Background: The thermophilic bacterium Caldanaerobius polysaccharolyticus can utilize mannan polysaccharides found in plant hemicellulose.
Results: The mannan degradation gene cluster contains a solute-binding protein with an unexpected tolerance for linear and branched manno-oligosaccharides.
Conclusion: Structural studies reveal a binding site optimized for linear and branched mannotriose.
Significance: The self-contained mannan-utilizing cluster can be utilized for engineering efforts for the conversion of mannan-containing hemicellulose into cellulosic biofuels.
Keywords: ABC Transporter, Biofuel, Cellulose, Crystal Structure, Plant Cell Wall
Abstract
Hemicelluloses, the polysaccharide component of plant cell walls, represent one of the most abundant biopolymers in nature. The most common hemicellulosic constituents of softwoods, such as conifers and cycads, are mannans consisting of a 1,4-linked β-mannopyranosyl main chain with branch decorations. Efforts toward the utilization of hemicellulose for bioconversion into cellulosic biofuels have resulted in the identification of several families of glycoside hydrolases that can degrade mannan. However, effective biofermentation of manno-oligosaccharides is limited by a lack of appropriate uptake route in ethanologenic organisms. Here, we used transcriptome sequencing to gain insights into mannan degradation by the thermophilic anaerobic bacterium Caldanaerobius polysaccharolyticus. The most highly up-regulated genes during mannan fermentation occur in a cluster containing several genes encoding enzymes for efficient mannan hydrolysis as well as a solute-binding protein (CpMnBP1) that exhibits specificity for short mannose polymers but exhibited the flexibility to accommodate branched polysaccharide decorations. Co-crystal structures of CpMnBP1 in complex with mannobiose (1.4-Å resolution) and mannotriose (2.2-Å resolution) revealed the molecular rationale for chain length and oligosaccharide specificity. Calorimetric analysis of several active site variants confirmed the roles of residues critical to the function of CpMnBP1. This work represents the first biochemical characterization of a mannose-specific solute-binding protein and provides a framework for engineering mannan utilization capabilities for microbial fermentation.
Introduction
Hemicellulose represents the second most abundant biopolymer on the planet and harbors a reservoir of polysaccharides that can be fermented to bioenergy products, such as ethanol (1). In softwoods (gymnosperms), mannans are the major constituents of hemicellulose (2) and are widely distributed throughout the plant tissues (3). Mannan polymers are heterogeneous and consist of a linear backbone of β-1,4-linked mannose sugars (mannans) or a combination of glucose and mannose sugars (glucomannans) that may be substituted with galactosyl groups (i.e. galactomannans or galactoglucomannans) (4). This structural complexity presents a challenge for both the import and subsequent enzymatic degradation of decorated mannans into constituent monosaccharides for fermentation into ethanolic biofuels (5).
Multiple enzymatic activities are required for mannan degradation, including β-mannanases (which cleave the 1,4-linked mannose backbone), β-mannosidases (which hydrolyze 1,4-β-d-mannopyranosides), and β-glucosidases (which cleave 1,4-linked glucopyranoses from the nonreducing ends of oligosaccharides released by β-mannanases) (see Fig. 1) (6–8). In addition, the debranching activities of acetyl esterases and α-galactosidases are required to process acetyl and galactosyl substituents, respectively. Multiple examples of each of these activities have been discovered from microbial sources, and the individual enzymes have been characterized in detail (9). Consequently, enzymatic mixtures of mannan-degrading enzymes have biotechnological applications for the in vitro saccharification of lignocellulosic biomass into fermentable sugars.
FIGURE 1.
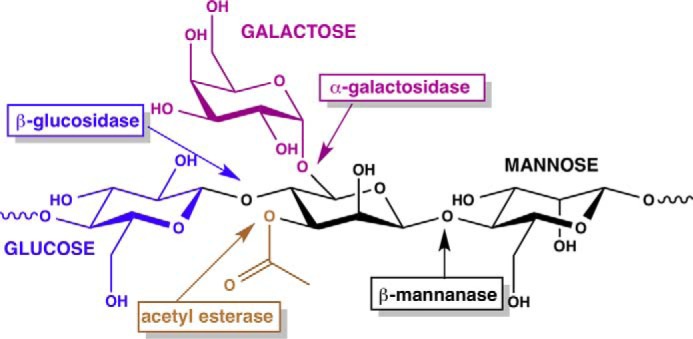
Chemical structure of acetylated galactoglucomannan demonstrating the complexity of the polysaccharide. The various enzymatic activities necessary for depolymerization into monomeric sugars are shown in shaded boxes.
Current efforts in industrial scale lignocellulosic biomass deconstruction are shifting away from the use of expensive enzyme mixtures for in vitro processing and toward engineering of microbial organisms for metabolic fermentation in vivo (10–12). In such efforts, ethanologenic organisms that can already ferment glucose are genetically transformed with the necessary genes for metabolism of other hexoses as well as pentose sugars (13). In some applications, the introduction of membrane-integral ATP-dependent oligosaccharide transporters allows facile import of short chain sugars, which can be further metabolized within the engineered strain (14, 15). We have previously characterized a xylan utilization gene cluster consisting of a xylan-specific transporter and the necessary hydrolytic enzymes for xylose metabolism, which may be used as a cassette to engineer xylan fermentative capacity into ethanologenic organisms (16). However, mannan-specific ATP-dependent transporters have not been characterized in detail, thus limiting the utility of cassette-based engineering approaches for the bioprocessing of mannan-rich softwoods. As mannans are decorated with branched substituents, enzymes for both the import and catabolism of manno-oligosaccharides must accommodate these decorations, a feature not commonly observed with the solute-binding components of similar transporters.
Here, we describe the identification of a mannan utilization cluster in the thermophilic bacterium Caldanaerobius polysaccharolyticus, the gene products of which target manno-oligosaccharide transport and degradation. The cluster encodes six genes, including man5b, a β-mannosidase predicted to be intracellularly located; a transcriptional regulator, which likely senses the presence of manno-oligosaccharides; a putative multicomponent ATP-dependent membrane-integral complex that is predicted to import the end products of extracellular mannan degradation; and a member of GH130, a newly described glycoside hydrolase family (17). Based on sequence comparison and genomic context, a putative solute-binding component (CpMnBP1), which may be a component of the ATP-binding cassette (ABC)4 transporter complex, was identified. We carried out calorimetric analyses on CpMnBP1 that reveal a tolerance for both linear and decorated manno-oligosaccharides. We also present the co-crystal structures of CpMnBP1 in complex with mannobiose (to 1.4-Å resolution) and mannotriose (to 2.2-Å resolution), which represent the first structural views of a manno-oligosaccharide-specific solute-binding protein. The structural data and previous biochemical analysis of gene products encoded in the mannan utilization gene cluster allow for an understanding of the specificity for manno-oligosaccharide binding by CpMnBP1.
EXPERIMENTAL PROCEDURES
Materials
Manno-oligosaccharides (mannobiose (M2), mannotriose (M3), mannotetraose (M4), mannopentaose (M5), mannohexaose (M6), and α-galactosyl mannotriose) and cello-oligosaccharides (cellotriose (G3), cellotetraose (G4), cellopentaose (G5), and cellohexaose (G6)) were purchased from Megazyme (Bray, Ireland). Cellobiose (G2) was purchased from Sigma-Aldrich. Protein molecular mass markers for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were obtained from Bio-Rad. High performance liquid chromatography (HPLC) grade sodium acetate trihydrate was purchased from EMD Chemicals (Darmstadt, Germany). All other reagents were purchased from Fisher Scientific and were of the highest purity available.
Growth of C. polysaccharolyticus and RNA Extraction
C. polysaccharolyticus was cultured anaerobically at 65 °C in butyl rubber-stoppered Balch tubes using a previously reported defined medium (16) in an atmosphere consisting of 5% H2 and 95% CO2. As sole carbohydrate source, glucose, mannose, glucomannan (Megazyme), or galactomannan (Megazyme) was added at 0.2% (w/v). The cells were adapted to the respective media by culturing for 24 h three times in succession. Subsequently, 0.2 ml of preculture was inoculated into 10 ml of fresh medium, and the absorbance at 600 nm (A600 nm) values were monitored every hour using a Spectronic 21D spectrophotometer (Milton Roy). To estimate growth rates, cells were cultured to their maximal A600 nm values. For RNA extraction experiments, the cells were harvested at early logarithmic phase (A600 nm of 0.15 for all carbon sources) by combining the culture with 2 volumes of RNAprotect® bacterial reagent (Qiagen) followed by centrifugation at 8,000 × g for 10 min at room temperature. The resulting cell pellets were stored at −80 °C until RNA extraction. In the subsequent steps, the cell pellets were treated with lysozyme and proteinase K for 30 min at room temperature, and the total RNA was extracted with the RNeasy Mini kit (Qiagen) with the optional on-column DNase treatment step. The RNA was eluted with diethyl pyrocarbonate-treated nuclease-free water and stored at −80 °C until sequencing.
