Genetic interaction analysis is used to identify new cytokinesis proteins involved in myosin II cleavage furrow accumulation and to demonstrate how different pathways collaborate to drive myosin II to the cleavage furrow. One of these proteins, RMD1, is required for myosin II cleavage furrow localization and acts in parallel with mechanical stress.
Abstract
How myosin II localizes to the cleavage furrow in Dictyostelium and metazoan cells remains largely unknown despite significant advances in understanding its regulation. We designed a genetic selection using cDNA library suppression of 3xAsp myosin II to identify factors involved in myosin cleavage furrow accumulation. The 3xAsp mutant is deficient in bipolar thick filament assembly, fails to accumulate at the cleavage furrow, cannot rescue myoII-null cytokinesis, and has impaired mechanosensitive accumulation. Eleven genes suppressed this dominant cytokinesis deficiency when 3xAsp was expressed in wild-type cells. 3xAsp myosin II's localization to the cleavage furrow was rescued by constructs encoding rcdBB, mmsdh, RMD1, actin, one novel protein, and a 14-3-3 hairpin. Further characterization showed that RMD1 is required for myosin II cleavage furrow accumulation, acting in parallel with mechanical stress. Analysis of several mutant strains revealed that different thresholds of myosin II activity are required for daughter cell symmetry than for furrow ingression dynamics. Finally, an engineered myosin II with a longer lever arm (2xELC), producing a highly mechanosensitive motor, could also partially suppress the intragenic 3xAsp. Overall, myosin II accumulation is the result of multiple parallel and partially redundant pathways that comprise a cellular contractility control system.
INTRODUCTION
Cytokinesis is a spectacular cell shape change in which a mother cell divides into two daughter cells in as little as 5 min. This process involves symmetry breaking as the cell and the mitotic spindle elongate, beginning at the metaphase-to-anaphase transition. The accumulation of a contractile meshwork then contributes to the equatorial mechanical changes that promote furrow ingression (Zhang and Robinson, 2005; Octtaviani et al., 2006; Reichl et al., 2008). During cytokinesis in Dictyostelium, the cell cortex becomes highly mechanosensitive and increases its deformability, particularly around the polar cortex (Effler et al., 2006; Reichl et al., 2008; Ren et al., 2009). This mechanosensitivity allows for mechanical stresses acting at the surface of the cortex to drive myosin II and cortexillin I accumulation in a cooperative manner that is dependent on myosin II mechanochemistry (Ren et al., 2009; Luo et al., 2012). This stress sensitivity acts independently of the mitotic spindle in postmetaphase cells and may help drive myosin II to the cleavage furrow cortex in three-dimensional culture and tissue environments, as well as in some asymmetric cell divisions (Cabernard et al., 2010; Ou et al., 2010; Surcel et al., 2010; Kee et al., 2012).
Other mechanisms for promoting myosin II cleavage furrow cortex accumulation are believed to involve signals directly emanating from the mitotic spindle. In metazoans, these signals originate from proteins that associate with the kinesin-6 family proteins, including MKLP1 (mammals), zen-4 (Caenorhabditis elegans), and kif12 (Dictyostelium). One class of kinesin-6–associated proteins includes MgcRacGAP, which constitutes the centralspindlin complex (Mishima et al., 2002; Pavicic-Kaltenbrunner et al., 2007). This complex, along with the ECT2 Rho GTP-exchange factor, maintains active RhoA at the cleavage furrow cortex (Yuce et al., 2005; Petronczki et al., 2007). Subsequently RhoA locally activates Rho kinase, which phosphorylates myosin II regulatory light chain, promoting a conformational change in the myosin II heavy chain, allowing it to assemble into bipolar thick filaments (BTFs) and enhancing the actin-activated myosin II ATPase activity. Other signals, such as from the heterotrimeric protein G0α, appear to promote cleavage furrow formation and myosin II accumulation in the absence of the centralspindlin complex in C. elegans (Bringmann and Hyman, 2005; Bringmann et al., 2007). However, this system of Rho kinase and MgcRacGAP appears to be absent from the Dictyostelium genome (Eichinger et al., 2005). Another major class of kinesin-6–associated proteins includes the chromosomal passenger complex proteins INCENP and Aurora kinase (Cooke et al., 1987; Adams et al., 2000). In Dictyostelium, aurora kinase is encoded by a single gene, appears to be essential, and accumulates measurably at the cleavage furrow only very late during furrow ingression (Li et al., 2008). INCENP accumulates at the cleavage furrow cortex in a myosin II–dependent manner, although myosin II can still accumulate at the cleavage furrow cortex of incenp-null cells (Chen et al., 2006; Chen et al., 2007). Finally, myosin II accumulation is modulated by its own set of kinases and phosphatases, which regulate heavy chain phosphorylation, thereby modulating myosin BTF assembly (Egelhoff et al., 1993; Yumura et al., 2005; Wang et al., 2011).
A clear signaling pathway has not yet been found that fully explains how myosin II accumulates at the cleavage furrow cortex. This is undoubtedly due to system redundancy, as multiple pathways can direct myosin II accumulation, including mechanical stress– and spindle-mediated pathways. Furthermore, myosin II can accumulate in several systems in the absence of the motor domain, apparently independent of actin polymers and in a manner that depends on the ability to form bipolar thick filaments rather than on a specific targeting sequence within the myosin II tail. However, in these instances, the myosin fails to incorporate tightly in the furrow cortex or only transiently associates before dissipating again (Zang and Spudich, 1998; Shu et al., 1999; Dean et al., 2005). These observations argue for other pathways and/or a cortical receptor (R) that can either help target the BTFs or retain the BTFs in the cleavage furrow cortex. 14-3-3 (encoded by a single gene in Dictyostelium) could be part of one such cortical receptor for myosin II (Robinson, 2010; Zhou et al., 2010). In metazoans, anillin is another candidate for such a cortical receptor (Straight et al., 2005; Piekny and Glotzer, 2008).
Here, to search for proteins involved in myosin II cleavage furrow cortex accumulation, we leveraged our genetic tools and the 3xAsp mutant myosin II heavy chain. This myosin II mutant has three critical threonines in the tail mutated to aspartic acids, mimicking the phosphorylated myosin heavy chain and driving myosin II toward the disassembled state (Egelhoff et al., 1993). The 3xAsp myosin II has a severely impaired ability to assemble into BTFs in vitro, remains largely disassembled in vivo, and typically fails to accumulate appreciably at the interphase or cleavage furrow cortices (Egelhoff et al., 1993; Sabry et al., 1997). Our experimental design is based on the assumption that myosin II exists in a minimum of four states: the assembly-incompetent monomer (M0, which is largely populated by the phosphorylated wild-type [WT] or 3xAsp myosin II), the assembly-competent monomer (M, which is believed to be largely populated by the unphosphorylated monomer), the BTFn(cytoplasm) (BTF of size-n myosin monomers in the cytoplasm), and the BTFn(cortex)⋅R (BTF of size n myosin monomers in the cortex, bound by any cortical receptors R). In this case, myosin II cleavage furrow recruitment will look like the following (Robinson, 2010):
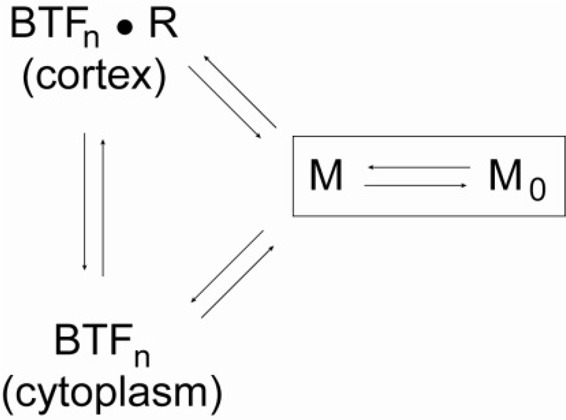
Myosin Recruitment Scheme
3xAsp alone is shifted toward M0, and upon expression of 3xAsp in a WT background (WT::3xAsp), normal myosin II assembly is altered, leading to cytokinesis defects. We subjected this strain to genetic selection and identified suppressors that could restore myosin II on the cortex and lead to improved growth (Robinson et al., 2002; Zhou et al., 2010). We found 11 genes that could suppress the growth deficiency of WT::3xAsp cells. For a secondary analysis, we tested the effect of these genes plus five additional ones of interest on the distribution of 3xAsp expressed in a myoII-null background, so that the only myosin II present is 3xAsp. Of these genes, six are able to significantly restore the ability of 3xAsp myosin II to accumulate at the cleavage furrow cortex. Initial characterization of one of these genes, regulator of microtubule dynamics 1 (RMD1), revealed that RMD1 contributes to cytokinesis fidelity, cortical tension, and myosin II cleavage furrow accumulation in a manner that is parallel to mechanical stress. We also found that the two major features of cytokinesis—furrow ingression dynamics and symmetry of daughter cell size—have different thresholds for myosin II activity, which RMD1 helps modulate. Finally, to test the relationship between myosin mechanosensitivity and heavy chain phosphorylation, we constructed and examined an engineered myosin with a long lever arm (2xELC) and the 3xAsp tail. The increased mechanosensitivity of the 2xELC motor allowed greater furrow accumulation than for 3xAsp alone, demonstrating cross-talk between the cooperative actin binding by the motor and thick filament assembly. Overall, myosin II cleavage furrow accumulation is a system-level function with many modulating determinants, stemming from both chemical and mechanical stimuli.
RESULTS
Genetic suppression of 3xAsp
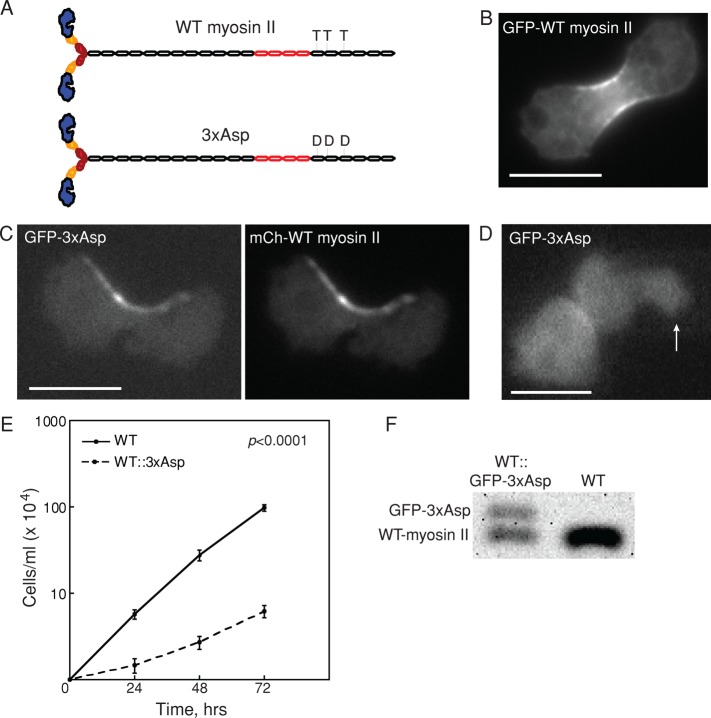
To establish a system for identifying proteins involved in novel pathways for myosin II cleavage furrow accumulation, we characterized the effects of 3xAsp myosin II on WT myosin II activity (Figure 1A). The 3xAsp mutant myosin II has the threonine residues at positions 1823, 1833, and 2029 mutated to aspartic acid residues, mimicking the phosphorylated WT myosin II and disrupting the ability of this myosin II to assemble into BTFs (Egelhoff et al., 1993). Compared to the uniform cleavage furrow distribution of WT myosin II (Figure 1B), when GFP-3xAsp was expressed along with mCh-WT myosin II, the two proteins colocalized at the cleavage furrow cortex, but the overall myosin II distribution was disrupted, with irregular, asymmetric accumulation often observed (Figure 1C). We also tested whether 3xAsp could accumulate in response to applied stress in the presence of WT myosin II. We found that the mechanosensory responsiveness of 3xAsp myosin II remained disrupted, indicating a severe impairment of myosin II dynamics (Figure 1D; Ren et al., 2009). Finally, although it is well characterized that myoII-null cells expressing 3xAsp are completely unable to grow in suspension culture (Egelhoff et al., 1993), WT cells expressing 3xAsp from an episomal plasmid were able to survive in shaking culture, albeit with greatly reduced growth rates as compared with WT cells (unpublished data).
