Abstract
Aqueous leachates prepared from natural antibacterial clays, arbitrarily designated CB-L, release metal ions into suspension, have a low pH (3.4–5), generate reactive oxygen species (ROS) and H2O2, and have a high oxidation-reduction potential. To isolate the role of pH in the antibacterial activity of CB clay mixtures, we exposed three different strains of Escherichia coli O157:H7 to 10% clay suspensions. The clay suspension completely killed acid-sensitive and acid-tolerant E. coli O157:H7 strains, whereas incubation in a low-pH buffer resulted in a minimal decrease in viability, demonstrating that low pH alone does not mediate antibacterial activity. The prevailing hypothesis is that metal ions participate in redox cycling and produce ROS, leading to oxidative damage to macromolecules and resulting in cellular death. However, E. coli cells showed no increase in DNA or protein oxidative lesions and a slight increase in lipid peroxidation following exposure to the antibacterial leachate. Further, supplementation with numerous ROS scavengers eliminated lipid peroxidation, but did not rescue the cells from CB-L-mediated killing. In contrast, supplementing CB-L with EDTA, a broad-spectrum metal chelator, reduced killing. Finally, CB-L was equally lethal to cells in an anoxic environment as compared to the aerobic environment. Thus, ROS were not required for lethal activity and did not contribute to toxicity of CB-L. We conclude that clay-mediated killing was not due to oxidative damage, but rather, was due to toxicity associated directly with released metal ions.
Introduction
We have identified a natural clay mixture, arbitrarily termed CB, that displays broad-spectrum, in vitro bactericidal activity in hydrated suspensions [1]–[4]. Previously, we prepared aqueous extracts from the natural clay mixtures, termed leachates, that are devoid of physical particles. These leachates, designated CB-L, maintain in vitro antibacterial activity, but do not cause cell lysis [2]. Thus, killing is dependent on the abiotic chemical, not physical, properties of hydrated clay suspensions [1]–[3]. When in hydrated suspension, the pH range of CB clay mixtures is 3–4 [1]. While we have previously shown that CB killing activity is dependent on a low pH environment, the acidic environment generated by CB minerals does not solely mediate toxicity [3]. Moreover, supplementation of a non-antibacterial leachate containing lower concentrations of Fe, Co, Ni, Cu, and Zn to final ion concentrations and a pH equivalent to that of the antibacterial leachate generated antibacterial activity against Escherichia coli and methicillin-resistant Staphylococcus aureus (MRSA), confirming the role of these ions in the antibacterial clay mixture leachates [1].
The first goal of the present study was to determine whether the acidic environment itself is exerting a degree of toxicity or whether it is simply necessary to maintain the desorbed metal ions in their more toxic speciation. For example, Fe is more toxic in acidic environments due to the increased solubility and reactivity of its reduced form, whereas Cu, Zn, and Cd are more toxic in elevated pH environments due to less competition with protons for cellular binding sites and less efflux [5]–[7]. E. coli O157:H7 was used as the model organism during low pH experimentation as specific strains offer extreme acid resistance resulting from cross protection between three well-characterized acid-response systems [8], [9]. The first acid-resistance system (AR1) is induced upon entry into stationary phase and involves activation of the RpoS general stress response [10]–[12]. AR2 transports glutamate into the cell via the antiporter GadC, converts glutamate to γ-amino butyric acid (GABA) by glutamate decarboxylase (GadA and GadB), and transports GABA out of the cell via GadC. During this continuous glutamate-to-GABA conversion, intracellular protons are consumed, thus raising the cytoplasmic pH and creating a pH gradient [13]–[15]. The second (AR2) and third (AR3) acid-resistance systems are functionally similar to one another, allowing the cytoplasmic pH to remain higher than the extracellular environment [11], [13], [14], [16], [17].
Chemical simulation modeling of the ions in CB-L revealed increased concentrations of soluble Cu2+ and Fe2+ in the antibacterial leachates, compared to non-antibacterial leachates, suggesting that these ionic species are modulating the antibacterial activity of the leachates [1]. Iron and copper are known to participate in the Fenton and Fenton-like reactions, respectively, whereby the metal reacts with H2O2 to produce a hydroxyl radical [18]. Current dogma asserts that reactive oxygen species (ROS)-driven oxidative damage to macromolecules is directly responsible for killing bacteria whereby H2O2 diffuses into the cells, damages DNA, and kills the cells [19], [20]. For example, Lloyd et al. [20] showed that salmon sperm DNA exposed to H2O2 along with at least one of nine different transition-metal ions showed significant increases in 8-hydroxydeoxyguanosine (8-OHdG), a biomarker of DNA damage. DNA damage has been shown to be iron-dependent, suggesting that the damage is actually from hydroxyl radicals or ferryl radical intermediates produced by the Fenton reaction [21], [22]. However, hydroxyl radicals are so reactive that they can only diffuse 5-10 molecular diameters before reacting nonspecifically with any reactive molecule in its vicinity [23]. Much of the current literature associated with ROS-driven oxidative damage to macromolecules is related directly to intracellular oxidative stress [24], [25]. Considering the high reactivity of hydroxyl radicals, superoxide, peroxynitrite, and hydrogen peroxide, oxidative damage to biological molecules is diffusion limited, and thus, exogenous ROS may not act on intracellular targets.
Here, we report evidence that metal-catalyzed oxidative damage is not responsible for antibacterial clay leachate-mediated killing. We also demonstrate that EDTA, not ROS scavengers or carbon supplements, disrupt antibacterial leachate killing and that bactericidal activity occurs in an anaerobic environment, thus negating the hypothesis that metal-catalyzed oxidative stress causes clay leachate antibacterial activity in vitro.
Materials and Methods
Bacterial Strains and Inoculum Preparation
E. coli ATCC 25922, E. coli O157:H7 ATCC 43889 (acid-tolerant), ATCC 43890 (acid-sensitive), and ATCC 43895 (acid-resistant, CDC EDL 933) were obtained from the American Type Culture Collection [26]. The methicillin-resistant Staphylococcus aureus (MRSA) strain, a clinical isolate obtained from Sonora Quest Laboratories (Tempe, AZ, USA), was grown as previously described [1]–[4]. For non-induced experiments, E. coli ATCC 25922 and E. coli O157:H7 were grown overnight in Luria-Bertani (LB) broth and trypticase soy broth (TSB), respectively, for 13–16 h at 37°C with gentle rotary mixing. Stationary-phase cultures were prepared by diluting saturated, overnight cultures into fresh growth medium to a starting cell density of 107 or 108 CFU/ml. Exponential phase cultures were prepared by diluting overnight cultures to a concentration of 107 CFU/ml into fresh growth medium and continuing growth at 37°C until the cultures reached mid-logarithmic phase (OD600 0.4–0.6). Prior to exposure, 1 ml aliquots of the cells were pelleted via centrifugation for 5 min at 13,000×g, washed with either 0.85% saline or 0.1× phosphate-buffered saline (PBS), pelleted via centrifugation for 5 min at 13,000×g, and suspended in the appropriate experimental medium. Non-induced stationary-phase E. coli O157:H7 (ATCC 43889) cells were suspended in EG medium, a minimal salts medium (73 mM K2HPO4, 17 mM NaNH4HPO4, 0.8 mM MgSO4) with 10 mM citrate and 0.4% glucose, and chemically-induced cells were suspended in EG medium with the addition of 2 mM glutamate [16]. All experiments were accompanied by cell viability assays, and CFU/ml was determined following overnight incubation at 37°C.
Clay Mixtures and Clay Mixture Leachates
Clay mixture leachates were prepared as previously described [5], [6] with minor modifications. Briefly, CB-L was obtained by continuously stirring clay mixtures (1 g/10 ml) in sterile, ultrapure, deionized H2O (dH2O) for 18–24 h, followed by centrifugation (31,000×g) for 3 h at 4°C. The aqueous supernatant (leachate) was then sterilized by passage through a 0.22 µm filter.
