Abstract
The circulating recombinant form (CRF) 07_BC is the most prevalent HIV-1 strain among injection drug users (IDUs) in Taiwan. It contains a 7 amino-acid deletion in its p6gag. We conducted a cohort study to compare viral loads and CD4 cell count changes between patients infected with subtype B and CRF07_BC and to elucidate its mechanism. Twenty-one patients infected with CRF07_BC and 59 patients with subtype B were selected from a cohort of 667 HIV-1/AIDS patients whom have been followed up for 3 years. Generalized estimated equation was used to analyze their clinical data and the results showed that patients infected with CRF07_BC had significantly lower viral loads (about 58,000 copies per ml less) than patients with subtype B infection (p = 0.002). The replicative capacity of nine CRF07_BC and four subtype B isolates were compared and the results showed that the former had significantly lower replicative capacity than the latter although all of them were CCR5- tropic and non-syncytium inducing viruses. An HIV-1-NL4-3 mutant virus which contains a 7 amino-acid deletion in p6gag (designated as 7d virus) was generated and its live cycle was investigated. The results showed that 7d virus had significantly lower replication capacity, poorer protease-mediated processing and viral proteins production. Electron microscopic examination of cells infected with wild-type or 7d virus demonstrated that the 7d virus had poorer and slower viral maturation processes: more viruses attached to the cell membrane and higher proportion of immature virions outside the cells. The interaction between p6gag and Alix protein was less efficient in cells infected with 7d virus. In conclusion, patients infected with CRF07_BC had significantly lower viral loads than patients infected with subtype B and it may due to the deletion of 7 amino acids which overlaps with Alix protein-binding domain of the p6gag.
Introduction
Understanding the factors affecting AIDS disease progression is very important for clinical management and counseling. It has been reported that HIV-1 patients infected with different subtypes have different rates of disease progression [1]–[3]. In Kenya, patients infected with subtype D recombinant virus had significantly faster disease progression than patients infected with subtype A in spite that they had similar viral loads [2]. A meta-analysis indicated that the trend of disease progression among different HIV-1 subtype in a descending order was subtype C>D>AE>G>A [4].
By the end of 2013, 27,366 individuals (including 891 foreigners) were reported as infected with HIV-1 by the Taiwan's Centers for Disease Control (CDC). Risk factor analyses showed that more than 50% of the HIV-1/AIDS patients were men who have sex with men (MSM) and about 25% were injection drug users (IDUs) [5]. In terms of subtype distribution, subtype B, CRF01_AE and CRF07_BC were predominant in MSM, heterosexuals, and IDUs respectively [6]–[10]. There are an estimated 60,000 to 100,000 IDUs in Taiwan and about 15% of them are infected with HIV-1 [9]. Since 83% of those HIV-1-infected IDUs may be infected with CRF07_BC [10], the numbers of IDUs in Taiwan who may be infected with CRF07_BC are between 7,470–12,450. Therefore, it is very important to understand the natural history and disease progression of CRF07_BC infection.
Previously, our full-length sequencing results indicated that all the Taiwanese CRF07_BC strains contain a signature 7 amino-acid deletion in p6gag [8]. The p6gag contains two motifs- PTAP and YPXnL (X can vary in sequence) which are important for viral assembling and budding. YPXnL motif is located between amino acid residues 36 and 44 at its C-terminal region and it interacts with AIP1 (apoptosis-linked gene 2-interacting protein, also known as Alix) [11], [12]. Since a 7 amino-acid deletion signature is overlapping with this motif, we hypothesized that such deletion may affect the viral life cycle, especially during virus assembly. In this study, 21 patients infected with CRF07_BC and 59 patients with subtype B were selected from a cohort of 667 HIV-1/AIDS patients whom have been followed up for more than 3 years. A GEE model was used to analyze multiple time points data and demonstrated that patients with CRF07_BC infection had significantly lower viral loads than patients with subtype B infection and it was mainly associated with the viral subtypes. Subsequently, we used both clinical isolates and molecular clones with specific deletion of those 7 amino-acid from p6gag to elucidate the mechanism and the results indicated that the lower replication capacity, poorer protease-mediated processing and viral proteins production of CRF07_BC were due to a 7 amino-acid deletion in its p6gag domain.
Methods
Patient Cohort and Study Design
In 2010, we established Taiwan HIV-1 Observational Database (TwHOD) to study the natural history and clinical aspects of Taiwanese HIV-1/AIDS patients. The TwHOD is a collaborative cohort study that involves the following hospitals located in the northern region (Taipei Veterans' General Hospital, Far Eastern Memorial Hospital, and Taipei City Hospital), central region (Taichung Veterans' General Hospital) and southern region (Kaohsiung Veterans' General Hospital) of Taiwan. The study was approved by the Institutional Review Boards (IRB) of all the participating hospitals. Written informed consent was obtained from patients who agreed to participate in this study. The procedure of data collection was similar to that reported by TREAT Asia HIV-1 Observational Database [13]. By the end of 2012, 667 HIV-1/AIDS patients enrolled in this study and the HIV-1 subtypes of 272 patients were determined. For the nested case control study, we selected 21 male treatment naïve patients infected with CRF07_BC and matched them with 59 subtype B-infected patients by gender, age and risk factor. Eleven patients who were infected with CRF07_BC were excluded due to gender (3 were female patients), previous antiretroviral therapy (2 patients) or lack of CD4 cell count or viral loads data (6 patients).
Subtyping and Isolation of HIV-1 viruses
HIV-1 subtypes were determined as described previously [14]. Nine CRF07_BC and 4 subtype B strains from treatment-naïve IDUs were obtained by using standard peripheral blood mononuclear cells (PBMCs) co-culture methods [15]. We enrolled treatment naïve HIV-1-infected IDU patients with CD4 cell counts of more than 500 cells/mm3. Initially, we selected 10 treatment naïve patients with CRF07_BC and 5 patients with subtype B infection for the virus isolation experiments. Eventually, 9 CRF07_BC and 4 subtype B isolates were obtained.
Determination of Co-receptor Usage and Syncytium Inducing (SI) Ability
Chemokine co-receptor usage and the SI ability were determined as described previously [16], [17]. In terms of sequence analysis of V3 region of env gene, co-receptor usage was determined as described previously [18]–[20].
