Dictyostelium Rap1 is dynamically activated during cytokinesis and drives cytokinesis progression by coordinating the three major cytoskeletal components: microtubules, actin, and myosin II. Importantly, mutated forms of Rap also affect cytokinesis in other organisms, suggesting a conserved role for Rap in cell division.
Abstract
Cytokinesis is the final step of mitosis when a mother cell is separated into two daughter cells. Major cytoskeletal changes are essential for cytokinesis; it is, however, not well understood how the microtubules and actomyosin cytoskeleton are exactly regulated in time and space. In this paper, we show that during the early stages of cytokinesis, in rounded-up Dictyostelium discoideum cells, the small G-protein Rap1 is activated uniformly at the cell cortex. When cells begin to elongate, active Rap1 becomes restricted from the furrow region, where the myosin contractile ring is subsequently formed. In the final stages of cytokinesis, active Rap1 is only present at the cell poles. Mutant cells with decreased Rap1 activation at the poles showed strongly decreased growth rates. Hyperactivation of Rap1 results in severe growth delays and defective spindle formation in adherent cells and cell death in suspension. Furthermore, Rap mutants show aberrant regulation of the actomyosin cytoskeleton, resulting in extended furrow ingression times and asymmetrical cell division. We propose that Rap1 drives cytokinesis progression by coordinating the three major cytoskeletal components: microtubules, actin, and myosin II. Importantly, mutated forms of Rap also affect cytokinesis in other organisms, suggesting a conserved role for Rap in cell division.
INTRODUCTION
Cell division is a fundamental process that is required for cell proliferation and differentiation of cell types. In anaphase, formation of the spindle apparatus pulls the chromosomes toward the poles of the dividing cell and triggers the beginning of cytokinesis, the final step in the separation of a mother cell into two daughter cells. Following the assembly of microtubule filaments in the expanding mitotic spindle, bundles of actin and non–muscle myosin filaments create a contractile ring that constricts the plasma membrane at the furrow region, while actin filaments are formed at the poles of the cells. This temporal and spatial regulation of the cytoskeleton is essential for the separation of the daughter cells (Glotzer, 2005; Kanada et al., 2005; Wang, 2005). Most of the current models therefore incorporate these global, polar, and equatorial processes in orchestrating the division process (Wang, 2005; Surcel et al., 2010); it is, however, not well understood how they are coordinated in time and space. To investigate this in more detail, we used the model organism Dictyostelium discoideum.
Cell division is one example of cell polarization that depends on the organization of the cytoskeletal elements. Establishment and maintenance of eukaryotic cell polarization can be induced by small fluctuations of an asymmetric stimulus. A stimulus can be either intracellular, like signals from the dividing nucleus during cell division, or extracellular, such as chemical gradients during chemotaxis. Similar processes are involved in establishing polarity during chemotaxis and in regulating asymmetry during cytokinesis (Wang, 2005). Myosin II filaments and cortexillin are localized at the back of chemotactic cells and at the furrow of dividing cells, whereas PIP3 (phosphatidylinositol (3,4,5)-triphosphate), dynacortin, coronin, Scar, and active Ras and Rac are localized at the leading edge and poles during chemotaxis and cell division, respectively (Janetopoulos et al., 2005; Sasaki et al., 2007; King et al., 2010; Surcel et al., 2010). Dictyostelium Rap1 is a well-known component in establishing cell polarity and regulating cytoskeletal rearrangements during chemotaxis (Lee and Jeon, 2012). During chemotaxis, Dictyostelium Rap1 is involved in the regulation of adhesion, myosin II disassembly, and PI3K (phosphatidylinositol-3-kinase) activation (Kortholt and van Haastert, 2008; Lee and Jeon, 2012), all processes that are also critical for cytokinesis. Consistently, knockdown of rap1 in Dictyostelium results in decreased growth rate and cell viability (Kang et al., 2002). In this paper, we show that proper Rap1 activation is important for regulating cytokinesis progression in Dictyostelium. Rap1 is dynamically activated during cytokinesis; in the early stages of cytokinesis, Rap1 is activated uniformly at the cell cortex, where it regulates adhesion and contractile force, while at later stages Rap1 regulates adhesion and cytoskeleton dynamics at the cell poles. We propose a model in which Rap1 drives cytokinesis progression by coordinating global, polar, and equatorial changes of the three major cytoskeletal components: microtubules, actin, and myosin II.
RESULTS
Rap1 regulates several processes in moving cells, such as adhesion and cytoskeletal rearrangements, that are also important during cell division (Jeon et al., 2007b; Kortholt and van Haastert 2008; Lee and Jeon, 2012). To investigate whether Rap1 plays a role during cytokinesis, we analyzed 1) Ras and Rap1 localization and activation in dividing Dictyostelium cells; 2) growth and cytokinesis of mutants with decreased or increased Rap1 activation; and 3) the role of Rap1 in coordinating microtubules, actin, and myosin II during cell division.
Dynamic Rap1 activation during Dictyostelium cytokinesis
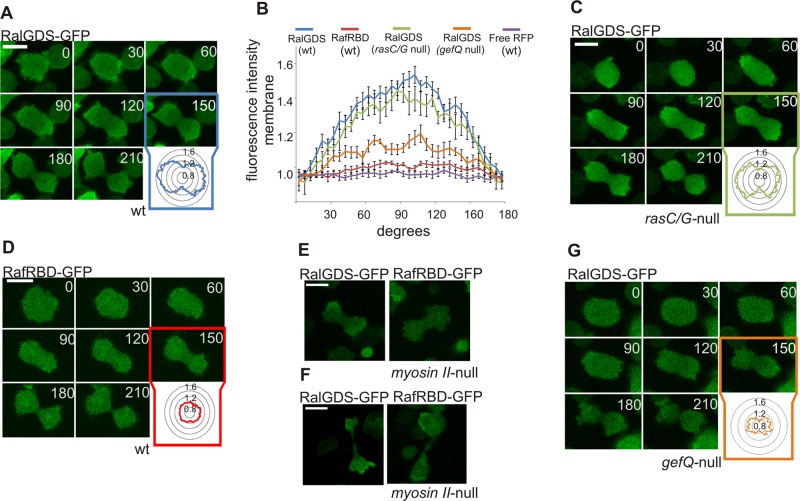
Supplemental Figure S1A shows that N-terminal green fluorescent protein (GFP)-fused Rap1 is localized uniformly at the cell membrane during both growth and cytokinesis of Dictyostelium cells. To monitor spatial activation of the protein, rather than its localization, we used the previously described molecular probe for active Rap1, RalGDS-GFP (Jeon et al., 2007b; Kortholt and van Haastert, 2008). In unstimulated vegetative cells, Rap1 activation was observed at the cell cortex and in patches at the cell membrane that may correspond to macropinosomes (Supplemental Figure S1B). At the moment cells rounded up and entered cytokinesis, these RalGDS patches disappeared and Rap1 became uniformly activated at the cell cortex. Once the cell started to elongate, Rap1 activation was restricted to the sites that later became the poles of the dividing cell (Figure 1A). In dumbbell-shaped cells, the RalGDS marker was depleted from the furrow region, and strongly localized to the poles (Figure 1, A and B). RalGDS-GFP intensity at the membrane was measured around the circumference of cells (Figure 1B). The average fluorescence of RalGDS-GFP at the poles was 1.48 ± 0.12 (n = 10) times the cytosolic fluorescence, while the fluorescence at the furrow region (1.01 ± 0.12, n = 10) was similar to that in the cytosol (Figure 1, A and B). This asymmetric Rap1 activation persisted until the moment the two daughter cells separated from each other.