Anaerobic Incubations and Viability Assays
CB leachate and exponential phase E. coli ATCC 25922 cells were prepared as described above. Anaerobic dH2O and CB-L were prepared by sparging the solutions for 30 min with a steady stream (8 PSI) of compressed nitrogen (99.97% pure, Airgas, Riverside, CA) through a 1 L bottle of water at room temperature [27]. Bacterial cells were transferred to the anaerobic chamber, resuspended in the anaerobically-prepared solutions, and incubated under anoxic conditions at 37°C. Cell viability was determined by plating in duplicate on LB plates either directly from the experimental samples or following appropriate 10-fold dilutions at the specified times.
pH Experiments
Initial screening experiments were performed to observe viability of acid-sensitive, non-induced acid-tolerant, and naturally acid-resistant E. coli O157:H7 when exposed to 10% suspensions of CB mixtures compared to a low-pH phosphate buffer. The pH of a 10% CB suspension is 3.1–3.3, so a low-pH 100 mM phosphate buffer (pH 3.1) was used for control experiments. Acid resistance was chemically induced in acid-tolerant E. coli O157:H7 by the addition of 2 mM glutamate to EG medium just prior to addition of 1% CB minerals. During acid-resistance induction, the pH of experimental media was adjusted to pH 2.5 with 1 M HCl.
H2O2 Detection Assays
H2O2 measurements were performed using the Fluorescent Hydrogen Peroxide/Peroxidase Detection Kit following the manufacturer's protocol (Cell Technology, Inc., Mountain View, CA). Diluted cultures of stationary-phase E. coli ATCC 25922 (107 CFU/ml) were prepared as described above and exposed to either a 10% CB mixture or CB-L. Briefly, after 0, 3, 6, and 9 h incubations, 150 µl aliquots were removed and centrifuged for 5 min at 13,000×g, and 50 µl of the supernatant was transferred to a 96-well plate in duplicate. The samples were mixed with the reaction cocktail and incubated for 15 min in the dark at room temperature prior to generating fluorescence readings on a Molecular Devices SpectraMax M2 microplate reader (Sunnyvale, CA) at Ex/Em 570/600 nm. Background H2O2 levels of E. coli ATCC 25922 incubated in dH2O were subtracted from reported results. A standard curve using different H2O2 concentrations was generated as a positive control and to quantify H2O2 concentration in experimental samples.
ROS Detection
Prior to ROS detection, cells were incubated in 1 mM dihydrorhodamine (DHR) for 30 min, washed in 0.85% saline solution, and suspended in the appropriate experimental medium. E. coli ATCC 25922 cells exposed to 5 mM diamide were used as a positive control for ROS production. A Cytomics FC 500 flow cytometer (Beckman Coulter, Inc., Brea, CA) fitted with a 488 nm excitation laser was used to measure ROS production in the intracellular environment during CB-L exposure. Exponential-phase E. coli (108 CFU/ml) was exposed to CB-L and incubated at 37°C with gentle rotary mixing for the specified time. Fluorescence was detected with the flow cytometer using channel FL1 with a 525 nm bandpass filter. A Molecular Devices SpectraMax M2 microplate reader was used to measure ROS production in CB-L without cells and of the E. coli intracellular environment while in the presence of CB-L. For these experiments, diluted cultures of stationary-phase E. coli (107 CFU/ml) were exposed to CB-L and incubated at 37°C with gentle rotary mixing for the specified time. Fluorescence was measured at an excitation of 485 nm and emission of 525 nm.
Carbon, Chelator, and ROS Scavenger Supplementation
E. coli ATCC 25922 cells were grown to mid-logarithmic phase, diluted to a starting density of 107 CFU/ml, and washed with sterile 0.85% saline. Glucose (40 mM), EDTA (10 mM), DMSO (1%), and thiourea (150 mM) were added to the antibacterial CB leachate and incubated for 1 h at room temperature prior to exposure to cells. Washed cell pellets were resuspended in dH2O or CB-L with and without the addition of glucose, EDTA, DMSO, or thiourea. At designated times, 100 µl aliquots were removed and appropriate 10-fold dilutions were performed to determine viable CFU/ml.
ELISA to Detect 8-OHdG and Oxidatively-damaged Nucleic Acids
E. coli ATCC 25922 cells were grown to mid-logarithmic phase, diluted to a 40 ml volume at a density of 5×108 CFU/ml, washed with sterile 0.85% saline, and resuspended in experimental conditions (dH2O, CB-L, 2 mM H2O2 in dH2O, 1% DMSO in dH2O, 1% DMSO in CB-L, 10 mM EDTA in dH2O, or 10 mM EDTA in CB-L). At the designated times, 100 µl aliquots were removed and appropriate 10-fold dilutions were performed to determine viable CFU/ml. In addition, 20 ml samples were removed, pelleted at 4,000 RPM for 10 min, and subjected to DNA extraction. To coat the wells, 100 µl aliquots of 0.1% poly-l-lysine were added to 96-well plates (Corning, high bind EIA) for 5 min at room temperature and removed via aspiration prior to subsequent experimental use. Plates were loaded with quadruplicate 50 µl samples of 100 ng/ml nucleic acids and allowed to dry overnight in a 37°C incubator. Once dry, the plates were incubated without agitation in blocking buffer (1×PBS, 1.5% FBS) for 1 h at 37°C and subsequently washed 3× with wash buffer (1×PBS, 0.5% Tween 20). 100 µl aliquots of the mouse anti-8-OHdG antibody (1∶20,000 in blocking buffer) were added to each well of the 96-well plate, and the plate was incubated without agitation for 1 h at 37°C. After washing 3× with wash buffer, 100 µl of rabbit anti-mouse enzyme-linked secondary antibody (1∶10,000 in blocking buffer) (Sigma Aldrich) was added to each well. Incubation was performed at room temperature for 1 h and subsequently washed 3× with wash buffer. Finally, to allow for color development, 100 µl of 4-methylumbelliferyl phosphate (Sigma Aldrich) solution was added to each well, and the plate was incubated without agitation for 30–60 min at 37°C. Fluorescence was measured with a microtiter plate reader (Molecular Devices SpectraMax M2) using wavelengths of 360 and 440 for excitation and emission, respectively.
Protein Oxidation
E. coli ATCC 25922 cells were grown to mid-logarithmic phase, diluted to a 20 ml volume at a density of 1×108 CFU/ml, washed with sterile 0.85% saline, and resuspended in either dH2O, 2 mM H2O2, or CB-L. At the designated times, 100 µl aliquots were removed and appropriate 10-fold dilutions were performed to determine viable CFU/ml. In addition, 10 ml samples of the culture were removed, pelleted at 4,000 RPM for 5 min, and resuspended in 500 µl of lysis buffer (10 mM HEPES, pH 7, 6% SDS). To collect cell extracts containing the total protein population, the pellets were resuspended in 500 µl of lysis buffer and boiled for 5 min. Carbonyl derivatization was completed according to methods described by Shacter et al. [28]. Briefly, 200 µl of 2,4-dinitrophenylhydrazine in 2 M HCl was added to 1 mg of protein, gently mixed, and incubated at room temperature for 30 min. Proteins were then precipitated by the addition of 200 µl of 20% trichloroacetic acid and pelleted at 13,000 RPM for 5 min. The pellets were washed three times in 1∶1 ethanol:ethyl acetate for 10 min each, dried for 10–20 min, and resuspended in 600 µl of 6M guanidine hydrochloride, 20 mM NaPO4, pH 8. Duplicate 200 µl samples were transferred to a 96-well plate, and the A280 and A360 readings were measured for each sample with a Molecular Devices Spectramax M2 spectrophotometer. Carbonyl concentration was calculated with a molar extinction coefficient of 22,000 M−1 cm−1. Reported carbonyl values were normalized across the samples based on the respective measured protein concentration.
