Background: Tim23 mediates protein translocation into mitochondria.
Results: Tim23 binds to mitochondria-like membranes through a hydrophobic anchor at its N terminus, with cardiolipin enhancing the interaction.
Conclusion: The intermembrane space domain of Tim23 can interact with both inner and outer mitochondria-like membranes.
Significance: Tim23 provides the central element for formation of the translocation contact.
Keywords: Lipid, Membrane, Mitochondria, Nuclear Magnetic Resonance (NMR), Protein Translocation, Protein Import
Abstract
Tim23 mediates protein translocation into mitochondria. Although inserted into the inner membrane, the dynamic association of its intermembrane space (IMS) domain with the outer membrane promotes protein import. However, little is known about the molecular basis of this interaction. Here, we demonstrate that the IMS domain of Tim23 tightly associates with both inner and outer mitochondrial membrane-like membranes through a hydrophobic anchor at its N terminus. The structure of membrane-bound Tim23IMS is highly dynamic, allowing recognition of both the incoming presequence and other translocase components at the translocation contact. Cardiolipin enhances Tim23 membrane attachment, suggesting that cardiolipin can influence preprotein import.
Introduction
The majority of mitochondrial proteins are synthesized in the cytosol and then imported and sorted into specific mitochondrial compartments. Protein import is governed by the coordinated interplay of highly dynamic hetero-oligomeric molecular machines: the translocases of the outer (TOM) and inner (TIM) mitochondrial membranes (1–5). Whereas the TOM40 complex works as a general mitochondrial entry gate, the inner membrane translocase TIM23 regulates the delivery of proteins, equipped with the proper presequence signal, into the mitochondrial matrix and the inner mitochondrial membrane (IMM)3 (6–9). During protein translocation across the mitochondrial membranes, TIM23 and TOM complexes transiently associate in supramolecular complexes at translocation contact sites (10–12).
The presequence translocase TIM23 is a very plastic assembly in which many subunits dynamically interact to favor preprotein import (8, 13, 14). The TIM23 core components are Tim17, Tim50, and Tim23, with Tim23 playing a crucial role in translocase activity (5). The Tim23 C-terminal domain (residues 97–222) anchors the protein within the IMM and forms the protein-conducting pore across the membrane, whereas the N-terminal domain (Tim23IMS, residues 1–96) is located in the intermembrane space (IMS) and works as a presequence receptor (15–18). The first 50 residues of Tim23 were shown to be required for yeast growth at 37 °C (15). In addition, removal of this region resulted in mitochondrial morphological aberrations (15), although it did not significantly affect protein import (10).
How could Tim23 contribute to the formation of translocation contact sites? One potential mechanism might be that Tim23 engages in a two-membrane topology whereby Tim23IMS crosses the outer mitochondrial membrane (OMM), with the first 20 residues being exposed on the outer surface, and thereby tethers the OMM to the IMM (15). Such an association of Tim23 with the OMM was shown to be highly dynamic and reversible (19) and to be regulated by the interaction with Tim50 and dependent on TIM23 translocation activity (13, 14). Despite the importance of the translocation contact, however, little is known about the molecular details that regulate the interaction of Tim23IMS with membranes. Here, we provide atomic resolution insights into the mechanisms underlying the membrane interaction of Tim23IMS. Our study reveals that a hydrophobic cluster at the N terminus of Tim23 works as a molecular hook to dynamically anchor Tim23 to membranes and thus enables Tim23 to contribute to the establishment of translocation contact sites.
EXPERIMENTAL PROCEDURES
Protein Preparation
The IMS domain of Saccharomyces cerevisiae Tim23 (residues 1–96, Tim23IMS) was expressed in Escherichia coli and purified as described previously (16). Due to cloning and tobacco etch virus nuclear inclusion a endopeptidase cleavage, two residues (Gly-Ser) were present at the N terminus (see Fig. 1A). 15N-Labeled samples were obtained by protein expression in M9 minimal medium with 15NH4Cl as the unique source of nitrogen. Tim23(1–13), Tim23(1–24), Tim23(1–24)W3S/L4S/F5S, and the 22-residue presequence peptide from retinal aldehyde dehydrogenase (rALDH) were obtained by standard Fmoc (N-(9-fluorenyl)methoxycarbonyl) solid-phase synthesis using an ABI 433A synthesizer (Applied Biosystems) and purified by reversed-phase HPLC.
FIGURE 1.