Generation of Recombinant Viruses
To generate HIV-1 infectious recombinant viruses with or without a 7 amino-acid deletion at its p6gag, MT2 cells were co-transfected with a linear marker plasmid pNL43HIVΔPR.RTBstEIInef-GFP and one of two linear plasmids pGEM-NCRT or pGEM-NCRT-7d using an electroporation method [21]. Briefly, a 1,652-bp fragment encompassing the coding regions of HIV-1 nucleocapsid protein-p6-protease-reverse transcriptase was amplified from plasmid pNL4-3 (corresponding to nucleotides 1827 to 3649 of the HIV-1 NL4-3 sequence) using PCR. It was sub-cloned into pGEM-T vector (Promega, Madison, Wisconsin) to generate a plasmid designated as pGEM-NCRT. Subsequently, we used PCR-based site-directed mutagenesis to delete nucleotide sequences 2220–2240 of HIV-1 NL4-3 to generate a plasmid containing a 7 amino-acid deletion in its p6gag (pGEM-NCRT-7d).
Growth Kinetic Assay
Growth kinetic of primary isolates and infectious recombinant viruses were measured in PBMCs and MT2 cells, respectively and described with some modifications [17]. Cells were plated in 24-well plates at 106 cells/well in 1 ml of RPMI 1640 medium, and 3,000 50% tissue culture infective dose (TCID50) of HIV-1 viruses were added. The cultures were split every 3-4 days by replacing 50% of the culture with the same volume of fresh medium and p24 quantified as a measure of ongoing virus replication. Growth kinetic of infectious recombinant viruses was measured in MT2 cells, as described elsewhere [17] with modification. A total of 2×106 cells were infected with 2,000 TCID50 of viruses. After incubation for 2 hours at 37°C, cells were washed twice with phosphate-buffered saline (PBS) and resuspended in RPMI 1640 medium. Triplicate cultures were tested, and viral growth was determined by HIV-1 p24 levels on days 2, 4, 6, 8, 10, 12 and 14. HIV-1 p24 antigen determined by enzyme-linked immunosorbent assay (ELISA) (PerkinElmer, Waltham, USA) was considered an indicator of virus replication.
Western Blot (WB) Assay
The details of WB have been described previously [22]. HIV-1 Gag proteins were detected by anti-p24gag mouse monoclonal antibody (clone 183-H12-5C) [23]. RT was detected by anti-RT mouse monoclonal antibody [24]. Protease was detected by anti-HIV protease mouse monoclonal antibody (Abcam). The bound antibody was detected by horseradish peroxidase-conjugated anti-mouse immunoglobulin secondary antibody (Amersham Corp.). Image J software (version 1.47) was used to analyze the intensity of reactive bands in WB.
Electron microscopy
Infected MAGIC-5 cells [25] were fixed in 2.5% glutaraldehyde-0.2M sodium cacodylate solution overnight at 4°C, and then fixed with 1% OsO4 in PBS for 1.5 hours. Specimens were then dehydrated in graded ethanol solution and embedded in Epon. Ultrathin sections were stained with uranyl acetate and lead citrate, and images were obtained using Jeol JEM-2000EXII transmission electron microscope (TEM).
Indirect Immunofluorescent Antibody (IFA) Staining and Total Internal Reflection Fluorescence (TIRF)
To detect and quantify the interaction between p6gag and Alix, IFA staining with TIRF and super-resolution fluorescence localization imaging methods were used (Leica SR GSD) [26], [27] for immunostaining, Gag was detected by anti-p24Gag mouse monoclonal antibody. Alix was detected by anti-Alix rabbit polyclonal antibody. The secondary antibodies were anti-mouse and anti-rabbit fluorescence (Alexa 488 and Alexa 647)-conjugated antibodies. Phosphate buffered saline containing 100 mM β-mercaptoethylamine (MEA) was used for SR fluorescence localization imaging. Imaging fields were magnified using a 100× oil objective (Leica) with a 1.47 numerical aperture and 1.6× optical magnification. The penetration depth of the excitation laser source for TIRF and super-resolution imaging was 200 nm. TIRF fluorescence image stacks consisting of over 30,000 frames were used to calculate SR fluorescence images. A two-dimensional spatial histogram map in each fluorescence channel was calculated using the SR images with an effective pixel size of 20 nm. The co-localization coefficients of two proteins were quantified by a combination of Manders analysis and two-dimensional spatial histogram maps of two fluorescence channels, with the fluorescence background removed during intensity-based co-localization analysis.
Statistical Analysis
A multivariate linear generalized estimating equations (GEE) model was performed to identify factors associated with the changes of CD4 cell count or viral loads. One-way ANOVA and Tukey's post hoc test were used to compare the p24 antigen levels between different subtypes or infectious recombinant viruses. SAS statistic software (SAS version 9.1; SAS Institute, Cary, North Carolina, USA) was used with significance level set at p<0.05.
Results
Study Population Characteristics
In total, 667 HIV-1/AIDS patients were recruited in this study. The median follow-up period was 46 months. In terms of socio-demographic variables, the age of the participants ranged from 15 to 81 years at diagnosis of HIV-1 infection. The median age was 31 years and 94.0% were men. The majority of the patients were MSM (388/600 [64.7%]) and 22.5% were IDUs. The results of HBsAg and HCV antibody tests were available for 301 (45.1%) and 388 (58.2%) patients, respectively. Seventy-one (23.6%) patients were HBsAg positive and 121 patients (31.2%) had anti-HCV antibodies. There were 466 patients (69.9%) under ART (Table 1).
Table 1. Demographic data, risk factors and clinical characteristics of HIV-1-infected patients recruited in the study.
| HIV-1 infected population | ||||||
| Male (%) | Female (%) | Total (%) | ||||
| (N = 627) | (N = 40) | (N = 667) | ||||
| Age (yrs) | n = 612 | n = 37 | n = 649 | |||
| 15–29 | 287 | (46.9) | 12 | (32.4) | 299 | (49.2) |
| 30–50 | 283 | (46.2) | 19 | (51.4) | 302 | (42.5) |
| ≥50 | 42 | (6.9) | 6 | (16.2) | 48 | (8.3) |
| Mode of infection | n = 561 | n = 39 | n = 600 | |||
| Homosexual contact | 388 | (69.2) | 0 | (0) | 388 | (64.7) |
| Heterosexual contact | 51 | (9.1) | 20 | (51.3) | 71 | (11.8) |
| Injecting drug use | 116 | (20.7) | 19 | (48.7) | 135 | (22.5) |
| Receipt of blood products | 6 | (1.1) | 0 | (0) | 6 | (1.0) |
| HBV coinfection | n = 280 | n = 21 | n = 301 | |||
| HBsAg, positive | 68 | (24.3) | 3 | (14.3) | 71 | (23.6) |
| HCV coinfection | n = 361 | n = 27 | n = 388 | |||
| HCVAb, positive | 105 | (29.1) | 16 | (59.3) | 121 | (31.2) |
| Antiretroviral treatment | n = 627 | n = 40 | n = 667 | |||
| HAART | 444 | (70.8) | 22 | (55.0) | 466 | (69.9) |
| HIV-1 subtype | n = 262 | n = 10 | n = 272 | |||
| B | 215 | (82.1) | 4 | (40.0) | 219 | (80.5) |
| C | 3 | (1.1) | 0 | (0) | 3 | (1.1) |
| CRF01_AE | 14 | (5.3) | 3 | (30.0) | 17 | (6.3) |
| CRF07_BC | 29 | (11.1) | 3 | (30.0) | 32 | (11.8) |
| CRF08_BC | 1 | (0.4) | 0 | (0) | 1 | (0.4) |
Among 272 patients whose subtypes have been determined using nested multiplex PCR, 80.5% were subtype B, 11.8% CRF07_BC, and 6.3% CRF01_AE. Three (1.1%) patients infected with subtype C and one patient with CRF08_BC (0.4%) (Table 1). Subtype B was predominant in MSM (93.9%) and heterosexuals (69.9%). 74.3% of IDUs were infected with CRF07_BC. Notably, two MSMs were infected with CRF07_BC and they denied using intravenous drugs before. In addition, we found that there was one MSM infected with CRF08_BC and three MSM infected with subtype C (Table 2).
