Abstract
Drug efflux pumps confer resistance upon bacteria to a wide range of antibiotics from various classes. The expression of efflux pumps are also implicated in virulence and biofilm formation. Moreover, organisms can only acquire resistance in the presence of active drug efflux pumps. Therefore, efflux pump inhibitors (EPIs) are attractive compounds to reverse multidrug resistance and to prevent the development of resistance in clinically relevant bacterial pathogens. We investigated the potential of pure compounds isolated from plants to act as EPIs. In silico screening was used to predict the bioactivity of plant compounds and to compare that with the known EPI, phe-arg-β-naphthylamide (PAβN). Subsequently, promising products have been tested for their ability to inhibit efflux. Plumbagin nordihydroguaretic acid (NDGA) and to a lesser degree shikonin, acted as sensitizers of drug-resistant bacteria to currently used antibiotics and were able to inhibit the efflux pump-mediated removal of substrate from cells. We demonstrated the feasibility of in silico screening to identify compounds that potentiate the action of antibiotics against drug-resistant strains and which might be potentially useful lead compounds for an EPI discovery program.
Keywords: Antibiotic, efflux pump inhibitors, multidrug resistance, natural products
Introduction
Antibiotic resistance is one of the world's most pressing health problems (Gootz 2010; Bush et al. 2011; Wise 2011; Piddock 2012). Paradoxically, the increase in antibiotic-resistant bacteria coincides with a dramatic reduction in the number of pharmaceutical companies developing new antimicrobial agents (Cooper and Shlaes 2011).
In Gram-negative pathogens, drug efflux protein complexes such as AcrAB-TolC confer resistance to antibiotics from many different classes and are the systems predominantly responsible for innate and acquired multidrug resistance (MDR) (Zhang et al. 2000; Poole 2001; Piddock 2006a, 2006b; Du et al. 2013). One way of prolonging antibiotic efficiency or of potentiating the activity of antibiotics against drug-resistant pathogens is by blocking drug efflux pumps with efflux pump inhibitors (EPIs) (Lomovskaya et al. 2006; Abreu et al. 2012; Kourtesi et al. 2013; Ruggerone et al. 2013). Inhibiting bacterial drug efflux machinery has additional benefits apart from the reversal of drug resistance because drug efflux pumps also have crucial roles in bacterial pathogenesis, virulence, and biofilm formation (Piddock 2006b; Martinez et al. 2009; Baugh et al. 2012, 2014). In addition, functional efflux pumps are necessary for the selection of drug-resistant bacteria (Lomovskaya and Bostian 2006; Ricci et al. 2006; Zhang et al. 2011).
A large proportion of the drugs used to treat infectious diseases have come from natural sources. Plants have evolved to effectively fight off infections by producing an array of special chemicals. We were specifically interested in identifying and developing EPIs from plant sources. So far, the plant compounds that have been shown to act synergistically with antibiotics were predominantly effective against Gram-positive bacteria (Lewis 2001; Stavri et al. 2007), while EPIs for Gram-negative pathogens are still evading us as phe-arg-β-naphthylamide (PAβN) and its derivatives are not clinically used due to problems with stability and toxicity (Marquez 2005). Gram-negative bacteria have evolved a significant permeability barrier to amphipathic compounds by the combination of an outer membrane and the expression of drug efflux pumps. Due to their intrinsic high drug resistance and the increased incidence of drug-resistant Gram-negative infections, it is important that new treatments against drug-resistant Gram-negative bacteria are developed.
In this respect, AcrB the inner membrane protein from the AcrAB-TolC drug transport complex is an ideal model for testing compounds with EPI properties. The structure of AcrB from Escherichia coli with and without substrates/inhibitors bound has been solved (Murakami et al. 2006; Seeger et al. 2006; Nakashima et al. 2011; Eicher et al. 2012; Hung et al. 2013). In addition, the first structure of a tripartite drug efflux complex has recently been published (Du et al. 2014).
Furthermore, AcrB homologues are present in most Gram-negative pathogens; it can even substitute for MexB from Pseudomonas aeruginosa and form an active drug efflux complex with MexA and OprM, the natural partners of MexB (Krishnamoorthy et al. 2008; Welch et al. 2010). Hence, an inhibitor of AcrB has great potential to be an EPI in other Gram-negative pathogenic organisms.
AcrB forms an asymmetric homotrimer where the monomers cycle through three conformational stages designated the loose, tight, and open or access, binding and extrusion stages according to Seeger et al. (Seeger et al. 2006) or Murakami et al. (Murakami et al. 2006), respectively. Structures with bound drugs revealed two discrete multisite binding pockets separated by a switch loop; the distal pocket in the binding/tight state and a proximal pocket in the access/loose state (Nakashima et al. 2011; Eicher et al. 2012).
A structure of AcrB with an inhibitor bound demonstrated that the inhibitor binding site partially overlaps with the minocycline-binding site in the distal binding pocket, while another part of the inhibitor binds to a narrow hydrophobic pit lined by several phenylalanine residues (F136, F178, F610, F615, F628). Tight binding of the inhibitor to residues in the pit prevents the functional rotation through the three stages; hence, prevents the efflux of antibiotics (Nakashima et al. 2013).
In this study, we used in silico screening to select five plant-derived compounds on their predicted ability to bind and inhibit AcrB. The selected compounds were screened for their antibacterial effect on wild-type E. coli cells and E. coli cells with a genomic deletion of the gene coding for AcrB. The ability of the compounds to improve antibiotic sensitivity to efflux-mediated-resistant E. coli cells was assessed. Direct inhibition of substrate transport was observed by measuring the reduction in Nile Red efflux in the presence of the natural products.
Materials and Methods
Bacterial strains, media, and chemicals
Bacteria used in this study were E. coli strain BW25113 wild type and strain BW25113 with a deletion of acrB (a gift from Professor Martin Pos, Institute of Biochemistry, Goethe-University, Frankfurt am Main, Germany). All strains were maintained in 25% (v/v) glycerol at −80°C until required. LB media were obtained from Acumedia and Agar from Oxoid. All antibiotics, chemicals, and natural products were obtained from Sigma.
