Background: Invariant insulin residue PheB24 (a site of diabetes-associated mutation) contacts the insulin receptor.
Results: Hormonal function requires hydrophobicity rather than aromaticity at this site.
Conclusion: The B24 side chain provides a nonpolar anchor at the receptor interface.
Significance: Nonstandard aliphatic modification of residue B24 may enhance therapeutic properties of insulin analogs.
Keywords: Diabetes, Mutagenesis, Protein Design, Protein Structure, Receptor Tyrosine Kinase, Hormone, nonstandard mutagenesis
Abstract
Crystallographic studies of insulin bound to fragments of the insulin receptor have recently defined the topography of the primary hormone-receptor interface. Here, we have investigated the role of PheB24, an invariant aromatic anchor at this interface and site of a human mutation causing diabetes mellitus. An extensive set of B24 substitutions has been constructed and tested for effects on receptor binding. Although aromaticity has long been considered a key requirement at this position, MetB24 was found to confer essentially native affinity and bioactivity. Molecular modeling suggests that this linear side chain can serve as an alternative hydrophobic anchor at the hormone-receptor interface. These findings motivated further substitution of PheB24 by cyclohexanylalanine (Cha), which contains a nonplanar aliphatic ring. Contrary to expectations, [ChaB24]insulin likewise exhibited high activity. Furthermore, its resistance to fibrillation and the rapid rate of hexamer disassembly, properties of potential therapeutic advantage, were enhanced. The crystal structure of the ChaB24 analog, determined as an R6 zinc-stabilized hexamer at a resolution of 1.5 Å, closely resembles that of wild-type insulin. The nonplanar aliphatic ring exhibits two chair conformations with partial occupancies, each recapitulating the role of PheB24 at the dimer interface. Together, these studies have defined structural requirements of an anchor residue within the B24-binding pocket of the insulin receptor; similar molecular principles are likely to pertain to insulin-related growth factors. Our results highlight in particular the utility of nonaromatic side chains as probes of the B24 pocket and suggest that the nonstandard Cha side chain may have therapeutic utility.
Introduction
Insulin is a small globular protein critical to the hormonal regulation of vertebrate metabolism (Fig. 1A) (1). Containing two peptide chains, A (21 residues) and B (30 residues), the hormone binds to a receptor tyrosine kinase, designated the insulin receptor (IR)6 (2). The IR is a disulfide-linked homodimer with subunits (αβ)2; the extracellular α subunit binds insulin, whereas the transmembrane β subunit contains an intracellular tyrosine kinase domain (1). The crystal structure of the dimeric ectodomain (αβΔ)2 (where βΔ designates the extracellular portion of the β subunit) has revealed a Λ-shaped conformation (Fig. 1B), whose legs sit upon the plasma membrane (coral disk at bottom) (3). A single insulin molecule binds with high affinity to a tandem site spanning both α subunits (4). This site consists of the N-terminal leucine-rich repeat domain (L1) of one α subunit and the C-terminal segment of the other (αCT). The structure of insulin bound in a ternary complex with these structural elements (designated the “micro-receptor”; μIR) has recently been determined (Fig. 1C) (5). In the μIR complex, the classical position of the C-terminal segment of the insulin B chain (brown in Fig. 1C) (2) would intersect αCT (magenta) and so must be displaced (5, 6). In this study we have investigated a key feature of this complex, stabilization of the bound conformation of insulin by an invariant aromatic residue (PheB24) docked within a nonpolar pocket of the IR (6).
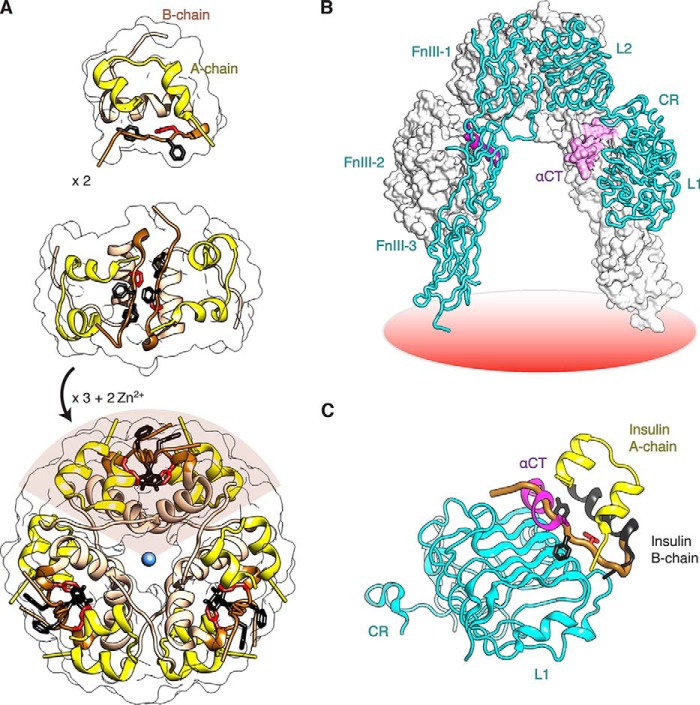
FIGURE 1.
Structure of insulin and receptor ectodomain. A, assembly of zinc insulin hexamer. The monomeric hormone (A and B chains, top panel) forms zinc-free dimers via anti-parallel association of B chain α-helices and C-terminal β-strands (brown, middle panel); two zinc ions then mediate assembly of three dimers to form the classical hexamer (T6, bottom panel). The A chain is shown in yellow (ribbon), and the B chain in light brown (B1–B19) or brown (B20–B30). Conserved aromatic residues PheB25 and TyrB26 are shown as black sticks, and PheB24 is shown in red. The Zn2+ ion is depicted in blue. B, Λ-shaped IR ectodomain homodimer. One protomer is shown as a ribbon (labeled), and the other as a molecular surface. Domains are as follows: L1, first leucine-rich repeat domain; CR, cysteine-rich domain; L2, second leucine-rich repeat domain; FnIII-1–3, respective first, second, and third fibronectin type III domains; and αCT, α-chain C-terminal segment. C, overlay illustrating insulin in its classical free conformation bound to site 1 of the microreceptor (L1-CR + αCT(704–719); designated μIR) (5). L1 and part of CR are shown in cyan, and αCT in magenta. PheB24, PheB25, and TyrB26 are as in A. The B chain is shown in dark gray (B6–B19); the position of the brown tube (residues B20–B30) would lead to a steric clash between B26–B30 and αCT. The figure was in part modified from Menting et al. (6), with permission of the authors. Coordinates were obtained from PDB entries 4INS, 2DTG, and 3W11.
Vertebrate insulin sequences are remarkable for an invariant phenylalanine (Phe) residue at position B24 (PheB24) and for the broad conservation of phenylalanine and tyrosine at B25 and B26 (PheB25 and TyrB26) (Fig. 2A) (7). A homologous triplet of aromatic residues is conserved among vertebrate insulin-related growth factors (IGF-I and IGF-II; Phe-Tyr-Phe) (8, 9). The contributions of these residues to the structure, stability, self-assembly, and activity of insulin have attracted long standing interest (10–12). The crystal structure of the zinc insulin hexamer, determined in 1969 by Hodgkin and co-workers (2, 13), revealed that residues B24–B26 participate in an aromatic-rich dimer interface (middle panel of Fig. 1A); this interface occurs three times in the hexamer (lower panel of Fig. 1A). The C-terminal deletion of this segment markedly impairs IR binding (14). The importance of these side chains was further highlighted by the discovery of clinical mutations associated with diabetes mellitus (DM) (2); effects of the substitution of PheB24 by Ser (clinical variant insulin Los Angeles) attracted detailed characterization (15, 16). Despite their intense study in the 45 years since the publication of the Hodgkin structure (13), how these aromatic side chains bind to the IR has, until recently, remained the subject of speculation. Photo-cross-linking studies suggested that in the holoreceptor complex PheB24 and TyrB26 primarily contact L1, whereas PheB25 primarily contacts αCT (17, 18).
FIGURE 2.
Sequence of insulin and its receptor engagement. A, sequence of insulin and sites of modification. Substitutions were introduced at position PheB24 (red with asterisk). Substitution of LysB29 by ornithine or the pairwise substitution of ProB28 and LysB29 by LysB28 and ProB29 facilitated semi-synthesis; the latter occurs in a prandial insulin analog (Humalog®). B, representation of residues B20–B27 (carbon atoms green, nitrogen atoms blue, and oxygen atoms red) packed between αCT and the L1-β2 sheet in a refined μIR complex (6). B chain residues B8–B19 are shown as a black ribbon and the A chain as a yellow ribbon; residues A1–A3 are concealed behind the surface of αCT. Key contact surfaces of αCT with B24–B26 are highlighted in magenta, and of the L1 domain with B24–B26 are highlighted in cyan; L1 and αCT surfaces that do not interact with B24–B26 are shown in lighter shades. C, orthogonal view to B, showing interaction of the side chain of PheB24 with the nonpolar surface of the L1-β2 sheet. TyrB26 is hidden below the surface of αCT. Engagement of conserved residues A1–A3 against the nonpolar surface of αCT is shown at top. D, environment of PheB24 within site 1 complex (stereo). Coordinates for B–D were obtained from PDB entry 4OGA.
The co-crystal structure of an insulin-μIR complex, recently refined at 3.5 Å resolution, has revealed that on receptor binding the B24–B27 segment is displaced from its position in the free hormone (Fig. 2B) (6). Such displacement, associated with reorganization of αCT on the surface of L1 (5), is critical to high affinity binding of the hormone. At this reorganized interface, the side chain of PheB24 plays a unique role as an “anchor” within a nonpolar pocket defined by the IR L1 surface, αCT, and central insulin B chain α-helix (Fig. 2C). The immediate environment of PheB24 in the μIR is delimited by L1 side chains Asn15, Leu37, and Phe39 by the αCT side chain Phe714 and by the insulin side chains LeuB15, TyrB16, and CysB19 (stereo model in Fig. 2D; for clarity B16 is omitted). Although past studies of selected insulin analogs suggested that aromaticity is required for such anchoring (12), the structure of the B24-related binding pocket in the μIR would seem compatible with aliphatic side chains of appropriate size and shape. To resolve this apparent discrepancy, we undertook systematic mutagenesis of the B24 position in human insulin. To facilitate trypsin-mediated semi-synthesis (19), our collection of analogs was prepared with ornithine (Orn) rather than lysine at position B29.7
Intriguingly, we observed that a nonaromatic side chain (MetB24) confers essentially native receptor-binding affinity and substantial biological activity in a rat model of DM. Molecular modeling of a simulated interface between [MetB24]insulin and the μIR suggested that this linear side chain might adopt a compact conformation within the confines of the B24-related pocket, intrinsically nonplanar due to the tetrahedral geometry of the side-chain carbon atoms and related sp3 configuration of the δ-sulfur atom. This model motivated design and characterization of a nonstandard insulin analog in which PheB24 was substituted by cyclohexanylalanine (Cha), an amino acid containing a cyclic and nonplanar aliphatic side chain. The ChaB24 insulin analog likewise exhibited high activity, and its crystal structure (determined at a resolution of 1.5 Å as an R6 zinc insulin hexamer) closely resembles that of wild-type (WT) insulin, including within the hexamer's three aromatic-rich B24-related dimer interfaces. The crystallographic analysis was enriched by the presence of an entire hexamer within the asymmetric unit (20). To our knowledge, the crystal structure of [ChaB24]insulin represents the first high resolution view of a cyclic aliphatic side chain in a globular protein (21).
In the crystal structure, the six ChaB24 side chains each exhibited a pair of overlapping chair conformations (oriented up or down) with variable occupancies. Although ChaB24 itself is unable to participate in weakly polar interactions (unlike PheB24) (22), neighboring aromatic-aromatic interactions among TyrB16, PheB25, TyrB26, and their dimer-related partners are preserved in the structure. Molecular modeling of the variant μIR complex likewise suggested that the side chain of ChaB24 would readily be accommodated within the aromatic-rich confines of the B24-related pocket. Nonetheless, the distinctive structure and flexibility of Cha's chair conformations may confer novel biophysical properties. In the context of a mealtime insulin analog in current clinical use ([LysB28,ProB29]insulin; named KP-insulin (23)), we observed that ChaB24 enhanced the rate of disassembly of the analog hexamer and delayed onset of protein fibrillation, properties of potential therapeutic advantage in the treatment of DM (24).
Together, our results have defined the structural requirements of an invariant anchor residue at a hormone-receptor interface (6). Conservation of PheB24 among vertebrate insulins and IGFs (2) may be enjoined by biological constraints (such as efficiency of folding and avoidance of toxic misfolding) unrelated to structure-activity relationships. Despite the broad conservation of PheB24 and its cognate IR binding pocket, nonstandard modifications at this site may be of translational interest.
EXPERIMENTAL PROCEDURES
Preparation of Insulin Analogs
Analogs were prepared by trypsin-catalyzed semi-synthesis using an insulin fragment, des-octapeptide (B23-B30)-insulin, and modified octapeptides as described (19). The fragment was made by tryptic cleavage of human insulin and purified by reverse phase HPLC. Modified octapeptides were prepared by solid phase synthetic methods (25). Trypsin-mediated formation of a peptide bond between ArgB22 and a synthetic octapeptide (in the series GXFYTPOT or GXFYTKPT, where X represents a standard or nonstandard amino acid and O designates ornithine) was favored by reaction in a mixed aqueous-organic solvent system containing 1,4-butanediol and dimethylacetamide as described (26); the resulting 51-residue insulin analogs were purified by preparative reverse phase C4 HPLC (Higgins Analytical Inc., Proto 300 C4 10 μm, 250 × 20 mm), and their purity was assessed by analytical reverse phase C4 HPLC (Higgins Analytical Inc., Proto 300 C4 5 μm, 250 × 4.6 mm). Predicted molecular masses were in each case verified using an Applied Biosystems 4700 proteomics analyzer MALDI-TOF.