FIGURE 1:
Dynamic Rap1 activation during cytokinesis. (A) Images of RalGDS-GFP (detecting active Rap) in dividing Dictyostelium wild-type cells. Inset: RalGDS-GFP fluorescence intensity was measured at the cell boundary around the circumference of the cell relative to the fluorescence intensity in the cytosol. (B) Quantification of the average fluorescence intensities of the indicated fluorescent markers along the cell membrane of dividing Dictyostelium cells presented as degrees from the cleavage furrow (see A, C, D, and G for representative images of the experiments). Error bars represent SEM. (C) Image of RalGDS-GFP in rasC/rasG-null cells. (D) Images of RafRBD-GFP (active Ras) in dividing Dictyostelium wild-type cells. Images of RalGDS-GFP or RafRBD-GFP in myosin II-null cells dividing by type B (E) and type C (F). (G) Image of RalGDS-GFP marker in gefQ-null cells. Scale bars: 10 μm.
Previous studies have shown that Rap activation at the leading edge of chemotactic Dictyostelium cells is completely dependent on heterotrimeric G-protein (Gα2βγ) and RasG signaling (Bolourani et al., 2008). In contrast, Rap1 activation during cytokinesis is not dependent on heterotrimeric G-protein signaling; dividing Gβ-null cells showed normal RalGDS localization at the poles (Supplemental Figure S1, C and D). Cells lacking rasC and rasG are unable to undergo chemotaxis and have severe growth defects in suspension culture (Tuxworth et al., 1997); however, when these cells are grown on substrates, the timing and progression of cytokinesis is essentially the same as that of wild-type cells (Figure 1C). Importantly, rasC/rasG-null cells showed normal RalGDS-GFP accumulation at the poles of dividing cells (Figure 1B), with average levels at the poles and furrow of 1.40 ± 0.04 and 0.96 ± 0.13, respectively (Figure 1, B and C; the numbers are not statistically different from wild-type cells).
To investigate whether Ras as well as Rap1 is activated during cytokinesis, we monitored Ras activation in vivo with the previously described GFP-RafRBD marker (Sasaki et al., 2004; Kortholt and van Haastert, 2008). Vegetative wild-type cells expressing RafRBD showed Ras activation at dynamic membrane patches (Supplemental Figure S1E). When cells rounded up to enter cell division, Ras activation completely disappeared for 2–3 min (Figure 1D). Importantly, during this stage of cell division, we already observed very strong activation of Rap1 at the poles (Figure 1A). The calculated average levels of GFP fluorescence for the poles and furrow region were 1.01 ± 0.04 and 0.95 ± 0.04 times that of cytosolic fluorescence (n = 10; Figure 1, B and D). Only during late stages of cytokinesis, when the two daughter cells were almost separated, did RafRBD-GFP become slightly enriched at the poles, as has been described before (Sasaki et al., 2007).
Rap and Ras activation during myosin-independent cytokinesis
Under normal conditions, wild-type Dictyostelium cells almost exclusively divide by a mechanism of cytokinesis called type A, which depends on the formation of a myosin contractile ring at the cell midzone (Fukui et al., 1990). This type of cytokinesis is characterized by evenly sized daughter cells, and the time to complete the scission is ∼3 min. Incidentally, wild-type cells grown on solid substrates divide without the formation of a myosin contractile ring (Uyeda et al., 2000). Such myosin-independent cytokinesis is critically dependent on substrate attachment (Nagasaki et al., 2002, 2009). Cells lacking myosin II are therefore only viable when grown on substrates. Analyses of RalGDS localization in myosin II-null cells revealed that Rap1 is strongly activated at the poles (Figure 1, E and F), indicating that spatial activation of Rap1 during cytokinesis is independent of myosin II.
There are two modes of this myosin-independent cytokinesis, called type B and type C (Neujahr et al., 1997; Nagasaki et al., 2009). Cytokinesis type B is spatiotemporally coupled to nuclear division, and the closing of the cleavage furrow is suggested to be passive and dependent on traction forces produced at the cell polar regions (Nagasaki et al., 2002). During cytokinesis B, Rap1 was strongly activated at the poles, while no activation of Ras could be detected (Figure 1E). This shows that during both cytokinesis type A and B, which are coordinated with karyokinesis, Rap1 activation is independent of Ras. Interestingly, during cytokinesis type C, both Ras and Rap were activated at the poles (Figure 1F), resembling Rap and Ras activation at the leading edge during chemotaxis (Kortholt and van Haastert, 2008). This is consistent with the observation that cytokinesis type C is not coupled to nuclear division and is the result of different parts of one multinucleate cell crawling away from each other. This process thus resembles directional movement of cells (Uyeda et al., 2000).
Together these data show that Rap1 but not Ras is activated at the poles of dividing Dictyostelium wild-type cells. In contrast to chemotaxis, this spatial activation of Rap1 does not depend on Ras activity. Similar results are obtained for cells lacking myosin II, except for the unusual cytokinesis type C. In the next sections, we discuss the growth and cytokinesis of mutants with decreased or increased Rap1 activation.
Decreased Rap1 activation results in severe growth defects
Expression of antisense rap1 mRNA in Dictyostelium cells results in decreased growth rate and cell viability (Kang et al., 2002). Multiple attempts to disrupt rap1 in Dictyostelium wild-type cells failed (Kang et al., 2002). Therefore we tried to generate a conditional knockout by disrupting rap1 in cells expressing Rap1 from an inducible extrachromosomal plasmid or by knocking in the inducible promoter sequence in the genomic rap1 promoter locus. Although a few clones were initially obtained, these cells were highly unstable and finally died.
GbpD and GefQ work together regulating Rap1 activation in vegetative cells (Plak et al., unpublished data). Mutants cells lacking or overexpressing GbpD have normal cytokinesis and growth rates (Bosgraaf et al., 2005). Cells overexpressing GefQ have severe cytokinetic defects in suspension and have limited ability to assemble myosin filaments (Mondal et al., 2008). We determined the growth rate of gefQ-null cells and observed that mutant cells grow much more slowly than the parental strain, with doubling times in shaking suspension of 30 and 13 h, respectively. These gefQ-null cells show an almost 50% reduction of Rap1 activation at the poles of dividing cells (Figure 1, B and G). Together these results suggest that GefQ-mediated Rap activation is important for regulating cytoskeleton rearrangements during cell division.