Accumulation of fluorescent compounds
LB-Broth was inoculated with an overnight culture of wild-type-resistant E. coli cells or cells with a genomic deletion of acrB (1:500 dilution) and incubated with shaking at 37°C until an OD660 of 0.5 was reached. The cells were harvested by centrifugation at 3000g for 10 min at 4°C, then washed three times by resuspension in 50 mmol/L potassium phosphate buffer (pH 7.0) containing 5 mmol/L MgSO4, and sedimented by centrifugation at 3000 g for 10 min at 4°C. The cells were then resuspended in the same buffer to an OD660 of 0.5 and incubated for 3 min at room temperature (25°C) in the presence of 25 mmol/L glucose to energize or 10 μmol/L carbonyl cyanide 3-chlorophenylhydrazone (CCCP) to deenergize the cells. The fluorescence measurement was started, and 60 sec later 2 μmol/L ethidium bromide, 0.125 μmol/L Hoechst 33342, or 0.25 μmol/L TMA-DPH [1-(4-trimethylammoniumphenyl)-6-phenyl-1,3,5-hexatriene p-toluenesulfonate] was added. The fluorescence was followed as a function of time in a PerkinElmer, Beaconsfield UK, LS 55B fluorimeter. Excitation and emission wavelengths and excitation and emission slit widths were 500, 580, 5, and 10 nm, respectively, for ethidium bromide; 355, 457, 10, and 4 nm, respectively, for Hoechst 33342; and 350, 425, 5, and 5 nm, respectively, for TMA-DPH.
Docking of natural products to AcrB
Docking of natural products was performed using the E. coli AcrB protein (Protein Data Bank: 4DX5, tight monomer) and AutoDock 4.2 (http://autodock.scripps.edu/) (Morris et al. 2009). Prior to docking, bound ligand (minocycline) was removed and the suitability of AutoDock was validated by blind cognate docking (Hetenyi and van der Spoel 2011). For this, the bound minocycline we first removed from the reference structure and minocycline (the structure of which was obtained from a separate source – the Zinc database, https://docking.org/) was redocked against the entire protein structure. The top-ranked pose of redocked minocycline largely superimposed (within ≤2Å) over the original pose of the drug present in the crystal structure (not shown). Since the putative interaction site(s) for the screened natural products were unknown, we used blind docking approach where the search space for the ligands comprised a grid that contained the entire protein structure. Ligands were downloaded from PubChem database (http://pubchem.ncbi.nlm.nih.gov/) and were energy minimized with MM2 force field using ChemBioOffice 2008 (http://www.cambridgesoft.com). Polar hydrogens and the Gasteiger partial atomic charges were then added to the protein and ligands using AutoDockTools (http://autodock.scripps.edu/resources/adt), and the prepared structures were used as input files for docking. From the estimated free energy of ligand interaction (ΔGbinding, kcal/mol), the inhibition constants (Ki) were calculated (Musa et al. 2011). Only the best pose (the one with the lowest ΔGbinding) is considered. Finally, the best ligand poses were analyzed using PoseViewWeb (http://poseview.zbh.uni-hamburg.de/). PyMol was used for figures.
Antimicrobial assays
Antimicrobial assays were carried out according to the 96-well microtiter broth dilution method (Ohene-Agyei et al. 2012). Briefly, the test compounds were added to the cell suspensions (105 cells/well in LB-broth) at increasing concentrations, and the cultures were incubated at 37°C with shaking. The A630 of the cultures were measured in a plate reader (BioTek, Potton, Bedfordshire, UK) after 18 h, and the lowest concentration of drug needed to prevent growth (no increase in A630 compared to the A630 at time zero) was determined by minimum inhibitory concentrations (MIC). Additionally, for some compounds where MIC determination with the plate reader was impaired due to their high optical densities bacterial growth was also determined using resazurin where changes of the color from blue to pink indicated a reduction of the compound caused by bacterial growth (Ndi et al. 2007). From this the MIC were determined as the lowest concentration of plant extract where no growth was observed (color remained blue).
All experiments employed basal levels of expression without induction. All MIC determinations were repeated at least three times in independent experiments. The antibiotics, compounds, and EPI were made up and used according to the manufacturer's instructions.
Checkerboard titration assay method
Interactions between antibiotics and natural products were assessed by a checkerboard titration assay (Lomovskaya et al. 2001; Orhan et al. 2005). The antibiotic was tested at seven concentrations, whereas the natural product was tested at five concentrations. A quantity of 100 μL of LB-broth was distributed into each well of the microdilution plates. The antibiotic was serially diluted along the ordinate, while the natural product was diluted along the abscissa. Each microtiter well was then inoculated with 100 μL of a bacterial inoculum to give 105cells/well. The plates were incubated at 37°C for 7 h under aerobic conditions.
The MIC were defined as the lowest concentration of antibiotic that completely inhibited the growth of the organism as detected by no increase in A630 over the 7 h. This was also confirmed with the use of resazurin to test cell viability (Ndi et al. 2007).
Nile red efflux assay
The ability of the natural products to reduce the efflux of efflux pump substrates was assessed by measuring the reduction in Nile Red efflux (Bohnert et al. 2010). LB-Broth (Acumedia from Cell Biosciences, Heidelberg, Victoria, Australia) was inoculated with an overnight culture of the required E. coli strain and incubated with shaking at 37°C until the OD660 did not increase any more. The cells were harvested by centrifugation at 3000g for 10 min at room temperature, then washed three times by resuspension in 50 mmol/L potassium phosphate buffer (pH 7.0) containing 1 mmol/L MgCl2 and sedimented by centrifugation at 3000g for 10 min at 4°C. The cells were then resuspended in the same buffer to an OD660 of 1 after which cells were allowed to rest for 15 min at room temperature. Aliquots (2 mL) were then transferred to Pyrex 15-mL conical centrifugation tubes and CCCP was added to a final concentration of 10 μmol/L for the AcrAB-overexpressing strain. After 15 min, the natural product was added to the desired final concentration and Nile Red added to a final concentration of 5 μmol/L after another 15 min. The cell suspension was incubated on a shaker (140 rpm; 37°C) for 3 h. The cells were left at room temperature for 60 min and then centrifuged for 5 min at 3000g. After the bulk of the supernatant was discarded, droplets of it clinging to the tube wall were thoroughly removed with absorbent tissues, and the cells were resuspended in 2-mL assay buffer. Immediately thereafter, 0.2 mL of this cell suspension was transferred to a quartz cuvette (PerkinElmer, UK) containing 1.8 mL of assay buffer, and the cuvette was placed in a PerkinElmer LS 55B fluorimeter. The excitation was at 552 nm, emission was at 636 nm, and excitation and emission slit widths were 10 and 5 nm, respectively. The fluorescence of the cell suspension was followed for 100 sec, after which Nile Red efflux was triggered by rapid energization with 50 mmol/L glucose. Fluorescence was monitored for another 200 sec.