Lipid Peroxidation
Lipid peroxidation measurements were generated using the Lipid Peroxidation (MDA) Assay kit (Abcam, Cambridge, MA) according to the manufacturer's guidelines. Briefly, E. coli ATCC 25922 cells were grown to mid-logarithmic phase of growth, diluted to 108 CFU/ml, and washed with sterile 0.85% saline prior to exposure to dH2O, 2 mM H2O2, or CB-L. At each time point, 400 µl of the sample was collected, pelleted, resuspended in the manufacturer's lysis buffer, and subjected to bead beating two times for 30-sec intervals. The lysates were pelleted, the supernatant was processed with thiobarbituric acid, and the fluorescent lipid peroxidation products were measured on a microtiter plate reader (Molecular Devices SpectraMax M2) using wavelengths of 532 and 553 for excitation and emission, respectively.
Preparation of Metal Ion Stock Solutions and Metal Salt Mixtures
Metal ion stock solutions were prepared by adding chloride salts to a final concentration of 100 mM in dH2O. Two different synthetic microbicidal mixtures (SMM3 and SMM16) were prepared by diluting metal ion stocks in dH2O to the concentrations found in CB07-L [1]. Elemental chemistry of the CB leachates was previously measured by inductively coupled plasma mass spectrometry (ICP-MS) (described in Otto et al. [1]), and ion concentrations from these ICP-MS measurements were used to prepare SMM3: Li (Sigma), 7.62 µM; Be(II) (Sigma), 0.80 µM; Na (Fisher), 171.61 µM; Mg(II) (Sigma), 773.11 µM; Al(III) (Sigma), 1079.6 µM; Ca(II) (Sigma), 6911.18 µM; Ti(IV) (Sigma), 0.01 µM; V(III) (Sigma), 0.49 µM; Mn(II) (Sigma), 99.31 µM; Fe(III) (Sigma), 229.88 µM; Co(II) (Sigma) 3.38 µM; Ni(II) (Sigma), 3.46 µM; Cu(II) (Sigma) 3.18 µM; Zn (Sigma) 14.52 µM [1]. The pH of the solution was adjusted to 3.5 with 1 M NaOH, and the solution was sterilized by passage through a 0.22 µM filter prior to further use. SMM16 was prepared in an identical manner as described above with the following metal ion concentrations: Fe(III), 200 µM; Co(II), 4 µM; Cu(II), 80 µM; Zn, 40 µM.
Statistical Analyses
Unpaired Student's t test and ANOVA were used to assess statistical significance. A p value of <0.05 was considered statistically significant.
Results
Low pH does not mediate clay antibacterial activity
To determine the role of pH in the antibacterial activity of CB clay mixtures, we exposed three acid-responsive strains of E. coli O157:H7 (acid resistant, acid sensitive, and acid tolerant) to a 10% suspension of CB clay. The clay suspension completely killed the acid-tolerant and acid-sensitive E. coli O157:H7 strains (ATCC 43889 and 43890, respectively) within 30 min, whereas a low-pH buffer resulted in a minimal decrease in viability after 2 h (Fig. 1A, B). Time course killing curves demonstrated that the more adept acid-response systems in acid-resistant E. coli O157:H7 (ATCC 43895) significantly slowed the rate of CB-mediated killing and enhanced survival over 2 h (Fig. 1C).
Figure 1. Stationary-phase, acid-tolerant (A), acid-sensitive (B), and acid-resistant (C) E. coli O157:H7 in 10% CB suspension (pH 3.1) and low-pH buffer (pH 3.1).
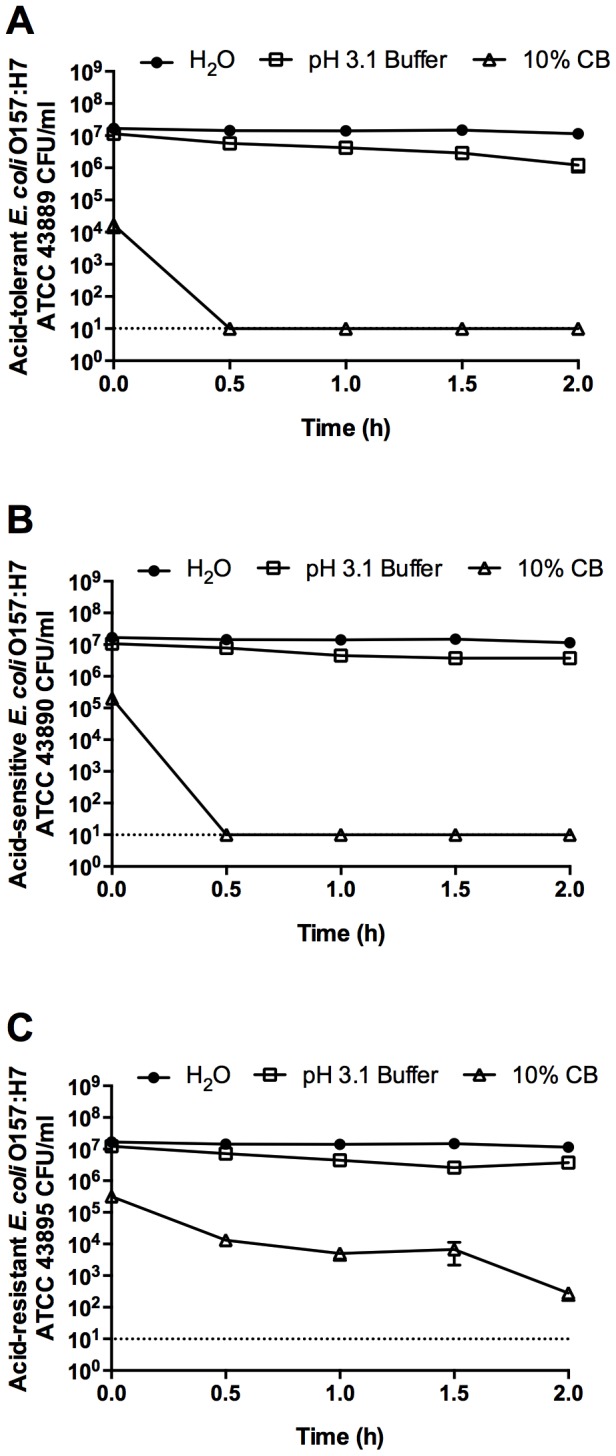
Stationary-phase cells were grown overnight in LB media. Data are presented as the mean ± standard deviation (SD) for three independent experiments. The dotted line represents the limit of detection.
Acid resistance was chemically induced in the acid-tolerant E. coli O157:H7 ATCC 43889 strain by supplementing the medium with glutamate during CB mineral exposure. Additionally, to eliminate cross protection between stationary-phase induced AR1 and glutamate-induced AR2, low pH experiments were also performed following overnight bacterial growth in glucose-supplemented medium which suppresses AR1 [11]. Induction of AR2 with glutamate significantly enhanced E. coli O157:H7 ATCC 43889 survival in the low pH EG medium (Fig. 2) Compared to a non-inducing (NI) environment with 1% CB, glutamate-induction of AR2 in acid-tolerant E. coli O157:H7 delayed the time required for the CB suspension to completely kill the cells (Fig. 2); however, after a 2 h exposure to 1% CB, chemically-induced, acid-resistant E. coli O157:H7 was completely killed (Fig. 2). Due to the rapid killing by CB-L, the viability of E. coli at the initial time point was often lower than the other control values. Further, we previously showed that when the pH of CB-L is increased, the killing activity is minimized [1]. Take altogether, these data (Figs. 1, 2) confirm that the low pH environment influences the rate at which CB clay mixtures kill, but does not directly mediate microbicidal activity.
