Background: USP28 is a deubiquitinating enzyme implicated in the DNA damage response, Myc stabilization, and cancer progression.
Results: USP28 activity is regulated by SUMO conjugation on the N-terminal region.
Conclusion: The N-terminal ubiquitin-binding domains of USP28 are not required for polyubiquitin processing.
Significance: Cross-talk exists between SUMO and ubiquitin in the regulation of USP28 enzymatic activity.
Keywords: Proteolytic Enzyme, SUMO-interacting Motif (SIM), Sumoylation, Ubiquitin, Ubiquitin-dependent Protease
Abstract
USP28 (ubiquitin-specific protease 28) is a deubiquitinating enzyme that has been implicated in the DNA damage response, the regulation of Myc signaling, and cancer progression. The half-life stability of major regulators of critical cellular pathways depends on the activities of specific ubiquitin E3 ligases that target them for proteosomal degradation and deubiquitinating enzymes that promote their stabilization. One function of the post-translational small ubiquitin modifier (SUMO) is the regulation of enzymatic activity of protein targets. In this work, we demonstrate that the SUMO modification of the N-terminal domain of USP28 negatively regulates its deubiquitinating activity, revealing a role for the N-terminal region as a regulatory module in the control of USP28 activity. Despite the presence of ubiquitin-binding domains in the N-terminal domain, its truncation does not impair deubiquitinating activity on diubiquitin or polyubiquitin chain substrates. In contrast to other characterized USP deubiquitinases, our results indicate that USP28 has a chain preference activity for Lys11, Lys48, and Lys63 diubiquitin linkages.
Introduction
Ubiquitin (Ub)4 and ubiquitin-like (Ubl) modifiers regulate many cellular processes (1, 2). This post-translation modification consists of the formation of an isopeptidic bond between the C terminus of the Ub or Ubl molecule and a lysine residue of the target protein. Ubiquitin tagging is the major cellular signal to promote the proteasomal degradation of protein targets, normally by the formation of Lys48-linked polyubiquitin chains. In addition to the protein degradation pathway, Ubls have also been implicated in multiple cellular processes, mainly by regulating protein-protein interactions, protein localization, or enzymatic activity (3, 4). Formation of the covalent isopeptidic bond between Ubls and the protein target occurs through a specific enzymatic cascade for each type of Ubl modification. This cascade involves the activating E1, the conjugating E2, and the ligating E3 enzymes (5). Substrate specificity of Ubl modification is determined primarily by the E3 ligase enzymes, which select the protein target and are comprised of a large and diverse family in mammals. Ub and Ubl modification can be reversed by a large family of proteases that remove Ub or Ubl from substrates and, in some instances, can protect them from degradation by the 26 S proteasome (6).
USP28 is a member of a family of deubiquitinating enzymes (DUBs). DUBs comprise a large class of intracellular proteases that can cleave ubiquitin from substrates. DUBs can be divided into five families: ubiquitin C-terminal hydrolases (UCH), ubiquitin-specific proteases (USPs), ovarian tumor proteases (OTU), MJD (Josephins), and MPN+/JAMM (JAB1/MPN/MOV34 metalloenzymes). All of these families are cysteine proteases except the MPN+/JAMM family, which are metalloproteases. USP28 belongs to the USP family, which is comprised of more than 50 members (7, 8). USP28 is highly homologous to USP25, which has been biochemically characterized. Searches in silico predicted that both USP28 and USP25 contain one ubiquitin-associated domain (UBA) and two ubiquitin interaction motifs (UIM) in the N-terminal region of the proteins.
The crystal structures of a number of DUBs in the USP class have been resolved, including USP7/HAUSP, USP14, USP2, USP21, and USP8 providing the basis for molecular recognition studies of these proteases in the apo form and in complex with ubiquitin (9–14). These structural studies demonstrated that the mechanism for ubiquitin recognition is similar in these proteins that are homologous only within their catalytic site regions, and it was hypothesized that this recognition mechanism is common to all DUBs of the USP/UBP class.
Genomic approaches have identified at least 530 human genes that putatively encode enzymes involved in the conjugation and deconjugation of ubiquitin. Of these, at least 79 are thought to encode functional DUBs, some of which have multiple isoforms (15, 16). Considerable progress has been made in the study of ubiquitin conjugation; however, the study of DUBs is still in its primary stages. Early research has been promising, implicating a number of DUBs, such as USP4 (UNP), USP6 (Tre-2), USP8 (UBPY), and USP28 and UCHL5 (UCH37) in neoplastic disease (17–22).
USP28 was identified through its homology to USP25 (24) and subsequently found as an interaction partner of 53BP1, a key regulator of DNA repair pathway choice (24). The catalytic activity of USP28 was reported to be required for IR-induced apoptosis and the stability of numerous DNA damage response regulators (25). Independently, USP28 was reported to stabilize the Myc proto-oncogene by antagonizing the activity of the SCFFBW7 ubiquitin ligase complex (17, 23). This function of USP28 was required for Myc induced transformation, and it was found to be up-regulated in human colon carcinoma and important to prevent differentiation. It was proposed that the dissociation of USP28 from Fbw7 in response to DNA damage provides a potential mechanism that coordinates Myc stability with the DNA damage response (23).
Subsequent work has confirmed the interaction between USP28 and 53BP1 but found only minor effects on the DNA damage response and no impairment in 53BP1-dependent processes, suggesting that it may not represent an attractive therapeutic target for chemosensitization (18, 26). However, its conditional depletion in a mouse model of colorectal cancer led to a significant increase in tumor latency, suggesting that in particular contexts, the modulation of its activity may influence cancer progression (22).
Although SUMO is not a direct tag for proteosomal degradation, there are several examples of cross-talk between the SUMO and the ubiquitin modification systems (27). For example, in the case of IκBα (inhibitor of transcription factor NF-κB) and proliferating cell nuclear antigen, SUMO or ubiquitin is conjugated on particular lysine residues, thus determining the protein fate in the cell (28, 29). In this context, of particular interest was the discovery of ubiquitin-dependent degradation by specific SUMO-target ubiquitin E3 ligases (STUbL) that can recognize substrates with polySUMO chains (30–33). Another example includes the ubiquitin E2 conjugating enzyme E2-25K, in which SUMO conjugation prevents interaction with the ubiquitin E1 enzyme (34). Finally, the DUB protease USP25 has been shown to be either SUMOylated or monoubiquitinated on Lys99. The ubiquitin modification of Lys99 enhanced USP25 deubiquitinating activity on the model substrate MyBPC1 (myosine-binding protein C), whereas SUMO modification had an inhibitory effect on its activity (35, 36). Interestingly, the ubiquitin-binding domains in the N-terminal region of USP25 seem to play a role in the regulation of its protease activity (35, 36).
In this study, we have investigated the catalytic properties of USP28 against different ubiquitin substrates and examined in particular the role of the N-terminal region in the regulation of protease activity. We have found that the SUMO modification of the N-terminal region inhibits the deubiquitinating activity of USP28, suggesting SUMO modification as a potential regulator of the activity of USP28 and adding another link to the cross-talk between these two post-translational modifications systems. However, in contrast to USP25, the N-terminal region of USP28 does not impair the deubiquitinating activity of the catalytic domain, and the analysis of the SUMO2-USP28 fusion proteins suggests a direct interaction of SUMO with the catalytic domain of USP28. Interestingly, USP28 displays a chain preference for Lys11, Lys48, and Lys63 diubiquitin linkages, in contrast to other nonspecific members of the USP deubiquitinase family. These results reveal unexpected diversity in the regulatory mechanisms and substrate preference of structurally similar USPs.
EXPERIMENTAL PROCEDURES
USP25/28 Constructs
The pENTR-USP25 construct was purchased from Open Biosystems (Human ORFeome Collection), and the pDZ-Flag-USP28 construct was purchased from Addgene (Plasmid 15665). The USP28 open reading frame was cloned into the pENTRC vector by PCR followed by TOPO cloning (pENTR directional TOPO cloning kits; Invitrogen). The USP28 constructs USP28FL, USP281–159, USP281–671, USP281–757, and USP28160–757 were generated by PCR amplification of the indicated residues and subcloned into the pET28-Smt3 vector (primers are shown in Table 1).