Table 2. Distribution of HIV-1 subtypes and circulating recombinant forms (CRFs) in different groups of patients recruited in this cohort study.
| Male | ||||||
| Subtype/CRF | Heterosexual | MSM | IDUs | Others | Femalea | Total |
| B | 16 (69.6) | 154 (93.9) | 9 (25.7) | 36 (90.0) | 4 (40) | 219 (80.5) |
| C | 0 (0) | 3 (1.8) | 0 (0) | 0 (0) | 0 (0) | 3 (1.1) |
| CRF01_AE | 7 (30.4) | 4 (2.4) | 0 (0) | 3 (7.5) | 3 (30) | 17 (6.3) |
| CRF07_BC | 0 (0) | 2 (1.2) | 26 (74.3) | 1 (2.5) | 3 (30) | 32 (11.8) |
| CRF08_BC | 0 (0) | 1 (0.6) | 0 (0) | 0 (0) | 0 (0) | 1 (0.4) |
| Total | 23 (100) | 164 (100) | 35 (100) | 40 (100) | 10 (100) | 272 (100) |
Includes 4 heterosexual female infected with subtype B, 3 heterosexual female infected with CRF01_AE, and 3 IDUs infected with CRF07_BC.
Factors Affecting Disease Progression
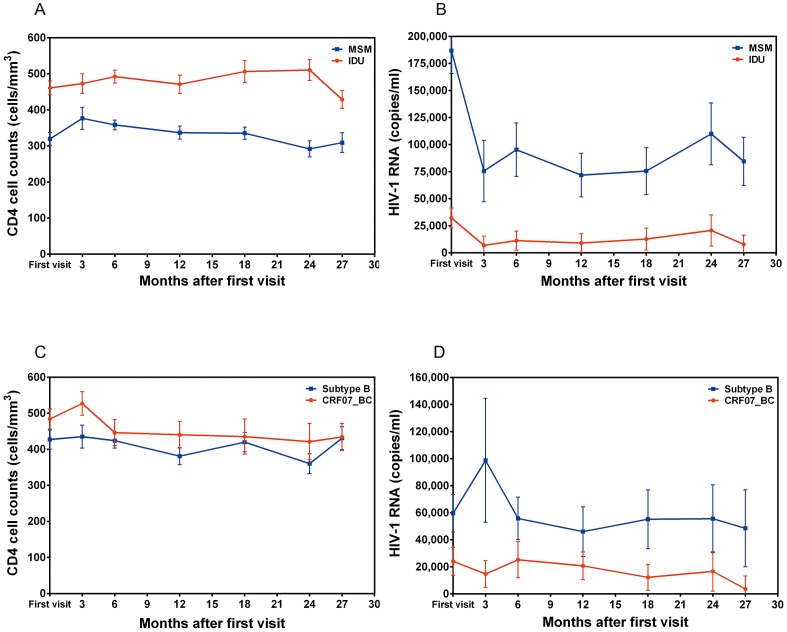
As shown in Figs. 1A and 1B, compared with MSM, IDUs had consistently higher CD4 cell count and lower HIV-1 viral loads over 2.5 years. A GEE model was used to identify factors associated with changes of CD4 cell count or viral loads. The variables which potentially may influence the CD4 cell count or HIV-1 viral loads were considered in this analysis. As shown in Table 3, regardless of whether “mode of infection” was added in the model (multivariate model I vs. model II), factors significantly associated with CD4 cell count changes included older age, subtypes (CRF01_AE and CRF08_BC), higher viral loads and no previous HAART. In contrast, when we analyzed factors associated with changes of viral loads, once we deleted “mode of infection” from the model (multivariate model II), we found that CRF07_BC infection became significantly associated with lower viral loads compared to subtype B infection (Table 4).
Figure 1. Comparisons of the changes of CD4 cell count and HIV-1 viral loads after the first clinical visit among the following four groups of treatment naïve patients.
Men who have sex with men (MSM, 357 patients for CD4 cell count and 371 patients for viral loads analysis) vs. injection drug users (IDUs, 129 patients for CD4 cell count and 128 patients for viral loads analysis) (Fig. 1A and 1B); patients infected with CRF07_BC vs. infected with subtype B (Figs. 1C and 1D). A generalized estimating equation model was used for the analyses.
Table 3. Univariate and multivariate generalized estimating equations models of factors associated with CD4 cell counts.
| Univariate | Multivariate model I | Multivariate model IIa | |||||
| No. | difference | p | difference | p | difference | p | |
| Gender | |||||||
| Male | 578 | ||||||
| Female | 37 | 20.8 | 0.541 | 43.0 | 0.187 | 52.2 | 0.082 |
| Age (yrs) | |||||||
| 15–29 | 271 | ||||||
| 30–50 | 281 | −31.1 | 0.084 | −43.6 | 0.012 | −36.4 | 0.040 |
| ≥50 | 46 | −153.2 | <0.001 | −140.6 | <0.001 | −170.7 | <0.001 |
| Mode of transmission | |||||||
| Homosexual contact | 357 | ||||||
| Heterosexual contact | 70 | −59.8 | 0.032 | −33.9 | 0.228 | - | - |
| Injecting drug use | 129 | 40.7 | 0.066 | 103.3 | 0.001 | - | - |
| Receipt of blood products | 6 | −164.4 | 0.115 | −118.5 | 0.222 | - | - |
| HIV-1 subtype | |||||||
| B | 201 | ||||||
| C | 3 | 85.6 | 0.317 | 68.2 | 0.384 | 70.5 | 0.367 |
| CRF01_AE | 15 | −147.7 | 0.004 | −103.3 | 0.041 | −118.0 | 0.020 |
| CRF07_BC | 31 | 9.9 | 0.746 | −24.4 | 0.510 | 53.1 | 0.084 |
| CRF08_BC | 1 | −225.6 | <0.001 | −132.3 | <0.001 | −135.5 | <0.001 |
| HIV viral loads (copies/ml) | |||||||
| <40 | 225 | ||||||
| 40–400 | 34 | −42.8 | 0.008 | −44.1 | 0.007 | −41.4 | 0.010 |
| 401–10,000 | 93 | −107.1 | <0.001 | −99.0 | <0.001 | −97.7 | <0.001 |
| >10,000 | 109 | −169.9 | <0.001 | −151.2 | <0.001 | −153.6 | <0.001 |
| Received ART | |||||||
| No | 261 | ||||||
| Yes | 354 | 118.3 | <0.001 | 50.0 | 0.005 | 41.6 | 0.020 |
The mode of transmission was removed from multivariate model II.