Transcriptional Analysis by RNA Sequencing
For RNA sequencing (RNA-seq) analyses, RNAs isolated from two biological replicates were used for each growth condition. Bacterial ribosomal RNAs were removed from 10 μg of total RNA with the MicrobExpress kit (Life Technologies). The enriched mRNA fraction was converted to RNA-seq libraries using the TruSeq Stranded RNA Sample Prep kit from Illumina, Inc. The barcoded libraries were pooled in equimolar concentration, and the pool was analyzed by quantitative PCR and sequenced on one lane for 101 cycles on a HiSeq2000 using a TruSeq SBS sequencing kit version 3. Fastq files were generated and demultiplexed with the bcl2fastq version 1.8.4 Conversion Software (Illumina, Inc.). RNA-seq library statistics are shown in Table 1.
TABLE 1.
RNA-seq mapping results
avg., average; nt, nucleotides.
| RNA-seq sample ID | Total reads (avg. length) | Reads after trimminga (avg. length) | Uniquely mapped readsb (percentage) | Nonspecifically mapped reads (percentage) | Unmapped reads (percentage) |
|---|---|---|---|---|---|
| Glu_1 | 27,144,132 (100 nt) | 27,142,479 (98.1 nt) | 21,896,756 (80.7%) | 146,706 (0.5%) | 5,099,017 (18.8%) |
| Glu_2 | 23,398,702 (100 nt) | 23,397,193 (98.1 nt) | 19,166,231 (81.9%) | 160,003 (0.7%) | 4,070,959 (17.4%) |
| Man_1 | 18,539,716 (100 nt) | 18,538,613 (97.9 nt) | 14,737,872 (79.5%) | 297,683 (1.6%) | 3,503,058 (18.9%) |
| Man_2 | 19,986,983 (100 nt) | 19,985,820 (98.1 nt) | 16,100,278 (80.6%) | 297,604 (1.5%) | 3,587,938 (18.0%) |
| GluMan_1 | 19,371,626 (100 nt) | 19,370,646 (98.1 nt) | 14,145,889 (73.0%) | 141,438 (0.7%) | 5,083,319 (26.2%) |
| GluMan_2 | 19,134,272 (100 nt) | 19,132,985 (98.3 nt) | 15,344,904 (80.2%) | 131,959 (0.7%) | 3,656,122 (19.1%) |
| GalMan_1 | 21,673,902 (100 nt) | 21,670,903 (98.0 nt) | 17,476,983 (80.6%) | 112,665 (0.5%) | 4,081,255 (18.8%) |
| GalMan_2 | 25,329,774 (100 nt) | 25,328,422 (98.0 nt) | 19,904,504 (78.6%) | 117,245 (0.5%) | 5,306,673 (21.0%) |
a Reads were trimmed using CLC Genomics Workbench version 5.5.1 with a quality score limit of 0.05 and a maximum number of ambiguities of 2.
b Reads were mapped to the C. polysaccharolyticus KMTHCJ genome using CLC Genomics Workbench version 5.5.1 with a minimum length fraction of 0.9, a minimum similarity fraction of 0.8, and a maximum number of hits for a read of 10.
The RNA-seq data were analyzed using CLC Genomics Workbench version 5.5.1 from CLC Bio (Cambridge, MA). The three genomic scaffold sequences of C. polysaccharolyticus DSM 13641 (GenBankTM accession numbers NZ_KE386493.1, NZ_KE386494.1, and NZ_KE386495.1) were used as the reference genome, and RNA-seq reads were mapped onto the reference sequences using the CLC software. Reads were only assembled if the fraction of the read that aligned with the reference genome was greater than 0.9 and if the read matched other regions of the reference genome at fewer than 10 nucleotide positions. The RNA-seq output files were analyzed for statistical significance by using the proportion-based test of Baggerly et al. (18). The RNA-seq data have been deposited in the NCBI server with accession data (SRP Number SRP048659).
Recombinant Protein Production and Purification
Escherichia coli BL21-CodonPlus(DE3)-RIPL cells harboring a recombinant plasmid bearing either the gene for CpMnBP1 (CalpoDRAFT_1212, GenBank accession number AGA35557.1) or variants were cultured in 10 ml of lysogeny broth medium containing 100 μg ml−1 sodium ampicillin and 50 μg ml−1 chloramphenicol for ∼8 h at 37 °C with vigorous shaking. The initial 10-ml cultures were used to inoculate lysogeny broth (1 liter) containing the two antibiotics at the same concentrations stated above in a 2.8-liter Fernbach flask. The cultures were incubated at 37 °C at 200 rpm until the absorbance at 600 nm reached ≈0.3 (after ∼2.5 h). Gene expression in E. coli cultures was then induced with the addition of isopropyl 1-thio-β-d-galactopyranoside (0.1 mm final concentration). The temperature was decreased to 16 °C, and the cultures were allowed to incubate for an additional 16 h with shaking at 200 rpm. Cell pellets were harvested from the cultures by centrifugation at 4,651 × g for 30 min at 4 °C, washed with 35 ml of binding buffer (50 mm Tris, 300 mm NaCl, pH 7.5), and frozen as cell pellets at −80 °C.
Frozen cell pellets were thawed and resuspended in 35-ml of ice-cold binding buffer and passed through an EmulsiFlex C-3 homogenizer (Avestin, Ottawa, Canada) to release the cell contents. The cell lysates were clarified by centrifugation at 12,857 × g for 20 min at 4 °C, and the supernatant containing the soluble fraction was recovered. The supernatant was incubated at 65 °C for 30 min and centrifuged at 20,000 × g for 15 min at 4 °C to pellet the precipitated heat-labile E. coli proteins. The supernatant was loaded onto an immobilized metal ion affinity resin (Talon resin, Novagen) pre-equilibrated with binding buffer (50 mm Tris-HCl, 300 mm NaCl, 20 mm imidazole, pH 7.5), and the bound proteins were washed with 10 column volumes of binding buffer and eluted with an elution buffer (50 mm Tris-HCl, 300 mm NaCl, 250 mm imidazole, pH 7.5). The eluted protein fractions were pooled, concentrated, and exchanged into 50 mm citrate HCl buffer, pH 5.5 for further purification by ion exchange chromatography. The protein was loaded onto a cation exchange column (5-ml HiTrap SP HP column, GE Healthcare) equilibrated with a citrate buffer (binding buffer, 50 mm citrate HCl, pH 5.5), and a gradient elution was applied by using the binding buffer supplemented with 1 m NaCl. Preparation of selenomethionine-labeled CpMnBP1 was carried out by repression of methionine synthesis in defined medium, and the resultant protein was purified as above but with the inclusion of reducing agent in all buffers.
Site-directed Mutagenesis
Site-directed mutagenesis was carried out with the QuikChange Multi Site-directed Mutagenesis kit according to the instructions of the manufacturer (Agilent Technologies). Oligonucleotide sequences are available upon request.
Substrate Binding Assay Using Isothermal Titration Calorimetry
The substrate binding activity of CpMnBP1 with different oligosaccharides was determined at 25 or 65 °C using a VP-ITC microcalorimeter (Microcal Inc., Northampton, MA) as described previously (16). Protein was purified by size exclusion chromatography using a SuperdexTM 200 HiloadTM 16/6 column and subsequently diluted to a concentration of 50 μm in citrate buffer (50 mm sodium citrate, pH 5.5). Mannose and manno-oligosaccharides (M1–M6) and glucose and cello-oligosaccharides (G1–G6) were prepared at a final concentration of 0.5 mm. The ligands were individually injected into the reaction cell containing CpMnBP1 in 24 or 28 successive 10-μl aliquots at 300-s intervals and 20-s duration. Binding data for CpMnBP1 variants utilized mannotetraose as the ligand. The resulting data were analyzed by non-linear regression with a single site model (MicroCal Origin), and thermodynamic parameters were calculated using the Gibbs free energy equation (ΔG = ΔH − TΔS) and the relationship ΔG = −RT lnKa.
Structure Determination of the CpMnBP1-Oligosaccharide Complexes
CpMnBP1 was purified as described above but with a size exclusion chromatographic step (Superdex 200 Hiload 16/60 size exclusion column) using a final buffer composed of 20 mm HEPES, pH 7.5, 100 mm KCl. Crystallization attempts with the full-length protein proved fruitless. Treatment of full-length CpMnBP1 with trypsin yielded a stable fragment from which the polyhistidine tag (and additional cloning artifacts) and the first 14 amino acids of the full-length protein were excised. This stable fragment was further purified by size exclusion chromatography prior to screening attempts for crystallization.
