Summary
Protein folding in the cell relies on the orchestrated action of conserved families of molecular chaperones, the Hsp70 and Hsp90 systems. Hsp70 acts early and Hsp90 late in the folding path, yet the molecular basis of this timing is enigmatic, mainly because the substrate specificity of Hsp90 is poorly understood. Here we obtained a structural model of Hsp90 in complex with its natural disease-associated substrate, the intrinsically disordered Tau protein. Hsp90 binds to a broad region in Tau that includes the aggregation-prone repeats. Complementarily, a 106 Å long substrate-binding interface in Hsp90 enables many low affinity contacts. It allows recognition of scattered hydrophobic residues in late folding intermediates that remain after early burial of the Hsp70 sites. Our model resolves the paradox of how Hsp90 specifically selects for late folding intermediates but also for some intrinsically disordered proteins – through the eyes of Hsp90 they look the same.
Keywords: molecular chaperones, protein folding, NMR spectroscopy, SAXS, protein-protein interactions, Alzheimer disease, intrinsically disordered proteins
INTRODUCTION
Protein folding in the cell is controlled by conserved families of molecular chaperones (Hartl et al., 2011). Crucial for the chaperone network in the cytoplasm are the ATP-driven Hsp70 and Hsp90 families (Bukau et al., 2006; Mayer, 2010). Both are complex systems that are controlled by co-chaperones and their activity is linked by the adaptor protein Hop (Li et al., 2012; Mayer, 2010; Pearl and Prodromou, 2006). One of the central questions in protein folding is how and when each of the chaperones assists the folding of a newly born protein in the cell.
Hsp70 chaperones recognise short hydrophobic sequences particularly enriched in leucine and are thought to interact with substrate proteins in the early stages of the folding path (Bukau et al., 2006; Rüdiger et al., 1997a). In contrast, Hsp90 acts during late folding stages, especially with proteins involved in signal transduction (Jakob et al., 1995; Li et al., 2012; Pearl and Prodromou, 2006). The molecular basis for the task distribution between Hsp90 and Hsp70, however, is unclear. This is predominantly due to the lack of mechanistic understanding of Hsp90 interaction with its client proteins (Mayer, 2010).
The substrate pool of Hsp90 does not share a common sequence or structural motif (Taipale et al., 2010; Taipale et al., 2012). Hsp90 interacts with a specific but structurally diverse subset of client proteins including kinases and steroid receptors (Echeverria and Picard, 2010; Jakob et al., 1995; Pearl and Prodromou, 2006; Street et al., 2011). Interestingly, Hsp90 also interacts with some intrinsically disordered proteins, including the microtubule-associated protein Tau (Dickey et al., 2007; Dou et al., 2003; Miyata et al., 2011; Thompson et al., 2012).
Tau normally stabilises microtubules which serve as tracks for axonal transport in neurons (Mandelkow and Mandelkow, 2012). It also accumulates abnormally in brain tissue of Alzheimer disease patients and is closely linked with neuronal toxicity in this most common neurodegenerative disease (Ittner et al., 2010; Roberson et al., 2007). Hsp90 facilitates microtubule association of Tau, but can also work with other (co-)chaperones, such as the ubiquitin ligase CHIP, to facilitate Tau degradation. In this way, Hsp90 can promote both stabilization and degradation of Tau (Dickey et al., 2007). Mutant Tau is particularly susceptible to inhibition of Hsp90, making it a promising lead for therapeutic strategies in Alzheimer disease and other Tau-related disorders (Dou et al., 2003; Luo et al., 2007). The molecular basis for Hsp90 interaction with Tau, however, is enigmatic. Tau is unfolded in its native conformation, which challenges the paradigm that chaperones support protein folding and maturation.
It is essential to determine Hsp90’s substrate binding site to reveal the molecular principles of its substrate recognition. This is a prerequisite to understand three central questions on chaperone action in the cell (i) How can the Hsp90 system take care of a diverse substrate pool, ranging from late folding states of kinases to proteins that never fold, such as Tau? (ii) How does the Hsp90 system share tasks with other chaperone systems, such as the Hsp70 machine? (iii) What determines the timing of chaperone action in the cell?
By combining a novel NMR approach with small-angle X-ray scattering (SAXS), we obtained a structural model of Hsp90 in complex with the Tau protein. A unique, extended binding interface in Hsp90 binds to Tau. Complementary, Hsp90 recognises a broad region that includes the aggregation-prone repeats in Tau. The long binding site predisposes Hsp90 to provide many low affinity contacts and hydrophobic residues as they scatter over the outer sphere of late folding intermediates. Thereby, Hsp90 can profit from the compaction that takes place in the early folding stages, after which Hsp70 sites are already buried. Our model resolves the paradox on how Hsp90 specifically selects for late folding intermediates but also some intrinsically disordered proteins.
RESULTS
Hsp90 binds to disordered Tau with low micromolar affinity
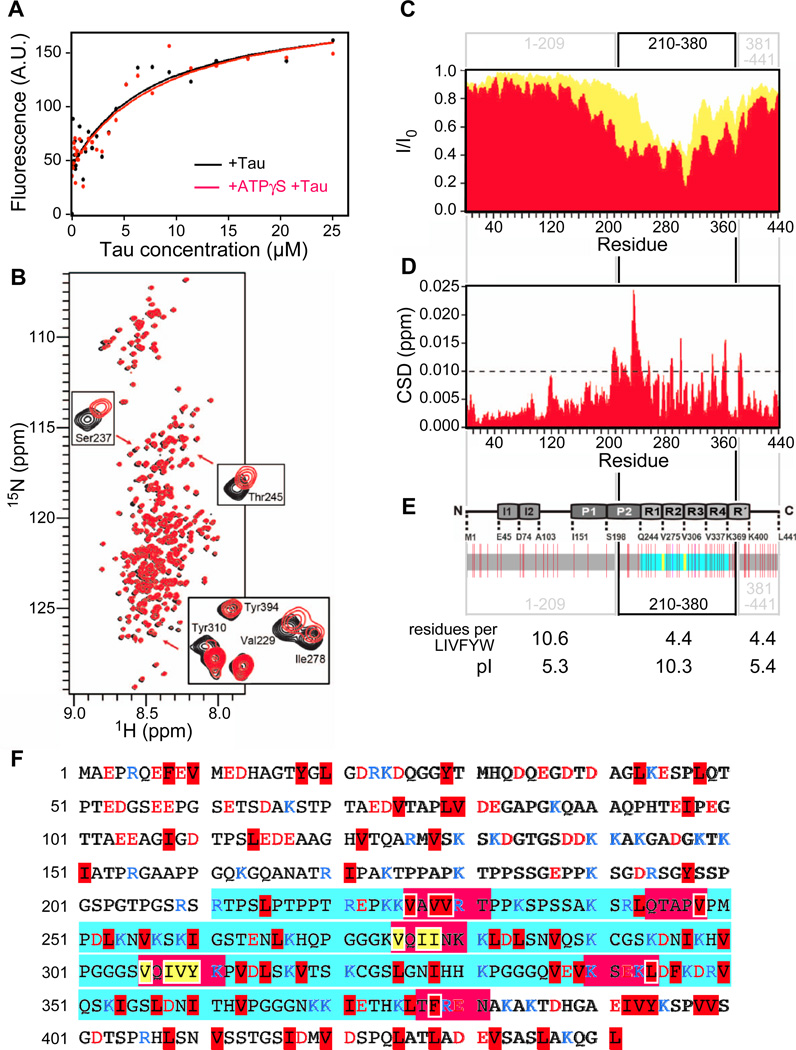
To study the affinity of Tau for Hsp90, we monitored intrinsic protein fluorescence (Fig. 1A). Titration of Hsp90 with tryptophan-free Tau revealed a binding affinity typical for chaperone-substrate interactions. Tau saturates Hsp90 and the affinity is sufficient for interaction of these abundant proteins at physiological levels (KD = 4.8 ± 1.0 µM; Fig. 1A) (Rüdiger et al., 1997a). As Hsp90 is an ATP-regulated machine, we used the ATP analogue ATPγS to measure Hsp90’s affinity for Tau in the ATP state (Karagöz et al., 2011). However, Hsp90’s affinity for Tau in the ATP state did not differ significantly from the affinity of Hsp90 in the nucleotide-free apo state (KD = 5.2 ± 3.0 µM; Fig. 1A). Thus, ATP binding does not trigger release of Tau nor induce extensive conformational changes that would influence affinity.
Fig. 1. Hsp90 binds to Tau’s repeat region.