Constitutive activation of Rap1 results in severe cytokinetic defects
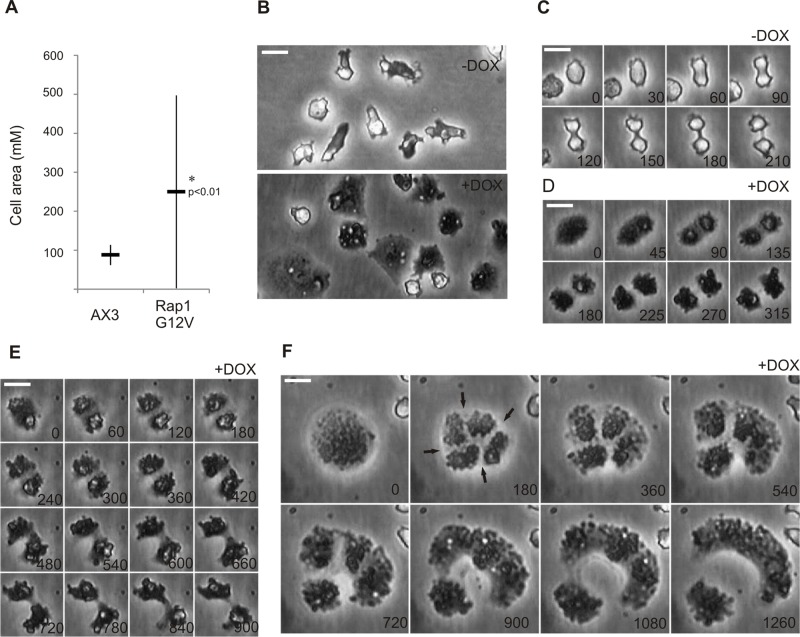
Most of the current knowledge about Rap-mediated signaling in Dictyostelium comes from studies using constitutively active Rap1G12V mutants. Cells expressing Rap1G12V are large and flattened (Rebstein et al., 1997), have strongly increased substrate adhesion, and exhibit chemotactic defects (Jeon et al., 2007b). Because of these severe defects, we used here a doxycycline-inducible plasmid system (pDM310) to study the effect of Rap1G12V on cell growth and cell division (Veltman et al., 2009). Seven hours after induction of Rap1G12V expression (Supplemental Figure S1F), cells showed the characteristic Rap1G12V phenotype and became larger and flattened, with an average cell area of 249 ± 247 μm2 (n = 180), compared with 87 ± 26 μm2 (n = 180) for uninduced control cells (Figure 2, A and B). The broad size distribution (Figure 2A) is caused by large variation in Rap1G12V expression levels within the cell population (Supplemental Figure S1G). This is the consequence of the variation in plasmid copy number per cell that is typically observed with the used inducible plasmid system (Veltman et al., 2009; Veltman and van Haastert, 2013).
FIGURE 2:
Hyperactivation of Rap1 leads to various cytokinetic defects on substrates. Dictyostelium Rap1G12V (+DOX) and control (−DOX) cells. Shown are (A) graph depicting the average cell size of vegetative grown cells and (B–F) images of cells undergoing cytokinesis on substrates; time in seconds is indicated. Arrows in F show the regions where the furrow is being formed. Scale bars: 10 μm. * indicates significantly different from wild type with p < 0.01.
In addition to the large cell surface area, Rap1G12V cells have strongly increased substrate attachment. Because precise spatiotemporal regulation of cellular adhesion strength has recently been proposed to be important in successful closure of the cleavage furrow in mitotic HeLa cells (Bastos et al., 2012), we investigated the closure of the cleavage furrow in Rap1G12V cells. Vegetative cells were observed during 30 h; wild-type cells did undergo approximately three division cycles, whereas 70% of the Rap1G12V cells did not manage to complete a single division cycle. The Rap1G12V cells that divided did so in a manner that was different from that of wild-type cells. As described in the next paragraph, two major types of cytokinetic defects are observed in the few Rap1G12V cells that completed cytokinesis: extended time for completing the scission and asymmetrical furrow ingression.
Wild-type cells dividing by myosin-dependent cytokinesis round up and slightly detach from the substrate at the moment they enter mitosis. The cells remain less attached throughout cytokinesis, as was observed by their bright appearance under phase-contrast microscopy (Nagasaki et al., 2002; Figure 2C). A subset of Rap1G12V cells showed only a moderate adhesion and cell-spreading phenotype (cell area of <250 μm). These cells rounded up before entering cell division, but in contrast to wild-type cells, they did not detach but remained strongly attached to the substrate during cytokinesis. Often, two daughter cells of equal size were formed (Figure 2D), but complete closure of the furrow did take significantly longer for the Rap1G12V cells (6.7 ± 0.8 min, n = 9) compared with wild-type cells (4.0 ± 0.3 min, n = 14). In several other cells, we observed that the closing of the furrow appeared to be asymmetrical, with one side of the furrow ingressing much more efficiently than the other side of the furrow. The cells that divided in this way did not exhibit the canonical dumbbell shape (Figure 2E) and needed more than 15 min, four times more than wild type, to complete division.
Hardly any of the cells with high levels of Rap1G12V expression (cell area of >250 μm) were able to divide within the given time period. The few cells that entered the division process initially rounded up and started to create a furrow, but this process suddenly stopped, and the cells returned back to the vegetative state. Only after a delay of several hours did these cells round up again and initially form multiple furrows, but they finally finished cytokinesis and divided asymmetrically into two daughter cells (Figure 2F).
Rap1G12V cells are unable to grow in shaking cultures
Rap1G12V cells were incubated with growth medium in a shaken culture. During the first 24 h, cells were growing but at a reduced rate compared with control cells; the doubling time of induced Rap1G12V cells is 29 h, while uninduced Rap1G12V cells or wild-type cells grown in the presence of doxycycline (DOX) have a doubling time of 15 h. In contrast to RapG12V cells grown on solid substrate (discussed previously), the size of Rap1G12V cells grown in suspension for 24 h did not increase (Supplemental Figure S2A). The DNA content of the Rap1G12V cells at this time point is not different from the uninduced control or wild-type cells (Supplemental Figure S2B), suggesting that Rap1 hyperactivation does not result in multinucleated cells. Nevertheless, after ∼48 h in shaken suspension, the Rap1G12V-expressing cells started to lyse.
Overexpression of Rap1G12V does not lead to multinucleated cell phenotype but affects nuclear shape and size
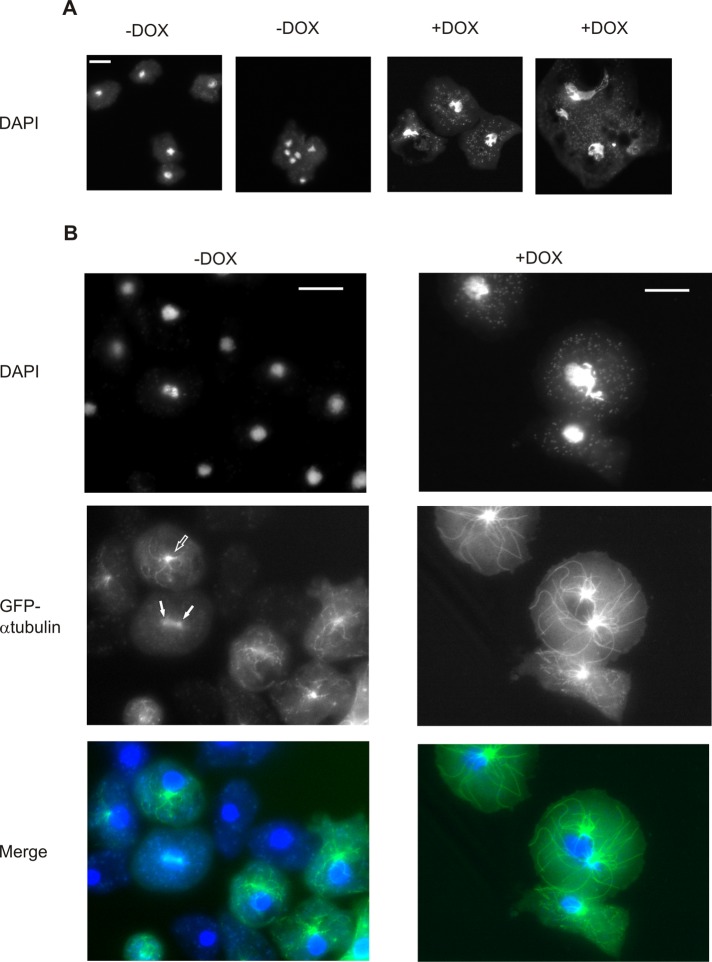
Staining of Rap1G12V cells grown on substrates in the presence or absence of DOX for 48 h with 4′,6-diamidino-2-phenylindole (DAPI) fluorescent dye showed no significantly (p > 0.01) different average number of nuclei in Rap1G12V-expressing cells (1.34, n = 114, SEM = 0.08) compared with the uninduced control cells (1.16, n = 90, SEM = 0.06). Although hyperactivation of Rap does not influence the number of nuclei, it dramatically affects nuclear size and shape. In control cells, the nuclei appeared small, round, and evenly sized both in single and in multinucleated cells (nuclear size of 8.6 ± 2 μm2; Figure 3A). In contrast, Rap1G12V cells had irregularly shaped and much larger nuclei (25.1 ± 13.6 μm2). The small fraction of multinucleated Rap1G12V cells had nuclei of various shapes and sizes, and some of them were elongated or appeared to be connected (Figure 3A).