Nitrocefin uptake assay
Nitrocefin uptake assay was carried out as described previously (Lomovskaya et al. 2001). E. coli cells with constitutive expression of chromosomal β-lactamase were grown in LB-broth, harvested, and washed in 50 mmol/L Potassium phosphate buffer. The cells were subsequently resuspended in the same buffer to OD660 of 0.5. The cell suspension was treated with 10 μmol/L CCCP and the plant compounds added to give the required final concentrations. Nitrocefin was then added to give a final concentration of 32 μg/mL. Hydrolysis of nitrocefin was monitored by measuring the increase in absorbance at 490 nm with a plate reader (BioTek).
Results and Discussion
Strains used in this study
As we wanted to identify plant products that could act as EPIs, it was important to distinguish between inhibition of AcrB-mediated antibiotic efflux and altered membrane permeability. In order to do this, we have compared the resistance profiles of wild type, drug-resistant E. coli cells with cells where the gene coding for the drug efflux protein AcrB has been deleted. We have used the uptake of fluorescent substrates to show that there is no intrinsic difference between the E. coli strains used in this study and that any effect observed would not be due to intrinsic differences in cell permeability.
Ethidium, Hoechst 33342 and TMA-DPH all share the property that they are virtually nonfluorescent in aqueous solution while undergoing a large increase in fluorescent quantum yield when in a hydrophobic environment such as when in a lipid bilayer or intercalated into DNA. Hence, the passive permeation of these compounds into bacterial cells can be followed by the increase in their fluorescence. It is clear from Figure 1A–C that the fluorescence (and hence passive permeation) of all three fluorescent compounds are identical in E. coli cells which have been deenergized by the addition of the protonophore CCCP. Therefore, there is no intrinsic difference in permeability between the cells used in this study. However, when the cells are energized by the addition of glucose, there is a marked decrease in fluorescence between the wild type (resistant) and ΔAcrB cells (Fig. 1D–F). This decrease in fluorescence is representative of AcrB-mediated efflux of the fluorescent compounds. Combined, the results mean that any effects on the MICs for the test compounds observed would be due to the active AcrB-mediated efflux of the compounds and not due to intrinsic differences in the membrane permeability between the strains used in the study.
Figure 1.
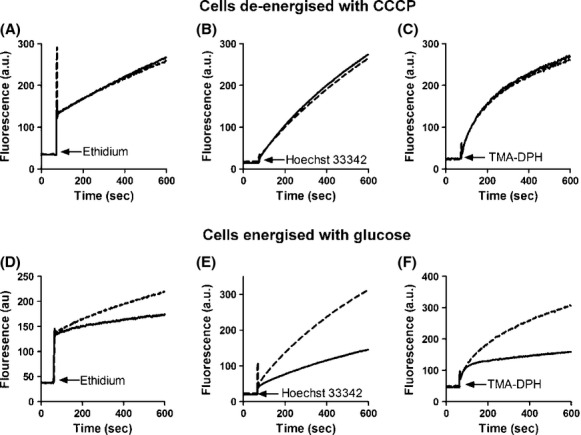
Accumulation of fluorescent compounds in the Escherichia coli strains used in this study. E. coli BW25113 (black line) and BW25113 ΔAcrB (broken line) cells were grown to an OD660 of 0.5. Cells were harvested, washed three times in 50 mmol/L potassium phosphate buffer (pH 7.0) containing 5 mmol/L MgSO4 and resuspended to a final OD660 of 0.5. (A–C). Cells were treated with carbonyl cyanide 3-chlorophenylhydrazone (CCCP) (10 μmol/L) for complete deenergization. (D–F). Cells were incubated in the presence of glucose (20 mmol/L) to generate metabolic energy before the fluorescence trace was started. Ethidium (2 μmol/L), Hoechst 33342 (0.125 μmol/L), or TMA-DPH (0.25 μmol/L) were added at the points indicated by the arrows and the increase in fluorescence was followed over time.
Plant compounds studied
In silico screening of an in-house database of 50 phytochemicals identified five different compounds that could be docked in the binding pocket of AcrB similar to the known EPI PAβN. These were the naphthoquinones plumbagin from Plumbago indica and shikonin from Lithospermum erythrorhizon; the flavonoid quercetin which is found in many fruits and vegetables; the xanthonoid mangiferin from Mangifera indica (mango), and the antioxidant nordihydroguaretic acid (NDGA) from creosote bush (Fig. 2).
Figure 2.
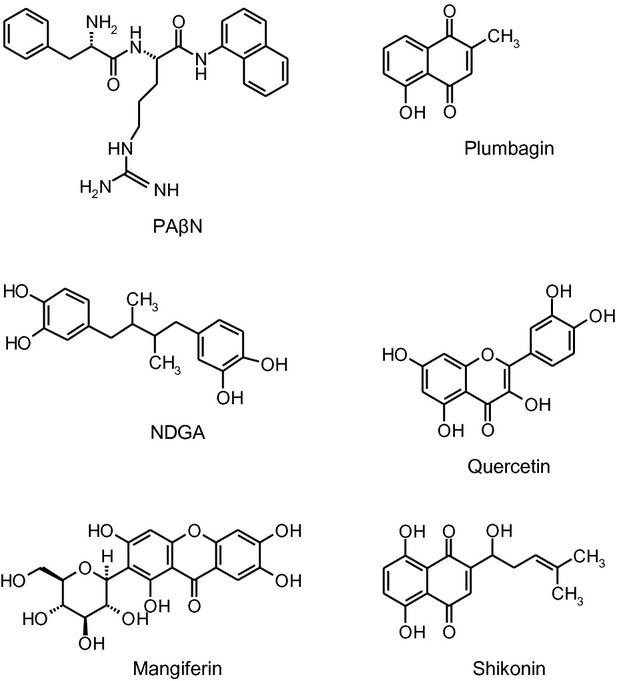
Structures of PAβN and the phytochemicals.
