Abstract
Reverse transcription-quantitative real-time PCR (RT-qPCR) is a widely used technique for gene expression analysis. The reliability of this method depends largely on the suitable selection of stable reference genes for accurate data normalization. Hypericum perforatum L. (St. John's wort) is a field growing plant that is frequently exposed to a variety of adverse environmental stresses that can negatively affect its productivity. This widely known medicinal plant with broad pharmacological properties (anti-depressant, anti-tumor, anti-inflammatory, antiviral, antioxidant, anti-cancer, and antibacterial) has been overlooked with respect to the identification of reference genes suitable for RT-qPCR data normalization. In this study, 11 candidate reference genes were analyzed in H. perforatum plants subjected to cold and heat stresses. The expression stability of these genes was assessed using GeNorm, NormFinder and BestKeeper algorithms. The results revealed that the ranking of stability among the three algorithms showed only minor differences within each treatment. The best-ranked reference genes differed between cold- and heat-treated samples; nevertheless, TUB was the most stable gene in both experimental conditions. GSA and GAPDH were found to be reliable reference genes in cold-treated samples, while GAPDH showed low expression stability in heat-treated samples. 26SrRNA and H2A had the highest stabilities in the heat assay, whereas H2A was less stable in the cold assay. Finally, AOX1, AOX2, CAT1 and CHS genes, associated with plant stress responses and oxidative stress, were used as target genes to validate the reliability of identified reference genes. These target genes showed differential expression profiles over time in treated samples. This study not only is the first systematic analysis for the selection of suitable reference genes for RT-qPCR studies in H. perforatum subjected to temperature stress conditions, but may also provide valuable information about the roles of genes associated with temperature stress responses.
Introduction
Gene expression analysis has been widely used as a method to study the complex signaling and metabolic pathways underlying cellular and developmental processes in biological organisms, including plants. Growing number of studies of expression levels of several genes in plants have been carried out in order to understand the cellular and molecular mechanisms involved in plant development and growth, as well as, in plant responses to biotic (pathogen infection) and abiotic (environmental) stresses [1]–[4].
The analysis of gene expression has been performed by using different methods such as, northern blotting, ribonuclease protection assay, reverse transcription-polymerase chain reaction (RT-PCR), reverse transcription-quantitative real-time PCR (RT-qPCR), DNA microarrays [5], and next generation sequencing (NGS) technologies [6]. These last three technologies in particular have gained a wider appeal for the quantification of gene expression. It is clear that microarrays and NGS are extremely popular due to the ability to perform high throughput analysis. It is also clear that because of their relative simplicity and portability, qPCR-based assays will continue to be in demand for some considerable time [6]. Moreover, NGS data is currently expanding in many plant species [7]–[9] and RT-qPCR provides a reliable method for validating such huge amount of RNA Sequencing (RNA-seq) data [10]. However, several variables need to be controlled to obtain reliable quantitative expression measures by RT-qPCR. These include variations in initial sample quantity, RNA recovery, RNA integrity, efficiency of cDNA synthesis, and differences in the overall transcriptional activity of the tissues or cells analyzed [11]. To overcome the problem of variability, a normalization step has to be used prior to gene expression analysis in order to minimize its effects. The most common approach to normalize RT-qPCR data is the introduction of reference genes (RG) [12]–[15]. A suitable reference gene is assumed to be unaffected by the experimental conditions and therefore should be expressed at a constant level among samples [16]. Consequently, any changes in its expression level are due only to technical variations which should be discounted from the variation of the target gene expression levels. Indeed, the purpose of a reference gene is to remove the technical variations, ending up with true biological changes [17]. The use of only one reference gene as well as the use of the most frequently used reference genes for normalization without a prior validation is no longer considered a good strategy [18]. It is widely recognized that the use of inappropriate RGs may result in misinterpretation of the expression pattern of a given target gene thereby introducing flaws in the understanding of the gene's role. Recently, efforts have been directed towards systematization and standardization of these type of analyses, and the MIQE (minimum information for publication of quantitative real-time PCR experiments) guidelines suggest the use of three reference genes to generate more reliable results [19]. The identification of stable RGs in various experimental designs will therefore contribute to have more accurate and reliable gene expression data.
Several algorithms such as GeNorm [20], NormFinder [11], BestKeeper [21], ΔCt [22], qBasePlus [23], as well as single-factor analysis of variance (ANOVA) and linear regression analysis [24] have been utilized. These algorithms have been applied in order to analyze the expression stability of candidate RGs among genotypes, organs, tissues, developmental stages and several biotic and abiotic stress conditions. Nowadays, a high diversity of plant species have been studied, including a number of important crops, vegetables and fruit plant species (Table 1).
Table 1. Previous studies on candidate reference genes selection for RT-qPCR data normalization in several crops, vegetables and fruit plants.
| Plant species | Experimental condition (validated reference genes) | References |
| Glycine max L. | Seed development (TUA5, UKN2) and seed germination (Glyma05g37470 and Glyma08g28550). | [25] |
| Different stresses (EF1B and UKN2). | [26] | |
| Zea mays L. | Different tissues, developmental stages, and stress treatments (CUL, FPGS, LUG, MEP and UBCP). | [27] |
| Respect to abiotic stresses, hormones across different tissue types (EF1a, b-TUB, and their combination - EF1a+b-TUB). | [28] | |
| Oryza sativa L. | Different stress conditions (EP (LOC_Os05g08980), HNR (LOC_Os01g71770), and TBC (LOC_Os09g34040)). | [29] |
| Solanum tuberosum L. | Compatibility of potato–nematode interactions (RPN7, UBP22, OXA1 and MST2). | [30] |
| Tuber tissues exposed to cold treatments during different time periods (ef1a and APRT). | [31] | |
| S. lycopersicum L. | Seed germination (SGN-U601022 (at2g20000), SGN-U580609 (at3g18780), SGN-U579915 (at4g02080), SGN-U569038 (at1g13320), SGN-U563892 (at5g25760), SGN-U568398 (at3g25800), SGN-U566667 (at5g46630), SGN-U567355, SGN-U584254 (at4g34270)). | [32] |
| Brassica napus L. | Different tissues and cultivars, and under different conditions. (GDI1, PPR, UBA, OTP80 and ENTH). | [33] |
| Lactuta sativa L. | Different abiotic stresses (MIR169, MIR171/170 and MIR172). | [34] |
| Fragaria x ananassa | Different tissues, cultivars, biotic stresses, ripening and senescent conditions, and SA/JA treatments (FaRIB413, FaACTIN, FaEF1a and FaGAPDH2). | [35] |
| Citrullus lanatus | Low temperature stress in leaves (ClYLS8 and ClPP2A). | [36] |
| Cucumis melo L. | Leaves and roots under various stresses and growth regulator treatments (CmRPL and CmADP). | [37] |
| Lycium sp. | Fruit developmental stages (combination of GAPDH and EF1a). | [38] |
| Ripening fruits (EF1a and ACTIN1) or plants under salt stress (H2B1 and H2B2). | [39] | |
| Coffea sp. | General assay (GAPHD, Cycl, and UBQ10), genotype (GAPDH, UBQ10, Ap47, and EF-1A), cold stress (UBQ10, GAPDH, ACT, and EF-1A), drought stress (GAPDH, ACT, EF1A, and Apt), multiple stress (UBQ10, GAPDH, ACT, and elf-4A). | [40] |
| First hours of interaction (12, 48 and 72 hpi) with C. kahawae (IDE). | [41] | |
| Vitis vinifera L. | First hours of interaction (0 h, 6, 12, 18 and 24 hpi) with P. viticola to study genotype and biotic stress effects (UBQ, EF1a and GAPDH (genotype effect), EF1a, SAND and SMD3 (data normalization) and EF1a, GAPDH and UBQ Biotic stress effect). | [42] |
| Citrus sp. | Different citrus tissues (18SrRNA, ACTB and rpII). | [43] |
| Musa acuminata | Evaluation of robustness under different conditions, and in different tissues and varieties (EF1, ACT, and TUB (normalization in expression in leaves of greenhouse plants), ACT and L2 (leaf discs), and combinations of TUB, ACT/ACT11, and EF1 (expression studies in meristems)). | [44] |
| Carica papaya L. | Different experimental conditions (EIF, TBP1 and TBP2). | [45] |
| Pyrus pyrifolia L. | Different environmental conditions, tissue types and developmental stages (HIS, SAND, TIP). | [10] |
| Prunus persica L. | Biotic stress treatments (miR5059 and miR5072). | [46] |
| Cocos nucifera L. | Abiotic stress and endosperm developmental (eEF1-α and UBC10). | [47] |
However, to the best of our knowledge, no studies were performed on the evaluation of the expression stability of candidate RGs in Hypericum perforatum L. (commonly named St. John's wort). Moreover, the few studies on gene expression by RT-qPCR performed in H. perforatum rely, even recently, on only one reference gene for data normalization [48]–[50]. Apart from these studies, no information is available on the most appropriate RGs to be used in data normalization in studies related with temperature stress response in this plant species.
H. perforatum is a widely known medicinal herb used mostly as a medication for depression [51] having also other broad pharmacological activities, such as anti-tumor, anti-inflammatory, antiviral, antioxidant, anti-cancer, and antibacterial properties [52], [53]. These properties are mainly due to the biosynthesis and accumulation of important secondary metabolites in its tissues. Naphtodianthrones, which include hypericin and pseudohypericins, and hyperforin, are the main biological active substances naturally present in H. perforatum [54]. H. perforatum, as a field growing plant, is frequently exposed to a variety of adverse environmental stresses which negatively affect both productivity and secondary metabolites content [55]. Environmental stress, such as extreme cold and heat stress (CS and HS, respectively), are two factors that greatly affect cultivated plants. The Mediterranean climate is characterized by high thermal amplitude, not only during a season, but also within a day, and therefore, plants need to suppress and respond quickly to the adverse effects of extreme temperature changes. It is important to understand how plants adapt to adverse temperatures in face of the current temperature changes that are occurring globally and that can critically affect plants behavior, such as physiological processes, crop production, and metabolite production.