X-ray Crystallography
Crystals of [ChaB24,OrnB29]insulin were obtained via hanging-drop vapor diffusion at 25 °C. 1-μl drops containing the protein at 10 mg/ml in 0.02 n HCl were mixed with a 1-μl drop of reservoir buffer containing 0.1 m sodium citrate, 0.08% zinc acetate, 2% phenol. Drops were suspended over 1 ml of reservoir buffer. A single crystal was transferred to a solution containing 20% (v/v) glycerol in the mother liquor for flash freezing. Diffraction data were collected at ∼100 K at a wavelength of 0.9537 Å at the Argonne National Laboratory, Structural Biology Center (beamline 19-ID; Argonne, IL). Data were processed using XDS, version January 10, 2014 (27), and the resolution limit was set according to the CC1/2 criterion (28) at the p = 0.001 level of significance. The structure was then determined by molecular replacement using PDB entry 1ZNJ as an initial search model and refined using PHENIX version 1.9.1692 (29). Hydrogen atoms were included in the final stages of refinement, which employed anisotropic B-factors for all non-hydrogen atoms. Within each insulin monomer, the segment GlyB23-ChaB24-PheB25 was modeled as having two alternate conformations to allow for gauche and anti-gauche chair conformations (defined with respect to the Cβ–Cγ bond). Occupancies of these paired segments were then refined independently across the protomers. The final model included residues A1–A21 and B1–B28 for each of the six insulin molecules in the asymmetric unit, 207 water molecules, two Zn2+ cations, two Cl− anions, and six phenol molecules. Full data processing and refinement, the statistics are given in Table 1.
TABLE 1.
X-ray data processing and refinement statistics
| Data processing | |
| Wavelength (Å) | 0.9537 |
| Resolution range (Å) | 50.0–1.50 (1.60–1.50)a |
| Space group | P21 |
| a (Å), b (Å), c (Å), β (°) | 46.04, 60.90, 59.30, 112.25 |
| Redundancy | 3.6 (3.5) |
| Completeness (%) | 98.9 (99.3) |
| Rmerge | 0.04 (0.84) |
| 〈I/σ(I)〉 | 14.8 (1.6) |
| CC1/2b | 0.999 (0.716) |
| Refinement | |
| Resolution range (Å) | 50.0–1.50 (1.60–1.50) |
| No. of reflections | 48189 (4823) |
| Rwork/Rfreec | 0.162/0.204 |
| No. protein atomsd | 2488 |
| No. of non-protein atoms | 248 |
| 〈Biso〉 protein atoms (Å2)d | 41.9 |
| 〈Biso〉 non-protein atoms (Å2) | 46.7 |
| σbonds (Å)/σangles (°) | 0.010/1.32 |
| Ramachandran plot | |
| Favored (%) | 100 |
| Outliers (%) | 0 |
a Numbers in parentheses refer to the outer resolution shell.
b Pearson correlation coefficient between merged intensities of two random halves of the diffraction data set (28).
c Free set contains 10% of total observed reflections.
d Atom counts and average B values exclude hydrogen atoms.
Circular Dichroism Spectroscopy
Far-ultraviolet (UV) spectra were obtained from 200 to 250 nm on an AVIV spectropolarimeter equipped with an automated syringe-driven titration unit. Insulin or analogs were made 50 μm in a buffer containing 10 mm potassium phosphate (pH 7.4) and 50 mm KCl. Helix-sensitive wavelength 222 nm was used as a probe of protein denaturation by guanidine hydrochloride. Thermodynamic parameters were obtained by application of a two-state model as described (30). In brief, data were fit by nonlinear least squares to a two-state-model as shown in Equation 1,
 |
where x is the concentration of guanidine hydrochloride, and θA and θB represent respective estimates of the baseline ellipticities of the protein in its native and unfolded states as extrapolated to a guanidine concentration of 0. Baseline values were approximated via pre- and post-transition lines represented by equations θA(x) = θAH2O + mAx and θB(x) = θBH2O + mBx. Such simultaneous fitting avoids artifacts of linear plots of ΔG versus concentration of denaturant.
NMR Spectroscopy
Spectra were acquired in aqueous D2O solution (pD 7.6, direct meter reading) at 700 MHz at 32 °C. A variant insulin B chain containing substitutions AspB10, LysB28, and ProB29 (designated the DKP template) was employed to facilitate high resolution studies of insulin as an engineered monomer (30, 31). Spectra of DKP-insulin and [ChaB24-DKP]insulin were each acquired at protein concentrations of 0.35 mm. Homonuclear two-dimensional nuclear Overhauser effect (NOE) spectroscopy (NOESY) and total correlation spectroscopy (TOCSY) were obtained with respective mixing times of 200 ms and 55 ms. Presumptive assignments were obtained by analogy to the assigned spectrum of [l-AlaB24-DKP]insulin, an analog that exhibits a native-like fold but without the aromatic ring current of PheB24 (30, 31). 1H NMR chemical shifts were calibrated to parts/million (ppm) relative to trimethylsilyl propionate as an internal standard, assumed to be 0.0 ppm.
Receptor Binding Assays
Affinities of WT insulin or semi-synthetic analogs for the IR (isoform B) were measured by a competitive-displacement assay (32). In brief, microtiter strip plates (Nunc Maxisorb) were incubated at 4 °C overnight with a stock solution containing 40 μg/ml anti-FLAG immunoglobulin G. Lysates of 293 PEAK cells transfected with cDNAs encoding the IR with a C-terminal FLAG tag were purified using wheat germ agglutinin chromatography as described previously (32). Partially purified receptors were then immobilized in the coated plates. Competitive binding assays employed human [3-125I-TyrA14]insulin as a tracer.
Receptor-based Screening of Insulin Analogs
Analogs were initially tested for their ability to displace pre-bound [125I-TyrA14]insulin from the IR at a single concentration of analog (0.75 nm), calibrated based on baseline displacement of 90% of receptor-bound [125I-TyrA14]insulin by control analog [OrnB29]insulin. The nondisplaced fraction of [125I-TyrA14]insulin (less than 20, 21–40, 41–70, and 71–100%) by a given B24 analog permitted its qualitative assignment to a corresponding class of relative affinities (designated high (I), intermediate (II), low (III), or very low (IV)).
Rodent Assays
Male Sprague-Dawley rats (mean body mass ∼300 g) were rendered diabetic by streptozotocin (33). To test the in vivo potency of insulin analogs relative to [OrnB29]insulin, the analogs were made 10 μg per 100 μl in a formulation buffer (16 mg/ml glycerin, 1.6 mg/ml meta-cresol, 0.65 mg/ml phenol, and 3.8 mg/ml sodium phosphate (pH 7.4)) and injected intravenously into tail veins. The normalized dose was 10 μg of insulin analog per 300-g rat with the actual dose (and so injection volume) being adjusted to each rat's body weight. Resulting changes in blood glucose concentration were monitored using a clinical glucometer (Hypoguard Advance Micro-Draw meter). WT insulin or analogs were each re-purified by HPLC, dried to powder, dissolved in diluent at the same maximum protein concentration, and re-quantitated by analytical C4 reverse phase HPLC to ensure uniformity of formulations; dilutions were made using the above buffer. Rats were injected at time t = 0. Blood was obtained from the clipped tip of the tail at time t = 0 and every 10 min for the 1st h, every 20 min for the 2nd h, every 30 min for the 3rd h, and every hour thereafter to a final time of 5 h. The efficacy of WT insulin or analog to reduce the blood glucose concentration was calculated using the following: (a) the change in blood glucose concentration over the first hour, and (b) the integrated area between the glucose time dependence and a horizontal line at the starting blood glucose concentration (area over the curve; AOC). Statistical significance was assessed using a Student's t test.
Assessment of Fibril Formation
To estimate lag times prior to onset of fibril formation, zinc-free WT insulin or analogs were made 60 μm in phosphate-buffered saline (pH 7.4) containing 0.01% sodium azide as an antimicrobial agent. The solutions were gently rocked at 37 °C in glass vials containing a liquid-air interface. Aliquots, taken at regular intervals, were frozen to enable later analysis of thioflavin T fluorescence. For a given tube, the assay was terminated on the 2nd day of the appearance of cloudiness in the solution (34).
Cobalt-coordinated Insulin Hexamers
Visual absorption spectroscopy was used to probe the formation and disassembly of phenol-stabilized R6 Co2+-substituted insulin hexamers. WT insulin or analogs were made 0.6 mm in a buffer containing 50 mm Tris-HCl (pH 7.4), 50 mm phenol, 0.2 mm CoCl2, and 1 mm NaSCN (35). Samples were incubated overnight at room temperature prior to the studies to ensure that a conformational equilibrium was reached. Spectra (450–700 nm) were obtained to monitor tetrahedral Co2+ coordination with its signature peak absorption band at 574 nm (35). To determine the rate of Co2+ release from the hexamers, metal ion sequestration was initiated at 25 °C by addition of an aliquot of EDTA (50 mm at pH 7.4) to a final concentration of 2 mm. Attenuation of the 574-nm absorption band was monitored on a time scale of seconds to hours. Kinetic data were consistent with a mono-exponential decay (36).
Molecular Dynamics Simulations
Molecular dynamics (MD) simulations were performed using GROMACS (version 4.5.5) (37), which employs the CHARMM all-atom additive force field (38, 39). Proteins were solvated in a cubic box of TIP3P water molecules. Ionizable residues and protein termini were assumed to be in their charged state. Sodium and chloride ions were added to neutralize the system at a final ionic strength of 0.10 m. Protein and solvent (including ions) were coupled separately to a thermal bath at 300 K employing velocity rescaling (40) applied with a coupling time of 1.0 ps. Pressure was maintained at 1 bar by coupling to a Berendsen barostat (41) with a coupling constant of 5.0 ps and compressibility of 4.5 × 10−5 bar. The time step was 2 fs. Simulations were performed with a single nonbonded cutoff of 10 Å and neighbor list update frequency of 10 steps (20 fs). The particle-mesh Ewald method was used to account for long range electrostatics (42), applying a grid width of 1.2 Å with fourth-order spline interpolation. Bond lengths were constrained using the LINCS algorithm (43). All simulations consisted of an initial minimization of water molecules, followed by 100 ps of MD with the protein restrained to permit equilibration of the solvent. Simulations of the WT, [MetB24]-, and [ChaB24]insulin monomers used as initial model coordinates the structure 2KJJ obtained from the Protein Data Bank (PDB). Calculations were continued for 120 ns from the geometries obtained after initial positionally restrained MD (below) at a temperature of 300 K.
Molecular Modeling of μIR Complexes
Comparative modeling of the MetB24- and ChaB24 variant μIR complexes was performed using MODELLER (44). The simulations employed as molecular templates the structure of the WT μIR-insulin complex (PDB entry 4OGA; Ref 6). The models included residues disordered in the crystal structure, viz. Cys159–Asn168 and Lys265–Gln276 of IR and AspB28–ThrB30 of insulin. An initial 50 models were created for each complex, and the structure with the lowest energy was selected for MD simulations. The μIR L1-CR fragment contains N-linked glycosylation sites at residues Asn16, Asn25, Asn111, Asn215, and Asn255 (45); N-acetylglucosamine carbohydrate was thus attached at each of these sites. Following positionally restrained MD, all restraints on the protein were removed, and MD continued for a further 50 ns. The surface area and volumes of protein cavities and crevices were estimated using programs VoroProt and PROVAT (46, 47), respectively. The surface areas of cavities were calculated based on rolling a sphere of radius 1.4 Å, whereas cavity volumes represent the difference between the sizes of cavities in the WT structure and rigid-body models wherein the residue of interest was substituted by Gly.
RESULTS
Receptor Binding Screen
Eighteen insulin analogs containing substitutions at B24 were prepared at small scale in the context of [OrnB29]insulin (Table 2), an active template chosen to facilitate trypsin-mediated semi-synthesis (6). To avoid disulfide interchange reactions, we did not prepare a CysB24 analog. To avoid introducing a tryptic site, LysB24 and ArgB24 analogs were also not prepared. A coarse receptor binding assay was designed to discriminate among insulin analogs based on displacement of pre-bound [125I-TyrA14]insulin at an analog concentration of 0.75 nm (Fig. 3A). At this concentration, [OrnB29]insulin was found to displace ≥80% of the bound [125I-TyrA14]insulin; a maximum of 20% of bound [125I-TyrA14]insulin was thus employed to define a high affinity group of analogs (class I in Fig. 3B). Intermediate, low, and very low affinity groups were similarly defined based on fractions of bound [125I-TyrA14]insulin in the presence of the 0.75 nm analog of 21–40, 41–70, and 71–100%, respectively (classes II–IV in Fig. 3B). This preliminary classification facilitated design of definitive binding assays.
TABLE 2.
Receptor-binding affinities of insulin analogs
Analogs were prepared in a template in which LysB29 was substituted by Orn. Assays employed the B isoform of the lectin-purified and detergent-solubilized IR.
| B24 residue | Kd | B24 residue | Kd |
|---|---|---|---|
| nm | nm | ||
| Phea | 0.05 ± 0.01 | Pro | VLb |
| Gly | 0.05 ± 0.01 | Ser | 3.9 ± 0.6c |
| Ala | 13.9 ± 2.3c | Thr | 2.1 ± 0.3c |
| Val | 0.65 ± 0.10 | Cys | NDd |
| Leu | 0.10 ± 0.02 | Asn | 6.2 ± 0.9c |
| Ile | 0.15 ± 0.02 | Asp | 3.7 ± 0.5c |
| Met | 0.06 ± 0.01 | Gln | VLb |
| Cha | 0.09 ± 0.01 | Glu | VLb |
| Tyr | 0.98 ± 0.14 | His | VLb |
| Trp | 0.93 ± 0.14 | Orn | >50e |
a This represents [OrnB29]insulin; its dissociation constant was indistinguishable from that of WT insulin.
b VL means very low affinity (class IV in coarse screen).
c Complete binding assays were performed on these representative members of class IV to obtain a quantitative calibration of the assay.
d ND means not determined.
e OrnB24 provided a model of a basic side chain as an isostere of Met; its Kd value was estimated to be 73 ± 17 nm. Lys and Arg were not included in the initial screen as these substitutions would have complicated semi-synthesis (see “Experimental Procedures”).
