Background: S-Acylation of hemagglutinin with stearate and palmitate is essential for influenza virus replication.
Results: Mass spectrometry showed that shifting a cysteine from the transmembrane region to a cytoplasmic position eliminates attachment of stearate.
Conclusion: The location of the acylation site is the decisive factor for site-specific acylation.
Significance: Similar differential acylation might occur in cellular transmembrane proteins.
Keywords: Fatty Acid, Glycoprotein, Influenza Virus, Mass Spectrometry (MS), Protein Acylation, Protein Palmitoylation, Transmembrane Domain, Hemagglutinin, Palmitate, Stearate
Abstract
S-Acylation of hemagglutinin (HA), the main glycoprotein of influenza viruses, is an essential modification required for virus replication. Using mass spectrometry, we have previously demonstrated specific attachment of acyl chains to individual acylation sites. Whereas the two cysteines in the cytoplasmic tail of HA contain only palmitate, stearate is exclusively attached to a cysteine positioned at the end of the transmembrane region (TMR). Here we analyzed recombinant viruses containing HA with exchange of conserved amino acids adjacent to acylation sites or with a TMR cysteine shifted to a cytoplasmic location to identify the molecular signal that determines preferential attachment of stearate. We first developed a new protocol for sample preparation that requires less material and might thus also be suitable to analyze cellular proteins. We observed cell type-specific differences in the fatty acid pattern of HA: more stearate was attached if human viruses were grown in mammalian compared with avian cells. No underacylated peptides were detected in the mass spectra, and even mutations that prevented generation of infectious virus particles did not abolish acylation of expressed HA as demonstrated by metabolic labeling experiments with [3H]palmitate. Exchange of conserved amino acids in the vicinity of an acylation site had a moderate effect on the stearate content. In contrast, shifting the TMR cysteine to a cytoplasmic location virtually eliminated attachment of stearate. Thus, the location of an acylation site relative to the transmembrane span is the main signal for stearate attachment, but the sequence context and the cell type modulate the fatty acid pattern.
Introduction
Hemagglutinin (HA) of influenza virus is a typical type I transmembrane glycoprotein with an N-terminal signal peptide, a large ectodomain, a single transmembrane region (TMR3; 27 amino acids) and an 11-amino acid-long cytoplasmic tail (1). HA from influenza A virus is S-acylated at three cysteine residues, two located in the cytoplasmic domain and one located at the end of the TMR (2–4). The hydrophobic modification of HA is essential for virus replication because (depending on the virus strain) virus mutants with more than one acylation site deleted either show drastically impaired growth or could not be created at all by reverse genetics (5–7). Acylation facilitates raft association of HA (8–10) and thus enrichment of the protein in small nanodomains of the plasma membrane (11), an observation that might explain why palmitoylation affects both assembly of virus particles (5, 7, 12) and the membrane fusion activity of HA (2, 6, 13–15). In accordance with the essential requirement of palmitoylation of HA for virus replication is the complete conservation of the acylation sites through all HA subtypes and variants, although HA is a highly variable molecule with very low amino acid conservation (16).
Cellular enzymes that acylate HA (or any other viral protein) have not been identified, but likely candidates are members of the Asp-His-His-Cys (DHHC) family, polytopic membrane proteins containing a DHHC motif within one of their cytoplasmic domains (17, 18). Most of the DHHC proteins are located in the endoplasmic reticulum or Golgi region where acylation of HA takes place (19). Alternatively, S-acylation of at least some proteins can occur by a non-enzymatic or autocatalytic mechanism (20, 21).
It has long been known that some “palmitoylated” proteins contain different acyl chains, indicating that the responsible enzyme(s) (in contrast to the N-myristoyltransferase) cannot strictly discriminate between long chain fatty acid species (22). However, advancements in mass spectrometry have allowed precise quantification of the type of fatty acid linked to an acyl protein or even to a single acylation site. The peripheral membrane protein GAP43 contains both palmitate (C16:0) and stearate (C18:0) at two N-terminal cysteine residues, but a preference of a fatty acid species for one of the cysteines was not observed (23). In contrast, our studies with HAs of influenza A virus revealed that stearate is exclusively attached to the cysteine positioned at the end of the transmembrane region, whereas the two cytoplasmic cysteines contain only palmitate (24). Site-specific acylation was also observed for the glycoproteins of influenza B and C viruses. HA of influenza B virus possesses two cytoplasmic cysteines that contain only palmitate, whereas the hemagglutinin esterase fusion protein HEF of influenza C virus has one transmembrane cysteine that is stearoylated (24), confirming previous studies performed with less sophisticated methodology (25). We also analyzed glycoproteins of other enveloped viruses and found that site-specific attachment of palmitate or stearate is a common feature of viral spike proteins (26) and (because viruses rely on the cellular acylation machinery) probably also of cellular transmembrane receptors.
Since this initial observation, we have analyzed more than 40 HA variants from 14 subtypes by MS (27, 28). The percentage of stearate in all HAs differs from 35% (suggesting that each of the TMR cysteines of the trimeric HA spike contains stearate) to 12% (indicating that only one of three TMR cysteines is stearoylated). Interestingly, HA present in virus strains initially isolated from humans contain less stearate compared with HA isolated from mammals and especially from birds. Because all our previous MS analyses were performed with viruses purified from the allantoic fluid of embryonated chicken eggs, it is not known whether cell type-specific differences in the fatty acid pattern might exist. Another reason for the different stearate content in the HA of human and avian viruses might be that other viral membrane proteins, especially the matrix protein M1, which is much less variable than HA but contains host-specific amino acid substitutions, affect acylation of HA. However, we could not detect variability in the stearate content of HA if internal proteins were exchanged between viruses (29).
Thus, the main molecular signal that determines preferential attachment of palmitate or stearate is likely to be located in HA either in the amino acid sequence around individual acylation sites or in their positioning relative to the membrane span. To analyze this question, we created recombinant viruses containing HA with point mutations adjacent to acylation sites or with a TMR cysteine shifted to a cytoplasmic location and analyzed the fatty acid content of HA by MS utilizing a new procedure for sample preparation that is more suitable for viruses grown in cell culture instead of embryonated eggs.
EXPERIMENTAL PROCEDURES
Cells and Plasmids
African green monkey kidney cells (CV1), human embryonic kidney cells (293T), DF-1 cells, a chicken fibroblast cell line, and Madin-Darby canine kidney (MDCK) II cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Pan Biotech) supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Chicken embryo fibroblasts were prepared from chicken embryos as described (30). All cells were cultured at 37 °C in 5% CO2 and passaged twice per week. The plasmid pHW2000 used for the rescue of recombinant influenza A/WSN/33 (H1N1) virus has been described (31).
Generation of Recombinant Virus
Recombinant influenza A/WSN/33 (H1N1) virus (WSN) was generated using the eight-plasmid reverse genetics system in which each plasmid contains cDNA of one viral RNA segment flanked by suitable promoters. In the HA-encoding cDNA segment, the codon for Ile-563 (ATA) was replaced by the isoleucine codon TTG, and Gly-557 (GGG) was replaced by the alanine codon GCT and the glutamic acid codon GAA, respectively. Gln-560 (CAG) was replaced by the glutamine codon GAA. Gly-547 (GGG) was replaced by the serine codon ACG. Cys-554 (TGT) and Leu-559 (TTG) were replaced by the serine codon (AGC) and the cysteine codon (TGC), respectively, using QuikChange mutagenesis (Stratagene) and confirmed by sequencing (GATC Biotech). 293T cells in 60-mm dishes were transfected with eight plasmids encoding WSN cDNA (1 μg each) using TurboFect (Fermentas/Thermo) in Opti-MEM (Invitrogen/Thermo). 4–6 h after transfection, medium was replaced by infection medium (DMEM containing 0.2% bovine serum albumin, 0.1% FBS, 2 mm glutamine, 100 units/ml penicillin/streptomycin, and 1 μg/ml TPCK-treated trypsin (Sigma-Aldrich), and incubation was continued at 37 °C. 72 h after transfection, supernatants were harvested and cleared of debris by low speed centrifugation (2,000 × g, 5 min, 4 °C). Recombinant viruses were first plaque-purified on MDCKII cells and then amplified and titrated in MDCKII cells to obtain a high titer virus stock.