Examples of genes which are associated with plant responses to abiotic stresses are, the alternative oxidase (AOX1 and AOX2), the chalcone synthase (CHS) and catalase (CAT). AOX is a mitochondrial membrane protein that functions as terminal oxidase in the alternative (cyanide-resistant) respiratory pathway, where it reduces oxygen to water [56]. AOX relieves oxygen species (OS) originating from environmental stresses by limiting mitochondrial reactive oxygen species (ROS) formation and preventing specific components of the respiration chain from over-reduction [57] and canalizing ROS signals [58]. In dicot plant species AOX is nuclear encoded by a small multigene family composed by two sub-family genes, AOX1 and AOX2 [59]. CAT is a nuclear encoded enzyme which depending on its isoform could be localized in both the cytosol and peroxisomes [60] and probably also in the mitochondria [61]. CAT, the major H2O2-scavenging enzyme in all aerobic organisms [62], performs the rapid removal of H2O2 from the cell by oxidation of H2O2 to H2O and O2 [61]. CHS, a key enzyme of the flavonoid/isoflavonoid biosynthesis pathway and a component of the plant developmental program, is induced in plants under stress conditions such as UV light, bacterial or fungal infection [63].
For further development of RT-qPCR studies in H. perforatum that analyze the expression of stress responsive genes upon temperature stress conditions, the present study aimed to determine the most suitable RGs for data normalization. Here, we report a systematic analysis of eleven candidate RGs of H. perforatum, out of which some are commonly used as such, like the 18S ribosomal RNA (18SrRNA), the glyceraldehyde-3-phosphate dehydrogenase A subunit (GAPDH), and the beta-tubulin (TUB) [28], [33], but others not, such as the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (RBCL), glutamate-1-semialdehyde 2,1-aminomutase (GSA), chamba phenolic oxidative coupling protein (HYP1), short-chain dehydrogenase/reductase (SDR), polyketide synthase 1 (PKS1) and polyketide synthase 2 (PKS2). The expression stabilities of these candidate RGs were evaluated using the three distinct statistical algorithms, GeNorm, NormFinder and BestKeeper, in order to determine the most stable and therefore most suitable genes for an accurate RT-qPCR normalization in H. perforatum upon temperature stress conditions.
Materials and Methods
Plant material and experimental conditions
Hypericum perforatum L. seeds were removed from the achenes of a mother plant growing under field conditions in the Alentejo region (Viana do Alentejo, Portugal, 38°21′37″N, 7°59′13″W). H. perforatum is not considered an endangered or protected species and no specific permissions were required for achene harvesting. After disinfection, seeds were in vitro inoculated (see details in [64]). Eight weeks after germination seedlings were transferred to 6.6×5.9 cm glass culture vessels (100 ml, Sigma-Aldrich, Sintra, Portugal) with fresh MS medium [65] (10 seedlings per flask). Cultures were maintained in a growth chamber at 16 h photoperiod, constant temperature of 25°C, and 80 µmol m−2s−1 of light intensity provided by day-light Philips fluorescent lamps. Nine-week-old seedlings were used for gene expression assays.
Two different experimental conditions were conducted in this study:
1) Cold stress (CS): cultures were transferred to 4°C. All the other parameters were maintained. Samples were collected at different time points: 0, 4, 8, 12, 24, 48 and 72 hours post incubation (hpi). Each biological sample consisted in a bulked sample of 10 seedlings. Three replicates were collected per time point.
2) Heat stress (HS): cultures were transferred to 35°C (the remaining parameters were maintained). Samples were collected at different time points: 0, 12, 24, 72 hpi and 7 days post incubation (dpi). Each biological sample consisted in a bulked sample of 3 seedlings. Three replicates were collected per time point.
RNA isolation and first-strand cDNA synthesis
Total RNA was isolated with the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany), according to the supplier's instructions and eluted in 30 µl volume of RNase-free water. To digest residual genomic DNA, RNA samples were treated on-column with DNase I (RNase-Free DNase Set, Qiagen, Hilden, Germany) following the manufacturer's instructions. The concentration of total RNA was determined with the NanoDrop-2000C spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and its integrity analyzed by agarose gel electrophoresis (prepared with DEPC-treated water) after visualization of the two ribosomal subunits in a Gene Flash Bio Imaging system (Syngene, Cambridge, UK). A maximum of 1.5 µg of total RNA was used for reverse transcription with the Maxima First Strand cDNA Synthesis Kit for RT-qPCR (Thermo Scientific, Wilmington, DE, USA). All the subsequent procedures were performed according to the manufacturer's instructions.
Quantitative real-time PCR
Relative quantification of gene expression was performed in the Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Real-time PCR reactions were carried out using 1× Maxima SYBR Green qPCR Master Mix, 300 nM of forward and reverse primers, and 1.25 ng of cDNA in a total volume of 18 µl. Primers for 11 candidate RGs and 4 target genes (Table 2) (designed from H. perforatum sequences deposited in National Center for Biotechnology Information - NCBI) were designed with the Primer Express v3.0 (Applied Biosystems, Foster City, CA, USA) using the default properties given by the software: amplicon length between 50 and 150 bp; primer length between 18 and 25 bp; melting temperature (Tm) of 60°C; guanine and cytosine (GC) content of 60%. All primer pairs were checked for their probability to form dimmers and secondary structures with the primer test tool of the software. cDNA samples were previously diluted in order to get the same concentration for all samples followed by a ten-fold dilution. The reactions were performed using the following thermal profile: 10 min at 95°C, and 40 cycles of 15 s at 95°C and 60 s at 60°C. No-template controls (NTCs) were used to assess contaminations and primer dimmers. A standard curve was performed using undiluted pool of all cDNA samples and three five-fold serial dilutions. All samples were run in duplicate. Melting curve analysis was done to ensure amplification of the specific amplicon. Quantification cycle (Cq) values were acquired for each sample with the Applied Biosystems 7500 software (Applied Biosystems, Foster City, CA, USA).
Table 2. Gene ontology of candidate reference genes and target genes.
| Gene | Accession Number | Complete name | Biological Process1 | Cellular Component1 | Molecular Function1 |
| Candidate reference genes | |||||
| Hp18SrRNA | AF206934 | 18S ribosomal RNA | Translation** | Cytosolic small ribossomal subunit** | Structural constituent of ribosome** |
| Hp26SrRNA | DQ110887 | 26S ribosomal RNA | N/A | N/A | N/A |
| HpGAPDH | EU301783 | glyceraldehyde-3-phosphate dehydrogenase A subunit | Calvin cycle* | Chloroplast, Membrane, Plastid* | oxidoreductase activity |
| HpGSA | KJ624985 | glutamate-1-semialdehyde 2,1-aminomutase | Chlorophyll biosynthesis, Porphyrin biosynthesis* | Chloroplast, Plastid* | Isomerase* |
| HpHYP1 | JF774163 | Chamba phenolic oxidative coupling protein | Plant defense | N/A | Pathogenesis-related protein |
| HpH2A | EU034009 | histone 2A | nucleosome assembly | nucleosome, nucleus | DNA binding |
| HpPKS1 | EF186675 | polyketide synthase 1 | biosynthetic process | N/A | Transferase, Acyltransferase |
| HpPKS2 | 186676 | polyketide synthase 2 | biosynthetic process | N/A | Transferase, Acyltransferase |
| HpRBCL | HM850066 | ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | reductive pentose-phosphate cycle, Photosynthesis, Carbon dioxide fixation, Calvin cycle | Plastid, Chloroplast | Oxidoreductase, Monooxygenase, Lyase |
| HpSDR | EU034010 | short-chain dehydrogenase/reductase | Abscisic acid biosynthesis* | Cytoplasm* | Oxidoreductase* |
| HpTUB | KJ669725 | beta-tubulin | microtubule-based process, protein polymerization* | Cytoplasm, Cytoskeleton, Microtubule* | GTPase activity, protein binding* |
| Target genes | |||||
| HpAOX1 | EU330415 | Alternative oxidase 1 | Electron transport Respiratory chain Transport | Membrane | Oxidoreductase |
| HpAOX2 | EU330413 | Alternative oxidase 2 | Electron transport Respiratory chain Transport | Membrane | Oxidoreductase |
| HpCAT1 | AY173073 | Catalase-1 | hydrogen peroxide catabolic process | Cytoplasm* | Oxidoreductase Peroxidase |
| HpCHS | AF461105 | chalcone synthase | biosynthetic process | endoplasmic reticulum, plant-type vacuole membrane, nucleus* | Transferase, Acyltransferase |
From http://www.uniprot.org, N/A: not available. *Information obtained from the Arabidopsis thaliana NCBI protein accessions. **Information obtained from the Arabidopsis thaliana and provided by TAIR (www.arabidopsis.org).
Determination of gene expression stability
To determine the expression stability for each of the candidate RGs, 3 different statistical algorithms were applied: GeNorm [20], NormFinder [11], and BestKeeper [21].