A, Hsp90 binds to Tau with high affinity. Titration of Tau increases intrinsic fluorescence of Hsp90 or Hsp90+ATPγS (Hsp90, 2 µM, black; Hsp90+ATPγS, red), fluorescence increase is plotted versus [Tau]. B, 2D 1H 15 N HSQC spectra of full-length Tau protein in the absence (black) and presence of Hsp90 (red). The insets highlight the broadenings and changes in the chemical shifts of selected resonances. (C) Intensity ratio broadening of Tau in the presence of Hsp90 (red). Yellow represents the signal intensity of Tau in the presence of Hsp72 (taken from Jinwal et al., 2013). (D) NMR chemical shift deviations of Tau in the presence of Hsp90. E, The domain organisation of Tau is indicated above (I = inserts, P = proline-rich region, R = pseudo-repeat), Hydrophobicity distribution of Tau underneath. Indicated are large hydrophobic and aromatic residues (LIVFYW; red lines), the Hsp70 binding sites (yellow lines) and the microtubule binding repeat region (cyan). For the section 1–209, 210–380 and 381–441 the content of LIVFYW residues and the pI are indicated. The LIVFYW content is given as occurrence per amino acid (aa) in the protein sequence of Tau. The average content of LIVFYW in the human proteome is 1 per 3.6 aa. (F) Color-coded primary sequence of Tau highlighting the regions affected upon interaction with Hsp90 (red boxes). LIVFYW residues are highlighted in red. The two Hsp70 binding stretches are indicated by yellow letters. Charged residues are indicated by blue (Arg and Lys) and red letters (Asp and Glu). The repeat region is indicated in cyan.
Hsp90 recognises Tau’s microtubule-binding repeats domain
As Hsp90 binds with reasonably high affinity to Tau, we set out to map its Hsp90 binding site by NMR spectroscopy. We acquired 2D HSQC spectra of 15N labelled Tau in the presence and absence of unlabelled Hsp90 (Fig. 1B). This method provides a fingerprint of all amide bonds of a protein, which allows mapping interaction sites by recording changes in position and/or intensity of individual peaks. A subset of signals shifted upon Hsp90 binding, including Ser237, Thr245, Ile278 and Tyr310 (Fig. 1B). The spectra also revealed a strong drop in intensity in a broad region between residues 210 and 380 upon Hsp90 binding (Fig. 1C). The reduction in signal intensity is a commonly observed phenomenon in protein NMR, which is typically caused by the combined effects of (i) the increase in molecular weight upon complex formation, which increases transverse relaxation rates and thereby line width, and (ii) the chemical exchange at the contact surface (Dekker et al., 1993; Sette et al., 1997; Wüthrich, 2003; Zuiderweg, 2002). The reduction in intensity is consistent with Hsp90-Tau binding on the intermediate exchange time scale, and also consistent with the affinity determined by fluorescence (Fig. 1A). Chemical shift changes in the boundaries of this region confirm that the loss of intensity is caused by direct interaction with Hsp90 (Fig. 1D).
The Hsp90 binding region contains the entire microtubule-binding repeat domain, including the 275VQIINK280 and 306VQIVYK311 hexapeptide motifs, which are crucial for pathological Tau aggregation in Alzheimer disease (Fig. 1EF) (Bergen et al., 2000). Within the broad Hsp90 binding region, some segments show dips in intensity, indicating particular importance for the interaction (226VAVVRT231, 244QTAPV248, 275VQIINK280, 306VQIVYK311, 340KSEKL344 and 377TFREN381; Fig. 1CF). These segments representing minima in intensity are characterised by large hydrophobic and aromatic side chains (LIVFYW residues; leucine, isoleucine, valine, phenylalanine, tyrosine and and tryptophane; tryptophane is not present in Tau; Fig. 1EF). They also have a positive net charge, although negatively charged residues are not excluded.
The Hsp90 binding region 210–380 has considerably higher content of LIVFYW residues compared to the N-terminal part 1–209 (1 per 4.4. vs 1 per 10.6 residues). Interestingly, the C-terminal segment 381–441 contains a similar fraction of LIVFYW (1 in 4.4), but it did not bind to Hsp90. The binding segment 210–380, however, is the only one that is characterised by a strong positive net charge (pI = 10.3; Fig. 1E). Thus, both LIVFYW residues and positive net charge characterise the Hsp90 binding regions in Tau. We conclude that it is not its disordered nature in itself that causes Tau binding to chaperones, but specific sequence properties.
We next compared the Hsp90 binding site in Tau to the known Hsp70 sites. Strikingly, the Hsp90 binding region is significantly broader compared to that of Hsp70 chaperones, which specifically bind to two short segments within this region (275VQIIN279 and 306VQIVY310; Fig. 1CF) (Jinwal et al., 2012; Sarkar et al., 2008). The binding regions of both chaperones are partially overlapping, despite dramatic differences in their width.
Tau binds to the N-terminal and middle domains of Hsp90
We then used an advanced NMR method to understand the chemical nature of Hsp90’s Tau binding site. Hsp90 is not accessible to standard NMR experiments due to the high molecular weight of the Hsp90 dimer (170 kDa). We recently overcame the typical NMR size limit for the Hsp90 system by employing specific isotope labelling of methyl groups in isoleucine side chains and methyl transverse relaxation optimised spectroscopy (methyl-TROSY) (Karagöz et al., 2011). Using this novel method, we were then able to monitor the interaction of isoleucine-labelled and NMR visible Hsp90 with unlabelled and, therefore, NMR-invisible Tau (Fig. 2A).
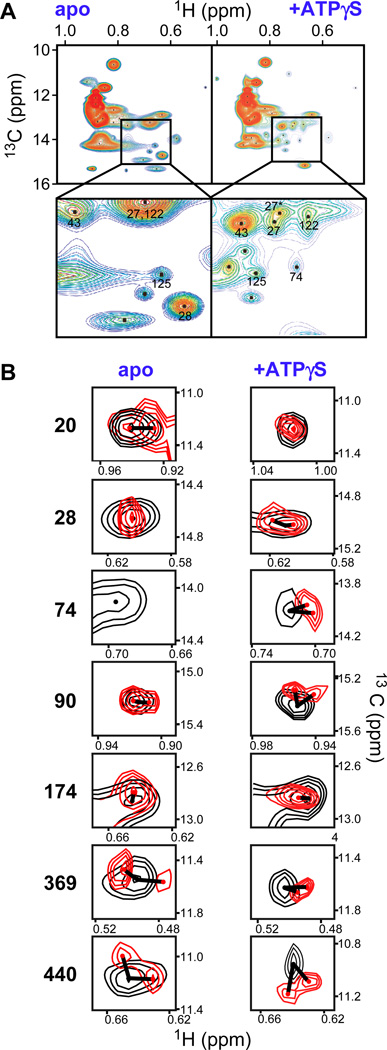
Fig. 2. Tau causes specific shift in Hsp90 NMR spectra.
A, ATP binding modulates the dynamics of the Hsp90-Tau complex. Rainbow representation of 1H-13C-Ile-methyl-TROSY cross-peaks of full-length Hsp90 in complex with Tau (left) and Tau+ATPγS (right). The inset magnifies peaks that sharpen upon ATPγS binding, numbers indicate assigned isoleucines (numbering without N-terminal methionine). B, Tau binding causes shifts and peak doubling in Hsp90 spectra (Hsp90, 125 µM; Tau, 287 µM; Hsp90, black; Hsp90+Tau, red), in the absence and presence of ATPγS (2 mM). The extent of each shift is indicated by a line. See also Fig. S1 and Table S1.
Compared to spectra of Hsp90 alone, the signal intensity of isoleucines in Hsp90-Tau complex is reduced, consistent with what would be expected from binding of a large, disordered protein in the fast to intermediate exchange NMR time scale (Fig. 2A, Fig. S1B) (Karagöz et al., 2011). A subset of Hsp90’s isoleucine signals shifted upon addition of Tau, including Ile20, Ile74, Ile174, Ile369 and Ile440 (Fig. 2B, Table S1). Some of the isoleucine signals split, including Ile90, Ile369 and Ile440 (Fig. 2B). NMR data of isoleucine side chains are particularly useful to map binding sites as they typically indicate local changes, as opposed to standard spectra monitoring the backbone. Isoleucines are not involved in hydrogen bonds, and therefore, less sensitive to long-range structural changes when compared to backbone amide signals, as observed in previous isoleucine-based NMR analyses of Hsp90 complexes (Didenko et al., 2011; Didenko et al., 2012; Karagöz et al., 2011). The locations of the shifting and splitting residues reveal Tau binding to both the N-terminal and the middle domains of Hsp90 (Fig. S1C).
