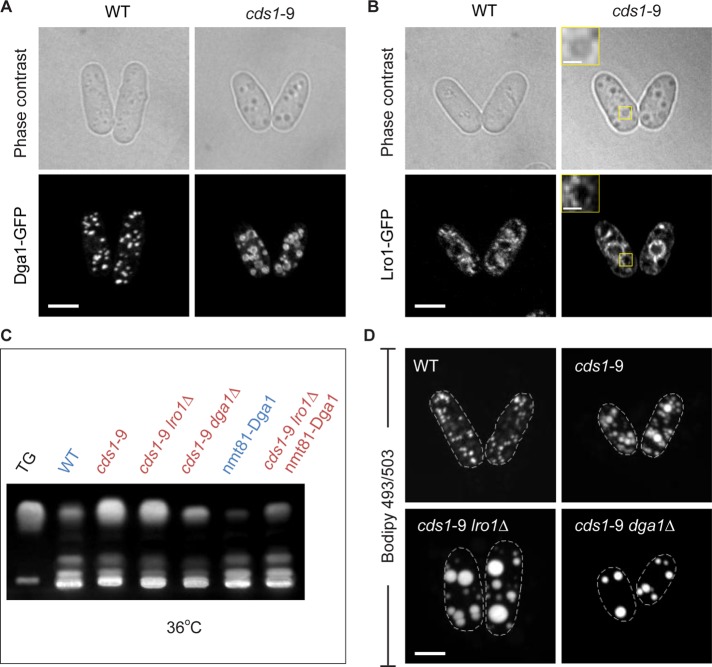
Fission yeast genomes encode orthologues of the two budding yeast TG synthases, Dga1 and Lro1. Both proteins contributed to TG synthesis in exponentially growing S. pombe (Supplemental Figure S7A; see also Zhang et al., 2003). These enzymes exhibited distinct patterns of subcellular localization. In wild type, the GFP-tagged Dga1, the DG acyltransferase that uses acyl-CoA as an acyl donor (Oelkers et al., 2002), was enriched at LDs (Figure 7A). In cds1-9 cells grown at the restrictive temperature of 36°C, Dga1-GFP also localized predominantly to the periphery of large LDs (Figure 7A). On the other hand, the acyl-CoA–independent diacylglycerol acyltransferase Lro1, which is believed to use PE or PC (Oelkers et al., 2000) as acyl donor, exhibited less-defined intracellular localization, with some enrichment at the peripheral ER (Figure 7B). Of interest, we observed enhanced Lro1-GFP signal at the ER membranes surrounding the abnormal LDs and around the NE in cds1-9 cells (Figure 7B). Thus the core machinery for TG biosynthesis appears to be concentrated in the vicinity of large, ER-associated LDs in cells lacking Cds1 activity.
FIGURE 7:
Both TG synthases Dga1 and Lro1 function at the large ER-associated lipid droplets in cds1-9 S. pombe cells. (A) Dga1-GFP localizes to the periphery of LDs in cds1-9 cells at the restrictive temperature of 36°C. (B) Lro1-GFP localizes to the ER, including the ER membranes associated with large LDs in cds1-9 cells, at the restrictive temperature of 36°C. Insets, magnified views. (C) TLC shows that deletion of dga1 massively reduces cellular TG content in cds1-9 genetic background at the restrictive temperature of 36°C. cds1-9 lro1Δ cells also show reduced total TG. (D) BODIPY 493/503–stained LDs in cds1-9 cells lacking the TG synthases Lro1 or Dga1 at the restrictive temperature of 36°C. (A–D) Cells grown in the chemically defined minimal medium. Scale bar, 5 μm.
We wondered which of the two TG synthases was essential for accumulation of TG in cds1-9 cells. The TLC analysis of cells grown in liquid minimal medium showed that cellular TG levels were considerably reduced in dga1Δ cds1-9 cells compared with the single cds1-9 mutant (Figure 7C and Supplemental Figure S7B). In line with this observation, the number of LDs decreased in dga1Δ cds1-9 cells (Figure 7D; LD number was 5 ± 2, n = 20 cells; see Supplemental Figure S7C for permissive temperature). Deletion of lro1 in cds1-9 genetic background also led to some reduction in TGs (Figure 7C and Supplemental Figure S7B), but, surprisingly, we observed formation of even larger LDs, which frequently occupied a large part of the intracellular volume at the restrictive temperature of 36°C (Figure 7D; LD number was 13 ± 3, n = 20 cells; see Supplemental Figure S7C for permissive temperature).
In spite of the highly abnormal morphology of lipid droplets, lro1Δ cds1-9 cells grew considerably better than the single cds1-9 mutant in minimal medium at 36°C (the doubling time was 5.3 vs. ∼10 h, respectively). On the other hand, dga1Δ cds1-9 double-mutant cells exhibited considerably increased doubling time even at the permissive temperature of 24°C (10.8 h) and virtually arrested growth at 36°C (>12 h). Thus activities of the two TG synthases appeared antagonistic to each other with respect to supporting growth of cds1-9 cells. We were unable to construct a strain in which both dga1 and lro1 were deleted in the cds1-9 mutant background, due to germination failure of the triple-mutant cells. However, replacement of the native dga1 promoter with a thiamine-repressible version allowed us to perform transcriptional shut-off of dga1 in exponentially growing lro1Δ cds1-9 cells, resulting in substantial suppression of TG production (Figure 7C and Supplemental Figure S7B). Of interest, under these conditions, the triple-mutant lro1Δ nmt81::dga1 cds1-9 cells grew similarly to wild type, with doubling time of just 2.8 h.
Taken together, our results suggest that both Lro1 and Dga1 participate in TG biosynthesis in Cds1-deficient S. pombe cells. Of interest, when the phospholipid-dependent acyltransferase Lro1 is inactivated, vegetatively growing cds1 mutant cells no longer require supplementation with exogenous lipid precursors. Conversely, it is possible that in the absence of Dga1, a higher relative activity of Lro1 results in PE/PC depletion inhibitory to growth of cds1 mutant cells deficient in the CDP-DG branch of phospholipid biosynthesis.
