We have demonstrated the potential of combining quantitative image features with advanced bioinformatics analysis to create radiogenomic maps providing molecular correlates of morphologic characteristics of glioblastoma multiforme.
Abstract
Purpose
To derive quantitative image features from magnetic resonance (MR) images that characterize the radiographic phenotype of glioblastoma multiforme (GBM) lesions and to create radiogenomic maps associating these features with various molecular data.
Materials and Methods
Clinical, molecular, and MR imaging data for GBMs in 55 patients were obtained from the Cancer Genome Atlas and the Cancer Imaging Archive after local ethics committee and institutional review board approval. Regions of interest (ROIs) corresponding to enhancing necrotic portions of tumor and peritumoral edema were drawn, and quantitative image features were derived from these ROIs. Robust quantitative image features were defined on the basis of an intraclass correlation coefficient of 0.6 for a digital algorithmic modification and a test-retest analysis. The robust features were visualized by using hierarchic clustering and were correlated with survival by using Cox proportional hazards modeling. Next, these robust image features were correlated with manual radiologist annotations from the Visually Accessible Rembrandt Images (VASARI) feature set and GBM molecular subgroups by using nonparametric statistical tests. A bioinformatic algorithm was used to create gene expression modules, defined as a set of coexpressed genes together with a multivariate model of cancer driver genes predictive of the module’s expression pattern. Modules were correlated with robust image features by using the Spearman correlation test to create radiogenomic maps and to link robust image features with molecular pathways.
Results
Eighteen image features passed the robustness analysis and were further analyzed for the three types of ROIs, for a total of 54 image features. Three enhancement features were significantly correlated with survival, 77 significant correlations were found between robust quantitative features and the VASARI feature set, and seven image features were correlated with molecular subgroups (P < .05 for all). A radiogenomics map was created to link image features with gene expression modules and allowed linkage of 56% (30 of 54) of the image features with biologic processes.
Conclusion
Radiogenomic approaches in GBM have the potential to predict clinical and molecular characteristics of tumors noninvasively.
© RSNA, 2014
Introduction
Glioblastoma multiforme (GBM) is the most frequent primary malignant brain tumor in adults. Despite decades of research and multimodality treatment with microsurgical resection followed by chemotherapy and radiation therapy, mean survival time is only 12–14 months (1). Genomic characterization has recently improved the clinical assessment of GBM with the description of distinct molecular gene expression profiles, underlying genomic abnormalities, and epigenetic modifications (2–5). The development of a radiogenomic map—a link between image features and underlying molecular data—holds the potential to address the clinical need for surrogate biomarkers that accurately predict underlying tumor biology and therapy response in GBM.
Our work focuses on extending radiogenomic analysis of GBM in two ways: Adding computational approaches to extract quantitative image features and utilizing integrated bioinformatics analysis that incorporates multiple modes of molecular data. In addition to manually annotated semantic features, computational methods can be used to generate quantitative image features. Previously, we developed a quantitative image feature pipeline to extract quantitative image features from computed tomographic (CT) and positron emission tomographic/CT data in non–small cell lung cancer and correlated these features with matched gene expression (6,7). Here, we extend these quantitative image features to GBM and magnetic resonance (MR) images. Additionally, we propose the derivation of quantitative image features for three distinct regions of interest (ROIs): necrosis and enhancement ROIs on T1-weighted postcontrast images and an edema ROI on T2-weighted fluid-attenuated inversion recovery (FLAIR) images.
We also propose to take advantage of the extensive molecular characterization, including gene expression, copy number, and DNA methylation status, available for the tumors in patients with GBM in the Cancer Genome Atlas (TCGA) by using a comprehensive integration strategy called Amaretto to summarize the molecular data as gene expression modules (8,9). A module is defined as a set of coexpressed genes together with a multivariate model of cancer driver genes predictive of the module’s expression pattern. The cancer driver genes are chosen on the basis of strong statistical evidence of genomic or epigenomic alterations. By correlating quantitative image features with Amaretto modules, we create richer hypotheses of how gene expression patterns are driving the morphologic manifestations captured by quantitative image features.