Initial crystallization conditions were obtained by a sparse matrix sampling method using commercial screens. Crystals of both complexes were grown using the hanging vapor drop diffusion method. Briefly, for crystallization of the CpMnBP1-mannotriose complex, 1 μl of protein at 20 mg/ml concentration was incubated with 5 mm oligosaccharide for 2 h on ice, and the complex was mixed with 1 μl of precipitant solution (30% polyethylene glycol 1500, 100 mm KCl, 20 mm HEPES, pH 7.5) and equilibrated over a well containing the precipitant solution at 9 °C. Crystals grew within 2 weeks and were briefly soaked in precipitant solution supplemented with 10% ethylene glycol prior to vitrification by direct immersion in liquid nitrogen. Crystallization of the CpMnBP1-mannobiose complex was carried out using selenomethionine-labeled protein.
Flash cooled crystals of native CpMnBP1 in complex with mannotriose diffracted x-rays to 2.2-Å resolution at an insertion device synchrotron beam line (Life Sciences Collaborative Access Team (LS-CAT) Sector 21 ID-F, Advanced Photon Source, Argonne, IL). These crystals occupy space group P21 with unit cell parameters a = 63.3 Å, b = 101.6 Å, c = 67.8 Å, and β = 116.7° with two molecules in the crystallographic asymmetric unit. A 4-fold redundant data set was collected to 2.2-Å resolution with an overall Rmerge of 9.0% and I/σ(I) of 2 in the highest resolution shell. All data were indexed and scaled using the HKL2000 package (19). Crystals of selenomethionine-CpMnBP1-mannobiose complex diffract to 1.4 Å and occupy space group P21 with unit cell parameters a = 53.7 Å, b = 62.3 Å, c = 60.0 Å, and β = 116.7°. For crystallographic phase determination, a 4-fold redundant data set was collected to 1.5-Å resolution near the selenium absorption edge (wavelength, 0.97856 Å) with an overall Rmerge of 5.5% and I/σ(I) of 6 in the highest resolution shell. A second data set was collected to a slightly higher resolution (1.4 Å) away from the selenium edge (wavelength, 1.07812 Å) and used for crystallographic refinement.
Crystallographic phases were determined by single wavelength anomalous scattering. The heavy atom substructure was determined using HySS, and refinement of heavy atom parameters using Phaser (20) as implemented in the PHENIX software package (21) yielded a figure of merit of 0.469. Solvent flattening and non-crystallographic symmetry averaging yielded experimental maps of exceptional quality, allowing nearly the entire polypeptide chain to be automatically traced using Buccaneer (22). Further manual fitting using Coot (23) was interspersed with rounds of refinement using REFMAC5 (24). Cross-validation using 5% of the data for the calculation of the free R factor was utilized throughout the model building process to monitor building bias (25). When a near complete model of the polypeptide was obtained, further crystallographic refinement was carried out against the 1.4-Å resolution data set. Clear density for the oligosaccharide could be observed in the initial experimental maps, and the ligand was manually built into the model only after the free R factor dropped below 30%.
Phases for the CpMnBP1-mannotriose complex were determined by the molecular replacement method using the atomic coordinates of the protein obtained from the CpMnBP1-mannobiose complex. Manual fitting using Coot was interspersed with rounds of REFMAC5 refinement until the free R factor dropped below 30%. The coordinates for the oligosaccharide were manually built and subjected to further rounds of refinement until convergence. The stereochemistry of all of the models was routinely monitored throughout the course of refinement using PROCHECK (26). Relevant data collection and refinement parameters are provided in Table 2. The refined coordinates have been deposited in the Protein Data Bank (27) with accession codes 4R9F (CpMnBP1-mannobiose complex) and 4R9G (CpMnBP1-mannotriose complex).
TABLE 2.
Data collection, phasing, and refinement statistics
| CpMnBP1-Mannobiose | CpMnBP1-Mannotriose | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| a, b, c (Å); β (°) | 53.7, 62.2, 60.0; 116.5 | 63.3, 101.7, 67.8; 116.7 |
| Resolution (Å)a | 50–1.4 (1.42–1.4) | 50–2.2 (2.28–2.2) |
| Rsym (%)b | 8.4 (19.9) | 8.3 (44.1) |
| I/σ(I) | 12.3 (4.8) | 14.3 (2.0) |
| Completeness (%) | 98.2 (96.1) | 96.2 (80.9) |
| Redundancy | 3.2 (3.0) | 2.7 (1.9) |
| Phasing statistics | ||
| FOMc (after DM) | 0.469 (0.659) | |
| Refinement | ||
| Resolution (Å) | 25.0–1.4 | 25.0–2.2 |
| No. reflections | 64,702 | 35,483 |
| Rwork/Rfreed | 18.3/21.3 | 20.1/26.5 |
| Number of atoms | ||
| Protein | 2,948 | 6,052 |
| Oligosaccharide | 23 | 66 |
| Water | 631 | 382 |
| B-factors | ||
| Protein | 14.2 | 32.5 |
| Oligosaccharide | 9.2 | 24.7 |
| Water | 28.1 | 36.8 |
| r.m.se deviations | ||
| Bond lengths (Å) | 0.004 | 0.01 |
| Bond angles (°) | 0.983 | 1.36 |
a Highest resolution shell is shown in parentheses.
b Rsym = Σ|(Ii − Ii|ΣIi where Ii is the intensity of the ith reflection and Ii is the mean intensity.
c Mean figure of merit (acentric and centric). DM, density modification.
d R factor = Σ(|Fobs| − k|Fcalc|)/Σ|Fobs|, and Rfree is the R value for a test set of reflections consisting of a random 5% of the diffraction data not used in refinement.
e Root mean square.
RESULTS
Identification of a Gene Cluster for Mannan Utilization
Multiple enzymatic activities are necessary to catabolize mannans (Fig. 1), and genes encoding several of these activities can be identified within a putative mannan utilization gene cluster in C. polysaccharolyticus (28). However, the gene coding for the extracellular enzyme (CpMan5A) that may be responsible for the initial hydrolysis of the polysaccharide (29) is located outside of this reported gene cluster elsewhere in the genome. This strategy is in agreement with our previous observation that in a gene cluster for xylan degradation by C. polysaccharolyticus the gene coding for the endoxylanase is located outside of the xylo-oligosaccharide metabolism gene cluster (16).
Growth of C. polysaccharolyticus with Monosaccharides and Polysaccharides
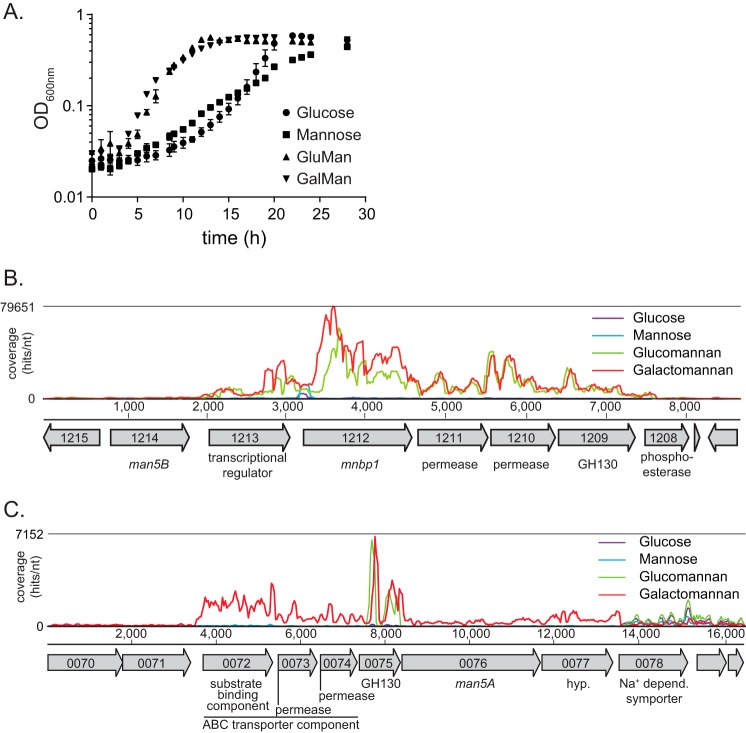
C. polysaccharolyticus was previously shown to ferment glucose and mannose (30) and to degrade mannan polysaccharides (28, 29) into oligosaccharides that can then be hydrolyzed into shorter oligosaccharides and the monomeric sugar mannose (28). The manno-oligosaccharide-cleaving enzyme Man5B characterized in this bacterium is likely located intracellularly because this protein seems to lack an identifiable signal peptide. To gain insight into the machinery utilized for manno-oligosaccharide transport into the cells of C. polysaccharolyticus, we cultured the cells on defined medium containing the polysaccharide glucomannan or galactomannan as carbohydrate source to identify the genes uniquely up-regulated (in the presence of these polysaccharides) compared with the monosaccharides glucose and mannose. Here, we show that the bacterium can utilize both polysaccharides (Fig. 2A). The doubling time during growth with glucomannan (GluMan) or galactomannan (GalMan) was 2.4-fold faster relative to mannose, the major monosaccharide component of mannan (Fig. 2A and Table 3). In contrast, the doubling time with glucose as the carbon source was comparable with that for the polysaccharides. These results indicate that although C. polysaccharolyticus can ferment free mannose the cells possess mechanisms optimized for energy capture from mannan polysaccharides. To understand the molecular mechanism for mannan utilization, we used an unbiased whole genome RNA sequencing approach.