ATP binding modulates dynamics of Hsp90-Tau complex
We then investigated the effect of ATP on the structure of the Hsp90-Tau complex, using ATPγS. The pattern of the NMR signals in ATP state differed significantly from its nucleotide free state. The ATP state induced additional shifts for some residues in Hsp90-Tau (e.g. Ile28 shifts, yet the nearby Ile20 does not), but also dramatically sharpened signals (region around 14 ppm (13C) and 0.7 ppm (1H); Fig. 2A, zoom). Sharpening of NMR signals typically indicates either changes in conformational flexibility or the chemical exchange rate. Given that the binding affinity of the ATP state does not change, we conclude that binding of the client and ATP jointly modulate the dynamics of Hsp90.
We anticipate that nucleotide-induced changes in Hsp90 dynamics in the presence of substrate is crucial for Hsp90’s chaperone activity. Notably, we observed splitting of NMR signals upon Tau binding both in the nucleotide-free and the ATP-bound states of Hsp90, indicating that Tau binding breaks the symmetry of the Hsp90 dimer (Fig. 2B). This may imply either that Tau bound Hsp90 adopts two specific conformations or that Tau breaks the symmetry of the Hsp90 dimer. While there is no evidence to support the first possibility, conformational asymmetry is characteristic for several previously described Hsp90 complexes (i.e. Hsp90-p23, Hsp90-Cdk4-Cdc37 and Hsp90-Aha1) (Karagöz et al., 2011; Retzlaff et al., 2010; Vaughan et al., 2006). Our NMR data are in line with a break of symmetry in the substrate cycle of Hsp90.
The Hsp90-Tau complex is extended
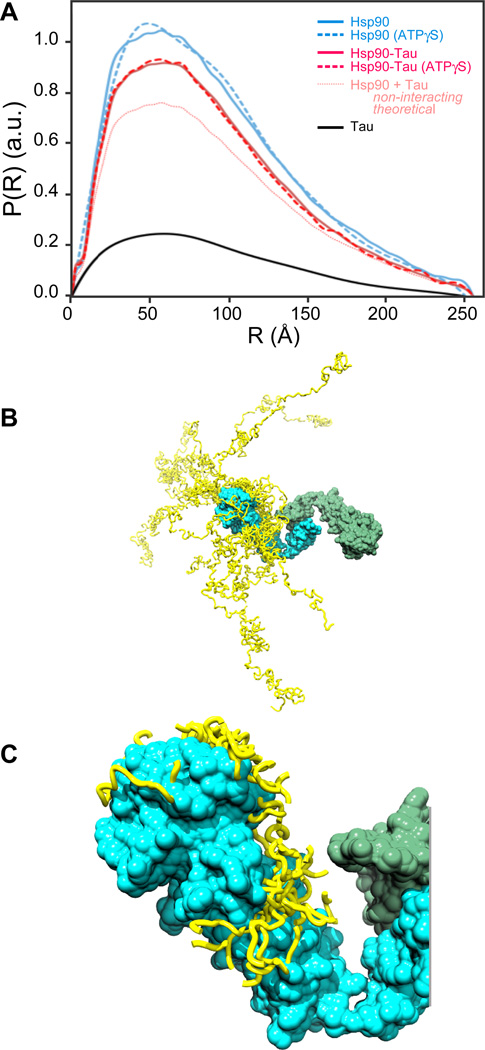
Next, we characterised the overall shape in solution of Hsp90, Tau and the Hsp90-Tau complex by small-angle X-ray scattering (SAXS). SAXS is a solution scattering method that had been successfully applied to several Hsp90 homologues to obtain structural models (Krukenberg et al., 2008; Krukenberg et al., 2009). In our SAXS experiments, Human Hsp90 appeared as an extended molecule, with a maximal extension (Dmax) of 25.5 nm and a radius of gyration (Rg) of 7.8 ± 0.5 nm (Fig. 3A, Fig. S2A, Table 1). This is consistent with previous SAXS studies on several Hsp90 homologues by the Agard group (Krukenberg et al., 2008; Krukenberg et al., 2009). Notably, we did not observe significant changes of Dmax and Rg values upon addition of ATP, indicating that ATP does not significantly affect the global shape of the conformational ensemble of human Hsp90 (Fig. S2B, Table 1). SAXS experiments of Tau revealed a significantly higher radius of gyration and a larger maximal extension than expected for globular proteins of the same size, consistent with its disordered nature and with previous studies (Rg = 6.8 ± 1.8 nm, Dmax = 24.9 nm; Fig. 3BC; Table 1) (Mylonas et al., 2008; Shkumatov et al., 2011).
Fig. 3. Convergence of Tau models on Hsp90 in NMR/SAXS models.
A, Hsp90 binds to Tau in an open conformation. SAXS curves of Tau (black), Hsp90 in apo (light blue, solid line) and ATP states (light blue, dashed line) and Hsp90+Tau in apo (red, solid line) and ATP states (red, dashed line) (red; dashed curve in the presence of ATPγS), showing P(R), the relative probability of interatomic scattering distances, normalised on the total intensity I0 and concentration of each sample. The theoretical P(R) assuming non-interacting proteins is shown as well. B, The ten best Tau models bound to Hsp90 (Tau models, yellow bands; Tau-binding Hsp90 protomer, cyan; other Hsp90 protomer, green). C, The ten best Tau models within 10 Å of Hsp90 indicate convergence on Hsp90 (colours as in B). See also Fig. S2 and S5.
Table 1.
Characteristics of Hsp90 and Tau in SAXS experiments
| Protein | Rg [nm] | Dmax [nm] |
|---|---|---|
| Hsp90 | 7.8 ± 0.5 | 25.5 |
| Hsp90 (AMP-PNP) | 7.6 ± 0.7 | 25.0 |
| Tau | 6.8 ± 1.8 | 24.9 |
| Hsp90-Tau | 7.5 ± 0.7 | 25.5 |
| Hsp90-Tau (ATPγS) | 7.2 ± 1.5 | 25.5 |
Tau binding to Hsp90 lead to a significant change in the P(R) values. The binding of Tau, however, did not modulate the shape of Hsp90. We found dimensions similar to Hsp90 alone in apo and ATP states (Rg = 7.5 ± 0.7 nm, Dmax = 25.5 nm in the absence of ATP; Rg = 7.2 ± 1.5 nm, Dmax = 25.5 nm in the presence of ATP; Fig. 3A, Table 1). This indicates that neither Tau, nor nucleotide-binding nor synergistic binding of both ligands caused an obligatory conformational rearrangement of Hsp90.
The Tau binding site maps to the N-terminal and middle domains of Hsp90
We next set out to obtain a structural model of Hsp90 in the Tau-bound state. We combined the structural information that could be extracted from our NMR restraints and scattering curves with previously published homology models based on published crystal structures of Hsp90 (Ali et al., 2006; Karagöz et al., 2011). The seven residues that shifted and/or split provided restraints that allowed us to precisely position the Tau binding site in structural models fitted to the SAXS curves (Fig. 2B).
Both NMR and SAXS are methods that result in an ensemble of structural models, which together provide a comprehensive description of the protein in solution. Tau-bound Hsp90 appeared as an open, dynamic ensemble, in which the N-terminal domains are the most distant points of the dimeric structure (Fig. 3B). In contrast to the converging Hsp90 ensemble, the part of the disordered Tau that is not bound to Hsp90 is highly heterogeneous (Fig. 3B). However, the part of Tau close to Hsp90 is strongly converging on the binding site (Fig. 3C). In this interacting region, the variation of the Hsp90-bound Tau ensemble has a minimal width of 19 Å and 17 Å at its most narrow side in Hsp90-N and Hsp90-M, respectively.