Plaque Assay
The plaque assay was performed on MDCKII cells in 6-well plates according to standard procedures. Briefly, cells were infected with serial 10-fold dilutions of the virus supernatants in infection medium, incubated for 1 h at 37 °C, washed twice with phosphate-buffered saline (PBS), and overlaid with 1.8% SeaPlaque agarose (Lonza) in 2× Eagle's minimum essential medium (Lonza) supplemented with 0.2% bovine serum albumin, 0.1% FBS, 2 mm glutamine, and 1 μg/ml TPCK-treated trypsin. After 2–3 days of incubation, cells were stained using neutral red (0.03% in PBS; Biochrom), and plaques were counted.
For isolation of virus clones, the overlaying agar of single, well separated plaques was excised, dissolved in 500 μl of PBS, incubated for 16 h at 4 °C, and then used to infect T25 cell culture flasks of MDCKII cells. After incubation for 1 h at 37 °C, the cells were washed twice with PBS, and infection medium was added. At 1–3 days postinfection when a cytopathic effect was visible, supernatants were harvested and cleared of debris by low speed centrifugation (2,000 × g, 5 min, 4 °C). Every virus sample was sequenced to confirm the presence of inserted mutations before titrating in the plaque assay and amplification in large scale in MDCKII cells.
Virus Preparation from Embryonated Eggs and MDCK Cells
Recombinant WSN virus (1,000 pfu; diluted in 0.9% NaCl) was inoculated with a 0.45 × 25-mm needle into the allantoic cavity of 11-day-old embryonated chicken eggs. For control experiments, natural virus strains A/WSN/33 (H1N1) and A/Puerto Rico/8/34 (H1N1) (PR8) were propagated in the 11-day-old embryonated chicken eggs via the same procedure. After 48–72 h at 37 °C, the embryos were euthanized at 4 °C for 16 h. The allantoic fluid was collected and centrifuged at 2,000 × g for 20 min at 4 °C to clarify from debris and erythrocytes.
To prepare virus from cell culture, the virus stock was used to infect MDCKII, chicken embryo fibroblast (CEF), or DF-1 cells with a multiplicity of infection of 0.01. After 1 h of adsorption, cells were washed with PBS, infection medium was supplied, incubation was continued until a cytopathic effect became evident, and then the supernatant was harvested.
The cleared supernatant or allantoic fluid was centrifuged at 100,000 × g for 2 h, and the viral pellet was resuspended in TNE (10 mm Tris/HCl, pH 7.4, 100 mm NaCl, and 1 mm EDTA). In some cases, the obtained pellet was centrifuged through 2 ml of a sucrose cushion (20% in TNE) solution and centrifuged at 100,000 × g for 2 h. The resulting virus pellet was resolved in TNE.
Reverse Transcription-Polymerase Chain Reaction (RT)-PCR and Sequencing
To check for correctness of HA sequences in the recombinant viruses, RNA was isolated from cell culture supernatants or virus preparations with an Invisorb Spin Virus RNA Mini kit (Stratec) followed by RT-PCR using HA-specific primers and a OneStep RT-PCR kit (Qiagen). PCR products were purified from agarose gels using the Fragment CleanUp kit (Stratec) and sequenced (GATC Biotech).
Expression of HA with the Vaccinia T7 System and Metabolic Labeling
HA was subcloned from pHW2000 constructs into the plasmid pTM1 behind the T7 RNA polymerase promoter. CV1 cells (2 × 106) in Petri dishes were infected with recombinant vaccinia virus expressing T7 polymerase (10 plaque-forming units/cell). At 2 h postinfection, the cells were washed with PBS prior to transfection with pTM1-HA plasmids using TurboFect. At 5 h postinfection, cells intended for [35S]methionine/cysteine labeling were starved in DMEM lacking both amino acids. Cells were labeled at 5.5 h postinfection with 0.5 mCi/ml [35S]methionine/cysteine (1,175 Ci/mmol; Easy Tag Express Protein Labeling Mix, PerkinElmer Life Sciences) or with 0.25 mCi/ml [3H]palmitic acid (30–60 Ci/mmol; 9,10-3H; PerkinElmer Life Sciences) for 3 h. Cells were lysed in radioimmune precipitation assay buffer (20 mm Tris-HCl, 150 mm NaCl, 10 mm EDTA, 10 mm iodoacetamide, 0.1% (w/v) SDS, 1% (v/v) Triton X-100, and 1% (w/v) sodium deoxycholate, pH 7.4), cell debris was pelleted, and the cleared supernatant was incubated at 4 °C for 16 h with 1 μl of anti-influenza H1N1 virus polyclonal rabbit antibody R309 (32) kindly provided by Y. Kawaoka. Following incubation with 30 μl of protein A-Sepharose, each lysate was washed with radioimmune precipitation assay buffer. The final pellet was analyzed by SDS-PAGE under non-reducing conditions and fluorography as described (33). Densitometric quantification of bands was carried out using Bio-1D software (Vilmer-Lourmat).
Mass Spectrometry: Sample Preparation by Proteolytic Digestion of Virus Particles
has been described (34). Briefly, the HA ectodomain was removed by digestion of gradient-purified virus particles with bromelain, shaved particles were pelleted, and the membrane-anchoring fragments of HA were extracted to the chloroform phase and analyzed directly by MALDI-TOF MS.
Sample Preparation by Blotting
Viral proteins were separated via non-reducing SDS-PAGE in a Mini-Protean (Bio-Rad) system and electroblotted onto a PVDF membrane (Merck), which was stained with Ponceau S (Fluka). The HA band was cut from the membrane, fragmented into small pieces, and stored at 4 °C in an Eppendorf tube with ∼ 50 μl of distilled water until further analysis. The membrane pieces were rinsed with 100–200 μl of 40% acetonitrile and 50 mm NH4HCO3, pH 7.5 and digested with 5–10 μl of trypsin (Promega; 15 ng/μl in 80% acetonitrile and 50 mm NH4HCO3, pH 7.5) for at least 4 h at 37 °C. The reaction was stopped by adding a 2-fold volume of 0.5% trifluoroacetic acid. The water-acetonitrile solution containing hydrophilic and slightly hydrophobic peptides was removed to a clean Eppendorf tube, and an aliquot (1–2 μl) was later used to confirm the protein identity by MALDI-TOF MS analysis. The strongly hydrophobic peptides were further extracted from the membrane pieces with 5–10 μl of hexafluoroisopropanol for 1 h. The water-acetonitrile and hexafluoroisopropanol extracts were combined with the membrane pieces and subjected to chloroform (or chloroform/methanol) extraction. The organic phase enriched with S-acylated peptides was analyzed by MALDI-TOF MS.
MALDI-TOF MS Analysis
Sample aliquots (1 μl) were mixed on a steel target with an equal volume of a 2,5-dihydroxybenzoic acid (Aldrich) in 30% acetonitrile and 0.5% trifluoroacetic acid water solution (10 mg/ml). Most mass spectra were recorded on an Ultraflextreme MALDI TOF-TOF mass spectrometer (Bruker Daltonik, Germany) equipped with a 355 nm neodymium laser. The MH+ molecular ions were measured in reflector mode; the accuracy of mass peak measuring was within 0.01%. The proportion of stearate/palmitate was calculated from the MS peak intensities using the method developed earlier (35).
Amino Acid Frequency Analysis
The influenza A virus HA amino acid sequences were extracted from the Influenza Research Database in April 2014 (33,254 entries of H1–H17 subtypes, excluding laboratory strains, with complete HA sequence). Among them, there were 17,311 unique sequences chosen for multiple alignments. The MAFFT program (36) working with very large samplings was used to align sequences within the subtype, whereas the MUSCLE server (37) was further used to align the subtype consensus sequences. The consensus sequence alignment was visualized using JalView. The frequency value F for every consensus amino acid X in position i in the multiple alignment was calculated as F = (#X/N) × 100% where #X is the number of sequences having amino acid residue X in position i, and N is the total number of sequences aligned.
RESULTS
Rationale for Mutagenesis in the Membrane-anchoring Domain of HA
In principle, the differential acylation pattern for HA might be influenced by the location of the acylated cysteine relative to the transmembrane span or (but mutually not exclusive) by the presence of specific amino acids near the acylation site. We first performed a bioinformatics comparison of all HA sequences of influenza A virus present in the NCBI database to reveal conserved residues in the TMR and cytoplasmic tail that might be part of a complex acylation signal (Fig. 1). We mutated residues in H1 subtype HA from the WSN strain that might interact with the active center of a DHHC protein, i.e. located in the inner part of the transmembrane region or in the cytoplasmic tail (Fig. 2).
FIGURE 1.