FIGURE 1:
Overexpression of 3xAsp in WT cells reduced growth rate. (A) Diagram of WT myosin II and 3xAsp myosin II. (B) GFP-WT myosin II accumulates at the furrow. Scale bar, 10 μm. (C) Myosin II distribution in myoII::GFP-3xAsp;mCh-WT myosin II was altered by expression of 3xAsp myosin II. GFP-3xAsp and mCherry-WT myosin II colocalized at the cleavage furrow in dividing cells but with an aberrant distribution. Scale bar, 10 μm. (D) WT cells expressing GFP-3xAsp (WT::3xAsp) did not show mechanosensitive myosin II accumulation in response to micropipette aspiration (white arrow). The background-corrected fluorescence intensity ratio of the cortex inside the micropipette (Ip) and the opposite cortex (Io) was measured and used to determine whether the myosin II undergoes mechanosensitive accumulation (Effler et al., 2006). Scale bar, 10 μm. (E) Suspension culture growth for WT control and WT::3xAsp cells. Average growth rate is 0.067 ± 0.008 h−1 for WT cells (n = 11) and 0.032 ± 0.009 h−1 for WT::3xAsp cells (n = 11; p value is on the graph). (F) Western analysis of WT and WT::GFP-3xAsp (where 3xAsp is integrated randomly in the genome) showed that 3xAsp is 40% and WT endogenous myosin II is 60% of the total myosin II in these cells.
This combination of defects in the uniformity of cleavage furrow accumulation, mechanosensitive localization, and growth rate were considered essential for our experimental design to identify genes that encode proteins involved in nonmechanosensitive myosin II accumulation. We stably integrated green fluorescent protein (GFP)–3xAsp into the genome of WT cells, creating WT::3xAsp cells. This insertion was integrated randomly. These WT::3xAsp cells displayed all of the phenotypes of the cells expressing the proteins from episomal plasmids, including the greatly reduced growth rates as compared with WT cells (Figure 1E). The mobility shift of the 3xAsp myosin II heavy chain due to the GFP fusion allowed us to perform Western analysis to determine the ratio of 3xAsp myosin II to WT endogenous myosin II. We found that 3xAsp represented 40% of the total myosin II in these cells (Figure 1F).
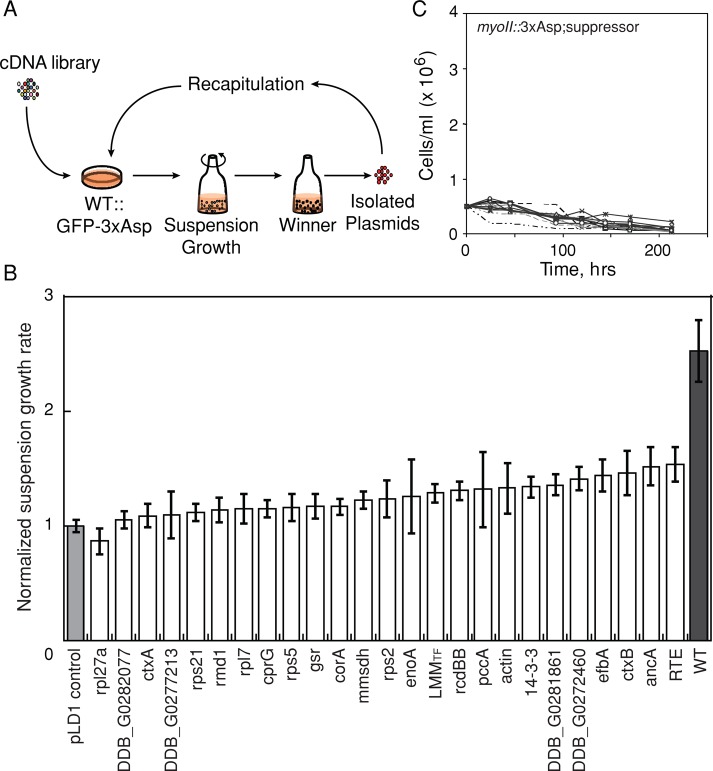
These WT::3xAsp cells were subjected to cDNA library suppression to select for genes that could rescue the WT::3xAsp cytokinesis defects in suspension growth (Figure 2A). A total of 77 pools of ∼1800 transformants/pool (140,000 total transformants) were generated and grown in suspension culture for ∼3–4 wk. Once a pool showed a growth rate increase of >30% over the empty vector control pool, the cDNAs were isolated, and individual cDNAs were identified (Robinson and Spudich, 2000; Zhou et al., 2010). After sequence analysis, we recovered 25 independent genes from the selection. One cDNA included the myosin-coding sequence spanning nucleotides 4459 to the poly A tail, which is essentially the coding sequence of the light meromyosin (LMM) region of the myosin heavy chain tail. However, the only in-frame ATG did not occur until nucleotide 5671. Thus LMMTF is predicted to encode only the myosin tail fragment (TF), which spans amino acids 1891–2116. The recovery of LMMTF demonstrated that our cDNA library suppression approach could identify genes involved in myosin II function.
FIGURE 2:
cDNA library complementation selection identified 3xAsp suppressors. (A) Strategy of cDNA library suppression. A Dictyostelium cDNA library was transformed into WT::3xAsp cells and subjected to selection using suspension growth. Plasmids were isolated and identified from winner pools based on growth rate. Recovered plasmids were reintroduced into WT::3xAsp cells, which were again subjected to suspension growth to identify strong suppressors. (B) Recapitulation results of “winner” plasmids were sorted according to mean growth rates. All growth rates were normalized over empty vector control (pLD1 control), as shown by the light gray bar. WT control (WT::GFP-myosin II) cells are shown by the dark gray bar. Error bars, SEM. (C) Suspension of growth of the myoII::3xAsp cells transformed with suppressor plasmids and several genes of interest, which included corA, rps2, RMD1, cortexillin I, and 14-3-3 hp. None of these genes was able to rescue the growth of myoII::3xAsp cells in suspension culture, a highly restrictive condition for myosin II–deficient growth.
To confirm that each of the genes could recapitulate the suppression, we reintroduced the purified clonal plasmids in duplicate in fresh WT::3xAsp cells. Culture growth rates were measured for up to five passages and normalized by the empty vector control (Figure 2B). Eleven of these genes recapitulated the suppression and rescued the suspension growth defect of WT::3xAsp mutant cells (p < 0.05 by Student's t test; Table 1). Note that cells were frequently imaged throughout the experiment to confirm that the genetic suppression was not due to the loss of 3xAsp expression. For these 11 suppressors, the cell lines continued to express GFP-3xAsp myosin II at initial levels. Because LMMTF includes WT myosin sequence, spanning the three mutated threonines in 3xAsp, it is possible that the cDNA recombined with the integrated 3xAsp sequence, correcting the residues to WT threonines; therefore, we focused our analysis on our other hits.
TABLE 1:
Recapitulation of 3xAsp suppressors from cDNA library suppression.
| Gene name | Gene ID | Mean ± SEM (n) | p |
|---|---|---|---|
| Wild-type controla | 2.5 ± 0.27 (9) | 0.0001 | |
| DDB_G0272460 | DDB_G0272460 | 1.4 ± 0.10 (10) | 0.0009 |
| DDB_G0293690_RTE (TRE5-A ORF1) | DDB_G0293690 | 1.5 ± 0.15 (10) | 0.0013 |
| DDB_G0281861 | DDB_G0281861 | 1.4 ± 0.094 (10) | 0.0016 |
| efbA | DDB_G0288373 | 1.4 ± 0.14 (8) | 0.002 |
| ancA | DDB_G0267454 | 1.5 ± 0.17 (9) | 0.0023 |
| rcdBB (random cDNA clone veg113) | DDB_G0274551 | 1.3 ± 0.080 (10) | 0.0026 |
| 14-3-3 | DDB_G0269138 | 1.3 ± 0.094 (11) | 0.0029 |
| ctxB (cortexillin II) | DDB_G0276893 | 1.5 ± 0.19 (9) | 0.013 |
| mmsdh | DDB_G0289085 | 1.2 ± 0.077 (9) | 0.016 |
| mhcA LMMTF | DDB_G0286355 | 1.3 ± 0.080 (6) | 0.019 |
| actin (e.g., act8) | DDB_G0269234 | 1.3 ± 0.22 (5) | 0.046 |
| corA (coronin) | DDB_G0267382 | 1.2 ± 0.071 (7) | 0.059 |
| rps2 (ribosomal protein S2) | DDB_G0293742 | 1.2 ± 0.16 (5) | 0.073 |
| cprG (cysteine proteinase 7) | DDB_G0279187 | 1.2 ± 0.077 (6) | 0.11 |
| pccA | DDB_G0275355 | 1.3 ± 0.33 (5) | 0.14 |
| gsr (gluthathione-SH reductase) | DDB_G0272754 | 1.2 ± 0.11 (10) | 0.15 |
| rps5 (ribosomal protein small subunit 5) | DDB_G0286075 | 1.2 ± 0.12 (9) | 0.18 |
| enoA | DDB_G0283137 | 1.3 ± 0.33 (4) | 0.19 |
| rmd1 | DDB_G0278793 | 1.1 ± 0.11 (10) | 0.21 |
| rpl7 (S60 ribosomal protein L7) | DDB_G0276441 | 1.2 ± 0.13 (8) | 0.23 |
| rpl27a (S60 ribosomal protein L27a) | DDB_G0292388 | 0.87 ± 0.11 (5) | 0.25 |
| rps21 (40S ribosomal protein S21) | DDB_G0293700 | 1.1 ± 0.073 (3) | 0.29 |
| ctxA (cortexillin I) | DDB_G0289483 | 1.1 ± 0.099 (7) | 0.37 |
| DDB_G0282077 | DDB_G0282077 | 1.1 ± 0.075 (4) | 0.51 |
| DDB_G0277213 | DDB_G0277213 | 1.1 ± 0.20 (7) | 0.54 |
| pLD1 vector controlb | 1 ± 0.049 (13) |
Plasmids recovered from the cDNA library suppression screen of WT::3xAsp cells (Ax3(Rep orf+)::pDRH:GFP-3xAsp) were isolated and transformed into fresh WT::3xAsp cells. These were subjected to suspension growth to determine growth rates.
aWild-type control cells were WT::GFP-WT-myosin II cells (Ax3(Rep orf+)::pDRH:GFP-myosin-II; pLD1).
bpLD1 vector control was Ax3(Rep orf+)::pDRH:GFP-3xAsp; pLD1 cells.
The primary selection was performed using WT::3xAsp cells, which expressed both endogenous WT myosin II and 3xAsp myosin II. For a more stringent test, we analyzed the ability of the suppressors to rescue myoII::3xAsp suspension growth (where 3xAsp is the only myosin II in these cells). None of the 15 genes tested was able to rescue the suspension growth of myoII::3xAsp cells (Figure 2C), which is not surprising, as this is a very rigorous test for myosin II function.