Figure 2. Stationary-phase, inducible-acid-tolerant E. coli O157:H7 challenged in EG medium (pH 2.5) with (I, induced) or without (NI, non-induced) 2 mM glutamate.
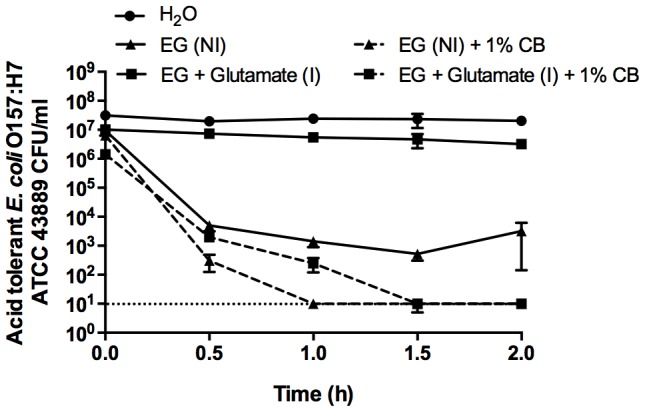
Experimental samples were exposed to a 1% CB suspension for 2 h. Data are presented as the mean ±SD for three independent experiments. The dotted line represents the limit of detection.
Hydrated antibacterial clays and derived leachates generate H2O2
We used a fluorescence detection system to measure H2O2 production in clay and leachate samples in the presence and absence of E. coli ATCC 25922 and observed continuous generation of 3–4 µM H2O2 in 10% suspensions of CB clay mixtures (Fig. 3). When the experimental medium was supplemented with up to 5 µM H2O2, the H2O2 was rapidly scavenged by the cells and was not detectable after 1 h (data not shown). H2O2 levels were slightly lower in CB-L with detected fluorescence correlating to 1–2 µM H2O2 when compared to a standard curve (Fig. 3). These lower H2O2 levels measured in CB-L could be due to the absence of CB clay mixtures required for surface-mediated production of H2O2 [29], [30]. Together, these data suggest that any H2O2 scavenged by the bacterial cells is rapidly regenerated by the hydrated CB clay mixtures or leachates, thus, resulting in a steady state concentration over time (Fig. 3). While there was continuous generation of 1–4 µM H2O2 in the clays and leachates, it is known that E. coli (strain AB1157) can produce and tolerate as much as 4 µM s−1 H2O2 during active growth [24].
Figure 3. Hydrogen peroxide concentration in 10% CB suspensions and CB-L.
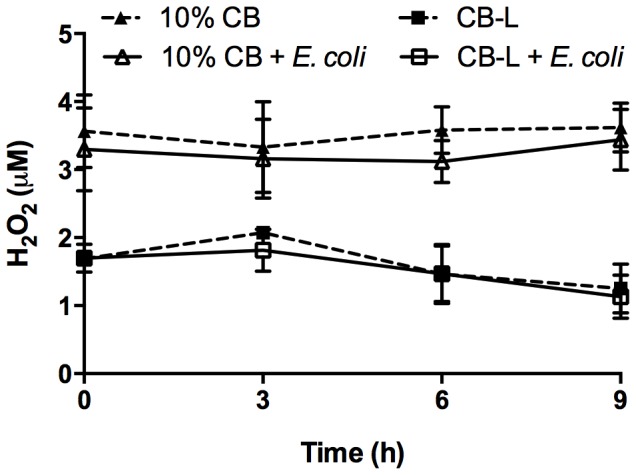
Hydrogen peroxide levels were measured in 10% CB suspensions and CB-L with or without the addition of stationary-phase E. coli ATCC 25922. Values represent the average H2O2 concentration ±SD for three independent experiments.
Hydrated antibacterial clays and derived leachates lead to intracellular and extracellular ROS
To determine the location of potential reactive oxygen toxicity, we assessed intracellular and extracellular ROS production. We measured 11-fold and 10-fold increases in ROS production in CB-L, as compared to a dH2O-only control, after 12 h and 24 h, respectively (Fig. 4). Following 12 h and 24 h CB-L exposures, flow cytometric analyses of E. coli ATCC 25922 cells revealed 3-fold and 2-fold increases in ROS production, respectively, as compared to the water-exposed control (Fig. 4). The oxidation-reduction potential (ORP) of CB10-L measured 593.3 mV [1], demonstrating that the solution is more oxidizing than the ultrapure, dH2O (432.6 mV) used in this study. Subsequent measurements over 24 h and thereafter revealed that the CB10-L ORP remains stable over time. Overall, these data demonstrated that CB-L generates an oxidizing environment and ROS. We note that our data conflicts with another study [31] which showed that the redox potential of antibacterial clay suspensions transitions to a reducing environment over time, however, the report did not include a methodological description for the ORP measurements.
Figure 4. CB-L-associated ROS production in the absence and presence of E. coli ATCC 25922.
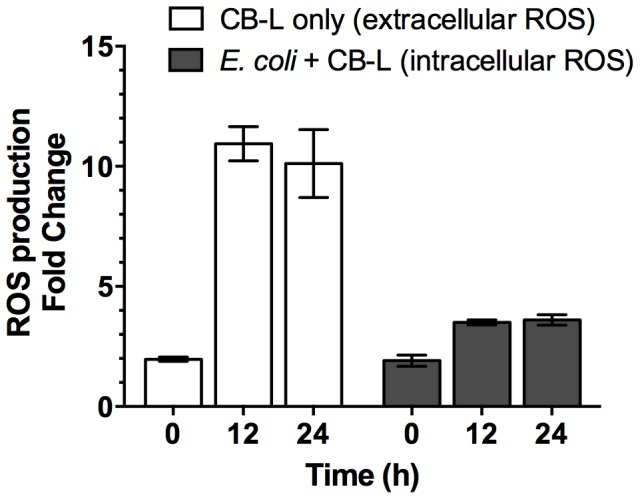
ROS was measured in CB-L without the presence of cells (extracellular; using a plate reader) and exclusively in the intracellular environment of exponential-phase E. coli following CB-L exposure (intracellular; using flow cytometry). Cells were grown overnight, transferred to fresh growth media and grown to mid-logarithmic phase, washed, and exposed to CB-L for 24 h. Reported values represent the ROS production fold change of CB-L compared to water and CB-L-exposed cells compared to water-exposed cells. Data are presented as the mean ± standard error of the mean (SEM) for three independent experiments.
Antibacterial leachates induce lipid peroxidation lesions in E. coli ATCC 25922
We measured the production of oxidative lesions in DNA, proteins, and lipids following leachate exposure. First, we used a carbonyl-derivative-based biomarker assay to measure direct oxidation of amino acid side chains in total protein samples of E. coli following CB-L exposure. Compared to water-exposed controls, E. coli exposure to CB-L decreased protein oxidative lesions, although the difference was not statistically significant (p = 0.0604) (Fig. 5A). Furthermore, addition of DMSO or EDTA to CB-L did not significantly affect protein oxidative damage (Fig. 5A). While it is possible that the water-exposed cells undergo osmotic shock during this exposure, the repeatable minimal change in viability suggested that influences of osmotic shock are negligible. Next, we used an ELISA-based method to measure changes in 8-oxo-2'-deoxyguanosine concentrations (8-OHdG), a well-characterized biomarker for DNA lesions induced by oxidative stress [32]. Compared to water-exposed controls, no significant change in 8-OHdG was detected following E. coli exposure to CB-L for 4 h (Fig. 5B). However, the addition of DMSO (hydroxyl radical scavenger) or EDTA (broad-spectrum chelator) to CB-L prior to E. coli exposure significantly decreased oxidative DNA lesions (Fig. 5B). Finally, we used a malonaldehyde (MDA)-based assay to quantify the extent of lipid peroxidation in E. coli cells following CB-L exposure. Data from these experiments demonstrated that after exposure of E. coli to CB-L, lipid peroxidation significantly increased by 12% over the water-exposed control cells (Fig. 5C). In summary, CB-L mediated bactericidal activity (Fig. 5D, Fig. 6) and significantly increased lipid peroxidation, but did not induce DNA or protein oxidative damage (Fig. 5A, B).