FIGURE 3:
Size and shape of the nucleus and microtubular cytoskeleton are affected by expression of hyperactivated Rap1. Images of Dictyostelium Rap1G12V (+DOX) and control (−DOX) cells. Shown are (A) cells stained with DAPI and (B) cells coexpressing GFP–α−tubulin. Interphase MTOC is marked with open arrows, while mitotic structures are indicated with closed arrows. Scale bars: 10 μm.
Together these results show that both reduced expression of Rap1 and hyperactivation of Rap1 lead to severe growth defects. Hyperactivation of Rap1 results in cytokinetic defects with misshapen nuclei. In the next sections, we describe the role of Rap1 in coordinating microtubules, actin, and myosin II during cell division.
Rap1 activation affects microtubule cytoskeleton organization
The above data show that Rap1G12V cells have severe growth and cytokinetic defects but are not multinucleated. This suggests that cells may fail to properly divide their nuclei at an early stage of cell division. Recently it was shown that an increased cellular area and decreased cell height can result in increased chromosome spreading after the breakdown of the nuclear envelope, which subsequently causes delays in chromosome capture and defects in spindle formation during the mitosis process (Lancaster et al., 2013). Since Dictyostelium cells undergo closed mitosis, during which the nuclear envelope does not disassemble (Gräf et al., 2000), we tested whether the increased size of the nucleus in Rap1G12V cells causes similar defects in spindle formation. Rap1G12V was coexpressed with GFP–α-tubulin, cells were stained with DAPI, and the distribution of nuclear DNA and the size of the microtubule cytoskeleton were analyzed. Wild-type interphase cells have microtubule-organizing centers (MTOCs) in close proximity to the nuclei, and in almost all cases (97%, n = 102), their number corresponds to the number of nuclei (Figure 3B, open arrow). Mitotic wild-type cells have clearly visible spindle microtubules, while nuclear DNA is condensed and starts to separate (Figure 3B, closed arrows). Rap1G12V cells have strong defects in microtubule cytoskeleton organization and abnormally shaped nuclei and MTOCs (Figure 3B). Furthermore, in 37% of the analyzed cells (n = 96), the number of MTOCs did not correspond to the number of nuclei present. Almost all of these Rap1G12V cells had a single, enlarged nuclei with two independent MTOCs attached to it, a phenotype that was never observed in wild-type cells. The two MTOCs are not connected by central-spindle microtubule filaments that are characteristic and essential for mitotic wild-type cells (Figure 3B).
Rap1 regulates myosin and actin rearrangements during cytokinesis
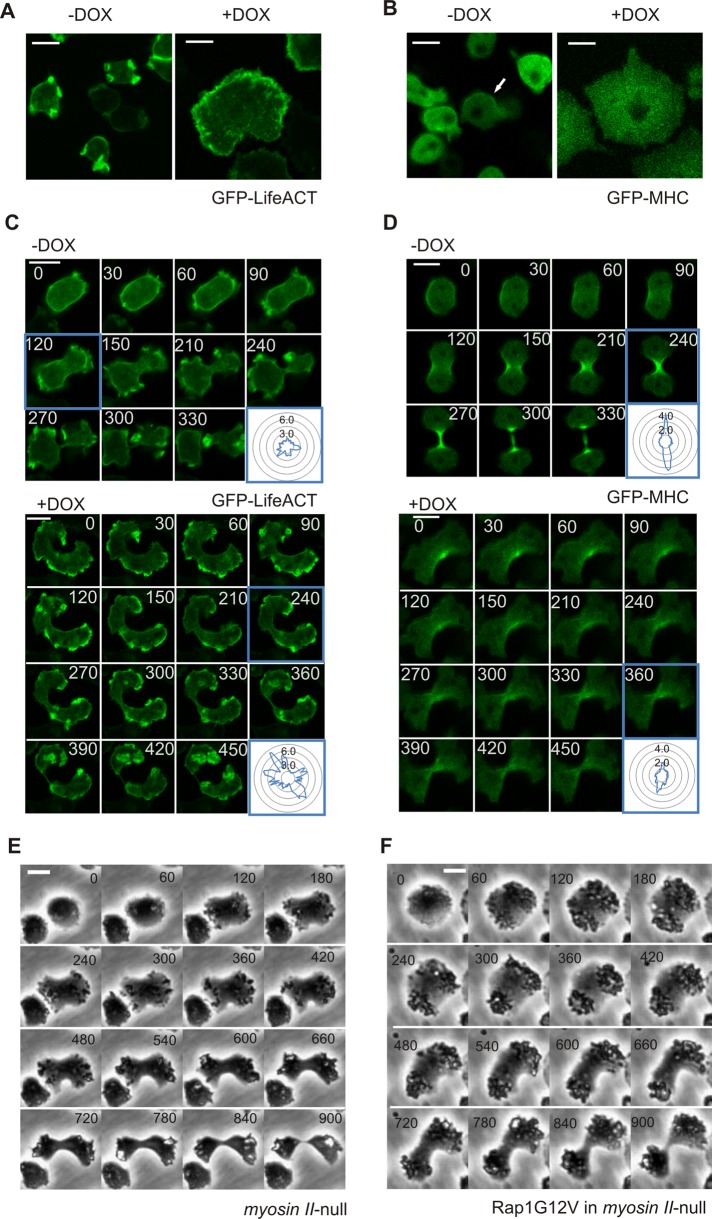
Dictyostelium Rap1 is an important regulator of both myosin and actin (Kortholt et al., 2006; Kortholt and van Haastert, 2008; Jeon et al., 2007b). Activation of Rap1 induces actin filaments when exposed to uniform chemoattractant stimulation (Mun and Jeon, 2012), while Rap1-mediated myosin heavy-chain phosphorylation results in myosin II filament disassembly at the leading edge of chemotactic cells (Jeon et al., 2007b). Proper regulation of actin and myosin dynamics is essential for successful closure of the cytokinetic furrow (Faix et al., 1996; O’Connell et al., 2001; Gerisch et al., 2004; Glotzer, 2005). Actin and myosin dynamics were monitored with the filamentous actin marker Lifeact (Riedl et al., 2008) and GFP-MHC (Moores et al., 1996), respectively. Vegetative control cells showed dynamic Lifeact patches at the cell boundary and weak cortical localization of myosin (Figure 4, A and B). After Rap1G12V expression was induced, the cortical myosin localization strongly diminished, and the cells showed a thick but dynamic actin-rich cortex around the whole cell boundary. During cell division, actin became enriched at the poles of both dividing wild-type and Rap1G12V cells, however, Rap1G12V cells showed larger amounts and more intense patches of F-actin at the cell boundary compared with the control cells (Figure 4C, bottom panels). Dividing control cells showed symmetrical localization of myosin at both sides of the cell furrow, which increased with time (Figure 4D, top panels). In contrast, dividing Rap1G12V cells showed an asymmetrical localization of myosin and even at late stages of cytokinesis the amount of myosin at the furrow was much lower than in control cells (Figure 4D, bottom panels). The side of the furrow with less myosin showed slower ingression, which consequently resulted in an asymmetrical cell shape during the division (Figure 4D, bottom panels). These data show that proper Rap1 activation is important for regulating the actomyosin cytoskeleton in response to nuclear division.