TABLE 1.
Primers used in this work
| Primers | Sequences (5′ → 3′) |
|---|---|
| USP25-BamHI-F | GGATCCATGACCGTGGAGCAGAACGT |
| USP28-BamHI-F | GGATCCATGACTGCGGAGCTGCAGCA |
| USP28-NT-Stop-NotI-R | GCGGCCGCTAAACCATCAACTCTCCTCCAGTCA |
| USP25-Stop-NotI-R | GCGGCCGCTTATCTTCCATCAGCAGGAG |
| USP28-Stop-NotI-R | GCGGCCGCTTATTTCACTGTCACAGTTG |
| U28-K64R-forw | CTCACTGATGAGAGAGTTAGGGAGCCCAGTCAAGACACT |
| U28-K64R-rev | AGTGTCTTGACTGGGCTCCCTAACTCTCTCATCAGTGAG |
| U28-K115R-forw | AGTCTACTGGAGTCTCCCAGAATTCAAGCTGATGGAAGA |
| U28-K115R-rev | TCTTCCATCAGCTTGAATTCTGGGAGACTCCAGTAGACT |
| MUT-U28NT-K99R-F | CCTTACTCATGATAACAGAGATGATCTTCAGGCTG |
| MUT-U28NT-K99R-R | CAGCCTGAAGATCATCTCTGTTATCATGAGTAAG G |
| U28CD757 ORI-F | CAAACACAGCCCGTGCCTATGAGAAGAGCGGTGTAG |
| U28CDΔNT-F | ATAGGATCCGATGGTTGGCCAGTTGGGCTG |
| U25FL-R | ATAGCGGCCGTTATCTTCCATCAGCAGGAGTTC |
| U28FL-R | ATAGCGGCCGCTTATTTCACTGTCACAGTTG |
| U25CD739E-F | GCAGCAGGAGACCCATAATATCTAGAGCAGCCATC |
| U25CD739E-R | GATGGCTGCTCTAGATATTATGGGTCTCCTGCTGC |
| U25CD757Y-F | CAAACACAGCCCGTGCCTAAGAGAAGAGCGGTGTAG |
| U25CD757Y-R | CTACACCGCTCTTCTCTTAGGCACGGGCTGTGTTTG |
Mutants of USP281–159 and USP281–757
The following mutations were introduced in the expression vectors: USP281–159 K64R, USP281–159 K115R, USP281–159 K99R, USP281–159 K64R/K115R, USP281–159 K64R/K115R/K99R, USP281–757 K115R, USP281–757 K99R, and USP281–757 K99R/K115R. These mutations were introduced into the USP281–159 and USP281–757 using QuikChange mutagenesis kit (Stratagene). SUMO2 fusion proteins were constructed by PCR and inserted into the USP28160–757 and USP281–757 constructs (primers are shown in Table 1).
SUMO Constructs
Plasmids containing Δ14-SUMO2-precursor (first 14-amino acid deletion of SUMO2 precursor) and Δ14-SUMO2 (first 14-amino acid deletion of the mature SUMO2) were constructed based on the results of Reverter and Lima (37). They are purified by the procedure mentioned above.
General Protein Purification Methods
cDNA encoding for the particular proteins were amplified by PCR and cloned into the vector pET28b-Smt3 to encode a polypeptide fused to a thrombin-cleavable N-terminal hexahistidine tag and followed by another SENP-cleavable Smt3 tag. Expression constructs were used to transform Escherichia coli BL21 (DE3) codon plus cells (Novagen). Bacterial cultures were grown by fermentation at 37 °C to A600 = 0.6, and isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.5 mm. Cultures were incubated for 4–5 h at 30 °C and harvested by centrifugation (6000 × g, 20 min), and the supernatant was discarded. Cell suspensions were equilibrated in 20% sucrose, 20 mm Tris-HCl (pH 8.0), 1 mm β-mercaptoethanol, 350 mm NaCl, 20 mm imidazole, 0.1% Igepal CA-630, and 10 mm MgCl2, and the cells were disrupted by sonication. Cell debris was removed by centrifugation (40,000 × g). Protein was separated from lysate by metal affinity chromatography using nickel-nitrilotriacetic acid-agarose resin (Qiagen); eluted with 20 mm Tris-HCl (pH 8.0), 350 mm NaCl, 300 mm imidazole, and 1 mm β-mercaptoethanol; and dialyzed against buffer containing 20 mm Tris-HCl (pH 8.0), 250 mm NaCl, and 1 mm β-mercaptoethanol with SENP2 at a 1:1000 ratio. After SENP2 cleavage, proteins were separated by gel filtration (Superdex 200; GE Healthcare). Fractions containing the protein of interest were pooled, diluted to 50 mm NaCl, applied to anion exchange resin (Mono Q; GE Healthcare), and eluted with a NaCl gradient from 0 to 50% of a buffer containing 20 mm Tris-HCl (pH 8.0), 1 m NaCl, and 1 mm β-mercaptoethanol in 15 column volumes. Fractions containing the protein of interest were pooled, concentrated, and snap-frozen in liquid nitrogen prior to storage at −80 °C.
USP28 SUMOylation Reactions
The small scale SUMOylation reactions of USP28 constructs were performed in a reaction mixture containing 20 mm Hepes (pH 7.5), 5 mm MgCl2, 0.1% Tween 20, 50 mm NaCl, 1 mm dithiothreitol, 1 mm ATP, 150 nm SAE1/SAE2 (E1)), 100 nm Ubc9 (E2), 10 nm IR1 (E3), 16 mm USPs, and 32 mm SUMO in MilliQ water. Samples are taken at 0, 30, and 60 min. The large scale SUMOylation of USP281–757 (containing the N-terminal and the catalytic domains) is 10 times the size of the small scale SUMOylation reaction. Products were verified by SDS-PAGE, purified by gel filtration (Superdex 75, GE Healthcare), concentrated to 1 mg/ml, and snap-frozen in liquid nitrogen prior to storage at −80 °C.
Mass Spectrometry
Mass spectrometry experiments were performed in the Institute for Research in Biomedicine Barcelona mass spectrometry core facility. Proteins were excised from polyacrylamide gels and subjected to in-gel digests with trypsin, chymotrypsin, or both enzymes. Digested peptides were diluted in 1% formic acid. The nano-LC-MS/MS set up was as follows. Samples were loaded on a 180 μm × 2 cm C18 Symmetry trap column (Waters) at a flow rate of 15 μl/min using a nano-Acquity Ultra Performance LCTM chromatographic system (Waters Corp., Milford, MA). Peptides were separated using a C18 analytical column (BEH130TM C18 75 μm × 25 cm, 1.7 μm; Waters Corp.) with a 90-min run, comprising three consecutive steps with linear gradients from 1 to 35% B in 30 min, from 35 to 50% B in 5 min, and from 50% to 85% B in 3 min, followed by isocratic elution at 85% B in 10 min and stabilization to initial conditions (A = 0.1% formic acid in water, B = 0.1% formic acid in CH3CN). The column outlet was directly connected to an Advion TriVersa NanoMate (Advion) fitted on an LTQ-FT Ultra mass spectrometer (Thermo). The mass spectrometer was operated in a data-dependent acquisition mode. Survey MS scans were acquired in the FT with the resolution (defined at 400 m/z) set to 100,000. Up to six of the most intense ions per scan were fragmented and detected in the linear ion trap. The ion count target value was 1,000,000 for the survey scan and 50,000 for the MS/MS scan. Target ions already selected for MS/MS were dynamically excluded for 30 s. Spray voltage in the NanoMate source was set to 1.70 kV. Capillary voltage and tube lens on the LTQ-FT were tuned to 40 and 120 V. Minimal signal required to trigger MS to MS/MS switch was set to 1000, and activation Q was 0.250. The spectrometer was working in positive polarity mode, and singly charged state precursors were rejected for fragmentation. At least one blank run before each analysis was performed to ensure the absence of cross-contamination from previous samples.