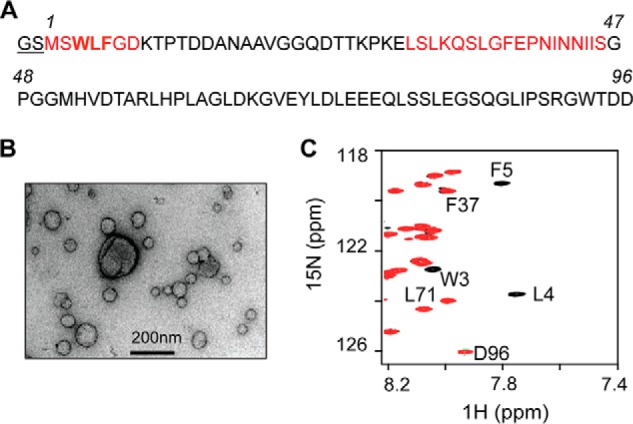
Tim23IMS and mitochondria-like membranes. A, amino acid sequence of Tim23IMS (residues 1–96). Two additional residues (Gly-Ser) are present due to His tag removal by tobacco etch virus nuclear inclusion a endopeptidase cleavage. The two membrane-binding regions are highlighted in red. B, electron micrograph of liposomes mimicking the phospholipid composition of the OMM. C, superposition of 1H-15N heteronuclear single quantum coherence (HSQC) spectra of 0.05 mm Tim23IMS without (black) and with (red) the addition of a 3-fold molar excess of total lipids in OMM-like liposomes. Selected resonance assignments are indicated.
1,2-Dihexanoyl-sn-glycero-3-phosphocholine (DHPC), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine), 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS), phosphatidylinositol (1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol 3′-phosphate), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol 4′,5′-bisphosphate)), and cardiolipin (heart, bovine) were purchased in lyophilized form from Avanti Polar Lipids. Lipid compositions corresponding to the OMM (48:28:10:10:4 DOPC/DOPE/DOPS/phosphatidylinositol/cardiolipin) and IMM (38:24:4:16:18 DOPC/DOPE/DOPS/phosphatidylinositol/cardiolipin) were dissolved in chloroform, mixed, and dried under N2 gas for 30 min to form a film. The semidried mixture of phospholipids was additionally lyophilized for 12 h to remove traces of organic solvent. NMR buffer was added to the dried phospholipid film and vortexed to homogeneously suspend the phospholipids. The resuspended phospholipid mixture was sonicated in a precooled water bath at 4 °C for 2 h with 5-min pulses alternating with 15-min delays to prepare the liposome stock. The liposome stock solution contained either 10 or 25 mg of total lipid. The stock solutions were diluted in NMR buffer prior to NMR measurements. Total lipid concentrations were estimated assuming that all lipids were rehydrated when forming liposomes.
Transmission Electron Microscopy
Samples were deposited on glow-discharged carbon-coated grids and stained with 1% uranyl acetate. EM pictures were taken with a Philips CM120 transmission electron microscope using a TemCam-224A slow scan CCD camera (TVIPS GmbH, Gauting, Germany).
Circular Dichroism
CD measurements were carried out with 0.01 mm Tim23IMS and OMM-like liposomes at 0.15 mm total lipid concentration in 50 mm sodium phosphate buffer (pH 7.0). CD spectra were recorded at room temperature on a Chirascan spectrometer (Applied Photophysics) using a 2-mm path length cuvette. Five repeats of each spectrum were obtained for the wavelength range of 190–260 nm.
Fluorescence Spectroscopy
Fluorescence emission spectra were recorded in the 285–395 nm range, using 280 nm as the excitation wavelength with a slit of 5 nm, on a Cary Eclipse fluorescence spectrophotometer (Varian). Measurements were done at 298 K in 20 mm Hepes (pH 7.2) and 50 mm NaCl.
NMR Spectroscopy
NMR spectra were recorded at 288 K on 600- and 700-MHz Bruker BioSpin spectrometers. For NMR studies, samples were prepared in 20 mm Hepes (pH 7.2), 50 mm NaCl, and 10% D2O.
NMR data were processed using NMRPipe (20) and analyzed with Sparky (T. D. Goddard and D. G. Kneller, University of California, San Francisco). Tim23IMS backbone assignment was obtained as described previously (16). For titration analysis, the normalized average chemical shift perturbation (ΔHN) was calculated as ΔHN = (((δN/5)2 + (δH)2)/2)1/2. Secondary chemical shift values were calculated as the differences between measured Cα chemical shifts and amino acid-specific random-coil values, which were taken from Refs. 21 and 22.
Steady-state 1H-15N hetero-NOE (HetNOE) experiments were measured using a recycle delay of 5 s. NOE values were calculated by the ratio of the peak intensities between saturated and reference spectra.