FIGURE 2.
Growth on mannan substrates and their sugar components and transcriptome map of the highly induced gene clusters. A, growth of C. polysaccharolyticus on mannan polysaccharides and monomeric sugars with time. Error bars represent S.D. for multiple measurements. B and C, transcriptional analysis showing RNA-seq coverage of two gene clusters (B, Man5B cluster; C, Man5A cluster) that contained the most highly up-regulated genes during growth of C. polysaccharolyticus with galactomannan, glucomannan, glucose, and mannose. Gene annotations and open reading frame numbers were determined by the Joint Genome Institute. nt, nucleotide; depend., dependent.
TABLE 3.
Bacterial growth characteristics under various substrates
| Growth substrate | A600 maximum | Growth ratea | Doubling timeb |
|---|---|---|---|
| h−1 | h | ||
| Glucose | 0.59 ± 0.009 | 0.37 ± 0.04 | 1.9 ± 0.2 |
| Mannose | 0.43 ± 0.007 | 0.19 ± 0.04 | 3.9 ± 0.8 |
| Glucomannan | 0.57 ± 0.02 | 0.46 ± 0.1 | 1.6 ± 0.5 |
| Galactomannan | 0.57 ± 0.008 | 0.45 ± 0.08 | 1.6 ± 0.3 |
a Growth rates were calculated using the following equation: μ = ln(At2/At1)/(t2 − t1).
b Doubling times (or generation times) were calculated using the following equation: G = ln(2)/μ.
The Genes Most Highly Induced by Mannan Map to the man5B-containing Mannan Utilization Cluster
The mannan utilization genes in C. polysaccharolyticus were identified by applying RNA-seq analysis to transcripts extracted from cells grown on the monosaccharide- and polysaccharide-amended minimal medium (i.e. with either glucose, mannose, glucomannan, or galactomannan). The RNA was extracted from cells at early log phase of growth, and the relative expression values for the genes were quantified by RNA-seq. The expression of four genes was induced greater than 100-fold during growth with glucomannan relative to mannose, and all of these genes mapped to the man5B-containing gene cluster (28). The cluster included the putative ABC transporter components (CpMnBP1, 126-fold; permease1, 286-fold; and permease2, 337-fold) and the putative GH130 manno-oligosaccharide phosphorylase (137-fold) (Table 4). The putative transcriptional regulator was also induced 11.9-fold (Table 4; CalpoDRAFT_1213). Interestingly, the magnitude of induction of man5B was rather low (2.46-fold), although the absolute expression values for this gene agreed well among the biological replicates, and the relative expression comparison yielded a very low p value (7.6 × 10−33). To evaluate the likelihood of these genes being co-transcribed on a single messenger RNA molecule (polycistronic mRNA), we performed read mapping on the entire genome and compared the coverage over the gene cluster for the glucomannan and mannose experiments. A large number of reads mapped to the ABC transporter genes in the case of the glucomannan-grown cells, whereas very few reads mapped to the ABC transporter in the mannose-grown cells (Fig. 2B), consistent with the expression data presented in Table 4. Although there were some fluctuations in coverage over the genes, overall the coverage was continuous over the entire region encompassing the transcriptional regulator (CalpoDRAFT_1213) through the GH130 gene (CalpoDRAFT_1209) (Fig. 2B). These observations suggest that the genes in this region are arranged in an operon, although further experiments are necessary to validate this hypothesis.
TABLE 4.
C. polysaccharolyticus genes expressed greater than 10-fold on galactomannan relative to mannose
JGI, Joint Genome Institute; CAZy, Carbohydrate-Active Enzymes Database.
A similar up-regulation of the man5B encoding gene cluster was observed during growth of C. polysaccharolyticus on galactomannan compared with growth on mannose. In fact, the -fold changes during growth on galactomannan compared with mannose were higher than those for growth on glucomannan compared with mannose (Table 4). Thus, the RNA-seq results identified this cluster of genes as being involved in mannan degradation or as part of a transcriptional program induced by C. polysaccharolyticus during growth on mannans.
CalpoDRAFT_1212 (MnBP1) Encodes a Substrate-binding Component of a Putative ABC Transport Complex That Is Specific for Manno-oligosaccharides
Up-regulation of the gene encoding CpMnBP1 was observed for growth on either glucomannan or galactomannan (Table 4 and Fig. 2B), suggesting that a mechanism exists whereby both the linear and decorated manno-oligosaccharides produced by the extracellular CpMan5A can be transported across the plasma membrane and into the cell for subsequent processing by the manno-oligosaccharide utilization genes, such as CpMan5B and the GH130 enzyme. The putative ABC composed of the three genes encoding a solute-binding protein and tandem repeats of the corresponding permease components is, therefore, the likely candidate for the transport of the manno-oligosaccharides. In agreement with this hypothesis, the components (solute-binding protein and permease) of a putative ABC transporter (CalpoDRAFT_0072, CalpoDRAFT_0073, and CalpoDRAFT_0074), located upstream of the Man5A-encoding gene, together with a GH130 gene (CalpoDRAFT_0075) were among the top 30 up-regulated genes during growth on galactomannan (Table 4 and Fig. 2C). The gene encoding Man5A was only slightly up-regulated during galactomannan utilization, suggesting that another mannanase may be present in the genome or the Man5A-encoding gene is mostly constitutively expressed.
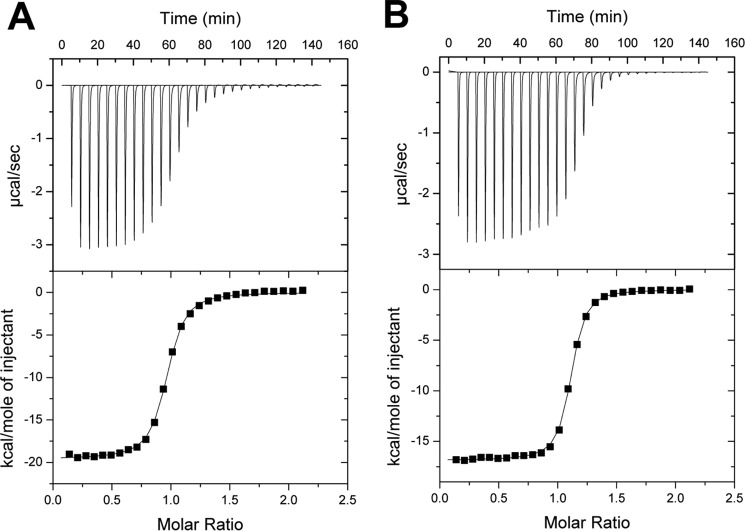
The analysis above shows that the gene encoding CpMnBP1 (CalpoDRAFT_1212; Table 4 and Fig. 2B) was up-regulated during growth on the two mannan polysaccharides used as energy sources in culturing C. polysaccharolyticus. To evaluate whether the ABC transporter is specific for manno-oligosaccharides, the gene encoding CpMnBP1 was cloned and expressed in E. coli, and the purified recombinant protein was tested for manno-oligosaccharide binding activity. The binding activity of purified CpMnBP1 (Fig. 3) was quantified against various substrates using isothermal titration calorimetry (ITC). CpMnBP1 did not show any appreciable binding to mannose (data not shown) but bound tightly to manno-oligosaccharides with degrees of polymerization ranging from 2 to 5 with increased binding correlated to increases in the oligosaccharide length (Table 5). In ITC experiments using glucose and cello-oligosaccharides, no significant binding was detected (data not shown), indicating that CpMnBP1 binds specifically to manno-oligosaccharides. Notably, CpMnBP1 demonstrated tight affinity for α-galactosyl mannotriose, a likely product generated extracellularly by the endo-acting degradation of galactomannan by a mannanase, such as CpMan5A. These results suggest that CpMnBP1 is part of an ABC transporter that is specific for both linear and branched manno-oligosaccharides.