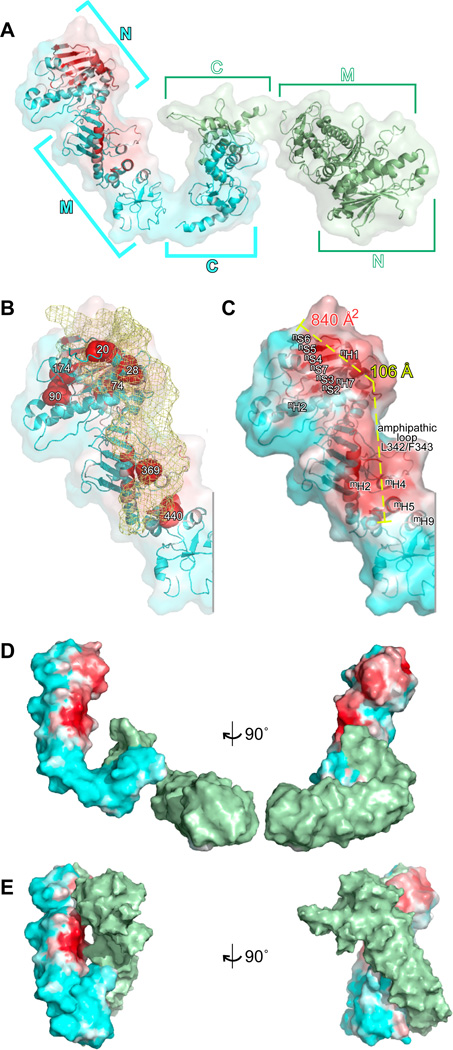
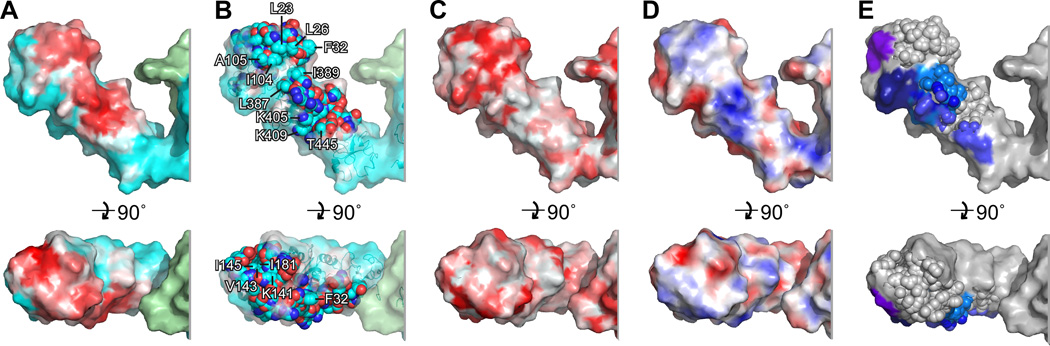
On Hsp90’s surface, the Tau binding site covers a remarkably extended surface running along Hsp90-N and Hsp90-M (Fig. 4AB). The binding site is formed by the Hsp90-N’s (3-sheet nS2-nS7 and alpha helix nH1 and the succeeding loop T30-K35 at the top, running down via the Hsp90-N’s helices nH7 and nH8 the N-terminal segment G102-K106 of the lid of the nucleotide binding pocket (Fig. 4C). In Hsp90-M it passes the helices mH1, mH5, mH4 and mH9. The border on one side is the long helix mH2 and on the other side it is the amphipathic loop around Leu342/Phe343, which has been earlier implicated in client interaction (Prodromou et al., 1997). The shifting isoleucine side chains are mostly buried in the structural model, which confirms that the Tau-dependent shifts are caused by significant interactions that can be sensed inside the molecule (Fig. S3AB).
Fig. 4. Structural model of the Hsp90-Tau complex.
A, NMR/SAXS models of Hsp90 bound to Tau as ribbon representation, the surface (80% transparent) of the molecule is overlaid (Tau-binding Hsp90 protomer, cyan; other Hsp90 protomer, green). The Tau binding site is indicated as white-red gradient. B, Localisation of Tau on its binding site in Hsp90-N and Hsp90-M. The approximate shape of the Hsp90-binding stretch of Tau is visualised by a yellow mesh (averaged from 5 best NMR/SAXS models, disordered parts outside Hsp90 binding site are, therefore, invisible; Hsp90 coloured as in A; isoleucines indicated in Fig. 2B are shown as red space fillings). C, The Tau binding site is extended. Dimensions of the binding site are indicated in Hsp90-N (white letters) and Hsp90-M (black letters; colours of Hsp90 as in A, surface is shown with 20% transparency). D, E, Structural models indicating the Tau binding site in different conformations, which represent the dynamic Hsp90 ensemble (D, NMR/SAXS model as in A; E, mapping the binding site as identified in D on a homology model of the closed conformation of Hsp90, as induced by ATP and p23; surface representations, colour code as in C; the models are vertically and horizontally aligned on Hsp90-M). See also Fig. S3.
The binding interface lacks a lid that would allow Hsp90 to temporarily bury crucial contact residues. The interface, however, is 106 Å long and 840 Å2 large (Fig. 4C). It is significantly more extended than that of the protomers of other chaperones that are known to be controlled by ATP (i.e. Hsp70, Hsp60 and Hsp100 classes). The large binding region allows for a high number of low affinity contacts to build up high-affinity binding energy despite the absence of a binding pocket or cleft.
We analysed how Hsp90 may be able to modulate the architecture of its binding site. Hsp90 functions as a dynamic dimer that is characterised by large inter-domain movements, which is also reflected by the crystal structures of Hsp90 homologues captured in different conformations (Ali et al., 2006; Dollins et al., 2007; Shiau et al., 2006). These states may reflect possible conformations in the active cycle of Hsp90 that could be crucial for regulation of Hsp90’s interaction with client proteins. To map the Tau binding site in those conformations, we compared structural models in which the interface between Hsp90-N and Hsp90-M is fixed to what was observed for ATP-state complexes of both yeast Hsp90 bound to p23 and Grp94 (Fig. 4DE; Fig. S3C). We noted that for the Grp94-based model (Fig. S3C) the binding site twists at the interface between Hsp90-N and Hsp90-M. Such rotations at the interface between both domains may allow Hsp90 to adapt to different substrates and to modulate affinity.
A key step in the active cycle of Hsp90 is closure of the Hsp90 dimer due to dimerisation of the N-terminal domains. Therefore, we mapped the Tau binding site onto the closed structure, as it would be induced by the pro-folding Hsp90 exclusive co-chaperone p23 (Fig. 4E). Remarkably, most of the substrate binding hotspots in Hsp90-N and Hsp90-M remained accessible, although they were not longer continuously connected. The extended nature of the binding site, however, may offer a regulated substrate release mechanism by inter-domain movement at the interface between Hsp90-N and Hsp90-M following ATP hydrolysis.
The Tau binding site in Hsp90 is hydrophobic and negatively charged
We next dissected the chemical nature of the interaction surface between Tau and Hsp90. Since Tau’s binding to Hsp90 is mediated by both basic and hydrophobic residues, we predicted that Hsp90’s surface would provide corresponding hydrophobic and acidic components. The most intense binding regions were two areas in Hsp90-N and Hsp90-M (Figs. 3, 4 and 5A). Although not particularly hydrophobic, the binding area contained some scattered hydrophobic patches including the residues Val143, Ile181 and Pro173 in the β-sheet S2-S7 in Hsp90-N (Fig. 5BC). Notably, the surface hydrophobicity was enhanced by combining large aliphatic side chains with other aliphatic groups, such as the methyl groups of e.g. alanine or threonine (Leu23, Leu26, Ile104 and Ala105 in Hsp90-N and, Leu387, Ile389, Lys409, Thr445 and Lys405 in Hsp90-M) as well as several surface exposed aromatic side chains (Phe31, Phe343) (Fig. 5B). The residues Phe31, Tyr32, Leu387 and Ile389 map to the Hsp90-N/Hsp90-M interface and domain movements might bring those in close proximity to provide a larger area with exposed hydrophobic regions. The scattered nature of Hsp90s surface hydrophobicity ensures that it has the ability to make a large number of low affinity contacts. This dictates that only larger proteins can make effective use of the interaction surface. The scattering also subverts aggregation of Hsp90 as it reduces the need to conceal its binding site.
Fig. 5. The Tau binding site has complex sequence properties.
Mapping of surface properties on Hsp90, close up highlighting Hsp90-N and Hsp90-M. A, Hsp90’s Tau binding site (colour code as in Fig. 3). B, Natural colour representation, indicating residues that expose hydrophobic groups (cyan, carbon; blue, nitrogen; red, oxygen). C, Surface hydrophobicity (gradient non-hydrophobic (white) to hydrophobic (red). D, Surface potential (gradient negative (red) to positive potential (blue). E, Interaction interfaces of the cochaperones Sgt-1, CHORD, p23, Cdc37 and Aha1, based co-crystal structures. Only the Cdc37 binding site overlaps partially with the Tau binding site (cochaperone binding sites are indicated in blue and purple shades p23 (blue), Aha1 (TV blue), Cdc37 (marine), CHORD (dark blue) and Sgt1 (deep blue; the Tau binding site is indicated by space fillings).