RESULTS
Tim23IMS Associates with Both IMM- and OMM-like Liposomes
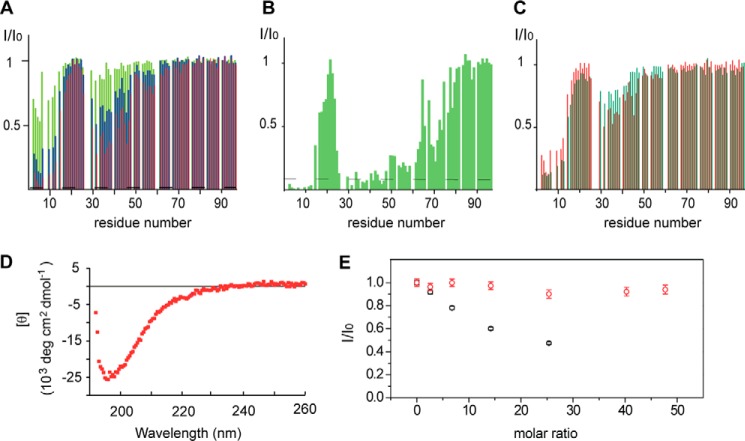
To obtain insight into the interaction of Tim23IMS with mitochondrial membranes, we used NMR spectroscopy. NMR is uniquely suited to study protein-membrane interactions, as it can provide structural information at high resolution even for dynamic protein-membrane systems. To this end, a variety of liposomes that mimic both the IMM and OMM were prepared. First, we studied the association of Tim23IMS with liposomes prepared with a lipid composition mimicking the OMM (Fig. 1B). At increasing concentrations of OMM-like liposomes, NMR signals belonging to residues 1–12 and 29–46 of Tim23IMS were strongly attenuated (Figs. 1C and 2A). The residue-specific perturbation of the NMR signals demonstrates that the two regions directly bound to the membrane. Moreover, the finding that residues 1–12 were most strongly perturbed suggests that the very N terminus of Tim23IMS has the highest affinity for OMM-like liposomes and works as the prime interaction site. In contrast, NMR signals of residues 18–25 and 61–96 were not affected, suggesting that these regions remain highly flexible when Tim23IMS is bound to OMM-like membrane.
FIGURE 2.
Tim23IMS binds to both IMM- and OMM-like liposomes. A, NMR signal intensity changes in Tim23IMS upon addition of OMM-like liposomes as a function of residue number, with 0.5-fold (green), 1.5-fold (blue), and 3-fold (red) molar excesses of total lipids. The intensity ratio (I/I0) is calculated from the data height (I) of a resonance in the 1H-15N HSQC spectrum of Tim23IMS at a defined molar excess of lipids and the data height (I0) of the same resonance in the spectrum of free Tim23IMS. I/I0 values of prolines and overlapping residues were set to zero. B, NMR signal intensity changes in Tim23IMS upon addition of a 0.5-fold molar excess of total lipids in IMM-like liposomes. The dashed line indicates the estimated noise level in NMR spectra. C, 1H-15N signal intensity attenuation profiles of Tim23IMS in the presence of OMM-like (red) and IMM-like (green) liposomes without cardiolipin at an equimolar total lipid/protein concentration. The Tim23IMS concentration in A–C was 0.05 mm. D, far-UV CD spectrum of 0.01 mm Tim23IMS bound to OMM-like liposomes at 0.15 mm total lipid concentration. deg, degrees. E, normalized signal changes in Tim23(1–24) upon addition of increasing amounts of IMM-like liposomes. Relative intensity values are plotted versus the total lipid/protein molar ratio. Titrations were carried out with wild-type Tim23(1–24) (black) and the triple mutant Tim23(1–24)W3S,L4S,F5S (red). The initial peptide concentration was 0.1 mm. Note that intensity values in the presence (I) or absence (I0) of liposomes were determined by overall integration of 1H spectra between 10.5 and 6.5 ppm, thus including residues both involved and not involved in binding.
To address whether Tim23IMS exclusively interacts with the OMM or can also associate with the IMM, where the C-terminal domain of Tim23 is located, we analyzed the binding of Tim23IMS to liposomes mimicking the lipid composition of the IMM. At a total lipid/protein molar ratio of 0.5, NMR resonances of residues 1–12 and 29–46 were already broadened beyond detection (Fig. 2B), suggesting that these residues were fully bound to the membrane. Residues of neighboring regions were partially broadened, possibly as a result of weak electrostatic interactions between the negatively charged cardiolipin and positively charged residues in Tim23IMS (His52, Arg57, His59, and Lys66).