Table 4. Univariate and multivariate generalized estimating equations model of factors associated with HIV-1 viral loads.
| Univariate | Multivariate model I | Multivariate model IIa | |||||
| No. | difference | p | difference | p | difference | P | |
| Gender | |||||||
| Male | 603 | ||||||
| Female | 38 | −26767.8 | <0.001 | −20826.2 | 0.013 | −26658.9 | <0.001 |
| Age (yrs) | |||||||
| <30 | 286 | ||||||
| 30–50 | 290 | −4936.2 | 0.449 | 1520.8 | 0.823 | −3313.5 | 0.633 |
| ≥50 | 48 | −4747.7 | 0.627 | 2665.1 | 0.802 | 8246.7 | 0.404 |
| Mode of transmission | |||||||
| Homosexual contact | 371 | ||||||
| Heterosexual contact | 70 | −3605.2 | 0.706 | 10465.9 | 0.387 | - | - |
| Injecting drug use | 128 | −22329.5 | <0.001 | −73432.1 | <0.001 | - | - |
| Receipt of blood products | 6 | −7205.5 | 0.752 | −35456.4 | 0.227 | - | - |
| HIV-1 subtype | |||||||
| B | 217 | ||||||
| C | 3 | 1607.7 | 0.940 | 23058.5 | 0.269 | 19596.9 | 0.325 |
| CRF01_AE | 16 | −10620.5 | 0.291 | −43785.9 | 0.004 | −37612.3 | 0.004 |
| CRF07_BC | 30 | −21153.8 | 0.005 | −9074.4 | 0.472 | −58045.6 | <0.001 |
| CRF08_BC | 1 | −7883.3 | 0.086 | −113409.7 | <0.001 | −106777.7 | <0.001 |
| CD4 cell count (cells/mm3) | |||||||
| ≤50 | 22 | ||||||
| 51–250 | 82 | −263241.1 | <0.001 | −240696.2 | <0.001 | −246030.4 | <0.001 |
| 251–500 | 209 | −313911.1 | <0.001 | −290801.3 | <0.001 | −296881.6 | <0.001 |
| >500 | 160 | −334294.9 | <0.001 | −300993.4 | <0.001 | −309618.0 | <0.001 |
| Received ART | |||||||
| No | 270 | ||||||
| Yes | 371 | −97661.2 | <0.001 | −102574.2 | <0.001 | −88088.5 | <0.001 |
The mode of transmission was removed from multivariate model II.
To evaluate the effect of HIV-1 subtypes on CD4 cell count and HIV-1 viral loads, a nested case control study which consisted of 21 patients with CRF07_BC infection and 59 patients with subtype B infection was established. They were all treatment-naïve patients and matched by age and gender. The results showed that there was no significant difference in CD4 cell count (GEE model, p = 0.168) (Fig. 1C). In contrast, patients infected with CRF07_BC had significantly lower viral loads than patients infected with subtype B (GEE model, p = 0.002) (Fig. 1D).
CRF07_BC Isolates had Lower Replication Capacity than the Subtype B Isolates
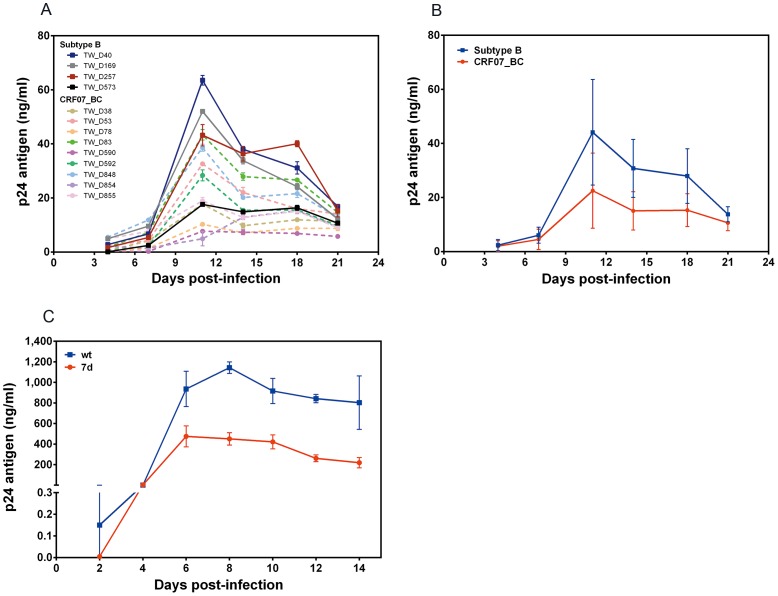
To confirm the findings mentioned above, we used co-culture methods to isolate CRF07_BC and subtype B strains from clinical specimens to compare their replication capacity. The results showed that the replication capacity of 9 CRF07_BC isolates was relatively lower than that of the 4 subtype B isolates (Fig. 2A). As shown in Fig. 2B, CRF07_BC isolates had significantly lower replication capacity than subtype B isolates.
Figure 2. Comparison of the replication kinetics of different HIV-1 isolates from patients infected with CRF07_BC or subtype B (A and B) as well as recombinant HIV-1 virus with or without a 7 amino-acid deletion in the p6gag protein.
(A) PBMCs were infected with fixed amounts of different HIV-1 subtype B (square) or CRF07_BC (circle) isolates and cultural supernatants were collected at days 4, 7, 11, 14, 18 and 21 post-infection. (B) The representative replicative curves of CRF07_BC and subtype B isolates deduced from Fig. 2A. (C) MT2 cells were infected with wild type (wt) or recombinant mutant virus with a 7 amino-acid deletion at the p6gag (7d). Supernatants were collected at days 2, 4, 6, 8, 10, 12, and 14 post-infection. Viral replication was monitored through p24 antigen production. One-way analysis of variance (ANOVA) and Tukey's post hoc test were used to estimate the differences between subtypes, or between the recombinant viruses.