Plumbagin has previously been reported as an antibacterial compound which became more potent in the presence of the EPI PAβN (Tegos et al. 2002; Kuete et al. 2011). In addition, plumbagin is also an inhibitor of ABCG2, the ATP-binding cassette transporter involved in the multidrug-resistant phenotype of cancer cells (Shukla et al. 2007). Many compounds inhibit drug transporters from both cancer cells and bacteria (Amaral et al. 2012) as the drug efflux proteins involved display similarities in their mechanism of action (Venter et al. 2003, 2005). Quercetin has previously been reported as a substrate for AcrB as deletion of AcrB from E. coli resulted in a more than 8-fold reduction in the MIC of quercetin (Al-Karablieh et al. 2009). Nordihydroguaretic acid is bioactive compound from creosote bush, a plant which has long been used for its medicinal properties by North American Indians (Ross 2005) while antimicrobial effects of NDGA have recently been reported (Martins et al. 2013). Mangiferin has many medicinal qualities (Matkowski et al. 2013) and limited antibacterial (Savikin et al. 2009) and biofilm inhibitory (Annapoorani et al. 2012) effects have also been reported. Shikonin is a compound isolated from herbs used in traditional Chinese medicine (Tse 2003; Andújar et al. 2013). It has antibacterial activity against Gram-positive organisms (Shen et al. 2002; Haghbeen et al. 2011) but not against Gram-negative bacteria such as P. aeruginosa and E. coli (Shen et al. 2002; Haghbeen et al. 2011).
In silico prediction of the binding of PAβN and phytochemicals to AcrB
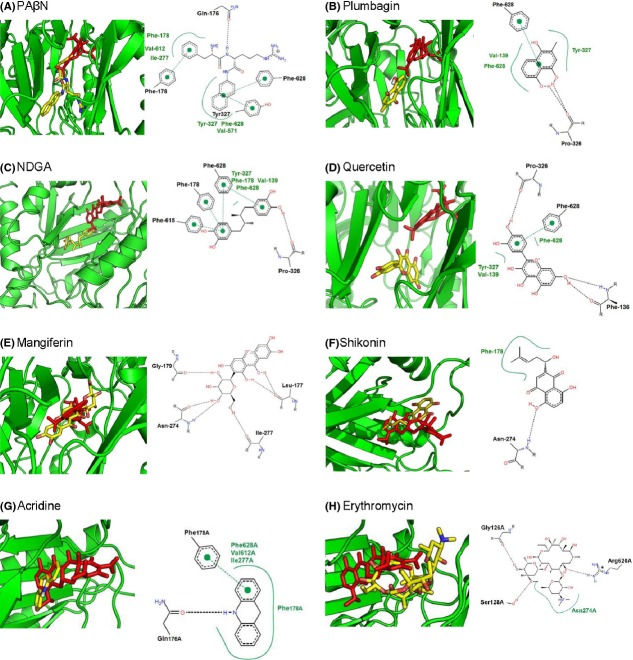
The binding of natural products to AcrB was predicted through docking experiments and compared with the docking of the known inhibitor PAβN and substrate minocycline. The T (tight or binding) monomer of AcrB (PDB code 4DX5) (Eicher et al. 2012) with minocycline bound to the deep binding pocket was used in the docking experiments. PAβN docked in a position closer to the lateral binding site of the T monomer compared to minocycline (Fig. 3A) and is stabilized by interactions with amino acids Phe-628, Phe-178, Tyr-287, and a hydrogen bond to the oxygen of Gln176 (Fig. 3A). The predicted docking site of PAβN correlates well with the binding of PAβN to AcrB as predicted by dynamic simulations where Phe-628, Phe-178, and Gln-176 were also some key residues for interaction with PAβN (Vargiu and Nikaido 2012). In addition, a recent crystal structure of AcrB and MexB revealed that Phe 178 contributes to the tight binding of an inhibitor molecule through π–π interactions (Nakashima et al. 2013). Incidentally, all the natural products which were predicted to be potential EPIs docked in a similar position as PAβN, while substrates such as minocycline, acridine, and erythromycin docked in the deep binding pocket (Fig. 3). Vargiu and Nikaido (Vargiu and Nikaido 2012) utilized molecular dynamics simulations which also predicted that residues from the deep binding pocket were much more involved in interaction with a range of substrates, while less involved in stabilizing the inhibitor PAβN. In our in silico binding predictions, all natural products were stabilized by hydrogen bonding and extensive hydrophobic interactions between phenylalanine side chains and aromatic groups within the compounds. This is not surprising as phenylalanine residues play an important role in substrate recognition and stabilization in RND-type drug transporters (Ohene-Agyei et al. 2012). According to the docking, mangiferin would act more like a substrate than an inhibitor and the predicted binding affinity was also the lowest of the natural products included in the assays. The free energy of interaction (ΔGbinding) determined from the docking of each compound was used to predict the Ki for the interactions (Table 1). Accordingly, the compounds could be arranged into an order of putative inhibition efficiency given as:
Figure 3.
Docking of phytochemicals and substrates onto AcrB (PDB: 4DX5, tight monomer). Positions of the examined ligands (yellow) compared to minocycline (red) are given in the left-hand pictures, while the interaction of the amino acids with the ligands is given on the right-hand side in each picture.
Table 1.
Free energy of binding and affinity constants calculated from the docking experiments
| Ligand | ΔGbinding (kcal/mol) | Predicted Ki (μmol/L) |
|---|---|---|
| PAβN | −7.95 | 1.5 |
| Plumbagin | −6.7 | 12.3 |
| NDGA | −6.66 | 13.1 |
| Quercetin | −7.15 | 5.7 |
| Mangiferin | −5.56 | 84 |
| Shikonin | −6.02 | 38.6 |
| Acridine | −7.8 | 1.9 |
| Erythromycin | −8.6 | 0.5 |
Subsequently, bioassays were performed to determine the validity of the in silico screening/modeling data.
Antibacterial activity of the plant compounds
The MIC of the natural products were determined so that subinhibitory concentrations could be used to check for efflux pump inhibition. For this purpose, wild-type-resistant E. coli BW25113 cells or cells with a genomic deletion of acrB were used to test the antibacterial activity of the natural products. Cells were grown in the presence of increasing concentrations of PAβN or in the presence of increasing concentrations of natural products. PAβN is toxic to cells at high concentrations and the presence of AcrB confers resistance to the cells against PAβN in accordance with the well-established role of PAβN as an EPI that is also an efflux pump substrate (Table 2).
Table 2.