FIGURE 4:
Rap1 regulates myosin and actin cytoskeleton dynamics. Images of vegetative induced (+DOX) or control (−DOX) cells expressing (A) Lifeact-GFP (detects filamentous actin) or (B) GFP-MHC. Localization of (C) Lifeact-GFP or (D) GFP-MHC during cytokinesis is shown. Inset: fluorescence intensity was measured at the cell boundary around the circumference of the cell relative to the fluorescence intensity in the cytosol. (E and F) Comparison of representative cell shape and timing of cells during cytokinesis. Shown are myosin II–null cells (E) and myosin II-null cells with Rap1G12V (F). Scale bars: 10 μm.
Both expression of Rap1G12V cells or disruption of myosin resulted in delayed closure of the cleavage furrow compared with wild-type cells (Figures 2, D–F, and 4E; Uyeda et al., 2000). To further study the importance of Rap1-mediated cytoskeletal rearrangements during cytokinesis, we expressed Rap1G12V in myosin II-null cells. In the time frame during which 74% (n = 86) of the myosin II-null control cells completed the cell cycle, only 26% (n = 98) of Rap1G12V myosin II-null cells divided. However, the timing and progression of the cytokinetic events were highly similar in Rap1G12V myosin II-null and myosin II-null cells (Figure 4, E and F). Interestingly, the defects of Rap1G12V myosin II-null cells were less severe than the defects of Rap1G12V cells, suggesting that the strong Rap1G12V phenotype partly depends on myosin II.
Rap1 affects cellular distribution of 14-3-3 in a RacE-dependent manner
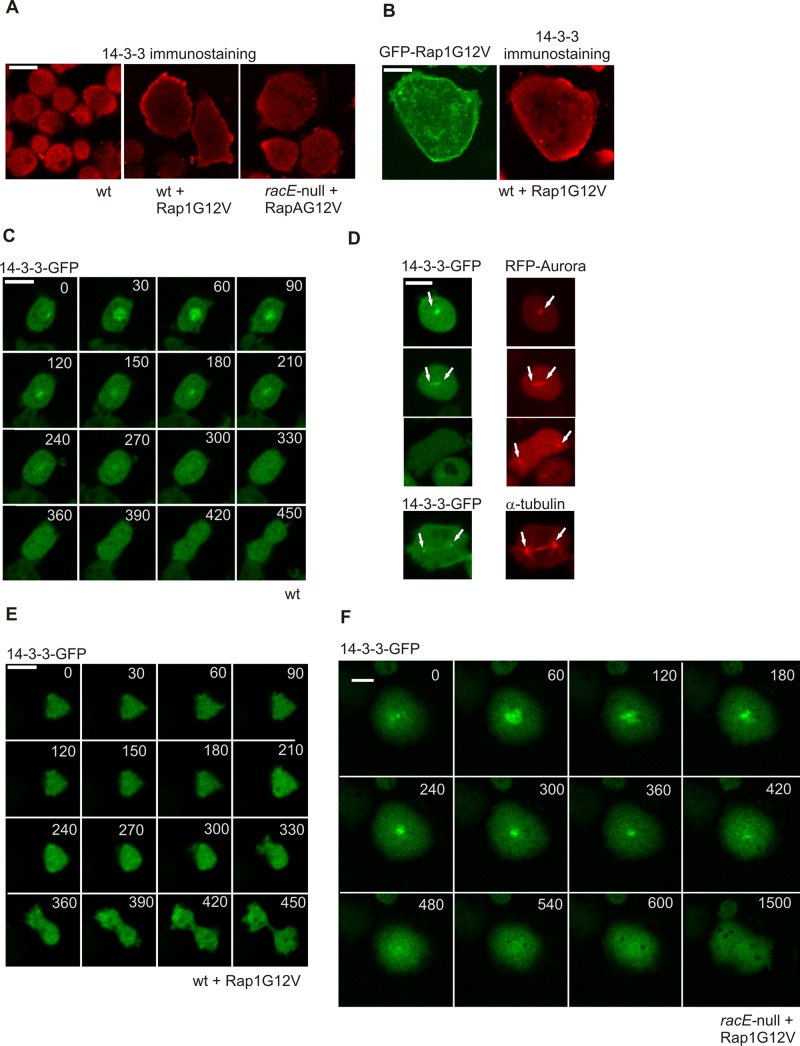
We showed that Rap1G12V expression affects a number of cellular processes during mitosis progression, including defects in myosin assembly, adhesion, actin polymerization, and microtubule formation. Recently a RacE/14-3-3–dependent pathway was identified in Dictyostelium that links microtubules and cortical myosin to actin dynamics at the cortex (Zhou et al., 2010). Although human 14-3-3 is an interaction partner for Rap proteins (Riou et al., 2013), we were unable to detect any direct binding between activated Dictyostelium Rap1 and 14-3-3. However, because Dictyostelium Rap1G12V cells have increased Rac activation (Plak et al., 2013), we hypothesized that Rap1 may cooperate with RacE and 14-3-3 in regulating Dictyostelium cytokinesis. Staining with 14-3-3–specific antibody, revealed weak cortical localization of 14-3-3 in nonmitotic wild-type cells (Figure 5A). Additionally, 14-3-3 and active Rap1 colocalized at the cell cortex (Figure 5B). A recent proteomic study suggested that Dictyostelium 14-3-3 is associated with centrosome during cell division (Reinders et al., 2006), whereas microscopy data suggest that the protein is enriched at the polar cortex region of the dividing cell in late cytokinesis (Zhou et al., 2010). Our data with both N-terminal or C-terminal GFP-fused 14-3-3 clearly show a dynamic cell cycle–dependent localization: in early mitotic stages, 14-3-3 is localized in the nucleus and subsequently at the centrosome (Figure 5C), where it colocalizes with Aurora kinase and, partially, α-tubulin (Figure 5D). Notably, we did not observe the previously described polar cortical localization of 14-3-3 (Zhou et al., 2010). As the cytokinesis progressed the centrosomal localization disappeared, and at the late stages of cytokinesis, appeared to be lost completely (Figure 5, C and D). This centrosomal localization appears to be specific, as all dividing cells observed(n = 22) showed the same localization pattern. In Rap1G12V-expressing cells, 14-3-3 is mislocalized; vegetative Rap1G12V cells have increased amounts of 14-3-3 at the cortex compared with wild-type cells (Figure 5A). Most importantly, and in contrast to control cells, the cells that expressed low amounts of Rap1G12V and that were capable of dividing did not show nuclear or centrosomal localization of 14-3-3 during their division (Figure 5E).
FIGURE 5:
Rap1G12V affects 14-3-3 localization in RacE-dependent manner. (A) Images of immunostained 14-3-3 protein in the indicated cell strains. (B) Images showing colocalization of GFP-Rap1G12V and 14-3-3 in fixed Dictyostelium cells. (C) 14-3-3-GFP localization in dividing wild-type cells. (D) Colocalization of 14-3-3 with Aurora-mRFP or α-tubulin. 14-3-3 localization in (E) dividing Rap1G12V and (F) racE-null/Rap1G12V cells. Time is indicated in seconds. Scale bars: 10 μm for all panels.