A database search was performed with Proteome Discoverer software v1.3 (Thermo) using the Sequest search engine and a custom database, which included N-terminal sequences of USP28 and USP25. Search parameters included no enzyme restriction, carbamidomethyl in cysteine as static modification and methionine oxidation, and +599.266 Da (QQQTGG) in lysine as dynamic modifications. Peptide mass tolerance was 10 ppm, and the MS/MS tolerance was 0.8 Da. Peptides with XCorr > 1.1 (z = 1), 1.25 (z = 2), and 1.68 (z = 3) were considered as positive identifications.
Deubiquitinating Assays against Different Types of Ubiquitin Substrates
Human polyubiquitin chain (Lys48, Lys63, and 3–7ubs) and diubiquitin (Lys48 and Lys63) were purchased from Boston Biochem. They were dissolved in the buffer containing 250 mm NaCl, 20 nm Tris 8.0, and 1 mm β-mercaptoethanol with a final concentration of 1 mg/ml. The polyubiquitin chains were diluted 10 times to 0.1 mg/ml and mixed with different concentrations of USP28 constructs (0.5, 5, 50, and 500 nm) at 37 °C in a buffer containing 25 mm Tris-HCl (pH 8.0), 150 mm NaCl, 0.1% Tween 20, and 2 mm dithiothreitol. Diubiquitin substrate was prepared with a narrow dilution of the USP28 constructs: 4, 20, 100, and 500 nm.
For the comparative experiment between USP281–757-SUMO2 and USP281–757 in the cleavage of polyubiquitin chains, SENP2 protease at 50 nm was incubated to the reaction mixture for 20 min at 37 °C, to release SUMO from the USP28-SUMO2 adduct. Reactions were stopped after 25 min with SDS loading buffer and analyzed by PAGE. Proteins were detected by staining with SYPRO (Bio-Rad). In the time course experiment with diubiquitin substrates, the concentrations of USP28 and USP28-SUMO2 were fixed at approximately 50 nm. A similar SENP2 incubation was prepared to release SUMO from the USP28-SUMO2 adducts.
For the analysis of the cleavage of the different diubiquitin substrates, eight types of diubiquitin linkages (linear, Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) were purchased from the UBPBio Company. All substrates were dissolved in the buffer containing 250 mm NaCl, 20 nm Tris (pH 8.0), and 1 mm β-mercaptoethanol at a final concentration of 1 mg/ml. A time course experiment was conducted with three USP28 constructs at fixed substrate (5 μm) and USP28 concentrations (120 nm).
RESULTS
Structural and Functional Characterization of USP28
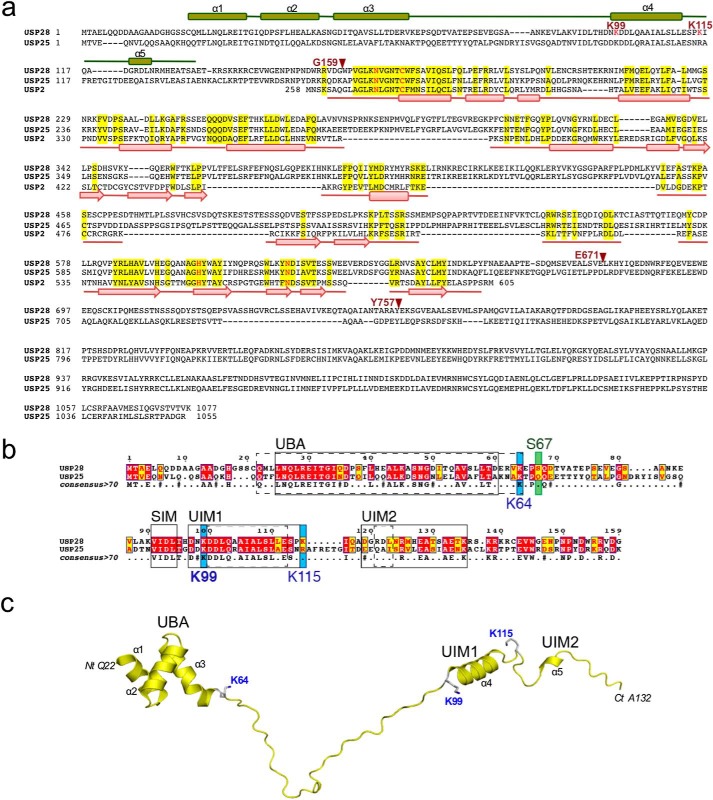
Based on structural alignments with other members of the USP family and on a previously published report on the homologous USP25, the USP28 full-length protein (1077 residues) can be divided in three domains: the N-terminal domain (∼160 residues long), the catalytic “conserved” USP domain (∼350–400 residues long), and the C-terminal extension domain (∼400 residues) (Fig. 1a).
FIGURE 1.
a, sequence alignment of USP2, USP25, and USP28. Secondary structure elements are based on USP2 structure (Protein Data Bank code 2HD5) and on the N-terminal USP28 NMR structure (Protein Data Bank code 2HD5) and shown below and above the sequences, respectively. USP28 active site residues are shown in red. Red arrows indicate the last residue of the USP28 constructs used in this work. b, Sequence alignment of the ubiquitin and SUMO binding motifs in USP28 and USP25. Secondary structure elements are labeled. The UBA, UIM, and SIM motifs predicted in USP25 are indicated by solid lines (34). Domains predicted by the NMR structure of USP28 are shown as dashed lines. Sequences were aligned using T-coffee, and the figure was generated using ESPript. Ser67, a target for DNA damage-induced phosphorylation, is indicated by a green box. Primary SUMOylation sites identified in Table 1 are indicated by blue boxes. c, NMR structure of the N-terminal domain of USP28. Ribbon representation of the deposited NMR structure (Protein Data Bank code 2LVA) of the N-terminal region of USP28 (residues 22–132) (37). Lys64, Lys99, and Lys115 side chains are labeled and shown in stick representation. Structural domains and secondary structure elements are labeled.
The N-terminal region of USP28 includes 159 residues (USP281–159), from Met1 to Gly159, the C-terminal residue corresponding with the beginning of the conserved USP catalytic domain (Fig. 1, a and b). In a recent report on USP25, in silico comparative searches predicted three different ubiquitin binding motifs in the N-terminal region: one UBA and two UIMs (35). The N-terminal regions of USP28 and USP25 are highly homologous, particularly in these ubiquitin-binding motifs (Fig. 1b).
Recently, the NMR structure of the USP28 N-terminal region (Protein Data Bank code 2LVA) was deposited by the Northeast structural genomics consortium (38) (Fig. 1c). The NMR structure confirms the presence of the predicted UBA domain in USP28, from Gln22 to Lys64, forming a characteristic three-helix bundle domain and constituting the only globular domain in the N-terminal region of USP28 (Fig. 1). The other regions of the N-terminal domain are disordered, with the exception of the formation of an α-helix, from Asp100 to Ser113, which would correspond to one of the predicted UIM domains in USP25 (Fig. 1) (35). The second predicted UIM domain, displaying a lower level of homology in USP28, only forms a short 310-helix in the NMR structure of USP28, from Arg121 to Leu123 (Fig. 1). Thus the NMR structure of the N-terminal region USP28 suggests that it is mainly disordered, with only a few secondary structure elements forming the 3-helix bundle UBA domain and an isolated α-helix corresponding to the first predicted UIM domain.
In addition to the presence of these ubiquitin-binding motifs elements, a SUMO interaction motif (SIM) was described in USP25 (36). This domain is highly conserved in USP28, corresponding to the region from Val91 to Leu94 (Fig. 1b). In USP25, this SIM was elegantly described to participate in the SUMO conjugation reaction through a novel conjugation mechanism (36). To examine the relative importance of the domains of USP28 to its enzymatic activity, we produced five different truncation variants of USP28 in E. coli: USP28FL, USP281–159, USP281–671, USP281–757, and USP28160–757 (the subindex indicates the first and last residues of the construct) (Fig. 1a).