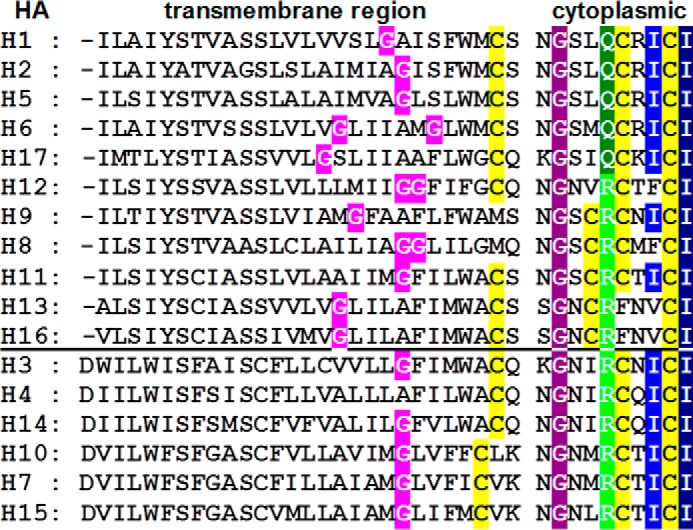
Consensus sequence of the TMR and cytoplasmic tail for each HA subtype. Acylation sites, the stearoylated cysteine at the end of the TMR, and the two palmitoylated cysteines in the cytoplasmic tail are highlighted in yellow. The C terminus contains the hydrophobic patch ICI where the C-terminal isoleucine is completely conserved (highlighted in dark blue) and the other isoleucine (highlighted in light blue) is in some subtypes replaced by a phenylalanine or valine. Also completely conserved is a glycine in the cytoplasmic tail (highlighted in purple). Another position in the cytoplasmic tail contains either a glutamine (dark green) or an arginine (light green). The location of the glycine(s) in the middle of the TMR present in all subtypes (except H4) is highlighted in pink. Above the horizontal bar are group 1 HAs; below are group 2 HAs (49).
FIGURE 2.
Mutations introduced into the TMR and cytoplasmic tail of HA. The amino acid sequence at the inner part of the transmembrane region and cytoplasmic tail of H1 subtype HA from the WSN strain is shown. Acylated cysteine residues are highlighted in gray; amino acid exchanges are in bold. The bars above the sequence depict the frequency of individual amino acids through all HA sequences present in the NCBI database determined as described under “Experimental Procedures.” + or − indicates whether infectious virus could be rescued by transfection of cells.
One of the latter is the hydrophobic sequence ICI at the C terminus where the terminal isoleucine 565 and the palmitoylated cysteine 564 are present in each HA variant. The adjacent isoleucine 563 is almost completely conserved through all HA subtypes; in some subtypes it is replaced by other large hydrophobic amino acids. To investigate whether the acylation pattern of HA is determined by the C-terminal hydrophobic patch, Ile-563 was exchanged by the polar residue glutamine (I563Q) or by the hydrophobic residue leucine (I563L).
Glycine at position 557 in the cytoplasmic tail is also completely conserved through all HA sequences. We exchanged it by the small residue alanine (G557A) or by the negatively charged glutamic acid (G557E). In another mutant, the cytoplasmic tail (including the two palmitoylated cysteines) was deleted by replacing the codon for glycine 557 with a stop codon (G557Stop). Position 560 located adjacent to a palmitoylated cysteine is not conserved through all HA sequences but accommodates only two consensus amino acids, either a glutamine or an arginine, depending on the subtype. The glutamine present in HA of the WSN strain was replaced by glutamic acid (Q560E).
Each HA subtype (except H4) contains in the middle of its TMR a glycine, which has a very low propensity to be present in an α-helical structure. It thus might induce a kink that presents the cysteine at the end of the TMR to an acyltransferase. We exchanged glycine 547 by serine (G547S), which is better compatible with an α-helical structure.
To determine whether the location of a cysteine relative to the transmembrane span is the main determinant for attachment of stearate, we shifted the cysteine at the end of the TMR to a cytoplasmic location. To do so, cysteine 554 was replaced by a serine, and leucine 559 was exchanged by cysteine (C554S/L559C). The resulting mutant thus contains an identical location of cysteines as in H8 and H9 subtype HAs from naturally occurring avian, equine, swine, and human virus strains.
Having verified the mutation by sequencing, the mutant HA plasmid together with seven plasmids encoding the other viral proteins were transfected into HEK 293T cells, and 3 days later the supernatant was tested for virus particle release by a plaque assay. For two mutants, G557Stop and I563Q, we never rescued infectious virus particles, although control experiments using wild type HA done in parallel showed that transfection was successful. Thus, the mutation might have destroyed one of the essential activities of HA, i.e. virus assembly and budding and/or membrane fusion during virus entry. All the other mutations in HA were not lethal: the nucleotide sequence was stable during several passages of replication, but viral titers tested in a growth curve were reduced by 1 or 2 orders of magnitude (data not shown).
Analyzing S-Acylation of HA with Lethal Mutations
The lethal replication defect of viruses containing the HA mutants I563Q and G557Stop might be due to a complete defect in acylation of HA, a modification essential for virus replication. To address whether the cytoplasmic tail is an essential part of an acylation signal, we expressed mutant HA with a vaccinia virus T7 system in CV1 cells and analyzed acylation by metabolic labeling with [3H]palmitate (Fig. 3). Labeling with [35S]methionine/cysteine done in parallel, immunoprecipitation, SDS-PAGE under non-reducing conditions, and fluorography showed two HA bands, which are likely to represent the mannose-rich precursor with higher SDS-PAGE mobility and the slower migrating product with complex glycans that has been described for HA for WSN (38). The intensity of the upper and lower bands differed between wild type HA and the mutants; especially HA with deleted tail exhibits more of the precursor relative to the mature form, indicating that its intracellular transport is retarded. The fluorogram of the [3H]palmitate-labeled samples revealed that each HA mutant is labeled but with varying intensity. Densitometric quantification of [35S]methionine/cysteine- and [3H]palmitate-labeled bands revealed no meaningful reduction in palmitoylation. The (non-lethal) HA mutant Q560E and the mutant I563Q are acylated with only slightly reduced efficiency (∼90%) relative to wild type HA (100%). Acylation of G557Stop in which two acylated cysteines are deleted is reduced to ∼60%, suggesting that the one remaining acylation site at the end of the TMR is efficiently used. Thus, the mutations introduced into HA did not prevent acylation, but subtle differences in the occupancy of individual acylation sites cannot be excluded.
FIGURE 3.
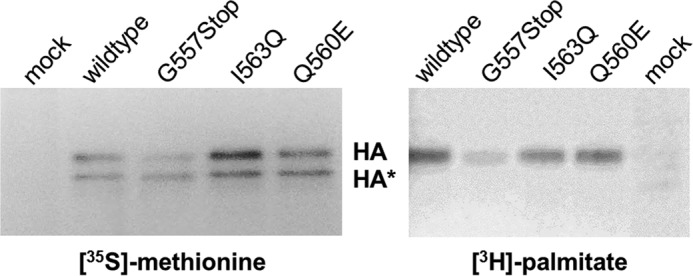
Expression and [3H]palmitoylation of non-functional HA mutants. HA mutants were expressed with a vaccinia virus T7 system in CHO cells. Transfected cells were labeled for 3 h with [35S]-methionine/cysteine and [3H]palmitate, respectively. HA was immunoprecipitated from cell extracts and subjected to non-reducing SDS-PAGE and fluorography. The lower HA band (HA*) is a mannose-rich precursor, and the upper HA band is the mature product (38). Densitometric quantification of both [35S]methionine/cysteine-labeled bands and the [3H]palmitate band revealed a 3H to 35S ratio of 90% for Q560E and 86% for I563Q relative to wild type HA (100%). The G557Stop mutant has a 3H to 35S ratio of 62%, but two acylation sites are deleted in this construct. mock, untransfected cells.