GeNorm determines the pairwise variation of every control gene with all other control genes as the standard deviation of the logarithmically transformed expression ratios, and defines the internal control gene-stability measure M as the average pairwise variation of a particular gene with all other control genes. GeNorm also determines the optimal number of genes required to calculate a reliable normalization factor [20]. The advantage of GeNorm is that it can find suitable combinations of reference genes required for normalization, which is more reliable than using only one reference gene. The main limitation of GeNorm is its sensitivity to co-regulation [66]–[68]. Co-regulated genes will have similar expression patterns having similar M values in the pairwise analysis of GeNorm, however, they might not have a stable expression in a specific tissue.
NormFinder enables estimation not only of the overall variation in the expression level of the candidate reference genes but also of the variation between sample subgroups of the sample set. It provides a direct measure for the estimated expression variation, enabling the user to evaluate the systematic error introduced when using the gene. The advantage of NormFinder is that it takes into account the inter- and intra-group variations and it shows less sensitivity to co-regulation of the candidate reference genes [11].
BestKeeper is an Excel based tool able to compare expression levels for up to ten reference genes together with ten target genes, each in up to hundred biological samples. It determines the ‘optimal’ reference genes employing the pair-wise correlation analysis of all pairs of candidate genes and calculates the geometric mean of the ‘best’ suited ones. The weighted index is correlated with up to ten target genes using the same pair-wise correlation analysis. While GeNorm software is restricted to the reference genes analysis only, in BestKeeper software, additionally up to ten target genes can be analyzed. Once a robust BestKeeper index is constructed, it can be applied as an expression standard in the same way like any single reference gene [21]. Both NormFinder and BestKeeper were used to avoid co-regulation and to compare the results obtained by these two software programs with those obtained by GeNorm, one of the most used software to determine suitable reference genes.
RefFinder is a user-friendly web-based comprehensive tool developed for evaluating and screening reference genes from extensive experimental datasets. It integrates the currently available major computational programs (GeNorm, NormFinder, BestKeeper, and the comparative ΔΔCt method) to compare and rank the tested candidate reference genes. Based on the rankings from each program, it assigns an appropriate weight to an individual gene and calculates the geometric mean of their weights for the overall final ranking (http://www.leonxie.com/referencegene.php). RefFinder was used here in order to obtain the expression stabilities values from GeNorm, NormFinder and BestKeeper in the same tool, instead of using each program separately.
The input data were the raw Cq values for each sample and each of the candidate RGs (these were organized in columns). For each candidate gene, the stability values from the GeNorm (or average expression stability value, M) and NormFinder algorithms were taken, as well as, the values of the standard deviation of Cq value and the Pearson coefficient of correlation (r) were taken from the BestKeeper algorithm. For each RG a weight was given which corresponds to the number of the position given by GeNorm, NormFinder and BestKeeper. That is, the most stable RG was assigned the number 1 and the least stable RG was assigned the number 11. By calculating the geometric mean of these values, a ranking of the RGs using the three algorithms together was obtained and designated in this work by geometric mean.
Reference genes validation
In order to validate the candidate RGs, four target genes, namely CHS, CAT1, AOX1 and AOX2, were selected from the literature based on their response to environmental stress conditions [63], [69]–[72]. For normalization of the target genes expression levels, Cq values were converted into relative quantities (RQ) by the delta-Ct method [20] using the formula RQ = EΔCq, where E is the amplification efficiency calculated for each primer pair and ΔCq = lowest Cq – sample Cq. Amplification efficiency (E) was calculated using the formula E = 10(-1/slope), where the slope was given by the Applied Biosystems (AB) software. A normalization factor was obtained by calculating the geometric mean between the relative quantities of the selected RGs (for each normalization strategy) for each sample. For each target gene, calculating the ratio between the relative quantities for each sample and the corresponding normalization factor, a normalized gene expression value was obtained. The graphs show the mean ± standard deviation of three biological replicates and correspond to the ratio between treated and untreated samples for each time point, with bars representing the fold-change related to control group (0 hpi), which was set to 1. Statistical significances (p≤0.05 and p≤0.01) between the two means were determined by the t-test using IBM SPSS Statistics version 22.0 (SPSS Inc., USA).
Results
Amplification specificity and efficiency
RNA samples were analyzed for their quantity and purity with a NanoDrop-2000C spectrophotometer (Thermo Scientific, Wilmington, DE, USA). All samples showed an absorbance ratio at 260/280 nm of above 1.8. Agarose gel electrophoresis revealed only the two rRNA subunits (18S and 28S) with well-defined bands and with no indication of RNA degradation (data not shown).
The amplification specificity of each gene was confirmed by performing a melting curve analysis after each PCR run. All the primer pairs used amplified the expected specific product and no formation of primer dimmers was observed (S1 Figure and S2 Figure). Amplification in NTCs was observed only for 18S and 26S rRNAs which was due to high transcript abundance and therefore confirmed by low Cq (Cq<15) values (data not shown). PCR efficiency of each primer pair was calculated using a standard curve with four points: the undiluted pool, containing all cDNA samples, and three five-fold serial dilutions of the undiluted cDNA pool. The slope, y-intercepts, correlation coefficient (r2), and efficiency values were given by the AB software. The slope values ranged between −3.234 and −3.674 for CS assays, and between −3.259 and −3.574 for HS assays (data not shown). The correlation coefficient (r2) ranged from 0.991 to 0.999 for CS assays and from 0.993 to 1.00 for HS assays (except for TUB which has a lower value of 0.985) (Table 3). PCR efficiency, it ranged from 1.90 (90.50%) to 2.04 (103.81%) in CS assays (except for PKS2 having only 87.15%), and from 1.90 (90.45%) to 2.03 (102.70%) in HS assays.
Table 3. Primers sequences for candidate reference genes and target genes and other parameters.
| Gene (Accession Number)* | Gene | Primer Sequence (5′ – 3′) | Amplicon Length (bp) | Amplicon Tm (°C) | R2/E (%)** | |
| Cold/Heat | Cold Stress | Heat Stress | ||||
| Candidate reference genes | ||||||
| AF206934 | Hp18SrRNA | Fw: CGTCCCTGCCCTTTGTACAC Rv: CGAACACTTCACCGGACCAT | 72 | 80.3/80.3 | 0.999/1.932 (93.18) | 0.999/1.905 (90.46) |
| DQ110887 | Hp26SrRNA | Fw: GCGTTCGAATTGTAGTCTGAAGAA Rv: CGGCACCCCCTTCCAA | 65 | 80.8/81.0 | 0.998/1.966 (96.57) | 0.999/1.924 (92.39) |
| EU301783 | HpGAPDH | Fw: GGTCGACTTCAGGTGCAGTGA Rv: CACCATGTCGTCTCCCATCA | 76 | 81.0/81.3 | 0.998/1.905 (90.49) | 0.999/1.928 (92.78) |
| KJ624985 | HpGSA | Fw: GCAATAATCCTTGAACCTGTTGTG Rv: CCTGCGGAGAGCGTTGA | 78 | 78.4/78.7 | 0.998/2.011 (101.05) | 0.997/1.982 (98.19) |
| JF774163 | HpHYP1 | Fw: GGAGGAAGCAAGGGTAAGATTACA Rv: CCCGATCTTGACTTCTTCTTCATT | 81 | 77.0/77.0 | 0.998/1.967 (96.69) | 0.999/1.968 (96.76) |
| EU034009 | HpH2A | Fw: CCGGTTGGGAGGGTTCA Rv: TGCACCGACCCTTCCATT | 63 | 79.7/79.7 | 0.997/1.988 (98.76) | 0.998/1.967 (96.72) |
| EF186675 | HpPKS1 | Fw: ACGGACGCTGCCATCAA Rv: ACATAACCGTGTACCTTGTCTTCACA | 76 | 79.2/78.9 | 0.991/2.018 (101.78) | 0.996/1.958 (95.79) |
| 186676 | HpPKS2 | Fw: GCCTGCGCGATTGTAGGA Rv: GCCCTTCACTAGCTCGAATATTG | 66 | 80.7/81.5 | 0.991/1.871 (87.14) | 0.997/1.965 (96.49) |
| HM850066 | HpRBCL | Fw: CGCGGTGGGCTTGATTT Rv: CGATCCCTCCATCGCATAAA | 71 | 76.9/76.9 | 0.999/1.935 (93.47) | 1.000/1.953 (95.30) |
| EU034010 | HpSDR | Fw: TCGACCAAGCCGACTTGTATC Rv: CACCAAATGCAATTCTTGAAAATATC | 79 | 76.7/76.7 | 0.991/1.943 (94.32) | 0.993/1.982 (98.23) |
| KJ669725 | HpTUB | Fw: GGAGTACCCTGACAGAATGATGCT Rv: TTGTACGGCTCAACAACAGTATCC | 80 | 78.0/78.1 | 0.999/1.991 (99.09) | 0.985/1.919 (91.92) |
| Target genes | ||||||
| EU330415 | HpAOX1 | Fw: TTGGACAATGGCAACATCGA Rv: GGGAGGTAGGCGCCAGTAGT | 69 | 80.5/80.5 | 0.995/1.934 (93.41) | 0.998/1.933 (93.33) |
| EU330413 | HpAOX2 | Fw: TCAACGCCTACTTTGTGATCTATCTC Rv: AATGGCCTCTTCTTCCAAATAGC | 80 | 78.6/78.7 | 0.998/2.022 (102.17) | 0.994/1.941 (94.13) |
| AY173073 | HpCAT1 | Fw: CGCTTCCTCAACAGATGGATTAG Rv: ACCCAGATGGCTCTGATTTCA | 71 | 79.1/79.1 | 0.998/2.018 (101.82) | 0.999/1.983 (98.27) |
| AF461105 | HpCHS | Fw: GCGCTGCATCGATCATCA Rv: CAGCTCGAACAAGGGCTTTT | 65 | 80.3/80.3 | 0.997/2.038 (103.81) | 0.999/2.027 (102.70) |
*NCBI accession number, R2: correlation coefficient, E: PCR efficiency.