It has previously been shown that cortex localization of 14-3-3 in vegetative Dictyostelium cells depends on the small G-protein RacE (Zhou et al., 2010). We tested vegetative racE-null cells expressing Rap1G12V for their ability to translocate 14-3-3 to the cell cortex and detected much weaker cortical localization of 14-3-3 compared with control wild-type cells expressing Rap1G12V (Figure 5A). Interestingly, 14-3-3 was shown to be present in the nucleus during cytokinesis of Rap1G12V racE-null cells. Despite this nuclear localization of 14-3-3, racE-null/Rap1G12V cells still had the strong spreading phenotype and showed difficulties in completing cytokinesis (Figure 5, A and F). During cell division, the racE-null/Rap1G12V cells appeared to acquire polarity and elongated in the direction that was in accordance with the spindle axis, but the furrow was either ingressing slowly and asymmetrically or never formed (Figure 5F), which suggests defects at later stages of cytokinesis.
DISCUSSION
Coordinated changes in cellular adhesion, microtubule organization, and actin and myosin filament rearrangements contribute to cell division (Wang, 2005; Surcel et al., 2010). We have shown here that the Rap1 activation pattern exhibits dynamical changes that allow for proper coordination of adhesion and cytoskeleton dynamics during the cell division process. Rap1 is specifically activated at the poles of dividing cells. Activation becomes detectable immediately after cells round up and persists while the mother cell elongates until the daughter cells are separated. The pathways that regulate activation of Rap1 during cytokinesis are different from those previously reported for chemotaxis. The heterotrimeric and monomeric G-protein signaling cascade that is indispensable for chemoattractant-induced Rap1 activation (Bolourani et al., 2008), is not necessary for activation of Rap1 during cell division. GefQ, one of the three known GEFs for Rap1 in Dictyostelium, appears to be important for regulating Rap1 during cytokinesis. Furthermore, a recent study suggests that RapGAP9 is also crucial for cytokinesis progression (Mun et al., 2014).
Knockdown of rap1 severely affects the growth of Dictyostelium cells (Kang et al., 2002), whereas expression of hyperactivated Rap1 results in multiple defects in progression of cytokinesis. Through comparison of the cytoskeleton in control and hyperactivated Rap1 cells, a picture is emerging on the complex role of Rap during cytokinesis.
At the mitotic entry, wild-type cells round up and detach from the substrate, which is associated with moderate and uniform Rap1 activation at the cell boundary. At this stage, the mitotic spindle assembles and designates the division plane, which allows for entry to cytokinesis. Previously it was shown that, in Dictyostelium, 14-3-3 links microtubules and cortical myosin to actin dynamics (Zhou et al., 2010). We showed here that, in early mitotic control cells, 14-3-3 is localized in the nucleus and subsequently at the centrosome. Hyperactivation of Rap1 leads to accumulation of 14-3-3 at the cell cortex and prevents it from localizing to the mitotic spindle during the interphase. Our data suggest that this aberrant 14-3-3 localization is mediated by the small G-protein RacE. We have not been able to demonstrate a direct interaction between 14-3-3 and Rap1; it is thus unlikely that Rap1 directly regulates 14-3-3 localization. It has been shown that 14-3-3 is localized to the cortex by binding to several actin cross-linking proteins (for a review, see Sluchanko and Gusev, 2010). RacE regulates cortical localization of several global actin cross-linkers and is important for 14-3-3 localization during cytokinesis (Zhou et al., 2010). Finally, we have shown that vegetative racE-null cells expressing Rap1G12V did not show the aberrant cortical localization of 14-3-3 when compared with wild-type cells expressing Rap1G12V, suggesting that RacE functions downstream of Rap1 and upstream of 14-3-3.
At the beginning of cytokinesis, activation of Rap1 is likely spatially regulated by cell cycle–activated GEFs (guanine exchange factors) and GAPs (GTPase-activating protein). Cells lacking gefQ showed reduced Rap1 activation at the poles of the dividing cells and reduced growth, while hyperactivation of GefQ resulted in decreased myosin assembly and defects in cytokinesis (Mondal et al., 2008). Consistently, the few Rap1G12V cells that did enter the cell cycle failed to properly regulate the actomyosin cytoskeleton, resulting in extended furrow ingression times and asymmetrical cell division. The timing and progression of cytokinesis in Rap1G12V myosin II-null cells is highly similar to that of myosin II-null cells, however, far fewer Rap1G12V/myosin II-null entered the cell cycle. Furthermore, expression of Rap1G12V in wild-type cells reduced the growth rate in shaking suspension but, in contrast with myosin II-null cells, did not result in multinucleated cells. Together these data suggest that Rap-mediated actin/myosin rearrangements are not involved in regulating the entry to mitosis but are important for proper progression of the process to separate the emerging daughter cells. We therefore propose that Rap1 activation at the poles promotes adhesion and cell polarization by inducing F-actin remodeling through pathways that most likely include PI3K and Rac proteins and by inhibiting myosin assembly at the poles through its effector Phg2 (Jeon et al. 2007a; Kortholt and van Haastert, 2008; Lee and Jeon, 2012). At the same time, low levels of Rap1 activation in the equatorial zone of the dividing cell cause decreased adhesion and allow for myosin filament assembly. Those two processes promote contractile ring formation and closing of the cytokinetic furrow. Consistent with this model, Lancaster et al. (2013) recently showed that the cortical actin cytoskeleton does not contribute to spindle formation in mitotic human cells but is crucial for progression of cytokinesis and for maintaining proper cortical tension in cells grown in physically constrained tissues.
Previously it was shown that levels of activated Rap are tightly controlled during the progression of cell division in HeLa cells (Dao et al., 2009). Furthermore, Rap hyperactivation in both Drosophila embryonic cells and human cell lines causes, as in Dictyostelium, defective spindle positioning (Carmena et al., 2011; Lancaster and Baum, 2014). This strongly suggests a conserved and important role for Rap in regulating cell cycle progression and cytokinesis.
MATERIALS AND METHODS
Strains and constructs
AX3 Dictyostelium cells were used as the wild-type strain. Cells were grown in HL5-C media (Formedium, Norfolk, UK) either on Nunclon-coated Petri dishes or in shaking flasks. For selection, the medium was supplemented with the appropriate antibiotics; 10 μg/ml G418, 50 μg/ml hygromycine B, 10 μg/ml DOX, or 10 μg/ml blasticidine S. Knockout strains of gβ (DBS0236531), rasC/rasG (DBS0236858), myosin (DBS0236387), and racE (DBS0235413) genes were provided by the Dictyostelium stock center (Fey et al., 2013). Rap1G12V was amplified by PCR, using the previously described pVEII Rap1G12V plasmid as a template (Rebstein et al., 1997). The obtained PCR fragments was digested with BamHI and ligated into the BglII site of the previously characterized Dictyostelium extrachromosomal Tet-ON plasmids pDM310 or pDM334 (N-terminal GFP fusion; Veltman et al., 2009). The 14-3-3, aurora, and α-tubulin genes were amplified from Dictyostelium cDNA by PCR, digested with BamHI, and cloned into the BglII site of a pDM317 (N-terminal GFP fusion), pDM318 (N-terminal red fluorescent protein [RFP]), or pDM323 (C-terminal GFP fusion) (Veltman et al., 2009). A rap1 knockout construct was created by amplifying the genomic sequence of Rap1 and ligating it to pBlue vector. Subsequently a blasticidin resistance cassette (bsr) was inserted within the rap1 gene, and the final construct was amplified by means of PCR. Dictyostelium cells were transformed by means of electroporation.