Large lipid droplets in Cds1-deficient cells arise from persistent growth rather than fusion
It was previously proposed that deficiency in the Cds1 function in budding yeast could lead to LD fusion and emergence of supersized LDs caused by a rise in intracellular PA levels (Fei et al., 2011). Strong enrichment of TG synthases at abnormal lipid droplets in cds1-9 cells, together with the fact that they were closely associated with the ER, raised a possibility that these structures arose as a result of progressive enlargement rather than fusion of the preexisting, normally sized LDs. To evaluate this possibility, we performed time-lapse imaging of cds1-9 cells expressing the LD marker Tgl4-GFP or the artificial luminal ER marker GFP-ADEL (Zhang et al., 2010). Cells were shifted to the restrictive temperature of 36°C, and imaging started 2 h after the shift. We observed emergence of highly refractile bodies that recruited Tgl4-GFP and expanded over time (Figure 8A; n = 3 cells). Imaging using GFP-ADEL showed initial clustering of the ER membranes, followed by their outward expansion and eventual formation of large, “bubble”-like structures (Figure 8B; n = 14 cells). Thus abnormally large LDs in cells lacking the ER-localized CDP-DG synthase activity form through continuous growth rather than fusion between the preexisting LDs.
FIGURE 8:
ER-associated lipid droplets in cds1-9 S. pombe cells grow by expansion. (A) A time-lapse sequence showing emergence and expansion of the Tgl4-GFP–marked lipid droplet in cds1-9 mutant cell (indicated by yellow arrowhead) at the restrictive temperature of 36°C. Also shown are the corresponding phase-contrast images. (B) A time-lapse sequence of GFP-ADEL– expressing cds1-9 mutant cell documents emergence of the ER-associated LD at the restrictive temperature of 36°C. Insets, magnified views of an indicated area. (A, B) Cells grown in rich yeast extract–based medium. Time is in hours and minutes. Scale bar, 5 μm; inset, 2 μm.
DISCUSSION
The evolutionarily conserved enzyme CDP-DG synthase catalyzes a key rate-limiting step in phospholipid biosynthesis by converting PA into the liponucleotide CDP-DG (Carter, 1968; Raetz and Kennedy, 1973; Ganong and Raetz, 1982; Saito et al., 1997). In yeast, CDP-DG is an intermediate for synthesis of major structural phospholipids, including PC, PE, PI, and PS, and the mitochondrial phospholipid species phosphatidylglycerol and cardiolipin (reviewed in Carman and Han, 2009). PA can be alternatively dephosphorylated to DG, which is then used for synthesis of PE and PC through the Kennedy pathway or for production of the storage lipid TG (reviewed in Carman and Han, 2009).
This branched architecture of cellular PA allocation might explain the pronounced shift toward high TG content in fission yeast deficient in CDP-DG synthase function. In cds1 mutants, PA is presumably channeled to DG, thus increasing TG production. High DG levels would also enable phospholipid biosynthesis through the Kennedy pathway. Indeed, our results suggest that significant down-regulation of Cds1 activity in fission yeast does not impede vegetative growth as long as the mutant cells are supplemented with choline or ethanolamine, the two exogenous substrates required for this phospholipid production route (Figure 5D).
Of interest, the fatty acid composition of cds1 mutant cells undergoes profound changes, depending on the external supply of lipid precursors. Without added choline, Cds1-deficient cells exhibit significant enrichment in lipid species containing stearic acid. This is likely related to the increased TG production in the mutant cells, since the addition of choline to the growth medium brings both TG and stearate contents close to their wild-type values (Table 1 and Figure 5E). Cycling of FAs through TG is believed to affect both cellular acyl chain composition and acyl-CoA availability (Petschnigg et al., 2009; Mora et al., 2012). Therefore it is possible that stearate accumulation may reflect insufficient FA recycling and lipid remodeling when the rate of phospholipid production is dramatically reduced. In line with this possibility, choline-stimulated phospholipid synthesis through the Kennedy pathway appears to promote FA turnover in Cds1-deficient cells (Table 1).
We observed a marked increase in cellular DG content in cells deficient in Cds1 function (Figure 4B and Supplemental Figure S4). Consistent with rapid utilization of PA in an alternative pathway, the amount of PA remained comparable to control (Figure 4A). This contrasts with the previously reported data from budding yeast showing an approximately twofold increase in total cellular PA upon Cds1 inactivation (Fei et al., 2011). Possible reasons for this apparent inconsistency include differences in experimental setup (e.g., transcriptional shut-off of Cds1 in budding yeast versus conditional loss-of-function mutant in fission yeast) and divergent lipid signaling and metabolism in budding and fission yeasts (Raychaudhuri et al., 2012). As an example, the PA-responsive gene regulation circuitry controlling inositol production and phospholipid biosynthesis is not conserved between budding and fission yeast; in fact, the fission yeasts are naturally auxotrophic for inositol (Fernandez et al., 1986; Gaynor and Greenberg, 1992; Ingavale and Bachhawat, 1999).
Cds1 has been reported as essential in both S. cerevisiae and S. pombe (Shen et al., 1996; Kim et al., 2010). Although we show that cycling fission yeast cells may tolerate a severe decline in Cds1 activity, the CDP-DG–dependent route of phospholipid biosynthesis could be indispensable in specific physiological scenarios, such as nutrient starvation, mating, or germination. Of interest, we observed an abnormally high number of spores that failed to germinate after mating of cds1 mutant cells in both S. japonicus and S. pombe (unpublished data).