SUMO Conjugation Analysis of the N-terminal Region of USP28
The N-terminal domain of USP28, from Met1 to Gly159, can be produced in high levels amounts in E. coli. Gel filtration purification was consistent with the presence of a dimeric protein, although we cannot discard the possibility that the disordered nature of the USP28 N-terminal region could result in an irregular elution profile in size exclusion chromatography. An in vitro SUMO conjugation reaction using purified E1, E2, and IR1 E3 SUMO ligase resulted in efficient attachment of SUMO to USP281–159 (Fig. 2a). MS analysis of the SUMO-modified USP28 revealed Lys99 as the major SUMOylation site in the N-terminal domain, followed to a lesser degree by modification on Lys64, Lys85, Lys115, and Lys135 (Table 2). Remarkably, Lys99 was also found to correspond to the major site for SUMOylation and monoubiquitination of USP25 in previous works (35, 36).
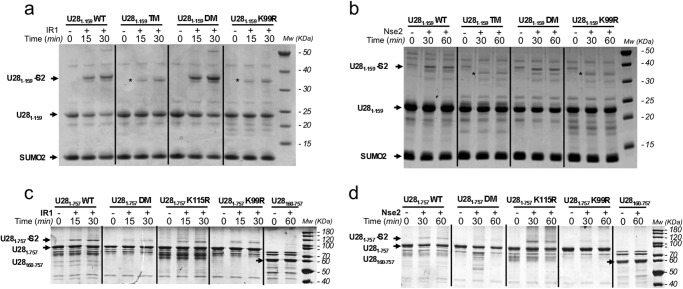
FIGURE 2.
SUMO conjugation reaction with USP constructs. a, time course SUMO conjugation reaction with IR1 E3 ligase using different point mutants of the N-terminal region construct of USP28 (USP281–159). b, similar reaction as in a but using Nse2 as SUMO E3 ligase. c, time course SUMO conjugation reaction with IR1 E3 ligase using different point mutants of the catalytic domain construct of USPS28 (USP281–757). d, similar reaction as in a but using Nse2 as SUMO E3 ligase. Reactions were run at 37 °C and stopped with SDS-PAGE loading buffer at marked times. USP281–159 K64R/K115R, USP281–159 K64R/K115R/K99R, and USP281–757 K99R/K115R are shown. DM, double mutant; TM, triple mutant.
TABLE 2.
Identification of SUMOylated residues in USP28
Shown are the sites identified by mass spectrometry in SUMOylated fragments of the USP281–159 and USP281–673 proteins. The number of peptide spectrum matches for the non-SUMOylated or SUMOylated fragments and the motif compared with the linear consensus motif are shown for each. Underline residues are hydrophobic and italic residues are acidic.
| Residue | Peptide spectrum matches |
Motif (ΨKXE) | |
|---|---|---|---|
| Nonmodified | SUMOylated | ||
| SUMOylation sites identified in USP281–159 | |||
| Lys64 | 12 | 1 | VKEP |
| Lys85 | 198 | 4 | NKEV |
| Lys99 | 177 | 257 | NKDD |
| Lys115 | 3 | 7 | PKIQ |
| Lys135 | 67 | 23 | TKRS |
| Lys138 | 0 | 1 | RKRK |
| Lys140 | 0 | 1 | RKRC |
| SUMOylation sites identified in USP281–673 | |||
| Lys64 | 47 | 1 | VKEP |
| Lys85 | 82 | 1 | NKEV |
| Lys99 | 82 | 23 | NKDD |
| Lys115 | 101 | 4 | PKIQ |
| Lys210 | 0 | 2 | EKRN |
| Lys305 | 75 | 1 | GKPF |
| Lys385 | 62 | 1 | NKLE |
To confirm the MS results, we mutated lysine to arginine on several sites within or near the UBA or UIM1 domains that appeared to be the most structured domains. These included single (K99R), double (K115R and K64R), and triple (K115R, K64R and K99R) point mutant constructs of the USP28 N-terminal domain. The SUMO conjugation reaction was conducted using two different E3 ligases, IR1 and Nse2, and despite the different abilities of SUMO conjugation, both ligases confirmed Lys99 as the major residue for SUMO conjugation in the N-terminal region of USP28 (Fig. 2, a and b). Whereas the USP28 double mutant (K115R and K64R) was conjugated to a comparable degree as the wild-type form, the addition of K99R in the USP28 triple mutant strongly decreases conjugation to levels comparable with the K99R single mutant. Interestingly, a SUMO conjugation reaction with a single (K99R) and triple mutant (K115R/K64R/K99R) yielded a faint band in the gel in a slightly different position than the Lys99 conjugate, probably indicating residual SUMO conjugation on another lysine residue such as Lys85 or Lys135 (Fig. 2, a and b, asterisks).
Based on the deposited NMR structure of the N-terminal domain of USP28 (Protein Data Bank code 2LVA), Lys99 is located at the beginning of the α-helix corresponding to the first predicted UIM domain (Fig. 1c). Although Lys99 is not located in a linear SUMO consensus motif (ΨKXE), two reasons might favor this lysine as the major conjugation site: the spatial conformation of Lys99 (together with other E2 interacting residues) in the UIM α-helix, as observed in examples of SUMO conjugation on lysines located in nonconsensus regions (34), and the SUMO conjugation enhancement produced by the interaction with SUMO E3 ligases, in our case with IR1 and Nse2. In the case of the homologous USP25, the nearby SIM region was proposed to enhance SUMO conjugation by favoring interaction with the charged E2-SUMO thioester. In our in vitro assays, SUMO conjugation of USP28 in the absence of E3 ligases was not observed, although we cannot discard the possibility that a similar conjugation mechanism occurs with USP28, as described for USP25 (36).
SUMO Conjugation Analysis of the Catalytic Domain of USP28
We next examined SUMO conjugation of the USP28 catalytic domain to determine whether the N-terminal region contained the major SUMOylation sites. MS analysis of an excised band of an in vitro SUMO conjugation reaction of a construct including the N-terminal region and the conserved catalytic domain of USP28 (USP281–757) also confirmed Lys99 as the major SUMOylation site (Table 2). The analysis also revealed the presence of other low level SUMOylation sites, including Lys115, Lys385, Lys511, and Lys513. To confirm the MS results, in vitro SUMO conjugation reactions were also conducted with two different SUMO E3 ligases, IR1 and Nse2, and with two point mutants of USP28, K99R and K115R. Again, Lys99 emerges as the major SUMOylation site, although SUMO conjugation can also occur to a lesser degree in Lys115 (Fig. 2, c and d). Double point mutations on K99R and K115R practically eliminate SUMO conjugation of USP28 in the two separate reactions using different E3 SUMO ligases. Interestingly, in our SUMO conjugation assays, the absence of the N-terminal region (USP28160–757) also reduces the formation of SUMO conjugates, indicating that the N-terminal region of USP28, and in particular Lys99, is the major site for SUMO conjugation in our in vitro assays (Fig. 2, c and d, far right lanes).
Functional Characterization of the USP28 Truncation Domains
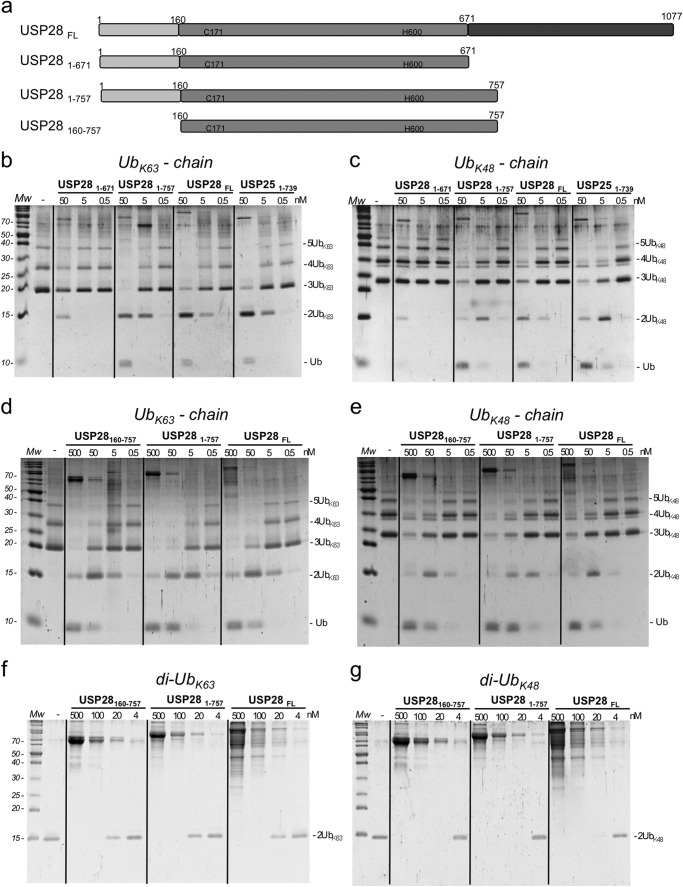
To understand the role of the N-terminal domain in the regulation of USP28 activity, we established deubiquitination assays using synthetic ubiquitin chain substrates with two different ubiquitin linkages (Lys48 and Lys63). We examined the activities of several USP28 constructs including the full-length protein (USP28FL), the N-terminal domain plus the catalytic domain (USP281–671), the N-terminal domain plus a longer catalytic domain (USP281–757), and only the catalytic domain (USP28160–757) (Fig. 3a). Based on a comparative sequence analysis with other USP family members, we first designed a construct for the catalytic domain ending at position Glu671 (USP281–671); however, despite good purification yields in bacteria, this construct had very low activity in our deubiquitinating assays (Fig. 3b). As has been described for USP25 (35), an extension of the C-terminal catalytic domain to Tyr757 yielded a recombinant protein with deubiquitinating activities comparable with the full-length USP28 and USP25 (Fig. 3, b and c). Thus in both USP25 and USP28, the conserved catalytic domain is longer in comparison with other USP family members.