In this study, we applied these two approaches to create radiogenomic maps in patients with GBM and to detail the insights they provide about underlying biologic mechanisms. In summary, the purpose of this study was to derive quantitative image features from MR images that characterize the radiographic phenotype of GBM lesions and to create radiogenomic maps associating these features with various molecular data.
Materials and Methods
Image Selection and Annotation Processing
We obtained clinical, molecular, and MR imaging GBM data from TCGA and the Cancer Imaging Archive after local ethics committee and institutional review board approval. The MR image data sets were downloaded from the Cancer Imaging Archive in July 2012 (www.cancerimagingarchive.net) and originated from four centers (Henry Ford Hospital, University of California San Francisco, M.D. Anderson Cancer Center, and Emory University). We extracted the imaging protocol descriptions from the Digital Imaging and Communications in Medicine, or DICOM, header and standardized the annotations for the axial images. The inclusion criteria for this study were presurgical axial T1-weighted images obtained before and after the administration of gadolinium-based contrast material, T2-weighted FLAIR images, and treatment-naive gene expression data. We included 55 of 75 patients. Eleven patients were excluded because MR images were obtained after surgery, and nine patients were excluded because no gene expression data were available. Next, a board-certified neuroradiologist (L.A.M., with 5 years of experience) outlined ROIs in two dimensions on the single imaging section that had the largest volume of tumor corresponding to the enhanced (“enhancement ROI”) and necrotic (“necrotic ROI”) tumor portions on T1-weighted postcontrast images by using the electronic Physician Annotation Device (ePAD), an Osirix plug-in allowing annotation of ROIs (10). ROIs were confirmed by an expert board-certified neurosurgeon (A.S.A., with 5 years of experience in neuroimaging and neurosurgical oncology). Enhancement was confirmed by comparison with the T1-weighted precontrast images. The signal abnormality seen on the T2-weighted FLAIR image was similarly outlined and defined as the “edema ROI,” which included both peritumoral edema and any nonenhancing tumor. These ROIs were then used to compute features that characterized the intensities within the ROIs, the sharpness of lesion boundaries, and the boundary shapes, as previously described (6). We used our quantitative image feature pipeline to generate 79 computational image features for each of 321 regions corresponding to 55 patients, including multiple ROIs in cases of multicentric or multifocal GBM. Image features for multicentric or multifocal cases where averaged for each type of ROI.
Image Feature Robustness Analysis
We estimated the robustness of the image features in two ways: digital algorithmic modification and test-retest analysis. For the digital algorithmic modification, we investigated the effect of variations in the ROIs by modifying the ROIs as follows: (a) horizontal translation by 2 pixels; (b) horizontal and vertical translation by 2 pixels; (c) 1° rotation; (d) 5° rotation; (e) moving each point on the outline along the horizontal and vertical axes by a zero-mean random number with a 0.1-pixel standard deviation; (f) same as e with a 0.5-pixel standard deviation; (g) combining modifications a, c, and e; (h) combining modifications b, d, and f; (i) enlarging by 1 pixel along radial lines; and (j) shrinking by 1 pixel along radial lines. Next, we performed a test-retest analysis based on 21 cases. For each of these cases, ROIs were drawn twice, either in the same imaging section (n = 18) or in a neighboring section (n = 3), and were subjected to the quantitative image feature pipeline to generate quantitative image features.