Figure 5. Assessment of E. coli ATCC 25922 oxidative damage to proteins (A), DNA (B), and lipids (C) and killing (D) upon exposure to CB-L and ROS scavengers or metal chelators.
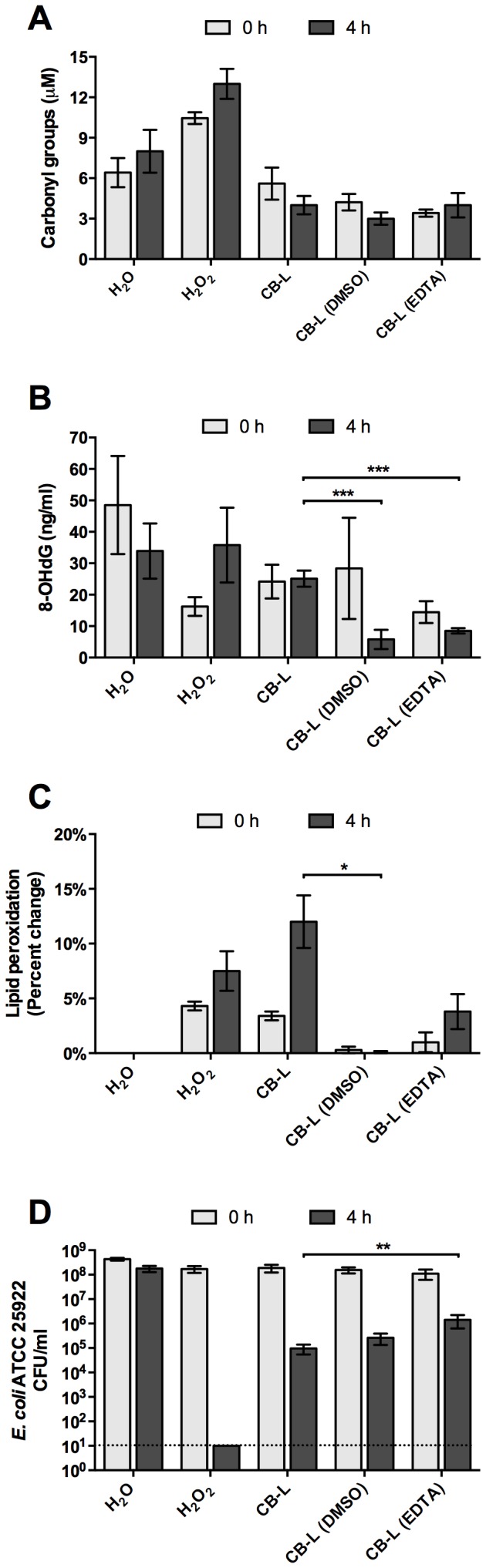
(A) Oxidative damage to protein samples collected from E. coli exposed to dH2O, H2O2 (2 mM), and CB-L with and without the addition of DMSO (1%) or EDTA (10 mM). Values represent the concentration of total carbonylated protein lesions formed as a result of protein oxidation during control and experimental exposures. Error bars represent the SEM for at least three independent experiments. (B) 8-OHdG levels measured in E. coli following CB-L exposure with and without the addition of DMSO or EDTA. Error bars represent the SEM for at least three independent experiments. (C) Lipid peroxidation levels measured in E. coli exposed to dH2O, H2O2, and CB-L with and without the addition of DMSO or EDTA. Values represent the percent change in lipid peroxidation over the water-exposed control, and the SEM for at least three independent experiments. (D) Viability of logarithmic phase E. coli following CB-L exposure with and without the addition of DMSO or EDTA. Error bars represent the SEM for at least three independent experiments. The dotted line represents the limit of detection. *, p<0.05; **, p<0.01; ***, p<0.002.
Figure 6. Comparison of log10 change in E. coli viability following exposure to CB-L with and without the addition of glucose (40 mM), EDTA (10 mM), DMSO (1%), and thiourea (150 mM).
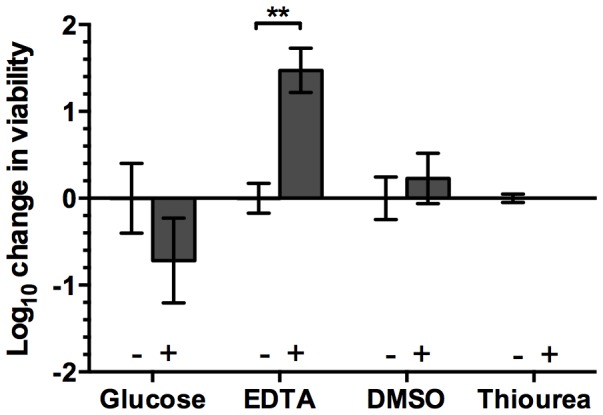
**, p<0.01.
Supplementation with glucose does not rescue cells from clay-mediated killing
E. coli K-12 (strain JM101) metabolite profiling studies using 13C-glucose labeling demonstrated that conserved post-transcriptional rerouting of the central carbon metabolic fluxome may be instrumental in combating oxidative stress and works to counteract oxidative shifts in the cytoplasmic redox state [33]–[36]. Collectively, the metabolic redistribution inactivates specific enzymes in the glycolytic pathway resulting in an increased flux through the pentose phosphate pathway (PPP). Induction of the PPP results in increased production of nicotinamide adenine dinucleotide phosphate-oxidase (NADPH) which is essential in maintaining the function of key antioxidant systems and other cellular processes [33], [34], [36]. We supplemented the antibacterial CB clays with glucose and assessed bacterial viability following clay exposure. With the addition of glucose, CB-mediated killing was maintained (Fig. 6), indicating that the bacterial cells were rapidly killed via mechanisms other than oxidative stress.
Supplementation with metal chelators rescues cells from clay-mediated killing
We supplemented CB-L suspensions with EDTA and assessed subsequent changes in E. coli ATCC 25922 viability. EDTA is a broad-spectrum chelator with high affinity for divalent and trivalent metal ions. The formation constants of EDTA-metal complexes vary depending on the pH of the solution and generally increase in affinity with decreasing pH [37]. EDTA is a hydrophilic molecule that does not penetrate cell membranes, thus, it will only chelate metals outside the cell. In a previous study, Jayasena et al. [38] exposed human neuroblastoma cells to Fe and H2O2 to induce hydroxyl radical stress. In these conditions, subsequent supplementation with EDTA did not significantly reduce hydroxyl radical production [38]. When CB-L was supplemented with EDTA, E. coli survival was significantly increased (p = 0.0094), indicating that the metals, not hydroxyl radicals, are killing the cells (Fig. 5D, Fig. 6). When CB-L was supplemented with DMSO, a hydroxyl radical scavenger, E. coli cells exhibited the same degree of killing as compared to CB-L alone (Fig. 5D, Fig. 6). Supplementation with DMSO during CB-L exposure eliminated lipid peroxidation (Fig. 5C). However, despite supplementation-based restoration of lipid integrity to levels comparable to the water-exposed control, CB-L maintained the same, or more, killing activity (Fig. 5D, Fig. 6). Further, when 150 mM thiourea, another hydroxyl radical scavenger, was added to the CB clay leachate or water immediately prior to contact with E. coli ATCC 25922, antibacterial activity remained unchanged, resulting in complete killing of E. coli within 24 h [3]. In summary, supplementation with ROS scavengers did not restore viability, whereas supplementation with metal chelators reduced CB-L-mediated killing.