Effect of suppressors on 3xAsp distribution during cytokinesis
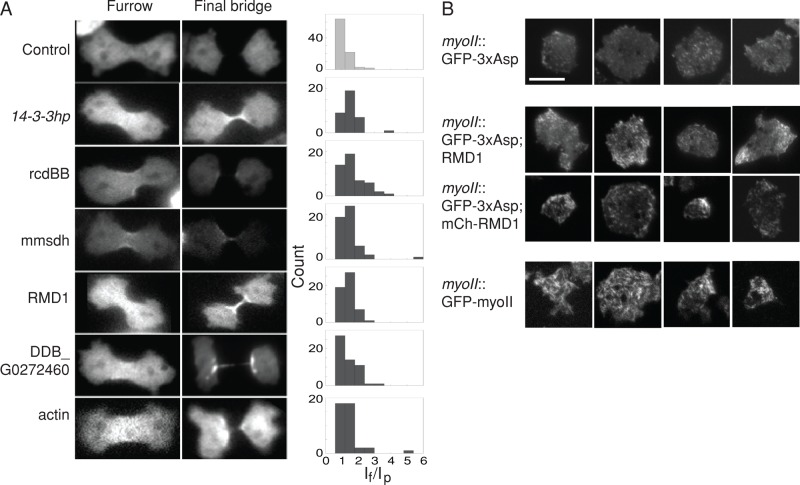
We then tested whether the suppressors could promote 3xAsp cleavage furrow localization in the myoII::3xAsp cells. When expressed alone, 3xAsp seldom showed accumulation at the cleavage furrow cortex (Figure 3A and Table 2). Based on our experimental design, if a gene plays a pivotal role in modulating myosin II assembly or targeting myosin II to the site of cell division, expression of this suppressor may help drive 3xAsp myosin II toward the cleavage furrow. We expressed each of the recapitulated suppressors in myoII::3xAsp cells, as well as five others that were just above the p < 0.05 threshold. These five included two actin cross-linking proteins (cortexillin I and coronin), RMD1, rps2, and 14-3-3, which we previously showed is involved in the myosin II–RacE pathway that controls myosin II cortical accumulation and dynamics (Zhou et al., 2010). From our conception of how 14-3-3 modulates myosin II assembly, we also tested the 14-3-3hp, which reduces 14-3-3 expression by 70% and also leads to overassembled myosin II in an otherwise WT background (Zhou et al., 2010). We quantified the myosin II levels at the cleavage furrow cortex by measuring the myosin II intensity at the furrow (If) and polar (Ip) cortices and then calculating the intensity ratio (If/Ip) for each cell. Histograms of the distribution of 3xAsp If/Ip ratios were plotted for each cell line and compared with the empty vector control (Figure 3A). Nine of 15 plasmids showed an enhancement of the 3xAsp furrow localization with p < 0.10. These plasmids were 14-3-3hp, rcdBB, mmsdh (methylmalonate-semialdehyde dehydrogenase), rmd1, a novel protein (gene ID: DDB_G0272460), actin, coronin, efbA, and cortexillin II (Table 2). As one example, the mean 3xAsp If/Ip ratio for the pLD1 empty vector control was 1.17 ± 0.034, whereas a strong suppressor plasmid rcdBB had a ratio of 1.64 ± 0.11 (Student's t test, p < 0.0001; Figure 3A and Table 2).
FIGURE 3:
3xAsp suppressors restored 3xAsp cleavage furrow accumulation. (A) Expression of 3xAsp suppressors increased furrow accumulation of GFP-3xAsp in myoII-null cells. Epifluorescence images of dividing cells at telophase and final bridge stages. Scale bar, 10 μm. Frequency histograms show the 3xAsp fluorescence intensity ratio of furrow/pole (If/Ip). (B) TIRF images show extent of myosin II assembly in the cortex. GFP-3xAsp myosin is imaged for myoII::GFP-3xAsp cells expressing empty vector control, RMD1, and mCherry-RMD1 (mCh-RMD1). mCh-RMD1 was used to confirm that RMD1 was expressed in these cells. GFP-WT myosin II (myoII::GFP-myosin II) was examined as a positive control. Four examples of each are shown. Scale bar, 10 μm.
TABLE 2:
GFP-3xAsp intensity ratio If/Ip in myoII nulls expressing 3xAsp suppressors.
| Gene | If/Ip ratio ± SEM (n) | p |
|---|---|---|
| 14-3-3hp | 1.51 ± 0.086 (37) | 0.0001 |
| rcdBB | 1.64 ± 0.11 (48) | 0.0001 |
| mmsdh | 1.46 ± 0.097 (52) | 0.0011 |
| rmd1 | 1.37 ± 0.051 (54) | 0.0012 |
| DDB_G0272460 | 1.38 ± 0.069 (54) | 0.0032 |
| actin | 1.41 ± 0.12 (41) | 0.011 |
| corA | 1.31 ± 0.074 (55) | 0.056 |
| efbA | 1.31 ± 0.086 (44) | 0.077 |
| ctxB | 1.28 ± 0.058 (53) | 0.099 |
| 14-3-3 | 1.24 ± 0.073 (48) | 0.32 |
| ctxA | 1.23 ± 0.058 (34) | 0.34 |
| Calcineurin-like | 1.22 ± 0.054 (57) | 0.40 |
| TRE5-A ORF | 1.20 ± 0.060 (52) | 0.69 |
| rps2 | 1.19 ± 0.061 (48) | 0.73 |
| ancA | 1.18 ± 0.053 (50) | 0.90 |
| pLD1 vector controla | 1.17 ± 0.034 (91) | |
| WT GFP myosin IIb | 2.1 ± 0.2 (29) | 0.0001 |
The background strain was the myoII:3xAsp cells (myoII::pDRH:GFP-3xAsp). n is the number of cells analyzed.
apLD1 vector control was the mutant transformed with the empty vector.
bData for late-stage furrows from Figure 6B.
One way in which the suppressors could rescue 3xAsp myosin II is by promoting assembly of the 3xAsp myosin II into BTFs. To test this, we performed total internal reflection fluorescence (TIRF) microscopy to examine the BTF assembly state of 3xAsp alone and with the suppressors and compared these to images of WT BTFs, which are readily visible by TIRF (Liang et al., 2002; Girard et al., 2006). Contrary to expectation, we found that 3xAsp can form some very short filaments and/or aggregations/bundles in the myoII-null cells. Actin and mmsdh did not significantly alter 3xAsp assembly compared with 3xAsp alone; however, RMD1 appeared to increase cortical 3xAsp levels, although not to the level of WT myosin (Figure 3B).
Characterization of 3xAsp suppressors for myosin II furrow recruitment
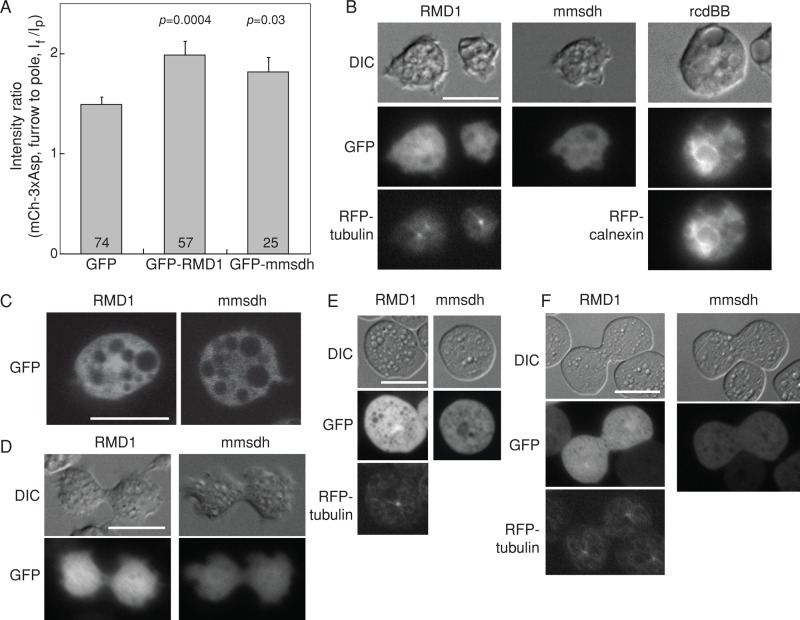
The genes that rescued 3xAsp myosin II furrow accumulation in the myoII-null background might play an important role in myosin II assembly dynamics and/or in pathways involved in myosin II localization and may include “hypothetical receptors” that might be involved in recruiting myosin II to the cleavage furrow during cytokinesis. We focused on three proteins—RMD1, rcdBB, and mmsdh—which are among the top hits in the 3xAsp furrow localization rescue and suspension growth assays. We analyzed the subcellular localizations of these proteins by live-cell imaging of WT cells expressing N-terminal GFP- or mCherry-tagged fusion proteins. Suspension growth and 3xAsp furrow localization assays were repeated to confirm that these fusions did not impair protein function. Both GFP-RMD1 and GFP-mmsdh increased suspension growth of WT::3xAsp cells and rescued the 3xAsp furrow accumulation in myoII-null cells (Figure 4A). These data are similar to those acquired for the non–fluorophore-tagged versions, confirming that the N-terminal GFP-fusion proteins were functional. We note that the C-terminal–labeled RMD1 protein lost its spindle localization, indicating that this fusion protein did not function normally.
FIGURE 4:
Localization of RMD1, mmsdh, and rcdBB proteins in cells. (A) Bar graph shows the cleavage furrow intensity ratio of mCherry-3xAsp myosin II in myoII::mCh-3xAsp cells when GFP-RMD1 and GFP-mmsdh were expressed. These data confirm that GFP-RMD1 and GFP-mmsdh are functional GFP-fusion proteins. Sample sizes and p values are displayed on the graph. (B) Epifluorescence images demonstrate subcellular localization of RMD1, mmsdh, and rcdBB. RMD1 is largely cytoplasmic, with some enrichment around the centrosome (RFP-tubulin is shown for comparison). mmsdh is only cytosolic, and rcdBB was found enriched in the endoplasmic reticulum (RFP-calnexin is shown for comparison). (C) Confocal imaging confirms the largely cytosolic distribution of RMD1, with weak enrichment around the centrosome and the cytosolic distribution of mmsdh. (D) During cytokinesis, RMD1 and mmsdh remain cytosolic, with RMD1 showing weak enrichment around the centrosome. (E) In interphase cells compressed by agarose overlay, which introduces mechanical stress to the cortex, RMD1 remains largely cytosolic, with weak enrichment around the centrosome (RFP-tubulin is shown for comparison). mmsdh also remains cytosolic. (F) In dividing cells compressed by agar overlay, RMD1 and mmsdh remain cytosolic, with RMD1 showing weak enrichment around the centrosome (RFP-tubulin is shown for comparison).
We found that RMD1 was primarily cytosolic, with some enrichment at the centrosome, whereas mmsdh was entirely cytosolic (Figure 4, B and C). RcdBB showed enrichment at the endoplasmic reticulum and colocalized with the endoplasmic reticulum marker protein calnexin (Figure 4B). None of these proteins showed furrow enrichment in dividing cells (Figure 4D). No significant change in subcellular localization of these proteins was observed when expressed in myoII-null cells (unpublished data). Owing to the extreme difficulty in expressing rcdBB, no further studies were pursued with this protein at this time.
Neither mmsdh nor RMD1 showed significant enrichment at the cell cortex, even though both promoted 3xAsp myosin II accumulation. Because myosin II undergoes mechanosensitive accumulation at the cortex and cleavage furrow in response to mechanical stress (Kee et al., 2012), we used agarose overlay to apply an external mechanical stress, which might direct greater cortical accumulation of mmsdh and/or RMD1. However, under these conditions, neither RMD1 nor mmsdh showed an increased cortical or cleavage furrow accumulation, confirming that these proteins do not appreciably accumulate at the cortex with or without applied stress (Figure 4, E and F).