Genomic Data Integration with Amaretto
All patients had extensive molecular data available in TCGA. We used Amaretto (8,9) to integrate gene expression, DNA methylation, and copy number data into gene expression modules, thereby creating 100 coexpressed gene expression modules. Amaretto is a two-step algorithm that we previously used to link gene expression modules with ovarian cancer subtypes (9). The first step of Amaretto identifies cancer driver genes by modeling the relationship between genomic and transcriptomic data on an individual-gene basis. The second step uses the cancer driver genes identified from the first step and takes a global approach by dissecting global gene expression data into modules of coexpressed genes. After model building, each module has an associated gene regulatory program that connects the cancer driver genes from the first step with their downstream targets. This gene regulatory program is modeled by using linear regression with elastic net regularization (11). Next, we selected modules that were significantly correlated with overall survival across all 426 TCGA cases (P < .05); this resulted in 35 modules. TCGA contains more cases than just those cases with imaging data, allowing us to indirectly infer the potential survival relationships of quantitative image features by correlating them with survival-correlated modules and thus focusing only on gene expression modules that are prognostic. The survival-correlated modules were represented by the mean expression of the genes in each module and were subsequently associated with the image features by using the Spearman correlation test, thereby creating a prognostic radiogenomic map for necrosis, enhancement, and edema ROIs.
Associating Quantitative Image Features with Molecular Pathways
We associated image features with molecular pathways by using the Amaretto modules as an intermediate (Fig E1 [online]). We started with the radiogenomic maps for necrosis, enhancement, and edema, providing an image feature–versus-module map. Next, we annotated all modules by using gene enrichment analysis, resulting in a module-versus-pathway association map (see below). Both association maps were defined by P values. We transformed these P values by taking their negative natural logarithms. Next, we used the matrix product of both matrices, creating an image feature–versus-pathway map (Fig E1 [online]). The matrix multiplication takes into account the multivariate association between an image feature and the modules and the association between a pathway and all modules. To evaluate the enrichment of Amaretto modules with biologic pathways, we used the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (12).
Statistical Analysis
We used the previously described 10 scenarios of digital algorithmic modification on all ROIs and assessed image feature robustness using the intraclass correlation coefficient (ICC) (13). Robust features were defined on the basis of an ICC of 0.6 for both the digital algorithmic modification and the test-retest analysis. We used hierarchic clustering with the Euclidean distance metric and average linkage to visualize the correlation structure of the robust quantitative image features. Next, we used Cox proportional hazards modeling to investigate univariate relationships between quantitative image features (n = 55), gene expression modules (n = 426), and overall survival (survival R package, version 2.36–10). Hazard ratios were used to report the direction of the survival effect, and the Wald test was used to determine the significance of Cox models. We applied the false discovery rate to correct for multiple hypothesis testing (14). Next, we used the Wilcoxon rank sum test to correlate quantitative image features with Visually Accessible Rembrandt Images (VASARI) features and with the GBM molecular subtypes. We used a hypergeometric test to check for enrichment of each KEGG pathway and all modules and corrected for multiple hypothesis testing using the false discovery rate (14).
Results
Quantitative Image Features Characterizing Necrotic, Enhancing, and Edema ROIs in GBM
Eighteen features had an ICC of 0.6 on the basis of digital algorithmic modification and the test-retest analysis (Table E1 [online]). Histogram and edge sharpness had high ICCs and were thus more robust to algorithmic and test-retest variation, whereas edge shape features had low ICCs and were less robust. We visualized the correlation structure between the robust quantitative image features using hierarchical clustering (Fig E2 [online]). This showed a cluster of correlated edema features where the mean edema intensity was highly correlated with the edge sharpness intensity difference (r = 0.81, P < .001). Overall, the correlation structure between the quantitative image features showed that, although clusters of correlated features exist, the quantitative image features captured unique characteristics of the lesion’s radiologic phenotype.