Establishment of a New Sample Preparation Procedure for Mass Spectrometry
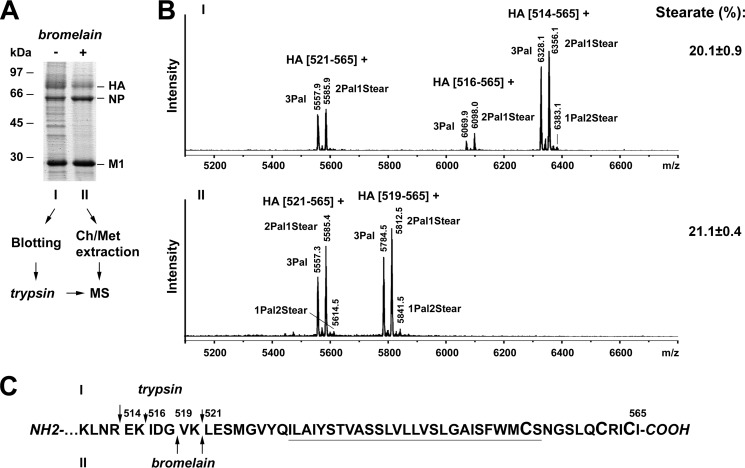
In our previous protocol of sample preparation for MS analysis, the HA ectodomain was removed by digestion of virus particles with bromelain, shaved particles were pelleted, and the membrane-anchoring fragments of HA were extracted to the chloroform phase and analyzed by MALDI-TOF MS. To remove impurities that might affect proteolytic cleavage of HA, virus particles were purified from the allantoic fluid of embryonated chicken eggs by sucrose gradient purification. Because the viral mutants were compromised in growth, we thought to develop an alternative procedure that does not require the materially demanding step of gradient purification and can thus be performed with viruses pelleted from the supernatant of MDCK cells. In the new approach, virus particles are separated by non-reducing SDS-PAGE, and proteins are blotted onto PVDF membranes and visualized by Ponceau S staining. The HA band is cut from the membrane and digested with trypsin, and the resulting peptides are eluted and analyzed by MALDI-TOF MS (Fig. 4A).
FIGURE 4.
Establishing a new method for sample preparation prior to MS. A, SDS-PAGE analysis under non-reducing conditions and Coomassie staining of A/WSN/33 (H1N1) virions grown in MDCK cells before (−) and after (+) digestion with bromelain and pelleting (100,000 × g, 1.5 h, 4 °C) of particles. The positions of viral proteins (HA, NP, and M1) and molecular mass markers in kDa are indicated. I, the new protocol includes electroblotting of undigested virus particles onto a PVDF membrane, “on-membrane” digestion of HA with trypsin, peptide elution, and MS analysis. II, the previous protocol is based on the proteolytic treatment of purified virions with bromelain followed by chloroform/methanol (Ch/Met) extraction of the anchoring peptides and their analysis with MS. B, MALDI-TOF MS analysis of S-acylated HA peptides obtained by methods I and II. Mass spectra obtained in reflector mode are depicted. Indicated are the m/z values of the peaks, the first and last amino acid residues of the corresponding peptides, and the number and type of HA-bound fatty acids. Amounts of HA-bound stearate (mean percentage ±S.D.) calculated from three to five spectra obtained for every of three blotting repeats (I; peaks of peptide HA(514–565) were taken for calculation) or from one bromelain digestion (II; both peptides HA(516–565) and HA(521–565) were used for calculation) are indicated. Minor peaks occasionally found between the peptides containing 3Pal and 2Pal1Stear and between 2Pal1Stear and 1Pal2Stear, respectively, probably result from methylation in acid medium (+14) or Met oxidation (+16). Note that non- or monoacylated HA peptides (peaks with m/z values around 4845 and 5083, respectively) were not detected in the spectra. C, amino acid sequence of the HA C-terminal region indicating sites of cleavage by trypsin (I) or bromelain (II). The arrow lengths approximately correspond to the respective peak heights in the spectra. Cysteines as potential acylation sites are enlarged, and the TMR is underlined.
To test whether the new approach yields reliable results, we compared both procedures with the same virus preparation, WSN wild type virus grown in MDCK cells. Each procedure utilizes a different protease, and therefore different peptides were observed in the MS spectra (Fig. 4B). According to its specificity, trypsin can cleave the polypeptide chain after arginine or lysine. We found that it cuts HA mainly after arginine 513 located 15 amino acid residues above the TMR, resulting in a peptide that matches to the complete membrane-anchoring plus linker region spanning amino acids 514–565. Two other cleavage sites after lysines 515 and 520 are also present in lower proportion (Fig. 4C). Bromelain possesses a wider substrate specificity compared with trypsin, but only glycine 518 and lysine 520 located in the linker region connecting the TMR with the well structured HA ectodomain are spatially accessible for the enzyme. Thus, the resulting peptides are shorter, spanning amino acids 519–565 and 521–565, respectively. Each of the mentioned peptides is present as two main peaks representing triple palmitoylated peptide and a peptide containing two palmitates and one stearate. A very small third peak representing a peptide containing one palmitate and two stearates also appears sometimes. Calculation of the amount of stearate revealed very similar results: 20.1% for the new blotting procedure and 21.1% for the previous extraction method. Thus, the new method is reliable and able to detect acylated peptides using ∼5–10 μg of protein, i.e. a Ponceau S-stained band.
Surprisingly, however, the stearate content from HA of the WSN virus was much higher than the amount previously determined for the closely related PR8 virus (12.6%; Ref. 24) that contains a very similar C-terminal amino acid sequence. Because the viruses were grown in MDCK cells and embryonated eggs, respectively, we hypothesized that the host cells might affect the acylation pattern of HA.
Comparison of Acylation Patterns of HA Synthesized in Avian and Mammalian Cells
All our previous MS analyses were performed with virus strains purified from an avian host, embryonated chicken eggs. In this system, HAs from human viruses have less stearate (12–18%) compared with HAs from avian viruses (18–35%; Refs. 24 and 27). To analyze whether HA from human viruses contains more stearate if grown in mammalian cells, we performed MS analysis with WSN using a variety of samples. We compared recombinant WSN (generated from transfected MDCK cells and then further amplified either in eggs or in MDCK cells) with a natural WSN virus strain amplified in eggs or in MDCK cells using both the previous extraction method and the new blotting procedure. In each case, WSN virus grown in eggs has less HA-bound stearate (8–12%) compared with WSN grown in MDCK cells (20.1–22.9%). Furthermore, to corroborate the surprising finding that avian cells apparently attach less stearate to HA compared with mammalian cells, we also analyzed WSN grown in CEFs, primary cells prepared from chicken embryos, and WSN grown in DF-1 cells, a cell line derived from CEFs. The amount of stearate attached to HA from CEF-derived WSN was identical (7.7%) to that attached to HA from egg-derived WSN (7.9%). DF-1 cells attach somewhat more stearate to HA (10.4%) but much less than MDCK cells (22.9%; results are summarized in Table 1). Thus, neither the sample preparation for MALDI-TOF MS nor the origin of the virus (from plasmid or from virus strain) significantly affected the result, indicating that mammalian cells attach more stearate to human viruses compared with cells of avian origin.
TABLE 1.
Comparison of amount of HA-bound stearate in virus grown in avian and mammalian cells determined by MALDI-TOF MS
Results are represented as mean ± S.D. The remainder is palmitate.
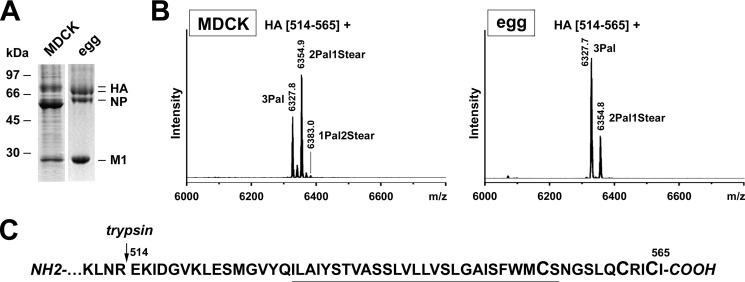
We then analyzed HA from the PR8 strain, which is of interest because it is used as a backbone to make seasonal flu vaccines; i.e. the glycoproteins of circulating human strains are reassorted into PR8, and the resulting virus strain is amplified in embryonated eggs for vaccine production. Identical to HA of WSN, we observed a tryptic peptide that matches to the complete membrane-anchoring region spanning amino acids 514–565 (Fig. 5). Calculation of the stearate amount in the two peaks revealed low stearate content in HA if the virus was grown in eggs (8.2%) compared with virus obtained from MDCK cells (21.2%). A low stearate amount was also previously determined with egg-grown PR8 virus using the previous extraction method (24). We conclude that HA from human strains is incompletely stearoylated if the virus is amplified in avian cells. However, no underacylated peptides were observed, indicating that the stearoylation site at the end of the TMR is occupied by palmitate. This was earlier confirmed by MALDI-TOF-TOF fragmentation analysis (35).
FIGURE 5.