An increase in TG synthesis in cds1 mutants did not translate into a higher number of LDs per cell. Instead, we observed emergence of a few expanding “supersized” droplets closely associated with the ER (Figure 8). We did not find any instances of LD fusion, and we did not detect elevated PA or lower PC levels (Figure 4A and Supplemental Figure S8), which are believed to promote fusion of LDs into larger entities (Guo et al., 2008; Fei et al., 2011; Krahmer et al., 2011). Consistent with persistent expansion of LDs in Cds1-deficient cells, the two terminal enzymes for TG biosynthesis, Lro1 and Dga1, were enriched at the LD-associated ER membranes (Figure 7, A and B). Once the “bubble”-like ER domains were established, they appeared continuous with the rest of the ER throughout LD expansion (Figures 1 and 8 and Supplemental Figure S1).
The emergence of large, TG-producing “factories” and, presumably, restructuring of the ER-LD contact sites could be potentially explained by an overall increase in the DG concentration in cds1 cells (Figure 4B and Supplemental Figure S4). Because DG is an immediate TG precursor, this is expected to promote TG synthesis through DG esterification. In a system functioning under higher synthetic load, the amount of phospholipids required to assemble LD shells may become limiting, especially in cells deficient in one of the two branches of phospholipid biosynthesis. In such situation, emergence of larger droplets would be favored. Of interest, overexpression of DGAT2, the orthologue of yeast Dga1, is sufficient for development of giant LDs in mammalian cells (Cases et al., 2001). An excess of DG in the ER membrane could additionally have a direct role in regulating TG packaging into LDs by changing membrane curvature or affecting recruitment of relevant proteins (reviewed in Carrasco and Merida, 2007). Of note, high local content of DG has been proposed to promote LD biogenesis in budding yeast by modifying ER membrane properties (Adeyo et al., 2011).
In conclusion, our data indicate that lowering the activity of the ER-resident CDP-DG synthase is sufficient to raise the cellular TG content. Of importance, studying unusual morphology of lipid droplets in Cds1-deficient cells may provide further insights into lipid droplet biogenesis and size control and aid in exploration of interactions between LDs and ER.
MATERIALS AND METHODS
Yeast strains
The original wild-type auxotrophic S. japonicus strains were kindly provided by H. Niki (National Institute of Genetics, Japan; Furuya and Niki, 2009). All strains constructed in the course of our study are listed in Supplemental Table S1. Genetic methods and culturing conditions for S. pombe and S. japonicus have been described (Gould, 2004; Furuya and Niki, 2009; Aoki et al., 2010). Typically, experiments have been performed during mid to late log phase of growth (typically at OD595 of 0.8). The S. japonicus open reading frames used in this study were tts1 (SJAG_05671), rlc1 (SJAG_01665), and cds1 (SJAG_00426). The S. pombe open reading frames used in this study were cds1 (SPBC13A2.03), tgl4 (SPAC1A6.05c), dga1 (SPCC1235.15), lro1 (SPBC776.14), and tts1 (SPBC1539.04).
S. japonicus mutagenesis screen
The S. japonicus Rlc1-GFP Tts1-mCherry–expressing strain (“parent”) was mutagenized using ultraviolet light and grown at 24°C on YES (yeast extract with supplements) medium agar plates until the appearance of individual colonies. Colonies were replica plated on YES plates containing phloxine B and grown at the restrictive temperature of 36°C to allow visualization of the temperature-sensitive mutant cells. After several rounds of screening, ∼900 temperature-sensitive mutants were isolated and tested for the ER and cell division abnormalities at the restrictive temperature.
Genome resequencing and analysis
We isolated high-quality genomic DNA from seven strains of S. japonicus (SOJ6, SOJ14, SOJ33, SOJ60, SO#20, SO#30, and SO#185) by our published method (Pomraning et al., 2009). Paired-end sequencing libraries for the Illumina platform were prepared as suggested by the manufacturer according to our modified methods (Pomraning et al., 2009). Each library prepared had a specific four-nucleotide barcode (SOJ33, ACGT; SO#185, GCTT; SO#20, CTGT; SO#30, GGGT). Libraries were pooled into four lanes and sequenced as single-end 40-nucleotide (nt) runs at the Oregon State University Center for Genome Research and Biocomputing core facility. We obtained 22,659,725 total 40-nt reads for SOJ33, of which 16,078,075 (or 71%) mapped to the reference genome, and 20,101,781 for the combination of all mutants, of which 13,857,423 (or 69%) mapped to the reference genome. The average sequencing depth in the four genomes analyzed was 26 reads/nt.
SOJ33 is the nonmutated parent strain most closely related to the S. japonicus strain from which the reference genome was derived (Rhind et al., 2011). By methods described elsewhere (Pomraning et al., 2011), we first mapped the reads obtained from SOJ33 to the reference genome and identified 445 single-nucleotide polymorphisms (SNPs) in coding regions that were present when compared with the relatively low-coverage reference genome. As a cutoff to call a high-quality SNP, we required 4 reads/nt and a variant frequency of >80%, as ∼99% of the genomes was covered at 5 reads/nt. We next compared the SOJ33 SNPs to SO#185, the originally mutated strain, and the two back-crossed siblings, SO#20 and SO#30, and enumerated SNPs encountered in these genomes. There were 456 SNPs found in all three mutant strains, 445 of which had been already found in SOJ33. This left 11 high-quality SNPs as primary candidates for the mutation causing the phenotype in the mutant strains. The 11 SNPs found by mapping were assigned to the underlying S. japonicus genes, the complete catalogue of which was downloaded from the Broad Institute website (www.broadinstitute.org/annotation/genome/schizosaccharomyces_group/MultiDownloads.html). We found that SJAG_00426.4 was the only mutated ORF out of the original list by directly resequencing all these ORFs in the “parent” and both the original #185 and the outcrossed sibling SO#20 and SO#30 strains.