Minimum inhibitory concentrations (MICs) of natural products and PAβN against Escherichia coli strains
| E. coli strain | MIC (mg/L) |
|||||
|---|---|---|---|---|---|---|
| Plumbagin | NDGA | Quercetin | Mangiferin | Shikonin | PAβN | |
| ΔAcrB drug-sensitive strain | 16 | 64 | >1024 | >512 | >256 | 128 |
| Wild-type drug-resistant strain | 128 | 512 | >1024 | >512 | >256 | 512 |
Of the compounds tested, only plumbagin and NDGA were antibacterial with a significant increase in the levels of susceptibility for the strains with the acrB deletion. This indicates the role of the AcrAB-TolC efflux pump in recognizing and transporting plumbagin and NDGA (Table 2). Quercetin, shikonin, and mangiferin were not toxic to bacterial cells at the maximum concentrations tested (Table 2).
According to our docking data, quercetin should be the best inhibitor of AcrB after PAβN. Cells lacking AcrB have also been shown to be hypersensitive to quercetin (Al-Karablieh et al. 2009). However, the E. coli strain we used was not susceptible to quercetin.
In order for an compound to qualify as an EPI, it must be able to potentiate the effect of antibiotics which are effluxed in strains containing an efflux pump and must not potentiate the activities of antibiotics that are not effluxed (Lomovskaya et al. 2001). We therefore tested the ability of the natural products to reverse drug resistance in AcrB expressing cells.
Some natural products are able to increase drug sensitivity in drug-resistant cells
The ability of the compounds to reverse antibiotic resistance conferred by the expression of the AcrAB-TolC efflux pump was determined. Standard checkerboard assays in which the MIC of the antibiotics were determined in the presence of different concentrations of the known EPI PAβN and the natural products were performed (Table 3). PAβN was synergistic with erythromycin, chloramphenicol, novobiocin, and tetracycline. NDGA was the most efficient potential EPI as it synergized with erythromycin, chloramphenicol, novobiocin, tetracycline, and Tetraphenylphosphonium (TPP) (Table 3). Plumbagin increased sensitivity to erythromycin, chloramphenicol, and TPP, while shikonin potentiated the antimicrobial activity of TPP (Table 3). Plumbagin, NDGA, and shikonin had no effect of the MIC on the tested antibiotics against the AcrB null strain, which confirmed that the observed synergism for the wild-type cells is specific to AcrB inhibition (Table 4). Quercetin and mangiferin failed to synergize with any of the antibiotics tested (results not shown). We also tested the ability of the natural products to synergize with fluoroquinolones such as levofloxacin and ciprofloxacin, but failed to observe any synergism with these antibiotics.
Table 3.
Minimum inhibitory concentration in the absence or presence of PAβN and natural products showing activity of natural products with different antibiotics and TPP against the drug-resistant (wild-type) strain of Escherichia coli
| MIC (mg/L) |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PAβN (mg/L) |
Plumbagin (mg/L) |
NDGA (mg/L) |
Shikonin (mg/L) |
|||||||||||||
| 0 | 50 | 100 | 200 | 0 | 16 | 32 | 64 | 0 | 64 | 128 | 256 | 0 | 64 | 128 | 256 | |
| Erythromycin | 256 | 128 | 64 | 32 | 256 | 256 | 128 | 64 | 256 | 256 | 256 | 64 | 256 | 256 | 256 | 256 |
| Chloramphenicol | 4 | 2 | 2 | 2 | 4 | 4 | 2 | 2 | 4 | 4 | 2 | 1 | 4 | 4 | 4 | 4 |
| Novobiocin | 512 | 256 | 128 | 128 | 512 | 512 | 512 | 512 | 512 | 512 | 512 | 64 | 512 | 512 | 512 | 512 |
| TPP | 1024 | 512 | 512 | 512 | 1024 | 512 | 512 | 256 | 1024 | 1024 | 128 | 64 | 1024 | 512 | 512 | 64 |
| Tetracycline | 1 | 1 | 1 | 0.5 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 0.25 | 1 | 1 | 1 | 1 |
| Nalidixic acid | 2 | 0.5 | 0.5 | 0.5 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
Table 4.
Minimum inhibitory concentration in the absence or presence of PAβN and natural products showing activity of natural products with different antibiotics and TPP against the drug-sensitive (ΔAcrB) strain of Escherichia coli
| MIC (mg/L) |
|||||
|---|---|---|---|---|---|
| No drug | PAβN (50 mg/L) | Plumbagin (10 mg/L) | NDGA (25 mg/L) | Shikonin (256 mg/L) | |
| Erythromycin | 16 | 16 | 16 | 16 | 16 |
| Chloramphenicol | 1 | 1 | 1 | 1 | 1 |
| Novobiocin | 16 | 16 | 16 | 16 | 16 |
| TPP | 16 | 16 | 16 | 16 | 16 |
| Tetracycline | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 |
| Nalidixic acid | 1 | 1 | 1 | 1 | 1 |
The fact that quercetin was unable to reverse the resistance of AcrAB-TolC-expressing cells against the antibiotics tested was unexpected as our docking data indicated that quercetin should be the best inhibitor of AcrB after PAβN. Quercetin has also been identified as a high-affinity substrate for TtgR the transcriptional repressor of TtgABC a major efflux pump of Pseudomonas putida (Alguel et al. 2007). The result obtained for the checkerboard assays can be interpreted in two ways, (1) quercetin could be a substrate, with no EPI activity or, (2) quercetin is unable to overcome the membrane permeability barrier of Gram-negative organisms and hence cannot access AcrB.
Further investigation would be needed to confirm which of the explanations apply. It is also worth to note that PAβN cannot reverse drug resistance for all drugs and has, for instance, no effect on efflux pump-mediated resistance to the dye ethidium (Lomovskaya et al. 2001). It also seems that similar to PAβN (Lomovskaya and Bostian 2006) the natural products act as substrates that compete with antibiotics and in this way reverse drug resistance. They are thus not trapping the efflux pump in an inactive conformation, but rather acting as high-affinity substrate inhibitors and would also explain why many inhibitors only synergize with some antibiotics.
The natural products do not permeabilize the outer membrane
In order to rule out false positives, we subsequently verified that the observed reduction in MIC in the presence of the natural products was not a result of nonspecific mechanisms such as increased membrane permeability. This was done in two ways: firstly by ascertaining that the natural products did not potentiate the activities of antibiotics that are not effluxed and secondly by determining the outer membrane permeability in the presence of the natural products using nicrocefin (Lomovskaya et al. 2001). Rifampicin is not effluxed by AcrB (MIC for wild type and ΔAcrB strains of 16 mg/L). We therefore determined the effect of the natural products at the maximum concentrations used in Table 1 on the MIC of rifampicin. With the exception of PAβN, known for its outer membrane permeability properties where the MIC was reduced from 16 mg/L to 0.5 mg/L, none of the natural products affected the MIC of rifampicin (results not shown).
