A method is introduced for optically imaging intracellular protein interactions at nanometer spatial resolution in live cells using photoactivatable complementary fluorescent (PACF) proteins. The PACF approach established here will enable the simultaneous visualization of homo- and heterodimeric interactions at the single live-cell level.
Abstract
Visualization of specific molecules and their interactions in real time and space is essential to delineate how cellular dynamics and the signaling circuit are orchestrated. Spatial regulation of conformational dynamics and structural plasticity of protein interactions is required to rewire signaling circuitry in response to extracellular cues. We introduce a method for optically imaging intracellular protein interactions at nanometer spatial resolution in live cells, using photoactivatable complementary fluorescent (PACF) proteins. Subsets of complementary fluorescent protein molecules were activated, localized, and then bleached; this was followed by the assembly of superresolution images from aggregate position of sum interactive molecules. Using PACF, we obtained precise localization of dynamic microtubule plus-end hub protein EB1 dimers and their distinct distributions at the leading edges and in the cell bodies of migrating cells. We further delineated the structure–function relationship of EB1 by generating EB1-PACF dimers (EB1wt:EB1wt, EB1wt:EB1mt, and EB1mt:EB1mt) and imaging their precise localizations in culture cells. Surprisingly, our analyses revealed critical role of a previously uncharacterized EB1 linker region in tracking microtubule plus ends in live cells. Thus PACF provides a unique approach to delineating spatial dynamics of homo- or heterodimerized proteins at the nanometer scale and establishes a platform to report the precise regulation of protein interactions in space and time in live cells.
INTRODUCTION
Cell migration is a dynamic process essential for various biological processes, including embryogenesis, tissue repair and regeneration, chemotaxis, and tumor metastasis. Microtubules determine the physical properties and functional plasticity of migrating cells (Watanabe et al., 2005). The plus ends of microtubules, extending into the peripheral regions of cells, orchestrate prompt switching between the growing and shortening phases (Vaughan, 2005). Microtubule dynamics are regulated by a group of associated proteins specially localized to “track” the growing microtubule plus ends; these are called plus end–tracking proteins (+TIPs; Honnappa et al., 2005; Slep and Vale, 2007; Akhmanova and Steinmetz, 2008; Komarova et al., 2009). However, the temporal and spatial dynamics at the plus ends of microtubules during cell migration have remained elusive.
Among the microtubule +TIPs, EB family proteins can track and regulate growing microtubules themselves and can act as scaffolds for other +TIPs to construct the highly complex plus end–tracking protein machinery (Vaughan, 2005). The EB family proteins include EB1, EB2, and EB3. These three proteins are highly conserved at their N-terminal calponin homology (CH) domains and their C-terminal domains (EBC). They form homo- or heterodimers with the EBC domain from three EB proteins (Komarova et al., 2009), which serves a main docking site of other +TIPs (Akhmanova and Steinmetz, 2008). Most of the +TIPs contain a SxIP motif for interaction with the hydrophobic cavity on a dimerized EBC domain. EB1 interacts with microtubules with its N-terminal CH domain (Slep and Vale, 2007). But its tracking property depends on dimerization (Honnappa et al., 2005; Slep and Vale, 2007; Komarova et al., 2009; Buey et al., 2011). It has further been demonstrated that the negatively charged EBC domain contributes to the dwell times of EB proteins on the microtubule lattice, and the dimerization of EB1 results in the spatial construction of two adjacent CH domains (Buey et al., 2011), but the intermolecular interaction and regulation of this structure have remained elusive. Visualization of specific molecules and their interactions in real space and time is essential to delineate how cellular plasticity and dynamics are achieved and orchestrated.
The discovery of photoactivatable fluorescence proteins and related analyses (Patterson and Lippincott-Schwartz, 2002; Betzig et al., 2006) provide a unique opportunity for studying and visualizing characters of protein dimerization at the nanometer scale in space and time in live cells using a photoactivatable complementary fluorescent (PACF) protein–based imaging strategy. In this study, we generated a complementary version of photoactivatable green fluorescence proteins and developed PACF, which reports protein dimerization in space and time. PACF proteins have the same photoactivatable character as their photoactivatable green fluorescent protein (PAGFP) parents, and that is confirmed by PACF fused to EB1 homodimer. We also performed superresolution imaging in fixed and live cells expressing PACF-EB1 proteins and found two different states of plus-end dynamics in migrating cells, consistent with the differences of precise EB1 localization in space in migrating cells. Our results provide an effective new method of detecting dimeric protein–protein interactions at the nanometer scale in live cells and further our understanding of the context-dependent organization and regulation of microtubule plus-end dynamics.
RESULTS
Generation of photoactivatable complementary green fluorescent proteins
The specificity of biological regulatory mechanisms relies on selective interactions between cognate partners in the right place at the right time in response to different extracellular signals. To visualize specific molecules and their interactions at the nanometer scale, we sought to establish an approach to visualize protein–protein interaction using the newly developed photoactivation localization microscope (Patterson and Lippincott-Schwartz, 2002; Betzig et al., 2006).