FIGURE 3.
Deubiquitinating activity of USP28 on Lys48 and Lys63 ubiquitin substrates. a, cartoon representation of the different constructs of USP28 used in the analysis. b, end point activities on Lys63-linked ubiquitin chains of three different dilutions of indicated USP28 constructs after a 30-min reaction. c, end point activities on Lys48-linked ubiquitin chains of three different dilutions of USP28 constructs after a 30-min reaction. d, end point activities on Lys63-linked ubiquitin chains of four different dilutions of indicated USP28 constructs after a 30-min reaction. e, end point activities on Lys48-linked ubiquitin chains of four different dilutions of indicated USP28 constructs after a 30-min reaction. f, end point activities on Lys63 diubiquitin substrate of four different dilutions of indicated USP28 constructs after a 30-min reaction. g, end point activities on Lys48 diubiquitin substrate of four different dilutions of the indicated USP28 constructs after a 30-min reaction. Mw, molecular mass markers.
In our deubiquitinating in vitro assays with the USP28 constructs, we do not observe substantial differences in the proteolytic activities between Lys48- and Lys63-linked polyubiquitin chains. Interestingly, our in vitro assays indicate that removal of the N-terminal region (USP28160–757) does not impair the deubiquitinating activity for either the Lys48- or Lys63-linked polyubiquitin chains substrates, and we cannot detect significant differences in comparison with the activities displayed by the USP28 constructs containing the N-terminal region, namely USP281–757 and USP28FL (Fig. 3, d and e). Thus our results indicate that despite the presence of ubiquitin-binding domains at the N-terminal region of USP28, the absence of such a region does not affect the deubiquitinating activity of USP28, at least in the activity against Lys48- and Lys63-linked polyubiquitin chains substrates in vitro.
We considered that substrate interaction with the N-terminal domain could lead to potential proteolytic differences that would only be observed when using shorter substrates, such as diubiquitin with a single ubiquitin linkage. However, deubiquitinating analysis using Lys48 or Lys63 diubiquitin substrates yielded results similar to those using polyubiquitin chains (Fig. 3, f and g), also indicating that the N-terminal region of USP28 is not a major determinant of activity in our in vitro deubiquitinating assays. In summary, our results indicate that there is not a particular preference for the most common Lys48 and Lys63 diubiquitin linkages and that the presence of the N-terminal region of USP28, which contains several ubiquitin-binding motifs, does not affect the deubiquitinating activity of the catalytic domain.
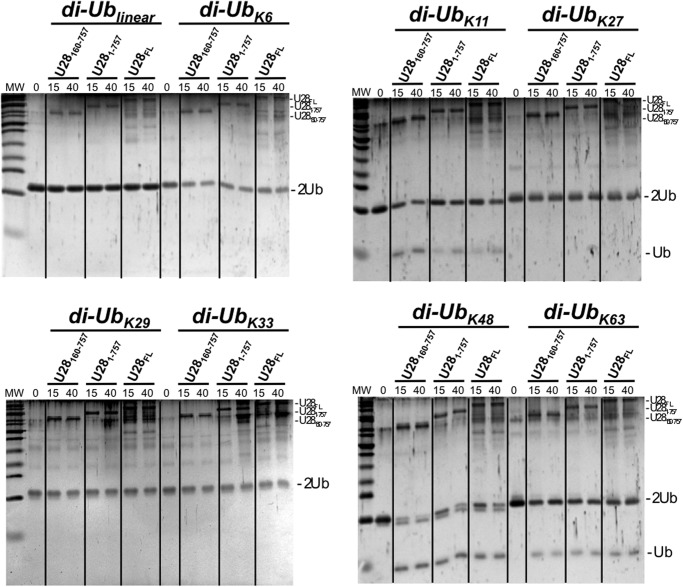
Diubiquitin Chain Specificity
To further investigate the specificity of our USP28 constructs, we have tested their deubiquitinating ability against all eight possible diubiquitin substrates, including the linear, Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63 diubiquitin (Fig. 4). Time course experiments were run at fixed substrate and USP28 concentrations, 5 μm and 120 nm, respectively. Under this experimental condition, only three diubiquitin substrates were cleaved by USP28, namely Lys11, Lys48, and Lys63 diubiquitin linkages, and in all instances with comparable activities between the three constructs tested, USP28160–757, USP281–757, and USP28FL. A previous report with several USP proteins, which includes the homologous USP25, showed only minor differences against all eight diubiquitin substrates (39). In contrast, the deubiquitinating activity of USP28 against diubiquitin linkages resembles the OTU DUB family, where members are specific for one or a small subset of diubiquitin linkages types (40). It is interesting that in addition to the most common Lys48 and Lys63 linkages, Lys11 is preferred by USP28 in our diubiquitin cleavage analysis (Fig. 4). Lys11 polyubiquitin chains have been recently implicated in anaphase-promoting complex-mediated tagging of proteins for an alternative mitotic degradation pathway (41).
FIGURE 4.
Diubiquitin linkage preference for USP28 constructs. USP28160–757, USP281-757, and USP28FL constructs were incubated with diubiquitin substrates of all linkage types (linear, Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63) for the indicated times and resolved in an SDS-PAGE stained with SYPRO-Ruby. All USP28 constructs were used at a fixed concentration (approximately 150 nm). MW, molecular mass markers.
Characterization of SUMO Modifications on USP28 Activity
We next examined the effects of SUMO modifications on USP28 activity using a deubiquitination assay. We first set up a large scale SUMO conjugation reaction using IR1 as a SUMO E3 ligase, followed by an ion exchange chromatography to separate nonmodified USP28 from the SUMO conjugated to USP28 (data not shown). This step is essential to reduce, as much as possible, any contamination of USP28 from the SUMO-USP28 preparation. As shown in the penultimate lane in Fig. 5a, the USP28 band is hardly observed in a Ruby-SYPRO-stained gel; however, we cannot discard some minor level of contamination of free USP28 in the SUMO-conjugated preparation.
FIGURE 5.