FIGURE 3.
Functional screening of insulin analogs and receptor binding assays. A, competitive binding assay of [OrnB29]insulin to the insulin receptor (black line, orange squares). For reference, predicted binding isotherms are shown corresponding to putative analogs with Kd values that are 10-fold (red), 100-fold (blue), or 1000-fold (green) greater than that of [OrnB29]insulin. Vertical axis, B/B0, where B is [125I-TyrA14]insulin bound by receptor at a given insulin analog concentration, and B0 is the baseline value ([125I-TyrA14]insulin bound in the absence of unlabeled insulin analog). Horizontal axis, log concentration of insulin analog. Filled black circles indicate approximate displacement of labeled [125I-TyrA14]insulin on binding of GluB24 (top), ValB24 (middle), and GlyB24 (bottom), each at a concentration of 0.75 nm. B, classification of insulin analogs based on fraction of remaining γ-counts as follows: high (I), ≤20%; intermediate (II), 21–40%; low (III), 41–70%; very low (IV), 71–100% relative to WT insulin. C, complete [125I-TyrA14]insulin displacement assays of selected B24 analogs as follows: [OrnB29]insulin (red squares); [GlyB24,OrnB29]insulin (blue diamonds); [IleB24,OrnB29]insulin (purple inverted triangles); [LeuB24,OrnB29]insulin (brown triangles); and [MetB24,OrnB29]insulin (green circles). D, corresponding receptor binding assays of ChaB24-substituted analogs in relation to respective controls as follows: [OrnB29]insulin (orange squares); [ChaB24,OrnB29]insulin (maroon x); [ChaB24-KP]insulin (purple stars) versus controls human insulin (black squares); and KP-insulin (green cross).
The high affinity group (class I) included Phe (the WT residue), an aliphatic residue (Met), and Gly; the anomalous activity of [GlyB24]insulin has been widely discussed in past studies (asterisk in Fig. 3B) (10, 48, 49). Class II contained Ile and Leu. Class III contained Val as well as the aromatic residues Tyr and Trp; the low affinity of [TyrB24,OrnB29]insulin is in accordance with prior studies (11, 12). Class IV (very low affinity) contained His (in accordance with Ref. 12), charged (Asp and Glu), constrained (Pro), polar (Asn, Gln, Ser, and Thr), and Ala (in accordance with Ref. 50). Following this screen, all analogs in classes I–III and representative analogs in class IV (Ala, Asn, Asp, Ser, and Thr) were prepared at larger scale to enable measurement of hormone-receptor dissociation constants (Kd) and, in four cases (Gly, Ile, Leu, and Met), measurement of thermodynamic stabilities (ΔGu).
Results of selected complete competitive displacement assays of receptor binding are given in Table 2; representative data are shown in Fig. 3C. The three analogs with highest affinities were GlyB24 (Kd 0.05 ± 0.01 nm), PheB24 (parent analog; Kd 0.05 ± 0.01 nm), and MetB24 (Kd 0.06 ± 0.01 nm) as compared with WT (Kd 0.05 ± 0.01 nm). To test whether the high affinity of the MetB24 analog depends on side-chain hydrophobicity, we prepared an OrnB24 analog (a basic isostere of Met); only negligible receptor binding was observed (Kd >50 nm; Table 2). The affinity of the LeuB24 variant was reduced by 2-fold (Kd 0.10 ± 0.02 nm) in accordance with past studies (19, 51); the affinity of the IleB24 variant was reduced by 3-fold (Kd 0.15 ± 0.02 nm). Such small differences are consistent with natural variation among mammalian insulins (species variants (52)). Aside from the special case of GlyB24 (which may reflect a shift in binding mode (6, 12, 53)),8 these findings suggest that the hormone-receptor interface selects for nonpolar side chains at B24 of suitable shape and size, irrespective of aromaticity. Avoidance of packing defects within the B24-related pocket presumably underlies the affinity order Phe, Met > Ile, Leu > Val > Ala.
Residues in class IV are likely to have imposed a combination of cavity-related and polarity penalties. Because of its clinical association with DM (15, 16), detailed binding studies were undertaken of [SerB24,OrnB29]insulin in relation to [AlaB24,OrnB29]insulin and other small polar or charged side chains in class IV (Thr, Asn, or Asp; Table 2). In accordance with previous studies, the affinity of the SerB24 analog (15, 54) was low (∼1% relative to [OrnB29]insulin) but 3-fold greater than that of the AlaB24 analog (50). Also preferred relative to AlaB24 were Thr, Asn, or Asp, which conferred affinities similar to that of [SerB24,OrnB29]insulin. Unlike Ala, each of these side chains would be able to participate in hydrogen bonding.
Folding and Stability
Overall α-helicity of the high affinity B24 insulin analogs (Met, Gly, Ile, and Leu) was monitored by far-UV CD (Fig. 4, A and C). Although the spectra indicated that a predominance of α-helix is maintained, attenuation of mean residue ellipticity at 222 nm and more negative values at 208 nm suggested overall α-helical destabilization. Such features were previously observed in the CD spectrum of KP-insulin in relation to WT insulin (49) and may reflect decreased dimerization at this protein concentration (50 μm) (55). In accordance with this interpretation, previous studies have shown that the spectrum of an AlaB24 analog of engineered monomer DKP-insulin9 was similar to that of DKP-insulin itself (30).
FIGURE 4.

Biophysical assays of structure and stability. A, CD spectra of [IleB24,OrnB29]insulin (amber), [LeuB24,OrnB29]insulin (purple, overlapping spectra), and [GlyB24,OrnB29]insulin (pink, overlapping spectra) in relation to control spectra of [OrnB29]insulin (black line) and WT insulin (open circles). B, guanidine-unfolding transitions as monitored by ellipticity at 222 nm; color code is as in A. C, CD spectra of [OrnB29]insulin (black line), [MetB24,OrnB29]insulin (blue), and [ChaB24,OrnB29]insulin (green). D, CD-detected guanidine unfolding transitions as monitored by ellipticity at 222 nm; color code is as in C. E, CD spectra of [LysB28,ProB29]insulin (black dot-dash) and [ChaB24,LysB28,ProB29]insulin (red). F, guanidine unfolding transitions as monitored by ellipticity at 222 nm; color code is as in E. Results of two-state modeling of the denaturation studies are given in Fig. 8A (histogram of ΔGu values) and Table 3 (ΔGu, Cmid, and m values). Control data for WT insulin in A and B were adapted from Pandyarajan et al. (62).
Helix-related ellipticity at 222 nm was exploited as a probe of fractional protein unfolding as a function of increasing concentrations of guanidine hydrochloride (Fig. 4, B, and D). Estimates of free energies of unfolding (ΔGu), based on a two-state model (30), demonstrated that the aliphatic substitutions at B24 were in each case associated with lower thermodynamic stability (Fig. 8A and Table 3). The B24 substitutions compatible with high affinity (Gly, Ile, Leu, and Met) exhibited decrements (ΔΔGu) in the range 0.7–1.9 kcal·mol−1 (including uncertainties) relative to the baseline stability of [OrnB29]insulin (ΔGu 3.5 ± 0.2 kcal/mol). The m values obtained in fitting the two-state model (i.e. the slope in a plot of ΔGu versus denaturation concentration; column 5 in Table 3) were also reduced. Because this slope correlates with extent of exposure of nonpolar surfaces on unfolding (30), this trend suggests that the nonaromatic side chains at B24 are associated with less efficient burial of such surfaces in the respective native states.
FIGURE 8.

Biophysical and self-assembly properties of B24 analogs. A, thermodynamic stabilities as inferred from two-state modeling of the guanidine denaturation assays shown in Fig. 4. B, time to fibril formation (lag time) at 37 °C for WT insulin (HI, purple circles), [OrnB29]insulin (black circles), [MetB24,OrnB29]insulin (blue circles), [ChaB24,OrnB29]insulin (green circles), [LysB28,ProB29]insulin (KP, open circles), and [ChaB24,LysB28,ProB29]insulin (ChaB24-KP, red circles). Circles indicate individual measurements. Onset of fibrillation was defined as a 2-fold enhancement of thioflavin T fluorescence. C and D, optical absorption spectra of Co2+-stabilized R6 insulin hexamers and kinetics of metal ion release. C, Co2+ d-d absorption bands of WT insulin (pink line), [OrnB29]insulin (black line), [ChaB24,OrnB29]insulin (green line), and [MetB24,OrnB29]insulin (blue line) near 550 nm provide a signature of the R state. The amplitudes of the [ChaB24,OrnB29]insulin and [MetB24,OrnB29]insulin are attenuated by 7 ± 1 and 37 ± 2% relative to [OrnB29]insulin. D, kinetic analysis based on Co2+ sequestration by EDTA. The color code is as in C. E, corresponding absorption spectra of Co2+-stabilized R6 insulin hexamers as follows: WT insulin (pink line); KP-insulin (black dot-dashed line); and [ChaB24-KP]insulin (red line). The amplitude of [ChaB24-KP]insulin is attenuated by 22% (±1) relative to KP-insulin. F, kinetic analysis based on Co2+ sequestration by EDTA. WT data were adapted from Ref. 62. The color code is as in E. Half-lives derived from the data in D and F are given in Table 5.
TABLE 3.
Properties of selected analogs
| Protein | Lag timea | ΔGub | Cmid | m |
|---|---|---|---|---|
| days | kcal·mol−1 | m | kcal·mol−1 | |
| human insulinc | 2.5 ± 1.2 | 3.4 ± 0.1 | 4.8 ± 0.1 | 0.69 ± 0.02 |
| LysB28, ProB29 (KP) | 3.3 ± 0.8 | 3.0 ± 0.1 | 5.1 ± 0.1 | 0.60 ± 0.01 |
| ChaB24-KP | 13 ± 5 | 2.4 ± 0.1 | 5.0 ± 0.2 | 0.48 ± 0.02 |
| OrnB29 | 6.4 ± 1.1 | 3.5 ± 0.2 | 4.8 ± 0.2 | 0.73 ± 0.03 |
| GlyB24, OrnB29 | NDd | 2.1 ± 0.2 | 4.1 ± 0.4 | 0.51 ± 0.05 |
| MetB24, OrnB29 | 2.0 ± 1.0 | 2.2 ± 0.1 | 4.1 ± 0.1 | 0.53 ± 0.02 |
| ChaB24, OrnB29 | 6.2 ± 3.5 | 2.7 ± 0.1 | 4.5 ± 0.2 | 0.59 ± 0.02 |
| LeuB24, OrnB29 | ND | 2.5 ± 0.1 | 4.2 ± 0.2 | 0.60 ± 0.02 |
| IleB24, OrnB29 | ND | 2.2 ± 0.1 | 4.2 ± 0.2 | 0.53 ± 0.02 |
a 2-Fold increase over baseline in thioflavin T fluorescence provided a criterion for onset of fibrillation. Fibrillation lag times pertain to zinc-free analogs; each protein was made 60 μm in phosphate-buffered saline (pH 7.4). For statistical analysis, one outlier in the OrnB29 trial was excluded due to it being more than two standard deviations greater than the mean.
b Thermodynamic parameters were inferred from CD-detected guanidine denaturation data by application of a two-state model.
c Data were adapted from Ref. 62.
d ND means not determined. A previous study has shown that lag time of [GlyB24-KP]insulin (1.4 ± 0.3 days) is 2-fold shorter than that of KP-insulin (3 ± 0.3 days).
Nonstandard Mutagenesis
The B24-related pocket, as visualized in the crystal structure of the WT μIR-complex (6), has a calculated volume of ∼200 Å3 in the absence of PheB24. Of this potential space, the side chain of PheB24 occupies only 117 Å3 (packing efficiency 58%); residual packing defects are predicted above, below, and to the sides of the aromatic ring (see “Discussion”). Although such quantitative analysis is limited by the low resolution of the structure (3.5 Å), modeling predicted that the planar aromatic ring of PheB24 could be replaced by a nonplanar aliphatic ring without steric clash. We therefore prepared nonstandard analogs [ChaB24,OrnB29]insulin (for comparison with our original set of B24 variants) and [ChaB24-KP]insulin (for assessment of its potential pharmacologic properties (56)); [ChaB24-DKP]insulin was also prepared to facilitate NMR studies (30, 31). These analogs exhibited a relative affinity for the insulin receptor (∼50–60%; class II) similar to that of the LeuB24 analog (Fig. 3D and Table 2). To test whether Phe and Cha are generally interchangeable at protein-protein interfaces, we also prepared a ChaB25 analog (in the context of KP-insulin); at this site the nonstandard substitution impaired receptor binding by ∼50-fold relative to KP-insulin (Table 4) in accordance with the low activity of DM-associated clinical variant [LeuB25]insulin (19, 51). The substantial receptor-binding affinity of the ChaB24 analogs is therefore an intrinsic feature of the B24-related pocket.
TABLE 4.