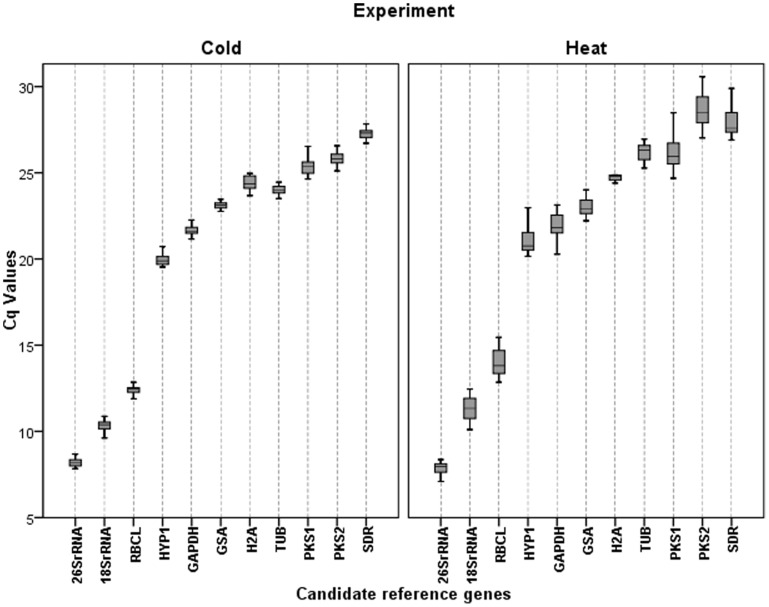
The expression levels of the candidate RGs were determined as Cq values. The 11 tested genes showed a wide range of transcript abundance in both CS and HS assays (Fig. 1). The candidate genes showed a relatively narrow variation in their expression levels across all samples, particularly in CS assays, as shown in box-plots where the boxes and whiskers are smaller than in HS assays. 18SrRNA, 26SrRNA and RBCL were the most highly expressed genes for both CS and HS with the lowest mean Cq (mCq) values (mCq between 7.97 and 13.91). SDR was the lowest expressed gene on both, CS and HS, showing the highest mCq (27.30 for CS and 28.02 for HS). With the exception of the ribosomal genes and the RBCL presenting mCq values around 8.0 and 14.0 for both assays, as mentioned above, the mCq values ranged between approximately 20.0 and 28.0 for all other candidate, being the HYP1 the most highly expressed gene with lower mCq values for both CS and HS assays (19.98 and 21.29, respectively), and SDR the less expressed gene.
Figure 1. Range of Cq values of the candidate reference genes for each experimental condition.
Each box corresponding to each candidate reference gene indicates the 25% and 75% percentiles. Whiskers represent the maximum and minimum values. The median is represented by the line across the box. 26SrRNA: 26S ribosomal RNA; 18SrRNA: 18S ribosomal RNA; RBCL: ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit; HYP1: Chamba phenolic oxidative coupling protein; GAPDH: glyceraldehyde-3-phosphate dehydrogenase A subunit; GSA: glutamate-1-semialdehyde 2,1-aminomutase; H2A: histone 2A; TUB: beta-tubulin; PKS1: polyketide synthase 1; PKS2: polyketide synthase 2; SDR: short-chain dehydrogenase/reductase.
Analysis of gene expression stability data
From the overall analysis, differences can be observed in the expression stability ranking of the candidate RGs between the two experimental conditions for each tested algorithm (Table 4). Additionally, differences can be also seen, although less significant, among the three algorithms applied within each assay. For CS experiments, GeNorm ranked GAPDH and GSA (M = 0.147) simultaneously as the most stable genes, and the 18SrRNA (M = 0.166) as the third best gene. For NormFinder, the most stable gene was the TUB (SV = 0.104) followed by 18SrRNA together with 26SrRNA (SV = 0.121 for both genes) and GAPDH (SV = 0.124). The BestKeeper algorithm ranked RBCL (SD = 0.10, r = 0.213) first, followed by TUB (SD = 0.12, r = 0.641) and 26SrRNA (SD = 0.12, r = 0.704) as the second and third ranked genes, respectively. From the geometric mean of all three algorithms, TUB, GSA, and GAPDH, were ranked in first, second and third positions, respectively.
Table 4. Candidate reference genes for cold and heat stresses determined by GeNorm, NormFinder, BesteKeeper, and by the combination of the 3 algorithms.
| COLD | COLD | COLD | COLD | HEAT | HEAT | HEAT | HEAT | |
| Rank | GeNorm M | NormFinder SV | BestKeeper SD/r | Geometric Mean | GeNorm M | NormFinder SV | BestKeeper SD/r | Geometric Mean |
| 1 | GAPDH | GSA (0.147) | TUB (0.104) | RBCL (0.10/0.213) | TUB (2.154) | 26S | H2A (0.174) | TUB (0.139) | 26S (0.28/0.798) | 26S (1.442) |
| 2 | 18S (0.121) | TUB (0.12/0.641) | GSA (2.714) | H2A (0.207) | H2A (0.34/0.826) | H2A (1.587) | ||
| 3 | 18S (0.166) | 26S (0.121) | 26S (0.12/0.704) | GAPDH (2.884) | GSA (0.336) | 26S (0.262) | 18S (0.35/0.494) | TUB (2.884) |
| 4 | 26S (0.179) | GAPDH (0.124) | GSA (0.16/0.697) | 26S (3.302) | SDR (0.425) | RBCL (0.338) | TUB (0.42/0.912) | 18S (4.932) |
| 5 | TUB (0.189) | GSA (0.152) | HYP1 (0.17/0.001) | 18S (3.476) | HYP1 (0.478) | 18S (0.473) | RBCL (0.54/0.760) | GSA (5.013) |
| 6 | SDR (0.199) | SDR (0.156) | GAPDH (0.17/0.705) | RBCL (3.826) | TUB (0.528) | GSA (0.561) | PKS1 (0.56/0.638) | RBCL (5.192) |
| 7 | H2A (0.208) | RBCL (0.169) | 18S (0.21/0.925) | SDR (6.604) | RBCL (0.584) | PKS1 (0.595) | GSA (0.57/0.634) | SDR (7.368) |
| 8 | RBCL (0.219) | H2A (0.240) | SDR (0.23/0.925) | HYP1 (7.399) | 18S (0.617) | GAPDH (0.648) | GAPDH (0.67/0.805) | HYP1 (7.399) |
| 9 | HYP1 (0.250) | HYP1 (0.339) | H2A (0.26/0.897) | H2A (7.958) | GAPDH (0.650) | HYP1 (0.716) | HYP1 (0.75/0.814) | PKS1 (7.489) |
| 10 | PKS2 (0.283) | PKS2 (0.384) | PKS2 (0.29/0.407) | PKS2 (10.00) | PKS1 (0.686) | SDR (0.764) | SDR (0.76/0.666) | GAPDH (8.320) |
| 11 | PKS1 (0.328) | PKS1 (0.486) | PKS1 (0.30/0.365) | PKS1 (11.00) | PKS2 (0.828) | PKS2 (1.416) | PKS2 (0.87/0.001) | PKS2 (11.00) |
M: expression stability average; SV: stability value; SD: standard deviation of Cq value; r: Pearson coefficient of correlation.
For HS tolerance assays, 26SrRNA and H2A were ranked in the two first positions with GeNorm (M = 0.174 for both genes) and BestKeeper (SD = 0.28, r = 0.798, SD = 0.34, r = 0.826, respectively). NormFinder ranked TUB in the first position (SV = 0.139) and H2A and 26SrRNA were selected as the second and the third best genes, respectively (SV = 0.207 and SV = 0.262, respectively). The third ranked RG by GeNorm was the GSA (M = 0.336) and by BestKeeper was the 18SrRNA (SD = 0.35, r = 0.494). For the overall final ranking, obtained by calculating the geometric mean of the ranked genes by the three algorithms, the three top RGs for the HS assay were the 26SrRNA, H2A, and TUB.
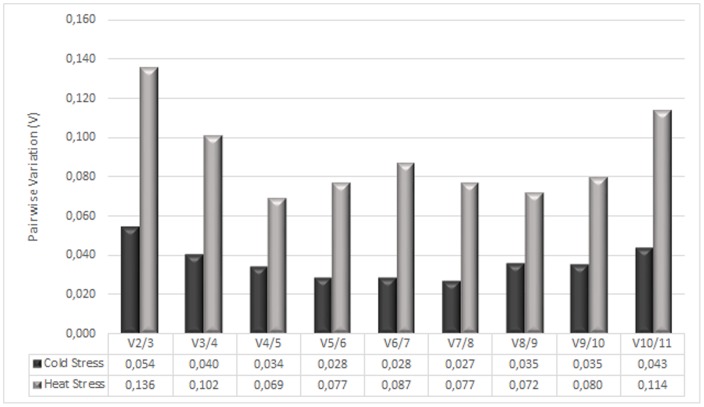
Determination of the optimal number of reference genes for normalization by GeNorm
The Genorm pairwise variation (V) values were determined for the candidate RGs in both experimental designs (CS and HS) using a cut-off value of 0.15, below which the inclusion of an additional RG is not required for normalization (Fig. 2). Interestingly, the optimal number of RGs used for normalization was only two for both CS assay (V2/3 = 0.054) and HS assay (V2/3 = 0.136), being both V values below the 0.15 cut-off value.
Figure 2. Determination of the optimal number of reference genes for normalization by pairwise variation using GeNorm.