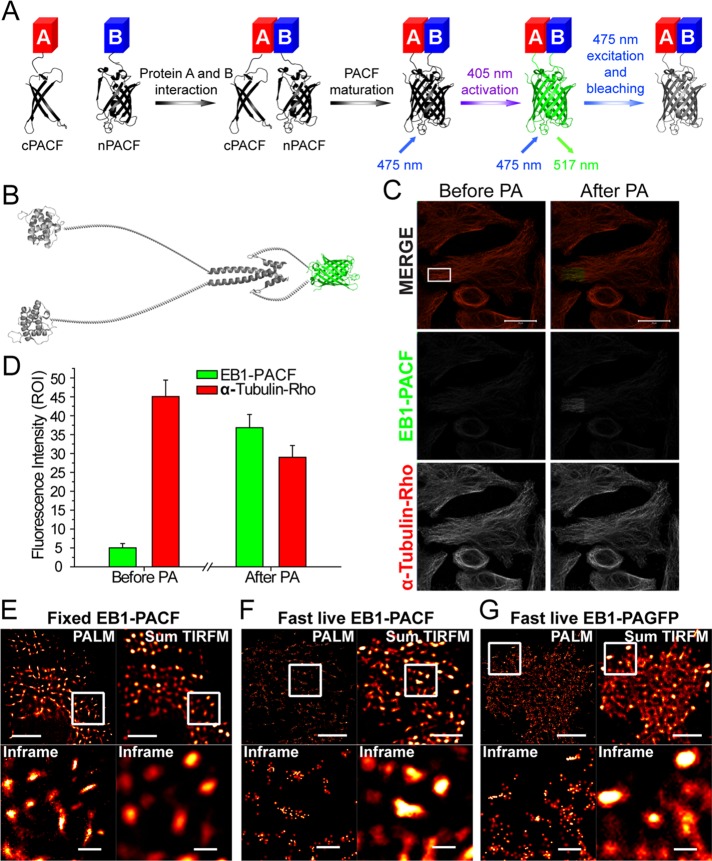
To this end, N-terminus/C-terminus (n/c) PACF was designed to fuse with different proteins to generate fluorescent protein (Figure 1A and Supplemental Figure 1). The interaction of target proteins provides the opportunity for PACF protein capture and maturation. After maturation, the green PACF proteins can be photoactivated with a 405-nm laser. To test the photoactivation property of PACF, we fused n/cPACF to EB1 to detect homodimerized EB1 proteins (Figure 1B). The expression of EB1-PACF in cells was confirmed by Western blot analyses of the right size (Supplemental Figure 2A). PACF can be activated with a 405-nm laser in cold methanol–fixed HeLa cells (Figure 1, C and D). As shown in Supplemental Figure 2, B–D, native gel analyses also confirm a functional activation of PACF and EB1-PACF by UV light, suggesting that PACF indeed possesses the property of being photoactivatable after its complementation. Photobleaching properties of EB1-PACF and EB1-PAGFP were also examined, and the parameters of PACF are comparable with those of PAGFP (Supplemental Figure 3, A and B).
FIGURE 1:
Characterization of photoactivatable complementary green fluorescent proteins. (A) Protein A and B were conjugated with nPACF or cPACF, respectively. On interaction of protein A with protein B, nPACF and cPACF were programmed to form a functional fluorescent protein under photoactivation. The photoactivation of PACF proteins was achieved with a 405-nm laser and excited with a 475-nm laser. The emission spectrum peaks at 517 nm. (B) Schematic illustration of engineered constructs containing EB1 dimer fused to nPACF and cPACF proteins. (C) HeLa cells transiently transfected to express EB1-PACF proteins were fixed and stained; this was followed by imaging under a Zeiss LSM 710 laser confocal microscope. After PA under UV irradiation, the EB1-PACF signal intensity increased (in frame), while the signal intensity of α-tubulin-rhodamine decreased (by photobleaching). Scale bars: 20 μm. (D) Statistical analyses of fluorescence intensity changes of EB1-PACF and α-tubulin marked by rhodamine in C before and after PA. (E) Comparative analyses of PALM and sum of TIRFM images of fixed MCF7 cells transiently transfected to express EB1-PACF proteins. Scale bar (top panels): 5 μm. Bottom panels are magnified views of insets mentioned above. Plus-end localization of EB1-PACF dimerized proteins was readily apparent and clearly defined under superresolution imaging. Scale bars: 1 μm. (F) Comparative analyses of live PALM and sum of TIRFM images of MCF7 cells expressing EB1-PACF. Scale bars: 5 μm (top panels); 1 μm (bottom panels). (G) Comparative analyses of live PALM and sum of TIRFM images of MCF7 cells expressing EB1-PAGFP. Scale bars: 5 μm (top panels); 1 μm (bottom panels).
To quantify the comparative levels of spontaneous and inducible complementation of PACF, we constructed a chemically inducible EB1 dimerization system in which the C-terminal of EB1 (EBC) was replaced by the homodimerization domain FK506-binding protein (FKBP; strategy illustrated in Supplemental Figure 3C). The FKBP-driven dimerization is initiated by a rapamycin derivative, AP20187 (Komarova et al., 2009). As shown in Supplemental Figure 3, D and E, the fluorescence signal in the cell was successfully detected after the addition of AP20187, which was significantly different from control groups, in which only a minimal increase in intensity could be measured after photoactivation and excitation, indicating the spontaneous complementation of PACF is insignificant in our experimental system. The low efficiency of spontaneous complementation seen in our PACF is similar to what was recently reported for complementary photoactivatable mCherry (Nickerson et al., 2014). Thus we conclude that the engineered PACF proteins bear the photoactivatable property of their parent PAGFP and can be used to accurately report the precise localization of the homo- or heterodimerized proteins.
Superresolution imaging with EB1-PACF in fixed cells
After completion of biochemical characterization of PACF, we next examined whether EB1-PACF can be utilized to perform superresolution imaging using the photoactivated localization microscopy (PALM) approach, as previously described (Betzig et al., 2006). Basically, MCF7 cells were transiently transfected to express PCAF-EB1; this was followed by chemical fixation. In general, 15,000 frames with an exposure time of 50 ms per frame were collected to generate a single PALM image, as detailed in Materials and Methods. Briefly, images were taken in total internal reflection fluorescence (TIRF) mode, which detects single photons with minimal autofluorescence and detection noise (Betzig et al., 2006). The microtubule plus-end localization of EB1-PACF was detected at the single-molecule level, with a localization precision of 23 nm (Figure 1E and Supplemental Figure 4A). These results presented the possibility of using PACF to picture the EB1 molecule distribution map with nanometer-scale precision.