Correlation of Quantitative Image Features with Survival, VASARI Features, and Molecular Subtypes
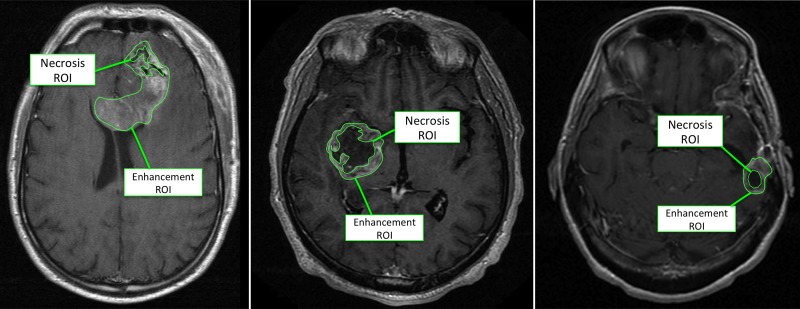
We investigated the correlation of robust quantitative image features with overall and progression-free survival and found three significant correlations for enhancement image features (P < .05; false discovery rate, <0.05; Table E2 [online]). For example, the variance of the radial distance signal of the enhancement ROI was the feature most highly correlated with overall survival (P = .017; hazard ratio, 0.67). This feature characterizes the irregularity of the border of the ROI, and a low value was correlated with good prognosis. Next, the edge sharpness of the enhancement was correlated with overall survival, with a smooth edge being characteristic of poor prognosis compared with a sharp edge (P = .023; hazard ratio, 1.43). Similarly, a low value for the blurriness of the edge sharpness was correlated with good progression-free survival (P = .028; hazard ratio, 0.48). Figure 1 shows examples of necrosis and enhancement ROIs in three different patients and displays the most extreme cases corresponding to the highest and lowest values for the irregularity of the border of the enhancement ROI. We could not study recurrence-free survival because of the limited number of patients with this outcome.
Figure 1:
Example axial T1-weighted postcontrast MR images show necrosis and enhancement ROIs.
Next, we found 77 significant correlations between quantitative features and the VASARI feature set, a community-developed visual guide to help standardize the assessment of GBM (Table E3 [online]). For example, the VASARI feature “well-defined enhancing margin” is correlated with several quantitative image features, including the variance of the intensity of the necrosis and enhancement ROIs and the enhancement edge sharpness. Similarly, the VASARI feature “ependymal extension” is correlated with two enhancement features that define the maximum intensity difference between the inside and the outside of the enhancement ROI. Edema edge sharpness features, on the other hand, are correlated with the VASARI features “proportion of edema” and “proportion of necrosis.” For example, when the average intensity difference between the inside and the outside of the edema ROI is low, most cases have less edema (<5%) than when it is high. Similarly, the VASARI feature “proportion of necrosis” is correlated with a high kurtosis of the edema intensity histogram, corresponding to a homogeneous edema ROI. Overall, we found a significant correlation with 12 distinct VASARI features. All significant associations between quantitative image features and VASARI image features are reported in Table E3 (online).
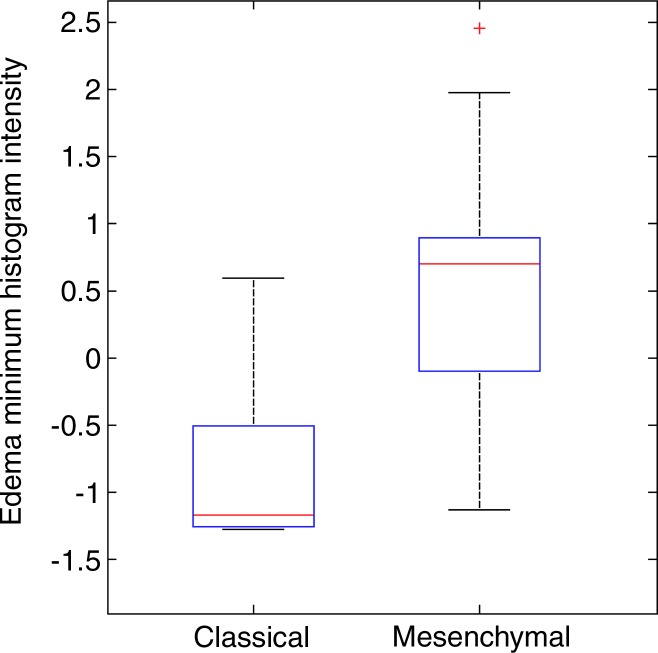
Next, we investigated correlations of quantitative image features with molecular subtypes and important molecular aberrations. TCGA described four molecular subtypes on the basis of gene expression analysis: classic, neural, proneural, and mesenchymal (2). We found that four features had a correlation with the TCGA molecular subgroups (P < .05). The mesenchymal subtype was correlated with one edema feature: the minimum intensity. The classic subtype was correlated with one necrosis and two edema image features related to edge sharpness and intensity. More specifically, edema ROIs in classic tumors have lower intensity than those in mesenchymal tumors (Fig 2).