The outer membrane permeabilizing effect of the natural products was assessed by examining the rate of hydrolysis of a chromogenic β-lactam, nitrocefin by intact cells of E. coli. Hydrolysis of nitrocefin by the β-lactamase releases a colored compound that can be measured at 490 nm. The rate of nitrocefin hydrolysis is limited by the rate of diffusion across the outer membrane, hence an increased rate of hydrolysis of nitrocefin would be indicative of outer membrane permeabilization (Lomovskaya et al. 2001). The effect of the natural products on the rate of nitrocefin hydrolysis was determined at concentrations, where they did not interfere with the A490 measurements. We observed no increase in the rate of nitrocefin hydrolysis in the presence of the natural products. We could therefore conclude that the synergism with known antibiotics observed was not due to secondary mechanisms such as membrane permeabilization (Fig. 4).
Figure 4.
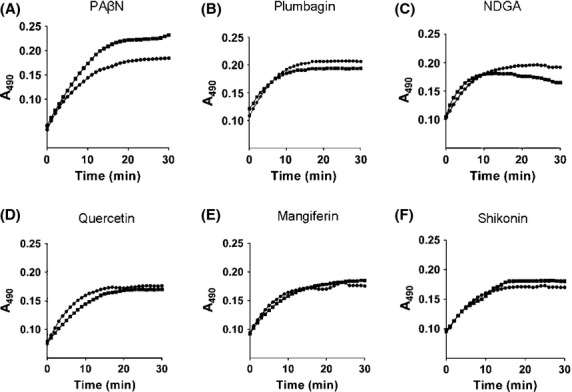
The phytochemicals do not permeabilize the outer membrane. The permeabilizing activity of compounds was measured as an increase in initial rates of nitrocefin hydrolysis by Escherichia coli wild-type cells treated with 10 μmol/L CCCP. Dots represent no treatment and squares represent treatment with compounds at (A) PAβN at 128 μg/mL, (B) plumbagin at 100 μg/mL, (C) NDGA at 100 μg/mL, (D) quercetin at 100 μg/mL, (E) mangiferin at 500 μg/mL, and (F) shikonin at 250 μg/mL. Nitrocefin hydrolysis was monitored over time by monitoring the increase in absorbance at 490 nm. Representative fluorescent traces are shown for experiments with different cells batches done on three different days.
The natural products inhibit drug transport
An EPI must also be able to reduce the level of extrusion of efflux pump substrates (Lomovskaya et al. 2001). We used the inhibition of Nile Red efflux to determine the ability of the natural products to reduce efflux pump activity (Bohnert et al. 2010). Nile Red is only weakly fluorescent in aqueous solutions, but undergoes a significant increase in fluorescent quantum yield when in nonpolar environments such as the cell membrane. Cells were preloaded with Nile Red in the absence or presence of the natural products. Once the cells were energized by the addition of glucose, the efflux of Nile Red could be observed as a drop in fluorescence. All the compounds tested were able to inhibit Nile Red efflux to an extent (Fig. 5). Plumbagin was the most efficient and complete inhibition of Nile Red efflux was observed using 50 μmol/L of plumbagin. At much higher concentrations (500 μmol/L), mangiferin was also able to inhibit the efflux, presumably through competition for the binding site, which would again be supportive of mangiferin being a low-affinity substrate rather than an inhibitor (Fig. 5).
Figure 5.
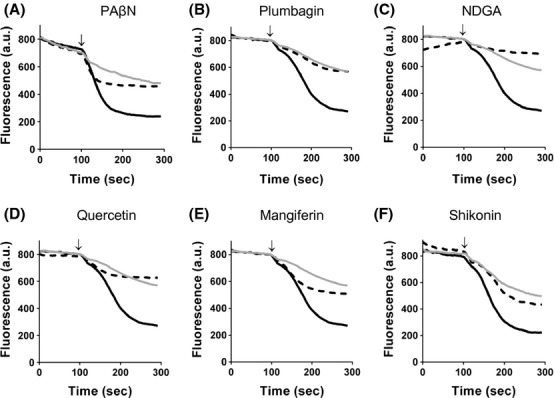
Inhibition of Nile Red efflux by the natural products. Wild-type-resistant cells (black line), ΔAcrB drug-sensitive cells (gray line) or wild-type cells in the presence of the natural products (dashed line) with (A) PAβN at 200 μmol/L, (B) plumbagin at 50 μmol/L, (C) NDGA at 100 μmol/L, (D) quercetin at 200 μmol/L, (E) mangiferin at 500 μmol/L, and (f) shikonin at 25 μmol/L were preloaded with Nile Red before the start of fluorescence measurements. Efflux was triggered at 100 sec by the addition of 50 mmol/L glucose (indicated by arrow). Representative fluorescent traces are shown for experiments with different cells batches done on three different days.
Conclusion
The obtained results of bioassays showed a good correlation with the molecular docking analysis, indicating that in silico screening could be a powerful tool to preselect compounds with potential EPI activity. To our knowledge, this is the first report of potential EPIs being identified with in silico screening and verified by bioassays.
Plumbagin, NDGA, and shikonin were able to increase susceptibility to antibiotics and toxic compounds and were also the most efficient in inhibiting AcrB-mediated substrate efflux; hence, they may be good lead compounds for a future drug discovery program. Given the similarity between AcrAB-TolC and the MexAB-OprM drug efflux system from pathogenic P. aeruginosa, future studies on MexAB-OprM using in silico screening and the methods described here have a high potential to deliver tangible outcomes in stemming the tide of drug-resistant infections.
Acknowledgments
This work was supported by the Royal Society (grant number 45605 to H. V. and UF110479 to T. R.) and the University of South Australia. TO is the recipient of a Cambridge Trust scholarship, an Adam Glinsman award and a Faculty for the Future Fellowship from the Schlumberger Foundation. R. M. is a recipient of an Australian Postgraduate Award.
Conflict of Interest
None declared.