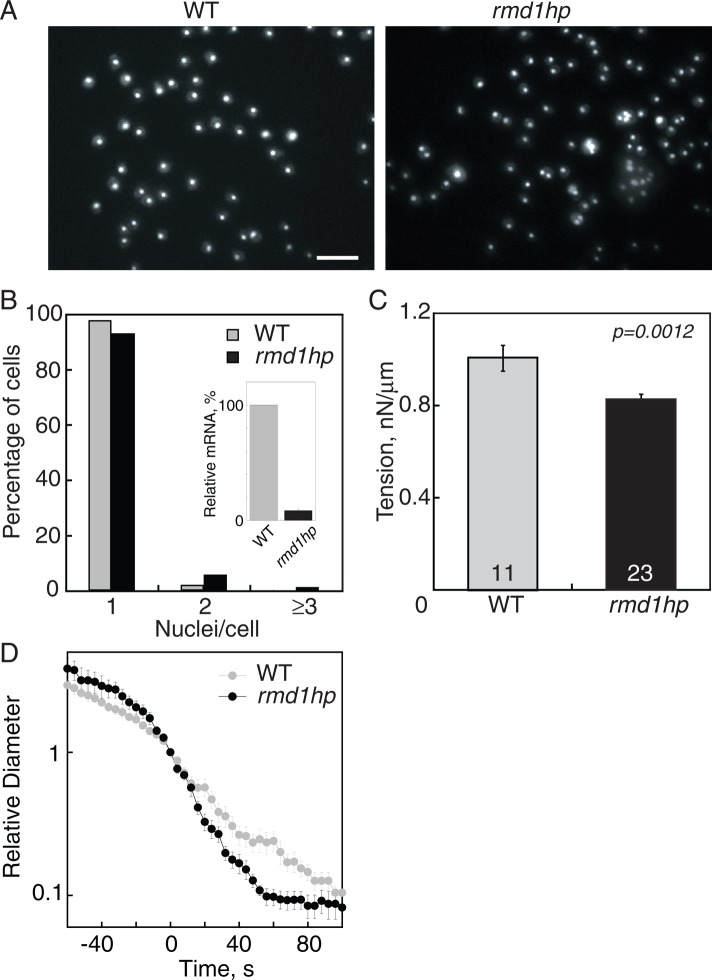
To test the role of RMD1 using loss-of-function analysis, we depleted rmd1 via small interference RNA (siRNA) using a hairpin construct (rmd1hp), which yielded ∼90–95% reduction of mRNA levels (Figure 5B, inset). Silencing of rmd1 in WT cells induced more binucleated and multinucleated cells (Figure 5, A and B) and reduced the cortical tension of cells (Figure 5C). Commensurate with the mild cytokinesis defect and reduction in cortical tension, rmd1hp cells also showed faster furrow ingression dynamics than the WT control, which had the stereotypical, near-exponential furrow ingression trajectory (Figure 5D). Of note, changes in the furrow ingression trajectory of rmd1hp cells are similar to what we observed previously for 14-3-3hp, myoII-null, and several other cell mechanics mutants (e.g., Zhang and Robinson, 2005; Reichl et al., 2008; Zhou et al., 2010).
FIGURE 5:
Depletion of rmd1 mRNA led to cytokinesis and cortical tension defects. (A) Micrographs of 4′,6-diamidino-2-phenylindole (DAPI)-stained cells show that rmd1hp cells have more multinucleated cells than WT control cells. Scale bar, 50 μm. (B) Frequency histogram reveals an increase in the fraction of multinucleated cells in rmd1hp cells. Inset demonstrates the 91% depletion of rmd1 mRNA in rmd1hp cells. (C) The rmd1hp cells had a 20% reduction in cortical tension. (D) Semilog plot of the furrow ingression dynamics of WT vs. rmd1hp cells, showing that the rmd1hp cells had a faster, more linear furrow ingression dynamic than WT control (n = 8–10 cells/genotype).
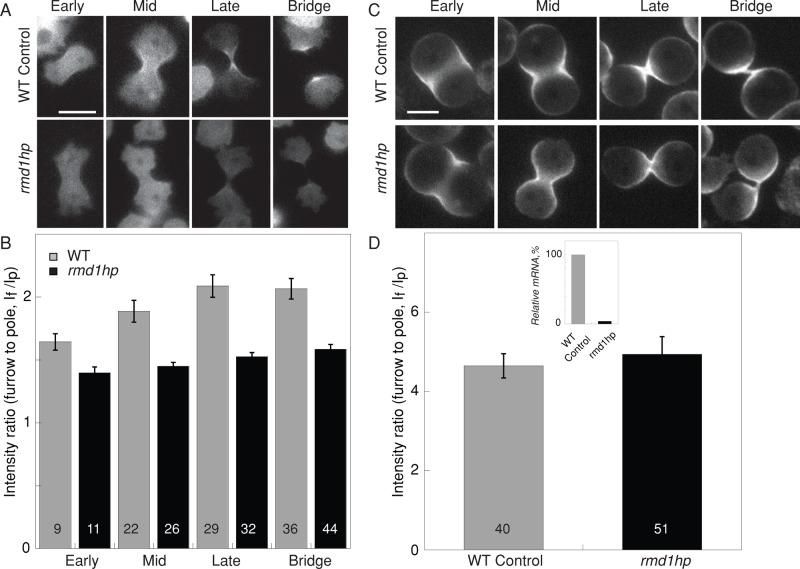
We then examined myosin II accumulation at the cleavage furrow cortex of rmd1hp cells and found that myosin II requires RMD1 for normal cleavage furrow accumulation (Figure 6, A and B). Because mechanical stress can also direct myosin II localization, including to the cleavage furrow (Effler et al., 2006; Ren et al., 2009; Kee et al., 2012), we tested whether mechanical stress could still enhance myosin II accumulation at the cleavage furrow of rmd1hp cells. Using agarose overlay (Yumura et al., 1984; Kee et al., 2012), we found that myosin II accumulated to comparable levels in WT control and rmd1hp cells (Figure 6, C and D). This result implies that mechanical stress acts in parallel with RMD1 in mediating myosin II accumulation.
FIGURE 6:
Depletion of rmd1 mRNA reduced GFP-myosin II cleavage furrow accumulation. (A) Micrographs show GFP-myosin II localization at the cleavage furrow cortex of WT control (myoII::GFP-myosin II; pLD1) and rmd1hp (myoII::GFP-myosin II; rmd1hp) cells. Scale bar, 10 μm. (B) Bar graph shows that the GFP-myosin II intensity at the cleavage furrow cortex is reduced in the rmd1hp cells compared with control (cell lines are the same as in A). Sample sizes are displayed in the bar graphs. Measurements are derived from 36 total control cells and 47 total rmd1hp cells. The differences between control and rmd1hp are significant (ANOVA, p < 0.0001). (C) Micrographs show myosin II accumulation at the cleavage furrow of cells compressed by agarose overlay. Scale bar, 10 μm. (D) WT control and rmd1hp cells showed comparable cleavage furrow accumulation when mechanical stress was applied. Because the data were similar across all stages of cytokinesis, the data were combined into a single group for each genotype. The inset shows the relative rmd1 mRNA levels in WT control and rmd1hp cells, which were used for the analyses in A–D.
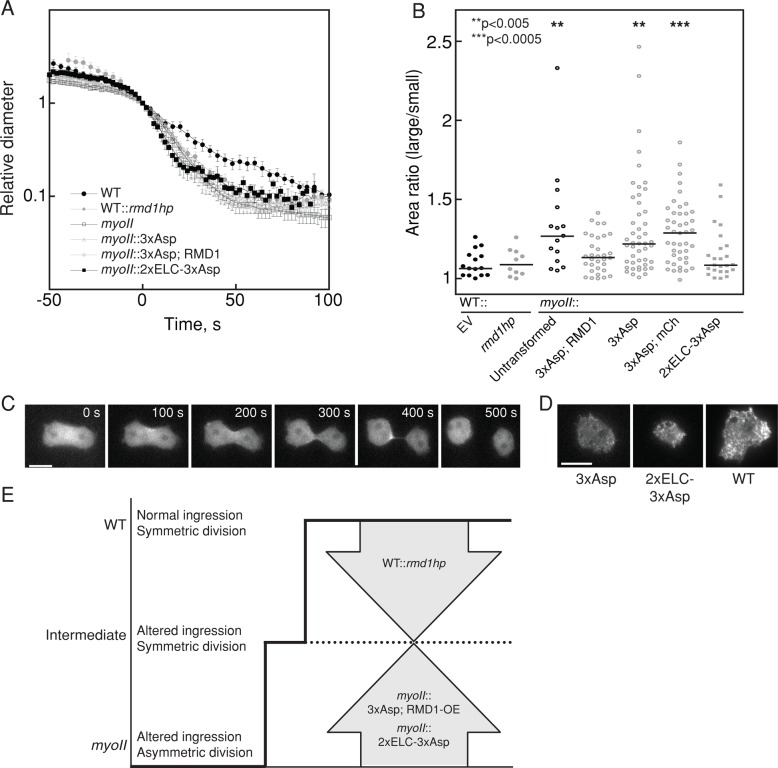
To ascertain the functional state of myosin II in the rmd1hp cells as well as in myoII::3xAsp cells rescued by RMD1, we measured the furrow ingression dynamics and symmetry of daughter cell sizes for these cells. None of the mutants was able to recover the near-exponential WT furrow ingression dynamics (Figure 7A). The products of cell division are two daughter cells, which for WT cells are highly symmetrical in size. To quantify this, we measured the two-dimensional cross-sectional area of the daughter cells and calculated the ratio of the larger cell to the smaller cell (Figure 7B). For WT, this ratio was 1.10 ± 0.021 (mean ± SEM), whereas for myoII-null cells, the ratio increased to 1.34 ± 0.084 and was more broadly distributed. As compared with WT cells, depletion of rmd1 did not alter the daughter cell symmetry despite altering furrow ingression kinetics (Figures 5D and 7, A and B). Of interest, RMD1 did not improve the furrow ingression kinetics of myoII::3xAsp, which was identical to myoII-null kinetics (Figure 7A), but it did increase the symmetry of the resulting daughter cells (Figure 7B). Thus daughter cell symmetry and furrow ingression kinetics appear to be established by different thresholds of myosin II activity: depletion of rmd1 from WT cells or expression of RMD1 in myoII::3xAsp cells causes the phenotype to converge to an intermediate level of myosin II function, where daughter cell symmetry is normal but furrow ingression kinetics is not (Figure 7E).
FIGURE 7:
Two thresholds of myosin II activity determine daughter cell symmetry and furrow ingression kinetics. (A) Furrow-thinning dynamics of WT, WT::rmd1hp, myoII-null, myoII::3xAsp, myoII::3xAsp, RMD1, and myoII::2xELC-3xAsp show that only WT cells have the characteristic exponential thinning dynamics. All other cell strains show a transition to more rapid thinning at a relative diameter of 1 (furrow diameter was rescaled by Dx, which is the point at which furrow diameter and length are equal; Zhang and Robinson, 2005). (B) Dot plot shows the distributions of area ratios of the large-/small-daughter cells, and the bars represent the medians. The WT class of cells (WT::EV [EV, empty vector], WT::rmd1hp, myoII::3xAsp,RMD1, and myoII::2xELC-3xAsp) produce symmetrically sized daughter cells (p = 0.12–1). The myoII-null class (myoII null, myoII::3xAsp, and myoII::3xAsp; mCh) produces highly asymmetrically sized daughter cells (p = 0.69–0.92). The two classes were significantly different (p = 0.0001–0.039). Asterisks denote significance relative to WT with empty vector. The p values were determined by Kruskal–Wallis test (p < 0.0001 for the entire data set), followed by pairwise Wilcoxon test. (C) Time series of a dividing cell expressing GFP-2xELC-3xAsp. Scale bar, 5 μm. (D) TIRF images of myoII-null cells expressing GFP-3xAsp, GFP-2xELC-3xAsp, and GFP-WT myosin II. Scale bar, 5 μm. (E) Cartoon depicts two thresholds of myosin II activity. Daughter cell symmetry requires intermediate level of function. WT furrow ingression and daughter cell symmetry require highest level of function. The myoII-null scenario has altered ingression dynamics and produces highly asymmetrically sized daughter cells.
Long lever arm partially suppresses 3xAsp
Finally, we asked what the hierarchical relationship is between heavy chain phosphorylation regulation and myosin mechanosensing. To do this, we engineered a super-mechanoresponsive myosin II, which has a long lever arm (2xELC), combined with the assembly-deficient 3xAsp tail. 2xELC myosin II accumulates very readily in response to low applied stress and accumulates normally to the cleavage furrow, whereas 3xAsp does neither of these (Figure 1; Ren et al., 2009; Kee et al., 2012; Luo et al., 2013). We then examined whether this 2xELC-3xAsp myosin behaved more like 3xAsp or more like WT. Indeed, myoII::2xELC-3xAsp had an intermediate phenotype in which more myosin accumulated at the cleavage furrow and cytokinesis resulted in greater daughter cell symmetry (Figure 7, B and C). 2xELC-3xAsp also showed greater filament formation by TIRF (Figure 7D). However, 2xELC-3xAsp could not rescue suspension growth (unpublished data) or furrow ingression kinetics (Figure 7A). Thus 2xELC could partially compensate for the 3xAsp lesions, demonstrating that myosin II function is governed by both heavy chain phosphorylation and cooperative binding of the motor to actin filaments (Luo et al., 2012).