The substrate-binding site of Hsp90 provides both positively and negatively charged patches. Negatively charged patches include surface areas around Glu80, Glu392, Asp366 and Asp517, which complement the positively charged segment of Tau involved in Hsp90 binding. In particular Hsp90-M’s long helix mH2 is positively charged (Fig. 5D). The 3D-isopotential surface analysis revealed that Hsp90-N and Hsp90-M exhibit a predominantly negative potential (Fig. S3D). The binding site itself, however, has a mixed potential, which may reduce unspecific charge-dominated interactions in the binding site itself.
In addition to substrates, Hsp90’s ATPase activity is regulated by a plethora of co-chaperones. Therefore, we mapped the binding sites of several key co-chaperones, including Aha1, p23, Cdc37, Sgt-1 and CHORD, which have previously been found in co-crystals with Hsp90 (Fig. 5E) (Li et al., 2012; Pearl et al., 2008). Strikingly, this revealed an elongated surface stretch that is almost free of co-chaperone interaction. Aha1, p23, CHORD and Sgt-1 bind on a surface stretch running parallel to the Tau binding site, which is enriched in positively charged residues (Fig. 5DE). Many of the positive charges are provided by the long helix mH2 that runs in N-C direction through the upper middle domain and separates the Aha1/p23 interaction area from the Tau binding region. The Cdc37 binding site overlaps partially with the Tau binding site, in between the main binding areas in Hsp90-N and Hsp90-M (Fig. 5E). As (i) the main substrate binding sites are still available for the client and (ii) Cdc37 is likely to bind to the other protomer (Vaughan et al., 2006), thus we do not expect significant infringements for Hsp90 substrate interaction by Cdc37. We conclude that the Tau binding site allows substrate interaction while simultaneously ensuring co-factor regulation.
Together, the ATP-dependent changes, its affinity and specific interaction site as determined from both SAXS and NMR experiments show that intrinsically disordered Tau behaves as a bona fide Hsp90 client.
Tau’s Hsp90 binding site resembles folding intermediates
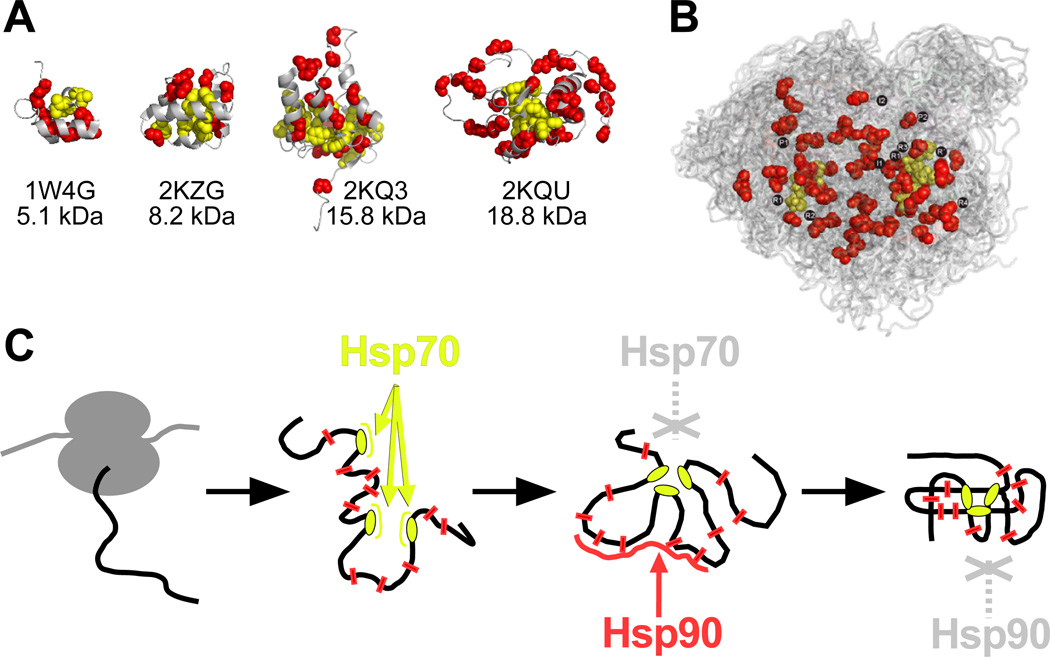
We then thought to determine whether Tau mimics the molecular features of folding intermediates. We analysed, therefore, structures of folding intermediates for hydrophobicity and potential Hsp70 binding sites. We predicted these sites using an algorithm developed for the homologue DnaK (Figs. 6A, S4) (Rüdiger et al., 1997a; Rüdiger et al., 1997b). We made two observations: (i) for all folding intermediates, the Hsp70-binding sites mapped to the protein nucleus, around which the protein folds, and outside the nucleus Hsp70 sites were absent; (ii) the structures showed hydrophobic residues outside the nucleus, but more scattered compared to their concentrated appearance in Hsp70 sites.
Fig. 6. Partitioning of chaperone action.
A, Predicted Hsp70 binding sites (side chains of leucine, isoleucine, valine, phenylalanine and tyrosine shown as yellow spheres) form the nucleus of folding intermediates (four examples, Dihydrolipoyllysine-residue acetyltransferase (1W4G), Pre-mRNA-processing factor 40 homolog A (2KZG), Thermonuclease (2KQ3) and Flavodoxin (2KQU)), leaving scattered hydrophobic residues in the periphery (LIVFYW residues; red spheres). (B) The Tau ensemble, color coded as in (A). The repeat regions are indicated as in Figure 1E. C, Model for partitioning of chaperone action between the Hsp70 and Hsp90 systems. Hsp70 binds early when its binding sites are accessible. During folding, the Hsp70 binding sites disappear, leaving scattered hydrophobics that are recognised by Hsp90. The folded protein will bury also those residues. See also Fig. S4.
At present, structures are only available for a few small, mono-domain intermediates, which are typically not Hsp90 clients. Nevertheless, even those small intermediates contain some exposed stretches with scattered hydrophobics that are buried in the folded protein. Interestingly, the largest folding intermediate exposed three large loop regions that have a similar degree of moderate, scattered hydrophobicity as the Tau repeat region (Fig. 6A, Fig. 2KQU). The distribution of hydrophobic residues of Tau’s repeat region in fact resembles that of the exposed regions of a folding intermediate (Fig. 6B).
DISCUSSION
We mapped the binding site of the natural Hsp90 substrate protein Tau, a protein that plays an important role in normal neuronal function as well as in neurodegenerative disease progression (Fig. 4). The complex we studied consisted of two full-length proteins. This is beneficial because (i) only full length Hsp90 could represent the complex architecture of the dimeric chaperone, and (ii) only a large protein substrate such as the 441 residue Tau protein can make full use of the extended Hsp90 binding site that runs over two domains. The bipartite nature of the binding site allows adaptation to the structural properties of the substrate. It is tempting to speculate that this bipartite nature may allow a potential substrate release mechanism due to domain movement at the Hsp90-N/Hsp90-M interface. Client binding may support transient conformational rearrangements of the Hsp90 dimer, which may affect the ATP hydrolysis rate via modulating the energy barrier for Hsp90 closure (Fig. 4E).
Overlap of Hsp90 binding sites for Tau and kinase
It is remarkable that another full-length protein, the late intermediate of kinase Cdk4 in complex with the co-chaperone Cdc37 partially overlaps with the Tau binding site (Vaughan et al., 2006). Hsp90 associates selectively with intrinsically unstable kinases independent of the sequence motif (Taipale et al., 2012). Cdk4 contacts the β-sheet in Hsp90-N and the amphipathic loop around Leu342 and Phe343 in Hsp90, both Tau contact sites (Fig. 4C). Interestingly, the Tau binding site also covers the proposed binding site of a small charged model substrate in Hsp90-M that does not overlap with the Cdk4 site and residues in Hsp90-M that had been implied in substrate interaction of E. coli Hsp90 (Fig. 4)(Genest et al., 2013; Street et al., 2011). This overlap suggests that the location of Hsp90’s substrate-binding site itself is not dependent on the type of substrate and co-chaperone repertoire.
Hsp90’s activity on chaperoning this diverse range of clients depends on ATP binding and hydrolysis. We did not find a significant difference in Tau in the presence and absence of ATP analogues (Fig. 1A). We relate this to the fact that ATP binding alone does not induce stable conformational changes in human Hsp90, in contrast to the yeast and E. coli homologues (Fig. S2) (Krukenberg et al., 2008; Krukenberg et al., 2009). As the lack of ATP-dependent closure in the absence of co-chaperones is an inherent property of human Hsp90, it is most likely independent from the nature of the bound substrate.