Fluorescence microscopy and image analysis
Fluorescence microscopy images were acquired using a Nikon TiE system (CFI Plan Apochromat VC 100XH 1.4 numerical aperture [NA] objective; Nikon, Melville, NY) equipped with a Yokogawa CSU-X1-A1 spinning disk unit (Yokogawa, Tokyo, Japan), a Photometrics CoolSNAP HQ2 camera (Photometrics, Tucson, AZ), and a DPSS 491-nm, 100-mW and DPSS 561-nm, 50-mW laser illumination unit under the control of MetaMorph Premier, version 7.7.5 (Universal Imaging, Sunnyvale, CA). Cells were cultured overnight at 24 and 30°C, respectively. Cells cultured at 30°C were shifted to 36°C for 4 h before harvest together with the cells cultured at 24°C; these were respectively imaged at 24 and 36°C. Cells were maintained on freshly sealed 2% agarose YES medium pads.
Time-lapse fluorescence microscopy images were acquired using a Zeiss Axiovert 200 M microscope (Plan Apochromat 100×, 1.4 NA objective; Carl Zeiss, Jena, Germany) equipped with an UltraView RS-3 spinning disk confocal system (PerkinElmer, Boston, MA) including a CSU21 confocal optical scanner, 12-bit digital cooled Hamamatsu Orca-ER camera (OPELCO, Sterling, VA), and a krypton–argon triple-line laser illumination source (DPSS 491 nm, 100 mW and DPSS 561 nm, 50 mW), under the control of MetaMorph Premier version 7.7.5. Cells were cultured overnight at 30°C and were kept at 36°C during imaging. Cells were maintained in Onix Microfluidic Perfusion System (CellASIC, Merck Millipore, Billerica, MA).
For quantification of LDs, fission yeast cells were stained by BODIPY 493/503 and subsequently imaged in both fluorescence and phase contrast modes. Quantifications were performed with ImageJ (Schneider et al., 2012), with only interphase cells counted (n = 50 cells). To quantify volume of S. japonicus cells, the width (W) and height (H) of each cell were measured in ImageJ. The cell volume was calculated as VC (volume) = ¾π(½W)3 + π(½W)2(H − W). We used a method described by Murphy et al. (2010) to quantify LD volume. Briefly, the diameter (D) of each LD was measured in ImageJ, and the volume was calculated as VL (volume) = πD3/6.
Transmission electron microscopy
We followed a previous protocol (Bauer et al., 2001) with some modifications. Cells were washed in phosphate-buffered saline (PBS) at 4°C and fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) with 1 mM calcium chloride for 30 min at 4°C. After rinsing in 0.1 M sodium cacodylate buffer, cell walls were digested by 0.15 mg/ml Zymolyase in 50 mM Tris-HCl (pH 7.5) with 5 mM magnesium chloride, 1.4 M sorbitol, and 0.5% (vol/vol) β-mercaptoethanol for 10 min. For a second fixation, cells were fixed in 0.5% osmium tetroxide and 0.8% potassium ferrocyanide in distilled water two times for 5 min at 4°C and washed in 0.1 M sodium cacoyldate buffer. Cells were mixed with a 2.5% solution of agarose (type VII) at 37°C. The solidified pellet were cut and washed in distilled water. Specimens were en bloc stained with 1% uranyl acetate for 90 min, dehydrated, and embedded in Epon. Hardened blocks were sectioned using an ultramicrotome (Leica ultra cut UCT; Leica, Wetzlar, Germany), and 80-nm (on average) ultrathin sections were collected on copper grids (300 mesh) and stained using lead citrate. Grids were imaged using a JEOL JEM-1230 transmission electron microscope (JEOL, Peabody, MA) at 120 kV, and digital images were taken using a Gatan SC200 charge-coupled device camera (2k × 2k, bottom mount) and Digital Micrograph 2.10.1282.0 (Gatan, Pleasanton, CA).
Isolation of lipids and lipid analyses
Cells were cultured overnight at 24 and 30°C, respectively, and the cells cultured at 30°C were shifted to 36°C for 4 h before being harvested together with the cells cultured at 24°C. Lipids were prepared according to protocols described in Fei et al. (2011). Briefly, the harvested cells were washed with ice-cold 1.2 M sorbitol in PBS, digested by enzyme mixture (MP Zymolyase 20T, 3 mg/ml; lysing enzyme, 5 mg/ml [Sigma-Aldrich, St. Louis, MO]), and sonicated. The lysates were subjected to methanol-chloroform extraction. The solvent of the chloroform phase was evaporated under nitrogen.
For TLC, harvested cells were resuspended in 150 μl of ice-cold methanol/chloroform (2:1) and broken by bead beating in a 1.5-ml Eppendorf tube. A tube was typically perforated with the needle and inserted into a 2-ml tube, and the lysate was separated from glass beads by centrifugation at 2000 × g for 2 min. The extracted lipids were dissolved in 200 μl of methanol/chloroform (2:1) and separated on silica gel 60 TLC plates (EMD, San Diego, CA) with hexane/diethyl/acetic acid (90:9:1). We loaded lipids from 10 OD595 units of cells/lane. Lipid spots were detected by 0.02% primuline in 20% acetone. Bands corresponding to fission yeast TGs were identified by comparing with the TG standard, triolein.
The molecular species of DG, TG, and phospholipids were analyzed by ESI triple quadrupole mass spectrometry (API 4000; Applied Biosystems, Foster City, CA), as described in Lee et al. (2011). Five replicates were analyzed for each genotype/condition combination. Statistical significance was determined by Student's t test (p < 0.05). Fatty acid composition was determined by gas–liquid chromatography on a Supelco 30-m Omegawax 250 column (Sigma-Aldrich) after derivitization of the fatty acids to methyl esters in 1.5 M methanolic HCl, followed by addition of water and extraction with pentane. Phospholipids were identified on the basis of the head group fragment and the mass/charge of the intact ion. Quantification was performed via comparison with a series of internal standards for each head group class. DGs and TGs are identified by the presence of one fatty acyl component, which is lost during tandem mass spectrometry, and the total number of acyl carbons and acyl double bonds in the other two fatty acyl chains. TGs are quantified by comparison with internal standards, but no response factor is calculated due to the variability in the ionization of individual TG species.