MS result from PR8 virus grown in avian and mammalian cells. A, non-reducing SDS-PAGE and Coomassie staining of A/Puerto Rico/34 (H1N1) virions grown in MDCKII cells or in embryonated eggs. The positions of viral proteins (HA, NP, and M1) and molecular mass markers in kDa are indicated. The strong band below NP is probably albumin from the serum used to grow virus in cell culture. HA was analyzed with the new blotting-MS protocol. B, MALDI-TOF MS analysis. Mass spectra obtained in reflector mode are depicted. Indicated are the m/z values of the peaks, the first and last amino acid residues of the corresponding peptides, and the number and type of HA-bound fatty acids. For simplicity, additional minor peaks producing the peptides HA(516–565) and HA(521–565) with lower m/z values are neither shown nor used for stearate calculation. The calculated stearate content found for the HA(514–565) peptides from several spectra is listed in Table 1. C, amino acid sequence of the HA C-terminal region indicating the prevailing site of cleavage by trypsin. Cysteines as potential acylation sites are enlarged, and the TMR is underlined.
Exchange of Amino Acids Modestly Affects the Acylation Pattern
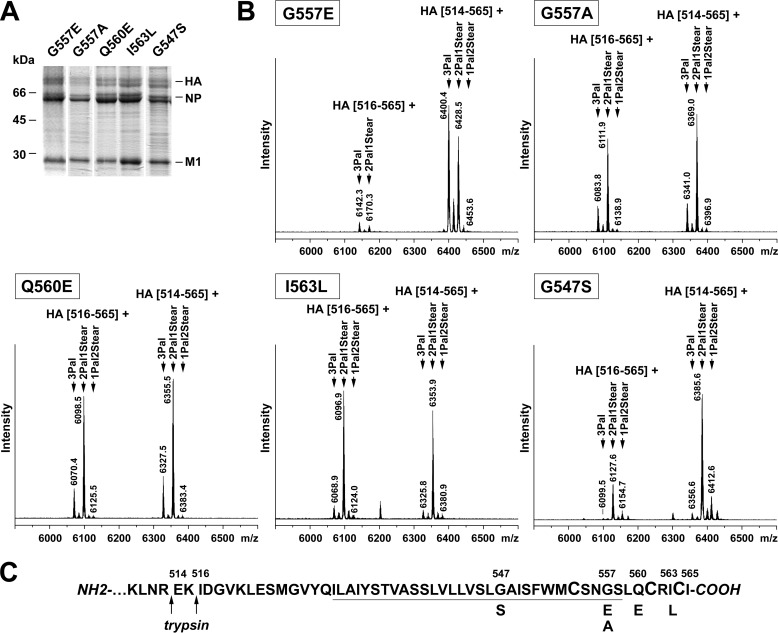
Next we determined the fatty acid pattern of mutant HAs from recombinant WSN viruses grown in MDCK cells using the blotting procedure. Because the error rate of the viral polymerase is high and virus with nucleotide reversions to the wild type sequence rapidly outgrows the mutant virus, we sequenced the genomic RNA encoding the C terminus of HA, and only virus preparations with the desired sequence were analyzed by MALDI-TOF MS. Similar to the wild type HA, trypsin cleaves the polypeptide chain after arginine 514 but also after arginine 516 such that two peptides appear in the spectra that encompass the cleavage site until the C terminus of HA (Fig. 6). The observed peptide masses are also consistent with the expected sequences shown in Fig. 2, indicating that no reversion to wild type (or any other amino acid) occurred.
FIGURE 6.
MS results from WSN mutants with exchange of amino acids. A, non-reducing SDS-PAGE and Coomassie staining of A/WSN/33(H1N1) recombinant virus particles grown in MDCKII cells. The mutation introduced into HA, the positions of viral proteins (HA, NP, and M1), and molecular mass markers in kDa are indicated. HA was analyzed with the new blotting-MS protocol. B, MALDI-TOF MS analysis. Mass spectra obtained in reflector mode are depicted. Indicated are the m/z values of the peaks, the first and last amino acid residues of the corresponding peptides, and the number and type of HA-bound fatty acids. A third peak with m/z values around 5557 corresponding to the triple acylated peptide HA(521–565) was also observed but is not shown because it overlaps with the trypsin autolysis peak at 5561 m/z. The stearate amount calculated for the peptides HA(516–565) and/or HA(514–565) of each mutant is depicted in Fig. 9. The m/z region corresponding to non- and underacylated peptides is not included in the spectra, but such peptides were never observed. C, amino acid sequence of the HA C-terminal region indicating the amino acid exchange and the prevailing site of cleavage by trypsin. Cysteines as potential acylation sites are enlarged, and the TMR is underlined.
Each of the mentioned peptides is present as two main peaks representing triple palmitoylated peptide and a peptide containing two palmitates and one stearate. However, the intensities of the peaks clearly differed between the HA mutants, indicating that the introduced amino acid replacements changed the acylation pattern. We calculated the stearate content for this and usually one other virus preparation; the results are graphically shown in Fig. 9.
FIGURE 9.
Graphical summary of MS results with recombinant WSN virus from MDCK cells. Stearate content was calculated from one (I563L and C554S/L559C), two (G557E, G557A, Q560E, and G547S) or three (WT) virus preparations. For each virus preparation, at least two blotting repeats were performed, and at least four mass spectra were recorded. The remainder is palmitate; the results are represented as mean percentage ±S.D. (error bars).
A reduction in the stearate content was observed only for G557E, which contains 15% of the longer acyl chain in comparison with 23% stearate calculated for wild type HA. Because the conservative replacement of the completely conserved glycine 557 by an alanine has no significant effect (24% stearate), it is rather the insertion of the acidic glutamic acid that is responsible for the observed stearate reduction in G557E. Exchange of glutamine by glutamic acid in position 560 caused a very minor increase in the stearate proportion (26%), indicating that the insertion of a negatively charged residue into the cytoplasmic tail does not reduce stearoylation per se. However, the conservative replacement I563L at the hydrophobic C terminus of HA significantly raised the stearate proportion to 31%. The largest increase in the stearate content to 33% was observed with the HA mutant that contains a serine instead of the glycine located in the middle of the TMR.
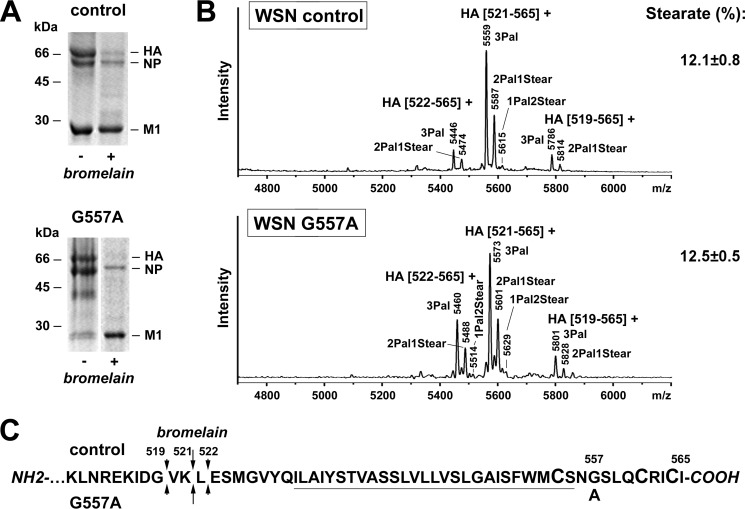
We also analyzed one of the HA mutants (G557A) with egg-grown virus using the previous, better established procedure for sample preparation (Fig. 7). Around 12% stearate was determined; this is identical to the wild type HA analyzed with the same procedure but half as much as was calculated for the same mutant virus grown in MDCK cells. This result reinforces our notion that avian cells attach less stearate to the HA of human viruses compared with mammalian cells. In addition, non- or underacylated peptides were not detected, suggesting that the mutation did not alter the stoichiometry of S-acylation. In summary, point mutations introduced into the TMR or cytoplasmic tail of HA modestly affected the fatty acid pattern of HA but did not abolish stearoylation or S-acylation per se.
FIGURE 7.
MS results from WSN wild type and G557A mutant grown in avian cells. A, non-reducing SDS-PAGE and Coomassie staining of A/WSN/33(H1N1) virions (control) and WSN G557A recombinant viral particles grown in eggs before (−) and after (+) digestion with bromelain and pelleting (100,000 × g, 1.5 h, 4 °C). The bromelain-digested viral particles were further analyzed with the previous extraction-MS protocol. The positions of viral proteins (HA, NP, and M1) and molecular mass markers in kDa are indicated. B, MALDI-TOF MS analysis. Mass spectra obtained in reflector mode are depicted. Indicated are the m/z values of the peaks, the first and last amino acid residues of the corresponding peptides, and the number and type of HA-bound fatty acids. Indicated is the percentage of HA-bound stearate (mean ± S.D.) calculated from four to six spectra obtained for each of one to three digestions performed for three (control) or one (G557A) virus preparations. Note that non- or monoacylated HA peptides (peaks with m/z values around 4845 and 5083, respectively) were not detected in the spectra, and only very tiny peaks of doubly acylated peptides (m/z around 5321) were observed. C, amino acid sequence of the HA C-terminal region indicating the amino acid exchange and the site of cleavage by bromelain. The arrow lengths approximately correspond to the respective peak heights in the spectra. Cysteines as potential acylation sites are enlarged, and the TMR is underlined.