Studies that used isolated domains and/or worked at subphysiological salt conditions also implied other surface elements involved in substrate binding (Didenko et al., 2012; Hagn et al., 2011; Park et al., 2011). It remains to be seen whether the relevance of those sites can be confirmed in the full-length protein. We noted, however, that the Tau binding sites in both Hsp90-N and Hsp90-M have been implicated in early crystal structures of both domains as potential substrate binding sites (Fig. 4)(Meyer et al., 2003; Prodromou et al., 1997). For the E. coli Hsp90 some mutations in Hsp90-M and Hsp90-C were recently implicated in substrate binding, and in fact the mutations in Hsp90-M are close to the Tau site (Genest et al., 2013). Together, our findings provide new context to the notion that the elongated site on Hsp90 that binds to Tau may also be used by other natural clients.
Since Tau is an intrinsically disordered protein (IDP), it is surprising that it interacts as a natural ligand of a molecular chaperone (Mukrasch et al., 2009). However, the microtubule-binding domain of Tau may define how Tau can become an Hsp90 client. This domain differs in sequence composition from the typical IDP by a higher content in hydrophobics, a typical trigger for chaperone interaction (Fig. 1E) (Dunker and Gough, 2011; Hartl et al., 2011; Uversky et al., 2009). In fact, the N-terminal segment of Tau, which is low in hydrophobic residues, does not interact with Hsp90. In this way exposed hydrophobicity may also drive a specific subset of disordered clients such as Tau onto Hsp90. This may ensure that the Hsp90 system is not poisoned by the large fraction of disordered proteins present in the eukaryotic cytoplasm.
The Hsp90-Tau interaction and therapeutic treatment of Alzheimer disease
Aggregation of the Tau microtubule binding region causes Alzheimer’s disease (Haass and Selkoe, 2007; Kosik et al., 1986). Hsp90’s association with Tau at this particular region allows the formation of a relay team, together with the Hsp70 system, that allows control of the fate of this potentially toxic stretch (Dickey et al., 2007; Jinwal et al., 2012; Sarkar et al., 2008). It demonstrates that the chaperonome is adapted to act selectively on toxic protein segments while ignoring the Tau regions that have no aggregation propensity (Fig. 1E).
The unusual sequence composition of Tau and its ability to interact with chaperones could be part of its pathogenic potential. Perhaps by trying to chaperone the microtubule binding domains of Tau, Hsp90 and other chaperones are extending the half-life of the disordered component in the cell, increasing its potential for toxicity (Dickey et al., 2007; Jinwal et al., 2012). Recent work suggests that Tau amyloid formation may actually be a protective mechanism that allows potentially toxic Tau intermediates to self-chaperone and sequester themselves into less toxic entities within long-lived neurons (Eisenberg and Jucker, 2012; Oddo et al., 2004; Santacruz et al., 2005). This feature is not reserved for Tau alone, but for other disease-associated proteins as well, such as α-synuclein (Daturpalli et al., 2013). Therefore, the association of Tau with chaperones like Hsp90 and Hsp70 within its amyloidogenic region may provide a nexus for Tau toxicity. As recently described, Hsp90 and the co-chaperone FKBP51 can prevent Tau from forming dense amyloids, and rather promote the formation of more toxic Tau intermediates (Blair et al., in the press). This demonstrates that the chaperonome can be a double-edged sword in the brain. This is the rationale for developing therapies aimed at chaperones, which in light of this recent evidence may be very beneficial for sufferers of these diseases.
Hsp90 can act on both folding intermediates and disordered proteins
Hsp90 is unique from other ATP-regulated chaperone systems in a number of ways. While Hsp90’s Tau binding site provides hydrophobic interaction and negative charges, similar to the other chaperone systems, it also provides a dramatically more extended substrate-binding site than any of the other chaperones (Fig. 5)(Buckle et al., 1997; Rüdiger et al., 1997a; Weibezahn et al., 2005; Xu et al., 1997). Moreover, the Hsp90 binding site for the substrate Tau is neither a cleft nor covered by a lid (Fig. 4). In fact, it is permanently open and accessible. This is made possible because Hsp90 distributes the hydrophobic contacts over a large surface, such that each individual contact constitutes only a fraction of the total binding energy (Fig. 5). This is confirmed by the fact that as of yet no single point mutations in Hsp90 have been identified to significantly alter its substrate affinity.
The architecture of the substrate-binding site of Hsp90 allows for sequence specific selection of some intrinsically disordered proteins such as Tau (Fig. 4). The Hsp90 substrate-binding site does not demand any specific structural features for a client to interact with, thus the exposed sequence pattern of the client will be decisive for binding. Folding intermediates indeed expose sequence patterns compatible with the properties of the binding site (Fig. 6A). This can explain why Hsp90 recognises a structurally diverse substrate pool with the same binding site (Fig. 4)(Echeverria and Picard, 2010; Jakob et al., 1995; Pearl and Prodromou, 2006; Street et al., 2011). We expect, therefore, that intermediates of client classes such as kinases and steroid receptors may bind via the same principles as disordered clients.
Hsp90 contributes to folding and maturation in concert with other chaperone systems, in particular Hsp70 (Wegele et al., 2004). Based on the comparison of the binding modes of Hsp90 and Hsp70, we propose a model for the temporal partitioning between both chaperones (Fig. 6C). Since Hsp70 binds to short, highly hydrophobic sequences it interacts with the folding protein early on (Bukau et al., 2006; Hartl et al., 2011; Rüdiger et al., 1997a). During the folding process these short hydrophobic stretches will most likely form the nucleus around which the protein condenses (Daggett and Fersht, 2003). Outside the folding nucleus are still less ordered stretches with scattered hydrophobics. Such stretches are not recognised by Hsp70, which may trigger its release from the late folding intermediate. The unique, 106 Å long binding surface of Hsp90, however, allows many low affinity contacts with the outer sphere of the late folding intermediates. Hsp90 profits from the compaction of the substrate during the nucleation process, yet co-chaperones will further specify substrate selection. Our model resolves the paradox on how Hsp90 specifically selects for late folding intermediates of regulatory proteins and but also some intrinsically disordered proteins – through the eyes of Hsp90 they look the same.
EXPERIMENTAL PROCEDURES
Proteins
Human proteins Hsp90β and hTau40 (isoform F, the longest isoform of Tau) were produced and purified as described (Barghorn et al., 2004; Karagöz et al., 2011).
Fluorescence spectroscopy
Fluorescence spectra were acquired on a Varian Cary Eclipse (excitation 295 nm, emission averaged 350–360 nm) at 25°C in 25 mM Na-phosphate, pH 7.2, 300 mM NaCl and 1 mM DTT, in microcuvettes with 3 mm pathlength. For ATPγS measurements, samples were measured again after addition of 1 mM ATPγS and 5 mM MgCl2 (final concentrations) and incubation for 10 min. The data were subjected to non-linear least square fitting using the Solver module in Excel (Kemmer and Keller, 2010), according to the equation F = Fmin + A *[Tau]n/(Kn+[Tau] n), in which F is the fluorescence intensity, Fmin is the minimal fluorescence intensity, A is the amplitude (difference maximal and minimal fluorescence intensity) and K is a constant value.
NMR spectroscopy
1H-13C-methyl TROSY spectra were recorded at 25 °C on a Bruker AvanceIII 900 MHz spectrometer (TCI cryoprobe) and referenced against DSS. Spectra processing, analysis and representations were done as described (Karagöz et al., 2011; Tugarinov et al., 2003). NMR samples of Hsp90 contained between 125 and 200 µM Hsp90 in D2O, 25 mM potassium phosphate, pH 7.2, 300 mM NaCl and 1 mM DTT. Spectra of Hsp90-Tau complexes were acquired with 136 µM and 125 µM Hsp90 and 1.2 to 2.3 molar equivalents of Tau (monomer concentrations).
Two-dimensional 1H–15N heteronuclear single quantum coherence (HSQC) NMR experiments were recorded at 5°C on a Bruker Avance 900 MHz spectrometer equipped with a cryogenic probe. Spectra were acquired using 600 complex points and 48 scans per increment with spectral widths of 8992 Hz and 2580 Hz in the 1H and 15N dimensions, respectively. Spectra were processed with NMRPipe (Delaglio et al., 1995) and analysed using CcpNMR Analysis (Vranken et al., 2005). NMR intensity ratio plots were reported with a 3–residue averaging window. Full-length 15N-labeled human Tau protein (hTau40) was expressed and purified as described previously (Mukrasch et al., 2009). NMR samples contained 50 µM 15N single labelled monomeric Tau protein in 50 mM phosphate buffer and 10 mM NaCl, pH 6.8, 1 mM DTT and 10 % (v/v) D2O. Spectra of Hsp90-Tau complexes were acquired using 21.7 µM and 41.7 µM of unlabelled Hsp90β (Overview isoleucine shifts in Table S1; The combined chemical shift difference Δv was calculated as follows: Δv= ((0.25ΔvC)2+ΔvH2)1/2).