Cardiolipin Enhances the Membrane Association of the N Terminus of Tim23
The IMM is particularly rich in cardiolipin, which has been shown to be crucial for mitochondrial morphology and the functional reconstitution of proteins into the IMM (23–25). At comparable total lipid/protein ratios, binding to IMM-like liposomes was more effective than to those mimicking the OMM (Fig. 2, A and B). We therefore tested the role of cardiolipin in Tim23IMS-IMM binding. To this end, we prepared liposomes lacking cardiolipin. Compared with IMM-like liposomes containing cardiolipin, the new IMM membrane systems showed lower affinity for Tim23IMS (Fig. 2, B and Fig. C). The data demonstrate that Tim23IMS can bind to both OMM- and IMM-like membranes. The membrane interaction is mediated by two distinct regions located within the N-terminal 50 residues: residues 1–12 and 29–46. Cardiolipin further tightens the membrane association of Tim23IMS by expanding the lipid-binding region outside these two major interaction sites.
Membrane-bound Tim23IMS Has a Highly Dynamic Structure
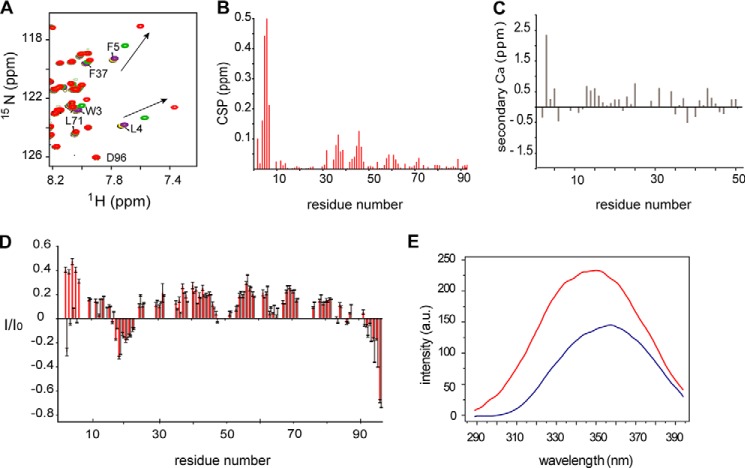
The membrane association/dissociation process of Tim23IMS is slow on the NMR time scale. Protein signals from the slowly tumbling complex are broadened beyond detection, and direct detection of membrane-bound Tim23IMS is not possible. To overcome this obstacle and obtain information on the membrane-bound structure of Tim23IMS, we used DHPC micelles, a membrane-mimicking system that has been proven to be highly useful for membrane proteins (26). In agreement with a specific interaction of Tim23IMS with DHPC micelles, no changes in the NMR signals of Tim23IMS were observed when the detergent was below its critical micelle concentration (∼16 mm). Above the critical micelle concentration, increasing amounts of DHPC most strongly perturbed residues 1–7, followed by residues 33–46 (Fig. 3, A and B). Thus, Tim23IMS contacts DHPC micelles through the same regions that mediate its binding to OMM- and IMM-like liposomes, supporting the choice of DHPC to mimic the membrane-bound conformation of Tim23IMS.
FIGURE 3.
Interaction of Tim23IMS with DHPC micelles. A, superposition of 1H-15N HSQC spectra of 0.05 mm Tim23IMS without DHPC (black) and with 8 mm (yellow), 16 mm (purple), 24 mm (green), and 32 mm (red) DHPC. Progressive changes in the resonance positions of Leu4 and Phe5 are marked. B, average chemical shift perturbation (CSP) of Tim23IMS resonances upon addition of 32 mm DHPC. C, secondary Cα chemical shifts of Tim23IMS in the presence of DHPC micelles (50 mm DHPC). For clarity, only the first 50 residues of Tim23IMS are shown. D, 1H-15N HetNOE analysis of Tim23IMS. Isat/Iref = I/I0 values in Tim23IMS in the presence (50 mm; red) and absence (gray) of DHPC. Error bars were calculated on the basis of the signal/noise ratio. E, fluorescence emission spectra of 0.02 mm Tim23(1–13) in the free state (blue) and in the presence of 100 mm DHPC (red). a.u., arbitrary units.