Shifting a Cysteine from the TMR to the Cytoplasm Eliminates Attachment of Stearate
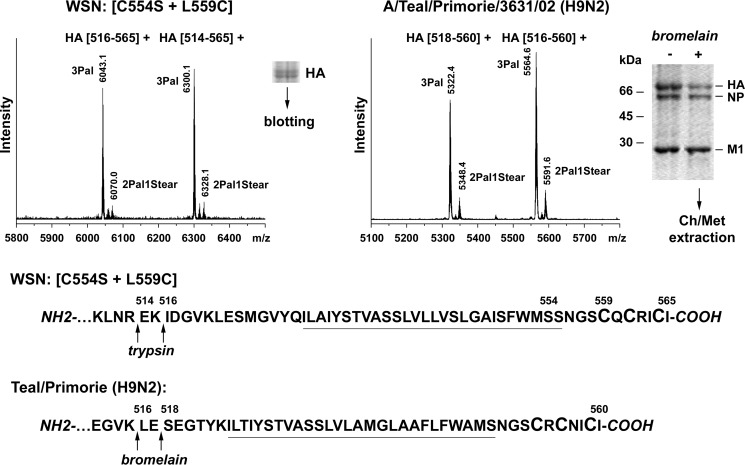
Finally, we determined the fatty acid pattern of the C554S/L559C mutant grown in MDCK cells; in this mutant the usually stearoylated transmembrane cysteine was shifted to a cytoplasmic location. Only very small peaks representing peptides with two palmitates and one stearate are detectable; the by far predominant peptide contains three palmitates (Figs. 8 and 9). Calculation of the stearate content revealed that it is reduced to 3.3%.
FIGURE 8.
MS results from a WSN mutant and a natural H9 subtype HA with shifted cysteine. The A/WSN/33 (H1N1) recombinant virus mutant (C554S/L559C) was grown in MDCK cells and analyzed using the blotting approach, whereas the A/Teal/Primorie3631/02 (H9N2) virus was grown in embryonated chicken eggs and analyzed using the previous protocol based on bromelain digestion of virions. Mass spectra obtained in reflector mode are depicted. Indicated are the m/z values of the peaks, the first and last amino acid residues of the corresponding peptides, and the number and type of HA-bound fatty acids. The stearate content was calculated as 3.3% for the H1 subtype mutant and 4.5% for H9 subtype HA. The peaks of both H1 subtype HA(516–565) and HA(514–565) and H9 subtype HA(518–560) and HA(516–560) were used for calculations. Depicted below the spectra are the C-terminal amino acid sequences of H1 and H9 subtype HAs indicating the cleavage sites by trypsin and bromelain, respectively. Cysteines as potential acylation sites are enlarged, and the TMR is underlined.
We also analyzed the fatty acid pattern of an H9 subtype HA from a teal virus grown in embryonated eggs that has the same spacing of cysteines embedded in a different amino acid context (SCRCNICI (H9) versus SCQCRICI (H1)). Despite these differences in the amino acid sequence, the same small peak intensities were detected for peptides with two palmitates and one stearate compared with those with three palmitates in the spectra of H9 and H1 HAs. Thus, the location of an acylated cysteine relative to the end of the transmembrane span is the decisive factor for the attachment of palmitate versus stearate as demonstrated both for an egg-grown avian virus and for a human virus from MDCK cells.
DISCUSSION
In this study, we used reverse genetics and mass spectrometry to determine the molecular signals that determine site-specific attachment of palmitate and stearate to HA of influenza virus. We first developed a new sample preparation procedure for MS analysis of acylated proteins that uses non-reducing SDS-PAGE of a virus preparation, blotting to PVDF membranes, identification of HA on the membrane by Ponceau S staining, in situ proteolysis, and elution of peptides from the membrane by organic solvents prior to MALDI-TOF MS. We have shown that the procedure produces the same result as the previous approach using proteolysis of gradient-purified virus particles (Fig. 4). The variation of the results between different virus preparations was also low (1–6%), albeit somewhat higher compared with the previous procedure (1–3%). Both protocols (proteolytic digestion of virions/MS or protein blotting/MS) demonstrated similar ∼1% (or less) variation when repeated for the same virus preparation. Nevertheless, the new approach might require only ∼10 μg of acylated protein to calculate the stearate/palmitate ratio and probably less just to detect the acylated peptides and is thus quite suitable to identify acylation patterns of abundantly expressed cellular membrane proteins. Purification of the protein is not even required as long as the protein of interest can be identified in a protein mixture as an individual (well represented) band by Ponceau S staining.
Using both the new and previous procedures, we found that HA from two human H1N1 strains (WSN and PR8) contains more stearate (20–23%) if grown in mammalian MDCK cells compared with the same virus amplified in avian cells (8–12%; Fig. 5 and Table 1). Together with the observation that HA from avian viruses already contains a high proportion of stearate (22–35%) when the virus is grown in eggs (24, 27), we speculate that viruses have adapted to their natural host cells such that they receive a high stearate proportion. However, before any firm conclusions can be drawn, more experiments with avian, porcine, equine, and human influenza viruses grown in natural and non-natural host cells should be performed. Nevertheless, this observation already indicates that there are cell type-specific differences in the acylation pattern of HA and thus probably also in cellular proteins. In addition, our results add to the very recent finding that influenza virions produced by mammalian and avian cells have distinct protein compositions, indicating that the host cell influences the architecture of virus particles (39).
We also showed that none of the introduced mutations prevented S-acylation of HA. The deleterious mutations in HA that did not allow generating infectious virus particles, i.e. I563Q and G557Stop, are acylated as efficiently as wild type HA as deduced from metabolic labeling with [3H]palmitate (Fig. 3). Thus, the functional defect in HA introduced by the mutation is not due to a significant blockade of acylation. In the MS spectra from HA mutants with which we could generate infectious virions, we never detected peaks that correspond to non-acylated or underacylated peptides (Figs. 4–8). Thus, the conserved glycine residues located in the cytoplasmic tail (Gly-557) and in the TMR (Gly-547) and Gln-560 in the cytoplasmic tail are apparently essential for acylation of HA. It is even possible to shift a cysteine from the TMR to a cytosolic location without compromising attachment of fatty acids at any of the three acylation sites.
However, a word of caution regarding the quantitative nature of MS data is necessary. Although in principle we could detect non- and underacylated peptides with our MALDI-TOF setting (35), we presently cannot exclude that non- or underacylated peptides containing exchange of crucial amino acids “do not fly” or are lost during sample preparation. Thus, we cannot rule out that some peptides are underacylated, i.e. that the mutations reduced the efficiency of acylation at certain sites. Nevertheless, it is fair to conclude that a putative acyltransferase does not need to recognize and interact with amino acids in the vicinity of acylation sites as an essential step of its catalytic cycle. Most likely, a reactive cysteine, i.e. reduced and deprotonated, that is accessible to the active site of an enzyme, i.e. exposed near the cytoplasmic face of the membrane, is sufficient for acylation to occur (40).
We also conclude that the location of a cysteine relative to the end of the TMR is the decisive factor for attachment of stearate. Shifting the cysteine to a cytoplasmic location reduced attachment of the longer chain fatty acid to 3%. The same proportion of stearate was found in the HA of a natural H9 subtype virus strain (Fig. 8), in the HA of H7 subtype if the TMR cysteine was exchanged for a serine (24), and in the HA of influenza B virus that evolutionarily lacks a cysteine residue at the end of the TMR (24). It is now obvious that the length of the cytoplasmic tail originally proposed to be important for stearoylation (41) plays no role at all because the fusion protein of Newcastle disease virus is stearoylated at a TMR cysteine despite containing a long (27-amino acid) cytoplasmic tail (26).
Nevertheless, an exchange of amino acids subtly influences the proportion of stearate attached to HA (Fig. 6). The most prominent effect was calculated for the mutant G547S, which showed an increase in the stearate proportion from 23 (wild type HA) to 33%. We hypothesize that the exchange of the conserved glycine in the TMR by a serine, which is more compatible with an α-helical conformation, straightens the transmembrane span and places the TMR cysteine in a better position for stearoylation. Alternatively, it was proposed that glycine residues in the transmembrane region are oriented toward the helix-helix interface to mediate interactions (42). Indeed, molecular modeling of the closely related H6 subtype HA implies that this is the case for the HA TMR (27). Substitution of glycine by serine might alter these interactions and/or rotate one TMR helix against the other. In all these cases, the cysteine at the end of the TMR might become better accessible by a putative stearate-specific acyltransferase.