Figure 2:
Boxplots show edema minimum histogram intensity and edema edge shape variability according to their differences within GBM molecular subgroups. The scales for both image features were standardized to have a mean of 0 and a standard deviation of 1.
Radiogenomic Map with Quantitative Image Features Revealed Novel Associations
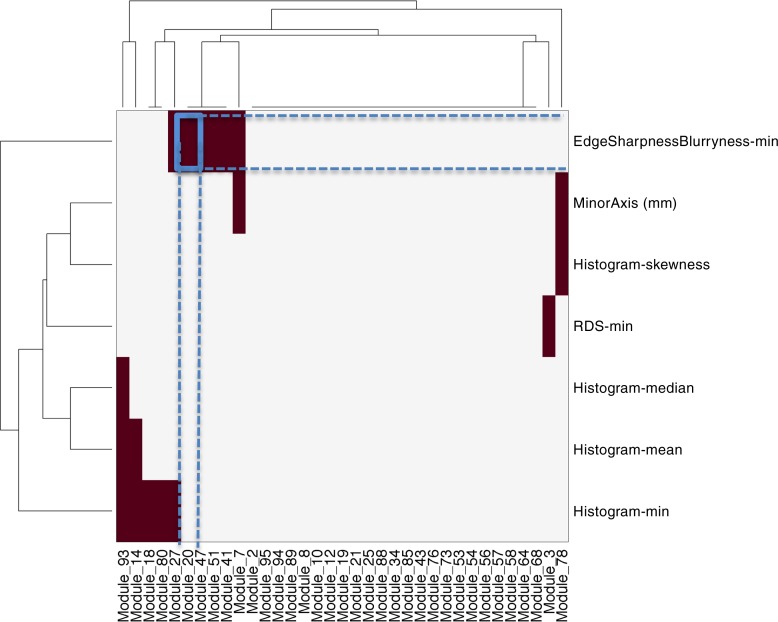
We used Amaretto to build 100 gene expression modules based on data in 426 patients in TCGA. Figure E3 (online) shows the gene expression modules together with important biologic pathway annotations for key modules. Next, we created radiogenomic maps by correlating only prognostic modules with quantitative image features. Figure 3 displays the radiogenomic maps between the necrosis quantitative image features and 35 prognostic modules separately for all ROIs. Figure E4 (online) illustrates the remaining radiogenomic maps for enhancement and edema. Each map shows several significant associations between modules and image features.
Figure 3:
Radiogenomic maps for necrosis features. Image features are in rows and modules are in columns. Each colored square = significant correlation between an image feature and a module. The association highlighted in blue in the necrosis map is presented in Figure 4. min = Minimum, RDS = radial distance signal.