Expression analysis of target genes for reference genes validation
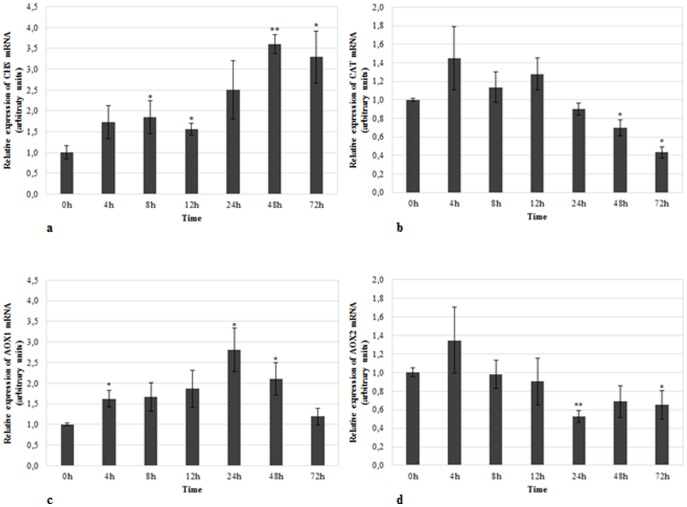
In order to validate the suitability of the selected candidate RGs, the expression profile of 4 target genes was analyzed in the present work. The target genes, selected from the literature based on their reaction to temperature stress response and oxidative stress [63], [69]–[72], were CHS, CAT1, AOX1 and AOX2. A single normalization strategy was applied to both assays, based on the geometric mean of the three algorithms by using the 3 top-ranked RGs. TUB, GSA and GAPDH were used as RGs for data normalization in CS whilst TUB together with 26S and H2A were used to normalize the expression results in HS assays. In CS assays, an accumulation of the CHS transcript was observed until 48 hpi with a 3.6 fold-change (p≤0.01) (Fig. 3a). From 24 hpi to 72 hpi, a down-regulation in the CAT1 mRNA expression for roughly half (p≤0.05) of the corresponding expression in the control group was observed (Fig. 3b). AOX1 transcript expression showed a gradual up-regulation in cold-treated samples until 24 hpi of about 2.8-fold (p≤0.05) with a following expression recovery until 72 hpi (Fig. 3c). AOX2 mRNA showed a reduction in its expression after 24 hpi in 1.9-fold (p≤0.01), with a further slight tendency to recover (Fig. 3d).
Figure 3. Relative mRNA expression of target genes in cold-treated samples.
Expression of a) CHS, b) CAT, c) AOX1, and d) AOX2 in cold-treated samples using TUB, GSA and GAPDH as reference genes in data normalization. The relative expression values are depicted as the mean ± standard deviation of three biological replicates and correspond to the ratio between treated and untreated samples for each time point. The bars represent the fold-change related to control group (0 hours) which was set to 1. Statistical significances (*p≤0.05 and **p≤0.01) between the two means were determined by the t-test using IBM SPSS Statistics version 22.0 (SPSS Inc., USA).
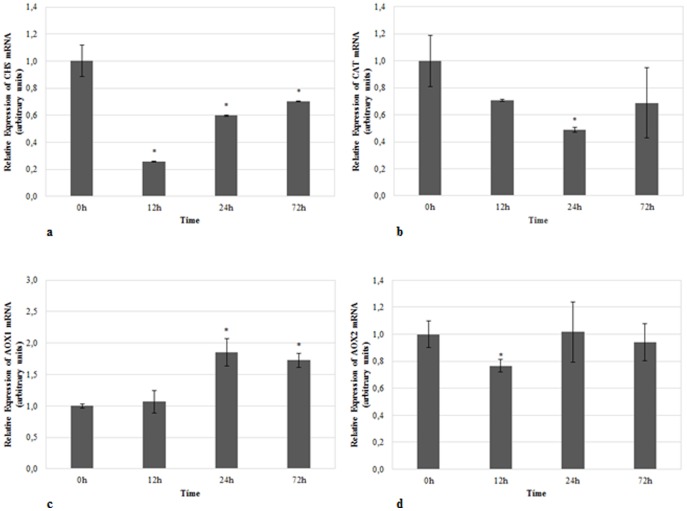
In HS assays, our results showed a down-regulation in the CHS at 12 hpi of 3.8 fold-change (p≤0.01) with a subsequent gradual recovery until 72 hpi (Fig. 4a). Similarly in CS, the mRNA expression of CAT1 had a tendency to decrease in heat-treated samples and after 24 hpi the value was around half (p≤0.05) of the expression in the control group (Fig. 4b). For AOX1, a slight increase in the mRNA expression of 1.8 fold-change (p≤0.05) was observed after 24 hpi, and this expression was maintained until 72 hpi (Fig. 4c). Although statistically significant (p≤0.05), the reduction in the AOX2 mRNA expression after 12 hpi was very modest demonstrating, in general, a high stability of this gene over time (Fig. 4d).
Figure 4. Relative mRNA expression of target genes in heat-treated samples.
Expression of a) CHS, b) CAT, c) AOX1, and d) AOX2 in heat-treated samples using TUB, 26S and H2A as reference genes in data normalization. The relative expression values are depicted as the mean ± standard deviation of three biological replicates and correspond to the ratio between treated and untreated samples for each time point. The bars represent the fold-change related to control group (0 hours) which was set to 1. Statistical significances (*p≤0.05 and **p≤0.01) between the two means were determined by the t-test using IBM SPSS Statistics version 22.0 (SPSS Inc., USA).
Discussion
In H. perforatum, studies on the expression stability of candidate RGs in a variety of experimental contexts has received no attention, and even recently only one RG is being used in RT-qPCR data normalization in this plant species [48]–[50] neglecting the knowledge that such a strategy is no longer admissible [19], [20]. Despite the pharmacological interest of the compounds produced by H. perforatum, very limited genomic information is available in NCBI. This is reflected by the absence of sequence data on a large number of frequently used genes. Consequently, for this study, a significant number of the genes selected had been recently isolated by members of our research group (GAPDH: EU301783, GSA: KJ624985, H2A: EU034009, SDR: EU034010, TUB: KJ669725, AOX1: EU330415, AOX2: EU330413). This study is the first detailed evaluation of the expression stability of several candidate RGs to be used for normalization in RT-qPCR studies in H. perforatum upon different stressful temperature conditions. Our analysis was based on the three most frequently used mathematical softwares, GeNorm, NormFinder and BestKeeper. Instead of using each software separately, the RefFinder tool was used here, since it integrates all three algorithms. We tested the expression stability of 11 candidate RGs, including commonly used RGs (TUB, GAPDH, and 18SrRNA), as well as, less frequently used RGs (HYP1, GSA, H2A, PKS1, PKS2, RBCL, SDR, and 26SrRNA). Within each algorithm, differences in the expression stability of the candidate genes were found when comparing the two experimental conditions of our study. These results reinforce previous conclusions, achieved by several authors using different plant species and experimental conditions, stating that the selection of the most stable RGs is highly dependent on the plant species and the experimental context [37], [73]–[75] and, therefore, assuming the existence of general RGs which can be used in different experimental situations could be of great risk. For example GAPDH, a commonly used RG, is ranked in first position by GeNorm in CS experiments, together with GSA. However, in heat treatment, this gene appeared to be an unsuitable RG ranking only in ninth position by the same algorithm. GAPDH has been also reported by others as a suitable RG, for example in Chlamydomonas during freezing acclimation [76] and in coffee upon CS [40]. However, it is not recommended for normalization of gene expression data in several studies involving temperature stress response and acclimatization performed in other plant species [29], [45], [77]. Another example from our study is the H2A, which is in final rankings in CS. However, in contrast, this gene is identified as one of the most stable RGs in HS, using the three algorithms.