For fatty acid content analysis by gas-chromatography mass spectrometry (GCMS), 10 ml of cells for each genotype and culture condition were grown overnight at 24 and 30°C, respectively, and the cells cultured at 30°C were shifted to 36°C for 4 h before being harvested together with the cells cultured at 24°C. The harvested cells were washed with ice-cold 1.2 M sorbitol in PBS and broken by beating with glass beads. The lysates were subjected to methanol-chloroform extraction. The solvent from the chloroform phase was evaporated under nitrogen. For transesterification of lipids, 100 μl of methanolic HCl (Supelco Analytical; Sigma-Aldrich) was added to each vial containing the dried total lipids and incubated for 1 h at 60°C with occasional vortexing. After the acid-based catalysis, the reactions were cooled on ice, followed by the addition of 60 μl of hexane and brief vortexing. The hexane layers (which contain the fatty acid methyl esters) were transferred into new vials and evaporated under nitrogen. The vials were then kept at −20°C until GCMS analysis. For GCMS, extracts were redissolved in 60 μl of hexane spiked with 10 μg/ml hexacosane (Sigma-Aldrich) and transferred into GCMS vials (Supelco Analytical). Analysis was performed with a GCMS QP2010 system (Shimadzu, Kyoto, Japan) equipped with a 5% phenyl-methylpolysiloxane (DB-5, 30-m length, 0.32-mm inner diameter, 0.25-μm film thickness; Agilent, Santa Clara, CA) column. The parameters were set at an initial column temperature of 50°C and increased to 210°C at a rate of 35°C/min, followed by an increment of 3°C/min to 280°C in splitless mode. Peaks corresponding to fatty acid methyl esters were identified based on retention times and electron ionization spectra of known standards. The absolute quantities of individual fatty acid methyl esters were obtained by normalizing the area under the corresponding signals by the area of an internal standard (hexacosane) and then multiplying each ratio by a response factor. The response factor corrects for different ionization properties of the individual fatty acid methyl ester species. To determine response factors, a standard curve was established for each fatty acid methyl ester at three concentrations (20, 100, and 500 ng/μl). The slope of the curve was used to calculate the ratio of the fatty acid methyl ester signal to the hexacosane signal for the same quantity in the samples. To lower the possibility of false positives, results were evaluated for significance using Student's t test with Holm–Sidak correction for multiple comparisons.
Enzyme activity assay
Cds1 activity assays were based on previous protocols (Ganong et al., 1980; Carman and Kelley, 1992). Fission yeast cells were precultured overnight at 30°C and shifted to 36°C for 4 h before being harvested. The harvested cells were washed with ice-cold 1.2 M sorbitol in PBS, digested by enzyme mixture (MP Zymolyase 20T, 3 mg/ml; Sigma-Aldrich lysing enzyme, 5 mg/ml) with complete protease inhibitor tablet (Roche, Indianapolis, IN) and 1 mM phenylmethylsulfonyl fluoride and sonicated. Lysates were centrifuged at 2000 × g at 4°C for 1 min. Protein concentration in supernatant was determined using the Bradford method. Total protein, 400 μg of S. japonicus and 600 μg of S. pombe, was mixed with 1.6 mM PA, 10 mM magnesium chloride, 0.2% Triton X-100 in 0.1 M potassium phosphate (pH 7.0) and 800 Ci/mmol [α-32P]CTP/5 mM cold CTP. The reactions took place at 30°C for 5, 10, 15, 25, and 60 min, respectively, and were terminated by the addition of 1 M sodium chloride in 200 μl HCl (pH 2) with 400 μl chloroform/methanol (1:1). The phases were separated by centrifugation at 9000 × g at 4°C for 5 min. A 150-μl sample of the chloroform phase was deposited onto a piece of filter paper. Samples were dried at room temperature, and their radioactivity was determined by liquid scintillation spectrometry. For measurements of kinetic parameters (KmPA and Vmax), the foregoing assay was performed for three concentrations of PA (1.6, 2.4, and 4.8 mM) for S. japonicus and four concentrations of PA (0.8, 1.6, 2.4, and 4.8 mM) for S. pombe. The reactions took place at 30°C for 15 min. V0 was determined by dividing amount of incorporated CTP by the reaction time (15 min). A plot was drawn with x-axis [PA]−1 and y-axis V0−1. A trend line of the plot was calculated using Excel (Microsoft, Redmond, WA) according to the formula V0−1 = KmPAVm−1[PA]−1 + Vm−1 The value of KmPA equaled the slope of this trend line divided by its ordinate, and Vmax equaled the reciprocal of the ordinate of the trend line.
Supplementary Material
Acknowledgments
We are grateful to D. Zhang, Y. Gu, and M. Makarova for discussions throughout this work and to E. Makeyev for suggestions on the manuscript. The lipid profile data were acquired at the Kansas Lipidomics Research Center, where instrument acquisition and method was supported by National Science Foundation Grants MCB 0455318, MCB 0920663, and DBI 0521587, Kansas INBRE (National Institutes of Health Grant P20 RR16475 from the INBRE program of the National Center for Research Resources), National Science Foundation EPSCoR Grant EPS-0236913, the Kansas Technology Enterprise Corporation, and Kansas State University. This work was supported by the Singapore Millennium Foundation to S.O., the Singapore National Research Foundation (RF2010-06) to J.Y., and start-up funds from the Oregon State University Computational and Genome Biology Initiative to M.F.