Clay leachate sustains antibacterial activity in an anoxic environment
We exposed E. coli to CB-L under anoxic conditions, whereby ROS are not formed due to the lack of molecular oxygen. Data from time-course exposures of E. coli ATCC 25922 to CB-L demonstrated parallel killing rates over 24 h, with complete killing occurring in the anoxic-exposed samples after 24 h (Fig. 7). Therefore, CB-L was equally lethal to cells in an anaerobic chamber as to cells incubated in an aerobic environment. Thus, reactive oxygen species were not required for lethal activity and did not contribute to toxicity of CB-L.
Figure 7. Assessment of E. coli viability during exposure to CB-L under aerobic and anoxic conditions.
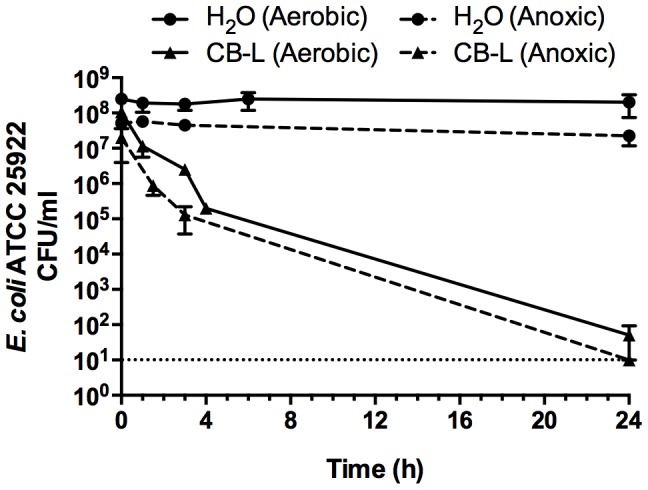
Error bars represent the SEM for three independent experiments. The dotted line represents the limit of detection.
Solutions of metal ion salts completely kill E. coli and MRSA
We previously used ICP-MS to analyze concentrations of metal cations present in the natural CB leachates [1]. Using the concentrations presented in Otto et al. [1], we prepared solutions of metal chloride salts, termed synthetic microbicidal mixtures (SMMs), that contained these metals at the same concentration and pH as in the natural leachate and tested these solutions for antibacterial activity against E. coli ATCC 25922 and MRSA. We should note, however, that the concentrations of metals in the natural leachates substantially vary [1] and, therefore, the metal concentrations in the SMMs only approximate those of natural leachates. SMM3, which was composed of all metals through Zn on the periodic table (see Materials and Methods for a complete list), completely killed E. coli and MRSA after a 24 h exposure (Fig. 8). Previous experiments in our lab demonstrated that E. coli exhibits increased survival in CB07-L supplemented with desferrioxamine [3], which readily chelates Fe, Co, Cu, and Zn [39]. Therefore, we prepared SMM16, which contained only Fe, Co, Cu, and Zn metal salts. The SMM16 solution completely killed E. coli and exhibited bactericidal properties against MRSA after a 24 h exposure (Fig. 8). These data support the conclusion that metal ions contribute to the antibacterial activity of the leachates.
Figure 8. Assessment of E. coli ATCC 25922 (A) and MRSA (B) viability following exposure to SMM3 and SMM16 synthetic microbicidal mixtures of metal salts.
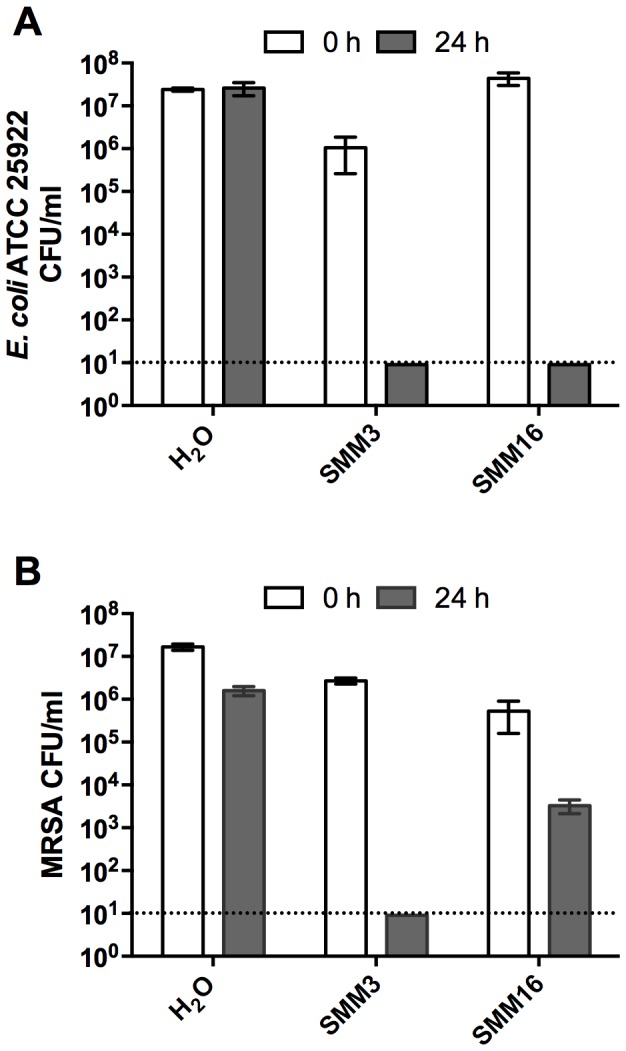
The dotted line represents the limit of detection.
Discussion
Some metals are known to exhibit toxicity in humans and some bacterial species. Previous literature reports that redox-active and -inactive metals increase production of hydroxyl radicals, superoxide, or hydrogen peroxide [40]–[42] and deplete cellular antioxidant defenses, particularly thiol-containing antioxidants and enzymes [43]. The exhaustion of antioxidant defenses is hypothesized to lead to oxidative stress and subsequent oxidative lesions in cellular lipids, proteins, and DNA. The toxic effects of metal exposure are often attributed to these lesions, as supplementation with extrinsic antioxidants ameliorates damage to the cellular macromolecules [44]. Here, we corroborate these previous data by demonstrating that antibacterial CB clay mixtures and leachates exhibit a strongly oxidizing environment, as revealed by high oxidation-reduction potential values [1], release redox-active metal ions [1], and generate H2O2 and ROS. We tested the hypothesis that metals released from the antibacterial CB clays generate H2O2 and ROS, and, in turn, oxidatively damage cellular macromolecules leading to eventual death. However, we did not observe a significant increase in oxidatively-damaged proteins or DNA. These data are consistent with previous literature demonstrating that Cu-exposed E. coli did not exhibit oxidatively-damaged DNA [45]. While Cu-supplemented cells exhibited substantial hydroxyl radical formation, the authors showed that the majority of H2O2-oxidizable Cu was located in the periplasm. Therefore, most of the Cu-mediated hydroxyl radical formation occurs in this compartment and away from the DNA [45]. While copper has previously been shown to cause oxidative damage to DNA and proteins, the experiments were conducted with purified DNA and protein samples and, thus, may not be relevant for living cell systems [46].
Supplementation of the leachate with thiourea and DMSO (ROS scavengers) failed to rescue the cells from killing. While catalase and superoxide dismutase are commonly used ROS scavengers, the enzymes were not used in this study because the low pH and high metal content of the leachates and clay suspensions would cause rapid loss of enzymatic activity. Rather, we supplemented the antibacterial CB leachate with thiourea and DMSO, which maintain ROS scavenging abilities in low pH environments [47], [48].
Exposure of E. coli ATCC 25922 to clay and leachate under anoxic conditions resulted in complete killing, demonstrating that oxygen was dispensable for toxicity. Iron toxicity is often explained by its participation in the Fenton reaction (Fe2+ + H2O2 → Fe3+ + HO· + OH−) [5], [49], [50]; however, this mechanism depends on the presence of O2 and thus cannot account for anaerobic toxicity [51].