Affinities of insulin analogs
These analogs were prepared in templates in which ProB28 and LysB29 were inverted in sequence ([LysB28,ProB29]insulin; KP-insulin) and with the additional substitution HisB10 → Asp (DKP-insulin) as an NMR model (30, 31). Binding studies employed the B isoform of the IR.
| Analog | Kd |
|---|---|
| nm | |
| KP-insulin | 0.09 ± 0.01 |
| [ChaB24-KP]insulin | 0.17 ± 0.02 |
| [ChaB25-KP]insulin | 5.1 ± 0.8 |
| DKP-insulin | 0.02 ± 0.01 |
| [ChaB24-DKP]insulin | 0.05 ± 0.01 |
CD Studies
Overall conformations of [ChaB24,OrnB29]insulin and [ChaB24-KP]insulin were assessed by far-UV CD (Fig. 4, C and E). CD spectra of [ChaB24,OrnB29]insulin and [ChaB24-KP]insulin more closely match that of KP-insulin (Fig. 4E), which is monomeric under these conditions (57). Guanidine denaturation studies (Fig. 4, D and F) indicated that in either the OrnB29 or KP contexts, substitution of PheB24 by Cha resulted in similar reductions in stability relative to their respective parent templates (ΔΔGu = 0.8 ± 0.3 and 0.6 ± 0.2 kcal·mol−1; Table 3). These values are similar to the decrement caused by LeuB24 and less severe than those imposed by IleB24 or MetB24.
1H NMR Spectroscopy
Spectra of [ChaB24-DKP]insulin were obtained in D2O at pD 7.6 and 32 °C in relation to baseline spectra of [DKP]insulin (30, 31). The absence of B24-related aromatic TOCSY and NOESY cross-peaks in the variant spectra were readily apparent (Fig. 5). Nevertheless resolved were the inter-residue NOEs involving the aromatic protons of TyrB26 (with presumptive assignments of B26-ValB12 and B26-LeuB15) and of TyrA19 (A19-IleA2 and A19-LeuB15). These qualitative NMR features indicate that the ChaB24 analog maintains a native-like conformation in solution, including with respect to the overall orientation of the C-terminal segment of the B chain relative to the α-helical core (30, 31). Overall 1H NMR chemical-shift dispersion among aliphatic resonances was reduced in accordance with the absence of the PheB24 ring-current diamagnetic field. Resonance assignments for ChaB24 were not obtained.
FIGURE 5.
1H NMR spectroscopy of DKP-insulin and [ChaB24-DKP]insulin. Two-dimensional spectroscopy of [ChaB24-DKP]insulin (A) and DKP-insulin (B) is shown. At left are TOCSY spectra of aromatic spin systems; at right are NOESY spectra in region containing cross-peaks between aromatic protons (vertical axis, ω2) and aliphatic protons, including upfield shifted methyl groups (horizontal axis, ω1). Brackets/arrows denote aromatic and methyl group resonances with labeled residues.
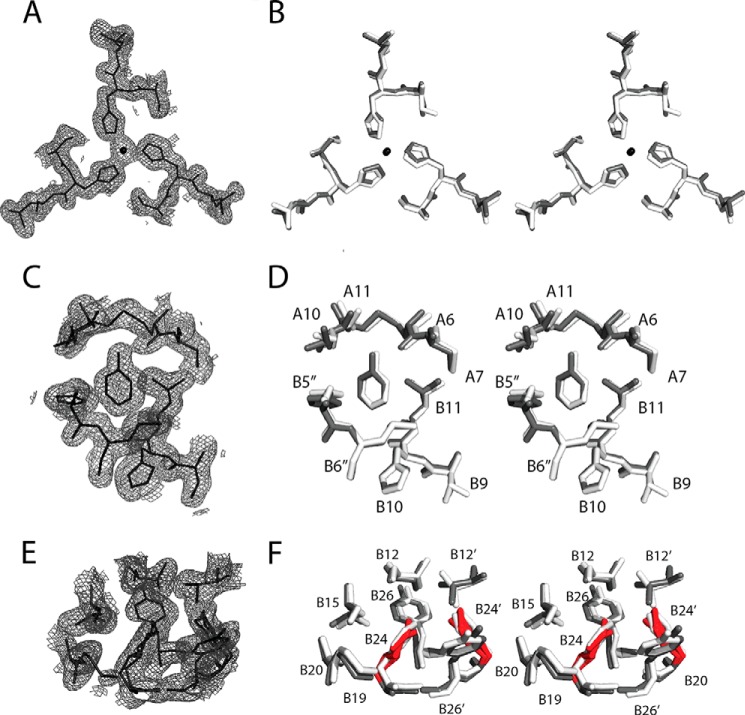
Crystal Structure of [ChaB24,OrnB29]Insulin
To obtain atomic level structural information, [ChaB24,OrnB29]insulin was crystallized under conditions that ordinarily lead to crystallization of WT insulin as a phenol-stabilized R6 hexamer (20). Indeed, a classical monoclinic lattice (Table 1) was observed in which one R6 hexamer (with protomers designated as having chains A and B, C and D, E and F, G and H, I and J, and K and L, respectively) defined the asymmetric unit.10 A ribbon model of the zinc insulin analog hexamer is shown in Fig. 6A; in this view, the six R-state-specific B1–B19 α-helices (green) are seen near the center, whereas the A chains (black) are near the periphery.
FIGURE 6.
Crystal structure of [ChaB24,OrnB29]insulin. A, R6 zinc hexamer. The A and B chains are shown in black and green; ChaB24 side chains are highlighted in red. The two axial zinc ions are aligned in the center (blue), coordinated by trimer-related HisB10 side chains (white). B, superposition of a representative analog protomer (gray) and WT protomer (white). The side chains of ChaB24 (red) and PheB24 (white) are shown as sticks. C, stereo view of aromatic residues (gray) and ChaB24 (red) at dimer interface. D, expanded view of the side chains near B24 in the analog structure (gray) and WT structure (white). Sulfur atoms of cysteine A20–B19 are shown as yellow spheres. Coordinates of the WT R6 hexamer were obtained from PDB entry 1ZNJ.
No significant differences were observed relative to the WT R6 hexamer in secondary structure, chain orientation, mode of assembly, or structures of the Zn2+- and phenol-binding sites. The six independent R-state protomers exhibited essentially identical conformations (average pairwise main-chain root-mean-square difference (r.m.s.d.) = 0.55 Å and average side-chain r.m.s.d. = 1.92 Å). Tetrahedral coordination of the two axial zinc ions (overlying blue spheres at center of Fig. 6A) by the side chains of HisB10 (three per R3 trimer; light gray side chains) was essentially identical to that observed in WT R6 hexamers (20). The fourth coordination site contained a presumed chloride anion. A representative (2Fobs − Fcalc) electron density map of one metal ion-binding site (Fig. 7A) is shown in relation to a superposition of the analog and WT coordination sites (dark and light gray in Fig. 7B; stereo stick models). Zn-N distances are 1.98, 1.98, and 2.00 Å in the zinc-binding site shown and 1.96, 2.01, 2.04 Å in the other trimer.
FIGURE 7.
(2Fobs − Fcalc) electron density maps. A, on-axis view of a representative zinc ion-binding site in R6 analog hexamer. B, superposition (stereo view) of zinc-binding sites in the analog hexamer (gray) and WT R6 hexamer (white). Zn2+ ion is shown as a black sphere. Not shown: Zn2+-coordinating Cl− anion. C, electron density of a representative bound phenolic ligand. Its para-OH group forms hydrogen bonds with the carbonyl oxygen and amide nitrogen of CysA6 and CysA11, respectively (cysteine A6–A11). The phenolic ring makes van der Waals contacts within the dimer-dimer interface of the hexamer; contacts include HisB5 of another dimer. D, stereo view corresponding to C. E, electron density of ChaB24 and surrounding residues. F, stereo view corresponding to E (stick representation). The two chair conformations of ChaB24 and ChaB24′ at this dimer interface are highlighted in red. Coordinates of the WT R6 hexamer were obtained from PDB entry 1ZNJ.
Superposition of a representative analog protomer and WT protomer is shown in Fig. 6B (dark and light gray ribbons, respectively). Average pairwise differences between individual protomers of [ChaB24,OrnB29]insulin and the WT R-state protomers (main-chain r.m.s.d. 0.65 = Å and side-chain r.m.s.d. = 2.03 Å) are only marginally different from the structural variation within a collection of WT protomers (main-chain r.m.s.d. = 0.54 Å and side-chain r.m.s.d. = 1.87 Å). Similarly, the variant dimer interface closely resembles that of WT insulin; a representative structure is shown in Fig. 6C. Side chains in close proximity to B24 are shown in Fig. 6D. Alignment of a representative analog dimer and WT dimer yielded main-chain r.m.s.d. = 0.29 Å and side-chain r.m.s.d. = 2.50 Å, the latter being inflated by the varying rotameric disposition of the side chain of residue B25 (and its 2-fold related partner) within and across both structures.
The analog hexamer contained six bound molecules of phenol, located at an interface between dimers as in the WT R6 hexamer (20). The six independent phenol-binding sites are essentially identical. The (2Fobs − Fcalc) electron density map volume associated with one such phenol is shown in Fig. 7C in relation to a superposition of variant and WT structures (dark and light gray; stereo stick models; Fig. 7D). In each case, a characteristic pair of hydrogen bonds from the phenolic –OH group was formed to the main-chain carbonyl oxygen (acceptor) and amide group (donor) of CysA6 and CysA11, respectively.
The positions of PheB24 (light gray side chain) and ChaB24 (red) closely overlap, with the side chain of ChaB24 (red in Fig. 7F) packing among the cluster of aromatic residues TyrB16, PheB25, TyrB26, and their dimer-related partners (black) (Fig. 6C). The six independent side chains of ChaB24 each exhibited the expected chair conformations but with varying relative occupancies of two overlapping conformations (with respective Cζ atoms oriented toward or away from LeuB15 of the same chain; defined as anti-gauche or gauche with respect to the B24 Cβ–Cγ bond). Occupancy refinement suggested that in four of the protomers the cyclohexanyl ring has a predominant anti-gauche conformation (anti-gauche occupancies 0.73 (chain B), 0.70 (chain D), 0.68 (chain F), and 0.82 (chain L), respectively); in the other two protomers the occupancy was split somewhat more evenly (anti-gauche occupancies 0.64 (chain H) and 0.54 (chain J), respectively). Dimer-related pairs of ChaB24 side chains thus exhibited adjoining anti-gauche occupancies of (0.73, 0.86), (0.68, 0.64), and (0.54, 0.82). Although these occupancies are consistent in each case with the appearance of the associated volumes of the final (2Fobs − Fcalc) electron density map (Fig. 7E), the diffraction data do not distinguish between dynamic disorder within a hexamer from static disorder among hexamers in the crystal lattice. Observation of nonequivalent occupancies across a given dimer interface nonetheless suggests that both anti-gauche/anti-gauche and anti-gauche/gauche packing schemes are allowed.
The environment of ChaB24 is similar but not identical to that of PheB24 at the dimer interface. A (2Fobs − Fcalc) electron density map of one such interface is shown in Fig. 7E in relation to a superposition of analog and WT structures (Fig. 7F). Although the four canonical inter-chain hydrogen bonds observed in the B24–B28/B28′-B24′ anti-parallel β-sheet (2) are indistinguishable from those observed in a collection of WT R6 structures (20), small adjustments occurred in the side-chain conformations of TyrB26′ (near ChaB24) and TyrB26 (near ChaB24′), where primed residues represent the dimer-related partners. Although the positions of the aromatic rings overlap, on average their respective χ1 and χ2 angles differ slightly (by 2° and 5° with respect to WT hexamer 1ZNJ). These adjustments are presumably related to the nonplanarity of ChaB24 and ChaB24′ and to changes in weakly polar interactions (22). The similarity of the variant and WT structures suggests that the essential features of the dimer interface do not depend on the asymmetric distribution of partial charges in the aromatic ring of PheB24 and associated pattern of aromatic-aromatic interactions.
Kinetic Analysis of Hexamer Disassembly
To gain further insight into native self-assembly, we next probed the rate of disassembly of [ChaB24,OrnB29]insulin and [ChaB24-KP]insulin hexamers in relation to their respective parent templates, WT insulin, and [MetB24,OrnB29]insulin. These studies employed Co2+ as an optical probe of the tetrahedral zinc-binding site in the R6 insulin hexamer (35).
Visible absorption spectra of [OrnB29]insulin and its derivatives are shown in Fig. 8C. The spectra of the variant complexes exhibited d-d absorption bands similar in shape to those of [OrnB29]insulin hexamers (black line) but reduced in magnitude. The respective absorption bands of the MetB24 and ChaB24 analogs were attenuated by 37 ± 2 and 7 ± 1% (blue and green lines in Fig. 8C). Such attenuation suggests either incomplete hexamer assembly at the protein concentration employed (0.6 mm) or a partial shift in the conformational equilibrium to subpopulations of T3Rf3 (and possibly T6) hexamers in which octahedral Co2+ coordination would attenuate or eliminate the d-d absorption bands. Corresponding studies of [ChaB24-KP]insulin are shown in Fig. 8E in relation to KP-insulin and WT insulin. Whereas the d-d absorption band in the spectrum of KP-insulin (black dot-dashed line in Fig. 8E) was similar in magnitude to those of WT insulin (pink line), the amplitude of [ChaB24-KP]insulin (red line) was attenuated by 22 ± 1% relative to KP-insulin.
The conformational equilibrium of insulin is characterized by rates of hexamer disassembly (with loss of bound metal ions) and reassembly (with restoration of metal ion binding). To probe the lifetimes of the tetrahedral Co2+-binding sites, we added an excess of EDTA to these protein solutions. The ensuing attenuation of the d-d absorption bands provided a kinetic probe for dissociation of the hexamer with release of the bound metal ion leading to its sequestration as an octahedral EDTA complex. In this assay, originally developed at the Lilly Research Laboratories (36), the time-course of d-d attenuation is essentially mono-exponential. Lifetimes are given in Table 5. In the context of [OrnB29]insulin substitution of PheB24 by Met led to a 10-fold reduction in lifetime, whereas substitution by Cha led to 3-fold reduction (Fig. 8D). The ChaB24 substitution had a similar effect in the context of KP-insulin (Fig. 8F). Accelerated disassembly of cobalt insulin hexamers is of potential pharmacologic relevance as the more rapid disassembly of KP-insulin (4-fold relative to WT) has been shown to correlate with its more rapid absorption from a subcutaneous depot in patients (23).