DISCUSSION
Cleavage furrow accumulation of myosin II
How myosin II localizes to the cleavage furrow cortex in dividing cells has been one of the central questions of cytokinesis research for many years. Originally, researchers believed that myosin II accumulated to the cleavage furrow through myosin motor–actin interactions. However, the observations that headless myosin II accumulates in the furrow region, although not fully integrated into the cortex in Dictyostelium cells (Zang and Spudich, 1998), and that inhibition of actin filaments with latrunculin still allowed transient cleavage furrow accumulation in Drosophila cells (Dean et al., 2005) indicate that other pathways help mediate cleavage furrow localization. The actin membrane–anchoring protein anillin has been proposed to provide one possible anchor (Straight et al., 2005), but structure–function analysis indicates that the putative myosin II–binding site in anillin is not essential for myosin II accumulation in C. elegans (Piekny and Maddox, 2010). Further, an anillin homologue is not found in Dictyostelium, implying that if this paradigm holds, it must be carried out by other proteins in this system. Owing to some functional similarities, cortexillins could be candidates; however, myosin II still accumulates to the cleavage furrow in cortexillin I/II double mutants, albeit to a lesser extent (Kee et al., 2012).
Important regulatory pathways for cleavage furrow cortex assembly include the Rho–Rho kinase pathway (not found in Dictyostelium, but implicated in metazoan cells), which phosphorylates myosin II on the regulatory light chain, and the chromosomal passenger complex proteins INCENP and Aurora kinase (implicated in Dictyostelium and metazoans). These cues are believed to be transported in part by microtubule-based motors such as the kinesin-6 family members (e.g., MKLP1, kif12; Mishima et al., 2002; Chen et al., 2007; Li et al., 2008). These regulators may then locally activate Rho kinase (in metazoans), which promotes local bipolar thick filament assembly (Jantsch-Plunger et al., 2000; Dean and Spudich, 2006; Loria et al., 2012). However, diffusion places limits on how signaling pathways alone can direct accumulation.
For a cell to build a contractile network, three fundamental questions must be addressed—When? Where? Which ones? The “When?” question is generally answered by signaling pathways, which often include Rho–Rho kinase, which leads to myosin II regulatory light chain phosphorylation, as well as other kinases and phosphatases responsible for heavy chain phosphorylation (Yumura et al., 2005; Breckenridge et al., 2009; Wang et al., 2011). However, from the measured cytoplasmic diffusion coefficient (∼1 μm2/s; Uehara et al., 2010) of an unassembled myosin II “functional monomer” (a hexamer of two heavy chains and four light chains), one can estimate that in ∼10 s, a myosin II monomer could diffuse across a small cell (∼8-μm diameter), and in ∼100 s, a myosin II monomer could travel ∼25 μm. Because myosin II bipolar thick filaments require ∼100 s to assemble (Mahajan et al., 1989; Mahajan and Pardee, 1996; Ren et al., 2009; Luo et al., 2012), diffusive signaling alone does not readily explain where myosin II assembly will occur in small cells (e.g., Dictyostelium, which are 8 μm in diameter), although it can help in larger cells (e.g., mouse oocytes, which are 80 μm in diameter; Larson et al., 2010; Evans and Robinson, 2011). For this reason, other pathways and processes, such as cortical receptors and mechanosensitive accumulation, must play a significant role in localizing myosin II. The questions “Where (in the cell)?” and “Which ones (actin filaments)?” are answered by myosin II–mediated mechanosensing, by which mechanical stresses guide where myosin II assembles on favorable actin filaments (Effler et al., 2007; Ren et al., 2009; Uyeda et al., 2011; Luo et al., 2012; Shutova et al., 2012). Track-associated proteins provide another strategy for motors to identify preferred actin filaments. Finally, cortical receptors can help localize myosin BTFs. For example, the septin-binding protein Bni5 and the IQGAP homologue Iqgap1 in budding yeast provide this type of function for myosin II accumulation to the bud neck (Fang et al., 2010).
Myosin mechanosensitive accumulation
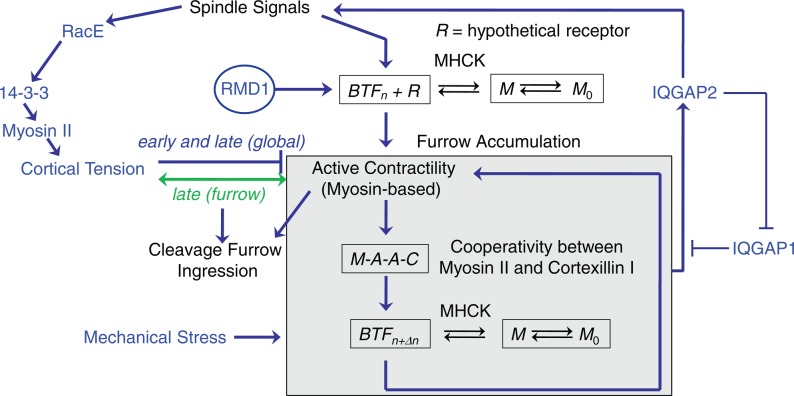
Mechanosensing provides an important mechanism for directing the localization of myosin II and is fundamental to a wide range of cellular and tissue functions (Engler et al., 2006; Luo and Robinson, 2011). We have shown that mechanosensitive accumulation plays a central role in tuning the cleavage furrow concentration of myosin II (Kee et al., 2012; Figure 8). This myosin mechanosensitive accumulation is performed by a three-part sensor, including force amplification through the myosin II lever arm, myosin BTF assembly/disassembly dynamics, and actin filament anchoring by cortexillin I (Ren et al., 2009). Multiscale modeling elucidates how myosin II force sensing and cooperative actin binding couple to myosin II BTF assembly (Luo et al., 2012), and this coupling is further demonstrated by the ability of 2xELC-3xAsp to accumulate at the cleavage furrow. This model quantitatively accounts for the amounts and kinetics of myosin II accumulation in response to applied stresses. However, this mechanosensory system is only part of a larger control system that tunes the total level of myosin II accumulation at the cleavage furrow under diverse mechanical constraints (Kee et al., 2012). In this control system (Figure 8), cortexillin I not only anchors the actin filaments, but it also links to signal transduction proteins (Faix et al., 1998; Mondal et al., 2010). These signaling proteins, including IQGAPs and the chromosomal passenger complex proteins, then reinforce signals that emanate from the mitotic spindle to direct myosin II accumulation. Overall this control system demonstrates that to decipher the recruitment pathways, these redundant systems must be taken into account.
FIGURE 8:
Myosin II accumulation and contractility are governed by a mechanosensitive control system. The flow diagram summarizes our view of how myosin II accumulates at the cleavage furrow and generates cortical tension and contractility, which together drive furrow ingression (Zhang and Robinson, 2005; Poirier et al., 2012). Cortical tension contributes in a complex manner, providing resistance early in furrow ingression and then assistance later in furrow ingression. The racE-14-3-3-myosin II pathway contributes to cell mechanics along the global/polar cortex (Zhou et al., 2010). The gray box encapsulates a cooperative module that includes myosin II and cortexillin I (Zhang and Robinson, 2005; Effler et al., 2006; Luo et al., 2012). The IQGAP1 and 2 proteins antagonize each other to modulate the cooperativity module. IQGAP2 further links back to the spindle signaling proteins, including kinesin 6 and INCENP proteins (Kee et al., 2012). Myosin II assembly is also modulated by myosin II heavy chain kinases (MHCKs), which maintain a free pool of myosin monomer (M0) so that myosin II can assemble where and when it is needed (Yumura et al., 2005; Ren et al., 2009). RMD1 also is required for myosin II accumulation at the cleavage furrow cortex. However, mechanical stress bypasses the RMD1 requirement.
Redundant pathways and multistate systems
To deal with these redundant systems and identify new factors involved in cytokinesis, we sensitized the cell by integrating 3xAsp myosin II into an otherwise WT background. 3xAsp myosin II was ideal for this because it does not readily assemble into BTFs (Egelhoff et al., 1993) and does not show mechanosensitive accumulation (Ren et al., 2009). This allowed us to use cDNA library suppression to identify important proteins involved in cytokinesis and in the proper function of the contractile machinery. We identified 11 proteins as suppressors of WT::3xAsp, which included several proteins previously known to be involved in cytokinesis in Dictyostelium (cortexillin II, actin, and 14-3-3; Faix et al., 1996, 2001; Zhou et al., 2010; Kee et al., 2012) and in other systems, namely C. elegans (RMD1; Oishi et al., 2007). Other known cytokinesis proteins, such as coronin and cortexillin I, were less significant during recapitulation but still offered some growth advantage, allowing them to survive the selection process (deHostos et al., 1993; Faix et al., 1996, 2001; Robinson and Spudich, 2000; Kee et al., 2012).
The identification of 14-3-3 highlights the multistate nature of the contractility machinery and confirms that the selection process could identify proteins that work directly on myosin II. Previously we discovered that 14-3-3 modulates the assembly of myosin II BTFs to control their distribution and ultimately the overall cortical tension of the cell (Zhou et al., 2010; Figure 7). Not surprisingly, we recovered 14-3-3 as a genetic suppressor of WT::3xAsp. Because 14-3-3 binds directly to myosin II heavy chains and inhibits BTF assembly (Zhou et al., 2010), it provides a buffer system for myosin BTF assembly. This buffering could rescue WT::3xAsp by increasing the turnover of the mixtures of WT and 3xAsp myosins. 14-3-3 did not rescue the localization of 3xAsp myosin II alone. However, reducing 14-3-3 expression levels using 14-3-3hp, which promotes the formation of larger myosin II assemblies (Zhou et al., 2010), allowed 3xAsp myosin II to accumulate at the cleavage furrow. Collectively these observations indicate that 3xAsp is not completely dead for assembly or localization, and the expression level of 14-3-3 shifts myosin populations between multiple states. By doing so, these factors can drive the system toward poor assembly and no localization, too much assembly and aberrant localization, or the proper balance of assembly and localization, depending on the combination of proteins expressed.
mmsdh and possible posttranslational modification of contractile machinery
mmsdh rescued the growth of WT::3xAsp and the cleavage furrow accumulation of 3xAsp in myoII-null cells. mmsdh catalyzes the degradation of valine to form propionyl- and acetyl-coA. Of interest, in mammalian cells, anillin and myosin IIA have been shown to be acetylated during mitosis, and histone deacetylase (HDAC) inhibitors cause mitotic failure (Chuang et al., 2010). Further, similar posttranslational modifications (PTMs) are found on the cardiac contractile machinery, where they modulate cardiac myosin II mechanochemistry, and HDAC associates with the cardiac sarcomeres (Samant et al., 2011). It is possible that mmsdh acts in the metabolic pathway that leads to PTM of the cleavage furrow contractile enzymes, including myosin II. TIRF results indicate that mmsdh does not increase the assembly of 3xAsp in the unstressed cortex. However, since the cleavage furrow cortex is a mechanically stressed environment, mmsdh-driven PTMs might make 3xAsp myosin II more active so that it is better able to accumulate at this stressed region. Of importance, we have also recovered mmsdh in a parallel proteomics effort searching for interactors with core mechanosensory proteins cortexillin and IQGAP2 (Srivastava, Chernysheva, Van Eyk, and Robinson, unpublished data), strongly supporting that the genetic interactions uncovered here are meaningful. An important future direction will be to determine which cytoskeletal proteins are propionylated or acetylated and whether these PTMs change the contractile properties of the dividing cell in a manner similar to what occurs in the cardiac sarcomere during heart failure (Samant et al., 2011).