Superresolution imaging of EB1-PACF molecules in live cells
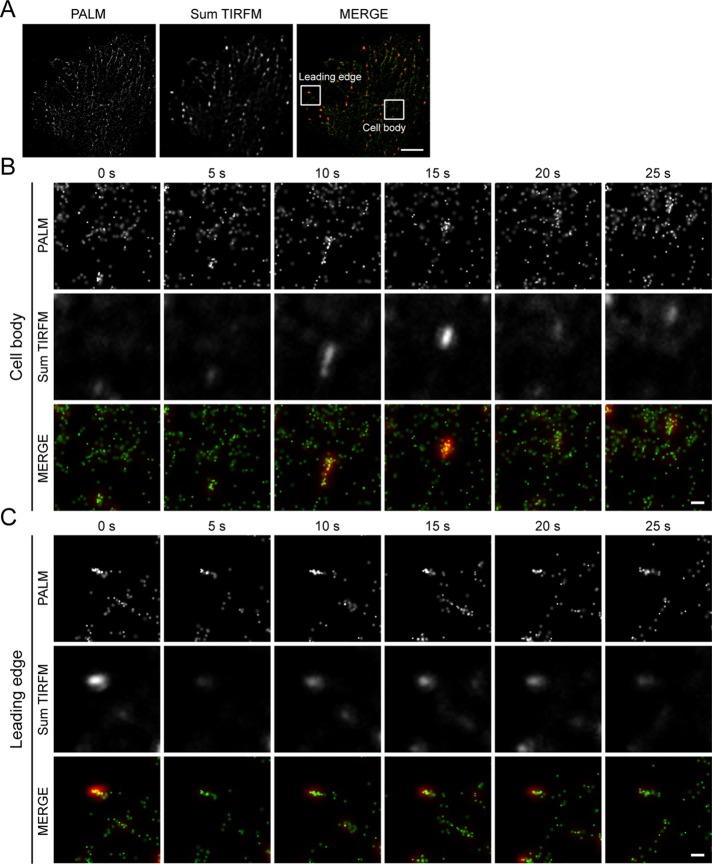
For probing microtubule plus-end dynamics at the nanometer scale in live cells, every single image was generated from 100 frames of 15 ms per exposure (Figure 1, F and G). The efforts to reduce frames and exposure time to obtain every single image resulted in a 10-nm loss in localization precision (Supplemental Figure 4B). However, this brief loss enabled us to collect superresolution images in 1.5 s and successfully trace the dynamic microtubule plus ends in live cells. Our results also demonstrated the advantage of EB1-PACF compared with EB1-PAGFP; the former has decreased background without perturbing the signal at the microtubule plus ends (Figure 1, F and G). Furthermore, we found that the EB1-PACF molecules at the leading edges of migrating cells exhibit a slow motion compared with those tracking the microtubule plus ends in the cell body (Figure 2, A–C, and Supplemental Figure 4C). We speculate that the growth rate of microtubule plus ends is reduced in the leading edge of migrating MCF7 cells, as these plus ends may dock into the cell cortex. This hypothesis was further confirmed by conventional live-cell imaging of MCF7 cells expressing EB1-eGFP (enhanced green fluorescent protein; Supplemental Figure 5).
FIGURE 2:
Superresolution imaging with EB1-PACF in live cells visualizes spatial dynamics of EB1 dimer. (A) Live PALM and TIRFM images of MCF7 cells transiently transfected to express EB1-PACF dimerized proteins. Frames indicate the regions of microtubule plus ends at the leading edge and in the cell body. Scale bar: 5 μm. (B) Time-lapse PALM and TIRFM images of the EB1-PACF dimerized proteins on a microtubule plus end in the region of the cell body indicated in the frame in A. Images of every time point were collected with 100 cycles of photoactivation and excitation. The exposure time is 15 ms per cycle. Paired images collected from PALM were merged with those from TIRFM to compare the clarity of molecules visualized. Scale bar: 500 nm. (C) Time-lapse PALM and TIRFM images of the EB1-PACF dimerized proteins on a microtubule plus end in the region of the leading edge indicated in the frame in A. Images of every time point were collected with 100 cycles of photoactivation and excitation. The exposure time is 15 ms per cycle. Paired images collected from PALM and TIRFM were merged to compare the clarity of molecules visualized by PALM. Scale bar: 500 nm.