Abbreviations used:
- ADEL
ER retention sequence
- CDP-DG
cytidine diphosphate diacylglycerol
- DG
diacylglycerol
- EM
electron microscopy
- ER
endoplasmic reticulum
- FA
fatty acid
- LD
lipid droplet
- NE
nuclear envelope
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- SE
sterol ester
- SNP
single nucleotide polymorphism
- TG
triacylglycerol
- TLC
thin-layer chromatography.
Footnotes
*These authors contributed equally to this work.
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-03-0832) on October 15, 2014.
REFERENCES
- Adeyo O, Horn PJ, Lee S, Binns DD, Chandrahas A, Chapman KD, Goodman JM. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J Cell Biol. 2011;192:1043–1055. doi: 10.1083/jcb.201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Hayashi H, Furuya K, Sato M, Takagi T, Osumi M, Kimura A, Niki H. Breakage of the nuclear envelope by an extending mitotic nucleus occurs during anaphase in Schizosaccharomyces japonicus. Genes Cells. 2011;16:911–926. doi: 10.1111/j.1365-2443.2011.01540.x. [DOI] [PubMed] [Google Scholar]
- Aoki K, Nakajima R, Furuya K, Niki H. Novel episomal vectors and a highly efficient transformation procedure for the fission yeast Schizosaccharomyces japonicus. Yeast. 2010;27:1049–1060. doi: 10.1002/yea.1815. [DOI] [PubMed] [Google Scholar]
- Athenstaedt K, Zweytick D, Jandrositz A, Kohlwein SD, Daum G. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J Bacteriol. 1999;181:6441–6448. doi: 10.1128/jb.181.20.6441-6448.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartz R, Li WH, Venables B, Zehmer JK, Roth MR, Welti R, Anderson RG, Liu P, Chapman KD. Lipidomics reveals that adiposomes store ether lipids and mediate phospholipid traffic. J Lipid Res. 2007;48:837–847. doi: 10.1194/jlr.M600413-JLR200. [DOI] [PubMed] [Google Scholar]
- Bauer C, Herzog V, Bauer MF. Improved technique for electron microscope visualization of yeast membrane structure. Microsc Microanal. 2001;7:530–534. doi: 10.1017.S1431927601010522. [DOI] [PubMed] [Google Scholar]
- Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, Kuhnlein RP. Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics. 2006;5:1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol. 2006;173:719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhman KK, Chen HC, Farese RV., Jr The enzymes of neutral lipid synthesis. J Biol Chem. 2001;276:40369–40372. doi: 10.1074/jbc.R100050200. [DOI] [PubMed] [Google Scholar]
- Carman GM, Han GS. Regulation of phospholipid synthesis in yeast. J lipid Res. 2009;50(Suppl):S69–S73. doi: 10.1194/jlr.R800043-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman GM, Kelley MJ. CDPdiacylglycerol synthase from yeast. Methods Enzymol. 1992;209:242–247. doi: 10.1016/0076-6879(92)09030-7. [DOI] [PubMed] [Google Scholar]
- Carrasco S, Merida I. Diacylglycerol, when simplicity becomes complex. Trends Biochem Sci. 2007;32:27–36. doi: 10.1016/j.tibs.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Carter JR., Jr Cytidine triphosphate: phosphatidic acid cytidyltransferase in Escherichia coli. J Lipid Res. 1968;9:748–754. [PubMed] [Google Scholar]
- Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, Farese RV., Jr Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J Biol Chem. 2001;276:38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- Connerth M, Grillitsch K, Kofeler H, Daum G. Analysis of lipid particles from yeast. Methods Mol Biol. 2009;579:359–374. doi: 10.1007/978-1-60761-322-0_18. [DOI] [PubMed] [Google Scholar]
- Czabany T, Wagner A, Zweytick D, Lohner K, Leitner E, Ingolic E, Daum G. Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J Biol Chem. 2008;283:17065–17074. doi: 10.1074/jbc.M800401200. [DOI] [PubMed] [Google Scholar]
- Daum G, Wagner A, Czabany T, Grillitsch K, Athenstaedt K. Lipid storage and mobilization pathways in yeast. Novartis Found Symp. 2007;286:142–151. doi: 10.1002/9780470985571.ch12. discussion, 151–154, 162–163, 196–203. [DOI] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, Yang H. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol. 2008;180:473–482. doi: 10.1083/jcb.200711136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Shui G, Zhang Y, Krahmer N, Ferguson C, Kapterian TS, Lin RC, Dawes IW, Brown AJ, Li P, et al. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 2011;7:e1002201. doi: 10.1371/journal.pgen.1002201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez S, Homann MJ, Henry SA, Carman GM. Metabolism of the phospholipid precursor inositol and its relationship to growth and viability in the natural auxotroph Schizosaccharomyces pombe. J Bacteriol. 1986;166:779–786. doi: 10.1128/jb.166.3.779-786.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto Y, Itabe H, Kinoshita T, Homma KJ, Onoduka J, Mori M, Yamaguchi S, Makita M, Higashi Y, Yamashita A, Takano T. Involvement of ACSL in local synthesis of neutral lipids in cytoplasmic lipid droplets in human hepatocyte HuH7. J Lipid Res. 2007;48:1280–1292. doi: 10.1194/jlr.M700050-JLR200. [DOI] [PubMed] [Google Scholar]
- Fujimoto T, Parton RG. Not just fat: the structure and function of the lipid droplet. Cold Spring Harbor Perspect Biol. 2011;3:a004838. doi: 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya K, Niki H. Isolation of heterothallic haploid and auxotrophic mutants of Schizosaccharomyces japonicus. Yeast. 2009;26:221–233. doi: 10.1002/yea.1662. [DOI] [PubMed] [Google Scholar]
- Ganong BR, Leonard JM, Raetz CR. Phosphatidic acid accumulation in the membranes of Escherichia coli mutants defective in CDP-diglyceride synthetase. J Biol Chem. 1980;255:1623–1629. [PubMed] [Google Scholar]