In addition to the differences encountered in the selection of the most stable genes between the two experimental conditions, we also found some slight differences amongst the distinct algorithms tested. For example, RBCL was ranked in the first position by BestKeeper and therefore considered a good candidate in CS. However, it showed high expression variability using both NormFinder and GeNorm, staying only in seventh and eighth positions, respectively. One example, in HS, is the case of SDR, which is ranked in fourth position by GeNorm, but ranked only in tenth position by both NormFinder and BestKeeper. Similarly, results from other authors also demonstrate the existence of minor differences among distinct algorithms [13], [33], [38], and that are most probably due to the differences in the statistical algorithms. Nevertheless, taking into account the existence of some differences between algorithms, and additionally to overcome different limitations of each algorithm studied here, the stability of candidate RGs was also determined based on the geometric mean of all three statistical algorithms. Indeed, depending on the software used, the ranking of candidate reference genes can be slightly different [40], [78], [79]. Although many statistical approaches exist to determine both the stability of gene expression and to select the most appropriate RGs, to date, there is no consensus on which approach gives valid results. Thus, some authors consider the best approach is to combine the distinct algorithms for the selection of the most reliable reference genes in order to reduce the risk of artificial selection [80]. In fact, the simultaneous use of more than one algorithm has been also noted by other authors [67], [81] as producing highly correlated results and thus represents a good strategy for the selection of RGs for RT-qPCR normalization. In this study, the normalization strategy adopted to analyze the expression levels of the target genes to validate the RGs was based on the geometric mean of all three algorithms. Indeed, the software programs such as GeNorm, NormFinder and BestKeeper exist to determine the stability of the candidate RGs. However, the suitability of these genes should be evaluated with target genes associated with the experimental conditions in order to obtain reliable results [34]. The target genes used here were the CHS, CAT1, AOX1 and AOX2, which encode for proteins, documented in the literature, as associated with plant stress responses and oxidative stress. From our study, AOX1 transcripts were revealed to be affected by both cold and heat stressful temperatures, presenting an up-regulation in both conditions. In recent years, transgenic plants overexpressing AOX1 genes have provided molecular evidence that AOX improves cold tolerance [69], [71]. AOX was proposed, in a hypothesis-driven approach, as target to develop functional markers for general stress tolerance across species and stresses [82], [83]. Extensive reports have shown that the expression of the AOX1 gene is highly responsive to environmental stress, such as CS and HS, whilst AOX2 members are generally not responsive, or are much less responsive (see reviews [84], [85]). Indeed, from our results, AOX2 showed high expression stability in heat-treated samples. However, in contrast, this gene showed differential expression, being down-regulated in cold-treated samples. Both AOX and CAT are two antioxidative enzymes already described as being highly involved in ROS detoxification [86]. Our results demonstrate that despite a slight tendency for CAT1 transcript to be up-regulated (not statistically significant) in CS for the first time points, CAT1 mRNA is down-regulated both in CS and HS. Previous reports on the effects of stress on CAT activities vary depending on experiment and plant species or cultivars. Decreases in CAT activity have been reported upon HS in various plant species [87]–[90], in contrast with other studies which reported increases in CAT activity [91]–[98]. An increase in the CAT activity has also been reported upon CS [72], [99]–[103]. Nevertheless, the majority of studies available have focused on protein content and activity, with very few studies analyzing the link between transcript accumulation and protein. Locato et al. [97] have reported that in Nicotiana tabacum there is a decrease in transcript accumulation upon HS starting at 4 h after HS exposition. These authors suggest that under those experimental conditions, the regulation of CAT occurs with a mechanism acting downstream gene expression. In the same way, Watanabe and co-workers [104] revealed that in Arabidopsis thaliana a decrease in CAT1 transcript level was detected 20 h after CS exposure. Mhamdi and co-workers [105] suggested that a transcriptional down-regulation of CAT could be important to induce or sustain increased H2O2 availability necessary for certain environmental responses or developmental processes. Our data are in agreement with these previous results, suggesting that the molecular mechanisms associated with temperature stress response involving differential expression of CAT mRNA encountered both in N. tabacum and in A. thaliana might be also found in H. perforatum. In contrast, previous studies from Skyba et al. [49] demonstrated, in H. perforatum, an increase in CAT transcript accumulation upon a low-temperature associated stressor, being the results genotype-dependent. The similar expression profile of CAT1 and AOX2 in CS, shown by our study, in which both are down-regulated, supports current knowledge of these proteins regarding their common involvement in ROS detoxification [86]. Worth a notice, our results suggest an earlier involvement of AOX2 than CAT1 in this process, as shown by the AOX2 having the lowest mRNA expression at a time point earlier than CAT1.
We observed an increase in the CHS transcript accumulation upon CS. It is known that CHS can be induced in response to a diversity of biotic and abiotic stress factors, thus resulting in enhanced production of secondary metabolites (see review in [63]). Interestingly, we found a similar expression profile for CHS and AOX1 in CS, and even for HS, with the exception of the first time point (12 hpi) for which a marked reduction was observed for CHS, from this time point on, both genes are similarly up-regulated. These findings corroborate the results obtained by Fiorani and co-workers [106] who reported a relationship between AOX1a and CHS, demonstrating that, at low temperature, transgenic A. thaliana overexpressing AOX1a shows enhanced transcription of CHS, a key gene of the polyketide phenylpropanoids biosynthesis [63].
In summary, to the best of our know ledge, this is the first study in which a set of candidate reference genes are analyzed in terms of their expression stability, by three distinct mathematical algorithms, in order to be used in RT-qPCR data normalization with H. perforatum submitted to stressful temperature conditions. Our results demonstrate considerable differences in the gene expression stability between cold and heat stress treatments, highlighting the importance of performing a selection of the most suitable reference genes prior to the RT-qPCR studies for each experimental condition. Our data also show that the reference gene ranking obtained by GeNorm, NormFinder and BestKeeper were overall very similar and presenting only slight differences. The present study pointed to TUB as a suitable RG in RT-qPCR data normalization in temperature-treated H. perforatum plants. Additionally, GSA and GAPDH are recommended for data normalization in studies of cold treatment, whilst 26SrRNA and H2A are suggested for studies of heat treatment. The expression of the target genes CHS, CAT1, AOX1 and AOX2 were analyzed in order to emphasize the importance of validating the reference genes. They showed differential expression in both CS/HS experimental conditions, showing consistent results with what is known about the involvement of these genes in plant response to stressful temperatures, allowing the validation of the identified candidate reference genes.
Supporting Information
Melting curves of the 11 candidate reference genes and 4 target genes tested in cold assays.
(PDF)
Melting curves of the 11 candidate reference genes and 4 target genes tested in heat assays.
(PDF)
Acknowledgments
The authors are very grateful to Tânia Nobre for valuable suggestions, critical comments and careful proofreading of the manuscript. The authors especially thank Mary Albury (School of Biological Sciences, University of Sussex, UK) for great help in the English-language correction of the manuscript. We also thank to Amaia Nogales for helpful suggestions, Vera Valadas for the help in sample preparation and to Virginia Sobral for technical assistance. We also thank the reviewers for their valuable comments on the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Portuguese Foundation for Science and Technology within the frame of the project PTDC/AGR-GPL/099263/2008 and the program POPH – Operational Program for Human Potential (Ciência 2007 to BA and Ciência 2008 to HC: C2008-UE/ICAM/06). This work is funded by FEDER Funds through the Operational Program for Competitiveness Factors – COMPETE, and National Funds through FCT under the Strategic Projects PEst-C/AGR/UI0115/2011 and PEst-OE/AGR/UI0115/2014. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, et al. (2010) Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329:1306–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yamaguchi H, Fukuoka H, Arao T, Ohyama A, Nunome T, et al. (2010) Gene expression analysis in cadmium-stressed roots of a low cadmium-accumulating solanaceous plant, Solanum torvum. J Exp Bot 61:423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang D, Pan Y, Zhao X, Zhu L, Fu B, et al. (2011) Genome-wide temporal-spatial gene expression profiling of drought responsiveness in rice. BMC Genomics 12:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dechorgnat J, Patrit O, Krapp A, Fagard M, Daniel-Vedele F (2012) Characterization of the Nrt2.6 gene in Arabidopsis thaliana: A link with plant response to biotic and abiotic stress. PLoS One 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Guénin S, Mauriat M, Pelloux J, Van Wuytswinkel O, Bellini C, et al. (2009) Normalization of qRT-PCR data: The necessity of adopting a systematic, experimental conditions-specific, validation of references. J Exp Bot 60:487–493. [DOI] [PubMed] [Google Scholar]