Both Subtype B and CRF07_BC Isolates were CCR5-tropic and Non-syncytia Inducing
Both genotypic and phenotypic assays were employed to compare the tropism between subtype B and CRF07_BC. In the genotypic assay, the C2-V5 regions of the env from 10 isolates were amplified using RT-PCR and the deduced amino acid sequences were aligned. Results indicated that the V3 loop sequences of all CRF07_BC isolates contained the typical GPGQ motif. Two of the three subtype B isolates had an amino acid deletion at position 25 of the V3 loop and all subtype B strains had the GPGR/K motif. Based on the 11/25 rule (presence of basic amino acid at positions 11 and 25 are X4 viruses) and the results from online tropism prediction (Geno2pheno and PSSM prediction programs), we concluded that all isolates were CCR5 tropic viruses. In the phenotypic prediction assay, results indicated that all isolates were R5 tropism and non-syncytium inducing (NSI) (Table 5).
Table 5. V3 amino acid sequences and predicted phenotypes of different HIV-1 isolates in Taiwan.
| Isolate | Consensus V3 region amino acid sequences 11 1819 232425 CTRPNNNTRKSIHIGPGRAFYTTGEIIGDIRQAHC | Genetic subtype | Biotype | MT-2 assay | Predictions | ||
| Sequencea | Geno2phenob | PSSMc | |||||
| TW_D78 | CTRPGNNTRKSIRIGPGQTFYATGDIIGDIRQAHC | CRF07_BC | R5 | NSI | R5 | R5 | R5 |
| TW_D83 | CTRPGNNTRKSIRIGPGQTFYATGDIIGDIRQAHC | CRF07_BC | R5 | NSI | R5 | R5 | R5 |
| TW_D590 | CTRPGNNTRKSIRIGPGQTFYATGDIIGDIRQAHC | CRF07_BC | R5 | NSI | R5 | R5 | R5 |
| TW_D592 | CTRPGNNTRKSIRIGPGQTFYATGDIIGDIRQAHC | CRF07_BC | R5 | NSI | R5 | R5 | R5 |
| TW_D848 | CTRPGNNTRKSIRIGPGQTFYATGEIIGNIRQAHC | CRF07_BC | R5 | NSI | R5 | R5 | R5 |
| TW_D854 | CTRPGNNTRKSIRIGPGQTFYATGDIIGDIRQAHC | CRF07_BC | R5 | NSI | R5 | R5 | R5 |
| TW_D855 | CTRPGNNTRKSIRIGPGQTFYATGDIIGDIRQAHC | CRF07_BC | R5 | NSI | R5 | R5 | R5 |
| TW_D40 | CTRPNNNTRRSIPIGPGRAFYTSE-IIGDIRKAHC | B | R5 | NSI | R5 | R5 | R5 |
| TW_D257 | CTRPNNNTRKSISMGPGKAFFATGDIIGDIRAAYC | B | R5 | NSI | R5 | R5 | R5 |
| TW_D573 | CTRPNNNTRKSIPIGPGRAFYTTN-IIGDIRKAHC | B | R5 | NSI | R5 | R5 | R5 |
A dash indicated a deletion or lack of an insertion.
The phenotype prediction based on 2 amino acid insertion/deletion between position 14 and 15, as well as variable amino acid positions (11, 18, 19, 23, 24 and 25) of V3 regions.
Geno2pheno (http://coreceptor.bioinf.mpi-inf.mpg.de/index.php), false-positive rate of 0.01.
Position-Specific Scoring Matrix (PSSM) (http://indra.mullins.microbiol.washington.edu/webpssm/).
The Effects of a 7 Amino-Acid Deletion of p6gag on Viral Life Cycle
Since co-receptor usage and SI properties of subtype B and CRF07_BC isolates are similar, we further studied the effect of a 7 amino-acid deletion of p6gag on the viral life cycle. We generated HIV-1 NL4-3 recombinant virus containing a 7 amino-acid deletion in the p6gag (7d virus). The replication capacity was analyzed by infecting MT-2 cells with wild-type (wt) and 7d viruses. Results showed that 7d virus replicated significantly slower than the wt virus (Fig. 2C).
To compare the efficiency of protease-mediated Gag processing and viral protein production in the wt and 7d viruses, we analyzed the reactive intensity of different protein bands in the viral lysates at 12, 24, 36 and 48 hours post-infection using WB assay. As shown in Fig. 3, compared to the wt virus, the level of viral proteins of 7d virus including p24, RT and PR appeared much lower in the cell lysates (24, 36 and 48 hours post-infection) and viral lysates (36 and 48 hours post-infection) (Fig. 3A). In addition, the relative expression levels of RT and PR of 7d virus in the viral lysates was significantly lower than those of the wt virus at 48 hours post-infection (Fig. 3B). Furthermore, we calculated the viral maturation index (the ratio of p24 to Pr55 in viral lysates) and found that 7d virus had significantly lower maturation index than the wt virus (Fig. 3C).
Figure 3. Characterization of the effects of a 7 amino-acid deletion in p6gag to the HIV-1 proteins expression, release and maturation.
MT2 cells were infected with wild type (wt) or deleted-type (7d) recombinant viruses. After 12, 24, 36 and 48 hours, supernatant was collected and pelleted by ultracentrifugation. (A) Western blot analysis of the cell lysates (left panel) and viral lysates (right panel) from cells infected with wt or 7d viruses. (B) The relative expression levels of PR and RT in the viral lysates of cells infected with wt or 7d virus. The total arbitrary densitometer units of PR and RT were standardized by p24 and normalized to those of wt in parallel experiments. The images were analyzed with Image J software. (C) The ratios of p24 vs. Pr55 (maturation index) in the viral lysates at different time points after infection were calculated. The total arbitrary densitometer units of each hours post infection were normalized to those of wt in parallel experiments. All results were representative of two independent experiments. (D) Electron microscopic (EM) examination of the viral particles of cells infected with wt or 7d recombinant viruses. MAGIC-5 cells were fixed and processed for transmission EM at different time points after they were infected with wt or 7d viruses. Scale bar indicates 200 nm. (E) Quantification of relative proportions of mature vs. immature virions released at different time points in the cells infected with wt or 7d viruses using EM. The method of virion quantification has been described previously [43].
We also used electron microscopy to compare the morphogenesis between wt and 7d virions. We collected MAGIC-5 cells at different time points after infection with equal amounts of wt and 7d viruses and performed TEM. Results showed that more budding virions were observed in cells infected with wt virus than 7d virus (Fig. 3D). Besides, a higher percentage of immature virus particles released from cell membranes was found in cells infected with 7d virus at different time points (Fig. 3E).