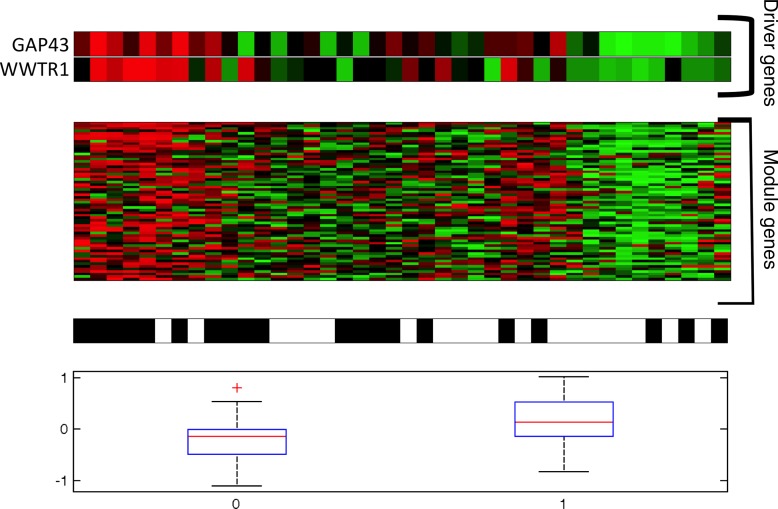
For example, we observed an anticorrelation between module 20 and the edge sharpness of the necrosis (Fig 4). Module 20 is highly expressed when the necrotic portion of the tumor has a blurry edge, whereas its expression is low when a tumor has a sharp necrosis edge. Module 20 is defined by GAP43 and WWTR1 as regulators, suggesting that these two genes are driving necrosis processes. WWTR1 is markedly hypermethylated and GAP43 is markedly deleted, suggesting that when both genes are switched off, sharp necrosis boundaries are created. GAP43 is associated with growth in neuronal development, and WWTR1 has been shown to be an oncogene in colorectal cancer cells that inhibits proliferation (15). Module 20 is further enriched with genes located in the cell membrane and genes in the IL4 pathway involved in T-cell differentiation and proliferation.
Figure 4:
Association between module 20 and the LAIIR2-var feature, defined as the variance of the necrosis edge shape. Top: Cancer driver genes of module 20. Red = high expression, green = low expression. Second-from-top: Expression of the genes in module 20. Second-from-bottom: LAIIR2-var feature. Black = high variance, white = low variance. Bottom: Boxplot of the LAIIR2-var feature for high versus low variance of the LAIIR2-var feature, corresponding to high or low expression of the module genes, respectively.
Radiogenomic Maps Associate Quantitative Image Features with Biologic Processes and Pathways
Although direct correlation of quantitative image features and modules provides insight into molecular biology, we also indirectly correlated each image feature with biologic pathways by leveraging the modules (Fig E1 [online]; see Materials and Methods). We were able to link 56% (30 of 54) of the image features with biologic processes (Fig E5 [online]). For example, the variance of the edema intensity was related to the KEGG cell cycle pathway. Similarly, image features related to edge sharpness were enriched with the KEGG focal adhesion pathway. The focal adhesion pathway involves genes related to cell motility, proliferation, and invasion. In normal tissues, genes in this pathway form the contact point with the extracellular matrix, and, when aberrantly regulated, this may lead to invasion and metastasis. Overall, we found several associations between image features and pathways, providing a rich resource for future experimental validation (Fig E5 [online]).
Discussion
In this study, we have shown exploratory results of two extensions to radiogenomic analysis of GBM cases from TCGA. We demonstrated the use of quantitative image features in GBM and reported significant correlations with survival, VASARI image features, and molecular data. We also showed the power of creating radiogenomic maps using Amaretto and using the gene expression modules to indirectly associate image features with underlying biologic processes. Our results demonstrate that building radiogenomic maps with quantitative image features and Amaretto is a promising complementary strategy toward noninvasive management of GBM.
The potential of radiogenomic maps was first demonstrated in hepatocellular carcinoma by Segal et al (16) and in GBM by Diehn et al (17). More recently, an editorial in Radiology reviewed recent efforts and concluded that “future research engagement will be most productive if it focuses on populating a three-domain Venn diagram intersection made up of genetics, clinical data, and imaging features” (18). A major enabling effort, TCGA focuses on extensively characterizing and making publicly available the molecular properties of several cancers, including GBM (19). At the same time, the Cancer Imaging Archive effort is underway to gather the medical images in these and other patients. These efforts provide enormous opportunities for the continued development of radiogenomic analytic techniques. For GBM, the TCGA glioma phenotype group has the most comprehensive data set, ranging from preoperative MR images to molecular analyses of resected tissue (20–23). A panel of eight neuroradiologists from multiple institutions has annotated a subset of the MR images by using the VASARI feature set (24), comprising 30 distinct manually annotated semantic features that describe GBM (20). Additionally, studies in other cohorts (17,25,26) have shown that molecular aberrations are associated with morphologic changes detectable on MR images and suggest that meaningful associations can be made between MR images and molecular data.