- Ganong BR, Raetz CR. Massive accumulation of phosphatidic acid in conditionally lethal CDP-diglyceride synthetase mutants and cytidine auxotrophs of Escherichia coli. J Biol Chem. 1982;257:389–394. [PubMed] [Google Scholar]
- Gaspar ML, Hofbauer HF, Kohlwein SD, Henry SA. Coordination of storage lipid synthesis and membrane biogenesis: evidence for cross-talk between triacylglycerol metabolism and phosphatidylinositol synthesis. J Biol Chem. 2011;286:1696–1708. doi: 10.1074/jbc.M110.172296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor PM, Greenberg ML. Regulation of CDP-diacylglycerol synthesis and utilization by inositol and choline in Schizosaccharomyces pombe. J Bacteriol. 1992;174:5711–5718. doi: 10.1128/jb.174.17.5711-5718.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman JM. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J Lipid Res. 2009;50:2148–2156. doi: 10.1194/jlr.R001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL. Protocols for experimentation with Schizosaccharomyces pombe. Methods. 2004;33:187–188. doi: 10.1016/j.ymeth.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Gu Y, Yam C, Oliferenko S. Divergence of mitotic strategies in fission yeasts. Nucleus. 2012;3:220–225. doi: 10.4161/nucl.19514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingavale SS, Bachhawat AK. Restoration of inositol prototrophy in the fission yeast Schizosaccharomyces pombe. Microbiology. 1999;145:1903–1910. doi: 10.1099/13500872-145-8-1903. [DOI] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci. 2011;124:2424–2437. doi: 10.1242/jcs.076836. [DOI] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, Simons K. Flexibility of a eukaryotic lipidome—insights from yeast lipidomics. PLoS One. 2012;7:e35063. doi: 10.1371/journal.pone.0035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein SD, Veenhuis M, van der Klei IJ. Lipid droplets and peroxisomes: key players in cellular lipid homeostasis or a matter of fat—store ‘em up or burn ‘em down. Genetics. 2013;193:1–50. doi: 10.1534/genetics.112.143362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Guo Y, Wilfling F, Hilger M, Lingrell S, Heger K, Newman HW, Schmidt-Supprian M, Vance DE, Mann M, et al. Phosphatidylcholine synthesis for lipid droplet expansion is mediated by localized activation of CTP:phosphocholine cytidylyltransferase. Cell Metab. 2011;14:504–515. doi: 10.1016/j.cmet.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuerschner L, Moessinger C, Thiele C. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 2008;9:338–352. doi: 10.1111/j.1600-0854.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem. 2006;281:491–500. doi: 10.1074/jbc.M508414200. [DOI] [PubMed] [Google Scholar]
- Kurat CF, Wolinski H, Petschnigg J, Kaluarachchi S, Andrews B, Natter K, Kohlwein SD. Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol Cell. 2009;33:53–63. doi: 10.1016/j.molcel.2008.12.019. [DOI] [PubMed] [Google Scholar]
- Lee J, Welti R, Schapaugh WT, Trick HN. Phospholipid and triacylglycerol profiles modified by PLD suppression in soybean seed. Plant Biotechnol J. 2011;9:359–372. doi: 10.1111/j.1467-7652.2010.00562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long AP, Manneschmidt AK, VerBrugge B, Dortch MR, Minkin SC, Prater KE, Biggerstaff JP, Dunlap JR, Dalhaimer P. Lipid droplet de novo formation and fission are linked to the cell cycle in fission yeast. Traffic. 2012;13:705–714. doi: 10.1111/j.1600-0854.2012.01339.x. [DOI] [PubMed] [Google Scholar]
- Mora G, Scharnewski M, Fulda M. Neutral lipid metabolism influences phospholipid synthesis and deacylation in Saccharomyces cerevisiae. PLoS One. 2012;7:e49269. doi: 10.1371/journal.pone.0049269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S, Martin S, Parton RG. Lipid droplet-organelle interactions; sharing the fats. Biochim Biophys Acta. 2009;1791:441–447. doi: 10.1016/j.bbalip.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Murphy S, Martin S, Parton RG. Quantitative analysis of lipid droplet fusion: inefficient steady state fusion but rapid stimulation by chemical fusogens. PLoS One. 2010;5:e15030. doi: 10.1371/journal.pone.0015030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy DJ, Vance J. Mechanisms of lipid-body formation. Trends Biochem Sci. 1999;24:109–115. doi: 10.1016/s0968-0004(98)01349-8. [DOI] [PubMed] [Google Scholar]
- Niki H. Schizosaccharomyces japonicus: the fission yeast is a fusion of yeast and hyphae. Yeast. 2014;31:83–90. doi: 10.1002/yea.2996. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem. 2002;277:8877–8881. doi: 10.1074/jbc.M111646200. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Tinkelenberg A, Erdeniz N, Cromley D, Billheimer JT, Sturley SL. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J Biol Chem. 2000;275:15609–15612. doi: 10.1074/jbc.C000144200. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Furuya K, Nozaki S, Aoki K, Niki H. Synchronous activation of cell division by light or temperature stimuli in the dimorphic yeast Schizosaccharomyces japonicus. Eukaryot Cell. 2013;12:1235–1243. doi: 10.1128/EC.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penno A, Hackenbroich G, Thiele C. Phospholipids and lipid droplets. Biochim Biophys Acta. 2013;1831:589–594. doi: 10.1016/j.bbalip.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Petschnigg J, Wolinski H, Kolb D, Zellnig G, Kurat CF, Natter K, Kohlwein SD. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J Biol Chem. 2009;284:30981–30993. doi: 10.1074/jbc.M109.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploegh HL. A lipid-based model for the creation of an escape hatch from the endoplasmic reticulum. Nature. 2007;448:435–438. doi: 10.1038/nature06004. [DOI] [PubMed] [Google Scholar]