Regulator of microtubule dynamics 1
RMD1 protein is a conserved protein with multiple coiled-coil domains that is found in species from Dictyostelium to humans. RMD1 was previously identified in C. elegans as important for chromosome segregation, maintenance of normal spindle microtubule–kinetochore attachments, and resistance to osmotic shock, and it was shown to bind microtubules (Oishi et al., 2007). The Dictyostelium RMD1 showed some enrichment at the centrosomes, which is a similar distribution to that observed in C. elegans embryos. The osmotic sensitivity in the C. elegans embryos suggests that RMD1 might have a role in maintaining cortical tension. In Dictyostelium, myosin II contributes to cortical tension, and myoII-null mutants are sensitive to osmotic stress (Pasternak et al., 1989; Kuwayama et al., 1996; Reichl et al., 2008). We showed here that depletion of RMD1 leads to a reduction in cortical tension. Thus our identification of RMD1 as a genetic suppressor of 3xAsp suggests that RMD1 plays a role in the pathway between microtubules and myosin II localization, and loss-of-function analysis confirmed that RMD1 is required for normal myosin II cleavage furrow accumulation. This linkage would be analogous to 14-3-3, which we also showed links the microtubule network to myosin II function (Zhou et al., 2010). Finally, RMD1 acts in parallel to mechanical stress, because mechanical stress rescued the defect in myosin II cleavage furrow accumulation in rmd1hp cells (Figure 8). Future analysis may identify how RMD1 interacts with myosin II, and possibly 14-3-3, and may clarify how the microtubule network and the cleavage furrow contractility machinery are integrated.
The genetic data presented here indicate that different thresholds of myosin II activity exist for ensuring daughter cell symmetry and normal furrow ingression kinetics (Figure 7C). Expression of RMD1 in the myoII::3xAsp cells was able to restore daughter cell symmetry but not furrow ingression dynamics. However, depletion of rmd1 from WT cells similarly caused a defect in furrow ingression kinetics while maintaining normal daughter cell symmetry. Of interest, RMD1 is the first mutant that we have identified that allows these two features of cytokinesis shape change to be resolved. Furthermore, the broadened distribution of daughter cell symmetry observed across multiple mutants (e.g., myoII and myoII::3xAsp) suggests an inability of the cells to compensate for force imbalances that may arise during cytokinesis (Figure 7B). Under more stringent, mechanically stressful situations, force imbalances may be uncorrectable by the suppressed mutants (e.g., myoII::3xAsp, RMD1, and myoII::2xELC-3xAsp), which may explain the inability of these cells to grow in suspension culture (Figure 2C).
Finally, it is worth noting that in addition to the 3xAsp mutations in the myosin II heavy chain, rmd1 and iqgap2 are the two strongest mutants with specific defects in myosin II cleavage furrow accumulation. As described earlier, mechanical stress can bypass the requirement for RMD1, and IQGAP2 is integrally linked to the mechanical feedback system (Kee et al., 2012; Figure 8). Deciphering the functions of these two proteins will likely hold the key for determining how myosin II accumulates at the cleavage furrow cortex.
Conclusion
The genetic selection described here identified several genes, some previously known in Dictyostelium, some implicated in other systems, and some completely novel, that point to the complexity of how myosin II accumulates at the cleavage furrow cortex (Figure 8). Further studies must decipher the biochemical mechanisms, and experimental designs must be used that separate mechanosensitive localization from pathway-directed mechanisms. Nevertheless, the data presented here show that myosin II functions as part of a multistate system in which an ideal balance of assembly and localization must be achieved for proper accumulation and cleavage furrow contractility. Either too much or too little assembly leads to a poisoned system. Thus cytokinesis continues to provide an elegant readout for exploring biochemical pathways and mechanical feedback, which tune the contractile machinery to ensure robust cell shape change.
MATERIALS AND METHODS
Dictyostelium strains, constructs, and cell culture
Dictyostelium discoideum strains used in this study are wild-type Ax3 (Rep orf+ cells; Robinson and Spudich, 2000), cortexillin-I–null strain (HS1151; Robinson and Spudich, 2000), and myoII heavy chain–null mutant (mhcA [HS1]; Ruppel et al., 1994; Table 3). Cells were cultured in Hans’ enriched HL-5 medium (1.4× HL-5, containing 8% FM (ForMedium, Hunstanton, Norfolk, UK), 60 U/ml penicillin, and 60 μg/ml streptomycin sulfate) and selected for plasmid transformations with 15–30 μg/ml G418, 4 μg/ml blasticidin, and/or 10–60 μg/ml hygromycin as appropriate. Cells were propagated at 22°C on 10-cm Petri dish plates. For suspension growth, cells were cultured in 10-ml culture volumes in 125-ml Erlenmeyer flasks at 180 rpm, 22°C. Cell densities were determined by counting cells on a hemacytometer.
TABLE 3:
Dictyostelium strains used in this study.
| Strain | Genotype | Experimental applications |
|---|---|---|
| WT::GFP-3xAsp | KAX3::pBIG:GFP-3xAsp | Suspension |
| WT::GFP-myoII | KAX3::pBIG:GFP-myoII | Suspension |
| myoII null | mhcA(HS1) null | Suspension, cytokinesis |
| WT::GFP-3xAsp; mCh-tub | KAX3::pBIG:GFP-3xAsp; pDRH:mCh-tub | 3xAsp localization, MPA, suspension |
| myoII::GFP-3xAsp; mCh-myoII | mhcA(HS1)::pBIG:GFP-3xAsp; pDRH:mCh-myoII | 3xAsp/myoII localization, MPA, suspension |
| myoII::GFP-myoII; mCh-myoII | mhcA(HS1)::pBIG:GFP-myoII; pDRH:mCh-myoII | myoII localization, suspension |
| WT::GFP-3xAsp | Ax3(Rep orf+)::pDRH:GFP-3xAsp | cDNA library suspension selection, recapitulations |
| WT::GFP-3xAsp control | Ax3(Rep orf+)::pDRH:GFP-3xAsp; pLD1A15SN | Suspension |
| myoII::GFP-3xAsp | mhcA(HS1)::pDRH:GFP-3xAsp | 3xAsp localization |
| WT::GFP-myoII | Ax3(Rep orf+)::pDRH:GFP-myoII; pLD1A15SN | Suspension |
| WT control | Ax3(Rep orf+)::pLD1A15SN | Suspension, DAPI, cortical tension, cytokinesis |
| WT::rmd1hp | Ax3(Rep orf+)::pLD1A15SN:rmd1hp | DAPI, cortical tension, cytokinesis |
| myoII::GFP-3xAsp; suppressor | mhcA(HS1)::pDRH:GFP-3xAsp; pLD1A15SN:suppressor | 3xAsp localization, suspension |
| myoII::GFP-3xAsp control | mhcA(HS1)::pDRH:GFP-3xAsp; pLD1A15SN | 3xAsp localization, TIRF, suspension, cytokinesis |
| myoII::mCh-3xAsp; GFP-RMD1 | mhcA(HS1)::pDRH:mCh-3xAsp; pDM181:GFP-RMD1 | 3xAsp localization |
| myoII::mCh-3xAsp; GFP-mmsdh | mhcA(HS1)::pDRH:mCh-3xAsp; pDM181:GFP-mmsdh | 3xAsp localization |
| myoII::mCh-3xAsp control | mhcA(HS1)::pDRH:mCh-3xAsp; pDM181:GFP | 3xAsp localization |
| myoII::GFP-3xAsp; rmd1hp | mhcA(HS1)::pDRH:GFP-3xAsp; pLD1A15SN:rmd1hp | 3xAsp localization |
| WT::GFP-RMD1; RFP-tub | Ax3(Rep orf+)::pDM181:GFP-RMD1; pDRH:RFP-tub | Protein localization |
| WT::GFP-RMD1 | Ax3(Rep orf+)::pDM181:GFP-RMD1 | Protein localization, agar overlay |
| WT::GFP-mmsdh | Ax3(Rep orf+)::pDM181:GFP-mmsdh | Protein localization, agar overlay |
| WT::GFP control | Ax3(Rep orf+)::pDM181:GFP | Protein localization, agar overlay |
| myoII::GFP-RMD1 | mhcA(HS1)::pDM181:GFP-RMD1 | Protein localization, agar overlay |
| myoII::GFP-mmsdh | mhcA(HS1)::pDM181:GFP-mmsdh | Protein localization, agar overlay |
| myolI::GFP control | mhcA(HS1)::pDM181:GFP | Protein localization, agar overlay |
| WT::mCh-rcdBB; cnx-GFP | Ax3(Rep orf+)::pDRH:mCh-rcdBB; pDEXRH:cnx-GFP | Protein localization |
| WT::RMD1-GFP | Ax3(Rep orf+)::pDM181:RMD1-GFP | Protein localization |
| myoII::GFP-3xAsp, RMD1 | mhcA(HS1)::pDRH:GFP-3xAsp; pLD1A15SN:RMD1 | TIRF |
| myoII::GFP-3xAsp, mmsdh | mhcA(HS1)::pDRH:GFP-3xAsp; pLD1A15SN:mmsdh | TIRF |
| myoII::GFP-3xAsp, actin | mhcA(HS1)::pDRH:GFP-3xAsp; pLD1A15SN:actin | TIRF |
| myoII::GFP-3xAsp, mCh-RMD1 | mhcA(HS1)::pBIG:GFP-3xAsp; pDRH:mCh-RMD1 | TIRF, agar overlay, cytokinesis |
| myoII::GFP-3xAsp, mCh-mmsdh | mhcA(HS1)::pBIG:GFP-3xAsp; pDRH:mCh-mmsdh | TIRF, agar overlay |
| myoII::GFP-3xAsp, mCh control | mhcA(HS1)::pBIG:GFP-3xAsp; pDRH:mCh | TIRF, agar overlay, cytokinesis |
| myoII::GFP-myoII, pLD1 | mhcA(HS1)::pDRH:GFP-myoII; pLD1A15SN | WT myosin II furrow accumulation, agar overlay |
| myoII::GFP-myoII, rmd1hp | mhcA(HS1)::pDRH:GFP-myoII; pLD1A15SN:rmd1hp | WT myosin II furrow accumulation, agar overlay |
| myoII::GFP-2xELC-3xAsp | mhcA(HS1)::pDM181:GFP-2xELC-3xAsp | Protein localization, TIRF, suspension, cytokinesis |
Myosin II was observed using pBIG:GFP-myosin II or pDRH:mCherry-myosin II (Moores et al., 1996; Zhou et al., 2010) and pBIG:GFP-3xAsp or pDRH:GFP-3xAsp constructs (Sabry et al., 1997; this work). Tubulin was observed using pDRH:mCherry-α-tubulin (Ren et al., 2009). RMD1 was observed using pDM181:GFP-RMD1 or pDM181:RMD1-GFP. mmsdh was observed using pDM181:GFP-mmsdh. RcdBB was observed using pDM181:GFP-rcdBB and pDM181:mCherry-rcdBB.
To prepare stably integrated WT::3xAsp cells, the plasmid pDRH:GFP-3xAsp was linearized by restriction enzyme digestion and then transformed into wild-type (Ax3(Rep orf+)) cells. To identify stably integrated clones, subclones of cells were subjected to suspension culture to select pools that had constant IC50 growth reduction even in the medium lacking the selective antibiotic. GFP-3xAsp expression was monitored throughout suspension culture to ensure that the fluorescence intensity remained at its original level even after many cycles of suspension culture. Finally, we extracted plasmid DNA and transformed the resulting eluent into DH5α and STBL2 (Life Technologies, Grand Island, NY) Escherichia coli cells to confirm that no plasmids could be recovered. We then performed Western analysis of WT and WT::3xAsp cells to measure the fraction of myosin II that was endogenous wild-type myosin II (60%) and GFP-3xAsp myosin II (40%; Figure 1F). These cells were then used for cDNA library suppression.