Precise localization of a single EB1-PACF molecule reveals its distinct spatial patterns in migrating cells
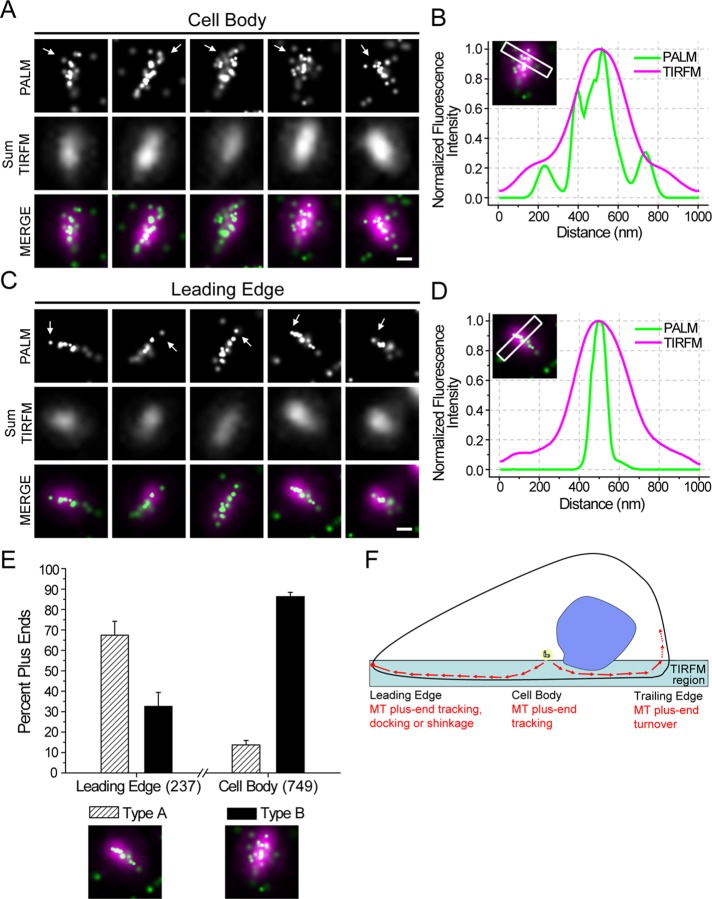
At the single-molecule level, we found that EB1-PACF tracking the growing microtubule plus ends in the cell body exhibits a complex construction, creating a profile shaped like a curving sheet (Figure 3A). These structures are consistent with the hypothetic model of microtubule plus ends (Vitre et al., 2008). Interestingly, we found many EB1-PACF molecules localized at the plus ends of the leading edge aligned in a narrow cone-shaped row (Figure 3C), distinctly different from the molecules tracking the microtubule plus ends in the cell body. In addition, this kind of difference could not be distinctly defined under TIRF microscopy (TIRFM) analyses at conventional resolution but were readily apparent with PACF imaging (Figure 3, B and D). We then confirmed that the difference seen between EB1-PACF signals at the cell body and leading edge is not because of blurring of the artifact; we collected 100 consecutive exposure frames (150 ms/frame) and separated them into two time series (frames 1–50 and frames 51–100). As shown in Supplemental Figure 6, A and B, the characteristics of each kind of plus end can be recognized, even with half-time exposure, although the integrity of images was obviously decreased (Supplemental Figure 6, C and D). We further classified the narrow cone-shaped EB1-PACF localization to type A and the complex curving sheet to type B (Figure 3E). Statistical analyses of type A and B plus ends at the leading edge or in the cell body exhibit significant differences. In the leading edge, 67.42 ± 6.86% plus ends are type A, while only 32.58 ± 6.86% are type B. However, in the cell body, only 13.72 ± 2.17% plus ends are type A, while 86.28 ± 2.17% plus ends are type B (Figure 3E). To illustrate the localization characteristics of EB1-PACF at the microtubule plus ends, we carried out superresolution images of EB1-PACF in fixed cells double-stained with tubulin and EB1 antibodies, respectively. The superresolution images of EB1-PACF were then merged with diffraction-limited images of tubulin and EB1 immunofluorescence. Careful examination reveals that EB1-PACF signals are superimposed onto the microtubules and microtubule plus ends (Supplemental Figure 6, C and D, respectively). As shown in Supplemental Figure 6E, statistical analyses demonstrate that the type A comet of EB1 dimeric molecules distributes preferentially at the leading edge of migrating cells, while the type B comet of EB1 dimers is enriched in the cell body. These precise localization analyses indicate that EB1 dimer molecules exhibit distinct distribution patterns on the microtubule plus ends in the leading edge, cell body, and trailing edge of a migrating cell (Figure 3F).
FIGURE 3:
EB1 molecules at the microtubule plus ends from the leading edge and cell body of migrating cells exhibit distinct distribution patterns. (A) Precise localization of single EB1-PACF molecules in the cell bodies of live MCF7 cells. Arrows indicate the plus ends of microtubules. Scale bar: 250 nm. (B) Fluorescence intensity distribution for PALM and TIRFM imaging of a rectangular region of 1 μm × 200 nm in the front of a plus end in the cell body was calculated and plotted. The fluorescence intensity was normalized to the highest value in the region to facilitate the comparison. (C) Precise localization of EB1-PACF proteins in the leading edge of live MCF7 cells. Arrows indicate the plus ends. Scale bar: 250 nm. (D) Fluorescence intensity distribution for PALM and TIRFM imaging of a rectangular region of 1 μm × 200 nm in the front of a plus end in the leading edge was calculated and plotted. The fluorescence intensity was normalized to the highest value in the region to facilitate the comparison. (E) The percentage of plus ends classified as type A and type B are plotted for the leading edge and cell body. Data were collected from six individual experiments. The total sample size (number of plus ends classified) is indicated in the parentheses on the abscissa. (F) A working model of plus-end tracking in a migrating cell was proposed and presented. PACF-based imaging demonstrates that EB1 molecules exhibit a context-dependent distribution pattern at the leading edge, cell body, and trailing edge of migrating cells. The distribution pattern of EB1 molecules reports the characteristics of microtubule plus-end dynamics in the aforementioned regions.