SAXS measurements and modelling
Small angle X-ray scattering data were collected from solutions of Hsp90, Tau, Hsp90-Tau, and the nucleotide-bound states for several solute concentrations and analysed using the package ATSAS and standard procedures. SAXS-based structural models were generated using the program CORAL (Petoukhov and Svergun, 2005), known crystal structures, a homology model for Hsp90 (Karagöz et al., 2011), and additional NMR data for the binding interfaces in case of the Hsp90-Tau complexes as input. The surface of the models obtained was coloured accordingly to increasing distance to Tau using a cyan-white-red gradient. Structural models are displayed using PyMOL 1.5, Schrödinger, LLC, and Chimera (UCSF). See supplementary information for details.
Supplementary Material
Highlights.
A model for temporal control of early (Hsp70) and late (Hsp90) chaperoning
Structural model of Hsp90 in complex with its client Tau
Hsp90 recognises clients by spreading weak contacts over an extended interface
Hsp90 binds to Tau’s aggregation-prone microtubule-binding repeat region
ACKNOWLEDGEMENTS
We are grateful to Ineke Braakman for continuous support. We thank Assaf Friedler, Sander van den Heuvel and Paul van Bergen en Henegouwen for comments on the manuscript. S.G.D.R. was supported by Marie-Curie Actions of the 6th and 7th Framework programme of the EU (Excellence Grant “chaperoning cascades” (No. 25651), Innovative Doctoral Programme “ManiFold” (No. 317371) and Initial Training Network “WntsApp” (No. 608180)), a VIDI career development grant by the Nederlands Organisation for Scientific Research NWO and a High Potential Grant of Utrecht University. A.M.S.D. was supported by of a grant from Fundação para a Ciência e a Tecnologia, Portugal (SFRH/BPD/78075/2011). R.B. was supported by NWO-CW for the 900 MHz NMR and the TCI cryoprobe. R.B., H.I. and T.M. were supported by the EU project “BioNMR” (No. 261863). M.Z. was supported by the German Research Foundation (ZW71/7-1). T.M. was supported by the Bavarian Ministry of Sciences, Research and the Arts (Bavarian Molecular Biosystems Research Network), the Austrian Academy of Sciences (APART-fellowship), and the German Research Foundation (Emmy Noether program MA 5703/1-1). The authors gratefully acknowledge the Leibniz Supercomputing Centre (LRZ, www.lrz.de) for providing computing time on the Linux Cluster.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCE LIST
- Ali MMU, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440(7087):1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barghorn S, Davies P, Mandelkow E. Tau paired helical filaments from Alzheimer's disease brain and assembled in vitro are based on beta-structure in the core domain. Biochemistry (N. Y.) 2004;43:1694–1703. doi: 10.1021/bi0357006. [DOI] [PubMed] [Google Scholar]
- Bergen Mv, Friedhoff P, Biernat J, Heberle J, Mandelkow EM, Mandelkow E. Assembly of tau protein into Alzheimer paired helical filaments depends on a local sequence motif ((306)VQIVYK(311)) forming beta structure. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5129–5134. doi: 10.1073/pnas.97.10.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair L, Nordhues BA, Hill SE, Scaglione KM, O'Leary JC, Fontaine SN, Breydo L, Zhang B, Li P, Wang L, et al. Accelerated neurodegeneration through chaperone-mediated oligomerization of tau. Journal of Clinical Investigation. doi: 10.1172/JCI69003. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckle AM, Zahn R, Fersht AR. A structural model for GroEL-polypeptide recognition. Proc Natl Acad Sci U S A. 1997;94:3571–3575. doi: 10.1073/pnas.94.8.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Daggett V, Fersht A. The present view of the mechanism of protein folding. Nat. Rev. Mol. Cell Biol. 2003;4:497–502. doi: 10.1038/nrm1126. [DOI] [PubMed] [Google Scholar]
- Daturpalli S, Wang S, Buell A, Waudby C, Meehan S, Jackson SE. Hsp90 Inhibits alpha-Synuclein Aggregation by Interacting with Soluble Oligomers. J. Mol. Biol. 2013 doi: 10.1016/j.jmb.2013.08.006. [DOI] [PubMed] [Google Scholar]
- Dekker N, Cox M, Boelens R, Verrijzer CP, van der Vliet PC, Kaptein R. Solution structure of the POU-specific DNA-binding domain of Oct-1. Nature. 1993;362:852–855. doi: 10.1038/362852a0. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dickey CA, Kamal A, Lundgren K, Klosak N, Bailey RM, Dunmore J, Ash P, Shoraka S, Zlatkovic J, Eckman CB, et al. The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Invest. 2007;117:648–658. doi: 10.1172/JCI29715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didenko T, Boelens R, Rüdiger SGD. 3D DOSY-TROSY to determine the translational diffusion coefficient of large protein complexes. Protein Eng. Des. Sel. 2011;24:99–103. doi: 10.1093/protein/gzq091. [DOI] [PubMed] [Google Scholar]
- Didenko T, Duarte AM, Karagöz GE, Rüdiger SGD. Hsp90 structure and function studied by NMR spectroscopy. Biochim. Biophys. Acta. 2012;1823:636–647. doi: 10.1016/j.bbamcr.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Dollins DE, Warren JJ, Immormino RM, Gewirth DT. Structures of GRP94-nucleotide complexes reveal mechanistic differences between the hsp90 chaperones. Mol. Cell. 2007;28:41–56. doi: 10.1016/j.molcel.2007.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou F, Netzer WJ, Tanemura K, Li F, Hartl FU, Takashima A, Gouras GK, Greengard P, Xu H. Chaperones increase association of tau protein with microtubules. Proc. Natl. Acad. Sci. U. S. A. 2003;100:721–726. doi: 10.1073/pnas.242720499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Gough J. Sequences and topology: intrinsic disorder in the evolving universe of protein structure. Curr. Opin. Struct. Biol. 2011;21:379–381. doi: 10.1016/j.sbi.2011.04.002. [DOI] [PubMed] [Google Scholar]
- Echeverria PC, Picard D. Molecular chaperones, essential partners of steroid hormone receptors for activity and mobility. Biochimica Et Biophysica Acta-Molecular Cell Research. 2010;1803:641–649. doi: 10.1016/j.bbamcr.2009.11.012. [DOI] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest O, Reidy M, Street TO, Hoskins JR, Camberg JL, Agard DA, Masison DC, Wickner S. Uncovering a region of heat shock protein 90 important for client binding in E. coli and chaperone function in yeast. Mol. Cell. 2013;49:464–473. doi: 10.1016/j.molcel.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Hagn F, Lagleder S, Retzlaff M, Rohrberg J, Demmer O, Richter K, Buchner J, Kessler H. Structural analysis of the interaction between Hsp90 and the tumor suppressor protein p53. Nature Struct. Mol. Biol. 2011;0:1–8. doi: 10.1038/nsmb.2114. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Ittner LM, Ke YD, Delerue F, Bi M, Gladbach A, van Eersel J, Wolfing H, Chieng BC, Christie MJ, Napier IA, et al. Dendritic function of tau mediates amyloid-beta toxicity in Alzheimer's disease mouse models. Cell. 2010;142:387–397. doi: 10.1016/j.cell.2010.06.036. [DOI] [PubMed] [Google Scholar]
- Jakob U, Lilie H, Meyer I, Buchner J. Transient interaction of Hsp90 with early unfolding intermediates of citrate synthase. Implications for heat shock in vivo. J Biol Chem. 1995;270:7288–7294. doi: 10.1074/jbc.270.13.7288. [DOI] [PubMed] [Google Scholar]
- Jinwal UK, Akoury E, Abisambra JF, O'Leary JC, 3rd, Thompson AD, Blair LJ, Jin Y, Bacon J, Nordhues BA, Cockman M, et al. Imbalance of Hsp70 family variants fosters tau accumulation. FASEB J. 2012 doi: 10.1096/fj.12-220889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagöz GE, Duarte AM, Ippel H, Uetrecht C, Sinnige T, van Rosmalen M, Hausmann J, Heck AJ, Boelens R, Rüdiger SGD. N-terminal domain of human Hsp90 triggers binding to the cochaperone p23. Proc. Natl. Acad. Sci. U. S. A. 2011;108:580–585. doi: 10.1073/pnas.1011867108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmer G, Keller S. Nonlinear least-squares data fitting in Excel spreadsheets. Nat. Protoc. 2010;5:267–281. doi: 10.1038/nprot.2009.182. [DOI] [PubMed] [Google Scholar]