2xELC-3xAsp was engineered by combining myosin 2xELC and 3xAsp cDNAs using standard molecular techniques. The rmd1hp was constructed in the pLD1A15SN (pLD1) vector, using PCR to amplify nucleotides 188–690 with a SalI site at the 3′ end and a NotI site at the 5′ end. When subcloned into the SalI and NotI sites of pLD1, the fragment was in the antisense orientation, yielding pLD1:rmd1AS (690–188). Nucleotides 388–690 were amplified with NotI and MluI sites introduced at the 5′ and 3′ ends, respectively. On subcloning into the NotI and MluI sites in pLD1:rmd1AS, the resulting plasmid encoded pLD1:rmd1hp with the stem composed of 388–690 and the loop composed of 388–188.
cDNA library suppression
A Dictyostelium cDNA library constructed in the pLD1A15SN expression vector was transformed into WT::3xAsp cells by electroporation as previously described (Robinson and Spudich, 2000). A 1.8-μg amount of cDNA library plasmids was transformed per 350-μl cells. We combined 280 transformations (500 colonies/transformation) into 77 distinct pools and subjected them to suspension growth. Cultures were started at ∼2 × 105 cells/ml and allowed to grow to >1 × 106 cells/ml. At this point, cells were split into new flasks with fresh medium and a density of 2 × 105 cells/ml. Because WT::3xAsp cells grew slowly in suspension, winners were identified as pools with at least a 30% increase in growth rate. More than six cycles of suspension for each pool were monitored by counting cell densities daily throughout the selection, allowing enough time for the winners to emerge from the population. Plasmid cDNAs of “winner” pools were extracted using the Wizard Plus Miniprep DNA Purification System (Promega, Madison, WI) and clonally isolated by transformation into STBL2 cells. After purification and restriction endonuclease digestion, appropriate clones were chosen for sequencing, and the cDNA was identified using Dictybase BLAST. The plasmids were then transformed into fresh WT::3xAsp cells to confirm the suppression.
Growth rate determination
Relative growth rate was determined by plotting cell densities versus time, and the resulting log- phase curve was fitted to a single-exponential equation using KaleidaGraph (Synergy Software, Reading, PA). Because of the time lag required for the winners to populate the cultures, only the growth rates of later suspension cycles were used for data analysis. The mutant growth rate was normalized to 1, and the growth rate of each transformant pool was normalized to the mutant control.
In the recapitulation experiment, 25 suppressor plasmids were independently transformed into the WT::3xAsp cells in duplicate, yielding a total of 50 pools with recovered plasmids. The suspension growth rate of each rescued pool was monitored for at least five cycles. The vector control Ax3(Rep orf+)::pDRH:GFP-3xAsp;pLD1A15SN and wild-type control Ax3(Rep orf+)::pDRH:GFP-myosin II; pLD1A15SN were analyzed in parallel. The rescued cells adapted to the suspension culture faster than the cDNA library transformants, so we used the mean growth rates of the first three cycles and individual growth rates of further cycles in the data analysis. The mutant growth rate was normalized to 1, and the growth rate of each complemented pool was normalized to the mutant control.
Microscopy and mechanical manipulation
Cells were imaged using an Olympus IX81 microscope (Olympus America, Center Valley, PA) with a 40× (numerical aperture [NA] 1.3; for epifluorescence imaging) or a 60× (NA 1.49; for total internal reflection fluorescence imaging) objective with 1.6× Optovar. Images were acquired using MetaMorph software (Molecular Devices, Sunnyvale, CA). For live-cell fluorescence imaging, cells were plated on imaging chambers for at least 30 min, and then the medium was replaced with MES buffer (50 mM 2-(N-morpholino)ethanesulfonic acid [MES], pH 6.8, 2 mM MgCl2, 0.2 mM CaCl2) right before imaging to reduce background fluorescence. Unless otherwise noted, cells were imaged in the differential interference contrast (DIC; 10-ms exposure) and GFP (40-ms exposure) channels. For micropipette aspiration, images were collected at 10-s intervals for DIC (10-ms exposure) and GFP (20-ms exposure) and at 40-s intervals for red fluorescent protein (RFP; 50-ms exposure). The magnitude of loaded responses was determined following a previously described protocol (Effler et al., 2006).
Fluorescence imaging of GFP3xAsp was used to confirm that each pool maintained 3xAsp expression throughout the suspension growth experiments during the initial cDNA library suppression selection and in the recapitulation studies. Any cell lines that lost 3xAsp expression were discarded, and their corresponding growth rates were not included in any data analysis.
For mechanical manipulation, we used micropipette aspiration and agarose overlay. For micropipette aspiration (MPA; our system and methods are described in full in Kee and Robinson, 2013), we use a custom-built motorized water manometer system to generate aspiration pressure. Aspiration pressure is applied to the surface of the cell using capillary pipettes that are pulled to a diameter of 5 μm and polished on a microforge. Pressure is applied to the cell in steps until the length of the portion of the cell pulled into the micropipette (Lp) equals the pipette radius (Rp). This critical pressure is maintained for ∼1 min to ensure that this deformation is stable. The cell is released and the measurement is repeated, generally three times for each cell to ensure that the critical pressure has been properly identified. The cortical tension is then calculated based on Laplace's law. For mechanosensory responses, cells are also aspirated, and the GFP-fusion protein is monitored using epifluorescence imaging as described previously (Effler et al., 2006; Ren et al., 2009; Kee and Robinson, 2013). For agarose overlay, cells were compressed by thin sheets of agarose using established methods (Yumura et al., 1984; Kee et al., 2012).
To quantify GFP-myosin II intensities, images were background corrected. The corrected intensities in the cleavage furrow and polar cortices were measured, and the ratios were calculated for normalization. The ratios were then used for statistical analysis. Image analysis was performed using ImageJ software (National Institutes of Health, Bethesda, MD), and statistical analysis was performed using KaleidaGraph (Synergy Software).
Cytokinesis analysis
Log-phase cells were screened under the microscope, and both DIC and GFP images of dividing cells were collected. Images were processed with ImageJ software. The furrow concentration of GFP-3xAsp was quantified using the ratio of background-subtracted mean fluorescence intensity at the cleavage furrow (If) and the polar cortex (Ip). The intensity ratio If/Ip was calculated and used for statistical analysis. Furrow ingression dynamics was measured as described previously (Zhang and Robinson, 2005). Relative diameter was determined by normalizing the measured diameter by the crossover distance, Dx, and the time axis was shifted so that the time point at which Dx was reached was reset to 0. Dx is the point in which the furrow length and diameter are equal.
To determine daughter cell symmetry, we measured the two dimensional cross-sectional area and calculated the ratio of the large cell to the small cell as described previously (Zhou et al., 2010). The data sets were analyzed by one-way analysis of variance (ANOVA) with a Fisher least significant difference test and Kruskal–Wallis and Wilcoxon nonparametric tests using KaleidaGraph. Similar results were obtained with the two methods of analysis.
Acknowledgments
We thank the members of the Robinson lab and Pablo Iglesias for helpful discussions and comments on the manuscript. This work was supported by National Institutes of Health Grant GM066817 and support to the Summer Academic Research Experience program.
Abbreviations used:
- 3xAsp
mutant myosin with three key threonines mutated to aspartic acids
- BTF
bipolar thick filament
- ELC
essential light chain
- If
intensity at the furrow
- Ip
intensity at the pole
- LMM
light meromyosin
- M
assembly-competent myosin monomer
- M0
assembly-incompetent myosin monomer
- mmsdh
methylmalonate semialdehyde dehydrogenase
- R
myosin cortical receptor
- RMD1
regulator of microtubule dynamics 1
- TF
myosin tail fragment
- TIRF
total internal reflection fluorescence
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-08-1322) on October 15, 2014.
REFERENCES
- Adams RR, Wheatley SP, Gouldsworthy AM, Kandels-Lewis SE, Carmena M, Smythe C, Gerloff DL, Earnshaw WC. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- Breckenridge MT, Dulyaninova NG, Egelhoff TT. Multiple regulatory steps control mammalian nonmuscle myosin II assembly in live cells. Mol Biol Cell. 2009;20:338–347. doi: 10.1091/mbc.E08-04-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H, Cowan CR, Kong J, Hyman A. LET-99, GOA-1/GPA-16, and GPR-1/2 are required for aster-positioned cytokinesis. Curr Biol. 2007;17:185–191. doi: 10.1016/j.cub.2006.11.070. [DOI] [PubMed] [Google Scholar]
- Bringmann H, Hyman A. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436:731–734. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- Cabernard C, Prehoda KE, Doe CQ. A spindle-independent cleavage furrow positioning pathway. Nature. 2010;467:91–94. doi: 10.1038/nature09334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Lakshmikanth GS, Spudich JA, De Lozanne A. The localization of inner centromeric protein (INCENP) at the cleavage furrow is dependent on Kif12 and involves interactions of the N terminus of INCENP with the actin cytoskeleton. Mol Biol Cell. 2007;18:3366–3374. doi: 10.1091/mbc.E06-10-0895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Li H, De Lozanne A. Contractile ring-independent localization of DdINCENP, a protein important for spindle stability and cytokinesis. Mol Biol Cell. 2006;17:779–788. doi: 10.1091/mbc.E05-08-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang C, Lin SH, Huang F, Pan J, Josic D, Yu-Lee LY. Acetylation of RNA processing proteins and cell cycle proteins in mitosis. J Proteome Res. 2010;9:4554–4564. doi: 10.1021/pr100281h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Heck MM, Earnshaw WC. The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J Cell Biol. 1987;105:2053–2067. doi: 10.1083/jcb.105.5.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean SO, Rogers SL, Stuurman N, Vale RD, Spudich JA. Distinct pathways control recruitment and maintenance of myosin II at the cleavage furrow during cytokinesis. Proc Natl Acad Sci USA. 2005;102:13473–13478. doi: 10.1073/pnas.0506810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean SO, Spudich JA. Rho kinase's role in myosin recruitment to the equatorial cortex of mitotic Drosophila S2 cells is for myosin regulatory light chain phosphorylation. PLoS One. 2006;1:e131. doi: 10.1371/journal.pone.0000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deHostos EL, Rehfuess C, Bradtke B, Waddell DR, Albrecht R, Murphy J, Gerisch G. Dictyostelium mutants lacking the cytoskeletal protein coronin are defective in cytokinesis and cell motility. J Cell Biol. 1993;120:163–173. doi: 10.1083/jcb.120.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effler JC, Iglesias PA, Robinson DN. A mechanosensory system controls cell shape during mitosis. Cell Cycle. 2007;6:30–35. doi: 10.4161/cc.6.1.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effler JC, Kee Y-S, Berk JM, Tran MN, Iglesias PA, Robinson DN. Mitosis-specific mechanosensing and contractile protein redistribution control cell shape. Curr Biol. 2006;16:1962–1967. doi: 10.1016/j.cub.2006.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhoff TT, Lee RJ, Spudich JA. Dictyostelium myosin heavy chain phosphorylation sites regulate myosin filament assembly and localization in vivo. Cell. 1993;75:363–371. doi: 10.1016/0092-8674(93)80077-r. [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Evans JP, Robinson DN. The spatial and mechanical challenges of female meiosis. Mol Reprod Dev. 2011;78:769–777. doi: 10.1002/mrd.21358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Clougherty C, Konzok A, Mintert U, Murphy J, Albrecht R, Mühlbauer B, Kuhlmann J. The IQGAP-related protein DGAP1 interacts with Rac and is involved in the modulation of the F-actin cytoskeleton and control of cell motility. J Cell Sci. 1998;111:3059–3071. doi: 10.1242/jcs.111.20.3059. [DOI] [PubMed] [Google Scholar]