FIGURE 3.
ITC of linear and branched manno-oligosaccharide binding by CpMnBP1. Binding isotherms and representative curves for CpMnBP1 show that the affinity for linear mannotriose (A) is similar to that for the branched ligand α-galactosyl mannotriose (B).
TABLE 5.
Binding parameters for wild-type and mutant CpMnBP1 interaction with oligosaccharides
ND, could not be determined as isotherms did not indicate any significant binding. Data for mutants were collected using mannotetraose as the ligand.
| N | Ka (×106) | ΔG | ΔH | ΤΔS | |
|---|---|---|---|---|---|
| kJ/mol | kJ/mol | kJ/mol | |||
| Mannobiose | 0.94 ± 0.002 | 3.30 ± 0.184 | −37.2 | −81.9 ± 0.34 | −44.66 |
| Mannotriose | 1.08 ± 0.001 | 6.39 ± 0.275 | −38.9 | −70.6 ± 0.17 | −31.69 |
| Mannotetraose | 1.14 ± 0.003 | 8.77 ± 0.975 | −39.8 | −83.7 ± 0.47 | −43.91 |
| Mannotetraose (65 °C) | 0.862 ± 0.008 | 1.86 ± 0.39 | −40.6 | −138.9 ± 2.2 | −98.33 |
| Mannopentaose | 0.861 ± 0.002 | 9.32 ± 0.882 | −39.6 | −79.5 ± 0.41 | −39.92 |
| W272A | ND | ND | ND | ND | ND |
| W25A | 0.751 ± 0.05 | 0.028 ± 0.003 | −25.3 | −78.6 ± 7.58 | −53.27 |
| Q191A | 0.835 ± 0.003 | 6.44 ± 0.685 | −38.8 | −72.3 ± 0.47 | −33.43 |
| N307A | 0.680 ± 0.015 | 0.116 ± 0.009 | −28.9 | −79.5 ± 2.52 | −50.65 |
The Molecular Basis for Manno-oligosaccharide Recognition by CpMnBP1
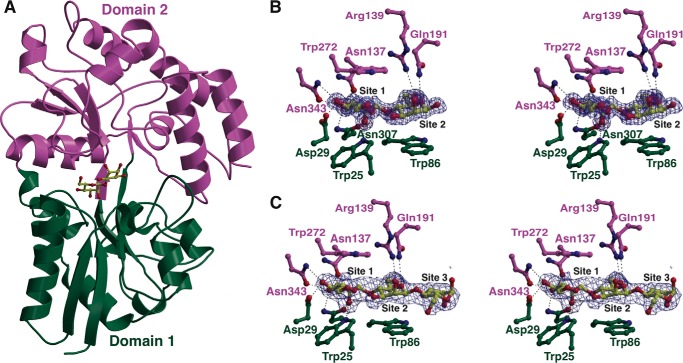
To elucidate the molecular rationale for manno-oligosaccharide specificity, we determined the co-crystal structures of CpMnBP1 in complex with mannobiose (1.4-Å resolution) and with mannotriose (2.2-Å resolution,). The overall structure of CpMnBP1 consists of the two-domain architecture that is common to oligopeptide- and oligosaccharide-specific solute-binding components of ABC transporters (Fig. 4A). The first domain consists of residues Lys-15 through Val-136 and residues Leu-303 through Asn-358, and the second domain includes Asn-137 to Ser-302 and Asp-359 to the C terminus. Both structures lack the first 14 amino acids, which were excised using trypsin to facilitate crystallization. Based on the structure-based classification of solute-binding proteins, CpMnBP1 belongs to the carbohydrate- and peptide-specific B cluster.
FIGURE 4.
Co-crystal structures of CpMnBP1-mannooligosaccharide complexes. A, ribbon diagram of the overall structure of CpMnBP1 showing the two-domain architecture common to all solute-binding proteins colored in green and purple. The mannobiose ligand is shown as a yellow ball-and-stick figure. B, difference Fourier electron density maps (contoured at 3σ over background) calculated with coefficients Fobs − Fcalc at the ligand-binding site for the CpMnBP1-mannobiose complex. The map was calculated from model phases with the coordinates of the ligand removed prior to a round of simulated annealing crystallographic refinement. The coordinates of the final refined model are superimposed. The color scheme for the protein residues follows that in A, and residues that interact with the oligosaccharide are shown as stick figures. C, difference Fourier electron density maps (contoured at 3σ over background) at the ligand-binding site for the CpMnBP1-mannotriose complex.
A DALI search (31) against the Protein Data Bank (27) reveals several maltose- and trehalose-specific solute-binding proteins to be the closest homologs, including the trehalose/maltose-binding protein from Thermococcus litoralis (Protein Data Bank code 1EU8; Z-score = 37.1, r.m.s.d. of 2.9 Å over 370 aligned Cα atoms), the maltodextrin-binding component from Thermoactinomyces vulgaris (Protein Data Bank code 2ZYM; Z-score = 36.9, r.m.s.d. of 2.4 Å over 355 aligned Cα atoms), and the acarbose solute receptor from Streptomyces glaucescens (Protein Data Bank code 3K00; Z-score = 36.5, r.m.s.d. of 2.5 Å over 355 aligned Cα atoms). As with other solute-binding proteins, the homology exists only at the structural level as CpMnBP1 shares less than 23% identity in primary sequences with these other proteins.
In contrast with other solute-binding proteins, CpMnBP1 engages the manno-oligosaccharide ligand through the non-reducing end (Fig. 4, B and C). The orientation of the mannobiose molecule in the 1.4-Å resolution co-crystal structure was unambiguous, and the hydrogen bonding interactions described below are consistent with the non-reducing end of the mannan as the major binding determinant in CpMnBP1 (Fig. 4B). The sugar-binding cleft is capped at one end by two α-helices that converge, and residue Val-341, located at the end of one of the helices, closes off where the terminal, non-reducing mannose is bound. This mannose residue is engaged between two aromatic residues (Trp-25 and Trp-272) that sandwich the pyranose. The penultimate mannose is similarly stacked against two aromatic residues (Trp-86 and Trp-272). These aromatic stacking interactions explain why CpMnBP1 requires a degree of polymerization of at least two manno-oligosaccharides for effective binding. Additional units of 1,4-β-linked mannose would only be engaged through minimal hydrogen bonding interactions as reflected by the lack of a large increase in binding affinity for polymers of lengths between 3 and 5.
The specificity of CpMnBP1 for manno-oligosaccharides and the inability to engage other sugar polymers can also be understood in the context of the co-crystal structures. In addition to the stacking interactions described above, a number of polar residues occupy the length of the oligosaccharide-binding cleft where they engage in specific interactions with mannose units. The first binding site, which harbors the non-reducing end of the oligosaccharide, O-2NR, of the terminal mannose, is within hydrogen bonding distance with the indole nitrogen of Trp-25 (2.9 Å) and with the carboxamide oxygen of Asn-307 (2.6 Å); O-3NR interacts with the carboxamide nitrogen of Asn-307 (2.9 Å) and Asn-343 (2.7 Å); and O-4NR is engaged by the oxygen of Asp-129 (2.8 Å) and Asn-343 (3.1 Å). These interactions likely dictate specificity for the non-reducing end of the chain. The next mannose unit is engaged through interactions between O-6 and Arg-139 (3.2 Å), between O-5 and Gln-191 (3.1 Å), and of O-2 (2.7 Å) with the backbone carbonyl of Asn-251. Additional mannose units are engaged through minimal interactions, most of which are mediated through intervening solvent molecules. Although the binding cleft can accommodate mannose chains with a degree of polymerization up to 5 (Table 5), there are very few polar or aromatic residues in the vicinity of these additional sites.