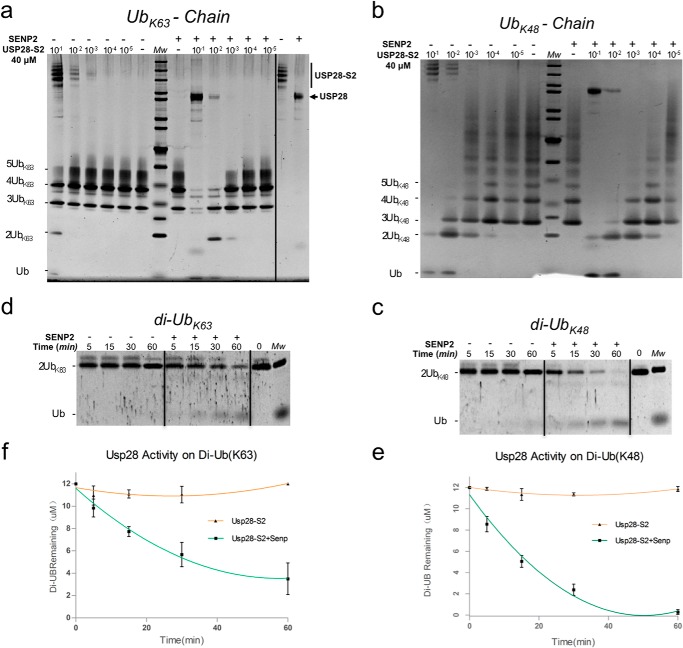
Inhibition of the deubiquitinating activity of USP28 by SUMO conjugation. a, end point deubiquitinating activity comparison on Lys48-linked ubiquitin chains using different dilutions of SUMO conjugated to the USP281–757 construct, before and after SENP2 treatment. b, end point deubiquitinating activity comparison on Lys63-linked ubiquitin chains using different dilutions of SUMO conjugated to the USP281–757 construct, before and after SENP2 treatment. c, time course reaction of deubiquitinating activity on Lys48 diubiquitin substrate using the SUMO-conjugated USP281–757 construct, before and after SENP2 treatment. d, time course reaction of deubiquitinating activity on Lys63 diubiquitin substrate using the SUMO-conjugated USP281–757 construct, before and after SENP2 treatment. e, graphic representation of the deubiquitination reaction shown in c. f, graphic representation of the deubiquitination reaction shown in d. The reactions were run in triplicate.
To assure equal amounts of USP28 protease in our comparative analysis, a serial dilution of USP28-SUMO conjugate was prepared with and without the presence of the SUMO protease SENP2. After proper cleavage of SUMO from USP28, the deubiquitinating activity was checked using the Lys48 and Lys63 polyubiquitin chains and diubiquitin substrates. Comparison of the processing of polyubiquitin chain substrates before and after SENP2 treatment clearly indicates that SUMO-conjugated USP28 has reduced activity (Fig. 5, a and b). Only at high protease concentrations can the SUMO-modified USP28 efficiently cleave the Lys48 or Lys63 polyubiquitin chains. This residual activity at high concentration could be a consequence of contamination of nonmodified USP28 in the SUMO-USP28 preparation or differences in the total SUMOylation levels of multiple lysines. We again did not observe significant differences in the proteolytic activity between the two polyubiquitin chains utilized, Lys48 or Lys63, indicating that in both cases, SUMO modification of the N-terminal region inhibits USP28 activity in our in vitro deubiquitinating assays.
These results with polyubiquitin chains can also be recapitulated using Lys48- and Lys63-linked diubiquitin substrates. To have a more quantitative assay, we performed a time course analysis for up to 60 min using a fixed concentration of USP28-SUMO, before and after treatment with SENP2 protease (Fig. 5, c and d). Similar to the results with polyubiquitin chains, we observed a diminished activity when USP28 was SUMO-conjugated, compared with the activity after treatment with SENP2. Interestingly we noted that after a digestion of 60 min, the Lys48 diubiquitin substrate is completely digested, whereas the proteolysis of the Lys63 linkage occurs at a slower rate (Fig. 5, e and f). Although differences in our in vitro assays are subtle, they might indicate a stronger interaction between USP28 and the Lys48 linkage, at least for the diubiquitin substrates.
In USP25, it was described that a covalent and a noncovalent interaction of SUMO with the N-terminal region of the protease resulted in an inhibition of the deubiquitinating activity of USP25 (36). To determine whether noncovalent SUMO interactions could inhibit USP28 activity, we conducted a competitive in vitro deubiquitinating activity assay in the presence of SUMO2 (Fig. 6, a and b). For this assay, increasing amounts of SUMO2 were added to a fixed concentration of the two truncated constructs of USP28: the N-terminal domain plus catalytic domain (USP281–757), and only the catalytic domain (USP28160–757). The processing of the Lys48- and Lys63-linked polyubiquitin chain substrates do not vary significantly after the addition of increasing amounts of SUMO2 (Fig. 6, a and b). We do not observe any inhibition of the USP28 deubiquitinating activity in the presence of the N-terminal domain, even when SUMO2 concentrations are several orders of magnitude higher. These results indicate that the covalent linkage formed between SUMO and the N-terminal region of USP28 (primarily through Lys99) is necessary for the inhibition of USP28 catalytic activity and that in our in vitro assays a noncovalent interaction of SUMO with the N-terminal region of USP28 does not affect the activity of the protease.
FIGURE 6.
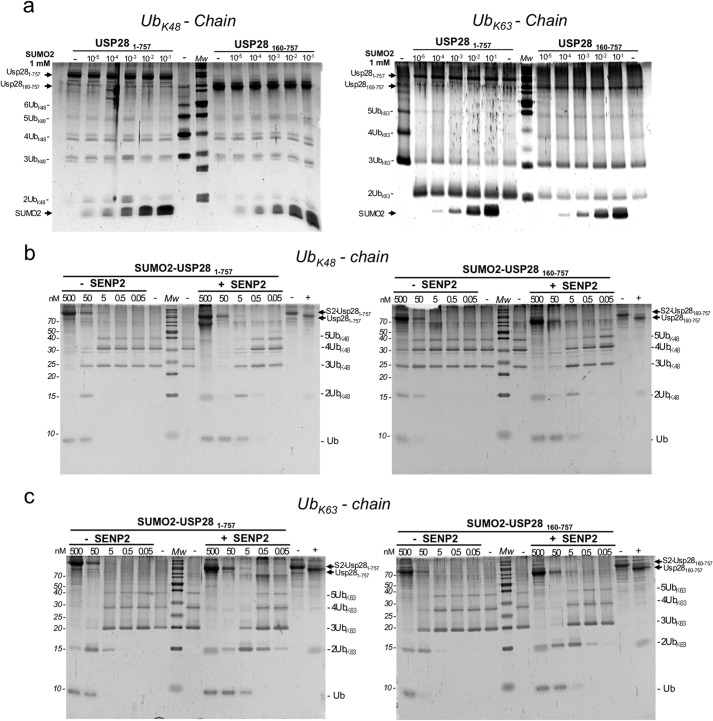
Noncovalent competitive analysis of SUMO2 and covalent SUMO2 fusion proteins analysis on the deubiquitinating activity of USP28. a, end point analysis of the deubiquitinating activity of the USP281–757 and USP28160–757 constructs on Lys48-linked (left panel) and K-63-linked (right panel) ubiquitin chains in the presence of increasing concentrations of SUMO2. b, end point deubiquitinating activity comparison on Lys48-linked ubiquitin chains using different dilutions of SUMO2-USP281–757 (left panel) and SUMO2-USP28160–757 (right panel) fusion constructs, before and after SENP2 treatment. Cleavage of the fusion protein is shown in the last two lanes of the gel. c, end point deubiquitinating activity comparison on Lys63-linked ubiquitin chains using different dilutions of SUMO2-USP281–757 (left panel) and SUMO2-USP28160–757 (right panel) fusion constructs, before and after SENP2 treatment. Cleavage of the fusion protein is shown in the last two lanes of the gel.
We next examined the relevance of the SUMO conjugation site in the inhibition of the USP28 proteolytic activity by using linear SUMO fusion constructs. We have conducted assays with Lys48 and Lys63 polyubiquitin chains using two different SUMO fusion constructs, SUMO2-USP281–757 and SUMO2-USP28160–757, in the presence and absence of SENP2 (Fig. 6, c and d). The results indicate that SUMO2 restrains the activity of USP28 in a similar fashion to the results shown in Fig. 5, in which SUMO was forming an isopeptidic bond to Lys99. We observed a similar inhibition in both SUMO fusion constructs, even in the absence of the N-terminal region, suggesting an inhibitory mechanism in which SUMO directly interacts with the catalytic domain of USP28, restraining its activity, and the N-terminal region of USP28 acts as a platform to promote SUMO conjugation.