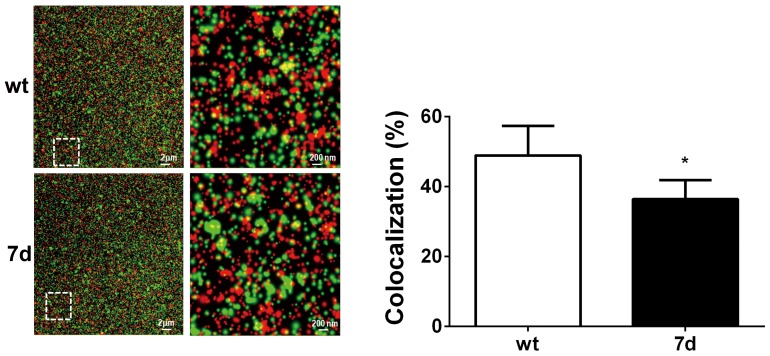
Since the 7 amino-acid deletion overlaps with the Alix binding domain of p6gag, we conducted IFA staining with anti- p24Gag and anti-Alix antibodies and analyzed their interaction using confocal microscopic exam with super-resolution program. The results showed that the co-localization coeffcicent of Gag and Alix was significantly lower in 7d virus than in wt virus (36.4% versus 48.88%, p<0.05) (Fig. 4).
Figure 4. The interaction between Alix protein and wild type/mutant Gag. MAGIC-5 cells were infected with wt or 7d recombinant virus for 48 hours.
The Alix and Gag proteins were analyze by TIRF-SR with rabbit anti-Alix polyclonal antibody and mouse anti-p24 monoclonal antibody. Red spots indicate Alix protein. Green spots indicate either wild type or 7d Gag protein. The proportion of co-localization of Alix and Gag protein was quantified using Volocity 3D Image Analysis Software.
Discussion
Previously, several studies demonstrated that compared with patients infected with subtype B, patients infected with CRF01_AE had faster disease progression [28], [29]. While studies have compared the viral loads of patients with different subtypes including CRF01_AE, this study was the first to conduct a comparison of viral loads between patients infected with subtype B and CRF07_BC. We conducted a nested case control study using an HIV-1/AIDS patient cohort which we have followed up since 2010 to demonstrate that patients infected with CRF07_BC had significantly lower viral loads (about 58,000 copies/ml in average) than patients with subtype B infection (Table 4 and Fig. 1D). We also found that gender, HIV-1 subtype, CD4 cell count and ART were strongly associated with viral loads. Furthermore, a GEE model was performed to compare viral load differences in treatment naïve patients infected with subtype B and CRF07_BC at multiple time points followed up for more than two years and matched by age, gender and initial CD4 cell count. The results showed that when we matched the cases by initial CD4 cell count, the viral loads of patients infected with CRF07_BC were consistently lower than those patients with subtype B (Fig. 1D). The GEE model has been widely used to estimate the parameters of a generalized linear model with a possible unknown correlation between outcomes [30]. Since the focus of the GEE is on estimating the average response over the population rather than the regression parameters that would enable prediction of the effect of changing one or more covariates on a given individual, it can be used to determine the association between multiple time points of plasma viral loads or CD4 cell counts in patients and model the association between factors [31]–[33]. Previous studies demonstrated that male, older age and fast CD4 cell count depletion were significantly associated with higher viral loads [34], [35]. Variable disease progression rates among individuals infected with HIV-1 have been recognized, and different factors influencing clinical outcome have been demonstrated, including host genetic, immunological and virological aspects [36]. Examples of host factors include human leukocyte antigens (HLA) and chemokine co-receptor genotype, as well as the age of the individual at the time of infection. Virological characteristics have also been shown to affect pathogenicity, such as HIV-1 subtypes, chemokine co-receptor use, syncytium-forming properties, and viral fitness [1]–[3], [37]. Since plasma viral loads have been shown to be the best prognostic marker for disease progression, our data suggests that patients infected with CRF07_BC may have a much slower disease progression rate than patients infected with subtype B [38].
We performed growth kinetic analysis to compare subtype B and CRF07_BC primary isolates. CRF07_BC, as well as infectious recombinant viruses carrying a 7 amino-acid deletion in p6gag, showed significantly reduced replication capacity (Fig. 2). The data presented here indicated that CRF07_BC was associated with slower clinical progression in treatment naïve HIV-1/AIDS patients. Previously, several studies have demonstrated that the different rates of disease progression strongly correlated with different HIV-1 subtypes [1], [2]. The different co-receptor usage of HIV-1 can affect replication rate. In general, NSI virus (or R5 virus) replicates more slowly than SI virus (or X4 virus) [39]. Our data indicated that the all the Taiwanese CRF07_BC isolates tested were NSI and replicated in CCR5-expressing cells. Furthermore, we found that CRF07_BC strains showed patterns of moderate-level replication compared with subtype B in PBMCs. We suggest that the reason may be that the genome in CRF07_BC strains was mostly subtype C with only five regions from subtype B [8]. However, subtype C infected patients are extreme rare in Taiwan. It is difficult to compare the disease progression between CRF07_BC and subtype C infected patients in Taiwan. Previous studies showed that R5-tropic subtype C strains replicate more slowly in PBMCs than R5-tropic subtype B strains in vitro growth competition assays [40].
The 7 amino-acid deletion (residues 30 to 36) is quite unique to CRF07_BC. We previously reported that the 7 amino-acid deletion is unique among almost all the CRF07_BC strains isolated in Taiwan. One Taiwanese CRF07_BC strain even had a 11 amino-acid deletion in the p6gag protein [8]. Subsequently, YM Shao's group sequenced 66 CRF07_BC strains from mainland China and found that the deletion was present in 25.8% of the cases [41]. As for whether other subtypes of CRFs have such deletion, according to our preliminary results using Los Alamos National Laboratory HIV-1 sequence database (http://hiv-web.lanl.gov) and data from YM Shao's paper, none of HIV-1 subtypes B, C CRF08_BC and other BC recombinants have this 7 amino-acid deletion [8]. In this study, we found that p6gag containing a 7 amino-acid deletion showed moderate to severe defects in Gag processing and fewer viral enzymatic proteins in infected cell and virions (Fig. 3). HIV-1 protease-mediated gag processing and gag protein interaction with host cell proteins are very important for virus assembly, budding and maturation [42]. Tsg101 binding domain of HIV-1 p6gag region (PTAP) was highly conserved in all the Taiwanese CRF07_BC strains, but they all have a 7 amino-acid deletion which overlaps with Alix binding domain (36-YPLASLRSL-44) at the residue 36Y [8]. A previous study demonstrated that Y36A mutation in p6gag protein was critical for Alix interaction and virus budding [43]. HIV-1 release forms a viral bud and the connection between the membrane bud and plasma membrane needs to be disassociated [44]. In this study, we found that co-localization coefficient between Gag and Alix was significantly lower in 7d virus (Fig. 4). We therefore suggest that such deletions in p6gag may affect its binding with Alix and subsequently affect virus release. In addition, Gag-Pol proteins are translated by -1 ribosomal frame shift during Gag translation [45]. Within the Gag-Pol, the p6gag is truncated and replaced by a trans-frame domain referred to as p6pol [46]. Deletions or mutations of p6pol affect p6pol-PR disassociation and further impair PR activity [47]. Chiu et al performed a single – cycle infection assay and demonstrated that p6pol deletion affected viral infectivity and reduced the p24/Pr55 protein ratio. [22]. A previous study demonstrated that truncation of p6gag reduced the amount of viral enzymatic proteins in the virions [48]. Furthermore, our data showed that the 7 amino-acid deletion in p6gag domain affected Gag processing efficiency and the amount of viral enzymatic proteins in HIV-1 virions. Further studies are needed to elucidate whether p6pol containing a 7 amino-acid deletion affects the incorporation of gag-pol into virus particle or virus assembly in the nearby cytoplasmic membrane.