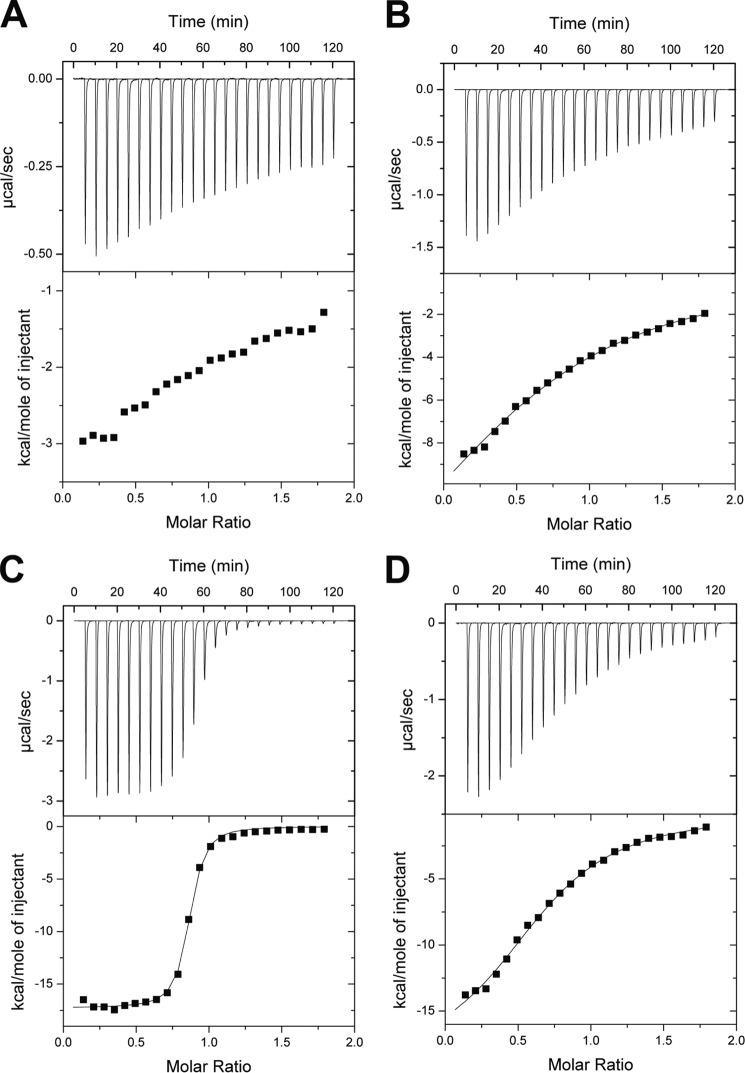
To delineate the contributions of residues to ligand binding, we generated Ala mutants of residues involved in stacking interactions with the non-reducing end (Trp-25 and Trp-272) as well as those involved in hydrogen bonding interactions (Gln-191 and Asn-307) and measured the binding of these variants to mannotetraose using ITC (Fig. 5 and Table 5). The W272A variant demonstrated the loss of all binding affinity, whereas the W25A bound manno-oligosaccharides but with a 100-fold loss in affinity. Deletion of Asn-307 resulted in a 17-fold loss in affinity, whereas the Q191A mutant was not significantly compromised.
FIGURE 5.
ITC of mannotetraose binding by variants of CpMnBP1. Binding isotherms and representative curves for CpMnBP1 show that the affinity for mannotetraose of the following CpMnBP1 variants: W272A (A), W25A (B), Q191A (C), and N307A (D).
As noted, CpMnBP1 can also engage decorated manno-oligosaccharides with high affinity, and the co-crystal structures provide a rationale for this tolerance. A superposition with various solute-binding proteins based on a structural alignment of secondary structural elements reveals that the CpMnBP1 ligand-binding site is flanked by a large loop spanning residues Gly-247 through Pro-258 (Fig. 6, A and B). In all other members of the oligosaccharide cluster B, the ligand-binding site is enclosed by a helix that runs parallel to the orientation of the binding cavity (Fig. 6, B, C, D, and E). This helix occludes the binding cavity and provides a hydrophobic cavity where the oligosaccharide or peptide is sequestered. The lack of this “capping helix” in CpMnBP1 results in a ligand-binding site that is open to solvent, which can presumably accommodate manno-oligosaccharides that contain branch decorations.
FIGURE 6.
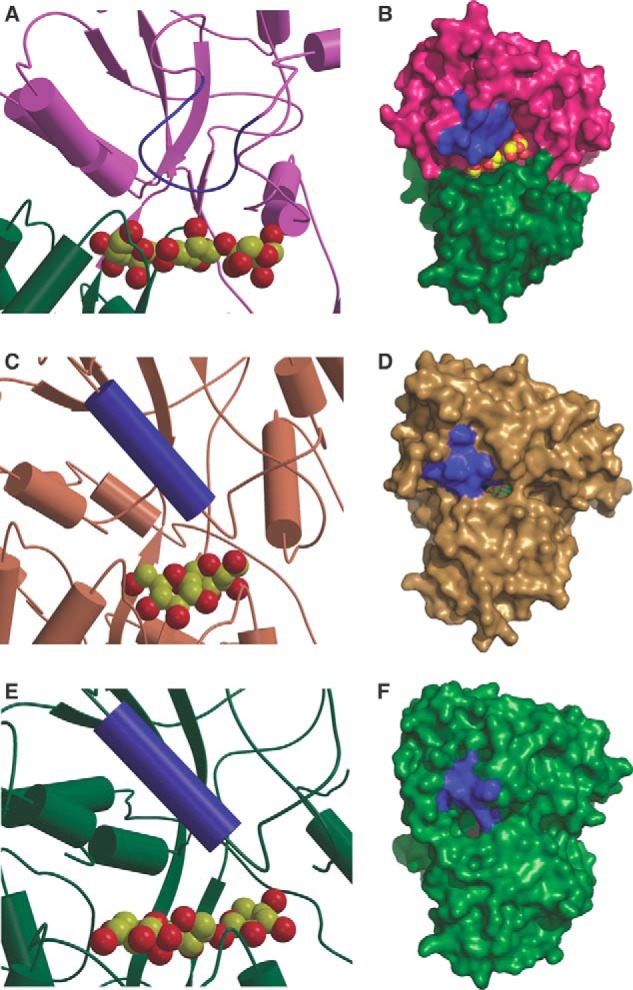
Comparison of the ligand pockets of various solute-binding proteins. Shown is a close-up view of the active site and a solvent-accessible surface representation of the solute-binding pockets of (A and B) CpMnBP1 (in purple and green) in comparison with those of the trehalose/maltose-binding protein from T. litoralis (in brown; Protein Data Bank code 1EU8) (C and D) and the xylan-binding protein from C. polysaccharolyticus (in green; Protein Data Bank code 4G68) (E and F). The bound oligosaccharides are shown in Corey-Pauling-Koltun representation, and the capping helix or equivalent structural element is colored in blue.
Despite the lack of this secondary structural element, the ligand-binding site is extensively defined, and a cavity that can accommodate branching decorations may be modeled into this cleft (Fig. 7). Lastly, we note that our calorimetric analysis demonstrates that CpMnBP1 engages polymeric mannose ligands with an affinity that is several orders of magnitude weaker (Ka around 106 m−1) than is observed with other solute-binding proteins and their cognate ligands (typically with Ka in the range of 109 m−1) (Table 5). Presumably, this weaker binding is a result of the fairly open nature of the ligand-binding cleft in CpMnBP1. Calorimetric analysis shows that CpMnBP1 binds to a branched substrate (α-galactosyl mannotriose) with an affinity similar to that of the undecorated counterpart (mannotriose) (Fig. 6), suggesting that tight ligand affinity is compromised to accommodate tolerance for decorated manno-oligosaccharides.
FIGURE 7.
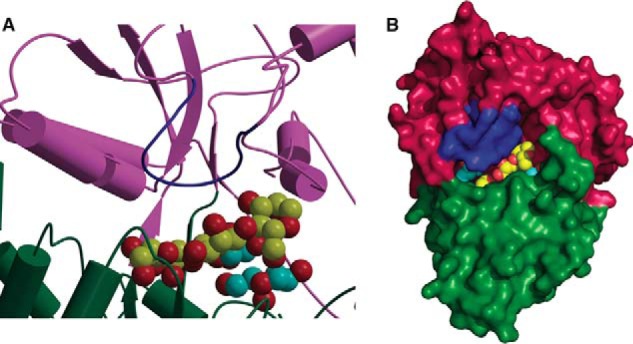
Model for binding of branched substrates by CpMnBP1. Shown is a close-up view of the active site (A) and the solvent-accessible surface (B) of a hypothetical model of a branched mannotriose ligand bound in the ligand-binding pocket of CpMnBP1. The oligosaccharide is shown in Corey-Pauling-Koltun representation with the branching carbon atoms colored in cyan.