TABLE 5.
Cobalt ion sequestration studies
| Protein | Co2+ dissociation half-lifea |
|---|---|
| min | |
| WT insulin | 12.9 ± 0.7 |
| KP-insulin | 2.9 ± 0.1 |
| [ChaB24-KP]insulin | 0.8 ± 0.1 |
| [OrnB29]insulin | 10.8 ± 0.1 |
| [ChaB24,OrnB29]insulin | 2.7 ± 0.1 |
| [MetB24,OrnB29]insulin | 1.2 ± 0.1 |
a Kinetic features of Co2+ release from protein hexamers were monitored by following the attenuation in absorbance at 574 nm after addition of excess EDTA; data were fitted to a mono-exponential decay.
Insulin Fibrillation
Whereas native state stabilities (summarized for the present analogs in Fig. 8A) are associated with relative rates of chemical degradation of clinical insulin formulations (58), the predominant mechanism of degradation above room temperature is through fibrillation (amyloid formation) (59). Accordingly, the susceptibility of the ChaB24 analogs to fibrillation was probed on gentle agitation at 37 °C in relation to WT insulin, KP-insulin, [OrnB29]insulin, and [MetB24,OrnB29]insulin (Table 3); the lag times were defined on the basis of a 2-fold enhancement of thioflavin T fluorescence (34). Because of the stochastic nature of a nucleation-propagation reaction (60), results are shown for each individual vial (Fig. 8B).
Whereas substitution of LysB29 by Orn was observed to prolong the lag time, this effect was mitigated by the destabilizing MetB24 substitution. By contrast, lag times of [OrnB29]insulin and [ChaB24,OrnB29]insulin were indistinguishable (p = 0.89). Furthermore, in the context of KP-insulin, ChaB24 appeared to provide a protective effect. Statistical analysis of the two groups (n = 6 and n = 5, respectively) indicated that the observed difference in mean lag times (3.3 and 13 days, respectively) was significant (p < 0.05). The unperturbed lag time of [ChaB24,OrnB29]insulin (relative to [OrnB29]insulin) and the prolonged lag time of [ChaB24-KP]insulin (relative to KP-insulin) were surprising given their reduced thermodynamic stabilities (above). Indeed, an [AlaB24]insulin analog, which is less stable than wild type insulin, exhibits accelerated fibrillation (30). Relative prolongation of the lag time of [ChaB24-KP]insulin seems remarkable as protective substitutions in insulin are uncommon in surveys of diverse insulin analogs (61).
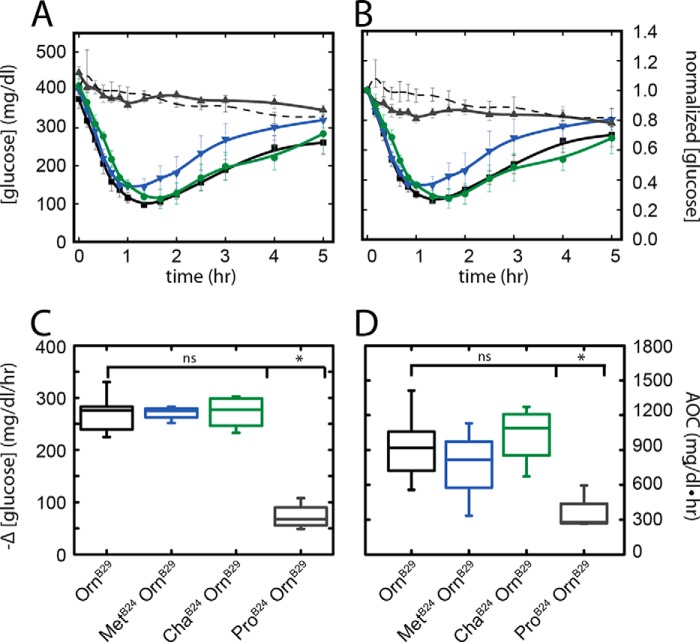
Biological Activity in Diabetic Rats
The potency and duration of action of [ChaB24,OrnB29]insulin were tested in a rat model of DM (33) in relation to control studies of the parent [OrnB29]insulin, class I analog [MetB24,OrnB29]insulin, and class IV analog [ProB24,OrnB29]insulin. The analogs were administered by i.v. bolus injection at a submaximal dose (10 μg insulin analog per 300 g rat). The activity of [OrnB29]insulin was similar to that described previously in rat studies of KP-insulin (62), whereas the negligible activity of the ProB24 analog was similar to that seen on injection of a buffer alone (the gradual linear fall in mean blood glucose concentration is likely to reflect progressive fasting). Because of variation in the initial blood glucose level between rats, data are shown both in relation to the actual blood glucose concentrations at time t = 0 (Fig. 9A) and after normalization with respect to their initial values (defined as 1.0; Fig. 9B). The pattern of fall and recovery of the blood glucose concentrations observed on injection of [ChaB24,OrnB29]insulin and [OrnB29]insulin (black and green lines in Fig. 9A) were indistinguishable. A trend toward a more rapid recovery phase was observed on injection of [MetB24,OrnB29]insulin (blue line), but the large errors precluded definitive assessment.
FIGURE 9.
Rat IV potency assay. A, time-dependent decrease and recovery of the blood glucose concentration (vertical axis) on i.v. bolus injection of insulin analogs as follows: [OrnB29]insulin (black, n = 8); [ChaB24,OrnB29]insulin (green, n = 4); [MetB24,OrnB29]insulin (blue, n = 4); [ProB24,OrnB29]insulin (gray, n = 4); or diluent (black dashed thin line, n = 3). Measurements were obtained at indicated times (horizontal axis) with vertical bars representing standard errors. B, data in A normalized to initial blood glucose concentrations; vertical scale provides fraction of initial value. C, initial rates of decrease in blood glucose concentration (defined over the 1st h). Box plots represent upper and lower quartiles; the central horizontal bar delineates the median, and the vertical bars represent minimum and maximum values. Although any differences among [OrnB29]insulin (black), [ChaB24,OrnB29]insulin (olive), and [MetB24,OrnB29]insulin (blue) were not significant (ns), each of these analogs was significantly more active than [ProB24,OrnB29]insulin (gray box and asterisk; p < 0.05). D, potencies of insulin analogs as measured by the integrated areas above the curves in A relative to a horizontal line at the starting blood glucose concentration. Of the analogs, only [ProB24,OrnB29]insulin exhibited a significant loss of potency (p < 0.05).
Initial rates of fall of the blood glucose concentration (calculated between 0 and 60 min) were similar among the three active analogs; any differences were not statistically significant (Fig. 9C). Although the MetB24 analog appeared to be less long-lived when mean values were plotted, quantitative analysis of total AOC demonstrated that any differences did not reach statistical significance (Fig. 9D).11
DISCUSSION
In a co-crystal structure of insulin bound to a domain-minimized model of the IR (6), insulin segment B24–B27 was displaced from the α-helical core of the hormone to insert between the tandem αCT-L1 element of the IR α subunit (4). The side chain of PheB24 inserts within a conserved pocket lined by L1 residues (Asn15, Leu37, and Phe39), αCT residue Phe714, and insulin residues LeuB15, TyrB16 and CysB19. The B24-binding cavity (with surface area 444 Å2 as defined by a 1.4-Å spherical probe) is predominantly nonpolar (∼80%). Whereas in the free insulin monomer one edge of the B24 aromatic ring is exposed to solvent, in the μIR complex this side chain is buried. Of the conserved aromatic residues in the B chain C-terminal segment (PheB24, PheB25, and TyrB26), only PheB24 inserts within a classical binding pocket to anchor the displaced segment; the B25- and B26 side chains are by contrast in contact with shallow depressions in otherwise exposed surfaces (6).
Structure-Activity Relationships
The long standing view that an aromatic side chain is required at B24 (10), recently reinforced by the studies of Brzozowski and co-workers (12), is broadly consistent with possible weakly polar interactions at the receptor interface, including potential π-π (Phe39 and Phe714), aryl-sulfur (cystine B19-A20), and aryl-amino (Asn15) interactions. Because such structural details were not well defined at the resolution of the refined co-crystal structure (3.5 Å), we undertook systematic mutagenesis of the B24 position. All possible amino acid substitutions were introduced with the exceptions of Cys (to avoid a free thiol group), Lys, and Arg; the latter were singly represented by Orn, a basic analog resistant to tryptic cleavage. In this collection of analogs, LysB29 was substituted by Orn.
The highest affinities were conferred by the native PheB24, MetB24, and GlyB24 (the latter being in accordance with past results (10, 49) as discussed separately below). Relative to these analogs, our results (Table 2) exhibited a trend in favor of aliphatic substitutions with substantial affinity requiring a side-chain volume greater than that of Ala (affinity <1% relative to WT insulin). Whereas alternative aromatic side chains (His, Tyr, and Trp) conferred low affinity, nonaromatic side chains at B24 exhibited the relationship Phe (WT) = ∼ Met > Leu > Ile > Val ≫ Ala. The affinity of [LeuB24,OrnB29]insulin was only 2-fold lower than that of WT insulin, whereas the affinity of [IleB24,OrnB29]insulin was reduced by 3-fold. Charged or polar side chains impaired hormone binding. This trend in structure-activity relationships suggests that the B24-related pocket of the IR selects for nonpolar side chains whose detailed size and shape are complementary to the borders of the pocket. The marked discrimination between PheB24 and TyrB24 suggests that at least one portion of this border is rigid in the hormone-receptor complex. Understanding how the para-OH group of TyrB24 impairs binding will require a higher resolution structure of the μIR complex. We imagine that the smaller size and increased polarity of His (relative to Phe) accounts for its essential inactivity (relative affinity <1%; see also Ref. 12), whereas the larger bicyclic structure of Trp may make unfavorable contacts with the cavity walls. [MetB24,OrnB24]Insulin also displayed native or near-native in vivo potency in male Lewis rats (Fig. 9).
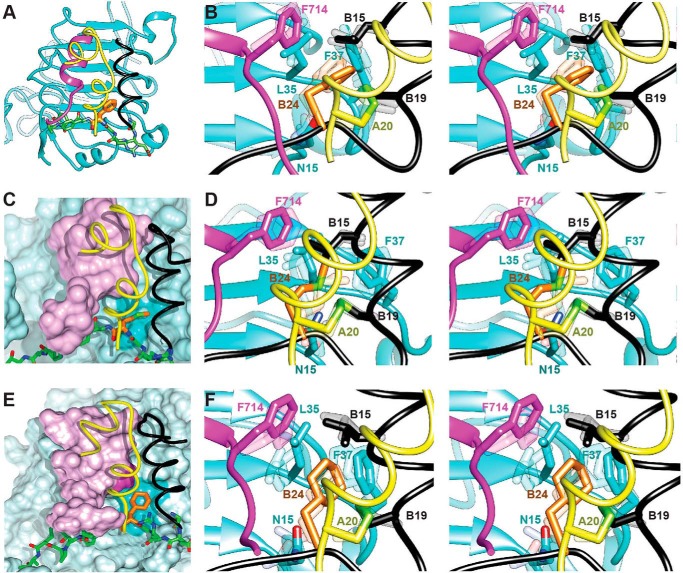
With the exception of GlyB24, the above structure-activity relationships are in general accordance with occupancy of a delimited nonpolar cavity (63). When expressed in terms of differences in free energies of association (RT ln(Ka/K′a)), limited discrimination exists between Phe, Met, Leu, and Ile (<0.5 kcal/mol at room temperature). Why might Met be preferred even to this small extent? The side chains of Met, Ile, and Leu are similar in volume (64) but differ in shape and repertoire of conformations. We speculate that whereas branched side chains (Leu and Ile) are restricted in potential conformations, the linear side chain of Met might be more adaptable (65) in conforming to the dimensions of the cavity. Molecular modeling (MD simulation followed by energy minimization) of MetB24 at the μIR interface supported the plausibility of this model (Fig. 10, C and D); essential features of the WT model (subjected to the same MD-energy minimization protocol) were retained (Fig. 10, A and B). The favorable bound conformation of MetB24 presumably compensates for its greater loss of conformational entropy relative to Leu or Ile.
FIGURE 10.
Models of variant hormone-μIR complexes. A, crystal structure of WT insulin complexed with the μIR. B, stereo view of the B24 binding pocket of the structure presented in A following MD simulation and energy minimization. Depicted in transparent shades are two intermediate and representative sets of side-chain orientations arising during the course of the MD simulation. This panel is included as a control for D and F. C and D, molecular models of [MetB24]insulin. C, representative position of MetB24 (orange carbon atom and light green sulfur atom) in the B24-related pocket. Molecular surfaces of L1 (blue) and αCT (lilac) are shown with respect to the insulin A chain (yellow ribbon) and B chain (residues B8–B19 in black and B20–B27 as green sticks). The darker blue patch on the L1 surface indicates key contact residues. D, corresponding stereo view with color code as in B. The side chains of MetB24 are shown with orange carbon atoms and light green sulfur atoms. E and F, molecular models of [ChaB24]insulin. E, representative position of ChaB24 (orange) in the B24-related pocket. The color code is as in C. F, corresponding stereo view of variant μIR complexes containing [ChaB24]insulin with color code as in E. Shown in D and F (transparent shades) are representative side-chain orientations arising in the respective MD simulations as depicted in B for WT insulin.
We imagine that increasing the size of an l-amino acid side chain at B24 would enhance the occupied percentage of the B24-related cavity and so would reduce the size of any potential cavity penalty. Similarly, increasing the hydrophobicity of the side chain would reduce the polarity penalty. These expectations are in accordance with the activities of ValB24 and ThrB24 analogs. Although these side chains are of similar shape and size, a preference is shown for Val versus Thr (respective affinities 8 and 2.5% relative to WT insulin). This further indicates that the B24-binding pocket displays a strong preference for the binding of nonpolar side chains relative to polar side chains.