Immunochemistry and DAPI staining
Cells were allowed to grow on glass-bottom dishes (MatTek) and were washed with PB buffer (10 mM KH2PO4/Na2HPO4, pH 6.5) before fixation. Cells were fixated by incubation for 15 min with PB containing 4% paraformaldehyde, washed with PB, and permeabilized by two 10-min incubations in PB containing 0.1% Triton X-100. Subsequently cells were blocked for 30 min in PB containing 1% wt/vol bovine serum albumin (BSA) and 0.1% Tween, incubated with 14-3-3 antibody (ab77187 [Abcam, Cambridge, UK], 1:200 diluted in 1% BSA, 0.1% Tween PB) or α-tubulin antibody (sc53029 [Santa Cruz, Heidelberg, Germany], 1:50 diluted in 1%BSA, 0.1% Tween PB) for 1 h, washed three times with PB buffer, and finally incubated for 1 h in the dark with a secondary antibody (ab150130 [Abcam] or sc2782 [SantaCruz], 1:200 dilution with 1% BSA, 0.1% Tween, PB). Staining of DNA was performed with 20 μg/ml DAPI solution in PB for 10 min.
Microscopy
A Motic AE31 (Motic, Wetzlar, Germany) bright-field inverted microscope with an LWD PH20×/0.4 NA objective was used to monitor growth of Dictyostelium on Nunclon-coated plates. The images were recorded every 8 s with a digital camera (ScopeTek DCM130) using VirtualDub software (www.virtualdub.org) and Indeo video 5.10 compression (Ligos). The field of observation was 550 × 440 μm.
Fluorescence in fixed or living Dictyostelium cells was monitored using a confocal laser-scanning microscope (LSM 510 META-NLO; Carl Zeiss Microimaging) equipped with a 63×/NA 1.4 objective (Plan-Apochromatic; Carl Zeiss Microimaging). For excitation of the fluorochromes GFP (S65T variant) and Alexa Fluor 555, a 488-nm argon/krypton laser and a 543-nm helium laser were used, respectively. The fluorescence was filtered through a BP500-530 IR and a LP560 filter and was detected by a photomultiplier tube. The field of observation was 202 × 202 μm, and for in vivo observation of dividing Dictyostelium cells, the images were recorded with a scan time of 15 s.
DAPI-stained cells were imaged with a wide-field fluorescence Zeiss Axio Observer Z1 microscope (Zeiss, Jena, Germany) equipped with an EC-Plan-Neofluar 100×/1.3 NA objective and CoolSNAP HQ2 Camera (Roper Scientific, Martinsried, Germany). A 365- to 395-nm band-pass excitation filter, a 420-nm dichroic mirror, and a 435- to 485-nm band-pass emission filter were used. The field of observation was 271 × 202 μm. All images were analyzed using ImageJ software (Abramoff et al., 2004).
Supplementary Material
Acknowledgments
A.K. is supported by a Vidi grant from the Netherlands Organisation for Scientific Research (NWO).
Abbreviations used:
- BSA
bovine serum albumin
- DAPI
4′,6-diamidino-2-phenylindole
- DOX
doxycycline
- GFP
green fluorescent protein
- MTOCs
microtubule-organizing centers
- RFP
red fluorescent protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-08-1285) on October 8, 2014.
REFERENCES
- Abramoff MD, Magalhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Bastos RN, Penate X, Bates M, Hammond D, Barr FA. CYK4 inhibits Rac1-dependent PAK1 and ARHGEF7 effector pathways during cytokinesis. J Cell Biol. 2012;198:865–880. doi: 10.1083/jcb.201204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolourani P, Spiegelman GB, Weeks G. Rap1 activation in response to cAMP occurs downstream of ras activation during Dictyostelium aggregation. J Biol Chem. 2008;283:10232–10240. doi: 10.1074/jbc.M707459200. [DOI] [PubMed] [Google Scholar]
- Bosgraaf L, Waijer A, Engel R, Visser AJ, Wessels D, Soll D, van Haastert PJ. RasGEF-containing proteins GbpC and GbpD have differential effects on cell polarity and chemotaxis in Dictyostelium. J Cell Sci. 2005;118:1899–1910. doi: 10.1242/jcs.02317. [DOI] [PubMed] [Google Scholar]
- Carmena A, Makarova A, Speicher S. The Rap1-Rgl-Ral signaling network regulates neuroblast cortical polarity and spindle orientation. J Cell Biol. 2011;195:553–562. doi: 10.1083/jcb.201108112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao VT, Dupuy AG, Gavet O, Caron E, de Gunzburg J. Dynamic changes in Rap1 activity are required for cell retraction and spreading during mitosis. J Cell Sci. 2009;122:2996–3004. doi: 10.1242/jcs.041301. [DOI] [PubMed] [Google Scholar]
- Faix J, Steinmetz M, Boves H, Kammerer RA, Lottspeich F, Mintert U, Murphy J, Stock A, Aebi U, Gerisch G. Cortexillins, major determinants of cell shape and size, are actin-bundling proteins with a parallel coiled-coil tail. Cell. 1996;86:631–642. doi: 10.1016/s0092-8674(00)80136-1. [DOI] [PubMed] [Google Scholar]
- Fey P, Dodson RJ, Basu S, Chisholm RL. One stop shop for everything Dictyostelium: dictyBase and the Dicty Stock Center in 2012. Methods Mol Biol. 2013;983:59–92. doi: 10.1007/978-1-62703-302-2_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y, De Lozanne A, Spudich JA. Structure and function of the cytoskeleton of a Dictyostelium myosin-defective mutant. J Cell Biol. 1990;110:367–378. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch G, Faix J, Kohler J, Muller-Taubenberger A. Actin-binding proteins required for reliable chromosome segregation in mitosis. Cell Motil Cytoskeleton. 2004;57:18–25. doi: 10.1002/cm.10150. [DOI] [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Graf R, Daunderer C, Schliwa M. Dictyostelium DdCP224 is a microtubule-associated protein and a permanent centrosomal resident involved in centrosome duplication. J Cell Sci. 2000;113:1747–1758. doi: 10.1242/jcs.113.10.1747. [DOI] [PubMed] [Google Scholar]
- Janetopoulos C, Borleis J, Vazquez F, Iijima M, Devreotes P. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev Cell. 2005;8:467–477. doi: 10.1016/j.devcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Jeon TJ, Lee DJ, Lee S, Weeks G, Firtel RA. Regulation of Rap1 activity by RapGAP1 controls cell adhesion at the front of chemotactic cells. J Cell Biol. 2007a;179:833–843. doi: 10.1083/jcb.200705068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon TJ, Lee DJ, Merlot S, Weeks G, Firtel RA. Rap1 controls cell adhesion and cell motility through the regulation of myosin II. J Cell Biol. 2007b;176:1021–1033. doi: 10.1083/jcb.200607072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanada M, Nagasaki A, Uyeda TQ. Adhesion-dependent and contractile ring-independent equatorial furrowing during cytokinesis in mammalian cells. Mol Biol Cell. 2005;16:3865–3872. doi: 10.1091/mbc.E05-03-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Kae H, Ip H, Spiegelman GB, Weeks G. Evidence for a role for the Dictyostelium Rap1 in cell viability and the response to osmotic stress. J Cell Sci. 2002;115:3675–3682. doi: 10.1242/jcs.00039. [DOI] [PubMed] [Google Scholar]