DISCUSSION
Mannan is a major form of hemicellulose, the second most abundant polysaccharide in nature after cellulose. Microbes, therefore, have developed sets of enzymes that allow them to release the monomeric sugars in mannans for fermentation to generate energy and cellular building blocks. C. polysaccharolyticus, formerly Thermoanaerobacterium polysaccharolyticum, is a versatile polysaccharide-degrading anaerobic bacterium. This thermophilic bacterium elaborates enzymes of potential biotechnological application, especially in the emerging biofuel industry. C. polysaccharolyticus was isolated from a waste pile from a corn canning factory in Illinois (30). Aside from C. polysaccharolyticus, only two other members of this genus (Caldanaerobius zeae and Caldanaerobius fijiensis) have been successfully cultured (32). Based on 16 S ribosomal DNA sequence phylogeny, C. polysaccharolyticus and its relatives are distantly related to the thermophilic and anaerobic genera Thermoanaerobacterium and Thermoanaerobacter (30). Growth on mannan polysaccharides and subsequent transcriptome sequencing allowed us to identify two gene clusters as the main loci up-regulated during mannan metabolism by C. polysaccharolyticus. We have named the two gene clusters the Man5A and Man5B gene clusters in recognition of the two hydrolytic enzymes previously characterized from these gene clusters (28, 29). Our previous biochemical analysis of the man5A gene product, a large modular polypeptide, suggests that it is anchored on the cell surface through three surface layer homology peptides located at the C terminus of the polypeptide (29).
We postulate that the gene product of CalpoDRAFT_1213 or the transcriptional regulator in the Man5B gene cluster (Fig. 2B) senses the manno-oligosaccharides released by Man5A or other endo-mannanases in the C. polysaccharolyticus genome and facilitates transcription of other genes located in the same cluster. The Man5B polypeptide lacks an obvious signal peptide and is likely located in the cytoplasm. The man5b gene is also located upstream of the transcriptional regulator, and based on the RNA-seq data, it may be regulated differently or expressed constitutively to catalyze the hydrolysis of the manno-oligosaccharides produced by CpMan5A. Our previous biochemical analysis demonstrated that CpMan5B does not hydrolyze mannobiose but does hydrolyze mannotriose, mannotetraose, and mannopentaose to mannose and mannobiose. Furthermore, CpMan5B released mannose from galactosyl mannotriose (28, 33). Interestingly, within each of the Man5A and Man5B gene clusters is located a putative GH130, a newly characterized glycoside hydrolase family. The two polypeptides encoded by the GH130 genes share 99% identity, and they lack identifiable signal peptides. The gene products, therefore, likely function together with Man5B in the cytoplasm to degrade the manno-oligosaccharides released from mannan by Man5A. We hypothesized that the GH130 polypeptides possess manno-oligosaccharide phosphorylase activity and thus release α-d-mannosyl 1-phosphate from manno-oligosaccharides. This metabolic pathway for mannan has been described for Bacteroides fragilis (34) and Ruminococcus albus 7 (35) where phosphomannose mutase and mannose-6-phosphate isomerase convert the α-d-mannosyl 1-phosphate to fructose 6-phosphate, which can then enter the glycolytic pathway for metabolism. Our preliminary biochemical analysis of the recombinant form of the GH130 located in the Man5B gene cluster confirmed that it possesses manno-oligosaccharide phosphorylase activity,5 and based on the high identity with the homolog in the Man5A cluster, the two GH130 proteins are likely to harbor the same enzymatic activity. Thus, the up-regulation of the GH130 alone (Fig. 2, B and C) may be sufficient for channeling the manno-oligosaccharides released by Man5A and other mannanases into the glycolytic pathway for metabolism.
It is also important to note that the RNA-seq analysis of the two GH130 proteins is not entirely clear in the present report because the two genes (GH130 located in Man5A and Man5B gene clusters) are 98% identical at the nucleotide level. Irrespective of the confounding data presented by their almost identical nucleotide sequence, the results suggest that the GH130 genes are critical for mannan utilization by C. polysaccharolyticus.
Aside from the hydrolytic enzymes, each gene cluster contains the components of a putative multiprotein membrane-integral ATP-dependent transporter, i.e. CalpoDRAFT_1210, CalpoDRAFT_1211, and CalpoDRAFT_1212 for the Man5B gene cluster and CalpoDRAFT_0072, CalpoDRAFT_0073, and CalpoDRAFT_0074 for the Man5A gene cluster (Table 4 and Fig. 2, B and C). A plausible periplasmic substrate-binding domain of the ABC transporter in the Man5B gene cluster (CpMnBP1) specifically binds manno-oligosaccharides and was up-regulated during fermentation of both glucomannan and galactomannan (Table 4). The results thus suggest that the ABC transport system is for the internalization of manno-oligosaccharides, and CpMnBP1 may be a cognate solute-binding protein of the transporter. This system is analogous to the mechanism for xylan utilization utilized by C. polysaccharolyticus, suggesting a conserved strategy for polysaccharide fermentation by this organism. Furthermore, other Gram-positive bacteria possessing surface layer homology repeat-associated polysaccharide utilization functions have been shown to use a similar strategy for xylan utilization (36). This observation suggests that the route described for sugar processing by C. polysaccharolyticus is widespread in bacteria.
The protein most similar to CpMnBP1 in the GenBank database is a putative extracellular solute-binding protein from Thermoanaerobacter mathranii (Tmath_1701) that shares just 37% amino acid sequence identity. To the best of our knowledge, the only solute-binding protein of an ABC transporter with manno-oligosaccharide binding properties is the MalE1 protein from Thermotoga maritima. MalE1 optimally binds mannotetraose, and its expression is induced by mannans. Despite the similar manno-oligosaccharide binding activity, TmMalE1 and CpMnBP1 share only 26% amino acid sequence identity. Unlike typical oligosaccharide-specific solute-binding proteins, CpMnBP1 engages mannan sugars through the non-reducing end. Although unexpected, such a binding mode is not without precedent as the family 32 carbohydrate-binding module from Clostridium thermocellum has been demonstrated to similarly engage manno-oligosaccharides through the non-reducing end (37). Likewise, structural studies of the family 6 carbohydrate-binding module reveal that β-1,3-glucan chains are targeted through the non-reducing end of the saccharide (38).
The inversion in binding mode provides a rationale for understanding why CpMnBP1 does not bind cello- or xylo-oligosaccharides with any appreciable affinity. Docking of the cellotriose model with the non-reducing end poised at the end of the cleft results in clashes with the Trp residues that form the aromatic sandwich. Likewise, docking of a xylotriose model results in similar clashes, providing an explanation of why CpMnBP1 precludes the binding of these sugars. One possible reason why CpMnBP1 binds to the non-reducing ends of manno-oligosaccharides may be to facilitate the import of structurally diverse mannan degradation products, such as branch decorations on the β-1,4-linked mannose backbone. Calorimetric studies demonstrate that decorated substrates bind with equal affinity to CpMnBP1, and the open nature of the binding site allows for capture of branched substrates. The results of the mutational data can be similarly reconciled in the context of the crystal structures. The severe loss in binding affinity with the W272A mutant compared with the 100-fold loss in the W25A variant is probably the consequence of Trp-272 forming one entire face of the aromatic stacking platform, whereas the role of Trp-25 in forming the other face is redundant with Trp-86. Likewise, the nearly 17-fold loss in activity with the N307A variant (compared with the insignificant loss with mutations at other hydrogen-bonding residues) may be due to Asn-307 engaging in interactions with multiple hydroxyls of the substrate.
The manno-oligosaccharide degradation gene cluster described here encodes the necessary constellation of activities required for manno-oligosaccharide hydrolysis and import of the resultant oligosaccharide products across the cell membrane. Moreover, the manno-oligosaccharide-specific response regulator is predicted to coordinate the expression of these genes in response to high extracellular concentration of mannans. Thus, the entire gene cluster can be utilized as a cassette for the transfer of manno-oligosaccharide degradation activity into ethanologenic organisms for bioenergy production.
Acknowledgments
We are grateful to Drs. Keith Brister, Joseph Brunzelle, and the staff at Life Sciences Collaborative Access Team (LS-CAT, Argonne National Laboratory, Argonne, IL) for facilitating data collection.
This work was supported by the Energy Biosciences Institute.
The atomic coordinates and structure factors (codes 4R9F and 4R9G) have been deposited in the Protein Data Bank (http://wwpdb.org/).