Conserved lysine/arginine residues in the flexible linker region of EB1 are essential for its plus end–tracking activity
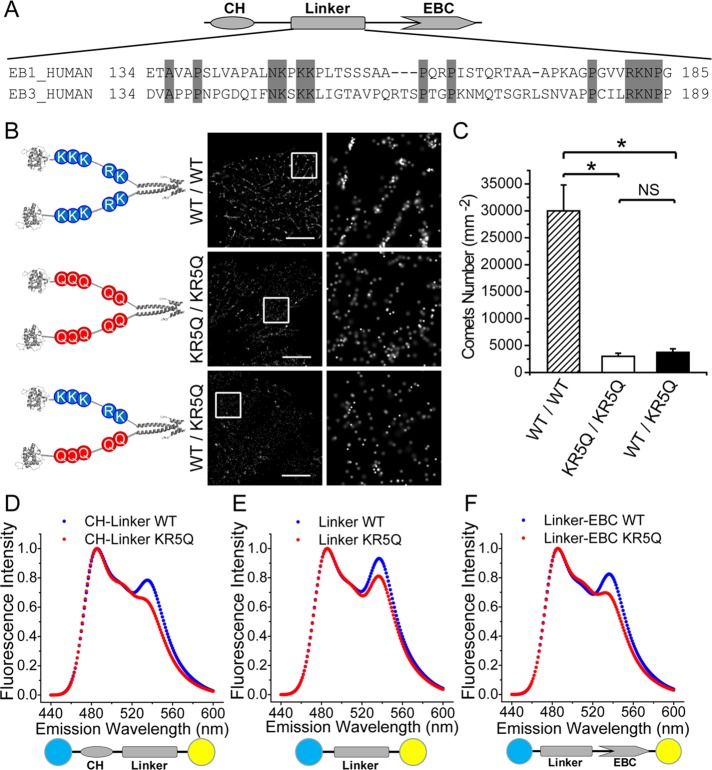
It is known that the EB family proteins possess highly conserved and rigid N-terminal CH domains (aa 1–130) and C-terminal domains (EBC: aa 191–268). The CH domain binds to the microtubule directly, while the EBC domains dimerize and serve as scaffolds for the other +TIPs. However, the structure–function relationship of the less-conserved flexible linker region between CH and EBC domains was unclear (Supplemental Figure 7; Akhmanova and Steinmetz, 2008). During our construction of the EB1-PACF system, we tried to introduce the fluorescent protein to engineer a fluorescent EB1 molecule without perturbing its microtubule plus end–tracking activity. To our surprise, insertion of the fluorescent protein either between the CH domain and the linker region or between the linker and the EBC impaired the plus-end tracking of EB1 (Supplemental Figure 8), suggesting that the less-conserved flexible linker region is also important for a stable plus-end loading.
To delineate the structure–function relationship of the linker region, we first performed sequence alignment of EB1 homologues and determined that several proline (P) residues and almost all the lysine (K)/arginine (R) residues were relatively conserved in this low homologue region (Figure 4A). Proline residue often serves at the turn or loop of a protein. However, mutation of proline residues to alanine or glycine did not alter the localization of EB1 to the plus end of the microtubule (unpublished data). We then examined whether the conserved and positively charged residues K/R modulate the localization of EB1. To this end, we took the advantage of EB1-PACF to design dimers with a combination of wild-type and mutant EB1 in which the five positively charged K/R residues were mutated to glutamine (Q; EB1KR5Q). In this regard, we can ascertain the respective contribution of each molecule of the engineered EB1 dimer to stable plus-end localization using EB1-PACF (Figure 4B, left panel). We first examined the protein expression of the PACF-fused EB1 proteins with Western blotting (Supplemental Figure 9). Consistent with our demonstration above, precise localization of EB1WT/WT-PACF exhibited a typical comet-like plus end–tracking character (Figure 4B, top right panel). However, expression of either EB1KR5Q/KR5Q-PACF or EB1WT/KR5Q-PACF failed to reach a stable localization to the plus ends, as evidenced by fewer comet-like structures (Figure 4, B and C, and Supplemental Figure 10). Thus the conserved K/R residues in the linker region are essential for the functional EB1 dimers, and their plus-end tracking property is impaired, even when the linker region of one molecule of the EB1 dimer is altered.
FIGURE 4:
The linker region of EB1 molecules governs the plus-end localization of functional EB1 dimers. (A) Schematic drawing of sequence alignment of the linker region of EB1 and EB3 molecules. The conserved residues are presented and highlighted with a gray background. (B) Schematic illustration and precise localization of PACF fused EB1WT/WT, EB1KR5Q/KR5Q, and EB1WT/KR5Q. Only EB1WT/WT dimer but not EB1KR5Q/KR5Q or even EB1WT/KR5Q mutant exhibits typical comet-like plus-end localization in transiently transfected HeLa cells. Scale bars: 5 μm. (C) Statistical analyses of histograms of comet numbers per square micrometer in live MCF7 cells expressing PACF fused EB1WT/WT, EB1KR5Q/KR5Q, and EB1WT/KR5Q. Our analyses show that the numbers of comets per square micrometer are decreased in cells expressing EB1KR5Q/KR5Q and EB1WT/KR5Q mutants compared with those of EB1WT/WT-expressing cells. Error bars indicate SD. *, p < 0.005; NS, no significance. (D) FRET spectra of EB1 CH linker (aa 1–190) wild type and KR5Q mutant. FRET efficiency decreases upon mutation of the conserved lysine and arginine, which demonstrates the importance of those residues in intramolecular interaction of EB1 molecules. (E) FRET spectra of EB1 linker (aa 131–190) wild type and KR5Q mutant. FRET efficiency decreases upon mutation of the conserved lysine and arginine. (F) FRET spectra of EB1 linker-EBC (aa 131–268) wild type and KR5Q mutant. FRET efficiency decreases upon mutation of the conserved lysine and arginine.
We next sought to examine how mutations in the linker region alter the microtubule plus end–tracking activity of EB1. Previous studies using electron microscopy and chemical cross-linking have suggested that the linker region is likely folded and may participate in coordinating the two CH domains of paired EB1 molecules (Buey et al., 2011). To probe whether the five K/R mutations would affect the folding of the linker region, we designed and performed fluorescence resonance energy transfer (FRET)-based analysis to assess the influence of mutations on the scale of the linker region. In addition, we inserted 1–190 (CH and linker), 131–190 (linker) or 131–268 (linker and EBC) between mTurquoise and mVenus to test whether the linker region would interact with the CH or the EBC domain or simply with itself. The expression and purification of the FRET-based sensor proteins was confirmed with SDS–PAGE (Supplemental Figure 11). Interestingly, the FRET efficiencies of mutant molecules all decreased compared with those of sensor containing the wild-type linker region (Figure 4, D–F), indicating that the linker region exhibits great flexibility as judged by its self-folding, and this folding is perturbed by the 5K/R mutation. Given the fact that the plus end–tracking property of EB1 is abolished when the linkage between linker and CH or linker and EBC is broken by insertion of GFP (Supplemental Figure 8), we propose that the linker region serves as a flexible lever by interacting with CH or EBC domain, which enables a stable loading of EB1 onto dynamic microtubule plus ends. In agreement with our rationale, structural analysis indicates that the surfaces of both CH and EBC domains are negatively charged (Supplemental Figure 12), which may exert electrostatic interaction with the positively charged K/R residues in the linker region.