The radiogenomic analyses demonstrated here extend previous radiogenomic work and are easily extendible. Quantitative image features have been successfully applied in non–small cell lung cancer, hepatocellular carcinoma, and now GBM. New features can be added to the computational pipeline and immediately assessed for prognostic relevance and molecular correlates. Additionally, Amaretto provides a framework to map image features to molecular data by integrating several molecular data sets in a comprehensive model.
Our study had limitations; the most important of these was that ROIs were manually annotated in a single section that contained the largest cross section of the lesion. Manually annotating ROIs is a tedious process. This process can be facilitated by using a dedicated software platform such as ePad (10) but introduces potential reader variability. By evaluating the robustness of quantitative image features to ROI definition and limiting our analysis to the most robust features, we mitigated this concern to some extent. We are actively developing semiautomatic approaches for outlining necrosis, enhancement, and edema ROIs in three dimensions to overcome this limitation. More importantly, this initial set of manually drawn ROIs allowed us and will allow others the possibility of investigating whether quantitative image features are a viable alternative and complement to semantic image features such as the VASARI feature set. We have provided all ROIs in Annotation and Image Markup, or AIM, format (27) at https://wiki.cancerimagingarchive.net/x/VwD-/.
In summary, we have demonstrated the potential of quantitative image features combined with advanced bioinformatics analysis to create radiogenomic maps providing molecular correlates with morphologic characteristics of GBM. Applying this framework in larger cohorts and in cohorts from different institutions will facilitate its value in advancing personalized treatment for patients with cancer.
Advances in Knowledge
■ Computational approaches allowed the extraction of 18 robust quantitative image features for each region of interest on MR images that captured meaningful properties of glioblastoma multiforme (GBM) tumors.
■ Three, 77, and seven robust quantitative image features were correlated with survival, the Visually Accessible Rembrandt Images features, and molecular subgroups, respectively (P < .05).
■ Robust quantitative image features can be used to build radiogenomic maps of GBM and allow the association of radiographic phenotype with underlying key biologic pathways for 56% (30 of 54) of the robust quantitative image features through a bioinformatics data integration algorithm.
Implication for Patient Care
■ Radiogenomic mapping in patients with GBM provides the potential for noninvasive assessment of underlying molecular processes in a tumor.
SUPPLEMENTAL FIGURES
SUPPLEMENTAL TABLES
Acknowledgments
Acknowledgments
We acknowledge the efforts of the Cancer Genome Atlas glioma phenotype group, the Cancer Imaging Archive, and the Cancer Genome Atlas glioblastoma project, without which this project would not have been possible.
Received August 14, 2013; revision requested November 6; revision received February 17, 2014; accepted March 12; final version accepted March 21.
From the 2013 RSNA Annual Meeting.
Supported by Information Sciences in Imaging at Stanford and the Center for Cancer Systems Biology at Stanford (grant U54 CA149145).
Funding: This research was supported by the National Institutes of Health (grants U54 CA149145 and R01 CA160251).
Disclosures of Conflicts of Interest: O.G. No relevant conflicts of interest to disclose. L.A.M. No relevant conflicts of interest to disclose. A.S.A. No relevant conflicts of interest to disclose. J.X. No relevant conflicts of interest to disclose. S.E. No relevant conflicts of interest to disclose. G.K.S. Financial activities related to the present article: none to disclose. Financial activities not related to the present article: is a consultant for Medtronic. Other relationships: none to disclose. S.H.C. No relevant conflicts of interest to disclose. S.N. Financial activities related to the present article: none to disclose. Financial activities not related to the present article: is on the advisory boards of Fovia and EchoPixel; is a consultant for Carestream. Other relationships: none to disclose. G.Z. Financial activities related to the present article: none to disclose. Financial activities not related to the present article: has received grant money from GE Healthcare. Other relationships: none to disclose. S.K.P. No relevant conflicts of interest to disclose.