J. R. Chekan, I. H. Kwon, V. Agarwal, D. Dodd, V. Revindran, R. I. Mackie, I. Cann, and S. K. Nair, unpublished data.
- ABC
- ATP-binding cassette
- M1–M6
- manno-oligosaccharides
- G1–G6
- cello-oligosaccharides
- RNA-seq
- RNA sequencing
- GluMan
- glucomannan
- GalMan
- galactomannan
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Scheller H. V., Ulvskov P. (2010) Hemicelluloses. Annu. Rev. Plant Biol. 61, 263–289 [DOI] [PubMed] [Google Scholar]
- 2. Popper Z. A., Fry S. C. (2003) Primary cell wall composition of bryophytes and charophytes. Ann. Bot. 91, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Popper Z. A. (2008) Evolution and diversity of green plant cell walls. Curr. Opin. Plant Biol. 11, 286–292 [DOI] [PubMed] [Google Scholar]
- 4. Ebringerová A., Hromádková Z., Heinze T. (2005) Hemicellulose. Adv. Polym. Sci. 186, 1–67 [Google Scholar]
- 5. Moreira L. R., Filho E. X. (2008) An overview of mannan structure and mannan-degrading enzyme systems. Appl. Microbiol. Biotechnol. 79, 165–178 [DOI] [PubMed] [Google Scholar]
- 6. Chauhan P. S., Puri N., Sharma P., Gupta N. (2012) Mannanases: microbial sources, production, properties and potential biotechnological applications. Appl. Microbiol. Biotechnol. 93, 1817–1830 [DOI] [PubMed] [Google Scholar]
- 7. Sánchez C. (2009) Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol. Adv. 27, 185–194 [DOI] [PubMed] [Google Scholar]
- 8. Wilson D. B. (2011) Microbial diversity of cellulose hydrolysis. Curr. Opin. Microbiol. 14, 259–263 [DOI] [PubMed] [Google Scholar]
- 9. Dhawan S., Kaur J. (2007) Microbial mannanases: an overview of production and applications. Crit. Rev. Biotechnol. 27, 197–216 [DOI] [PubMed] [Google Scholar]
- 10. Du J., Shao Z., Zhao H. (2011) Engineering microbial factories for synthesis of value-added products. J. Ind. Microbiol. Biotechnol. 38, 873–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hong K. K., Nielsen J. (2012) Metabolic engineering of Saccharomyces cerevisiae: a key cell factory platform for future biorefineries. Cell. Mol. Life Sci. 69, 2671–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee S. J., Lee S. J., Lee D. W. (2013) Design and development of synthetic microbial platform cells for bioenergy. Front. Microbiol. 4, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jang Y. S., Park J. M., Choi S., Choi Y. J., Seung do Y., Cho J. H., Lee S. Y. (2012) Engineering of microorganisms for the production of biofuels and perspectives based on systems metabolic engineering approaches. Biotechnol. Adv. 30, 989–1000 [DOI] [PubMed] [Google Scholar]
- 14. Galazka J. M., Tian C., Beeson W. T., Martinez B., Glass N. L., Cate J. H. (2010) Cellodextrin transport in yeast for improved biofuel production. Science 330, 84–86 [DOI] [PubMed] [Google Scholar]
- 15. Du J., Li S., Zhao H. (2010) Discovery and characterization of novel d-xylose-specific transporters from Neurospora crassa and Pichia stipitis. Mol. Biosyst. 6, 2150–2156 [DOI] [PubMed] [Google Scholar]
- 16. Han Y., Agarwal V., Dodd D., Kim J., Bae B., Mackie R. I., Nair S. K., Cann I. K. (2012) Biochemical and structural insights into xylan utilization by the thermophilic bacterium Caldanaerobius polysaccharolyticus. J. Biol. Chem. 287, 34946–34960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ladevèze S., Tarquis L., Cecchini D. A., Bercovici J., André I., Topham C. M., Morel S., Laville E., Monsan P., Lombard V., Henrissat B., Potocki-Véronèse G. (2013) Role of glycoside phosphorylases in mannose foraging by human gut bacteria. J. Biol. Chem. 288, 32370–32383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baggerly K. A., Deng L., Morris J. S., Aldaz C. M. (2003) Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics 19, 1477–1483 [DOI] [PubMed] [Google Scholar]
- 19. Otwinowski Z., Borek D., Majewski W., Minor W. (2003) Multiparametric scaling of diffraction intensities. Acta Crystallogr. A 59, 228–234 [DOI] [PubMed] [Google Scholar]
- 20. Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., Adams P. D. (2008) Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 [DOI] [PubMed] [Google Scholar]
- 21. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 22. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 23. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25. Kleywegt G. J., Brünger A. T. (1996) Checking your imagination: applications of the free R value. Structure 4, 897–904 [DOI] [PubMed] [Google Scholar]
- 26. Laskowski R. A., Rullmannn J. A., MacArthur M. W., Kaptein R., Thornton J. M. (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 [DOI] [PubMed] [Google Scholar]
- 27. Rose P. W., Bi C., Bluhm W. F., Christie C. H., Dimitropoulos D., Dutta S., Green R. K., Goodsell D. S., Prlic A., Quesada M., Quinn G. B., Ramos A. G., Westbrook J. D., Young J., Zardecki C., Berman H. M., Bourne P. E. (2013) The RCSB Protein Data Bank: new resources for research and education. Nucleic Acids Res. 41, D475–D482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Han Y., Dodd D., Hespen C. W., Ohene-Adjei S., Schroeder C. M., Mackie R. I., Cann I. K. (2010) Comparative analyses of two thermophilic enzymes exhibiting both β-1,4 mannosidic and β-1,4 glucosidic cleavage activities from Caldanaerobius polysaccharolyticus. J. Bacteriol. 192, 4111–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cann I. K., Kocherginskaya S., King M. R., White B. A., Mackie R. I. (1999) Molecular cloning, sequencing, and expression of a novel multidomain mannanase gene from Thermoanaerobacterium polysaccharolyticum. J. Bacteriol. 181, 1643–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cann I. K., Stroot P. G., Mackie K. R., White B. A., Mackie R. I. (2001) Characterization of two novel saccharolytic, anaerobic thermophiles, Thermoanaerobacterium polysaccharolyticum sp. nov., and Thermoanaerobacterium zeae sp. nov., and emendation of the genus Thermoanaerobacterium. Int. J. Syst. Evol. Microbiol. 51, 293–302 [DOI] [PubMed] [Google Scholar]
- 31. Holm L., Park J. (2000) DaliLite workbench for protein structure comparison. Bioinformatics 16, 566–567 [DOI] [PubMed] [Google Scholar]
- 32. Lee Y. J., Mackie R. I., Cann I. K., Wiegel J. (2008) Description of Caldanaerobius fijiensis gen. nov., sp. nov., an inulin-degrading, ethanol-producing, thermophilic bacterium from a Fijian hot spring sediment, and reclassification of Thermoanaerobacterium polysaccharolyticum and Thermoanaerobacterium zeae as Caldanaerobius polysaccharolyticus comb. nov., and Caldanaerobius zeae comb. nov. Int. J. Syst. Evol. Microbiol. 58, 666–670 [DOI] [PubMed] [Google Scholar]
- 33. Oyama T., Schmitz G. E., Dodd D., Han Y., Burnett A., Nagasawa N., Mackie R. I., Nakamura H., Morikawa K., Cann I. (2013) Mutational and structural analyses of Caldanaerobius polysaccharolyticus Man5B reveal novel active site residues for family 5 glycoside hydrolases. PLoS One 8, e80448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Senoura T., Ito S., Taguchi H., Higa M., Hamada S., Matsui H., Ozawa T., Jin S., Watanabe J., Wasaki J., Ito S. (2011) New microbial mannan catabolic pathway that involves a novel mannosylglucose phosphorylase. Biochem. Biophys. Res. Commun. 408, 701–706 [DOI] [PubMed] [Google Scholar]
- 35. Kawahara R., Saburi W., Odaka R., Taguchi H., Ito S., Mori H., Matsui H. (2012) Metabolic mechanism of mannan in a ruminal bacterium, Ruminococcus albus, involving two mannoside phosphorylases and cellobiose 2-epimerase: discovery of a new carbohydrate phosphorylase, β-1,4-mannooligosaccharide phosphorylase. J. Biol. Chem. 287, 42389–42399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stjohn F. J., Rice J. D., Preston J. F. (2006) Paenibacillus sp. strain JDR-2 and XynA1: a novel system for methylglucuronoxylan utilization. Appl. Environ. Microbiol. 72, 1496–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mizutani K., Fernandes V. O., Karita S., Luís A. S., Sakka M., Kimura T., Jackson A., Zhang X., Fontes C. M., Gilbert H. J., Sakka K. (2012) Influence of a mannan binding family 32 carbohydrate binding module on the activity of the appended mannanase. Appl. Environ. Microbiol. 78, 4781–4787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Bueren A. L., Morland C., Gilbert H. J., Boraston A. B. (2005) Family 6 carbohydrate binding modules recognize the non-reducing end of β-1,3-linked glucans by presenting a unique ligand binding surface. J. Biol. Chem. 280, 530–537 [DOI] [PubMed] [Google Scholar]