- 6. Bustin SA (2010) Why the need for qPCR publication guidelines?-The case for MIQE. Methods 50:217–226. [DOI] [PubMed] [Google Scholar]
- 7. Zenoni S, Ferrarini A, Giacomelli E, Xumerle L, Fasoli M, et al. (2010) Characterization of transcriptional complexity during berry development in Vitis vinifera using RNA-Seq. Plant Physiol 152:1787–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhargava A, Clabaugh I, To JP, Maxwell BB, Chiang Y-H, et al. (2013) Identification of cytokinin-responsive genes using microarray meta-analysis and RNA-Seq in Arabidopsis. Plant Physiol 162:272–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhai R, Feng Y, Wang H, Zhan X, Shen X, et al. (2013) Transcriptome analysis of rice root heterosis by RNA-Seq. BMC Genomics 14:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Imai T, Ubi BE, Saito T, Moriguchi T (2014) Evaluation of Reference Genes for Accurate Normalization of Gene Expression for Real Time-Quantitative PCR in Pyrus pyrifolia Using Different Tissue Samples and Seasonal Conditions. PLoS One 9:e86492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250 doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 12. Yoo WG, Kim TI, Li S, Kwon OS, Cho PY, et al. (2009) Reference genes for quantitative analysis on Clonorchis sinensis gene expression by real-time PCR. Parasitol Res 104:321–328. [DOI] [PubMed] [Google Scholar]
- 13. Demidenko N V, Logacheva MD, Penin AA (2011) Selection and validation of reference genes for quantitative real-time PCR in buckwheat (Fagopyrum esculentum) based on transcriptome sequence data. PLoS One 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Everaert BR, Boulet GA, Timmermans JP, Vrints CJ (2011) Importance of suitable reference gene selection for quantitative real-time PCR: Special reference to mouse myocardial infarction studies. PLoS One 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hoenemann C, Hohe A (2001) Selection of reference genes for normalization of quantitative real-time PCR in cell cultures of Cyclamen persicum. Electron J Biotechnol 14. doi: 10.2225/vol14-issue1-fulltext-8.. [DOI] [Google Scholar]
- 16. Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol 29:23–39 doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 17. Derveaux S, Vandesompele J, Hellemans J (2010) How to do successful gene expression analysis using real-time PCR. Methods 50:227–230 doi: 10.1016/j.ymeth.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 18. Gutierrez L, Mauriat M, Pelloux J, Bellini C, Van Wuytswinkel O (2008) Towards a systematic validation of references in real-time rt-PCR. Plant Cell 20:1734–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. [DOI] [PubMed] [Google Scholar]
- 20. Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515. [DOI] [PubMed] [Google Scholar]
- 22. Silver N, Best S, Jiang J, Thein SL (2006) Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol 7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol 8:R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brunner AM, Yakovlev IA, Strauss SH (2004) Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Q, Fan C-M, Zhang X-M, Fu Y-F (2012) Validation of reference genes for real-time quantitative PCR normalization in soybean developmental and germinating seeds. Plant Cell Rep 31:1789–1798. [DOI] [PubMed] [Google Scholar]
- 26. Ma S, Niu H, Liu C, Zhang J, Hou C, et al. (2013) Expression stabilities of candidate reference genes for RT-qPCR under different stress conditions in soybean. PLoS One 8:e75271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manoli A, Sturaro A, Trevisan S, Quaggiotti S, Nonis A (2012) Evaluation of candidate reference genes for qPCR in maize. J Plant Physiol 169:807–815. [DOI] [PubMed] [Google Scholar]
- 28. Lin Y, Zhang C, Lan H, Gao S, Liu H, et al. (2014) Validation of Potential Reference Genes for qPCR in Maize across Abiotic Stresses, Hormone Treatments, and Tissue Types. PLoS One 9:e95445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maksup S, Supaibulwatana K, Selvaraj G (2013) High-quality reference genes for quantifying the transcriptional responses of Oryza sativa L. (ssp. indica and japonica) to abiotic stress conditions. Chinese Sci Bull 58:1919–1930. [Google Scholar]
- 30. Castro-Quezada P, Aarrouf J, Claverie M, Favery B, Mugniéry D, et al. (2013) Identification of Reference Genes for Normalizing RNA Expression in Potato Roots Infected with Cyst Nematodes. Plant Mol Biol Report 31:936–945. [Google Scholar]
- 31. Lopez-Pardo R, Ruiz de Galarreta JI, Ritter E (2013) Selection of housekeeping genes for qRT-PCR analysis in potato tubers under cold stress. Mol Breed 31:39–45. [Google Scholar]
- 32. Dekkers BJW, Willems L, Bassel GW, van Bolderen-Veldkamp RPM, Ligterink W, et al. (2012) Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53:28–37. [DOI] [PubMed] [Google Scholar]
- 33. Yang H, Liu J, Huang S, Guo T, Deng L, et al. (2014) Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 538:113–122. [DOI] [PubMed] [Google Scholar]
- 34.Borowski JM, Galli V, da Silva Messias R, Perin EC, Buss JH, et al. (2014) Selection of candidate reference genes for real-time PCR studies in lettuce under abiotic stresses. Planta: 1–14. [DOI] [PubMed]
- 35. Amil-Ruiz F, Garrido-Gala J, Blanco-Portales R, Folta KM, Muñoz-Blanco J, et al. (2013) Identification and validation of reference genes for transcript normalization in strawberry (Fragaria × ananassa) defense responses. PLoS One 8:e70603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kong Q, Yuan J, Gao L, Zhao S, Jiang W, et al. (2014) Identification of suitable reference genes for gene expression normalization in qRT-PCR analysis in watermelon. PLoS One 9:e90612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kong Q, Yuan J, Niu P, Xie J, Jiang W, et al. (2014) Screening Suitable Reference Genes for Normalization in Reverse Transcription Quantitative Real-Time PCR Analysis in Melon. PLoS One 9:e87197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang L, Wang Y, Zhou P (2013) Validation of reference genes for quantitative real-time PCR during Chinese wolfberry fruit development. Plant Physiol Biochem 70:304–310. [DOI] [PubMed] [Google Scholar]
- 39. Zeng S, Liu Y, Wu M, Liu X, Shen X, et al. (2014) Identification and validation of reference genes for quantitative real-time PCR normalization and its applications in lycium. PLoS One 9:e97039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goulao LF, Fortunato AS, C. Ramalho J (2011) Selection of Reference Genes for Normalizing Quantitative Real-Time PCR Gene Expression Data with Multiple Variables in Coffea spp. Plant Mol Biol Report 30:741–759. [Google Scholar]
- 41. Figueiredo A, Loureiro A, Batista D, Monteiro F, Várzea V, et al. (2013) Validation of reference genes for normalization of qPCR gene expression data from Coffea spp. hypocotyls inoculated with Colletotrichum kahawae. BMC Res Notes 6:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Monteiro F, Sebastiana M, Pais MS, Figueiredo A (2013) Reference gene selection and validation for the early responses to downy mildew infection in susceptible and resistant Vitis vinifera cultivars. PLoS One 8:e72998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yan J, Yuan F, Long G, Qin L, Deng Z (2012) Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Mol Biol Rep 39:1831–1838. [DOI] [PubMed] [Google Scholar]
- 44. Podevin N, Krauss A, Henry I, Swennen R, Remy S (2012) Selection and validation of reference genes for quantitative RT-PCR expression studies of the non-model crop Musa. Mol Breed 30:1237–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu X, Li X, Chen W, Chen J, Lu W, et al. (2012) Evaluation of new reference genes in papaya for accurate transcript normalization under different experimental conditions. PLoS One 7:e44405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luo X, Shi T, Sun H, Song J, Ni Z, et al. (2014) Selection of suitable inner reference genes for normalisation of microRNA expression response to abiotic stresses by RT-qPCR in leaves, flowers and young stems of peach. Sci Hortic (Amsterdam) 165:281–287. [Google Scholar]
- 47. Xia W, Liu Z, Yang Y, Xiao Y, Mason AS, et al. (2014) Selection of reference genes for quantitative real-time PCR in Cocos nucifera during abiotic stress. Botany 92:179–186. [Google Scholar]
- 48. He M, Wang Y, Hua W, Zhang Y, Wang Z (2012) De novo sequencing of Hypericum perforatum transcriptome to identify potential genes involved in the biosynthesis of active metabolites. PLoS One 7:e42081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Skyba M, Petijová L, Kosuth J, Koleva DP, Ganeva TG, et al. (2012) Oxidative stress and antioxidant response in Hypericum perforatum L. plants subjected to low temperature treatment. J Plant Physiol 169:955–964. [DOI] [PubMed] [Google Scholar]
- 50. Košuth J, Hrehorová D, Jaskolski M, Čellárová E (2013) Stress-induced expression and structure of the putative gene hyp-1 for hypericin biosynthesis. Plant Cell, Tissue Organ Cult 114:207–216. [Google Scholar]
- 51. Butterweck V (2003) Mechanism of action of St John's wort in depression: what is known? CNS Drugs 17:539–562. [DOI] [PubMed] [Google Scholar]
- 52. Birt DF, Widrlechner MP, Hammer KDP, Hillwig ML, Wei J, et al. (2009) Hypericum in infection: Identification of anti-viral and anti- inflammatory constituents. Pharm Biol 47:774–782 doi: 10.1080/13880200902988645.Hypericum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Caraci F, Crupi R, Drago F, Spina E (2011) Metabolic drug interactions between antidepressants and anticancer drugs: focus on selective serotonin reuptake inhibitors and hypericum extract. Curr Drug Metab 12:570–577. [DOI] [PubMed] [Google Scholar]
- 54. Sirvent TM, Krasnoff SB, Gibson DM (2003) Induction of hypericins and hyperforins in Hypericum perforatum in response to damage by herbivores. J Chem Ecol 29:2667–2681. [DOI] [PubMed] [Google Scholar]
- 55. Gadzovska S, Maury S, Delaunay A, Spasenoski M, Hagège D, et al. (2013) The influence of salicylic acid elicitation of shoots, callus, and cell suspension cultures on production of naphtodianthrones and phenylpropanoids in Hypericum perforatum L. Plant Cell, Tissue Organ Cult 113:25–39. [Google Scholar]
- 56. Umbach AL, Gonzalez-Meler MA, Sweet CR, Siedow JN (2002) Activation of the plant mitochondrial alternative oxidase: insights from site-directed mutagenesis. Biochim Biophys Acta 1554:118–128. [DOI] [PubMed] [Google Scholar]
- 57. Popov V, Simonian R, Skulachev V, Starkov A. (1997) Inhibition of the alternative oxidase stimulates H2O2 production in plant mitochondria. FEBS Lett 415:87–90. [DOI] [PubMed] [Google Scholar]