- Kosik KS, Joachim CL, Selkoe DJ. Microtubule-associated protein tau (tau) is a major antigenic component of paired helical filaments in Alzheimer disease. Proc. Natl. Acad. Sci. U. S. A. 1986;83:4044–4048. doi: 10.1073/pnas.83.11.4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukenberg KA, Bottcher UM, Southworth DR, Agard DA. Grp94, the endoplasmic reticulum Hsp90, has a similar solution conformation to cytosolic Hsp90 in the absence of nucleotide. Protein Sci. 2009;18:1815–1827. doi: 10.1002/pro.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krukenberg KA, Forster F, Rice LM, Sali A, Agard DA. Multiple conformations of E. coli Hsp90 in solution: insights into the conformational dynamics of Hsp90. Structure. 2008;16:755–765. doi: 10.1016/j.str.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Soroka J, Buchner J. The Hsp90 chaperone machinery: Conformational dynamics and regulation by co-chaperones. Biochim. Biophys. Acta. 2012;1823:624–635. doi: 10.1016/j.bbamcr.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Luo W, Dou F, Rodina A, Chip S, Kim J, Zhao Q, Moulick K, Aguirre J, Wu N, Greengard P, Chiosis G. Roles of heat-shock protein 90 in maintaining and facilitating the neurodegenerative phenotype in tauopathies. Proc. Natl. Acad. Sci. U. S. A. 2007;104:9511–9516. doi: 10.1073/pnas.0701055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E. Biochemistry and cell biology of tau protein in neurofibrillary degeneration. Cold Spring Harb Perspect. Med. 2012;2:a006247. doi: 10.1101/cshperspect.a006247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP. Gymnastics of molecular chaperones. Mol. Cell. 2010;39:321–331. doi: 10.1016/j.molcel.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11:647–658. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- Miyata Y, Koren J, Kiray J, Dickey CA, Gestwicki JE. Molecular chaperones and regulation of tau quality control: strategies for drug discovery in tauopathies. Future Med. Chem. 2011;3:1523–1537. doi: 10.4155/fmc.11.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukrasch MD, Bibow S, Korukottu J, Jeganathan S, Biernat J, Griesinger C, Mandelkow E, Zweckstetter M. Structural Polymorphism of 441-Residue Tau at Single Residue Resolution. Plos Biology. 2009;7:399–414. doi: 10.1371/journal.pbio.1000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonas E, Hascher A, Bernado P, Blackledge M, Mandelkow E, Svergun DI. Domain conformation of tau protein studied by solution small-angle X-ray scattering. Biochemistry (N. Y.) 2008;47:10345–10353. doi: 10.1021/bi800900d. [DOI] [PubMed] [Google Scholar]
- Oddo S, Billings L, Kesslak JP, Cribbs DH, LaFerla FM. Abeta immunotherapy leads to clearance of early, but not late, hyperphosphorylated tau aggregates via the proteasome. Neuron. 2004;43:321–332. doi: 10.1016/j.neuron.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Park SJ, Kostic M, Dyson HJ. Dynamic Interaction of Hsp90 with Its Client Protein p53. J. Mol. Biol. 2011;411:158–173. doi: 10.1016/j.jmb.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C. Structure and mechanism of the hsp90 molecular chaperone machinery. Annu Rev Biochem. 2006;75:271–294. doi: 10.1146/annurev.biochem.75.103004.142738. [DOI] [PubMed] [Google Scholar]
- Pearl LH, Prodromou C, Workman P. The Hsp90 molecular chaperone: an open and shut case for treatment. Biochem. J. 2008;410:439–453. doi: 10.1042/BJ20071640. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Global Rigid Body Modeling of Macromolecular Complexes against Small-Angle Scattering Data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Roe SM, Piper PW, Pearl LH. A molecular clamp in the crystal structure of the N-terminal domain of the yeast Hsp90 chaperone. Nat Struct Biol. 1997;4:477–482. doi: 10.1038/nsb0697-477. [DOI] [PubMed] [Google Scholar]
- Retzlaff M, Hagn F, Mitschke L, Hessling M, Gugel F, Kessler H, Richter K, Buchner J. Asymmetric activation of the hsp90 dimer by its cochaperone aha1. Mol. Cell. 2010;37:344–354. doi: 10.1016/j.molcel.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Roberson ED, Scearce-Levie K, Palop JJ, Yan F, Cheng IH, Wu T, Gerstein H, Yu GQ, Mucke L. Reducing endogenous tau ameliorates amyloid beta-induced deficits in an Alzheimer's disease mouse model. Science. 2007;316:750–754. doi: 10.1126/science.1141736. [DOI] [PubMed] [Google Scholar]
- Rüdiger S, Buchberger A, Bukau B. Interaction of Hsp70 chaperones with substrates. Nat Struct Biol. 1997a;4:342–349. doi: 10.1038/nsb0597-342. [DOI] [PubMed] [Google Scholar]
- Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. Embo J. 1997b;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santacruz K, Lewis J, Spires T, Paulson J, Kotilinek L, Ingelsson M, Guimaraes A, DeTure M, Ramsden M, McGowan E, et al. Tau suppression in a neurodegenerative mouse model improves memory function. Science. 2005;309:476–481. doi: 10.1126/science.1113694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar M, Kuret J, Lee G. Two motifs within the tau microtubule-binding domain mediate its association with the hsc70 molecular chaperone. J. Neurosci. Res. 2008;86:2763–2773. doi: 10.1002/jnr.21721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette M, van Tilborg P, Spurio R, Kaptein R, Paci M, Gualerzi CO, Boelens R. The structure of the translational initiation factor IF1 from E.coli contains an oligomer-binding motif. EMBO J. 1997;16:1436–1443. doi: 10.1093/emboj/16.6.1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Shkumatov AV, Chinnathambi S, Mandelkow E, Svergun DI. Structural memory of natively unfolded tau protein detected by small-angle X-ray scattering. Proteins. 2011;79:2122–2131. doi: 10.1002/prot.23033. [DOI] [PubMed] [Google Scholar]
- Street TO, Lavery LA, Agard DA. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol. Cell. 2011;42:96–105. doi: 10.1016/j.molcel.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nature Reviews Molecular Cell Biology. 2010;11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell. 2012;150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AD, Scaglione KM, Prensner J, Gillies AT, Chinnaiyan A, Paulson HL, Jinwal UK, Dickey CA, Gestwicki JE. Analysis of the tau-associated proteome reveals that exchange of Hsp70 for Hsp90 is involved in tau degradation. ACS Chem. Biol. 2012;7:1677–1686. doi: 10.1021/cb3002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. Cross-correlated relaxation enhanced 1H-13C NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J. Am. Chem. Soc. 2003;125:10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]
- Uversky VN, Oldfield CJ, Midic U, Xie H, Xue B, Vucetic S, Iakoucheva LM, Obradovic Z, Dunker AK. Unfoldomics of human diseases: linking protein intrinsic disorder with diseases. BMC Genomics. 2009;(10 Suppl 1):S7. doi: 10.1186/1471-2164-10-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MM, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol. Cell. 2006;23:697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Wegele H, Muller L, Buchner J. Hsp70 and Hsp90--a relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 2004;151:1–44. doi: 10.1007/s10254-003-0021-1. [DOI] [PubMed] [Google Scholar]
- Weibezahn J, Schlieker C, Tessarz P, Mogk A, Bukau B. Novel insights into the mechanism of chaperone-assisted protein disaggregation. Biol. Chem. 2005;386:739–744. doi: 10.1515/BC.2005.086. [DOI] [PubMed] [Google Scholar]
- Wüthrich K. NMR studies of structure and function of biological macromolecules (Nobel lecture) Angew. Chem. Int. Ed Engl. 2003;42:3340–3363. doi: 10.1002/anie.200300595. [DOI] [PubMed] [Google Scholar]
- Xu Z, Horwich AL, Sigler PB. The crystal structure of the asymmetric GroEL-GroES-(ADP)7 chaperonin complex. Nature. 1997;388:741–750. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- Zuiderweg ER. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.