Next, we analyzed the structure and dynamics of micelle-bound Tim23IMS. To this end, we determined secondary Cα chemical shifts, a highly sensitive probe for protein backbone geometry. Secondary Cα chemical shifts were, with the exception of Ser2, smaller than 0.5 ppm (Fig. 3C), demonstrating that DHPC-bound Tim23IMS lacks rigid helical or β-strand secondary structure. The dynamic nature of micelle-bound Tim23IMS was further supported by analysis of 1H-15N HetNOE, which reports on pico-to-nanosecond backbone dynamics. For residues 2–6, HetNOE values increased from below 0.1 in the free form to values approaching 0.4 when bound to DHPC micelles (Fig. 3D). The data show that the N-terminal region of Tim23IMS, which forms the primary interaction site, was rigidified upon binding to DHPC micelles. HetNOE values downstream of residues 2–6 were very similar in the free and DHPC-bound forms, indicating that most of the Tim23IMS sequence remains highly flexible. The dynamic nature of membrane-bound Tim23IMS was further supported by CD. CD spectra of liposome-bound Tim23IMS had a shape that is typical of random-coil structure (Fig. 2D), indicating that liposome-bound Tim23IMS does not fold into a helical or β-strand conformation.
Tim23IMS Recognizes Presequences in the Presence of Membranes
The primary membrane-binding site of Tim23IMS contains the hydrophobic residues Trp3, Leu4, and Phe5. To test if the side chains of these residues anchor Tim23IMS to the membrane, we designed a 13-residue peptide, Tim23(1–13), which comprises the primary Tim23IMS membrane-binding region. The DHPC interaction of Tim23(1–13) was probed using fluorescence spectroscopy. Upon binding of Tim23(1–13) to DHPC micelles, the fluorescence emission spectrum strongly changed. The emission maximum shifted from 358 to 350 nm, and the intensity increased (Fig. 3E). These changes indicate that the solvent exposure experienced by Trp3 was changed and that Trp3 was buried within the hydrophobic core of the micelle. Thus, fluorescence provided direct experimental evidence for the insertion of the side chain of Trp3 into a hydrophobic environment. The crucial role of Trp3-Leu4-Phe5 in binding to mitochondrial membranes was supported by mutational analysis. Mutation of Trp3-Leu4-Phe5 to Ser3-Ser4-Ser5 abolished the ability of Tim23(1–24) to bind to IMM-like liposomes (Fig. 2E). Thus, the hydrophobic cluster formed by Trp3-Leu4-Phe5 acts as a molecular hook mediating the membrane attachment of Tim23IMS.
We then asked if Tim23IMS keeps its fundamental ability to work as a presequence receptor when bound to mitochondrial membranes. To this end, we titrated Tim23IMS bound to IMM-like liposomes (Fig. 4, A–C) with increasing concentrations of the presequence from rALDH. Analysis of Tim23IMS signal intensities (Fig. 4A, black bars) showed that before addition of presequence, Tim23IMS was in the membrane-bound state. Addition of presequence increased signal intensities in the second membrane-binding region, suggesting that its interaction with the membrane was attenuated. At the same molar ratio, however, no changes in signal intensity were observed for residues 1–12, demonstrating that the N-terminal hook remained bound to the membrane, whereas chemical shift changes were already present in the presequence-binding site (residues 71–84) (Fig. 4, B and C). The direction and magnitude of the chemical shift changes in the presequence-binding site were similar to those observed in the absence of liposomes (Fig. 4C). Residues 1–12 gained signal intensity only at higher presequence concentrations, suggesting that the N terminus of Tim23IMS was partially released from the membrane. The observed membrane release might arise from competition between Tim23IMS membrane-binding regions and the presequence peptide to bind to liposomes, consistent with the ability of several mitochondrial presequence peptides to interact with membranes (27–29). Taken together, the data suggest that membrane-bound Tim23IMS can recognize presequences.
FIGURE 4.

Tim23IMS recognizes presequences in the presence of membranes. A, intensity changes in 1H-15N resonances of Tim23IMS in the presence of IMM-like liposomes at a 0.5:1 total lipid/protein molar ratio without (black bars) or with 1-fold (red circles), 2-fold (cyan triangles), 4-fold (purple crosses), and 8-fold (blue circles) molar excesses of rALDH presequence peptide. B, 1H-15N chemical shift perturbation (CSP) in Tim23IMS in the presence of IMM-like liposomes at a 0.5:1 total lipid/protein molar ratio without (black squares) or with 1-fold (red squares), 2-fold (cyan triangles), 4-fold (purple crosses), and 8-fold (blue circles) molar excesses of rALDH presequence peptide. C, superposition of the Leu71 region from 1H-15N HSQC spectra of Tim23IMS in the presence (right) or absence (left) of IMM-like liposomes at a 0.5:1 total lipid/protein molar ratio and of increasing concentrations of rALDH presequence. Molar ratios of Tim23IMS to presequence are 1:0 (orange), 1:1 (red), 1:2 (cyan), 1:4 (purple), and 1:8 (blue). The Tim23IMS concentration was 0.125 mm.