Abbreviations:
- FLAIR
- fluid-attenuated inversion recovery
- GBM
- glioblastoma multiforme
- ICC
- intraclass correlation coefficient
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- ROI
- region of interest
- TCGA
- the Cancer Genome Atlas
- VASARI
- Visually Accessible Rembrandt Images
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2.Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bredel M, Scholtens DM, Harsh GR, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA 2009;302(3):261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 5.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gevaert O, Xu J, Hoang CD, et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data—methods and preliminary results. Radiology 2012;264(2):387–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nair VS, Gevaert O, Davidzon G, et al. Prognostic PET 18F-FDG uptake imaging features are associated with major oncogenomic alterations in patients with resected non-small cell lung cancer. Cancer Res 2012;72(15):3725–3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gevaert O, Plevritis S. Identifying master regulators of cancer and their downstream targets by integrating genomic and epigenomic features. Pac Symp Biocomput 2013:123–134. [PMC free article] [PubMed] [Google Scholar]
- 9.Gevaert O, Villalobos V, Sikic BI, Plevritis SK. Identification of ovarian cancer driver genes by using module network integration of multi-omics data. Interface Focus 2013;3(4):20130013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubin DL, Rodriguez C, Shah P, Beaulieu C. iPad: Semantic annotation and markup of radiological images. AMIA Annu Symp Proc 2008 Nov 6:626–630. [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 2014;42(Database issue):D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGraw KO, Wong S. Forming inferences about some intraclass correlation coefficients. Psychol Methods 1996;1(1):30–46. [Google Scholar]
- 14.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57(1):289–300. [Google Scholar]
- 15.Pan J, Li S, Chi P, Xu Z, Lu X, Huang Y. Lentivirus-mediated RNA interference targeting WWTR1 in human colorectal cancer cells inhibits cell proliferation in vitro and tumor growth in vivo. Oncol Rep 2012;28(1):179–185. [DOI] [PubMed] [Google Scholar]
- 16.Segal E, Sirlin CB, Ooi C, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol 2007;25(6):675–680. [DOI] [PubMed] [Google Scholar]
- 17.Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A 2008;105(13):5213–5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaffe CC. Imaging and genomics: is there a synergy? Radiology 2012;264(2):329–331. [DOI] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research Network . Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455(7216):1061–1068. [Published correction appears in Nature 2013;494(7438):506.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutman DA, Cooper LA, Hwang SN, et al. MR imaging predictors of molecular profile and survival: multi-institutional study of the TCGA glioblastoma data set. Radiology 2013;267(2):560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jain R, Poisson L, Narang J, et al. Genomic mapping and survival prediction in glioblastoma: molecular subclassification strengthened by hemodynamic imaging biomarkers. Radiology 2013;267(1):212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zinn PO, Mahajan B, Sathyan P, et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS ONE 2011;6(10):e25451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zinn PO, Sathyan P, Mahajan B, et al. A novel volume-age-KPS (VAK) glioblastoma classification identifies a prognostic cognate microRNA-gene signature. PLoS ONE 2012;7(8):e41522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.VASARI Research Project . https://wiki.cancerimagingarchive.net/display/Public/VASARI+Research+Project. Updated February 28, 2013. Accessed April 25, 2014.
- 25.Pope WB, Chen JH, Dong J, et al. Relationship between gene expression and enhancement in glioblastoma multiforme: exploratory DNA microarray analysis. Radiology 2008;249(1):268–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barajas RF, Jr, Hodgson JG, Chang JS, et al. Glioblastoma multiforme regional genetic and cellular expression patterns: influence on anatomic and physiologic MR imaging. Radiology 2010;254(2):564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abajian AC, Levy M, Rubin DL. Informatics in radiology: improving clinical work flow through an AIM database: a sample web-based lesion tracking application. RadioGraphics 2012;32(5):1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.