References
- Abreu AC, McBain AJ. Simoes M. Plants as sources of new antimicrobials and resistance-modifying agents. Nat. Prod. Rep. 2012;29:1007–1021. doi: 10.1039/c2np20035j. [DOI] [PubMed] [Google Scholar]
- Alguel Y, Meng C, Teran W, Krell T, Ramos JL, Gallegos MT, et al. Crystal structures of multidrug binding protein TtgR in complex with antibiotics and plant antimicrobials. J. Mol. Biol. 2007;369:829–840. doi: 10.1016/j.jmb.2007.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Karablieh N, Weingart H. Ullrich MS. Genetic exchange of multidrug efflux pumps among two enterobacterial species with distinctive ecological Niches. Int. J. Mol. Sci. 2009;10:629–645. doi: 10.3390/ijms10020629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral L, Spengler G, Martins A, Armada A, Handzlik J, Kiec-Kononowicz K, et al. Inhibitors of bacterial efflux pumps that also inhibit efflux pumps of cancer cells. Anticancer Res. 2012;32:2947–2957. [PubMed] [Google Scholar]
- Andújar I, Recio M, Giner R. Ríos J. Traditional chinese medicine remedy to jury: the pharmacological basis for the use of shikonin as an anticancer therapy. Curr. Med. Chem. 2013;20:2892–2898. doi: 10.2174/09298673113209990008. [DOI] [PubMed] [Google Scholar]
- Annapoorani A, Umamageswaran V, Parameswari R, Pandian SK. Ravi AV. Computational discovery of putative quorum sensing inhibitors against LasR and RhlR receptor proteins of Pseudomonas aeruginosa. J. Comput. Aided Mol. Des. 2012;26:1067–1077. doi: 10.1007/s10822-012-9599-1. [DOI] [PubMed] [Google Scholar]
- Baugh S, Ekanayaka AS, Piddock LJ. Webber MA. Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J. Antimicrob. Chemother. 2012;67:2409–2417. doi: 10.1093/jac/dks228. [DOI] [PubMed] [Google Scholar]
- Baugh S, Phillips CR, Ekanayaka AS, Piddock LJ. Webber MA. Inhibition of multidrug efflux as a strategy to prevent biofilm formation. J. Antimicrob. Chemother. 2014;69:673–681. doi: 10.1093/jac/dkt420. [DOI] [PubMed] [Google Scholar]
- Bohnert JA, Karamian B. Nikaido H. Optimized Nile Red efflux assay of AcrAB-TolC multidrug efflux system shows competition between substrates. Antimicrob. Agents Chemother. 2010;54:3770–3775. doi: 10.1128/AAC.00620-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K, Courvalin P, Dantas G, Davies J, Eisenstein B, Huovinen P, et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011;9:894–896. doi: 10.1038/nrmicro2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MA. Shlaes D. Fix the antibiotics pipeline. Nature. 2011;472:32. doi: 10.1038/472032a. [DOI] [PubMed] [Google Scholar]
- Du D, Venter H, Pos KM. Luisi BF. The Machinery and Mechanism of Multidrug Efflux in Gram-negative Bacteria. In: Yu EW, Zhang Q, Melissa H, Brown MH, editors; Microbial efflux pumps: current research. Poole, UK: Caister Academic Press; 2013. pp. 35–48. [Google Scholar]
- Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, et al. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eicher T, Cha HJ, Seeger MA, Brandstatter L, El-Delik J, Bohnert JA, et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl Acad. Sci. USA. 2012;109:5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootz TD. The global problem of antibiotic resistance. Crit. Rev. Immunol. 2010;30:79–93. doi: 10.1615/critrevimmunol.v30.i1.60. [DOI] [PubMed] [Google Scholar]
- Haghbeen K, Pourmolaei S, Mareftjo MJ, Mousavi A, Akbari Noghabi K, Hosseini Shirazi F, et al. Detailed investigations on the solid cell culture and antimicrobial activities of the Iranian Arnebia euchroma. J. Biomed. Biotechnol. 2011;2011:165852. doi: 10.1155/2011/165852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetenyi C. van der Spoel D. Toward prediction of functional protein pockets using blind docking and pocket search algorithms. Protein Sci. 2011;20:880–893. doi: 10.1002/pro.618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung LW, Kim HB, Murakami S, Gupta G, Kim CY. Terwilliger TC. Crystal structure of AcrB complexed with linezolid at 3.5 A resolution. J. Struct. Funct. Genomics. 2013;14:71–75. doi: 10.1007/s10969-013-9154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtesi C, Ball AR, Huang YY, Jachak SM, Vera DM, Khondkar P, et al. Microbial efflux systems and inhibitors: approaches to drug discovery and the challenge of clinical implementation. Open Microbiol. J. 2013;7:34–52. doi: 10.2174/1874285801307010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy G, Tikhonova EB. Zgurskaya HI. Fitting periplasmic membrane fusion proteins to inner membrane transporters: mutations that enable Escherichia coli AcrA to function with Pseudomonas aeruginosa MexB. J. Bacteriol. 2008;190:691–698. doi: 10.1128/JB.01276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuete V, Alibert-Franco S, Eyong KO, Ngameni B, Folefoc GN, Nguemeving JR, et al. Antibacterial activity of some natural products against bacteria expressing a multidrug-resistant phenotype. Int. J. Antimicrob. Agents. 2011;37:156–161. doi: 10.1016/j.ijantimicag.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Lewis K. In search of natural substrate and inhibitors ofMDR pumps. J. Mol. Microbiol. Biotechnol. 2001;3:247–254. [PubMed] [Google Scholar]
- Lomovskaya O. Bostian KA. Practical applications and feasibility of efflux pump inhibitors in the clinic–a vision for applied use. Biochem. Pharmacol. 2006;71:910–918. doi: 10.1016/j.bcp.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 2001;45:105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya O, Zgurskaya HI, Totrov M. Watkins WJ. Waltzing transporters and ‘the dance macabre’ between humans and bacteria. Nat. Rev. Drug Discov. 2006;6:56–65. doi: 10.1038/nrd2200. [DOI] [PubMed] [Google Scholar]
- Marquez B. Bacterial efflux systems and efflux pumps inhibitors. Biochimie. 2005;87:1137–1147. doi: 10.1016/j.biochi.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Martinez JL, Sánchez MB, Martínez-Solano L, Hernandez A, Garmendia L, Fajardo A, et al. Functional role of bacterial multidrug efflux pumps in microbial natural ecosystems. FEMS Microbiol. Rev. 2009;33:430–449. doi: 10.1111/j.1574-6976.2008.00157.x. [DOI] [PubMed] [Google Scholar]