DISCUSSION
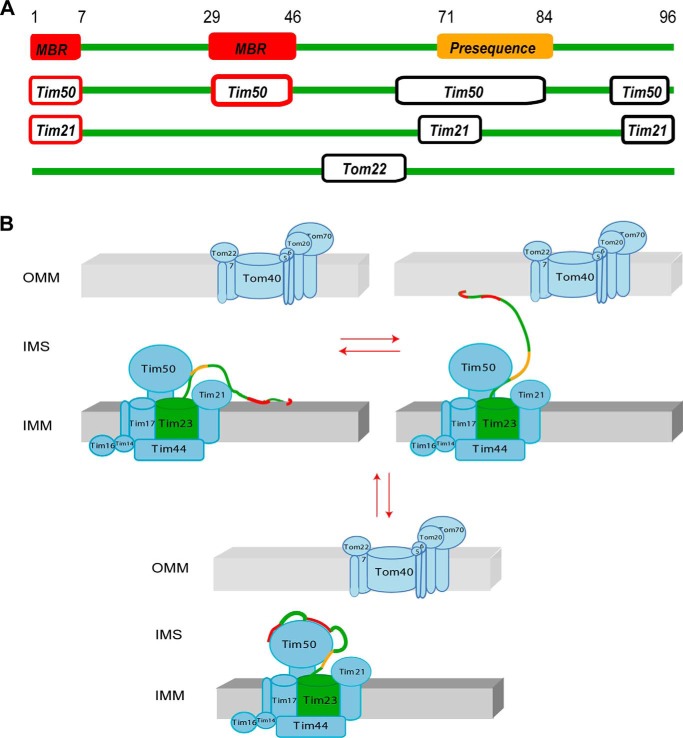
Transfer of presequence-carrying proteins into mitochondria requires the coordinated action of the TOM and TIM23 translocases (8, 9, 30). Tim23 is a key subunit of the TIM23 complex. It is inserted via its C-terminal domain into the IMM and can contact the OMM through the IMS domain (15). By contacting both mitochondrial membranes, Tim23 can promote preprotein import. However, little is known about the molecular basis of the dynamic association of the IMS domain of Tim23 with mitochondrial membranes. Our study demonstrated that together with residues 29–46 of Tim23, residues 1–7 directly bind to the membrane (Figs. 2 and 5A). The length of each of the two binding regions is not sufficient to transverse the lipid bilayer, in agreement with sequence analysis, which did not predict a helical transmembrane segment (15). Tim23IMS-membrane binding is enabled through a hydrophobic hook that is formed by the N-terminal residues Trp3-Leu4-Phe5, with the side chain of Trp3 inserted into the hydrophobic environment of the lipid bilayer (Figs. 2 and 3). Trp3-Leu4-Phe5 are conserved across many members of the fungus kingdom.
FIGURE 5.
Model of dynamic membrane interactions of Tim23. A, scheme of binding motifs used by Tim23IMS for contacting multiple partners within the IMS: OMM and IMM (membrane-binding region (MBR)), Tim21IMS, Tim50IMS, Tom22IMS, and presequences. B, model of alternative and dynamic binding of Tim23IMS to the OMM and IMM. The membrane- and presequence-binding regions are also involved in Tim23IMS binding to Tim50IMS (33).
We further showed that the N terminus of Tim23 not only anchors to OMM-like membranes, but also has a pronounced affinity for IMM-like environments (Fig. 2). This suggests that the N-terminal domain of Tim23 might contact dynamically both the IMM and OMM (Fig. 5B), in agreement with its high conformational flexibility (16). Cardiolipin, which is abundant in the IMM and is enriched at translocation contact sites (31), strongly promoted the membrane interaction of Tim23IMS through an enhanced interaction of residues 29–46 with cardiolipin-containing membranes. In the absence of other protein interactions from TIM or TOM proteins, IMM binding might therefore dominate over OMM binding. In this way, cardiolipin might influence preprotein import. Moreover, as TIM23-mediated protein import requires a membrane potential (5) and cardiolipin has been suggested to function as a proton trap (32), the presence of a membrane potential and proton-bound cardiolipin might further modulate membrane binding of Tim23IMS.