DISCUSSION
The PACF is based on photoactivation of complementary fluorescent proteins (Yang et al., 1996; Hu et al., 2002). On maturation, sparse subsets of PACF molecules were activated, localized, and then bleached (Betzig et al., 2006). This feature provides benefits for superresolution microscopic analyses of applying photoactivation localization and spatially matched molecular interaction. The aggregate position information from all PACF subsets was then assembled into a nanoscale map that enabled us to image a single-molecule copy of a specifically targeted protein–protein interaction in space and time in fixed and live cells. We imaged dimerized EB1 molecules at the microtubule plus ends of the leading edge and cell body in migrating cells and revealed the novel molecular function of the EB1 linker region, a previously uncharacterized structure in microtubule plus-end tracking.
This work provides a new method for observing and quantifying protein–protein interactions in live cells at the single-molecule level and permits easy manipulation of amino acids in a given copy of a testing dimer so the structure–function relationship can be visualized at high precision. For proof-of-principle purposes, in the current study, we successfully demonstrated the efficiency of a PACF system and observed the precise localization of dimerized EB1 proteins tracking the microtubule plus ends in live cells with a resolution of ∼35 nm. We further show that the plus ends at the leading edge and cell body have different motility, and the construction of the plus ends in these different cellular regions is also different. These different types of EB1 molecules were indistinguishable by conventional microscopy, but their distribution patterns are clearly defined by PACF (e.g., Figure 3). With the advantage of PACF system, we successfully distinguished the precise localization of EB1 dimers in defined combinations (e.g., two wild type, two KR5Q mutants, and one wild type plus one mutant). In addition, we demonstrated the important role of the conserved K/R residues in the flexible linker region of EB1 using PACF fusion proteins. On the basis of those results, we speculate that the K/R residues are important for remodeling of the linker region so the adjacent CH domain can be precisely localized and track the plus ends. These findings will further our understanding of the organization and regulation of microtubule plus ends underlying a variety of cellular processes in different model systems (Jiang et al., 2009; Huang et al., 2012; Zhang et al., 2013). Given the large collection of EB1-interacting proteins at the microtubule plus ends, nanometer-scale imaging of EB1-binding proteins at the single-molecule level will enable us to visualize and quantify the plus-end dynamics of individual microtubules in live cells. We have recently revealed that PCAF acetylates EB1 on K220 and disrupts the stability of a hydrophobic cavity on the dimerized EB1 C-terminal, which was previously reported to interact with +TIPs containing the SxIP motif (Xia et al., 2012; Ward et al., 2013). As determined with an EB1 acetyl-K220–specific antibody, K220 acetylation is dramatically increased in mitosis and is localized to the spindle microtubule plus ends. Thus it would be of great interest to use the PACF-based analyses established here to probe how acetylation of K220 alters EB1-binding protein dynamics at kinetochore microtubules and how different EB1-binding proteins function in mitosis at the single-molecule level to enable us to conduct single-molecule dynamics at a single-microtubule scale.
In the future, PACF should benefit from improvements of the palette of available photoactivatable fluorescent proteins, as well as from the discovery of means to improve photostability and brightness with minimized blinking. During the submission of this study, Nickerson and colleagues have demonstrated the feasibility of a complementary photoactivatable mCherry (Nickerson et al., 2014), which provides support of proof of principle of our PACF strategy reported here. In this regard, a parallel application of PACF with complementary photoactivatable mCherry approach will enable us to achieve precision localization of different subsets of dimeric proteins and measurement of their molecular dynamics in space and time. For example, we will be able to detect and quantify individual spindle microtubule plus ends at a given kinetochore and see how their dynamics differ during chromosome movements in mitosis. Given the fact that a large number of EB1-binding proteins possess the SxIP motif, PACF-based analyses will categorize the list of EB1-binding proteins and ascertain their specific roles in mitosis (Ward et al., 2013). A combination of PACF with functional protein interactome data will enable us to validate their interactions and build a high-resolution atlas of protein society with precise localization in eukaryotic cells in various contexts. The successful demonstration of EB1-PACF in tracking cellular dynamics presented here provides proof of concept.
It is worth noting that classic complementary fluorescent proteins exhibit some artificial activities when they are applied to proteins with unknown ability to dimerize or less-characterized dimerization activity (Honnappa et al., 2009). However, PACF exhibits greatly reduced activity with respect to spontaneous dimerization, as it requires large-scale preparation of PACF-expressing 293T cells in order for its fluorescence to be detected. In fact, this low-background characteristic was also observed in the PACF-cherry approach (Nickerson et al., 2014). Nevertheless, the use of both N- and C-terminal orientations for PACF, together with classic biochemical characterization of protein dimerization, will alleviate artifacts if trace amounts exist in cells.
In sum, PACF provides an excellent tool kit for delineating structural determinants underlying dimeric protein–protein interaction in space and time at a resolution of 35 nm. The PACF approach established here will enable the simultaneous visualization of interactions between one copy of wild-type protein and another copy of a protein with a disease-prone mutation and/or a single-nucleotide polymorphism (Ren et al., 2010). In this case, high-resolution correlation maps between genotype and phenotype can be established with ease. It is also expected that the PACF approach will enable a new generation of single molecule–based screening approaches aimed at modulating dimeric protein interactions.
MATERIALS AND METHODS
DNA constructs and chemicals
n/cPACF, segments were subcloned from PAGFP by PCR followed by insertion to PEGFP-N3/C1 vectors to replace GFP. Full-length EB1 was described previously (Xia et al., 2012) and was cloned to the nPACF-N3 and cPACF-N3, respectively. The homodimerization kit of FKBP was from ARIAD Phamaceuticals (Cambridge, MA). The working concentration of rapamycin-derived homodimer-induced AP20187 (ARIAD Pharmaceuticals) was 100 nM.