Peroxidative damage of lipids correlated with high levels of extracellular ROS, but did not cause lethality. Recent reports suggest that classic antibiotics kill bacteria by indirect formation of ROS [52]. However, additional studies revealed that antibiotic exposures do not produce ROS and that lethality likely resulted from the direct inhibition of cell wall assembly, protein synthesis, and DNA replication [53]. These latter data are analogous to our data whereby oxidative stress is secondary to the direct mode of killing.
To our knowledge, this work is the first analysis of a complex antibacterial metal ion mixture with a strong oxidizing environment [1]. Previous literature addressing individual metals provides insight into the possible mechanism of action of the CB leachate, although the mechanism likely involves a sequence of events that are more complex than those in systems only containing one type of toxic metal ion. For example, Cd exposure increases the generation of ROS during exposure to bacterial cells [54], [55]. However, Cd is not a redox-active metal, so the mechanism of superoxide generation was unclear. Wang et al. [54] and Pacheco et al. [56] demonstrated that Cd interferes with cellular respiration by directly binding to complexes II and III in the electron transport chain in liver, brain, and heart mitochondria of guinea pig. When the function of complex III is inhibited in mitochondria, superoxide is generated, leading to further site-specific damage; however, the oxidative damage is not the principle mechanism of Cd toxicity [54], [57], [58]. Pacheco et al. [56] demonstrated that fermenting E. coli K-12 cultures, grown in media streamed with nitrogen and capped with a rubber stopper, are less sensitive to Cd toxicity than actively respiring cultures. Wang and Crowley [59] reported that Cd affects E. coli K-12 expression of genes associated with protein synthesis, energy metabolism, and cell rescue following 5, 10, and 25 min exposures. The up-regulation of genes associated with anaerobic metabolism and the shutdown of all high-energy consumption processes such as the biosynthesis of amino acids suggests that, upon exposure to Cd, cells switch to an energy conservation mode [59]. In a similar manner, the mechanism of action of Ag has been linked to its interaction with thiol group compounds such as those found in the respiratory enzymes of bacterial cells [60]–[63].
In this report, we corroborate the historical correlation between oxidative stress and metal exposure. However, we negate the hypothesis that the oxidative environment, generated by antibacterial clays and leachates, is required for killing. We collectively conclude that clay-mediated killing is not due to oxidative damage, but rather via toxicity associated directly with released metal ions. Our ongoing transcriptomics and molecular genetics studies will be used to investigate direct molecular markers of oxidative and metal stress and define the molecular mechanism of action. A comparison of expression profiles from cells exposed to the leachates under oxic versus anoxic conditions will also be important to assist in differentiating between the mechanism of toxicity in these two environments.
Acknowledgments
We thank Anca Delgado and Sudeep Popat for assistance with the anaerobic chamber training and protocols and Ryan LaMarca for assistance performing the anaerobic viability studies.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This research was supported by Public Health Service grant AT004690 awarded to SEH from the NIH National Center for Complementary and Alternative Medicine and an Arizona Board of Regents Doctoral Research Grant awarded to CCO. CCO was also partially supported by a graduate research fellowship from Science Foundation Arizona. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Otto CC, Haydel SE (2013) Exchangeable ions are responsible for the in vitro antibacterial properties of natural clay mixtures. PLoS ONE 8:e64068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Otto CC, Cunningham TM, Hansen MR, Haydel SE (2010) Effects of antibacterial mineral leachates on the cellular ultrastructure, morphology, and membrane integrity of Escherichia coli and methicillin-resistant Staphylococcus aureus . Ann Clin Microbiol Antimicrob 9:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cunningham TM, Koehl JL, Summers JS, Haydel SE (2010) pH-Dependent metal ion toxicity influences the antibacterial activity of two natural mineral mixtures. PLoS ONE 5:e9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haydel SE, Remenih CM, Williams LB (2008) Broad-spectrum in vitro antibacterial activities of clay minerals against antibiotic-susceptible and antibiotic-resistant bacterial pathogens. J Antimicrob Chemother 61:353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Touati D (2000) Iron and oxidative stress in bacteria. Arch Biochem Biophys 373:1–6. [DOI] [PubMed] [Google Scholar]
- 6. Halliwell B, Gutteridge JM (1984) Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J 219:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sandrin TR, Maier RM (2002) Effect of pH on cadmium toxicity, speciation, and accumulation during naphthalene biodegradation. Environ Toxicol Chem 21:2075–2079. [PubMed] [Google Scholar]
- 8. Chung H, Bang W, Drake M (2006) Stress response of Escherichia coli . Comp Rev Food Sci Food Safety 5:52–64. [Google Scholar]
- 9. Lin J, Lee IS, Frey J, Slonczewski JL, Foster JW (1995) Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli . J Bacteriol 177:4097–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hengge R (2008) The two-component network and the general stress sigma factor RpoS (sigma S) in Escherichia coli . Adv Exp Med Biol 631:40–53. [DOI] [PubMed] [Google Scholar]
- 11. Castanie-Cornet MP, Penfound TA, Smith D, Elliott JF, Foster JW (1999) Control of acid resistance in Escherichia coli . J Bacteriol 181:3525–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin J, Smith MP, Chapin KC, Baik HS, Bennett GN, et al. (1996) Mechanisms of acid resistance in enterohemorrhagic Escherichia coli . Appl Environ Microbiol 62:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rowbury RJ, Goodson M (1999) An extracellular acid stress-sensing protein needed for acid tolerance induction in Escherichia coli . FEMS Microbiol Lett 174:49–55. [DOI] [PubMed] [Google Scholar]
- 14. Hersh BM, Farooq FT, Barstad DN, Blankenhorn DL, Slonczewski JL (1996) A glutamate-dependent acid resistance gene in Escherichia coli . J Bacteriol 178:3978–3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Waterman SR, Small PL (1998) Acid-sensitive enteric pathogens are protected from killing under extremely acidic conditions of pH 2.5 when they are inoculated onto certain solid food sources. Appl Environ Microbiol 64:3882–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Small PL, Waterman SR (1998) Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli . Trends Microbiol 6:214–216. [DOI] [PubMed] [Google Scholar]
- 17. Stim KP, Bennett GN (1993) Nucleotide sequence of the adi gene, which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli . J Bacteriol 175:1221–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Winterbourn CC (1995) Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol Lett 82–83:969–874. [DOI] [PubMed] [Google Scholar]
- 19. Morones-Ramirez JR, Winkler JA, Spina CS, Collins JJ (2013) Silver enhances antibiotic activity against gram-negative bacteria. Sci Transl Med 5:190ra181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lloyd DR, Carmichael PL, Phillips DH (1998) Comparison of the formation of 8-hydroxy-2′-deoxyguanosine and single- and double-strand breaks in DNA mediated by fenton reactions. Chem Res Toxicol 11:420–427. [DOI] [PubMed] [Google Scholar]
- 21. Fang FC (2004) Antimicrobial reactive oxygen and nitrogen species: concepts and controversies. Nat Rev Microbiol 2:820–832. [DOI] [PubMed] [Google Scholar]