This is the first study that combines a longitudinal clinical follow-up study and a virological characterization of CRF07_BC infection. The Taiwanese CRF07_BC strains have about 97% nucleotide sequence homology with the prototypic CRF07_BC strains in China [8]. According to our previous phylogenetic analysis using the env gene, the Taiwanese CRF07_BC strains collected in 2004 formed at least two clusters with bootstrap value of 71 [8]. This phenomenon was reconfirmed by analysis of a larger number of CRF07_BC strains collected between 2005-2008, which showed a bootstrap value of 80 [10]. Therefore, there were more than 1 wave of CRF07_BC infection being transmitted to Taiwan and we believe that the findings in this study can also be applied to the CRF07_BC strains in mainland China.
In conclusion, our results suggest that patients infected with CRF07_BC have slower rate of disease progression and the deletion of 7 amino acids in its p6gag region plays an important role in the assembly, budding and maturation processes of the viral life cycle.
Acknowledgments
We thank staffs from the Genome Research Center at National Yang-Ming University and colleagues at the CICAR, Kaohsiung Medical University for their technical assistance. We also thank patients and medical personnel in TwHOD network for their participation and supports. We thank the following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 p24 Monoclonal Antibody (183-H12-5C) from Dr. Bruce Chesebro and Kathy Wehrly; HIV-1 RT Monoclonal Antibody (MAb21) from Dr. Stephen Hughes.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This research was supported by Republic of China (ROC) National Health Research Institutes [grant number NHRI-EX103-10149SI] and partially by Kaohsiung Medical University “Aim for the Top Universities Grant [grant number KMU-TP103E02, KMU-TP103E20]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vasan A, Renjifo B, Hertzmark E, Chaplin B, Msamanga G, et al. (2006) Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin Infect Dis 42:843–852. [DOI] [PubMed] [Google Scholar]
- 2. Baeten JM, Chohan B, Lavreys L, Chohan V, McClelland RS, et al. (2007) HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis 195:1177–1180. [DOI] [PubMed] [Google Scholar]
- 3. Rangsin R, Piyaraj P, Sirisanthana T, Sirisopana N, Short O, et al. (2007) The natural history of HIV-1 subtype E infection in young men in Thailand with up to 14 years of follow-up. AIDS 21 Suppl 6 S39–46. [DOI] [PubMed] [Google Scholar]
- 4. Pant Pai N, Shivkumar S, Cajas JM (2012) Does genetic diversity of HIV-1 non-B subtypes differentially impact disease progression in treatment-naive HIV-1-infected individuals? A systematic review of evidence: 1996-2010. J Acquir Immune Defic Syndr 59:382–388. [DOI] [PubMed] [Google Scholar]
- 5.(2013) Center for Disease Control, ROC (Taiwan). HIV/AIDS data.
- 6. Chen YM, Huang KL, Jen I, Chen SC, Liu YC, et al. (2001) Temporal trends and molecular epidemiology of HIV-1 infection in Taiwan from 1988 to 1998. J Acquir Immune Defic Syndr 26:274–282. [DOI] [PubMed] [Google Scholar]
- 7. Chen YM, Lan YC, Lai SF, Yang JY, Tsai SF, et al. (2006) HIV-1 CRF07_BC infections, injecting drug users, Taiwan. Emerg Infect Dis 12:703–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin YT, Lan YC, Chen YJ, Huang YH, Lee CM, et al. (2007) Molecular epidemiology of HIV-1 infection and full-length genomic analysis of circulating recombinant form 07_BC strains from injection drug users in Taiwan. J Infect Dis 195:1283–1293. [DOI] [PubMed] [Google Scholar]
- 9. Chen YM, Kuo SH (2007) HIV-1 in Taiwan. Lancet 369:623–625. [DOI] [PubMed] [Google Scholar]
- 10. Chen YJ, Lee CM, Chen M, Chuang SY, Liu HF, et al. (2012) Molecular epidemiology of HIV-1 infection in Taiwan from 2005 to 2008: further spread of CRF07_BC and emergence of CRF07_BC/subtype B dual infection. J Acquir Immune Defic Syndr 59:438–446. [DOI] [PubMed] [Google Scholar]
- 11. Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO (2007) An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J Biol Chem 282:3847–3855. [DOI] [PubMed] [Google Scholar]
- 12. Strack B, Calistri A, Craig S, Popova E, Gottlinger HG (2003) AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689–699. [DOI] [PubMed] [Google Scholar]
- 13. Zhou J, Kumarasamy N, Ditangco R, Kamarulzaman A, Lee CK, et al. (2005) The TREAT Asia HIV Observational Database: baseline and retrospective data. J Acquir Immune Defic Syndr 38:174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei M, Guan Q, Liang H, Chen J, Chen Z, et al. (2004) Simple subtyping assay for human immunodeficiency virus type 1 subtypes B, C, CRF01-AE, CRF07-BC, and CRF08-BC. J Clin Microbiol 42:4261–4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xin X, Shioda T, Fukushima M, Hu H, Oka S, et al. (1998) Facilitation of HIV-1 isolation from patients by neuraminidase. Arch Virol 143:85–95. [DOI] [PubMed] [Google Scholar]
- 16. Trkola A, Paxton WA, Monard SP, Hoxie JA, Siani MA, et al. (1998) Genetic subtype-independent inhibition of human immunodeficiency virus type 1 replication by CC and CXC chemokines. J Virol 72:396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karlsson A, Parsmyr K, Sandstrom E, Fenyo EM, Albert J (1994) MT-2 cell tropism as prognostic marker for disease progression in human immunodeficiency virus type 1 infection. J Clin Microbiol 32:364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fouchier RA, Groenink M, Kootstra NA, Tersmette M, Huisman HG, et al. (1992) Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J Virol 66:3183–3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shioda T, Levy JA, Cheng-Mayer C (1992) Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 89:9434–9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cardozo T, Kimura T, Philpott S, Weiser B, Burger H, et al. (2007) Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res Hum Retroviruses 23:415–426. [DOI] [PubMed] [Google Scholar]