- Pomraning KR, Smith KM, Freitag M. Genome-wide high throughput analysis of DNA methylation in eukaryotes. Methods. 2009;47:142–150. doi: 10.1016/j.ymeth.2008.09.022. [DOI] [PubMed] [Google Scholar]
- Pomraning KR, Smith KM, Freitag M. Bulk segregant analysis followed by high-throughput sequencing reveals the Neurospora cell cycle gene, ndc-1, to be allelic with the gene for ornithine decarboxylase, spe-1. Eukaryot Cell. 2011;10:724–733. doi: 10.1128/EC.00016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CR, Kennedy EP. Function of cytidine diphosphate-diglyceride and deoxycytidine diphosphate-diglyceride in the biogenesis of membrane lipids in Escherichia coli. J Biol Chem. 1973;248:1098–1105. [PubMed] [Google Scholar]
- Rajakumari S, Daum G. Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol Biol Cell. 2010;21:501–510. doi: 10.1091/mbc.E09-09-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumari S, Rajasekharan R, Daum G. Triacylglycerol lipolysis is linked to sphingolipid and phospholipid metabolism of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2010;1801:1314–1322. doi: 10.1016/j.bbalip.2010.08.004. [DOI] [PubMed] [Google Scholar]
- Raychaudhuri S, Young BP, Espenshade PJ, Loewen C., Jr Regulation of lipid metabolism: a tale of two yeasts. Curr Opin Cell Biol. 2012;24:502–508. doi: 10.1016/j.ceb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Goto K, Tonosaki A, Kondo H. Gene cloning and characterization of CDP-diacylglycerol synthase from rat brain. J Biol Chem. 1997;272:9503–9509. doi: 10.1074/jbc.272.14.9503. [DOI] [PubMed] [Google Scholar]
- Savage MJ, Goldberg DJ, Schacher S. Absolute specificity for retrograde fast axonal transport displayed by lipid droplets originating in the axon of an identified Aplysia neuron in vitro. Brain Res. 1987;406:215–223. doi: 10.1016/0006-8993(87)90785-2. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Heacock PN, Clancey CJ, Dowhan W. The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. J Biol Chem. 1996;271:789–795. doi: 10.1074/jbc.271.2.789. [DOI] [PubMed] [Google Scholar]
- Sturmey RG, O’Toole PJ, Leese HJ. Fluorescence resonance energy transfer analysis of mitochondrial:lipid association in the porcine oocyte. Reproduction. 2006;132:829–837. doi: 10.1530/REP-06-0073. [DOI] [PubMed] [Google Scholar]
- Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J Biol Chem. 2002;277:44507–44512. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer HF, Wolinski H, Veenhuis M, van der Klei IJ, Kohlwein SD. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2014;25:290–301. doi: 10.1091/mbc.E13-08-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV., Jr The life of lipid droplets. Biochim Biophys Acta. 2009;1791:459–466. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte MA, Cermelli S, Griner J, Viera A, Guo Y, Kim DH, Gindhart JG, Gross SP. Regulation of lipid-droplet transport by the perilipin homolog LSD2. Curr Biol. 2005;15:1266–1275. doi: 10.1016/j.cub.2005.06.062. [DOI] [PubMed] [Google Scholar]
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, et al. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev Cell. 2013;24:384–399. doi: 10.1016/j.devcel.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinski H, Kolb D, Hermann S, Koning RI, Kohlwein SD. A role for seipin in lipid droplet dynamics and inheritance in yeast. J Cell Sci. 2011;124:3894–3904. doi: 10.1242/jcs.091454. [DOI] [PubMed] [Google Scholar]
- Xu N, Zhang SO, Cole RA, McKinney SA, Guo F, Haas JT, Bobba S, Farese RV, Jr, Mak HY. The FATP1-DGAT2 complex facilitates lipid droplet expansion at the ER-lipid droplet interface. J Cell Biol. 2012;198:895–911. doi: 10.1083/jcb.201201139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam C, Gu Y, Oliferenko S. Partitioning and remodeling of the Schizosaccharomyces japonicus mitotic nucleus require chromosome tethers. Curr Biol. 2013;23:2303–2310. doi: 10.1016/j.cub.2013.09.057. [DOI] [PubMed] [Google Scholar]
- Yam C, He Y, Zhang D, Chiam KH, Oliferenko S. Divergent strategies for controlling the nuclear membrane satisfy geometric constraints during nuclear division. Curr Biol. 2011;21:1314–1319. doi: 10.1016/j.cub.2011.06.052. [DOI] [PubMed] [Google Scholar]
- Yang H, Bard M, Bruner DA, Gleeson A, Deckelbaum RJ, Aljinovic G, Pohl TM, Rothstein R, Sturley SL. Sterol esterification in yeast: a two-gene process. Science. 1996;272:1353–1356. doi: 10.1126/science.272.5266.1353. [DOI] [PubMed] [Google Scholar]
- Yazawa H, Kumagai H, Uemura H. Characterization of triglyceride lipase genes of fission yeast Schizosaccharomyces pombe. Appl Microbiol Biotechnol. 2012;96:981–991. doi: 10.1007/s00253-012-4151-8. [DOI] [PubMed] [Google Scholar]
- Zhang D, Vjestica A, Oliferenko S. The cortical ER network limits the permissive zone for actomyosin ring assembly. Curr Biol. 2010;20:1029–1034. doi: 10.1016/j.cub.2010.04.017. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Chieu HK, Low CP, Zhang S, Heng CK, Yang H. Schizosaccharomyces pombe cells deficient in triacylglycerols synthesis undergo apoptosis upon entry into the stationary phase. J Biol Chem. 2003;278:47145–47155. doi: 10.1074/jbc.M306998200. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Peisker H, Weth A, Baumgartner W, Dormann P, Frentzen M. Extraplastidial cytidinediphosphate diacylglycerol synthase activity is required for vegetative development in Arabidopsis thaliana. Plant J. 2013;75:867–879. doi: 10.1111/tpj.12248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.