- 58. Amirsadeghi S, Robson C a, Vanlerberghe GC (2007) The role of the mitochondrion in plant responses to biotic stress. Physiol Plant 129:253–266. [Google Scholar]
- 59.Costa JH, McDonald AE, Arnholdt-Schmitt B, Fernandes de Melo D (2014) A classification scheme for alternative oxidases reveals the taxonomic distribution and evolutionary history of the enzyme in angiosperms. Mitochondrion: 1–12. [DOI] [PubMed]
- 60. McClung CR (1997) Regulation of catalases in Arabidopsis. Free Radic Biol Med 23:489–496. [DOI] [PubMed] [Google Scholar]
- 61. Møller IM, Jensen PE, Hansson A (2007) Oxidative modifications to cellular components in plants. Annu Rev Plant Biol 58:459–481. [DOI] [PubMed] [Google Scholar]
- 62. Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, et al. (2010) Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J Exp Bot 61:4197–4220. [DOI] [PubMed] [Google Scholar]
- 63. Dao TTH, Linthorst HJM, Verpoorte R (2011) Chalcone synthase and its functions in plant resistance. Phytochem Rev 10:397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ferreira AO, Cardoso HG, Macedo ES, Breviario D, Arnholdt-Schmitt B (2009) Intron polymorphism pattern in AOX1b of wild St John's wort (Hypericum perforatum) allows discrimination between individual plants. Physiol Plant 137:520–531. [DOI] [PubMed] [Google Scholar]
- 65. Murashige T, Skoog F (1962) A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol Plant 15:473–497. [Google Scholar]
- 66. Expósito-Rodríguez M, Borges AA, Borges-Pérez A, Pérez JA (2008) Selection of internal control genes for quantitative real-time RT-PCR studies during tomato development process. BMC Plant Biol 8:131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yeap W-C, Loo JM, Wong YC, Kulaveerasingam H (2013) Evaluation of suitable reference genes for qRT-PCR gene expression normalization in reproductive, vegetative tissues and during fruit development in oil palm. Plant Cell, Tissue Organ Cult 116:55–66. [Google Scholar]
- 69. Sugie A, Naydenov N, Mizuno N, Nakamura C, Takumi S (2006) Overexpression of wheat alternative oxidase gene Waox1a alters respiration capacity and response to reactive oxygen species under low temperature in transgenic Arabidopsis. Genes Genet Syst 81:349–354. [DOI] [PubMed] [Google Scholar]
- 70. Cansev A, Gulen H, Eris A (2011) The activities of catalase and ascorbate peroxidase in olive (Olea europaea L. cv. Gemlik) under low temperature stress. Hortic Environ Biotechnol 52:113–120. [Google Scholar]
- 71. Li CR, Liang DD, Xu RF, Li H, Zhang YP, et al. (2013) Overexpression of an alternative oxidase gene, OsAOX1a, improves cold tolerance in Oryza sativa L. Genet Mol Res. 12:5424–5432. [DOI] [PubMed] [Google Scholar]
- 72. Ma NL, Rahmat Z, Lam SS (2013) A Review of the “Omics” Approach to Biomarkers of Oxidative Stress in Oryza sativa. Int J Mol Sci 14:7515–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hruz T, Wyss M, Docquier M, Pfaffl MW, Masanetz S, et al. (2011) RefGenes: identification of reliable and condition specific reference genes for RT-qPCR data normalization. BMC Genomics 12:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhu J, Zhang L, Li W, Han S, Yang W, et al. (2013) Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PLoS One 8:e53196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu M, Jiang J, Han X, Qiao G, Zhuo R (2014) Validation of reference genes aiming accurate normalization of qRT-PCR data in Dendrocalamus latiflorus Munro. PLoS One 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu C, Wu G, Huang X, Liu S, Cong B (2012) Validation of housekeeping genes for gene expression studies in an ice alga Chlamydomonas during freezing acclimation. Extremophiles 16:419–425. [DOI] [PubMed] [Google Scholar]
- 77. Løvdal T, Lillo C (2009) Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal Biochem 387:238–242. [DOI] [PubMed] [Google Scholar]
- 78. Qi J, Yu S, Zhang F, Shen X, Zhao X, et al. (2010) Reference Gene Selection for Real-Time Quantitative Polymerase Chain Reaction of mRNA Transcript Levels in Chinese Cabbage (Brassica rapa L. ssp. pekinensis). Plant Mol Biol Report 28:597–604. [Google Scholar]
- 79. Rivera-Vega L, Mamidala P, Koch JL, Mason ME, Mittapalli O (2012) Evaluation of Reference Genes for Expression Studies in Ash (Fraxinus spp.). Plant Mol Biol Report 30:242–245. [Google Scholar]
- 80. Ayers D, Clements DN, Salway F, Day PJR (2007) Expression stability of commonly used reference genes in canine articular connective tissues. BMC Vet Res 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Reid KE, Olsson N, Schlosser J, Peng F, Lund ST (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol 6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Arnholdt-Schmitt B, Costa JH, De Melo DF (2006) AOX–a functional marker for efficient cell reprogramming under stress? Trends Plant Sci 11:281–287. [DOI] [PubMed] [Google Scholar]
- 83.Cardoso HG, Arnholdt-Schmitt A (2013) Functional Marker Development Across species in Selected Traits. In: Lübberstedt T, Varshney RK, editors. Diagnostics in Plant Breeding.Dordrecht: Springer Netherlands. pp.467–515. [Google Scholar]
- 84.Feng H, Guan D, Sun K, Wang Y, Zhang T, et al. (2013) Expression and signal regulation of the alternative oxidase genes under abiotic stresses. Acta Biochim Biophys Sin (Shanghai). doi: 10.1093/abbs/gmt094. [DOI] [PubMed]
- 85. Vanlerberghe GC (2013) Alternative Oxidase: A Mitochondrial Respiratory Pathway to Maintain Metabolic and Signaling Homeostasis during Abiotic and Biotic Stress in Plants. Int J Mol Sci 14:6805–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Møller IM (2001) P LANT M ITOCHONDRIA AND O XIDATIVE S TRESS: Electron Transport, NADPH Turnover, and Metabolism of Reactive Oxygen Species. [DOI] [PubMed]
- 87. Foyer CH, LopezDelgado H, Dat JF, Scott IM (1997) Hydrogen peroxide- and glutathione-associated mechanisms of acclimatory stress tolerance and signalling. Physiol Plant 100:241–254. [Google Scholar]
- 88. Dat J, Lopez-Delgado H, Foyer C, Scott I (1998) Parallel changes in H2O2 and catalase during thermotolerance induced by salicylic acid or heat acclimation in mustard seedlings. Plant Physiol 116:1351–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sato Y (2001) Heat shock-mediated APX gene expression and protection against chilling injury in rice seedlings. J Exp Bot 52:145–151. [PubMed] [Google Scholar]
- 90. Jiang Y (2001) Effects of calcium on antioxidant activities and water relations associated with heat tolerance in two cool-season grasses. J Exp Bot 52:341–349. [PubMed] [Google Scholar]
- 91. Sairam RK, Srivastava GC, Saxena DC (2000) Increased Antioxidant Activity under Elevated Temperatures: A Mechanism of Heat Stress Tolerance in Wheat Genotypes. Biol Plant 43:245–251. [Google Scholar]
- 92. Chaitanya K V, Sundar D, Masilamani S, Ramachandra Reddy A (2002) Variation in heat stress-induced antioxidant enzyme activities among three mulberry cultivars. Plant Growth Regul 36:175–180. [Google Scholar]
- 93. Orendi G, Zimmermann P, Baar C, Zentgraf U (2001) Loss of stress-induced expression of catalase3 during leaf senescence in Arabidopsis thaliana is restricted to oxidative stress. Plant Sci 161:301–314. [DOI] [PubMed] [Google Scholar]
- 94. He Y, Liu Y, Cao W, Huai M, Xu B, et al. (2005) Effects of Salicylic Acid on Heat Tolerance Associated with Antioxidant Metabolism in Kentucky Bluegrass. Crop Sci 45:988–995 doi: 10.2135/cropsci2003.0678. [DOI] [Google Scholar]
- 95. He Y, Huang B (2010) Differential Responses to Heat Stress in Activities and Isozymes of Four Antioxidant Enzymes for Two Cultivars of Kentucky Bluegrass Contrasting in Heat Tolerance. J Amer Soc Hort Sci 135:116–124. [Google Scholar]
- 96. Almeselmani M, Deshmukh PS, Sairam RK, Kushwaha SR, Singh TP (2006) Protective role of antioxidant enzymes under high temperature stress. Plant Sci 171:382–388. [DOI] [PubMed] [Google Scholar]
- 97. Locato V, Gadaleta C, De Gara L, De Pinto MC (2008) Production of reactive species and modulation of antioxidant network in response to heat shock: a critical balance for cell fate. Plant Cell Environ 31:1606–1619. [DOI] [PubMed] [Google Scholar]
- 98. Kumar RR, Goswami S, Sharma SK, Singh K, Gadpayle KA, et al. (2012) Protection against heat stress in wheat involves change in cell membrane stability, antioxidant enzymes, osmolyte, H2O2 and transcript of heat shock protein. Int J Plant Physiol Biochem 4:83–91. [Google Scholar]
- 99. Matsumura T, Tabayashi N, Kamagata Y, Souma C, Saruyama H (2002) Wheat catalase expressed in transgenic rice can improve tolerance against low temperature stress. Physiol Plant 116:317–327. [Google Scholar]
- 100. Kuk YI, Shin JS, Burgos NR, Hwang TE, Han O, et al. (2003) Antioxidative Enzymes Offer Protection from Chilling Damage in Rice Plants. Crop Sci 43:2109–2117. [Google Scholar]
- 101. Chen Y, Zhang M, Chen T, Zhang Y, An L (2006) The relationship between seasonal changes in anti-oxidative system and freezing tolerance in the leaves of evergreen woody plants of Sabina. South African J Bot 72:272–279. [Google Scholar]
- 102. Cansev A, Gulen H, Eris A (2011) The activities of catalase and ascorbate peroxidase in olive (Olea europaea L. cv. Gemlik) under low temperature stress. Hortic Environ Biotechnol 52:113–120. [Google Scholar]
- 103.Gong X-Q, Hu J-B, Liu J-H (2014) Cloning and characterization of FcWRKY40, A WRKY transcription factor from Fortunella crassifolia linked to oxidative stress tolerance. Plant Cell, Tissue Organ Cult: 1–14.
- 104. Watanabe CK, Hachiya T, Terashima I, Noguchi K (2008) The lack of alternative oxidase at low temperature leads to a disruption of the balance in carbon and nitrogen metabolism, and to an up-regulation of antioxidant defence systems in Arabidopsis thaliana leaves. Plant Cell Environ 31:1190–1202. [DOI] [PubMed] [Google Scholar]
- 105. Mhamdi A, Noctor G, Baker A (2012) Plant catalases: peroxisomal redox guardians. Arch Biochem Biophys 525:181–194. [DOI] [PubMed] [Google Scholar]
- 106. Fiorani F, Umbach AL, Siedow JN (2005) The Alternative Oxidase of Plant Mitochondria Is Involved in the Acclimation of Shoot Growth at Low Temperature. A Study of Arabidopsis AOX1a Transgenic Plants. Plant Physiol 139:1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Melting curves of the 11 candidate reference genes and 4 target genes tested in cold assays.
(PDF)
Melting curves of the 11 candidate reference genes and 4 target genes tested in heat assays.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.