- 21. Hu Z, Giguel F, Hatano H, Reid P, Lu J, et al. (2006) Fitness comparison of thymidine analog resistance pathways in human immunodeficiency virus type 1. J Virol 80:7020–7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chiu HC, Wang FD, Chen YM, Wang CT (2006) Effects of human immunodeficiency virus type 1 transframe protein p6* mutations on viral protease-mediated Gag processing. J Gen Virol 87:2041–2046. [DOI] [PubMed] [Google Scholar]
- 23. Toohey K, Wehrly K, Nishio J, Perryman S, Chesebro B (1995) Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology 213:70–79. [DOI] [PubMed] [Google Scholar]
- 24. Ferris AL, Hizi A, Showalter SD, Pichuantes S, Babe L, et al. (1990) Immunologic and proteolytic analysis of HIV-1 reverse transcriptase structure. Virology 175:456–464. [DOI] [PubMed] [Google Scholar]
- 25. Hachiya A, Aizawa-Matsuoka S, Tanaka M, Takahashi Y, Ida S, et al. (2001) Rapid and simple phenotypic assay for drug susceptibility of human immunodeficiency virus type 1 using CCR5-expressing HeLa/CD4(+) cell clone 1-10 (MAGIC-5). Antimicrob Agents Chemother 45:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Spira F, Mueller NS, Beck G, von Olshausen P, Beig J, et al. (2012) Patchwork organization of the yeast plasma membrane into numerous coexisting domains. Nat Cell Biol 14:640–648. [DOI] [PubMed] [Google Scholar]
- 27.Manders EMM, Verbeek FJ, Aten JA (1993) Measurement of co-localization of objects in dual-colour confocal images. Journal of Microscopy. pp. 8. [DOI] [PubMed]
- 28. Hu DJ, Vanichseni S, Mastro TD, Raktham S, Young NL, et al. (2001) Viral load differences in early infection with two HIV-1 subtypes. AIDS 15:683–691. [DOI] [PubMed] [Google Scholar]
- 29. Zhao K, Kang W, Liu Q, Li Y, Liu Q, et al. (2014) Genotypes and Transmitted Drug Resistance among Treatment-Naive HIV-1-Infected Patients in a Northwestern Province, China: Trends from 2003 to 2013. PLoS One 9:e109821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zeger SL, Liang KY, Albert PS (1988) Models for longitudinal data: a generalized estimating equation approach. Biometrics 44:1049–1060. [PubMed] [Google Scholar]
- 31. Raugi DN, Gottlieb GS, Sow PS, Toure M, Sall F, et al. (2013) HIV-1 outcompetes HIV-2 in dually infected Senegalese individuals with low CD4(+) cell counts. AIDS 27:2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kittikraisak W, van Griensven F, Martin M, McNicholl J, Gilbert PB, et al. (2009) Blood and seminal plasma HIV-1 RNA levels among HIV-1-infected injecting drug users participating in the AIDSVAX B/E efficacy trial in Bangkok, Thailand. J Acquir Immune Defic Syndr 51:601–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Morrison CS, Demers K, Kwok C, Bulime S, Rinaldi A, et al. (2010) Plasma and cervical viral loads among Ugandan and Zimbabwean women during acute and early HIV-1 infection. AIDS 24:573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gandhi M, Bacchetti P, Miotti P, Quinn TC, Veronese F, et al. (2002) Does patient sex affect human immunodeficiency virus levels? Clin Infect Dis 35:313–322. [DOI] [PubMed] [Google Scholar]
- 35. Nakagawa F (2014) Factors associated with short-term changes in HIV viral load and CD4+ cell count in antiretroviral-naive individuals. AIDS 28:1351–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pantaleo G, Fauci AS (1996) Immunopathogenesis of HIV infection. Annu Rev Microbiol 50:825–854. [DOI] [PubMed] [Google Scholar]
- 37. Tscherning C, Alaeus A, Fredriksson R, Bjorndal A, Deng H, et al. (1998) Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology 241:181–188. [DOI] [PubMed] [Google Scholar]
- 38. Mellors JW, Rinaldo CR Jr, Gupta P, White RM, Todd JA, et al. (1996) Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167–1170. [DOI] [PubMed] [Google Scholar]
- 39. Weber J, Piontkivska H, Quinones-Mateu ME (2006) HIV type 1 tropism and inhibitors of viral entry: clinical implications. AIDS Rev 8:60–77. [PubMed] [Google Scholar]
- 40. Ball SC, Abraha A, Collins KR, Marozsan AJ, Baird H, et al. (2003) Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J Virol 77:1021–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Song YH, Meng ZF, Xing H, Ruan YH, Li XP, et al. (2007) Analysis of HIV-1 CRF07_BC gag p6 sequences indicating novel deletions in the central region of p6. Arch Virol 152:1553–1558. [DOI] [PubMed] [Google Scholar]
- 42. Chen SW, Chiu HC, Liao WH, Wang FD, Chen SS, et al. (2004) The virus-associated human immunodeficiency virus type 1 Gag-Pol carrying an active protease domain in the matrix region is severely defective both in autoprocessing and in trans processing of gag particles. Virology 318:534–541. [DOI] [PubMed] [Google Scholar]
- 43. Fujii K, Munshi UM, Ablan SD, Demirov DG, Soheilian F, et al. (2009) Functional role of Alix in HIV-1 replication. Virology 391:284–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hurley JH, Hanson PI (2010) Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat Rev Mol Cell Biol 11:556–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jacks T, Power MD, Masiarz FR, Luciw PA, Barr PJ, et al. (1988) Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280–283. [DOI] [PubMed] [Google Scholar]
- 46. Partin K, Krausslich HG, Ehrlich L, Wimmer E, Carter C (1990) Mutational analysis of a native substrate of the human immunodeficiency virus type 1 proteinase. J Virol 64:3938–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Partin K, Zybarth G, Ehrlich L, DeCrombrugghe M, Wimmer E, et al. (1991) Deletion of sequences upstream of the proteinase improves the proteolytic processing of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 88:4776–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu XF, Dawson L, Tian CJ, Flexner C, Dettenhofer M (1998) Mutations of the human immunodeficiency virus type 1 p6Gag domain result in reduced retention of Pol proteins during virus assembly. J Virol 72:3412–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.