The conservative substitution of isoleucine with leucine at the hydrophobic patch at the C terminus of HA (and thus at a distance of nine amino acids from the TMR cysteine) increased the stearate proportion to 31%. Because of limited sample material, we could not identify the fatty acid pattern for each acylation site with MS/MS sequencing as we did in our previous analysis. However, in all our previous analyses, we never observed attachment of stearate at sites other than the TMR cysteine. Thus, it is likely that additional stearate (relative to the wild type HA) is also attached here and not at the C-terminal cysteine 564. One might speculate that the hydrophobic patch at the C terminus of the cytoplasmic tail interacts with the TMR, thereby contributing to a subtle increase of the stearate content at cysteine 554.
A reduction in the stearate content to 15% was observed in the mutant G557E. This is not due to replacement of the completely conserved, cytosolic glycine residue because its exchange by an alanine had no effect on the stearate content. It is also not caused by the presence of glutamic acid per se because in the mutant Q560E a slight increase in the stearate content was noticed. We suppose that insertion of a negatively charged residue at the membrane-cytosol interface pulls the cysteine out of the membrane and exposes it to the cytosol such that it is in a less favorable location for stearate attachment. Because the TMR of HA is very long (27 amino acids), it has a wide range to move perpendicular to the plane of the membrane, especially at the intracellular site of acylation (the late endoplasmic reticulum or early Golgi region) where the membrane bilayer is supposed to be thinner. In summary, conservative as well as drastic exchanges of amino acids in various regions at the C terminus of HA modestly alter the stearate proportion in a rather unpredictable manner. We hypothesize that the mutations alter the location of the cysteine within the transition region between the membrane bilayer and the cytosol, thereby affecting its accessibility for a putative stearate-specific acyltransferase.
Our last conclusion alludes to the mechanism of S-acylation of HA, i.e. whether attachment of palmitate and/or stearate requires an enzyme or whether only the presence of the lipid donor, i.e. Pal-CoA or Stear-CoA, is sufficient. Because acyl-CoAs are sequestered with very high affinity (∼0.1 nm) by the acyl-CoA-binding protein (43), their cytosolic concentration is presumably too low to facilitate spontaneous acylation. However, acyl-CoA partitions into lipid bilayers with a high partition constant (1–5 × 105/m) by insertion of its acyl chain. Using atomic force microscopy and supported lipid bilayers, it was found that acyl-CoA forms lateral aggregates in the plane of the bilayer (44) that might represent the lipid donor for non-enzymatic S-acylation if such clusters exist in living cells. One might imagine that a fatty acid is spontaneously transferred to a protein if acyl-CoA encounters a cysteine at the end of the TMR of a protein in transit along the exocytic pathway.
However, such a non-enzymatic reaction would not show any preference for the attachment of a particular fatty acid to a certain site but should rather reflect the concentration of individual acyl-CoAs at the intracellular site of acylation, the membranes of the late endoplasmic reticulum or early Golgi (19). Although very little is known about the distribution of acyl-CoAs in cellular membranes, there is no indication that the concentrations of Pal-CoA and Stear-CoA grossly differ in the endoplasmic reticulum/Golgi because both are also used for the synthesis of phospholipids that contain palmitate and stearate as the major type of fatty acids.
In addition, the cell type-specific heterogeneity we found in the acylation pattern of HA is also difficult to reconcile with a non-enzymatic mechanism of S-acylation. Moreover, although insect cells are able to acylate HA when expressed with the baculovirus system (45), attachment of stearate was observed neither for HA nor for endogenous insect proteins (46). Thus, stearoylation is apparently an evolutionary achievement of higher cells, and the responsible enzyme(s) might have then acquired slightly different substrate specificities in mammalian and avian cells.
In sum, it is more likely that (at least) two enzymes exist that differ in their acyl-CoA specificities as recently demonstrated for DHHC2 and -3 (47). One can imagine that the enzyme responsible for palmitoylation recognizes cysteine exposed to the hydrophilic milieu of the cytosol. In contrast, the active site of a DHHC protein with a strong preference for Stear-CoA might have a certain affinity for the unique biophysical environment of the border region between the hydrophobic and hydrophilic parts of the lipid bilayer. However, it cannot penetrate deeply into the lipid bilayer because H7 and H10 subtype HAs contain one cysteine and H3, H4, and H14 subtype HAs contain two cysteines in the middle of their transmembrane regions (Fig. 1) that are not acylated (4, 27). Identification of the DHHC proteins that acylate HA is a prime aim of our future investigations. Albeit 23 or 24 different DHHC proteins with distinct, partly overlapping substrate specificities are encoded by the mammalian genome (17, 18, 48), only a few might catalyze acylation of HA. These DHHC proteins are promising drug candidates because their inhibition might not compromise acylation of most cellular proteins.
Acknowledgments
We are grateful to Dr. S. G. Markushin and E. A. Kropotkina for providing us natural WSN and PR8 virus strain preparations. The MALDI MS facility was made available in the framework of Moscow State University Development Program PNG 5.13.
This work was supported by Deutsche Forschungsgemeinschaft Grant Ve 141/10 and Priority Program 1175 and Russian Foundation for Basic Research Grants 13-04-00290 and 14-04-01693.
- TMR
- transmembrane region
- DHHC protein
- polytopic membrane protein containing an Asp-His-His-Cys motif
- MDCK
- Madin-Darby canine kidney
- TPCK
- l-1-tosylamido-2-phenylethyl chloromethyl ketone
- NP
- nucleoprotein
- Pal
- palmitate
- Stear
- stearate
- CEF
- chicken embryo fibroblast.
REFERENCES
- 1. Skehel J. J., Wiley D. C. (2000) Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69, 531–569 [DOI] [PubMed] [Google Scholar]
- 2. Naeve C. W., Williams D. (1990) Fatty acids on the A/Japan/305/57 influenza virus hemagglutinin have a role in membrane fusion. EMBO J. 9, 3857–3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Steinhauer D. A., Wharton S. A., Wiley D. C., Skehel J. J. (1991) Deacylation of the hemagglutinin of influenza A/Aichi/2/68 has no effect on membrane fusion properties. Virology 184, 445–448 [DOI] [PubMed] [Google Scholar]
- 4. Veit M., Kretzschmar E., Kuroda K., Garten W., Schmidt M. F., Klenk H. D., Rott R. (1991) Site-specific mutagenesis identifies three cysteine residues in the cytoplasmic tail as acylation sites of influenza virus hemagglutinin. J. Virol. 65, 2491–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen B. J., Takeda M., Lamb R. A. (2005) Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J. Virol. 79, 13673–13684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wagner R., Herwig A., Azzouz N., Klenk H. D. (2005) Acylation-mediated membrane anchoring of avian influenza virus hemagglutinin is essential for fusion pore formation and virus infectivity. J. Virol. 79, 6449–6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zurcher T., Luo G., Palese P. (1994) Mutations at palmitylation sites of the influenza virus hemagglutinin affect virus formation. J. Virol. 68, 5748–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Engel S., Scolari S., Thaa B., Krebs N., Korte T., Herrmann A., Veit M. (2010) FLIM-FRET and FRAP reveal association of influenza virus haemagglutinin with membrane rafts. Biochem. J. 425, 567–573 [DOI] [PubMed] [Google Scholar]
- 9. Levental I., Grzybek M., Simons K. (2010) Greasing their way: lipid modifications determine protein association with membrane rafts. Biochemistry 49, 6305–6316 [DOI] [PubMed] [Google Scholar]
- 10. Melkonian K. A., Ostermeyer A. G., Chen J. Z., Roth M. G., Brown D. A. (1999) Role of lipid modifications in targeting proteins to detergent-resistant membrane rafts. Many raft proteins are acylated, while few are prenylated. J. Biol. Chem. 274, 3910–3917 [DOI] [PubMed] [Google Scholar]