DISCUSSION
In contrast to the other DUBs families, such as the OTU DUBs (40), most of the members of the USP family have been reported to display a promiscuous deubiquitinating activity preference in an assay against the eight possible diubiquitin linkages (39). An exception to this is the tumor suppressor CYLD, which possesses specific deubiquitinating activity for Lys63-linked ubiquitin chains synthesized in response to cytokine-mediated activation of TRAF2 and TRAF3 ubiquitin E3 ligases, thus antagonizing NF-κB signaling (13). Our in vitro assays indicate that USP28 does not display strict chain-type specificity for either Lys48- or Lys63-linked polyubiquitin chains (Fig. 3). In our experiments using all possible diubiquitin linkages, we observe comparable deubiquitinating activity against Lys48-, Lys63-, and Lys11-linked diubiquitin substrates with all of our truncated USP28 constructs (Fig. 4). Interestingly, these are the three main types of diubiquitin linkages reported to have a clear connection to cellular functions (42). In particular, Lys11-linked polyubiquitin chain has been recently described as an alternative degradation signal used to facilitate cell cycle progression (41). It will be of interest to define the structural determinants required for the cleavage of Lys11-linked chains by USP28, because they likely depend on specific interactions with the catalytic domain, in contrast to what has been described for CYLD and some OTU DUB members (13, 40).
Despite the substrate preference of USP28 in the diubiquitin array (Fig. 4), our assays using truncated USP28 constructs indicate that the role of the putative ubiquitin-binding domains in the N-terminal region is not immediately clear. The NMR structure indicates that the USP28 N-terminal region is mostly disordered, and only the three-helix bundle of the UBA domain appears to be a compact globular domain. In our assays, the removal of the N-terminal region, which includes the UBA, UIM, and SIM domains, does not impair the deubiquitinating activity of USP28. Thus the interaction of ubiquitin chains with USP28 during catalysis is not strictly controlled by the interaction with the ubiquitin binding domains in the N-terminal region. This could also explain the lack of discrimination between the Lys48- and Lys63-linked polyubiquitin chains in our deubiquitinating assays (Fig. 3).
Although loss of the N-terminal region did not affect deubiquitinating activity in our assays, our results indicate that the SUMO modification of the N-terminal region strongly compromises the activity of USP28. This inhibition of deubiquitinating activity would not depend on the type and length of the ubiquitin chain, because similar results are observed for both Lys48- and Lys63-linked substrates. SUMO conjugation has been reported to result in different outcomes, including the modification of the enzymatic activity of the target substrate that is modified. Another example of SUMO modification regulating enzymatic activity is the DNA mismatch repair protein; SUMO conjugation of thymine DNA glycosylase reduces its affinity for DNA and promotes its release from the abasic site (43). Remarkably, the linear SUMO-USP28 fusion proteins, with either the presence or absence of the N-terminal region, can also inhibit the USP28 activity in a similar fashion as the SUMO conjugation through Lys99, suggesting a direct interaction of SUMO with the catalytic domain of USP28. These results propose a role for the N-terminal region of USP28 as a platform for SUMO conjugation, with Lys99 being the major conjugation site. Structural studies of SUMO-conjugated USP28 would shed some light on this proposed mechanism of the negative regulation of USP28 activity by SUMO.
Our experiments indicate that the inhibition of USP28 catalytic activity only occurs when SUMO is covalently linked to the N-terminal region. Previous MS analysis also identified SUMO2 in pulldown experiments with USP28 (44), although whether this is covalent or not is not apparent in these experiments. In our assays we have not observed any noncovalent SUMO inhibition of USP28 activity, even at high concentrations of SUMO and in the presence of the putative SIM that is identical to that described for USP25. We have also not detected interactions between SUMO1 or SUMO2 and the N-terminal region of USP28 using size exclusion chromatography (data not shown), potentially because of weak affinity between the proteins. In USP25, this noncovalent interaction with the SIM was proposed to promote SUMO conjugation to the N-terminal region in the absence of any SUMO E3 ligase, by facilitating interaction and transfer from the E2-SUMO-thioester-conjugating enzyme (36). This SUMO conjugation mechanism in the absence of E3 ligase by means of the SIM domain has also been proposed for other proteins, including BLM (Bloom syndrome mutated RecQ helicase) (45). The presence of a SIM region in USP28 identical to that of USP25 suggested that a similar SUMO conjugation mechanism would be expected. However, we do not observe any SUMOylation of USP28 in the absence of an E3 ligase activity and have not seen an effect of high noncovalent SUMO levels on activity in our assays. Because the SIM is present in a region that appears disordered in the NMR structure, it is possible that this difference is due to sequence divergence between USP25 and USP28 in this region or is regulated by additional post-translational modifications. In USP28, serine 67, which is directly C-terminal to the UBA domain, is phosphorylated following DNA damage, and this residue, as well as the sequence between it and the SIM domain, is not conserved in USP25. We speculate that this phosphorylation event could affect the structure in a way that could influence SUMO or ubiquitin binding.
In summary, the SUMOylation of the N-terminal region of USP28 impairs its deubiquitinating activity, similarly to what has been reported for USP25. However, we find that in contrast to USP25, the UIM domains of USP28 are not critical for its activity on ubiquitin chains, and we do not find any evidence for non-E3-mediated SUMOylation of USP28 in vitro. The results suggest a regulatory mechanism in which SUMO2 can directly interact with the catalytic domain of USP28, as observed by the inhibition of the linear SUMO-USP28 fusion constructs. These results highlight regulatory differences between structurally similar USPs and add additional detail to the extensive cross-talk between SUMOylation and ubiquitination mechanisms. In addition, they suggest that active SUMOylation of USP28, and perhaps many other USPs, may influence the half-life of their substrates. USP28 has been implicated in the regulation of c-Myc stability, as well as the DNA damage response through its interactions with 53BP1 and its identification as a substrate of the damage induced kinases ATM and ATR (23, 25, 46). USP28 has been identified as polySUMOylated by SUMO2 following heat shock, suggesting that the regulatory mechanism described here in vitro may play a role in controlling USP28 activity in response to cellular stresses in vivo (47, 48). Consistent with this, extensive overlap between ATM/ATR kinase and SUMO2 substrates was revealed by functional annotation of a proteome wide analysis of SUMO2 substrates (47). Further work will be required to elucidate the precise roles of USP28 and the mechanism and significance of its enzymatic regulation by phosphorylation and SUMOylation.
Acknowledgments
We thank Marta Vilaseca and Marina Gay in the Institute for Research in Biomedicine Mass Spectrometry core facility for technical advice.
This work was supported by Grants BFU2012-37116 (to D. R.) and BFU2012-39521 (to T. H. S.) from the Ministerio de Economía y Competitividad of Spain.
- Ub
- ubiquitin
- Ubl
- ubiquitin-like
- SUMO
- small ubiquitin modifier
- DUB
- deubiquitinating enzyme
- UBA
- ubiquitin-associated domain
- UIM
- ubiquitin interaction motif
- SIM
- SUMO interaction motif.