Mutagenesis
For site-directed mutagenesis of EB1, the QuikChange Site-Directed Mutagenesis Kit (Stratagene, Santa Clara, CA) was used according to the manufacturer's instructions. Error-prone random mutagenesis and directed simultaneous mutagenesis of multiple sites were performed according to standard protocols and confirmed by sequencing.
Protein expression and purification
Proteins were expressed in the Escherichia coli strain BL21-CodonPlus (Stratagene) and purified by Ni-NTA affinity chromatography followed by a gel-filtration step on a Bio-Rad column using a Tris buffer (100 mM Tris-HCl, 150 mM NaCl, pH 7.5), as previously described (Xia et al., 2012).
For native PAGE and subsequent characterization of photoactivation, PACF and EB-PACF proteins from 293T cells (three Petri dishes for each protein) were isolated using a GFP-Trap A affinity matrix (Chromotek, Planegg-Martinsried, Germany) according to the user's manual. Aliquots of purified proteins (5 μg) were loaded onto a 10% polyacrylamide gel. After electrophoresis, the PAGE gel was stained with Coomassie blue to visualize PACF and EB-PACF proteins; this was followed by an examination of their excitabilities using Molecular Imager FX (Bio-Rad). The PAGE gel was first irradiated with a 413-nm light, which was followed by an excitation at 488 nm.
The identity of EB1-PACF was confirmed with anti-EB1 blotting analysis (1:1000; #610534, BD Biosciences, San Jose, CA).
Cell culture
HeLa and MCF7 cells from the American Type Culture Collection (Manassas, VA) were grown in DMEM (GIBCO, Grand Island, NY) culture medium with 10% fetal bovine serum (FBS). Cells grown on cover glasses were transfected 24 h before experiments. Live-cell experiments were performed in CO2-independent culture medium (Invitrogen, Grand Island, NY) containing 10% FBS.
Immunofluorescence imaging
Cells transiently transfected with related plasmids were fixed with cold methanol and stained with anti–α-tubulin mAb (1:2000; DM1A, Sigma Chemicals, St. Louis, MO) and a secondary antibody conjugated with rhodamine dyes (Jackson ImmunoResearch, West Grove, PA). Images were acquired with Plan-Apo 63×/1.42 |NA objective lens on a Zeiss LSM710 confocal microscope (Carl Zeiss, Jena, Germany). PACF-N/C or proteins were activated with a 405-nm laser and excited with a 475-nm laser, respectively.
Western blot analyses
Samples were subjected to SDS–PAGE and transferred onto a nitrocellulose membrane. For detection of EB1-nPACF and EB1-cPACF expression, proteins were probed by anti-EB1 monoclonal antibody (1:1000; #610534; BD Biosciences) and detected using a standard ECL protocol.
Protein structure modeling
GFP structure (PDB entry 1GFL; Yang et al., 1996), EB1 CH domain (PDB entry 1PA7; Hayashi and Ikura, 2003), and EB domain (PDB entry 3GJO; Honnappa et al., 2009) were used in structure modeling as previously described (Xia et al., 2012). Images were made with Pymol, Powerpoint, and Adobe Illustrator.
FRET spectroscopy
Static fluorescence emission spectra of purified FRET constructions were measured on a SHIMADZU RF-5301PC spectrofluorometer using excitation at 433 nm and measuring the emission fluorescence spectra between 460 and 560 nm, as previously described (Chu et al., 2011; Xia et al., 2012). Spectra were normalized and presented with OriginPro7.0.
Wide-field imaging
Time-lapse imaging of cultured cells expressing EB1-eGFP was accomplished with a 60×/1.4 NA objective on an Olympus IX71 microscope (Deltavision Seattle, WA), as previously described (Honnappa et al., 2009; Xia et al., 2012). Images were recorded at 30°C with a frame rate of 2 s each.
PALM imaging
All PALM experiments were performed on Zeiss ELYRA system (Carl Zeiss, Göttingen, Germany) with a 100×/1.42 NA Plan-Apo objective, as previously described (Betzig et al., 2006). For correction of sample drift, 100-nm polystyrene fluorescent beads (Invitrogen) were affixed to the cover-glass by incubation in the medium for 1 h at room temperature; this was followed by a wash with phosphate-buffered saline buffer to clean up the unbound beads. As previously described (Betzig et al., 2006), images were taken by a TIRF microscope coupled to an EMCCD camera that can detect single photons and minimize both autofluorescence and detector noise. Live-cell PALM experiments were performed at 30°C. PALM images of fixed cells were collected with 15,000 cycles of photoactivation (PA) and excitation. The exposure time of each cycle was ∼50 ms. PALM images of live cells were collected with 100 cycles of PA and excitation. The exposure time was 15 ms per cycle.
In general, initial image frames consisted of sparse fields of individually resolvable single molecules on a weaker background presumably dominated by the much larger population of PACF molecules still in the inactivated state, similar to those of PAGFP, as previously described (Betzig et al., 2006). When necessary, excitation and thus bleaching was maintained until such sparse fields were obtained. Additional image frames were then captured until single-molecule bleaching resulted in a mean molecular separation considerably larger than that required for isolation, as previously described (Betzig et al., 2006; Manley et al., 2008).
Data analyses
The fluorescence intensity of the photoactivated or control region and PALM imaging precision were analyzed with ImageJ and Zeiss Zen2011 software. The definition of two different EB1 comets at superresolution level was done based on their apparent shapes. The type A comet was defined by its appearance as a single line of EB1 molecules at the tip of microtubule. In general, typical type A comets possessed a width <70 nm on the microtubule tips. On the other hand, the type B comet was defined based on the unaligned and broad complex structures of PACF-EB1. Statistics were performed with OriginPro 7.0 software. Images were prepared with Adobe Photoshop and Illustrator.