Anomalous Activity of [GlyB24]Insulin
Whereas the low affinity of [AlaB24]insulin may be attributed to a cavity penalty (due to the predicted packing defect at the variant interface (63)), the high activity of the GlyB24 analog has long posed a seeming paradox (10, 11, 49, 66). How may an unanchored hormone-receptor interface be tolerated? This enigma is deepened by the enhanced affinity of [d-AlaB24]insulin and other d-analogs at B24 (10, 12, 30). Although destabilization of the B20–B30 segment (GlyB24) (6) or its frank detachment (d-AlaB24) (30) might reduce the cost of induced fit (and so enhance affinity), an alternative model has envisaged a one-residue shift in register between the C-terminal B chain β-strand and the site 1 surface.12 In this model, PheB25 would occupy the B24-binding pocket as an equivalent anchor; TyrB26 would occupy the B25 binding pocket (which would be expected to be well tolerated (53)), and ThrB27 would occupy the exposed B26-related surface observed in the μIR complex.
The register shift model may rationalize why small polar or charged side chains at position B24 (Ser, Thr, Asn, or Asp) confer a higher affinity (∼1%) than does the nonpolar side chain of l-Ala (< 0.5%). Although the differences are small, an intriguing idea is that higher observed affinities of [SerB24]insulin and related analogs reflect the coexistence of two binding modes, one resembling WT insulin in which the B24-related cavity is incompletely filled and the other resembling the register-shifted model envisaged for [GlyB24]insulin. The latter scheme may be favored (relative to AlaB24 analog) due to possible hydrogen bonding by the side chains of Ser, Thr, Asn, or Asp to the main chain as in turns. We imagine that such compensation is incomplete, i.e. the putative GERGX element (where X indicates the B24 substitution) Ser, Thr, Asn, or Asp is less compatible, relative to GlyB24 or d-AlaB24, with the novel pentaloop conformation. These possibilities may in the future be tested through photo-cross-linking studies of insulin analogs containing one or another of the B24 substitutions together with para-azido-Phe at position B25 or B26 (17, 18). A shift in register would be associated with a shift in pattern of domain-specific photo-cross-linking in respective holoreceptor complexes between L1 and αCT. Coexistence of native-like and register-shifted binding modes would in turn be implied by dual photo-cross-linking at each site to both L1 and αCT.
Evolutionary Constraints
To probe why neither Met nor Gly was selected by nature as an alternative to PheB24 among vertebrate insulin sequences (7), we evaluated three other aspects of structure. (i) Thermodynamic stability: in the zinc-free monomer MetB24 and GlyB24 are each associated with reduced free energies of unfolding (ΔΔGu = 2.2 ± 0.1 and 2.2 ± 0.2 kcal·mol−1, respectively). Such destabilization is likely to extend to the variant proinsulins and could be associated with impaired efficiency of disulfide pairing in pancreatic β-cells (48, 67). (ii) Susceptibility to fibrillation: just as a zinc-free GlyB24 variant exhibited a decreased lag time (in the context of KP-insulin (6)) relative to its parent monomer, analogous studies of [MetB24,OrnB29]insulin demonstrated marked foreshortening of the lag time relative to [OrnB29]insulin. (iii) Hexamer assembly: [GlyB24]insulin has previously been shown to impair R6 hexamer assembly (48). In this study, we observed analogous attenuation of the R6-specific Co2+-visible absorption band of the MetB24 analog with more rapid disassembly of the variant R6 hexamer relative to WT insulin or [OrnB29]insulin. Although insulin is stored in the secretory granules of pancreatic β-cells as micro-crystals of zinc insulin hexamers (68, 69), the structure of these hexamers is not known (i.e. whether R6, T3Rf3, T6, or in a novel allosteric state). It is possible that the perturbed spectroscopic and kinetic features of the [MetB24,OrnB29]insulin hexamer might be associated with perturbed packaging and storage in secretory granules.
Our findings highlight the multiple biological constraints that may have governed the evolution and divergence of vertebrate insulins. By analogy to the endoreticular stress previously described in studies of β-cell lines expressing [SerB24]proinsulin (48, 67), we speculate that Met and Gly have been excluded at position B24 due to structural requirements of biosynthesis and storage (70). The general conservation of insulin sequences among vertebrates, which is more stringent than that typically observed in globular domains (2), may reflect the multiple roles played by specific side chains in the course of a complex “conformational life cycle” from nascent folding to receptor binding. Of particular interest, we envisage that a combination of inefficient or unstable disulfide pairing, perturbed hexamer assembly, and heightened susceptibility to fibrillation might be associated with a risk of toxic protein deposition as an amyloidogenic disease (70). Proof of principle is provided by the selective deposition of insulin fibrils in the islet of Langerhans as observed in the South American rodent Octodon degus (71), whose insulin contains a divergent B chain sequence (72).
Insulin-like Growth Factor System
We anticipate that the homologous B domains of IGF-I and IGF-II undergo hinge-like detachment on binding to site 1 of the type 1 IGF receptor (IGF-1R) similar to that observed on binding of insulin to the μIR model (6). Indeed, insulin, IGF-I, and IGF-II are each capable of binding to IR (isoforms A and B) and IGF-1R (1). In structures of free IGFs (73–76), corresponding B22-B26 segments exhibit almost identical dispositions (with respect to the α-helical core) as in insulin; the structure of L1 and sequence of αCT are also conserved between IR and IGF-1R (5). We thus envision that Phe23 in IGF-I and Phe26 in IGF-II (homologs of PheB24 in insulin) function as corresponding anchor residues within homologous nonpolar pockets.
This proposal is supported by a detailed correspondence of structural features between these homologous signaling systems. The B20-B23 β-turn (sequence GERG in insulin and maintained in the μIR complex) is conserved among IGFs both in sequence (GDRG) and structure (75, 76), suggesting that the homologous turn is also maintained as IGFs engage IGF-1R. Furthermore, the aromatic triplet in insulin (PheB24–PheB25–TyrB26) is conserved in IGFs (as Phe-Tyr-Phe) as are the cognate binding residues in IR and IGF-1R (Fig. 11A). An apparent violation is at L1 residue Phe39 in the IR; this nonpolar side chain in the PheB24 binding pocket of the μIR is substituted by Ser in IGF-1R. However, this exception reflects a two-residue insertion in the L1 domain of IGF-1R that repositions the Ser outside of the predicted binding pocket (Fig. 11B).13
FIGURE 11.
Sequence and structural comparison of L1 domains in IR and IGF-1R. A, alignment of representative sequences of the insulin receptor and IGF-1R showing the conservation of L1 residues involved in interaction with PheB24, PheB25, and TyrB26 in the μIR complex with the exception of His32 (Phe or Tyr at corresponding position 28 in the L1 domain of IGF-1R) or Phe39 (Ser35 in IGF-1R) (6). The B24 binding pocket contains residues Asn15, Leu37, Phe39, and Phe714; the B25-binding surface contains residue Arg14, whereas the B26-binding surface contains residues Asp12, Arg14, and Val715. For brevity, residues N- and C-terminal to the aligned sequences have been omitted; the numbering refers to the top sequence. B, overlay of the structure of the insulin-complexed μIR (PDB entry 4OGA) with that the L1 domain of IGF-1R (PDB entry 1IGR). The domains of the μIR are (L1) as follows: cyan (insulin A chain); yellow (insulin B chain); black, and (αCT) magenta; the L1 domain of IGF-1R is shown in brown.
The detached model of the IGF-IGF-1R interface is broadly consistent with the structure of the μIR complex. On replacement of PheB25 by Tyr in IGFs, for example, the para carbon (Cζ) would be directed away from αCT and L1, and so Tyr could readily be accommodated. The Phe in IGFs corresponding to TyrB26 likewise poses no conflict as its contact residues (L1 residues Arg14 and Asp12) are identical in IGF-1R (Fig. 11A). In accordance with these expectations, substitution of PheB25 by Tyr is well tolerated in insulin (77), whereas substitution of TyrB26 by Phe results in only a small reduction in binding to the insulin receptor (78). Despite such similarities, a salient difference distinguishes the IGFs, their respective C domains linking B and A domains. Modeling suggests that, due to their length and flexibility, these linking peptides would not constrain segmental detachment of the respective B domain β-strands from the α-helical cores of these single-chain growth factors. Furthermore, the bound position of the B24–B27 segment (running anti-parallel to the central β-sheet (L1-β2) of L1) suggests that the IGF C domains would be directed toward the CR domain of the receptor (whether IR or IGF-1R). We speculate that the shorter C domain of IGF-II (relative to IGF-I) may explain its higher affinity for the A isoform of IR (1).
Clinical Pharmacology and Nonstandard Mutagenesis
A pioneering application of protein engineering in pharmacology was provided by the design of rapid-acting insulin analogs (79). The essential goal was to accelerate the disassembly of insulin hexamers in the subcutaneous depot, thereby facilitating the absorption of insulin into the bloodstream (80). This was accomplished by interchange of residues ProB28 and LysB29 (in Humalog® (23); KP-insulin in Table 3) and alternatively by substitution of ProB28 by Asp (in Novolog® (79)). These modifications lie at the edge of the dimer-related anti-parallel β-sheet (residues B24–B26 and B26′-B24′) in the hexamer (2). Although the ChaB24 substitution receptor binding led to a 2–3-fold attenuation of receptor-binding affinity in vitro (in both the OrnB29 and KP frameworks; Tables 2 and 4), full potency was retained in a rat model of DM. Molecular modeling suggested that this cyclic aliphatic side chain could fill the B24-related pocket without steric clash (Fig. 10, E and F).
Although our studies of ChaB24 analogs were motivated by the unexpected high affinity of [MetB24,OrnB29]insulin, it is possible that this analog might confer clinical benefits. Intriguingly, substitution of PheB24 by Cha led to accelerated disassembly of [OrnB29]insulin and to a further acceleration in the disassembly of KP-insulin. Although these assays employed cobalt insulin hexamers, we expect that these trends would extend to the zinc insulin hexamers as conventionally employed in insulin formations (81). Such ultra-rapid dissociation of insulin hexamers, if pertinent to the subcutaneous depot, is of current interest in relation to the safety and efficacy of “smart” insulin pumps in which a computer-based algorithm couples the output of a continuous monitor to insulin infusion rate (82). Such closed-loop systems promise to enhance glycemic control in type 1 DM. The utility of the ChaB24 derivative of KP-insulin in continuous pump-based infusion may be further enhanced by its augmented resistance to fibrillation relative to KP-insulin or WT insulin.
We speculate that the enhanced rate of disassembly of ChaB24 cobalt hexamers is related to the flexibility of the aliphatic ring as visualized in the present high resolution crystal structure. Indeed, the electron density of the ChaB24 side chain indicated the presence of at least two conformational substrates, ascribed to gauche and anti-gauche chair conformations. Their interconversion (with respect to the Cβ–Cγ bond), which seems to occur independently at the three dimer interfaces, may facilitate access to transition states for hexamer dissociation. Although mechanisms of WT hexamer disassembly are not well understood, the nature of such transition states may in principle be probed by MD simulations at elevated temperature and at prolonged time scales (83).
Concluding Remarks
PheB24, invariant among vertebrate insulins and IGFs (7), plays a key role in the three-dimensional structure of the hormone and its self-assembly (2). A recent co-crystal structure of insulin bound to a fragment of the insulin receptor has further demonstrated that the aromatic ring of PheB24 inserts into a nonpolar pocket at the interface between the L1 β-helix of the receptor α subunit, αCT residue Phe714, and the central B chain α-helix (6). Such side-chain insertion appears to anchor the displaced B24–B27 segment in a groove between L1 and αCT. The B24-related pocket in the μIR complex is lined by aromatic (TyrB16, Phe39, and Phe714), aliphatic (LeuB15 and Leu37), and polar (Asn15) residues. Like PheB24 itself, the lining residues are broadly conserved among vertebrate insulins, IGFs, and their respective receptors (5).
This study exploited systematic mutagenesis to define structure-activity relationships at position B24. Surprisingly, we have found that aromaticity (long thought to be central (10, 12)) is not required to achieve high affinity binding in vitro and native potency in a rat model of DM. Although the lining of the B24-related pocket in principle offers an opportunity for weakly polar interactions (which may be provided by aromatic-aromatic and aromatic-carboxyamide contacts (84)), the high affinity and biological activity of analogs containing MetB24 and ChaB24 provide evidence that any such directional interactions are replaceable by an aliphatic anchor. It is possible that the similar affinities of these analogs masks complex and compensating differences in underlying thermodynamic driving forces. The general resemblance of the B24-related pocket in the μIR complex (6) to a “druggable” binding pocket (as seen in a variety of pharmaceutical targets) and its tolerance to modified “ligands” (i.e. the present B24 modifications) suggest that nonstandard insulin analogs or IR agonists may be of therapeutic utility in the future.
Why has PheB24 been conserved within the vertebrate insulin family, an evolutionary history extending for 500 million years? Such invariance is particularly striking given the following: (i) the essentially native activities of GlyB24 and MetB24 analogs; (ii) the divergence generally observed among hydrophobic cores of globular proteins over this time scale (85); and (iii) the corresponding plasticity of core packing in model systems as revealed by random combinatorial mutagenesis (86). In light of the clinical association between SerB24 (16) and a monogenic form of DM (15) linked to endoreticular stress in pancreatic β-cells (67), we speculate that PheB24 confers a uniquely favorable combination of foldability, avoidance of misfolding, native assembly, stability, receptor binding and hormonal signaling. We thus envisage that, at this and other sites invariant among vertebrate insulins and IGFs, exploration of sequence space has become frozen at the narrow intersection of multiple biophysical and biological constraints.