- 22. McCormick ML, Buettner GR, Britigan BE (1998) Endogenous superoxide dismutase levels regulate iron-dependent hydroxyl radical formation in Escherichia coli exposed to hydrogen peroxide. J Bacteriol 180:622–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pryor WA (1986) Oxy-radicals and related species: their formation, lifetimes, and reactions. Annu Rev Physiol 48:657–667. [DOI] [PubMed] [Google Scholar]
- 24. Gonzalez-Flecha B, Demple B (1995) Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli . J Biol Chem 270:13681–13687. [DOI] [PubMed] [Google Scholar]
- 25. Slauch JM (2011) How does the oxidative burst of macrophages kill bacteria? Still an open question. Mol Microbiol 80:580–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Berry ED, Cutter CN (2000) Effects of acid adaptation of Escherichia coli O157:H7 on efficacy of acetic acid spray washes to decontaminate beef carcass tissue. Appl Environ Microbiol 66:1493–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Popat SC, Deshusses MA (2009) Reductive dehalogenation of trichloroethene vapors in an anaerobic biotrickling filter. Environ Sci Technol 43:7856–7861. [DOI] [PubMed] [Google Scholar]
- 28. Shacter E, Williams JA, Lim M, Levine RL (1994) Differential susceptibility of plasma proteins to oxidative modification: examination by western blot immunoassay. Free Radic Biol Med 17:429–437. [DOI] [PubMed] [Google Scholar]
- 29. Borda MJ, Elsetinow AR, Schoonen MA, Strongin DR (2001) Pyrite-induced hydrogen peroxide formation as a driving force in the evolution of photosynthetic organisms on an early earth. Astrobiology 1:283–288. [DOI] [PubMed] [Google Scholar]
- 30. Ahlberg E, Elfström BA (1996) Oxygen reduction at sulphide minerals. 1. A rotating ring disc electrode (RRDE) study at galena and pyrite. Int J Miner Process 46:73–89. [Google Scholar]
- 31. Williams LB, Metge DW, Eberl DD, Harvey RW, Turner AG, et al. (2011) What makes a natural clay antibacterial? Environ Sci Technol 45:3768–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Toyokuni S, Tanaka T, Hattori Y, Nishiyama Y, Yoshida A, et al. (1997) Quantitative immunohistochemical determination of 8-hydroxy-2′-deoxyguanosine by a monoclonal antibody N45.1: its application to ferric nitrilotriacetate-induced renal carcinogenesis model. Lab Invest 76:365–374. [PubMed] [Google Scholar]
- 33. Grant C (2008) Metabolic reconfiguration is a regulated response to oxidative stress. J Biol 7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ralser M, Wamelink M, Kowald A, Gerisch B, Heeren G, et al. (2007) Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J Biol 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Averill-Bates D, Przybytkowski E (1994) The role of glucose in cellular defenses against cytotoxicity of hydrogen peroxide in Chinese hamster ovary cells. Arch Biochem Biophys 312:52–58. [DOI] [PubMed] [Google Scholar]
- 36. Rui B, Shen T, Zhou H, Liu J, Chen J, et al. (2010) A systematic investigation of Escherichia coli central carbon metabolism in response to superoxide stress. BMC Syst Biol 4:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harris D (2010) Quantitative chemical analysis. New York, NY: W. H. Freeman and Company.
- 38. Jayasena T, Grant RS, Keerthisinghe N, Solaja I, Smythe GA (2007) Membrane permeability of redox active metal chelators: an important element in reducing hydroxyl radical induced NAD+ depletion in neuronal cells. Neurosci Res 57:454–461. [DOI] [PubMed] [Google Scholar]
- 39. Keberle H (1964) The biochemistry of desferrioxamine and its relation to iron metabolism. Ann N Y Acad Sci 119:758–768. [DOI] [PubMed] [Google Scholar]
- 40. Imlay JA (2003) Pathways of oxidative damage. Annu Rev Microbiol 57:395–418. [DOI] [PubMed] [Google Scholar]
- 41. Freeman JL, Persans MW, Nieman K, Salt DE (2005) Nickel and cobalt resistance engineered in Escherichia coli by overexpression of serine acetyltransferase from the nickel hyperaccumulator plant Thlaspi goesingense . Appl Environ Microbiol 71:8627–8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Geslin C, Llanos J, Prieur D, Jeanthon C (2001) The manganese and iron superoxide dismutases protect Escherichia coli from heavy metal toxicity. Res Microbiol 152:901–905. [DOI] [PubMed] [Google Scholar]
- 43. Ercal N, Gurer-Orhan H, Aykin-Burns N (2001) Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Curr Top Med Chem 1:529–539. [DOI] [PubMed] [Google Scholar]
- 44. Gaetke LM, Chow CK (2003) Copper toxicity, oxidative stress, and antioxidant nutrients. Toxicology 189:147–163. [DOI] [PubMed] [Google Scholar]
- 45. Macomber L, Rensing C, Imlay JA (2007) Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli . J Bacteriol 189:1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cervantes-Cervantes MP, Calderon-Salinas JV, Albores A, Munoz-Sanchez JL (2005) Copper increases the damage to DNA and proteins caused by reactive oxygen species. Biol Trace Elem Res 103:229–248. [DOI] [PubMed] [Google Scholar]
- 47. Arifoglu M, Marmer W, Dudley R (1992) Reaction of thiourea with hydrogen peroxide: 13C NMR studies of an oxidative/reductive bleaching process. Text Res J 62:94–100. [Google Scholar]
- 48. Crow JP, Spruell C, Chen J, Gunn C, Ischiropoulos H, et al. (1994) On the pH-dependent yield of hydroxyl radical products from peroxynitrite. Free Radic Biol Med 16:331–338. [DOI] [PubMed] [Google Scholar]
- 49. Cornelis P, Wei Q, Andrews SC, Vinckx T (2011) Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3:540–549. [DOI] [PubMed] [Google Scholar]
- 50. Dupont CL, Grass G, Rensing C (2011) Copper toxicity and the origin of bacterial resistance—new insights and applications. Metallomics 3:1109–1118. [DOI] [PubMed] [Google Scholar]
- 51. Bird LJ, Coleman ML, Newman DK (2013) Iron and copper act synergistically to delay anaerobic growth of bacteria. Appl Environ Microbiol 79:3619–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ (2007) A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130:797–810. [DOI] [PubMed] [Google Scholar]
- 53. Liu Y, Imlay JA (2013) Cell death from antibiotics without the involvement of reactive oxygen species. Science 339:1210–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang Y, Fang J, Leonard SS, Rao KM (2004) Cadmium inhibits the electron transfer chain and induces reactive oxygen species. Free Radic Biol Med 36:1434–1443. [DOI] [PubMed] [Google Scholar]
- 55. Lopez E, Arce C, Oset-Gasque MJ, Canadas S, Gonzalez MP (2006) Cadmium induces reactive oxygen species generation and lipid peroxidation in cortical neurons in culture. Free Radic Biol Med 40:940–951. [DOI] [PubMed] [Google Scholar]
- 56. Pacheco CC, Passos JF, Castro AR, Moradas-Ferreira P, De Marco P (2008) Role of respiration and glutathione in cadmium-induced oxidative stress in Escherichia coli K-12. Arch Microbiol 189:271–278. [DOI] [PubMed] [Google Scholar]
- 57. Muller FL, Liu Y, Van Remmen H (2004) Complex III releases superoxide to both sides of the inner mitochondrial membrane. J Biol Chem 279:49064–49073. [DOI] [PubMed] [Google Scholar]
- 58. Turrens JF (1997) Superoxide production by the mitochondrial respiratory chain. Biosci Rep 17:3–8. [DOI] [PubMed] [Google Scholar]
- 59. Wang A, Crowley DE (2005) Global gene expression responses to cadmium toxicity in Escherichia coli . J Bacteriol 187:3259–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bragg PD, Rainnie DJ (1974) The effect of silver ions on the respiratory chain of Escherichia coli . Can J Microbiol 20:883–889. [DOI] [PubMed] [Google Scholar]
- 61. Klasen HJ (2000) Historical review of the use of silver in the treatment of burns. I. Early uses. Burns 26:117–130. [DOI] [PubMed] [Google Scholar]
- 62. Liau SY, Read DC, Pugh WJ, Furr JR, Russell AD (1997) Interaction of silver nitrate with readily identifiable groups: relationship to the antibacterial action of silver ions. Lett Appl Microbiol 25:279–283. [DOI] [PubMed] [Google Scholar]
- 63. Feng QL, Wu J, Chen GQ, Cui FZ, Kim TN, et al. (2000) A mechanistic study of the antibacterial effect of silver ions on Escherichia coli and Staphylococcus aureus . J Biomed Mater Res 52:662–668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.