- 11. Simons K., Gerl M. J. (2010) Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 11, 688–699 [DOI] [PubMed] [Google Scholar]
- 12. Rossman J. S., Lamb R. A. (2011) Influenza virus assembly and budding. Virology 411, 229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Melikyan G. B., Jin H., Lamb R. A., Cohen F. S. (1997) The role of the cytoplasmic tail region of influenza virus hemagglutinin in formation and growth of fusion pores. Virology 235, 118–128 [DOI] [PubMed] [Google Scholar]
- 14. Sakai T., Ohuchi R., Ohuchi M. (2002) Fatty acids on the A/USSR/77 influenza virus hemagglutinin facilitate the transition from hemifusion to fusion pore formation. J. Virol. 76, 4603–4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ujike M., Nakajima K., Nobusawa E. (2004) Influence of acylation sites of influenza B virus hemagglutinin on fusion pore formation and dilation. J. Virol. 78, 11536–11543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Veit M., Serebryakova M. V., Kordyukova L. V. (2013) Palmitoylation of influenza virus proteins. Biochem. Soc. Trans. 41, 50–55 [DOI] [PubMed] [Google Scholar]
- 17. Greaves J., Chamberlain L. H. (2011) DHHC palmitoyl transferases: substrate interactions and (patho)physiology. Trends Biochem. Sci. 36, 245–253 [DOI] [PubMed] [Google Scholar]
- 18. Linder M. E., Deschenes R. J. (2007) Palmitoylation: policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 19. Veit M., Schmidt M. F. (1993) Timing of palmitoylation of influenza virus hemagglutinin. FEBS Lett. 336, 243–247 [DOI] [PubMed] [Google Scholar]
- 20. Kümmel D., Heinemann U., Veit M. (2006) Unique self-palmitoylation activity of the transport protein particle component Bet3: a mechanism required for protein stability. Proc. Natl. Acad. Sci. U.S.A. 103, 12701–12706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rocks O., Gerauer M., Vartak N., Koch S., Huang Z. P., Pechlivanis M., Kuhlmann J., Brunsveld L., Chandra A., Ellinger B., Waldmann H., Bastiaens P. I. (2010) The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141, 458–471 [DOI] [PubMed] [Google Scholar]
- 22. Schmidt M. F. (1984) The transfer of myristic and other fatty acids on lipid and viral protein acceptors in cultured cells infected with Semliki Forest and influenza virus. EMBO J. 3, 2295–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liang X., Lu Y., Neubert T. A., Resh M. D. (2002) Mass spectrometric analysis of GAP-43/neuromodulin reveals the presence of a variety of fatty acylated species. J. Biol. Chem. 277, 33032–33040 [DOI] [PubMed] [Google Scholar]
- 24. Kordyukova L. V., Serebryakova M. V., Baratova L. A., Veit M. (2008) S acylation of the hemagglutinin of influenza viruses: mass spectrometry reveals site-specific attachment of stearic acid to a transmembrane cysteine. J. Virol. 82, 9288–9292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Veit M., Herrler G., Schmidt M. F., Rott R., Klenk H. D. (1990) The hemagglutinating glycoproteins of influenza B and C viruses are acylated with different fatty acids. Virology 177, 807–811 [DOI] [PubMed] [Google Scholar]
- 26. Kordyukova L. V., Serebryakova M. V., Baratova L. A., Veit M. (2010) Site-specific attachment of palmitate or stearate to cytoplasmic versus transmembrane cysteines is a common feature of viral spike proteins. Virology 398, 49–56 [DOI] [PubMed] [Google Scholar]
- 27. Kordyukova L. V., Serebryakova M. V., Polyansky A. A., Kropotkina E. A., Alexeevski A. V., Veit M., Efremov R. G., Filippova I. Y., Baratova L. A. (2011) Linker and/or transmembrane regions of influenza A/Group-1, A/Group-2, and type B virus hemagglutinins are packed differently within trimers. Biochim. Biophys. Acta 1808, 1843–1854 [DOI] [PubMed] [Google Scholar]
- 28. Serebryakova M. V., Kordyukova L. V., Semashko T. A., Ksenofontov A. L., Rudneva I. A., Kropotkina E. A., Filippova I. Y., Veit M., Baratova L. A. (2011) Influenza virus hemagglutinin spike neck architectures and interaction with model enzymes evaluated by MALDI-TOF mass spectrometry and bioinformatics tools. Virus Res. 160, 294–304 [DOI] [PubMed] [Google Scholar]
- 29. Serebryakova M. V., Kordyukova L. V., Rudneva I. A., Kropotkina E. A., Veit M., Baratova L. A. (2013) Mass spectrometry analysis of influenza virus reassortant clones does not reveal an influence of other viral proteins on S-acylation of hemagglutinin. Arch. Virol. 158, 467–472 [DOI] [PubMed] [Google Scholar]
- 30. Hernandez R., Brown D. T. (2010) Growth and maintenance of chick embryo fibroblasts (CEF). Curr. Protoc. Microbiol. Appendix 4, 4I [DOI] [PubMed] [Google Scholar]
- 31. Neumann G., Watanabe T., Ito H., Watanabe S., Goto H., Gao P., Hughes M., Perez D. R., Donis R., Hoffmann E., Hobom G., Kawaoka Y. (1999) Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. U.S.A. 96, 9345–9350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sugita Y., Noda T., Sagara H., Kawaoka Y. (2011) Ultracentrifugation deforms unfixed influenza A virions. J. Gen. Virol. 92, 2485–2493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Veit M., Ponimaskin E., Schmidt M. F. (2008) Analysis of S-acylation of proteins. Methods Mol. Biol. 446, 163–182 [DOI] [PubMed] [Google Scholar]
- 34. Kordyukova L. V., Ksenofontov A. L., Serebryakova M. V., Ovchinnikova T. V., Fedorova N. V., Ivanova V. T., Baratova L. A. (2004) Influenza A hemagglutinin C-terminal anchoring peptide: identification and mass spectrometric study. Protein Pept. Lett. 11, 385–391 [DOI] [PubMed] [Google Scholar]
- 35. Serebryakova M. V., Kordyukova L. V., Baratova L. A., Markushin S. G. (2006) Mass spectrometric sequencing and acylation character analysis of C-terminal anchoring segment from influenza A hemagglutinin. Eur. J. Mass Spectrom. 12, 51–62 [DOI] [PubMed] [Google Scholar]
- 36. Katoh K., Toh H. (2008) Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 9, 286–298 [DOI] [PubMed] [Google Scholar]
- 37. Edgar R. C. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rodriguez-Boulan E., Paskiet K. T., Salas P. J., Bard E. (1984) Intracellular transport of influenza virus hemagglutinin to the apical surface of Madin-Darby canine kidney cells. J. Cell Biol. 98, 308–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hutchinson E. C., Charles P. D., Hester S. S., Thomas B., Trudgian D., Martínez-Alonso M., Fodor E. (2014) Conserved and host-specific features of influenza virion architecture. Nat. Commun. 5, 4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dietrich L. E., Ungermann C. (2004) On the mechanism of protein palmitoylation. EMBO Rep. 5, 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Veit M., Reverey H., Schmidt M. F. (1996) Cytoplasmic tail length influences fatty acid selection for acylation of viral glycoproteins. Biochem. J. 318, 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Javadpour M. M., Eilers M., Groesbeek M., Smith S. O. (1999) Helix packing in polytopic membrane proteins: role of glycine in transmembrane helix association. Biophys. J. 77, 1609–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Faergeman N. J., Knudsen J. (1997) Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem. J. 323, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cohen Simonsen A., Bernchou Jensen U., Faergeman N. J., Knudsen J., Mouritsen O. G. (2003) Acyl-coenzyme A organizes laterally in membranes and is recognized specifically by acyl-coenzyme A binding protein. FEBS Lett. 552, 253–258 [DOI] [PubMed] [Google Scholar]
- 45. Kuroda K., Veit M., Klenk H. D. (1991) Retarded processing of influenza virus hemagglutinin in insect cells. Virology 180, 159–165 [DOI] [PubMed] [Google Scholar]
- 46. Reverey H., Veit M., Ponimaskin E., Schmidt M. F. (1996) Differential fatty acid selection during biosynthetic S-acylation of a transmembrane protein (HEF) and other proteins in insect cells (Sf9) and in mammalian cells (CV1). J. Biol. Chem. 271, 23607–23610 [DOI] [PubMed] [Google Scholar]
- 47. Jennings B. C., Linder M. E. (2012) DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J. Biol. Chem. 287, 7236–7245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Roth A. F., Wan J., Bailey A. O., Sun B., Kuchar J. A., Green W. N., Phinney B. S., Yates J. R., 3rd, Davis N. G. (2006) Global analysis of protein palmitoylation in yeast. Cell 125, 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pica N., Palese P. (2013) Toward a universal influenza virus vaccine: prospects and challenges. Annu. Rev. Med. 64, 189–202 [DOI] [PubMed] [Google Scholar]