Acknowledgments
We thank Q.-X. Hua, W. Jia, and S. H. Nakagawa for advice regarding experimental procedures and P. Arvan, T. L. Blundell, P. De Meyts, P. G. Katsoyannis, M. Liu, D. F. Steiner, and C. Ward for discussion. The crystallographic results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by the UChicago Argonne, LLC, for the United States Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK04949 and DK079233 from NIDDK (to M. A. W.). This work was also supported by the Australian National Health and Medical Research Council Project Grant 1058233, the Hazel and Pip Appel Fund (to M. C. L.), and computational resources at the Victorian Life Sciences Computation Initiative. M. A. W. has equity in Thermalin Diabetes, LLC (Cleveland, OH), where he serves as Chief Scientific Officer; he has also been a consultant to Merck Research Laboratories and DEKA Research & Development Corp. N. B. P. and J. W. are consultants to Thermalin Diabetes, LLC.
The atomic coordinates and structure factors (code 4P65) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The terminal atoms of LysB29 are seldom ordered in crystal structures of insulin (2) and are not required for biological activity (6, 87). The affinities of KP-insulin and [OrnB29]insulin for the lectin-purified IR are indistinguishable from that of WT insulin (6).
DKP-insulin contains one substitution in the trimer-related surface of insulin (HisB10 → Asp) and two substitutions in or adjoining its dimer-related surface (ProB28 → Lys and LysB29 → Pro) (30, 31).
Crystal structures of WT insulin as R6 hexamers have also been obtained in which the asymmetric unit contains the full hexamer (20, 88) and alternatively in which the asymmetric unit contains an RR′ dimer (88).
Initial rates of fall of the blood glucose concentration were not statistically significant (Fig. 9C). Although [MetB24,OrnB29]insulin appeared to be less potent than [OrnB29]insulin when mean values were plotted (Fig. 9, A and B), quantitative analysis of total AOC demonstrated that any differences were not of statistical significance (Fig. 9D). The p value was 0.43. We cannot exclude that in a larger study (i.e. with a greater number of animals per group) statistically significant differences might be observed. If the small apparent differences in mean AOC in the present data should represent a true difference in potency, a power calculation suggests that a trial of >24 rats per group would be required to demonstrate statistical significance. In standard pharmaceutical formulations, any such differences in intrinsic potency would be compensated by redefinition of the formulation strength in relation to international units (IU) per mg of protein such that equivalent biological activity per ml is obtained.
Although certain NMR features of a GlyB24 insulin analog were interpreted by Kaarsholm and co-workers (53) as evidence for a stable register shift in the free hormone, re-investigation of the solution structures of [GlyB24-DKP]insulin (6) and a related d-AlaB24 analog of enhanced activity (30) lack 1H NMR features (strong inter-residue NOEs and aromatic ring-current shifts) of a register shift (at least not as a stable element of super-secondary structure). Flexibility of the GlyB24-destabilized C-terminal B chain segment in the free hormone may lead to weak inter-residue NOEs arising from transient contacts in an ensemble of partially folded conformations without a stable shift in register (6).
The two-residue insertion at Phe39 in IR with respect to IGF-1R is the major difference between the otherwise conserved structures of their respective L1 domains. The resultant alteration in backbone structure (Fig. 11B) rationalizes why an F39A substitution in IR leads to a marked reduction in its affinity for insulin. By contrast, no reduction in affinity for IGF-1 results from an S35A substitution in IGF-1R (89).
- IR
- insulin receptor
- αCT
- C-terminal segment of the IR α subunit
- μIR
- domain-minimized model of the insulin receptor
- Cha
- cyclohexanylalanine
- CR
- cysteine-rich domain of IR
- DM
- diabetes mellitus
- IGF
- insulin-like growth factor
- IGF-1R
- type 1 IGF receptor
- L1
- first leucine-rich repeat domains of IR
- L1-β2
- central β-sheet of the L1 domain
- Orn
- ornithine
- r.m.s.d.
- root-mean-square deviation
- TOCSY
- total correlation 1H NMR spectroscopy
- PDB
- Protein Data Bank
- AOC
- area over the curve
- MD
- molecular dynamics; Amino acids are designated by standard one- and three-letter codes.
REFERENCES
- 1. De Meyts P., Whittaker J. (2002) Structural biology of insulin and IGF1 receptors: implications for drug design. Nat. Rev. Drug Discov. 1, 769–783 [DOI] [PubMed] [Google Scholar]
- 2. Baker E. N., Blundell T. L., Cutfield J. F., Cutfield S. M., Dodson E. J., Dodson G. G., Hodgkin D. M., Hubbard R. E., Isaacs N. W., Reynolds C. D. (1988) The structure of 2Zn pig insulin crystals at 1.5 Å resolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 319, 369–456 [DOI] [PubMed] [Google Scholar]
- 3. McKern N. M., Lawrence M. C., Streltsov V. A., Lou M. Z., Adams T. E., Lovrecz G. O., Elleman T. C., Richards K. M., Bentley J. D., Pilling P. A., Hoyne P. A., Cartledge K. A., Pham T. M., Lewis J. L., Sankovich S. E., Stoichevska V., Da Silva E., Robinson C. P., Frenkel M. J., Sparrow L. G., Fernley R. T., Epa V. C., Ward C. W. (2006) Structure of the insulin receptor ectodomain reveals a folded-over conformation. Nature 443, 218–221 [DOI] [PubMed] [Google Scholar]
- 4. Smith B. J., Huang K., Kong G., Chan S. J., Nakagawa S., Menting J. G., Hu S.-Q., Whittaker J., Steiner D. F., Katsoyannis P. G., Ward C. W., Weiss M. A., Lawrence M. C. (2010) Structural resolution of a tandem hormone-binding element in the insulin receptor and its implications for design of peptide agonists. Proc. Natl. Acad. Sci. U.S.A. 107, 6771–6776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Menting J. G., Whittaker J., Margetts M. B., Whittaker L. J., Kong G. K., Smith B. J., Watson C. J., Záková L., Kletvíková E., Jirácek J., Chan S. J., Steiner D. F., Dodson G. G., Brzozowski A. M., Weiss M. A., Ward C. W., Lawrence M. C. (2013) How insulin engages its primary binding site on the insulin receptor. Nature 493, 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Menting J. G., Yang Y., Chan S. J., Phillips N. B., Smith B. J., Whittaker J., Wickramasinghe N. P., Whittaker L. J., Pandyarajan V., Wan Z. L., Yadav S. P., Carroll J. M., Strokes N., Roberts C. T., Jr., Ismail-Beigi F., Milewski W., Steiner D. F., Chauhan V. S., Ward C. W., Weiss M. A., Lawrence M. C. (2014) Protective hinge in insulin opens to enable its receptor engagement. Proc. Natl. Acad. Sci. U.S.A. 111, E3395–E3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Conlon J. M. (2001) Evolution of the insulin molecule: insights into structure-activity and phylogenetic relationships. Peptides 22, 1183–1193 [DOI] [PubMed] [Google Scholar]
- 8. Rinderknecht E., Humbel R. E. (1978) The amino acid sequence of human insulin-like growth factor I and its structural homology with proinsulin. J. Biol. Chem. 253, 2769–2776 [PubMed] [Google Scholar]
- 9. Rinderknecht E., Humbel R. E. (1978) Primary structure of human insulin-like growth factor II. FEBS Lett. 89, 283–286 [DOI] [PubMed] [Google Scholar]
- 10. Mirmira R. G., Tager H. S. (1989) Role of the phenylalanine B24 side chain in directing insulin interaction with its receptor: Importance of main chain conformation. J. Biol. Chem. 264, 6349–6354 [PubMed] [Google Scholar]
- 11. Mirmira R. G., Nakagawa S. H., Tager H. S. (1991) Importance of the character and configuration of residues B24, B25, and B26 in insulin-receptor interactions. J. Biol. Chem. 266, 1428–1436 [PubMed] [Google Scholar]
- 12. Záková L., Kletvíková E., Veverka V., Lepsík M., Watson C. J., Turkenburg J. P., Jirácek J., Brzozowski A. M. (2013) Structural integrity of the B24 site in human insulin is important for hormone functionality. J. Biol. Chem. 288, 10230–10240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Adams M. J., Blundell T. L., Dodson E. J., Dodson G. G., Vijayan M., Baker E. N., Hardine M. M., Hodgkin D. C., Rimer B., Sheet S. (1969) Structure of rhombohedral 2 zinc insulin crystals. Nature 224, 491–495 [Google Scholar]
- 14. Weitzel G., Bauer F. U., Eisele K. (1978) Structure and activity of insulin, XVI. Semisyntheses of desheptapeptide-(B24–30)- up to destripeptide-(B28–30)-insulin with lysine or alanine in place of arginine in position B22: influence on the three-step-increase of activity in positions B24–26 (Phe-Phe-Tyr). Hoppe-Seyler's Z. Physiol. Chem. 359, 945–958 [DOI] [PubMed] [Google Scholar]
- 15. Shoelson S., Fickova M., Haneda M., Nahum A., Musso G., Kaiser E. T., Rubenstein A. H., Tager H. (1983) Identification of a mutant human insulin predicted to contain a serine-for-phenylalanine substitution. Proc. Natl. Acad. Sci. U.S.A. 80, 7390–7394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shoelson S. E., Polonsky K. S., Zeidler A., Rubenstein A. H., Tager H. S. (1984) Human insulin B24 (Phe→Ser), secretion and metabolic clearance of the abnormal insulin in man and in a dog model. J. Clin. Invest. 73, 1351–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kurose T., Pashmforoush M., Yoshimasa Y., Carroll R., Schwartz G. P., Burke G. T., Katsoyannis P. G., Steiner D. F. (1994) Cross-linking of a B25 azidophenylalanine insulin derivative to the carboxyl-terminal region of the α subunit of the insulin receptor. Identification of a new insulin-binding domain in the insulin receptor. J. Biol. Chem. 269, 29190–29197 [PubMed] [Google Scholar]
- 18. Xu B., Huang K., Chu Y. C., Hu S. Q., Nakagawa S., Wang S., Wang R. Y., Whittaker J., Katsoyannis P. G., Weiss M. A. (2009) Decoding the cryptic active conformation of a protein by synthetic photoscanning: insulin inserts a detachable arm between receptor domains. J. Biol. Chem. 284, 14597–14608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Inouye K., Watanabe K., Tochino Y., Kobayashi M., Shigeta Y. (1981) Semisynthesis and properties of some insulin analogs. Biopolymers 20, 1845–1858 [DOI] [PubMed] [Google Scholar]
- 20. Derewenda U., Derewenda Z., Dodson E. J., Dodson G. G., Reynolds C. D., Smith G. D., Sparks C., Swenson D. (1989) Phenol stabilizes more helix in a new symmetrical zinc insulin hexamer. Nature 338, 594–596 [DOI] [PubMed] [Google Scholar]
- 21. Lachenmann M. J., Ladbury J. E., Qian X., Huang K., Singh R., Weiss M. A. (2004) Solvation and the hidden thermodynamics of a zinc finger probed by nonstandard repair of a protein crevice. Protein Sci. 13, 3115–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burley S. K., Petsko G. A. (1988) Weakly polar interaction in proteins. Adv. Protein Chem. 39, 125–189 [DOI] [PubMed] [Google Scholar]
- 23. Hirsch I. B. (2005) Insulin analogues. N. Engl. J. Med. 352, 174–183 [DOI] [PubMed] [Google Scholar]
- 24. Brange J., Andersen L., Laursen E. D., Meyn G., Rasmussen E. (1997) Toward understanding insulin fibrillation. J. Pharm. Sci. 86, 517–525 [DOI] [PubMed] [Google Scholar]
- 25. Barany G., Merrifield R. B. (1980) in The Peptides (Gross E., Meienhofer J., eds) pp. 273–284, Academic Press, New York [Google Scholar]
- 26. Kubiak T., Cowburn D. (1986) Enzymatic semisynthesis of porcine despentapeptide (B26–30) insulin using unprotected desoctapeptide (B23–30) insulin as a substrate. Int. J. Pept. Protein Res. 27, 514–521 [DOI] [PubMed] [Google Scholar]
- 27. Kabsch W. (2010) Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr. D. Biol. Crystallogr. 66, 133–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karplus P. A., Diederichs K. (2012) Linking crystallographic model and data quality. Science 336, 1030–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hua Q. X., Xu B., Huang K., Hu S. Q., Nakagawa S., Jia W., Wang S., Whittaker J., Katsoyannis P. G., Weiss M. A. (2009) Enhancing the activity of insulin by stereospecific unfolding. Conformational life cycle of insulin and its evolutionary origins. J. Biol. Chem. 284, 14586–14596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hua Q. X., Hu S. Q., Frank B. H., Jia W., Chu Y. C., Wang S. H., Burke G. T., Katsoyannis P. G., Weiss M. A. (1996) Mapping the functional surface of insulin by design: structure and function of a novel A-chain analogue. J. Mol. Biol. 264, 390–403 [DOI] [PubMed] [Google Scholar]
- 32. Whittaker J., Whittaker L. (2005) Characterization of the functional insulin binding epitopes of the full-length insulin receptor. J. Biol. Chem. 280, 20932–20936 [DOI] [PubMed] [Google Scholar]