A detailed analysis of the change in NMR signal position and intensity with increasing concentrations of the presequence from rALDH (Fig. 4) showed that Tim23IMS is able to bind to a presequence in the presence of membranes and therefore recognize the incoming protein. In addition, as large parts of membrane-bound Tim23IMS remain highly flexible, it is likely that the flexible residues, which are not membrane-bound, are able to interact with other components of the TOM/TIM translocation machinery. Consistent with this hypothesis, in vivo experiments showed that Tom22 can be cross-linked to residue 41 of Tim23 (14).
The TIM23 complex is a highly complex system that is constantly remodeled in vivo and involves multiple components. Supporting a complex interaction network, we have recently shown that Tim23IMS contains multiple sites to efficiently interact with the IMS domains of Tim21, Tim50, and Tom22 (33). Both membrane-binding regions identified in the present study overlap with sites that bind to Tim50IMS, whereas only the N-terminal membrane-binding region is involved in binding to Tim21IMS (Fig. 5A). This suggests that binding of the N terminus of Tim23IMS to membranes can influence binding of Tim23IMS to Tim50IMS (and vice versa), as well as to Tim21IMS. The combined data also indicate that the Tim23IMS binding site for Tom22 (residues 53–61) (33) remains accessible when the N terminus of Tim23 is bound to membranes. Thus, Tim23IMS might contribute to the translocation contact site both by binding to Tom22IMS and by directly associating with the OMM (Fig. 5B). Notably, the binding site in Tim23IMS for presequence is distinct from the membrane-binding region and from the binding site of Tom22, whereas the Tom22-binding site partially overlaps with one of the regions involved in binding to Tim50IMS (33), providing another level of regulation of the TIM23 complex.
Acknowledgments
We thank Kerstin Overkamp for peptide synthesis and Dietmar Riedel for electron micrographs.
This work was supported by Deutsche Forschungsgemeinschaft Collaborative Research Center 860 Project B2 (to M. Z.).
- IMM
- inner mitochondrial membrane
- IMS
- intermembrane space
- OMM
- outer mitochondrial membrane
- rALDH
- retinal aldehyde dehydrogenase
- DHPC
- 1,2-dihexanoyl-sn-glycero-3-phosphocholine
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DOPS
- 1,2-dioleoyl-sn-glycero-3-phospho-l-serine
- HetNOE
- hetero-NOE
- HSQC
- heteronuclear single quantum coherence.
REFERENCES
- 1. Bauer M. F., Hofmann S., Neupert W., Brunner M. (2000) Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol. 10, 25–31 [DOI] [PubMed] [Google Scholar]
- 2. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., Pfanner N. (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ryan K. R., Jensen R. E. (1995) Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell 83, 517–519 [DOI] [PubMed] [Google Scholar]
- 4. Schatz G. (1996) The protein import system of mitochondria. J. Biol. Chem. 271, 31763–31766 [DOI] [PubMed] [Google Scholar]
- 5. Neupert W., Herrmann J. M. (2007) Translocation of proteins into mitochondria. Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 6. Glick B. S., Brandt A., Cunningham K., Müller S., Hallberg R. L., Schatz G. (1992) Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell 69, 809–822 [DOI] [PubMed] [Google Scholar]
- 7. Pfanner N., Geissler A. (2001) Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2, 339–349 [DOI] [PubMed] [Google Scholar]
- 8. van der Laan M., Hutu D. P., Rehling P. (2010) On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim. Biophys. Acta 1803, 732–739 [DOI] [PubMed] [Google Scholar]
- 9. van der Laan M., Meinecke M., Dudek J., Hutu D. P., Lind M., Perschil I., Guiard B., Wagner R., Pfanner N., Rehling P. (2007) Motor-free mitochondrial presequence translocase drives membrane integration of preproteins. Nat. Cell Biol. 9, 1152–1159 [DOI] [PubMed] [Google Scholar]
- 10. Chacinska A., Rehling P., Guiard B., Frazier A. E., Schulze-Specking A., Pfanner N., Voos W., Meisinger C. (2003) Mitochondrial translocation contact sites: separation of dynamic and stabilizing elements in formation of a TOM-TIM-preprotein supercomplex. EMBO J. 22, 5370–5381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Horst M., Hilfiker-Rothenfluh S., Oppliger W., Schatz G. (1995) Dynamic interaction of the protein translocation systems in the inner and outer membranes of yeast mitochondria. EMBO J. 14, 2293–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schülke N., Sepuri N. B., Gordon D. M., Saxena S., Dancis A., Pain D. (1999) A multisubunit complex of outer and inner mitochondrial membrane protein translocases stabilized in vivo by translocation intermediates. J. Biol. Chem. 274, 22847–22854 [DOI] [PubMed] [Google Scholar]