REFERENCES
- 1. Kerscher O., Felberbaum R., Hochstrasser M. (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 [DOI] [PubMed] [Google Scholar]
- 2. Ciechanover A., Schwartz A. L. (1998) The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc. Natl. Acad. Sci. U.S.A. 95, 2727–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 4. Hochstrasser M. (2009) Origin and function of ubiquitin-like proteins. Nature 458, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 6. Gareau J. R., Lima C. D. (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and deconjugation. Nat. Rev. Mol. Cell Biol. 11, 861–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Komander D., Clague M. J., Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 8. Reyes-Turcu F. E., Ventii K. H., Wilkinson K. D. (2009) Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78, 363–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu M., Li P., Li M., Li W., Yao T., Wu J. W., Gu W., Cohen R. E., Shi Y. (2002) Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell 111, 1041–1054 [DOI] [PubMed] [Google Scholar]
- 10. Hu M., Li P., Song L., Jeffrey P. D., Chenova T. A., Wilkinson K. D., Cohen R. E., Shi Y. (2005) Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J. 24, 3747–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Renatus M., Parrado S. G., D'Arcy A., Eidhoff U., Gerhartz B., Hassiepen U., Pierrat B., Riedl R., Vinzenz D., Worpenberg S., Kroemer M. (2006) Structural basis of ubiquitin recognition by the deubiquitinating protease USP2. Structure 14, 1293–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Avvakumov G. V., Walker J. R., Xue S., Finerty P. J., Jr., Mackenzie F., Newman E. M., Dhe-Paganon S. (2006) Amino-terminal dimerization, NRDP1-rhodanese interaction, and inhibited catalytic domain conformation of the ubiquitin-specific protease 8 (USP8). J. Biol. Chem. 281, 38061–38070 [DOI] [PubMed] [Google Scholar]
- 13. Komander D., Lord C. J., Scheel H., Swift S., Hofmann K., Ashworth A., Barford D. (2008) The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Mol. Cell 29, 451–464 [DOI] [PubMed] [Google Scholar]
- 14. Ye Y., Akutsu M., Reyes-Turcu F., Enchev R. I., Wilkinson K. D., Komander D. (2011) Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep. 12, 350–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 16. Wong B. R., Parlati F., Qu K., Demo S., Pray T., Huang J., Payan D. G., Bennett M. K. (2003) Drug discovery in the ubiquitin regulatory pathway. Drug Discov Today 8, 746–754 [DOI] [PubMed] [Google Scholar]
- 17. Popov N., Herold S., Llamazares M., Schülein C., Eilers M. (2007) Fbw7 and Usp28 regulate myc protein stability in response to DNA damage. Cell Cycle 6, 2327–2331 [DOI] [PubMed] [Google Scholar]
- 18. Jacq X., Kemp M., Martin N. M., Jackson S. P. (2013) Deubiquitylating enzymes and DNA damage response pathways. Cell Biochem. Biophys. 67, 25–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu Y., Wang Y., Yang X. H., Kang T., Zhao Y., Wang C., Evers B. M., Zhou B. P. (2013) The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep. 5, 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shah S. P., Morin R. D., Khattra J., Prentice L., Pugh T., Burleigh A., Delaney A., Gelmon K., Guliany R., Senz J., Steidl C., Holt R. A., Jones S., Sun M., Leung G., Moore R., Severson T., Taylor G. A., Teschendorff A. E., Tse K., Turashvili G., Varhol R., Warren R. L., Watson P., Zhao Y., Caldas C., Huntsman D., Hirst M., Marra M. A., Aparicio S. (2009) Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature 461, 809–813 [DOI] [PubMed] [Google Scholar]
- 21. Flügel D., Görlach A., Kietzmann T. (2012) GSK-3β regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1α. Blood 119, 1292–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Diefenbacher M. E., Popov N., Blake S. M., Schülein-Völk C., Nye E., Spencer-Dene B., Jaenicke L. A., Eilers M., Behrens A. (2014) The deubiquitinase USP28 controls intestinal homeostasis and promotes colorectal cancer. J. Clin. Invest. 124, 3407–3418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Popov N., Wanzel M., Madiredjo M., Zhang D., Beijersbergen R., Bernards R., Moll R., Elledge S. J., Eilers M. (2007) The ubiquitin-specific protease USP28 is required for MYC stability. Nat. Cell Biol. 9, 765–774 [DOI] [PubMed] [Google Scholar]
- 24. Valero R., Bayés M., Sánchez-Font M. F., González-Angulo O., Gonzàlez-Duarte R., Marfany G. (2001) Characterization of alternatively spliced products and tissue-specific isoforms of USP28 and USP25. Genome Biol. 2, RESEARCH0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang D., Zaugg K., Mak T. W., Elledge S. J. (2006) A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 126, 529–542 [DOI] [PubMed] [Google Scholar]
- 26. Knobel P. A., Belotserkovskaya R., Galanty Y., Schmidt C. K., Jackson S. P., Stracker T. H. (2014) USP28 is recruited to sites of DNA damage by the tandem BRCT domains of 53BP1 but plays a minor role in double-strand break metabolism. Mol. Cell. Biol. 34, 2062–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Praefcke G. J., Hofmann K., Dohmen R. J. (2012) SUMO playing tag with ubiquitin. Trends Biochem. Sci. 37, 23–31 [DOI] [PubMed] [Google Scholar]
- 28. Desterro J. M., Rodriguez M. S., Hay R. T. (1998) SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell. 2, 233–239 [DOI] [PubMed] [Google Scholar]
- 29. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 30. Prudden J., Pebernard S., Raffa G., Slavin D. A., Perry J. J., Tainer J. A., McGowan C. H., Boddy M. N. (2007) SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 26, 4089–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sun H., Leverson J. D., Hunter T. (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xie Y., Kerscher O., Kroetz M. B., McConchie H. F., Sung P., Hochstrasser M. (2007) The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 282, 34176–34184 [DOI] [PubMed] [Google Scholar]
- 33. Uzunova K., Göttsche K., Miteva M., Weisshaar S. R., Glanemann C., Schnellhardt M., Niessen M., Scheel H., Hofmann K., Johnson E. S., Praefcke G. J., Dohmen R. J. (2007) Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 282, 34167–34175 [DOI] [PubMed] [Google Scholar]
- 34. Pichler A., Knipscheer P., Oberhofer E., van Dijk W. J., Körner R., Olsen J. V., Jentsch S., Melchior F., Sixma T. K. (2005) SUMO modification of the ubiquitin-conjugating enzyme E2–25K. Nat. Struct. Mol. Biol. 12, 264–269 [DOI] [PubMed] [Google Scholar]
- 35. Denuc A., Bosch-Comas A., Gonzàlez-Duarte R., Marfany G. (2009) The UBA-UIM domains of the USP25 regulate the enzyme ubiquitination state and modulate substrate recognition. PLoS One 4, e5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meulmeester E., Kunze M., Hsiao H. H., Urlaub H., Melchior F. (2008) Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell 30, 610–619 [DOI] [PubMed] [Google Scholar]
- 37. Reverter D., Lima C. D. (2004) A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure 12, 1519–1531 [DOI] [PubMed] [Google Scholar]
- 38. Lemak A., Yee A., Houliston S., Garcia M., Dhe-Paganon S., Montelione G. T., Arrowsmith C.; Northeast Structural Consortium (NESG); Structural Genomics Consortium (SGC) (2012) NMR solution structure of the N-terminal domain of human USP28. Northeast Structural Genomics Consortium, in press [Google Scholar]
- 39. Faesen A. C., Luna-Vargas M. P., Geurink P. P., Clerici M., Merkx R., van Dijk W. J., Hameed D. S., El Oualid F., Ovaa H., Sixma T. K. (2011) The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 18, 1550–1561 [DOI] [PubMed] [Google Scholar]
- 40. Mevissen T. E., Hospenthal M. K., Geurink P. P., Elliott P. R., Akutsu M., Arnaudo N., Ekkebus R., Kulathu Y., Wauer T., El Oualid F., Freund S. M., Ovaa H., Komander D. (2013) OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154, 169–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wickliffe K. E., Williamson A., Meyer H. J., Kelly A., Rape M. (2011) K11-linked ubiquitin chains as novel regulators of cell division. Trends Cell Biol. 21, 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Komander D., Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 43. Baba D., Maita N., Jee J. G., Uchimura Y., Saitoh H., Sugasawa K., Hanaoka F., Tochio H., Hiroaki H., Shirakawa M. (2005) Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature 435, 979–982 [DOI] [PubMed] [Google Scholar]
- 44. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu J., Zhu S., Guzzo C. M., Ellis N. A., Sung K. S., Choi C. Y., Matunis M. (2008) SUMO binding determines substrate recognition and paralog-selective SUMO modification. J. Biol. Chem. 283, 29405–29415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 47. Tammsalu T., Matic I., Jaffray E. G., Ibrahim A. F., Tatham M. H., Hay R. T. (2014) Proteome-wide identification of SUMO2 modification sites. Sci. Signal 7, rs2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bruderer R., Tatham M. H., Plechanovova A., Matic I., Garg A. K., Hay R. T. (2011) Purification and identification of endogenous polySUMO conjugates. EMBO Rep. 12, 142–148 [DOI] [PMC free article] [PubMed] [Google Scholar]