Sequence-alignment analyses
Sequence alignment of the EB1 homologue was performed by ClustalX2 and modified with BioEdit software. Designed vectors were illustrated with Vector NTI software (see Supplemental Figure 1).
Supplementary Material
Acknowledgments
We thank Yunyu Shi for support and Martin Chalfie for input. This work was supported by Chinese 973 projects 2014CB964803, 2012CB945002, and 2010CB912103; Chinese Natural Science Foundation grants 31320103904, 314300054, 31271518, and 91313303; Chinese Academy of Science grant KSCX1-YW-R-65; MOST grant 2009DFA31010; Ministry of Education grants 20113402130010 and PCSIRT IRT13038; Anhui Project 08040102005; National Institutes of Health grants CA164133, DK56292, and G12RR03034; and Central University grants WK2340000032 and WK2340000021.
Abbreviations used:
- CH
calponin homology
- EBC
C-terminal domains of EB proteins
- FBS
fetal bovine serum
- FRET
fluorescence resonance energy transfer
- PACF
photoactivatable complementary fluorescence
- +TIPs
plus end–tracking proteins
- TIRF
total internal reflection fluorescence
- TIRFM
TIRF microscopy.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1133) on October 29, 2014.
REFERENCES
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Buey RM, Mohan R, Leslie K, Walzthoeni T, Missimer JH, Menzel A, Bjelic S, Bargsten K, Grigoriev I, Smal I, et al. Insights into EB1 structure and the role of its C-terminal domain for discriminating microtubule tips from the lattice. Mol Biol Cell. 2011;22:2912–2923. doi: 10.1091/mbc.E11-01-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Yao PY, Wang W, Wang D, Wang Z, Zhang L, Huang Y, Ke Y, Ding X, Yao X. Aurora B kinase activation requires survivin priming phosphorylation by PLK1. J Mol Cell Biol. 2011;3:260–267. doi: 10.1093/jmcb/mjq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi I, Ikura M. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1) J Biol Chem. 2003;278:36430–36434. doi: 10.1074/jbc.M305773200. [DOI] [PubMed] [Google Scholar]
- Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Honnappa S, John CM, Kostrewa D, Winkler FK, Steinmetz MO. Structural insights into the EB1-APC interaction. EMBO J. 2005;24:261–269. doi: 10.1038/sj.emboj.7600529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell. 2002;9:789–798. doi: 10.1016/s1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- Huang Y, Wang W, Yao P, Wang X, Liu X, Zhuang X, Yan F, Zhou J, Du J, Ward T, et al. CENP-E interacts with SKAP to orchestrate accurate chromosome segregation in mitosis. J Biol Chem. 2012;287:1500–1509. doi: 10.1074/jbc.M111.277194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Wang J, Liu J, Ward T, Wordeman L, Davidson A, Wang F, Yao X. TIP150 interacts with and targets MCAK to the plus-end of microtubules. EMBO Rep. 2009;10:857–865. doi: 10.1038/embor.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova Y, De Groot CO, Grigoriev I, Gouveia SM, Munteanu EL, Schober JM, Honnappa S, Buey RM, Hoogenraad CC, Dogterom M, et al. Mammalian end binding proteins control persistent microtubule growth. J Cell Biol. 2009;184:691–706. doi: 10.1083/jcb.200807179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- Nickerson A, Huang T, Lin L, Nan X. Photoactivated localization microscopy with bimolecular fluorescence complementation (BiFC-PALM) for nanoscale imaging of protein-protein interactions in cells. PLoS One. 2014;9:e100589. doi: 10.1371/journal.pone.0100589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- Ren J, Jiang C, Gao X, Liu Z, Yuan Z, Jin C, Wen L, Zhang Z, Xue Y, Yao X. PhosSNP for systematic analysis of genetic polymorphisms that influence protein phosphorylation. Mol Cell Proteomics. 2010;9:623–634. doi: 10.1074/mcp.M900273-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT. TIP maker and TIP marker: EB1 as a master controller of microtubule plus ends. J Cell Biol. 2005;171:197–200. doi: 10.1083/jcb.200509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B, Coquelle FM, Heichette C, Garnier C, Chretien D, Arnal I. EB1 regulates microtubule dynamics and tubulin sheet closure in vitro. Nat Cell Biol. 2008;10:415–421. doi: 10.1038/ncb1703. [DOI] [PubMed] [Google Scholar]
- Ward T, Wang M, Liu X, Wang Z, Xia P, Chu Y, Wang X, Yu H, Yan M, Wang J, et al. Regulation of a dynamic interaction between two microtubule-binding proteins, EB1 and TIP150, by the mitotic PCAF orchestrates kinetochore microtubule plasticity and chromosome stability during mitosis. J Biol Chem. 2013;288:15771–15785. doi: 10.1074/jbc.M112.448886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Noritake J, Kaibuchi K. Regulation of microtubules in cell migration. Trends Cell Biol. 2005;15:76–83. doi: 10.1016/j.tcb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Xia P, Wang Z, Liu X, Wu B, Wang J, Ward T, Zhang L, Ding X, Gibbons G, Shi Y, Yao X. EB1 acetylation by P300/CBP-associated factor (PCAF) ensures accurate kinetochore-microtubule interactions in mitosis. Proc Natl Acad Sci USA. 2012;109:16564–16569. doi: 10.1073/pnas.1202639109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Moss LG, Phillips GN., Jr The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- Zhang L, Shao H, Zhu T, Xia P, Wang Z, Liu L, Yan M, Hill DL, Fang G, Chen Z, et al. DDA3 associates with microtubule plus ends and orchestrates microtubule dynamics and directional cell migration. Sci Rep. 2013;3:1681. doi: 10.1038/srep01681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.