- 13. Popov-Celeketić D., Mapa K., Neupert W., Mokranjac D. (2008) Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 27, 1469–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamura Y., Harada Y., Shiota T., Yamano K., Watanabe K., Yokota M., Yamamoto H., Sesaki H., Endo T. (2009) Tim23-Tim50 pair coordinates functions of translocators and motor proteins in mitochondrial protein import. J. Cell Biol. 184, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donzeau M., Káldi K., Adam A., Paschen S., Wanner G., Guiard B., Bauer M. F., Neupert W., Brunner M. (2000) Tim23 links the inner and outer mitochondrial membranes. Cell 101, 401–412 [DOI] [PubMed] [Google Scholar]
- 16. de la Cruz L., Bajaj R., Becker S., Zweckstetter M. (2010) The intermembrane space domain of Tim23 is intrinsically disordered with a distinct binding region for presequences. Protein Sci. 19, 2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bauer M. F., Sirrenberg C., Neupert W., Brunner M. (1996) Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell 87, 33–41 [DOI] [PubMed] [Google Scholar]
- 18. Truscott K. N., Kovermann P., Geissler A., Merlin A., Meijer M., Driessen A. J., Rassow J., Pfanner N., Wagner R. (2001) A presequence- and voltage-sensitive channel of the mitochondrial preprotein translocase formed by Tim23. Nat. Struct. Biol. 8, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 19. Vogel F., Bornhövd C., Neupert W., Reichert A. S. (2006) Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 175, 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 21. Schwarzinger S., Kroon G. J., Foss T. R., Chung J., Wright P. E., Dyson H. J. (2001) Sequence-dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 123, 2970–2978 [DOI] [PubMed] [Google Scholar]
- 22. Berjanskii M. V., Wishart D. S. (2005) A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 127, 14970–14971 [DOI] [PubMed] [Google Scholar]
- 23. Jiang F., Ryan M. T., Schlame M., Zhao M., Gu Z., Klingenberg M., Pfanner N., Greenberg M. L. (2000) Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275, 22387–22394 [DOI] [PubMed] [Google Scholar]
- 24. Schlame M., Ren M. (2009) The role of cardiolipin in the structural organization of mitochondrial membranes. Biochim. Biophys. Acta 1788, 2080–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang M., Mileykovskaya E., Dowhan W. (2002) Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 277, 43553–43556 [DOI] [PubMed] [Google Scholar]
- 26. Fernández C., Wüthrich K. (2003) NMR solution structure determination of membrane proteins reconstituted in detergent micelles. FEBS Lett. 555, 144–150 [DOI] [PubMed] [Google Scholar]
- 27. Wieprecht T., Apostolov O., Beyermann M., Seelig J. (2000) Interaction of a mitochondrial presequence with lipid membranes: role of helix formation for membrane binding and perturbation. Biochemistry 39, 15297–15305 [DOI] [PubMed] [Google Scholar]
- 28. Hammen P. K., Gorenstein D. G., Weiner H. (1996) Amphiphilicity determines binding properties of three mitochondrial presequences to lipid surfaces. Biochemistry 35, 3772–3781 [DOI] [PubMed] [Google Scholar]
- 29. Swanson S. T., Roise D. (1992) Binding of a mitochondrial presequence to natural and artificial membranes–role of surface potential. Biochemistry 31, 5746–5751 [DOI] [PubMed] [Google Scholar]
- 30. Chacinska A., Lind M., Frazier A. E., Dudek J., Meisinger C., Geissler A., Sickmann A., Meyer H. E., Truscott K. N., Guiard B., Pfanner N., Rehling P. (2005) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120, 817–829 [DOI] [PubMed] [Google Scholar]
- 31. Ardail D., Privat J. P., Egret-Charlier M., Levrat C., Lerme F., Louisot P. (1990) Mitochondrial contact sites. Lipid composition and dynamics. J. Biol. Chem. 265, 18797–18802 [PubMed] [Google Scholar]
- 32. Haines T. H., Dencher N. A. (2002) Cardiolipin: a proton trap for oxidative phosphorylation. FEBS Lett. 528, 35–39 [DOI] [PubMed] [Google Scholar]
- 33. Bajaj R., Jaremko L., Jaremko M., Becker S., Zweckstetter M. (2014) Molecular basis of the dynamic structure of the TIM23 complex in the mitochondrial intermembrane space. Structure 22, 1501–1511 [DOI] [PubMed] [Google Scholar]