- King JS, Veltman DM, Georgiou M, Baum B, Insall RH. SCAR/WAVE is activated at mitosis and drives myosin-independent cytokinesis. J Cell Sci. 2010;123:2246–2255. doi: 10.1242/jcs.063735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortholt A, Rehmann H, Kae H, Bosgraaf L, Keizer-Gunnink I, Weeks G, Wittinghofer A, van Haastert PJ. Characterization of the GbpD-activated Rap1 pathway regulating adhesion and cell polarity in Dictyostelium discoideum. J Biol Chem. 2006;281:23367–23376. doi: 10.1074/jbc.M600804200. [DOI] [PubMed] [Google Scholar]
- Kortholt A, van Haastert PJ. Highlighting the role of Ras and Rap during Dictyostelium chemotaxis. Cell Signal. 2008;20:1415–1422. doi: 10.1016/j.cellsig.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Lancaster OM, Baum B. Shaping up to divide: coordinating actin and microtubule cytoskeletal remodelling during mitosis. Semin Cell Dev Biol. 2014 doi: 10.1016/j.semcdb.2014.02.015. [DOI] [PubMed] [Google Scholar]
- Lancaster OM, Le Berre M, Dimitracopoulos A, Bonazzi D, Zlotek-Zlotkiewicz E, Picone R, Duke T, Piel M, Baum B. Mitotic rounding alters cell geometry to ensure efficient bipolar spindle formation. Dev Cell. 2013;25:270–283. doi: 10.1016/j.devcel.2013.03.014. [DOI] [PubMed] [Google Scholar]
- Lee MR, Jeon TJ. Cell migration: regulation of cytoskeleton by Rap1 in Dictyostelium discoideum. J Microbiol. 2012;50:555–561. doi: 10.1007/s12275-012-2246-7. [DOI] [PubMed] [Google Scholar]
- Mondal S, Bakthavatsalam D, Steimle P, Gassen B, Rivero F, Noegel AA. Linking Ras to myosin function: RasGEF Q, a Dictyostelium exchange factor for RasB, affects myosin II functions. J Cell Biol. 2008;181:747–760. doi: 10.1083/jcb.200710111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores SL, Sabry JH, Spudich JA. Myosin dynamics in live Dictyostelium cells. Proc Natl Acad Sci USA. 1996;93:443–446. doi: 10.1073/pnas.93.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun H, Jeon TJ. Regulation of actin cytoskeleton by Rap1 binding to RacGEF1. Mol Cells. 2012;34:71–76. doi: 10.1007/s10059-012-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun H, Lee MR, Jeon T. RapGAP9 regulation of the morphogenesis and development in Dictyostelium. Biochem Biophys Res Commun. 2014;446:428–433. doi: 10.1016/j.bbrc.2014.01.196. [DOI] [PubMed] [Google Scholar]
- Nagasaki A, de Hostos EL, Uyeda TQ. Genetic and morphological evidence for two parallel pathways of cell-cycle-coupled cytokinesis in Dictyostelium. J Cell Sci. 2002;115:2241–2251. doi: 10.1242/jcs.115.10.2241. [DOI] [PubMed] [Google Scholar]
- Nagasaki A, Kanada M, Uyeda TQ. Cell adhesion molecules regulate contractile ring-independent cytokinesis in Dictyostelium discoideum. Cell Res. 2009;19:236–246. doi: 10.1038/cr.2008.318. [DOI] [PubMed] [Google Scholar]
- Neujahr R, Heizer C, Gerisch G. Myosin II-independent processes in mitotic cells of Dictyostelium discoideum: redistribution of the nuclei, re-arrangement of the actin system and formation of the cleavage furrow. J Cell Sci. 1997;110:123–137. doi: 10.1242/jcs.110.2.123. [DOI] [PubMed] [Google Scholar]
- O’Connell CB, Warner AK, Wang Y. Distinct roles of the equatorial and polar cortices in the cleavage of adherent cells. Curr Biol. 2001;11:702–707. doi: 10.1016/s0960-9822(01)00181-6. [DOI] [PubMed] [Google Scholar]
- Plak K, Veltman D, Fusetti F, Beeksma J, Rivero F, van Haastert PJ, Kortholt A. GxcC connects Rap and Rac signaling during Dictyostelium development. BMC Cell Biol. 2013;14:6. doi: 10.1186/1471-2121-14-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebstein PJ, Cardelli J, Weeks G, Spiegelman GB. Mutational analysis of the role of Rap1 in regulating cytoskeletal function in Dictyostelium. Exp Cell Res. 1997;231:276–283. doi: 10.1006/excr.1996.3466. [DOI] [PubMed] [Google Scholar]
- Reinders Y, Schulz I, Graf R, Sickmann A. Identification of novel centrosomal proteins in Dictyostelium discoideum by comparative proteomic approaches. J Proteome Res. 2006;5:589–598. doi: 10.1021/pr050350q. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riou P, Kjaer S, Garg R, Purkiss A, George R, Cain RJ, Bineva G, Reymond N, McColl B, Thompson AJ, et al. 14-3-3 proteins interact with a hybrid prenyl-phosphorylation motif to inhibit G proteins. Cell. 2013;153:640–653. doi: 10.1016/j.cell.2013.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki AT, Chun C, Takeda K, Firtel RA. Localized Ras signaling at the leading edge regulates PI3K, cell polarity, and directional cell movement. J Cell Biol. 2004;167:505–518. doi: 10.1083/jcb.200406177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki AT, Janetopoulos C, Lee S, Charest PG, Takeda K, Sundheimer LW, Meili R, Devreotes PN, Firtel RA. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol. 2007;178:185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluchanko NN, Gusev NB. 14-3-3 proteins and regulation of cytoskeleton. Biochemistry (Mosc) 2010;75:1528–1546. doi: 10.1134/s0006297910130031. [DOI] [PubMed] [Google Scholar]
- Surcel A, Kee YS, Luo T, Robinson DN. Cytokinesis through biochemical-mechanical feedback loops. Semin Cell Dev Biol. 2010;21:866–873. doi: 10.1016/j.semcdb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuxworth RI, Cheetham JL, Machesky LM, Spiegelmann GB, Weeks G, Insall RH. Dictyostelium RasG is required for normal motility and cytokinesis, but not growth. J Cell Biol. 1997;138:605–614. doi: 10.1083/jcb.138.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda TQ, Kitayama C, Yumura S. Myosin II-independent cytokinesis in Dictyostelium: its mechanism and implications. Cell Struct Funct. 2000;25:1–10. doi: 10.1247/csf.25.1. [DOI] [PubMed] [Google Scholar]
- Veltman DM, Keizer-Gunnink I, Haastert PJ. An extrachromosomal, inducible expression system for Dictyostelium discoideum. Plasmid. 2009;61:119–125. doi: 10.1016/j.plasmid.2008.11.002. [DOI] [PubMed] [Google Scholar]
- Veltman DM, van Haastert PJ. Extrachromosomal inducible expression. Methods Mol Biol. 2013;983:269–281. doi: 10.1007/978-1-62703-302-2_14. [DOI] [PubMed] [Google Scholar]
- Wang YL. The mechanism of cortical ingression during early cytokinesis: thinking beyond the contractile ring hypothesis. Trends Cell Biol. 2005;15:581–588. doi: 10.1016/j.tcb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Kee YS, Poirier CC, Jelinek C, Osborne J, Divi S, Surcel A, Will ME, Eggert US, Muller-Taubenberger A, et al. 14-3-3 coordinates microtubules, Rac, and myosin II to control cell mechanics and cytokinesis. Curr Biol. 2010;20:1881–1889. doi: 10.1016/j.cub.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.