- 33. Yang Y., Petkova A., Huang K., Xu B., Hua Q. X., Ye I. J., Chu Y. C., Hu S. Q., Phillips N. B., Whittaker J., Ismail-Beigi F., Mackin R. B., Katsoyannis P. G., Tycko R., Weiss M. A. (2010) An Achilles' heel in an amyloidogenic protein and its repair. Insulin dynamics, misfolding, and therapeutic design. J. Biol. Chem. 285, 10806–10821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang K., Dong J., Phillips N. B., Carey P. R., Weiss M. A. (2005) Proinsulin is refractory to protein fibrillation. Topological protection of a precursor protein from cross-β assembly. J. Biol. Chem. 280, 42345–42355 [DOI] [PubMed] [Google Scholar]
- 35. Roy M., Brader M. L., Lee R. W., Kaarsholm N. C., Hansen J. F., Dunn M. F. (1989) Spectroscopic signatures of the T to R conformational transition in the insulin hexamer. J. Biol. Chem. 264, 19081–19085 [PubMed] [Google Scholar]
- 36. Birnbaum D. T., Kilcomons M. A., DeFelippis M. R., Beals J. M. (1997) Assembly and dissociation of human insulin and LysB28ProB29-insulin hexamers: a comparison study. Pharm. Res. 14, 25–36 [DOI] [PubMed] [Google Scholar]
- 37. Hess B., Kutzner C., Van Der Spoel D., Lindahl E. (2008) GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435–447 [DOI] [PubMed] [Google Scholar]
- 38. MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiórkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 39. Guvench O., Mallajosyula S. S., Raman E. P., Hatcher E., Vanommeslaeghe K., Foster T. J., Jamison F. W., 2nd, Mackerell A. D., Jr. (2011) CHARMM additive all-atom force field for carbohydrate derivatives and its utility in polysaccharide and carbohydrate-protein modeling. J. Chem. Theory Comput. 7, 3162–3180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bussi G., Donadio D., Parrinello M. (2007) Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 [DOI] [PubMed] [Google Scholar]
- 41. Berendsen H. J., Postma J. P., van Gunsteren W. F., DiNola A., Haak J. (1984) Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 [Google Scholar]
- 42. Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G. (1995) A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 [Google Scholar]
- 43. Hess B. (2008) P-LINCS: A parallel linear constraint solver for molecular simulation. J. Chem. Theory Comput. 4, 116–122 [DOI] [PubMed] [Google Scholar]
- 44. Fiser A., Sali A. (2003) Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374, 461–491 [DOI] [PubMed] [Google Scholar]
- 45. Elleman T. C., Frenkel M. J., Hoyne P. A., McKern N. M., Cosgrove L., Hewish D. R., Jachno K. M., Bentley J. D., Sankovich S. E., Ward C. W. (2000) Mutational analysis of the N-linked glycosylation sites of the human insulin receptor. Biochem. J. 347, 771–779 [PMC free article] [PubMed] [Google Scholar]
- 46. Olechnovic K., Margelevicius M., Venclovas C. (2011) Voroprot: an interactive tool for the analysis and visualization of complex geometric features of protein structure. Bioinformatics 27, 723–724 [DOI] [PubMed] [Google Scholar]
- 47. Gore S. P., Burke D. F., Blundell T. L. (2005) PROVAT: a tool for Voronoi tessellation analysis of protein structures and complexes. Bioinformatics 21, 3316–3317 [DOI] [PubMed] [Google Scholar]
- 48. Nakagawa S. H., Tager H. S. (1993) Importance of main-chain flexibility and the insulin fold in insulin-receptor interactions. Biochemistry 32, 7237–7243 [DOI] [PubMed] [Google Scholar]
- 49. Shoelson S. E., Lu Z. X., Parlautan L., Lynch C. S., Weiss M. A. (1992) Mutations at the dimer, hexamer, and receptor-binding, surfaces of insulin independently affect insulin-insulin and insulin-receptor interactions. Biochemistry 31, 1757–1767 [DOI] [PubMed] [Google Scholar]
- 50. Kobayashi M., Ohgaku S., Iwasaki M., Maegawa H., Watanabe N., Takada Y., Shigeta Y., Inouye K. (1984) Changes in receptor binding, biological activity and immunoreactivity of insulin caused by replacing the residues B23-B26 with alanine. Biomed. Res. 5, 267–272 [Google Scholar]
- 51. Tager H., Thomas N., Assoian R., Rubenstein A., Saekow M., Olefsky J., Kaiser E. T. (1980) Semisynthesis and biological activity of porcine [LeuB24]insulin and [LeuB25]insulin. Proc. Natl. Acad. Sci. U.S.A. 77, 3181–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Steiner D. F., Chan S. J., Welsh J. M., Kwok S. C. (1985) Structure and evolution of the insulin gene. Annu. Rev. Genet. 19, 463–484 [DOI] [PubMed] [Google Scholar]
- 53. Ludvigsen S., Olsen H. B., Kaarsholm N. C. (1998) A structural switch in a mutant insulin exposes key residues for receptor binding. J. Mol. Biol. 279, 1–7 [DOI] [PubMed] [Google Scholar]
- 54. Haneda M., Kobayashi M., Maegawa H., Watanabe N., Takata Y., Ishibashi O., Shigeta Y., Inouye K. (1985) Decreased biologic activity and degradation of human [SerB24]-insulin, a second mutant insulin. Diabetes 34, 568–573 [DOI] [PubMed] [Google Scholar]
- 55. Pocker Y., Biswas S. B. (1980) Conformational dynamics of insulin in solution. Circular dichroic studies. Biochemistry 19, 5043–5049 [DOI] [PubMed] [Google Scholar]
- 56. Pandyarajan V., Weiss M. A. (2012) Design of non-standard insulin analogs for the treatment of diabetes mellitus. Curr. Diab. Rep. 12, 697–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bakaysa D. L., Radziuk J., Havel H. A., Brader M. L., Li S., Dodd S. W., Beals J. M., Pekar A. H., Brems D. N. (1996) Physicochemical basis for the rapid time-action of LysB28ProB29-insulin: dissociation of a protein-ligand complex. Protein Sci. 5, 2521–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brange J., Langkjoer L. (1993) Insulin structure and stability. Pharm. Biotechnol. 5, 315–350 [DOI] [PubMed] [Google Scholar]
- 59. Brange J., Langkjaer L. (1997) in Protein Delivery: Physical Systems (Sanders L. M., Hendren R. W., eds) pp. 343–410, Plenum Press, New York [Google Scholar]
- 60. Waugh D. F., Wilhelmson D. F., Commerford S. L., Sackler M. L. (1953) Studies of the nucleation and growth of selected types of insulin fibrils. J. Am. Chem. Soc. 75, 2592–2600 [Google Scholar]
- 61. Nielsen L., Frokjaer S., Brange J., Uversky V. N., Fink A. L. (2001) Probing the mechanism of insulin fibril formation with insulin mutants. Biochemistry 40, 8397–8409 [DOI] [PubMed] [Google Scholar]
- 62. Pandyarajan V., Phillips N. B., Cox G. P., Yang Y., Whittaker J., Ismail-Beigi F., Weiss M. A. (2014) Biophysical optimization of a therapeutic protein by nonstandard mutagenesis: Studies of an iodo-insulin derivative. J. Biol. Chem. 289, 23367–23381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Eriksson A. E., Baase W. A., Zhang X. J., Heinz D. W., Blaber M., Baldwin E. P., Matthews B. W. (1992) Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science 255, 178–183 [DOI] [PubMed] [Google Scholar]
- 64. Janin J. (1979) Surface and inside volumes in globular proteins. Nature 277, 491–492 [DOI] [PubMed] [Google Scholar]
- 65. Gellman S. H. (1991) On the role of methionine residues in the sequence-independent recognition of nonpolar protein surfaces. Biochemistry 30, 6633–6636 [DOI] [PubMed] [Google Scholar]
- 66. Hua Q. X., Shoelson S. E., Kochoyan M., Weiss M. A. (1991) Receptor binding redefined by a structural switch in a mutant human insulin. Nature 354, 238–241 [DOI] [PubMed] [Google Scholar]
- 67. Liu M., Haataja L., Wright J., Wickramasinghe N. P., Hua Q. X., Phillips N. F., Barbetti F., Weiss M. A., Arvan P. (2010) Mutant INS-gene induced diabetes of youth: proinsulin cysteine residues impose dominant-negative inhibition on nonmutant proinsulin transport. PLoS One 5, e13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Greider M. H., Howell S. L., Lacy P. E. (1969) Isolation and properties of secretory granules from rat islets of Langerhans. II. Ultrastructure of the β granule. J. Cell Biol. 41, 162–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Michael J., Carroll R., Swift H. H., Steiner D. F. (1987) Studies on the molecular organization of rat insulin secretory granules. J. Biol. Chem. 262, 16531–16535 [PubMed] [Google Scholar]
- 70. Weiss M. A. (2013) Diabetes mellitus due to the toxic misfolding of proinsulin variants. FEBS Lett. 587, 1942–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hellman U., Wernstedt C., Westermark P., O'Brien T. D., Rathbun W. B., Johnson K. H. (1990) Amino acid sequence from degu islet amyloid-derived insulin shows unique sequence characteristics. Biochem. Biophys. Res. Commun. 169, 571–577 [DOI] [PubMed] [Google Scholar]
- 72. Nishi M., Steiner D. F. (1990) Cloning of complementary DNAs encoding islet amyloid polypeptide, insulin, and glucagon precursors from a New World rodent, the degu, Octodon degus. Mol. Endocrinol. 4, 1192–1198 [DOI] [PubMed] [Google Scholar]
- 73. Sato A., Nishimura S., Ohkubo T., Kyogoku Y., Koyama S., Kobayashi M., Yasuda T., Kobayashi Y. (1993) Three-dimensional structure of human insulin-like growth factor-I (IGF- I) determined by 1H NMR and distance geometry. Int. J. Pept. Protein Res. 41, 433–440 [DOI] [PubMed] [Google Scholar]
- 74. Torres A. M., Forbes B. E., Aplin S. E., Wallace J. C., Francis G. L., Norton R. S. (1995) Solution structure of human insulin-like growth factor II. Relationship to receptor and binding protein interactions. J. Mol. Biol. 248, 385–401 [DOI] [PubMed] [Google Scholar]
- 75. Vajdos F. F., Ultsch M., Schaffer M. L., Deshayes K. D., Liu J., Skelton N. J., de Vos A. M. (2001) Crystal structure of human insulin-like growth factor-1: detergent binding inhibits binding protein interactions. Biochemistry 40, 11022–11029 [DOI] [PubMed] [Google Scholar]
- 76. Brzozowski A. M., Dodson E. J., Dodson G. G., Murshudov G. N., Verma C., Turkenburg J. P., de Bree F. M., Dauter Z. (2002) Structural origins of the functional divergence of human insulin-like growth factor-I and insulin. Biochemistry 41, 9389–9397 [DOI] [PubMed] [Google Scholar]
- 77. Mirmira R. G., Tager H. S. (1991) Disposition of the phenylalanine B25 side chain during insulin-receptor and insulin-insulin interactions. Biochemistry 30, 8222–8229 [DOI] [PubMed] [Google Scholar]
- 78. Gauguin L., Klaproth B., Sajid W., Andersen A. S., McNeil K. A., Forbes B. E., De Meyts P. (2008) Structural basis for the lower affinity of the insulin-like growth factors for the insulin receptor. J. Biol. Chem. 283, 2604–2613 [DOI] [PubMed] [Google Scholar]
- 79. Brange J., Ribel U., Hansen J. F., Dodson G., Hansen M. T., Havelund S., Melberg S. G., Norris F., Norris K., Snel L. (1988) Monomeric insulins obtained by protein engineering and their medical implications. Nature 333, 679–682 [DOI] [PubMed] [Google Scholar]
- 80. DeFelippis M. R., Chance R. E., Frank B. H. (2001) Insulin self-association and the relationship to pharmacokinetics and pharmacodynamics. Crit. Rev. Ther. Drug Carrier Syst. 18, 201–264 [PubMed] [Google Scholar]
- 81. Brange J. (ed) (1987) Galenics of Insulin: The Physico-chemical and Pharmaceutical Aspects of Insulin and Insulin Preparations, Springer, Berlin [Google Scholar]
- 82. Hovorka R. (2006) Continuous glucose monitoring and closed-loop systems. Diabet. Med. 23, 1–12 [DOI] [PubMed] [Google Scholar]
- 83. Shaw D. E., Maragakis P., Lindorff-Larsen K., Piana S., Dror R. O., Eastwood M. P., Bank J. A., Jumper J. M., Salmon J. K., Shan Y., Wriggers W. (2010) Atomic-level characterization of the structural dynamics of proteins. Science 330, 341–346 [DOI] [PubMed] [Google Scholar]
- 84. Burley S. K., Petsko G. A. (1985) Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science 229, 23–28 [DOI] [PubMed] [Google Scholar]
- 85. Chen J., Stites W. E. (2001) Packing is a key selection factor in the evolution of protein hydrophobic cores. Biochemistry 40, 15280–15289 [DOI] [PubMed] [Google Scholar]
- 86. Bowie J. U., Reidhaar-Olson J. F., Lim W. A., Sauer R. T. (1990) Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science 247, 1306–1310 [DOI] [PubMed] [Google Scholar]
- 87. Katsoyannis P. G., Ginos J., Cosmatos A., Schwart G. (1973) Synthesis of destetrapeptide B27–30 human (porcine) insulin. A biologically active insulin analog. J. Am. Chem. Soc. 95, 6427–6434 [DOI] [PubMed] [Google Scholar]
- 88. Smith G. D., Ciszak E., Magrum L. A., Pangborn W. A., Blessing R. H. (2000) R6 hexameric insulin complexed with m-cresol or resorcinol. Acta Crystallogr. D Biol. Crystallogr. 56, 1541–1548 [DOI] [PubMed] [Google Scholar]
- 89. Lou M., Garrett T. P., McKern N. M., Hoyne P. A., Epa V. C., Bentley J. D., Lovrecz G. O., Cosgrove L. J., Frenkel M. J., Ward C. W. (2006) The first three domains of the insulin receptor differ structurally from the insulin-like growth factor 1 receptor in the regions governing ligand specificity. Proc. Natl. Acad. Sci. U.S.A. 103, 12429–12434 [DOI] [PMC free article] [PubMed] [Google Scholar]