- Martins S, Teixeira JA. Mussatto SI. Solid-state fermentation as a strategy to improve the bioactive compounds recovery from Larrea tridentata leaves. Appl. Biochem. Biotechnol. 2013;171:1227–1239. doi: 10.1007/s12010-013-0222-2. [DOI] [PubMed] [Google Scholar]
- Matkowski A, Kuss P, Goralska E. Wozniak D. Mangiferin – a bioactive xanthonoid, not only from mango and not just antioxidant. Mini. Rev. Med. Chem. 2013;13:439–455. [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Nakashima R, Yamashita E, Matsumoto T. Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- Musa MA, Cooperwood JS, Khan MO. Rahman T. In-vitro antiproliferative activity of benzopyranone derivatives in comparison with standard chemotherapeutic drugs. Arch. Pharm. (Weinheim) 2011;344:102–110. doi: 10.1002/ardp.201000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima R, Sakurai K, Yamasaki S, Nishino K. Yamaguchi A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature. 2011;480:565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- Nakashima R, Sakurai K, Yamasaki S, Hayashi K, Nagata C, Hoshino K, et al. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500:102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- Ndi CP, Semple SJ, Griesser HJ. Barton MD. Antimicrobial activity of some Australian plant species from the genus Eremophila. J. Basic Microbiol. 2007;47:158–164. doi: 10.1002/jobm.200610262. [DOI] [PubMed] [Google Scholar]
- Ohene-Agyei T, Lea JD. Venter H. Mutations in MexB that affect the efflux of antibiotics with cytoplasmic targets. FEMS Microbiol. Lett. 2012;333:20–27. doi: 10.1111/j.1574-6968.2012.02594.x. [DOI] [PubMed] [Google Scholar]
- Orhan G, Bayram A, Zer Y. Balci I. Synergy tests by E test and checkerboard methods of antimicrobial combinations against Brucella melitensis. J. Clin. Microbiol. 2005;43:140–143. doi: 10.1128/JCM.43.1.140-143.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 2006a;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock LJ. Multidrug-resistance efflux pumps? not just for resistance. Nat. Rev. Microbiol. 2006b;4:629–636. doi: 10.1038/nrmicro1464. [DOI] [PubMed] [Google Scholar]
- Piddock LJ. The crisis of no new antibiotics—what is the way forward? Lancet. Infect. Dis. 2012;12:249–253. doi: 10.1016/S1473-3099(11)70316-4. [DOI] [PubMed] [Google Scholar]
- Poole K. Multidrug efflux pumps and antimicrobial resistance in Pseudomonas aeruginosa and related organisms. J. Mol. Microbiol. Biotechnol. 2001;3:255–264. [PubMed] [Google Scholar]
- Ricci V, Tzakas P, Buckley A, Coldham NC. Piddock LJ. Ciprofloxacin-resistant Salmonella enterica serovar Typhimurium strains are difficult to select in the absence of AcrB and TolC. Antimicrob. Agents Chemother. 2006;50:38–42. doi: 10.1128/AAC.50.1.38-42.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross I. Medicinal plants of the world. New York City, NJ: Humana Press; 2005. [Google Scholar]
- Ruggerone P, Murakami S, Pos KM. Vargiu AV. RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr. Top. Med. Chem. 2013;13:3079–3100. doi: 10.2174/15680266113136660220. [DOI] [PubMed] [Google Scholar]
- Savikin K, Menkovic N, Zdunic G, Stevic T, Radanovic D. Jankovic T. Antimicrobial activity of Gentiana lutea L. extracts. Z. Naturforsch. C. 2009;64:339–342. doi: 10.1515/znc-2009-5-606. [DOI] [PubMed] [Google Scholar]
- Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K. Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- Shen CC, Syu WJ, Li SY, Lin CH, Lee GH. Sun CM. Antimicrobial activities of naphthazarins from Arnebia euchroma. J. Nat. Prod. 2002;65:1857–1862. doi: 10.1021/np010599w. [DOI] [PubMed] [Google Scholar]
- Shukla S, Wu CP, Nandigama K. Ambudkar SV. The naphthoquinones, vitamin K3 and its structural analogue plumbagin, are substrates of the multidrug resistance linked ATP binding cassette drug transporter ABCG2. Mol. Cancer Ther. 2007;6:3279–3286. doi: 10.1158/1535-7163.MCT-07-0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavri M, Piddock LJ. Gibbons S. Bacterial efflux pump inhibitors from natural sources. J. Antimicrob. Chemother. 2007;59:1247–1260. doi: 10.1093/jac/dkl460. [DOI] [PubMed] [Google Scholar]
- Tegos G, Stermitz FR, Lomovskaya O. Lewis K. Multidrug pump inhibitors uncover remarkable activity of plant antimicrobials. Antimicrob. Agents Chemother. 2002;46:3133–3141. doi: 10.1128/AAC.46.10.3133-3141.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse T. Use of common Chinese herbs in the treatment of psoriasis. Clin. Exp. Dermatol. 2003;28:469–475. doi: 10.1046/j.1365-2230.2003.01322.x. [DOI] [PubMed] [Google Scholar]
- Vargiu AV. Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc. Natl Acad. Sci. USA. 2012;109:20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter H, Shilling RA, Velamakanni S, Balakrishnan L. van Veen HW. An ABC transporter with a secondary-active multidrug translocator domain. Nature. 2003;426:866–870. doi: 10.1038/nature02173. [DOI] [PubMed] [Google Scholar]
- Venter H, Shahi S, Balakrishnan L, Velamakanni S, Bapna A, Woebking B, et al. Similarities between ATP-dependent and ion-coupled multidrug transporters. Biochem. Soc. Trans. 2005;33:1008–1011. doi: 10.1042/BST20051008. [DOI] [PubMed] [Google Scholar]
- Welch A, Awah CU, Jing S, van Veen HW. Venter H. Promiscuous partnering and independent activity of MexB, the multidrug transporter protein from Pseudomonas aeruginosa. Biochem. J. 2010;430:355–364. doi: 10.1042/BJ20091860. [DOI] [PubMed] [Google Scholar]
- Wise R. The urgent need for new antibacterial agents. J. Antimicrob. Chemother. 2011;66:1939–1940. doi: 10.1093/jac/dkr261. [DOI] [PubMed] [Google Scholar]
- Zhang L, Li X-Z. Poole K. Multiple antibiotic resistance in Stenotrophomonas maltophilia: involvement of a multidrug efflux system. Antimicrob. Agents Chemother. 2000;44:287–293. doi: 10.1128/aac.44.2.287-293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Lambert G, Liao D, Kim H, Robin K, Tung CK, et al. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science. 2011;333:1764–1767. doi: 10.1126/science.1208747. [DOI] [PubMed] [Google Scholar]