- Faix J, Steinmetz M, Boves H, Kammerer RA, Lottspeich F, Mintert U, Murphy J, Stock A, Aebi U, Gerisch G. Cortexillins, major determinants of cell shape and size, are actin-bundling proteins with a parallel coiled-coil tail. Cell. 1996;86:631–642. doi: 10.1016/s0092-8674(00)80136-1. [DOI] [PubMed] [Google Scholar]
- Faix J, Weber I, Mintert U, Köhler J, Lottspeich F, Marriott G. Recruitment of cortexillin into the cleavage furrow is controlled by Rac1 and IQGAP-related proteins. EMBO J. 2001;20:3705–3715. doi: 10.1093/emboj/20.14.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Luo J, Nishihama R, Wloka C, Dravis C, Travaglia M, Iwase M, Vallen EA, Bi E. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J Cell Biol. 2010;191:1333–1350. doi: 10.1083/jcb.201005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard KD, Kuo SC, Robinson DN. Dictyostelium myosin II mechanochemistry promotes active behavior of the cortex on long time scales. Proc Natl Acad Sci USA. 2006;103:2103–2108. doi: 10.1073/pnas.0508819103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Gonczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M. CYK-4: A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol. 2000;149:1391–1404. doi: 10.1083/jcb.149.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee YS, Ren Y, Dorfman D, Iijima M, Firtel RA, Iglesias PA, Robinson DN. A mechanosensory system governs myosin II accumulation in dividing cells. Mol Biol Cell. 2012;23:1510–1523. doi: 10.1091/mbc.E11-07-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y-S, Robinson DN. Micropipette aspiration for studying cellular mechanosensory responses and mechanics. Methods Mol Biol. 2013;983:367–382. doi: 10.1007/978-1-62703-302-2_20. [DOI] [PubMed] [Google Scholar]
- Kuwayama H, Ecke M, Gerisch G, Van Haastert PJ. Protection against osmotic stress by cGMP-mediated myosin phosphorylation. Science. 1996;271:207–209. doi: 10.1126/science.271.5246.207. [DOI] [PubMed] [Google Scholar]
- Larson SM, Lee HJ, Hung PH, Matthews LM, Robinson DN, Evans JP. Cortical mechanics and meiosis II completion in mammalian oocytes are mediated by myosin-II and Ezrin-Radixin-Moesin (ERM) proteins. Mol Biol Cell. 2010;21:3182–3192. doi: 10.1091/mbc.E10-01-0066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Chen Q, Kaller M, Nellen W, Graf R, De Lozanne A. Dictyostelium Aurora kinase has properties of both Aurora A and Aurora B kinases. Eukaryot Cell. 2008;7:894–905. doi: 10.1128/EC.00422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W, Licate LS, Warrick HM, Spudich JA, Egelhoff TT. Differential localization in cells of myosin II heavy chain kinases during cytokinesis and polarized migration. BMC Cell Biol. 2002;3:19. doi: 10.1186/1471-2121-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria A, Longhini KM, Glotzer M. The RhoGAP domain of CYK-4 has an essential role in RhoA activation. Curr Biol. 2012;22:213–219. doi: 10.1016/j.cub.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Mohan K, Iglesias PA, Robinson DN. Molecular mechanisms of cellular mechanosensing. Nat Mater. 2013;12:1064–1071. doi: 10.1038/nmat3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Mohan K, Srivastava V, Ren Y, Iglesias, Pablo A, Robinson, Douglas N. Understanding the cooperative interaction between myosin II and actin cross-linkers mediated by actin filaments during mechanosensation. Biophys J. 2012;102:238–247. doi: 10.1016/j.bpj.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Robinson DN. The role of the actin cytoskeleton in mechanosensation. In: Kamkin A, Kiseleva I, editors. Mechanosensitivity in Cells and Tissues: Mechanosensitivity and Mechanotransduction. Vol. 4. New York: Springer-Verlag; 2011. pp. 25–65. [Google Scholar]
- Mahajan RK, Pardee JD. Assembly mechanism of Dictyostelium myosin II: Regulation by K+, Mg2+, and actin filaments. Biochemistry. 1996;35:15504–15514. doi: 10.1021/bi9618981. [DOI] [PubMed] [Google Scholar]
- Mahajan RK, Vaughan KT, Johns JA, Pardee JD. Actin filaments mediate Dictyostelium myosin assembly in vitro. Proc Natl Acad Sci USA. 1989;86:6161–6165. doi: 10.1073/pnas.86.16.6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- Mondal S, Burgute B, Rieger D, Muller R, Rivero F, Faix J, Schleicher M, Noegel AA. Regulation of the actin cytoskeleton by an interaction of IQGAP related protein GAPA with filamin and cortexillin I. PLoS One. 2010;5:e15440. doi: 10.1371/journal.pone.0015440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores SL, Sabry JH, Spudich JA. Myosin dynamics in live Dictyostelium cells. Proc Natl Acad Sci USA. 1996;93:443–446. doi: 10.1073/pnas.93.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Octtaviani E, Effler JC, Robinson DN. Enlazin, a natural fusion of two classes of canonical cytoskeletal proteins, contributes to cytokinesis dynamics. Mol Biol Cell. 2006;17:5275–5286. doi: 10.1091/mbc.E06-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi K, Okano H, Sawa H. RMD-1, a novel microtubule-associated protein, functions in chromosome segregation in Caenorhabditis elegans. J Cell Biol. 2007;179:1149–1162. doi: 10.1083/jcb.200705108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou G, Stuurman N, D’Ambrosio M, Vale RD. Polarized myosin produces unequal-size daughters during asymmetric cell division. Science. 2010;330:677–680. doi: 10.1126/science.1196112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak C, Spudich JA, Elson EL. Capping of surface receptors and concomitant cortical tension are generated by conventional myosin. Nature. 1989;341:549–551. doi: 10.1038/341549a0. [DOI] [PubMed] [Google Scholar]
- Pavicic-Kaltenbrunner V, Mishima M, Glotzer M. Cooperative assembly of CYK-4/MgcRacGAP and ZEN-4/MKLP1 to form the centralspindlin complex. Mol Biol Cell. 2007;18:4992–5003. doi: 10.1091/mbc.E07-05-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M, Glotzer M, Kraut N, Peters JM. Polo-like kinase 1 triggers the initiation of cytokinesis in human cells by promoting recruitment of the RhoGEF Ect2 to the central spindle. Dev Cell. 2007;12:713–725. doi: 10.1016/j.devcel.2007.03.013. [DOI] [PubMed] [Google Scholar]
- Piekny AJ, Glotzer M. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr Biol. 2008;18:30–36. doi: 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- Piekny AJ, Maddox AS. The myriad of roles of anillin in cytokinesis. Semin Cell Dev Biol. 2010 doi: 10.1016/j.semcdb.2010.08.002. 21, 881–891. [DOI] [PubMed] [Google Scholar]
- Poirier CC, Ng WP, Robinson DN, Iglesias PA. Deconvolution of the cellular force-generating subsystems that govern cytokinesis furrow ingression. PLoS Comput Biol. 2012;8:e1002467. doi: 10.1371/journal.pcbi.1002467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichl EM, Ren Y, Morphew MK, Delannoy M, Effler JC, Girard KD, Divi S, Iglesias PA, Kuo SC, Robinson DN. Interactions between myosin and actin crosslinkers control cytokinesis contractility dynamics and mechanics. Curr Biol. 2008;18:471–480. doi: 10.1016/j.cub.2008.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Effler JC, Norstrome M, Luo T, Firtel RA, Iglesias PA, Rock RS, Robinson DN. Mechanosensing through cooperative interactions between myosin II and the actin crosslinker cortexillin. Curr Biol. 2009;19:1421–1428. doi: 10.1016/j.cub.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN. 14-3-3, an integrator of cell mechanics and cytokinesis. Small GTPases. 2010;1:165–169. doi: 10.4161/sgtp.1.3.14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN, Ocon SS, Rock RS, Spudich JA. Dynacortin is a novel actin bundling protein that localizes to dynamic actin structures. J Biol Chem. 2002;277:9088–9095. doi: 10.1074/jbc.M112144200. [DOI] [PubMed] [Google Scholar]
- Robinson DN, Spudich JA. Dynacortin, a genetic link between equatorial contractility and global shape control discovered by library complementation of a Dictyostelium discoideum cytokinesis mutant. J Cell Biol. 2000;150:823–838. doi: 10.1083/jcb.150.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppel KM, Uyeda TQP, Spudich JA. Role of highly conserved lysine 130 of myosin motor domain. In vivo and in vitro characterization of site specifically mutated myosin. J Biol Chem. 1994;269:18773–18780. [PubMed] [Google Scholar]
- Sabry JH, Moores SL, Ryan S, Zang J-H, Spudich JA. Myosin heavy chain phosphorylation sites regulate myosin localization during cytokinesis in live cells. Mol Biol Cell. 1997;8:2647–2657. doi: 10.1091/mbc.8.12.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samant SA, Courson DS, Sundaresan NR, Pillai VB, Tan M, Zhao Y, Shroff SG, Rock RS, Gupta MP. HDAC3-dependent reversible lysine acetylation of cardiac myosin heavy chain isoforms modulates their enzymatic and motor activity. J Biol Chem. 2011;286:5567–5577. doi: 10.1074/jbc.M110.163865. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shu S, Lee RJ, LeBlanc-Straceski JM, Uyeda TQP. Role of myosin II tail sequences in its function and localization at the cleavage furrow in Dictyostelium. J Cell Sci. 1999;112:2195–2201. doi: 10.1242/jcs.112.13.2195. [DOI] [PubMed] [Google Scholar]
- Shutova M, Yang C, Vasiliev JM, Svitkina T. Functions of nonmuscle myosin II in assembly of the cellular contractile system. PLoS One. 2012;7:e40814. doi: 10.1371/journal.pone.0040814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Field CM, Mitchison TJ. Anillin binds nonmuscle myosin II and regulates the contractile ring. Mol Biol Cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surcel A, Kee Y-S, Luo T, Robinson DN. Cytokinesis through biochemical-mechanical feedback loops. Semin Cell Dev Biol. 2010;21:866–873. doi: 10.1016/j.semcdb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Goshima G, Mabuchi I, Vale RD, Spudich JA, Griffis ER. Determinants of myosin II cortical localization during cytokinesis. Curr Biol. 2010;20:1080–1085. doi: 10.1016/j.cub.2010.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda TQ, Iwadate Y, Umeki N, Nagasaki A, Yumura S. Stretching actin filaments within cells enhances their affinity for the myosin II motor domain. PLoS One. 2011;6:e26200. doi: 10.1371/journal.pone.0026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Steimle PA, Ren Y, Ross CA, Robinson DN, Egelhoff TT, Sesaki H, Iijima M. Dictyostelium huntingtin controls chemotaxis and cytokinesis through the regulation of myosin II phosphorylation. Mol Biol Cell. 2011;22:2270–2281. doi: 10.1091/mbc.E10-11-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuce O, Piekny A, Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J Cell Biol. 2005;170:571–582. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumura S, Mori H, Fukui Y. Localization of actin and myosin for the study of ameboid movement in Dictyostelium using improved immunofluorescence. J Cell Biol. 1984;99:894–899. doi: 10.1083/jcb.99.3.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumura S, Yoshida M, Betapudi V, Licate LS, Iwadate Y, Nagasaki A, Uyeda TQ, Egelhoff TT. Multiple myosin II heavy chain kinases: roles in filament assembly control and proper cytokinesis in Dictyostelium. Mol Biol Cell. 2005;16:4256–4266. doi: 10.1091/mbc.E05-03-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang J-H, Spudich JA. Myosin II localization during cytokinesis occurs by a mechanism that does not require its motor domain. Proc Natl Acad Sci USA. 1998;95:13652–13657. doi: 10.1073/pnas.95.23.13652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Robinson DN. Balance of actively generated contractile and resistive forces controls cytokinesis dynamics. Proc Natl Acad Sci USA. 2005;102:7186–7191. doi: 10.1073/pnas.0502545102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou QQ, Kee Y-S, Poirier CC, Jelinek C, Osborne J, Divi S, Surcel A, Tran ME, Eggert US, Müller-Taubenberger A, et al. 14-3-3 coordinates microtubules, Rac and myosin II to control cell mechanics and cytokinesis. Curr Biol. 2010;20:1881–1889. doi: 10.1016/j.cub.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]